Abstract
A longstanding goal in the field of intrinsically disordered proteins (IDPs) is to characterize their structural heterogeneity and pinpoint the role of this heterogeneity in IDP function. Here, we use multinuclear chemical exchange saturation (CEST) nuclear magnetic resonance to determine the structure of a thermally accessible globally folded excited state in equilibrium with the intrinsically disordered native ensemble of a bacterial transcriptional regulator CytR. We further provide evidence from double resonance CEST experiments that the excited state, which structurally resembles the DNA-bound form of cytidine repressor (CytR), recognizes DNA by means of a “folding-before-binding” conformational selection pathway. The disorder-to-order regulatory switch in DNA recognition by natively disordered CytR therefore operates through a dynamical variant of the lock-and-key mechanism where the structurally complementary conformation is transiently accessed via thermal fluctuations.
The intrinsically disordered cytidine repressor binds DNA via a folding-before-binding conformational selection mechanism.
INTRODUCTION
Between 16 and 45% of bacterial and over 50% of eukaryotic proteins contain long intrinsically disordered protein regions (IDPs), which do not have stable structure in isolation but instead exist as a heterogeneous ensemble of interconverting conformations (1). The structural plasticity of IDPs makes them particularly suited for participating in signal transduction cascades and regulatory networks, and disorder is prevalently found in kinases, transcription factors, and nucleic acid–binding proteins (2, 3). The amino acid sequences of IDPs are simpler than sequences of folded proteins and are characterized by low complexity coupled with a biased amino acid composition (1). While small (Gly and Ser) and charged amino acids (Lys, Arg, Asp, and Glu) are enriched in IDPs, there is a simultaneous depletion of large aliphatic (Leu, Ile, and Val) and aromatic (Phe, Tyr, and Trp) hydrophobic residues which promote order and folding (3). The conformational tendencies of an IDP are largely dictated by its amino acid sequence, and the conformational free energy surfaces of IDPs are believed to be flat and lacking pronounced minima (4–6).
Despite being unable to fold spontaneously into stable three-dimensional (3D) structures, IDPs have evolved distinctive mechanisms for performing their function. IDPs frequently undergo disorder-to-order transitions that result from interactions with binding partners (7, 8) or as a consequence of posttranslational modifications (9). In addition, disordered regions have been shown to nucleate liquid-liquid phase separation (10–12), thereby creating dynamic membrane-less organelles for compartmentalizing the localization and function of cellular components. The fundamental motif underlying IDP function is biomolecular recognition, which is enabled by the weak, multivalent, and often promiscuous interactions of IDPs with themselves and with other physiological binding partners (12, 13).
A central challenge in IDP biophysics has been the difficulty in obtaining an atomic-resolution description of the mechanisms of interaction between IDPs and their partner proteins. When recognition involves a disorder-to-order transition, the widely accepted mechanism of recognition is the folding-upon-binding pathway (14, 15), where transient encounter complexes stabilized by native or non-native interactions between IDPs and their partners mature into the final protein-protein or protein–nucleic acid complex without dissociation of the IDP from its binding partner (16–18). This pathway requires no prior folding of the IDP (19), and while IDPs have been known to have small amounts of preformed secondary structure (20), such residual structure is not required for binding (21) and often has to unfold before the binding event (17).
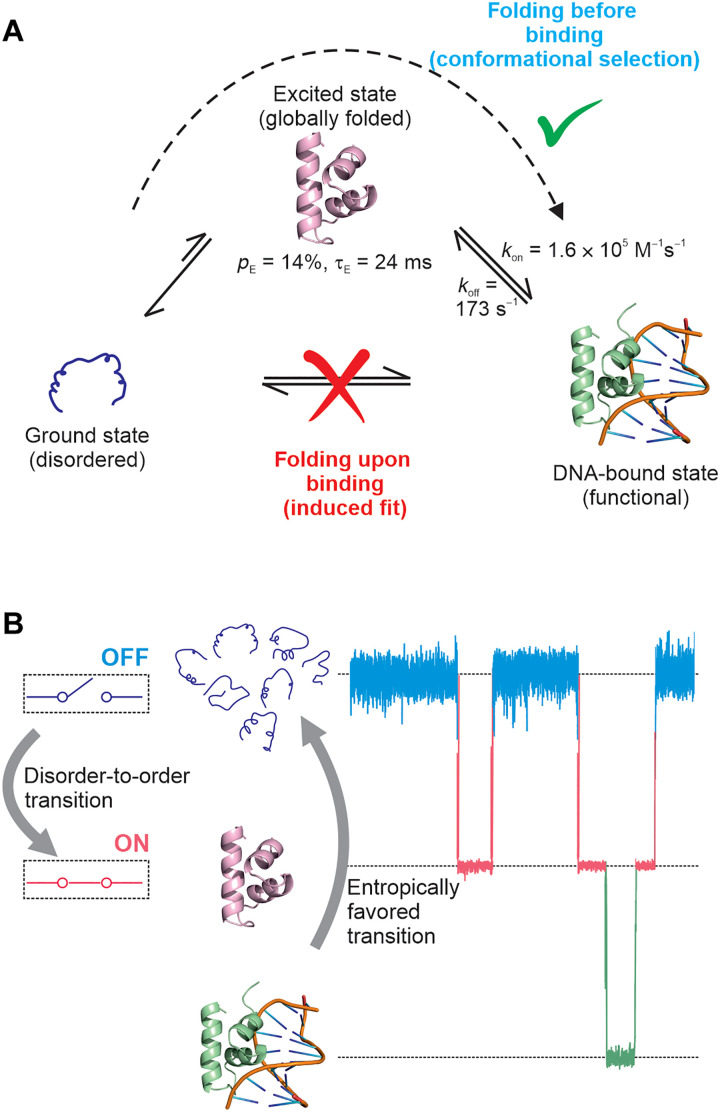
Here, we dissect the DNA binding mechanism of the natively disordered N-terminal domain of the cytidine repressor (CytR), belonging to the lactose repressor (LacR) family of transcriptional regulators. CytR undergoes a disorder-to-order transition in the presence of DNA and is folded in the DNA-bound state. We show using multinuclear chemical exchange saturation transfer (CEST) nuclear magnetic resonance (NMR) that the DNA binding domain (DBD) of CytR transiently populates a globally folded conformationally excited state. We use chemical shifts and residual dipolar couplings (RDCs) to determine the structure of the excited state, which is a well-organized three-helix bundle containing a helix-turn-helix (HTH) DNA recognition motif. We then use the recently developed multifrequency irradiation Double Resonance DANTE-CEST (DRD-CEST) to demonstrate an unconventional mode of IDP functional regulation, in which the excited state alone binds DNA through a folding-before-binding conformational selection (CS) mechanism.
RESULTS
CytRN is an intrinsically disordered DBD
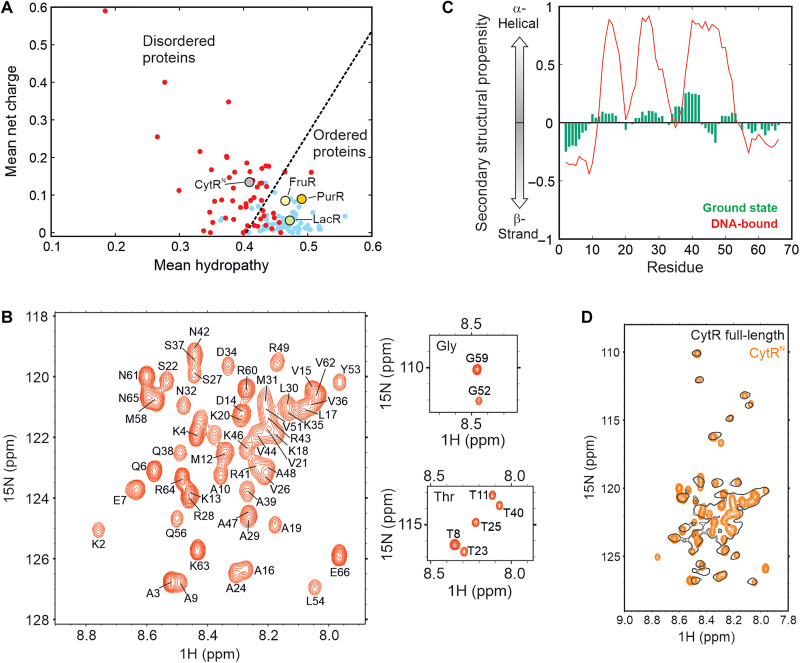
The Escherichia coli CytR (22) is a member of the LacR family of transcriptional repressors (23, 24). It is 341 amino acids long and consists of a 66-residue N-terminal DBD (referred to as CytRN), followed by a C-terminal region responsible for binding cytidine as well as dimerization (25). While the DBDs of members of the LacR family including LacR, FruR, and PurR fold into stable 3D structures in the absence of DNA (26–28), sequence-specific disorder prediction algorithms PONDR (29, 30) and IUPRED (31) classify CytRN as an IDP (fig. S1A). CytRN has higher mean net charge and lower hydrophobicity than its family members that places it in the disordered region of the Uversky charge-hydropathy plot (Fig. 1A). In addition, CytRN is depleted in hydrophobic residues that promote ordering and is instead rich in Ala and Lys, which are abundant in disordered proteins (fig. S1B). Figure 1B shows the 1H-15N heteronuclear single-quantum coherence (HSQC) spectrum of CytRN in its native state. The backbone amide resonances of CytRN are dispersed over a narrow chemical shift range between 7.9 and 8.8 ppm in the 1H dimension, and backbone (1HN, 15N, 13C′, 13Cα, 13Cβ, and 1Hα) chemical shifts correlate well with random coil values (fig. S2A), confirming that CytRN is intrinsically disordered. Residue-specific secondary structure propensity (SSP) (32) scores, calculated using backbone chemical shifts, indicate that native CytRN does not adopt stable secondary structure, although it has up to ~30% residual helicity in regions of the protein that form helices in the DNA-bound conformation (Fig. 1C) (33). This conclusion is consistent with residue-specific patterns in 13Cα and 13C′ secondary chemical shifts that suggest the presence of a low degree of helical content between residues M12-K18 and K35-E45 within an overall disordered ensemble (fig. S2B).
Fig. 1. CytRN is intrinsically disordered by itself and in full-length CytR.
(A) Charge versus hydropathy scores show that CytRN falls in the region occupied by disordered proteins (red dots), while LacR, FruR, and PurR fall in the region of ordered proteins (blue dots). (B) The 1H-15N HSQC spectrum of CytRN. The resonance assignments for each peak are indicated on the spectrum. (C) SSP of native disordered (green) and DNA-bound CytRN (red). (D) Overlay of 1H-15N HSQC spectra of CytRN (orange) and full-length CytR (black).
To determine whether native CytRN is unable to fold because of unsatisfied interactions with the C-terminal domain that are absent in the truncated version, we acquired a 1H-15N HSQC spectrum of full-length CytR which exists as a dimer of 38-kDa protomers. Most of the resonances visible in the HSQC spectrum of CytR (Fig. 1D, black contours) overlay very well with matching resonances from CytRN (Fig. 1D, orange contours), indicating that the N-terminal DBD is unstructured in full-length CytR also. Although resonances from structured regions of the 76-kDa CytR are expected to be broadened out in the HSQC spectrum because of slow tumbling and large transverse relaxation rate constants, peaks from the N terminus are sharp, demonstrating that CytRN behaves independently of the rest of the protein and retains considerable local mobility in full-length CytR. Together, NMR data confirm predictions based on the sequence of CytRN that native CytRN is intrinsically disordered in both the full-length and truncated versions of the protein.
CytRN transiently populates a higher free energy excited state
Proteins are dynamic and often use thermally accessible higher free energy conformations (referred to here as excited states) for performing function (34). However, our current understanding suggests that excited conformations of IDPs are markedly destabilized with respect to the native disordered ensemble, because the latter exists as a heterogeneous collection of microstates with comparable energies that increases its entropy (4). Therefore, the populations of excited states of IDPs are expected to be too low to be detectable or functional. Nevertheless, previous fluorescence-detected stopped-flow experiments provided evidence that CytRN may be undergoing compaction on the millisecond time scale (35). Hence, we next probed the slow dynamics of CytRN using CEST (36) NMR.
CEST experiments were carried out by irradiating 15N-labeled CytRN with a weak radiofrequency field of amplitude B1 over a range of 15N offsets for a fixed exchange duration (Tex), “searching” for chemical shifts of NMR spins in invisible excited states. These can be ascertained by quantifying the intensities (I) of peaks from the disordered state, which are graphs of the ratio of I with the corresponding intensity in a reference experiment where the exchange duration is not present (I0). The fingerprint of conformational exchange in CEST profiles is the appearance of two or more dips, the larger one arising from saturation of the major state and the smaller one, from perturbations of the minor state by the B1 field which are subsequently transferred to the major state. Since the minor dip in intensity occurs at the chemical shift of a spin in the excited state, CEST profiles provide an avenue for determining the chemical shifts of “invisible” conformations (36) that cannot be detected in routine NMR spectra.
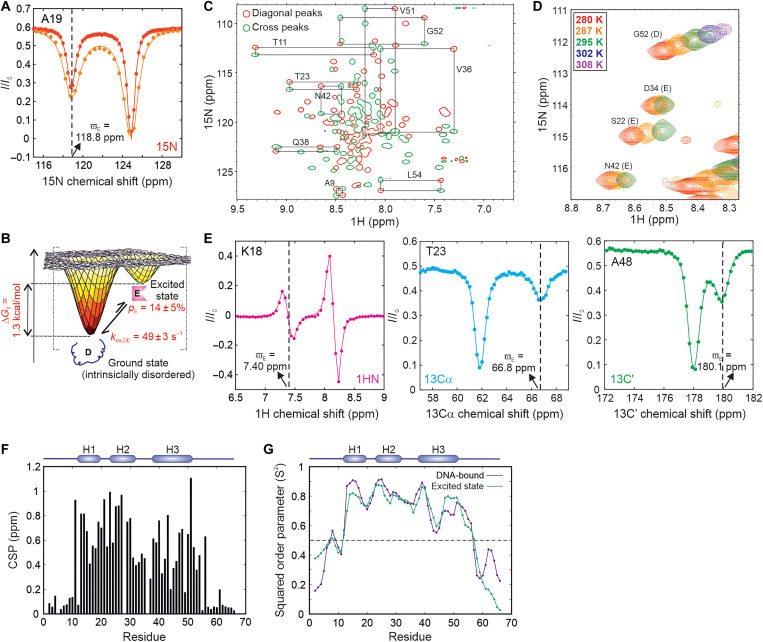
The presence of a kinetically distinct excited state in the thermal ensemble of CytRN can be discerned from the two dips in intensity in 15N CEST profiles of a number of residues (Fig. 2A and fig. S3). CEST data acquired at two B1 fields (14.7 and 28.6 Hz) for isolated residues showing two well-separated dips were globally modeled using the Bloch-McConnell equations. Fits of the CEST data reveal that the population of the excited state (E) is 14 ± 5% at 287 K (table S2, error determined from values for three independent samples, see fig. S4 and table S2 for sample-specific errors), which places this conformation at 1.04 kcal/mol (or 1.82 kT) higher in free energy than the native disordered form (D; Fig. 2B). The exchange rate constant between the two states (kex,DE) is 49 ± 3 s−1, which implies that the excited state is transiently populated with a lifetime of 24 ± 2 ms (table S2). A global analysis of 15N CEST profiles from 39 isolated residues gave very similar values of pE and kex,DE, confirming that CEST data across the entire CytRN molecule report on the formation of the same excited state (table S3). In accordance with this conclusion, pE and kex,DE obtained from single-residue fits cluster closely around the values extracted from a global analysis (fig. S5), strongly suggesting that D↔E interconversion conforms to a system in two-state exchange. Together, our data thus unequivocally demonstrate that, contrary to the currently accepted notions, IDPs can access excited conformations with millisecond lifetimes via thermal fluctuations.
Fig. 2. Intrinsically disordered CytRN populates a globally folded excited state.
(A) 15N CEST profiles of A19, acquired at 14.7 Hz (red) and 28.6 Hz (orange). Solid lines are global fits of the CEST data to the Bloch-McConnell equations. The dashed line shows the chemical shift position of the nucleus in the excited state. (B) Cartoon representation of the conformational free energy surface of CytRN, showing the disordered and excited conformations observed in NMR experiments. The relative population of the excited state (pE) and the rate constant for its exchange with the disordered state (kex,DE) are indicated on the plot. (C) Magnetization exchange experiment connecting peaks from the ground and excited states of CytRN. Only the diagonal (red) and cross-peaks (green) of residues with exchanging peaks resolved in both 1H and 15N dimensions remain in this spectrum. Black rectangles connect the diagonal peaks of the disordered and excited states with their respective cross-peaks and are labeled with the corresponding residue number. (D) Overlay of HSQC spectra of CytRN at different temperatures showing changes in excited (E) or disordered (D) peak intensities with temperature. (E) CEST profiles of the amide proton of K18 (left, magenta), alpha carbon of T23 (middle, cyan), and carbonyl carbon of A48 (right, green). The solid lines are fits to the Bloch-McConnell equations, while the dashed lines indicate the chemical shift position of the excited state dip. (F) CSPs between the excited and ground states of CytRN, calculated as described in Materials and Methods. (G) Residue-specific squared order parameters (S2) for the excited state (green) and DNA-bound CytRN (purple). The secondary structural elements of DNA-bound CytRN are shown at the top of the plot in (F) and (G).
To independently assess the existence of the excited state, we ran Carr-Purcell-Meiboom-Gill relaxation dispersion (CPMG) experiments, which are known to be effective in detecting and structurally characterizing sparsely populated conformations with millisecond lifetimes (37). CPMG profiles are plots of the effective transverse relaxation rate constant (R2,eff) as a function of the frequency of the CPMG pulse train (νCPMG), which consists of τCP - 180°- τCP repeats. Dephasing due to conformational exchange (exchange broadening) is quenched by the CPMG pulse train, so that micro-millisecond time scale exchange broadening decreases with increasing νCPMG. 15N CPMG profiles (fig. S6A) were acquired at two static magnetic field strengths [16.45 T (700 MHz) and 18.8 T (800 MHz)] and profiles of 16 nonoverlapping residues with Rex [=R2,eff(νCPMG = 33.3 Hz) − R2,eff(νCPMG = 1000 Hz), 800 MHz] > 3 s−1 were globally fit to a two-state conformational exchange model. Chemical shift differences between the disordered and excited states estimated from CEST and CPMG experiments match very well (fig. S6B), confirming that both experiments report on the same higher free energy conformation. In addition, pE and kex,DE values obtained from this analysis of 21 ± 1% and 47 ± 2 s−1 agree well with CEST-derived parameters (table S2), confirming the existence of a transiently populated excited state in equilibrium with intrinsically disordered CytRN.
The conformationally excited state of CytRN can be observed as a distinct set of resonances visible in the 1H-15N HSQC of CytRN at low contour levels (fig. S7A). Magnetization transfer experiments show cross-peaks between these low-intensity resonances and peaks from natively disordered CytRN, establishing that the two states are in millisecond time scale conformational exchange (Fig. 2C). The 1HN and 15N chemical shifts of the low-intensity resonances agree very well with the chemical shifts of the excited state measured by CEST (fig. S7, B and C), indicating that the sparsely populated alternate conformation seen in 1H-15N correlation spectra is the same excited state of CytRN detected in CEST profiles. The ratio of excited to disordered peak intensities estimated from 1H-15N HSQC spectrum of CytRN gives the population of the excited state as 12 ± 3% (fig. S7D), which agrees well with pE values from CEST experiments. 1H-15N correlation spectra, CEST, CPMG, and magnetization transfer data are therefore internally consistent with each other and demonstrate the independent existence of a transiently populated excited state in equilibrium with the disordered ensemble of CytRN.
To evaluate whether the CytRN disordered state seen at 287 K is a cold-denatured conformation, we first acquired 1H-15N HSQC spectra at temperatures ranging from 280 to 308 K (fig. S8A). HSQC spectra show that CytRN remains predominantly disordered at temperatures up to 302 K, indicating that the native disorder seen at 287 K is not a result of cold denaturation. Minor resonances from the CytRN excited state disappear beyond ~295 K, reflecting a destabilization of the excited conformation with increasing temperature (Fig. 2D and fig. S8, B and C). Subsequently, we used a combination of CEST and CPMG experiments (fig. S9) to obtain precise estimates of pE and kex,DE at different temperatures. The population of the disordered state increases from 59% at 280 K to 93% at 302 K (fig. S10), clearly showing that CytRN is not cold-denatured at 287 K. pE increases from 7% at 302 K to 41% at 280 K, indicating that the melting temperature (Tm) for the D↔E transition is below 280 K. kex,DE values also show a substantial increase from 13 s−1 at 280 K to 248 s−1 at 302 K. The excited state populations and lifetimes match well with those of a compact conformation detected in temperature-dependent stopped-flow fluorescence (fig. S10) (35), single-molecule Forster resonance energy transfer experiments (38), and statistical mechanical modeling (39), providing additional evidence for the existence for a compact excited state in the CytRN ensemble.
The excited state of CytRN is globally folded
IDPs have been known to undergo local folding as well as global disorder-to-order transitions in the presence of binding partners or upon posttranslational modifications. To determine the extent of structure in the excited state of CytRN, we used multinuclear CEST profiles to extract 15N (40), 1HN (41), 13Cα (42), and 13C′ (43) chemical shifts in the excited conformation (Fig. 2E and fig. S11). 15N, 1HN, 13Cα, and 13C′ chemical shifts were obtained for 53, 50, 40, and 44 residues respectively (tables S4 and S5), providing excellent coverage of the structural changes occurring in the entirety of CytRN.
Since chemical shifts are extremely sensitive probes of protein structure (44), regions of the protein experiencing large chemical shift perturbations (CSPs) will also be the regions that display large differences in conformation between the excited state and the native disordered ensemble. Figure 2F shows the residue-specific CSPs in CytRN resulting from the conformational interconversion. CSPs are small (<0.2 ppm) at the N (M1 to A10) and C termini (P57 to E66) and signify that the extremities of CytRN remain disordered in the excited state. While residues 50 to 57 in LacR form the hinge helix when the Lac repressor is bound to operator DNA (45), the corresponding region in CytR remains disordered in both the excited state (Fig. 2F) and the DNA-bound state (33), likely because of the helix-breaking Pro57 residue at position 2 of the putative hinge helix. In contrast, the rest of the protein from T11 to Q56 undergoes sizeable changes in chemical shift upon transitioning to the excited state with an average CSP of 0.59 ± 0.25 ppm over 44 residues, underscoring the fact that disordered native CytR acquires a globally folded structure in the excited state. The magnitude of CSPs plotted on the structure of DNA-bound CytRN (fig. S12) also illustrates that CSPs are not localized to a particular region but are instead distributed across the entire protein sequence from T11 to Q56.
Chemical shifts can also be used within the Random Coil Index framework to obtain estimates of generalized squared order parameter (S2) values (46) that are measures of the amplitude of motion occurring at backbone amide sites in the protein. Figure 2G shows the residue-specific S2 values of the excited state evaluated from 15N, 1HN, 13Cα, and 13C′ chemical shift information. S2 values are high in the interior of CytRN and range between 0.60 and 0.88, consistent with the rigidity expected from a globally folded excited state conformation. On the other hand, S2 values drop to less than 0.5 for the flexible terminal residues M1 to M12 and P57 to E66 (Fig. 2G). Taken in conjunction with the large spectral dispersion observed for the excited state resonances in the 1H-15N HSQC (fig. S7A), our data suggest that CytRN adopts a rigid globally folded conformation in the excited state.
The excited state is a three-helix bundle containing an HTH motif
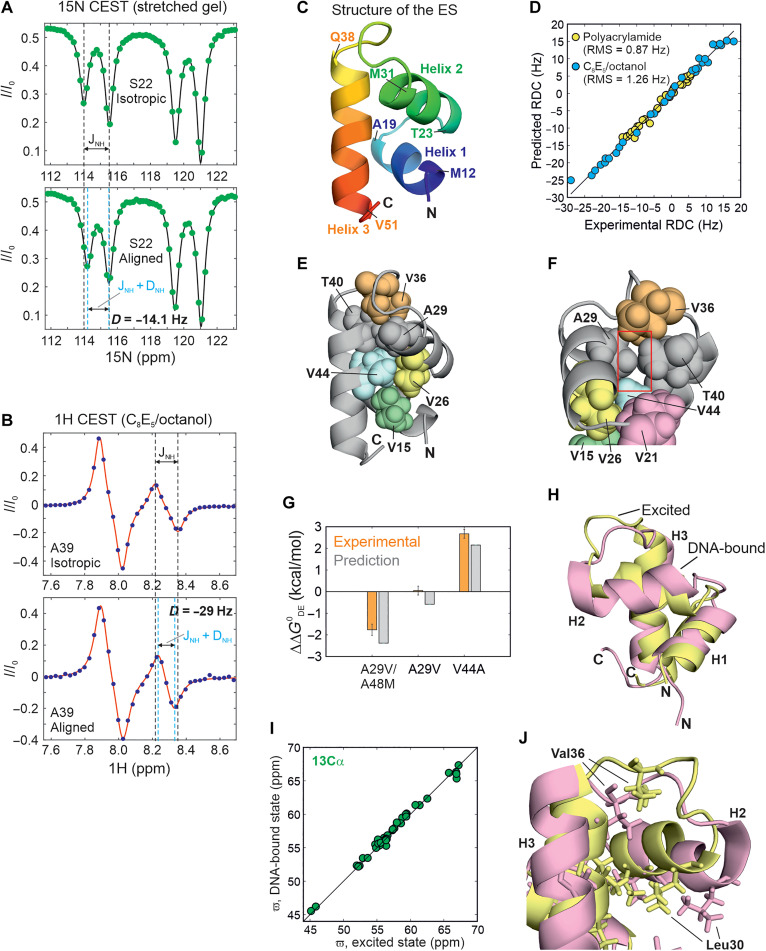
To elucidate the atomic-resolution structure of the excited state, we measured 15N-1H RDCs (47, 48) as additional structural restraints for use in a structural calculation algorithm. 15N-1H RDCs were measured using either 15N- (49) or 1HN-CEST pulse sequences (fig. S13). Briefly, in the 15N-CEST sequence, doublets are obtained by removing the 90x°-240y°-90x° 1H decoupling module during the exchange duration, and the spacing between the two components of the doublet corresponds to the scalar coupling (1JNH) in the isotropic sample (Fig. 3A, top) or the sum of 1JNH and the 15N-1H RDC in the aligned sample (Fig. 3A, bottom). Accordingly, the difference in doublet spacings between the two samples provides the value of the RDC (fig. S13). On the other hand, the 1HN-CEST sequence is already designed so that every CEST profile represents a difference of the TROSY (HzNβ) and anti-TROSY (HzNα) CEST profiles. Therefore, fitting each intensity dip to a difference of two Lorentzians directly furnishes 1JNH for data acquired on an isotropic sample (Fig. 3B, top), or 1JNH + 15N-1H RDC (Fig. 3B, bottom) in the case of an aligned sample (fig. S13). Simulations demonstrate that RDCs can be obtained reliably using this procedure, provided the magnitude of the 1HN chemical shift difference between the ground and excited states (|ΔDE|) is larger than 1JNH; in our case, fit 1JNH values match well with the inputs supplied in the simulation for |ΔDE| > 0.25 ppm (corresponding to 175 Hz on a 700-MHz NMR spectrometer), so that all four dips in intensity are clearly resolved in the 1HN CEST profile (Supplementary Text and fig. S14).
Fig. 3. Structure of the CytRN excited state.
1H-15N RDCs for S22 (A) and A39 (B) obtained via the 15N CEST (A) or the 1H CEST experiment (B) on CytRN aligned in stretched polyacrylamide gels (PAGs) (A) or in a lyotropic C8E5/octanol mixture (B). In both panels, CEST profiles in isotropic media are shown at the top for comparison. The black dashed lines indicate the position of the in-phase (A) or “anti-phase” (B) doublets separated by 1JNH in isotropic solution. Cyan lines reflect the positions of the same doublets in aligned media separated by 1JNH + 15N-1H RDC. (C) The three-helix bundle structure of the globally folded excited state. The amino acids marking each helix boundary are indicated on the structure. (D) A comparison of the experimental RDCs in PAG (yellow) and C8E5/octanol (blue) with the RDCs predicted for the lowest-energy CS-Rosetta conformer. (E and F) Different orientations of the excited state structure of CytRN shown in (C). The hydrophobic core of the ES is depicted as spheres. The cavity formed by the smaller A29 (compared to the larger Val in other family members) is indicated by a red box in panel (F). (G) Bar plot of the predicted [gray; FoldX (57)] and experimentally observed (orange) free energy differences between the disordered and excited states of CytRN mutants shown as differences from the WT value. Experimental values are from global fits of CEST profiles for A29V and V44A CytRN (fig. S21 and S23) and from volumes of HSQC spectral resonances for A29V/A48M CytRN (fig. S22). (H and J) A comparison of the structures of excited (yellow) and DNA-bound (pink) CytRN [Protein Data Bank ID: 2LCV (33)]. Leu and Val side chains are shown as sticks. (I) Comparison of the 13Cα chemical shifts in the excited state with those in the DNA-bound conformation (BMRB ID: 17419).
15N-labeled CytRN was aligned either in a 6% stretched polyacrylamide gel (PAG; Fig 3A) (50) or in a lyotropic phase made from bicelles containing a mixture of C8E5 and n-octanol (Fig. 3B) (51). Using a combination of the above 15N- and 1HN-CEST–based methods, 15N-1H RDCs were obtained for 35 residues in PAG and ranged from −14 to 6 Hz, while 32 RDCs varying between −29 and 18 Hz were extracted from CytRN aligned in C8E5/octanol (fig. S15 and tables S6 and S7).
We then calculated the structure of the excited state of CytRN using CS-Rosetta (52, 53) incorporating 119 backbone chemical shifts and 65 15N-1H RDCs as structural restraints (Fig. 3C and Supplementary Text). A well-defined funnel is observed in the energy versus root mean square deviation (RMSD) plot, confirming that the structure calculations have converged (fig. S16). Figure S16C shows the final ensemble of 10 excited state structures, defined by an all-atom RMSD of 0.84 Å and a Cα RMSD of 0.15 Å. There is good agreement between the input chemical shifts and the chemical shifts predicted by Sparta+ (54) for the excited state; the scatter in the correlations is of the same order of magnitude as the prediction accuracy of Sparta+ for each nucleus (fig. S17). Moreover, RDCs predicted with PALES (55) using the lowest-energy structure agree very well with the experimental RDCs and return Q values of 0.124 and 0.091 and RMSD values of 0.87 and 1.3 Hz for PAG and C8E5/octanol, respectively (Fig. 3D and fig. S18). Last, we also fit the experimental chemical shift and RDC data to ensembles of CytRN generated using Monte Carlo sampling (fig. S19) or molecular dynamics simulations (fig. S20) in which the helices are allowed to fluctuate starting from the excited state model. None of these sampled structures agrees better with the experimental data than the current excited state structure (Supplementary Text and figs. S19 and S20), reinforcing that the CS-Rosetta structure of the excited state reported here is a faithful representation of the NMR spectroscopic data collected on the CytRN excited state.
CytRN adopts a three-helix bundle topology in the excited state, in which helix 3 (H3) docks on to the HTH motif formed by helices 1 (H1) and 2 (H2) (Fig. 3C). The hydrophobic core is composed of five conserved valine residues V15, V21, V26, V36, and V44, supplemented by the methyl side chains from A29 and T40 (Fig. 3E). While T40 is conserved within the LacR family, the residue at position 29 is occupied by a valine in FruR, PurR, and LacR (56). The alteration of Val to Ala in CytRN causes a small cavity in the excited state hydrophobic core (Fig. 3F), and CEST data (fig. S21) show that the excited state of CytRN is slightly stabilized by the A29V mutation, in which the larger Val side chain can fill the hydrophobic core without generating cavities in the structure. When the core is further strengthened by replacing the small A48 side chain with Met (the corresponding residue in LacR) (56), there is a marked stabilization of the excited state which becomes the dominant state in A29V/A48M CytRN (pE = 78 ± 6%; fig. S22), while the population of the disordered state is 22 ± 6%. 15N and 1HN chemical shifts of the ground state of A29V/A48M CytRN match very well with the excited state chemical shifts of wild-type (WT) CytRN (fig. S22), establishing that this double mutant undergoes population switching. In contrast, disrupting the hydrophobic core by modifying V44 to the smaller Ala side chain considerably destabilizes the excited state, which disappears from the CEST profile altogether (fig. S23). The ΔΔG0,DE values obtained from HSQC and CEST data for all three mutants agree well with structure-based stability predictions from FoldX (Fig. 3G) (57). Overall, the structure of the excited state reveals that CytRN undergoes a disorder-to-order transition in the absence of binding partners, highlighting that IDP sequences with a high net charge and a small fraction of hydrophobic residues can encode rigid globally structured topologies while incurring only a small (ΔG0,DE = 1.04 kcal/mol, 287 K) free energy penalty.
There is a close resemblance between the backbone secondary structure of the excited state and the NMR structure of DNA-bound CytRN (33) (Cα backbone RMSD = 2.4 Å) (Fig. 3H and fig. S24). This is reflected in the excellent agreement between the 13Cα chemical shifts of the two states (Fig. 3I) which indicates that the helix boundaries are virtually identical in the two conformations and furnishes key evidence for the folded nature of the excited state. To evaluate whether the three helices are also oriented along the same directions in the two structures, we fit the excited state RDCs to the DNA-bound CytRN structure. The predicted RDCs for DNA-bound CytRN match well with the experimental RDCs (fig. S25), confirming that the relative orientations of the three helices are comparable, although not identical, in the two forms of CytRN. However, there is more scatter in the correlation between the 15N (fig. S24A) and 1HN (fig. S24B) chemical shifts of the excited and DNA-bound states; this is likely a consequence, not only of the charged DNA molecule containing aromatic nucleobases in the vicinity of DNA-bound CytRN but also of small differences in helix orientation and hydrophobic packing between the two structures. For example, in DNA-bound CytRN (33), the side chain of Leu30 rotates outward toward the DNA, while Val36 moves inward toward the hydrophobic core, consistent with the tertiary structure of CytRN adapting to the presence of the bound DNA molecule (Fig. 3J). In addition, the recognition helix H2 is slightly smaller in the excited state than in DNA-bound CytR and LacR.
CytRN binds DNA through the excited three-helix bundle state
The coexistence of the disordered ensemble of CytRN with the three-helix bundle excited state, which is structurally similar to the DNA-bound state, raises the intriguing question of whether this excited state is selected by DNA molecules for binding. While this would result in a CS mechanism of molecular recognition, the other possible models include the induced fit (IF) (58), which is known as the folding-upon-binding mechanism in IDP literature (15), and the triangular model, in which binding occurs through both the CS and IF pathways.
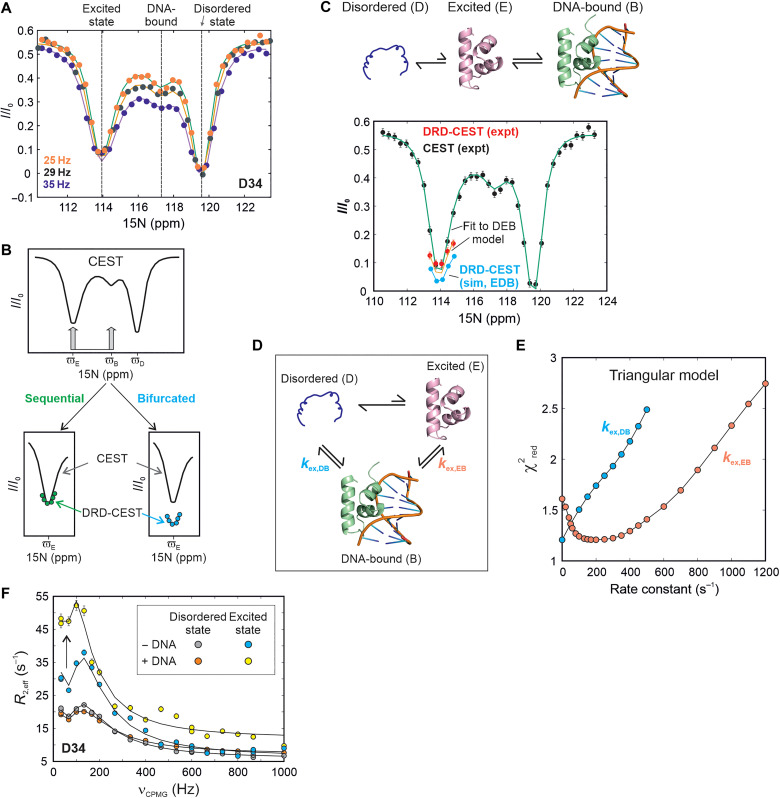
We addressed this question by first choosing D34 as a simultaneous NMR reporter of the disordered state (D), the excited conformation (E), and the DNA-bound form (B). D34 is located in the H2-H3 loop and has distinct chemical shifts of 119.6, 113.9, and 117.2 ppm in D, E, and B respectively. 15N CEST profiles of the D34 resonance in the absence of DNA show two intensity dips at the chemical shifts of the disordered (D) and excited (E) states (fig. S26). We then acquired CEST data on a sample containing 628 μM 15N-labeled CytRN and 150 μM uridine phosphorylase (udp) half-site double-stranded DNA, which is one of the operator regions specifically recognized by CytRN (33). In the presence of cognate DNA, a distinct third dip in intensity can be clearly discerned in the CEST profile of CytRN D34 at the 15N chemical shift of state B (117.2 ppm) which is the direct result of exchange between free and DNA-bound forms of CytRN (Fig. 4A).
Fig. 4. The excited state of CytRN is responsible for functional regulation.
(A) 15N CEST profiles of D34 acquired on a sample containing 628 μM CytRN and 150 μM DNA. The three dips in intensity at the chemical shifts of the disordered (119.6 ppm), excited (113.9 ppm), and DNA-bound (117.2 ppm) states are indicated with dashed lines. (B) Schematic representation of the DRD-CEST experiment (58) for a system undergoing three-state exchange between one visible (D) and two invisible states (E and B). The CEST profile for this system is shown at the top. Overlays of the minor dip at ϖE in CEST (black line) and DRD-CEST (colored circles) are shown at the bottom for a sequential (left) and a bifurcated model (right). (C) 15N CEST (black circles) and DRD-CEST (red circles) profiles of D34 acquired on the same sample as (A). Green and orange lines are global fits of the regular and DRD-CEST data at multiple B1 fields to the D↔E↔B model shown above the CEST profile. Cyan circles are simulated DRD-CEST data points assuming a bifurcated E↔D↔B model. Simulations were done using best-fit parameters obtained by fitting five CEST and three DRD-CEST datasets globally to the E↔D↔B model. (D) The triangular model for binding of CytRN to DNA, where rate constants describing the binding of D and E are kex,DB and kex,EB respectively. (E) distribution of kex,DB (cyan) and kex,EB (orange) generated from fits of CEST and DRD-CEST profiles to the triangular model. (F) 15N CPMG profiles of D34 in the disordered (gray and red) and excited states (blue and yellow) of CytRN acquired in the absence (gray and blue) and presence (red and yellow) of 5% DNA. The arrow indicates the increase in the size of excited state CPMG dispersions upon addition of DNA (blue to yellow).
Having obtained CEST profiles for D34 which report on the simultaneous exchange between D, E, and B, we turned to the recently developed DRD-CEST (59) for determining whether binding occurs via the excited state. DRD-CEST has established potential for unequivocally distinguishing between sequential and bifurcated models of chemical exchange (59). In the case of CytRN, viewed from the perspective of the NMR-visible disordered state D, the CS mechanism of binding (D↔E↔B) is a sequential model, while the IF pathway is a bifurcated model (E↔D↔B). DRD CEST builds upon D-CEST or DANTE CEST (60), in which the continuous wave irradiation that is typically used during the CEST exchange time is replaced by the DANTE selective excitation scheme (Fig. 4B) (61). When implementing DRD-CEST, a sufficiently large B1 field is first chosen so that the magnetization of molecules arriving at E is completely dephased with respect to the starting D magnetization (Fig. 4A). Then, the sample of CytRN containing DNA is simultaneously irradiated with this B1 field at the chemical shifts of both E and B, and the CEST profile of D34 is quantified. If the model of conformational exchange is a sequential one, then irradiation at E and B will not change the size of the dip compared to irradiation only at E. This is because dephasing is complete at E itself, so additional irradiation at D does not make a difference. On the other hand, if the model is bifurcated, then dephasing at E and dephasing at B occur on magnetization from different molecules, so even if dephasing is complete at E, simultaneous irradiation at E and B will increase the dip size compared to irradiation only at E. Therefore, if conformational exchange proceeds through a sequential model, the size of the dip at the chemical shift of state E remains unchanged between the regular and DRD-CEST profiles. On the other hand, the fingerprint of a bifurcated model is a pronounced increase in dip size in the DRD-CEST profile compared to regular CEST data (Fig. 4B).
Figure 4C and fig. S27 show the regular CEST profile of D34 (D state) in black and the DRD-CEST data in red. The size of the intensity dip for state E clearly remains the same between the CEST and DRD-CEST profiles, unambiguously establishing that DNA binding occurs via the excited state. The simulated dip for a bifurcated model is shown in cyan circles for comparison. To further evaluate the binding mechanism, we globally fit the CEST (five B1 fields) and DRD-CEST data (three B1 fields) separately to the sequential and bifurcated models. The value for the bifurcated model (= 1.6) is substantially higher than the sequential model ( = 1.2), signifying that the CEST data are better described by the CS mechanism (fig. S27).
Last, we quantitatively assessed whether both the CS and IF mechanisms are simultaneously operative in the DNA-CytRN interaction by fitting the CEST and DRD-CEST data globally to a triangular model (Fig. 4D). Four of the five global parameters, pE, pB, kex,DB (=kDB + kBD), and kex,EB (=kEB + kBE) were floated during the fit, while kex,DE was kept constant at the value obtained in the absence of DNA (table S2). Here, kex,DB and kex,EB are the exchange rate constants for the binding of states D and E to DNA, respectively, while kex,DE is the disordered-excited state interconversion rate constant and pD, pE, and pB are the fractional populations of states D, E, and B such that pD + pE + pB = 1. The best-fit values of kex,DB and kex,EB from the triangular fit are 0.00015 ± 0.3 and 194 ± 51 s−1 respectively, while pE = 16.5 ± 0.3% and pB = 2.1 ± 0.3%. The vast difference between kex,DB and kex,EB signifies that the disordered state cannot directly bind DNA to form the specific DNA-CytRN complex. surfaces (Fig. 4E) show that the optimal fit of the triangular model to the CEST data is obtained when kex,DB is very close to 0. In contrast, there is a heavy penalty in when kex,EB is forcibly set to 0 and the optimum value is around 200 s−1, which matches the kex,EB value of 194 s−1 obtained above. A flux-based analysis (62) of the triangular model (Supplementary Text) shows that the flux along the CS pathway is at least ~100-fold (and up to ~106-fold) larger than the flux along the IF pathway.
The CEST and DRD-CEST data together demonstrate that only the folded excited state (and not the disordered ground state) is kinetically coupled to the DNA-bound conformation. Since the disordered and excited states interconvert slowly (49 s−1), while DNA binding occurs on the millisecond time scale (kex,DB = 194 s−1 under our conditions), our model predicts that DNA binding will cause exchange broadening of the excited state but not the ground state, provided that ΔEB is large. To test this prediction, we acquired CPMG data on samples of CytRN lacking or containing 5% DNA and extracted CPMG profiles of both the disordered and excited states of D34 (ΔDE = −5.7 ppm, ΔEB = 3.3 ppm). In the absence of DNA, CPMG profiles of both states show dispersions of R2,eff with νCPMG, which originates from the D↔E interconversion (Fig. 4F, gray and blue). In the presence of DNA, the CPMG profile of the disordered state remains unchanged, indicating that DNA binding does not cause additional exchange broadening of the ground state (Fig. 4F, red). In notable contrast, the exchange broadening incurred by the excited increases by as much as 14 s−1, resulting in substantially larger dispersions in the CPMG profile (Fig. 4F, yellow). CPMG data thus provide a direct validation of the CS-based model, where only the CytRN excited state but not the disordered state binds DNA. In summary, the CEST, DRD-CEST, and CPMG experiments establish that the DNA recognition pathway of disordered CytRN predominantly follows a folding-before-binding mechanism, where molecules of CytRN in the structured excited state but not the disordered state bind to DNA (Fig. 5).
Fig. 5. The folding-before-binding model for DNA recognition by intrinsically disordered CytRN as a regulatory switch.
(A) CytRN is intrinsically disordered in its native state, and the molecular recognition event leading to the DNA-bound functional form involves a CS-based folding-before-binding mechanism. (B) A model for how the disorder-to-order transition in CytRN turns on the regulatory switch into the functional state and primes it for DNA binding, while the large entropy of the disordered state favors the dissociation of the complex and subsequent unfolding of the folded conformation, resetting the switch into the off mode. Molecular dynamics traces of CytRN are schematic representations and do not correspond to actual data.
DISCUSSION
How intrinsically disordered proteins function despite lacking stable secondary and tertiary structure remains an open question. The recognition of a cognate binding partner is often accompanied by the folding of the IDP; this is seen in the case of intrinsically disordered CytRN, belonging to the iconic LacR family of bacterial transcriptional repressors, which is folded in the DNA-bound state. In this work, we show that CytRN transiently adopts a globally folded structure that is accessible via thermal fluctuations in the absence of DNA. We have also provided evidence that this disorder-to-order transition functions as a regulatory switch by converting the binding-incompetent disordered ensemble into a three-helix bundle HTH structure capable of binding DNA. In stark contrast to the IF folding-upon-binding models that are routinely used to describe folding-coupled-binding by IDPs, the CytRN-DNA interaction follows a CS mechanism that we term “folding-before-binding” (Fig. 5).
The free energy landscape of an IDP controls its folding and molecular recognition properties and is often depicted as a flat surface with multiple shallow minima separated by very small barriers (4–6). Interconversion between these structurally similar conformations occurs on the picosecond-nanosecond time scales. Kinetically distinct higher free energy excited states in IDP landscapes are considered to be thermally inaccessible because they are substantially destabilized with respect to the high entropy disordered native ensemble (4). While this may generally be true, intrinsically disordered CytRN furnishes a notable counter-example by populating a globally ordered three-helix bundle that is a mere 1.04 kcal/mol (1.82 kT, 287 K) higher in free energy than the disordered ensemble. This excited state is not stabilized by interaction with partner ligands, osmolytes, crowding agents, or posttranslational modifications but is instead as much an intrinsic property of the free energy landscape of CytRN as the native disordered ensemble. Moreover, the detection of an excited state of CytRN with a millisecond lifetime confirms that IDPs can dynamically sample conformations over a hierarchy of time scales ranging from picoseconds to milliseconds. In the case of CytRN, this slow disorder-to-order transition has been leveraged for performing its DNA binding function.
The coexistence of the disordered and three-helix bundle conformations of CytRN is ultimately encoded in its amino acid sequence. IDP sequences are characterized by low hydrophobicity, a high net charge, and a paucity of large aromatic and aliphatic hydrophobic amino acids (1), and the sequence of CytRN meets all these criteria. Our results therefore clearly show that sequences classified as IDPs can nevertheless accommodate not only locally folded but also globally ordered domains at comparable free energies to the disordered state. This raises the question whether IDPs such as CytRN can be categorized as folding-competent proteins but with appreciably lower melting temperature. The data reported here (figs. S8 to S10) suggest that such a classification may be valid, although there is very little information in literature to support it (63).
The precise sequence determinants which allow the cytR gene to encode a thermally accessible excited state remain tantalizingly unknown. However, a comparison of the sequences of CytRN with its family members FruR, LacR, and PurR (56) shows that CytR retains most of the hydrophobic core necessary for forming the three-helix bundle while increasing the fraction of positively charged residues in the variable regions of the sequence. Simultaneously, two large hydrophobic residues at sites 29 and 48 are replaced with Ala in CytRN, resulting in a slight loosening of the hydrophobic core and concomitant destabilization (56). The synergistic combination of the above factors seems to assist CytRN in remaining intrinsically disordered without rendering the folded state thermally inaccessible. Our data clearly show that the key A29V/A48M mutant swaps the folds of the ground and excited states in CytRN, making the folded three-helix bundle conformation more stable than the intrinsically disordered ensemble.
Our CEST data show that the formation of the specific CytRN-DNA complex occurs primarily by the binding of DNA to the globally structured excited state. Since CytRN is disordered in its native state and folded in the bound form, the CytRN-DNA interaction can be classified as folding coupled to binding. However, unlike conventional IF-like folding-upon-binding situations that are prevalent in IDP literature (14, 15, 17), CytRN folds before binding DNA and therefore conforms to a classical CS mechanism. There are a number of points that are noteworthy in this regard. First, the function of the intrinsically disordered CytRN domain is to bind DNA. Since this function is carried out by the three-helix bundle folded conformation, the excited state is responsible for the functional regulation of the CytRN IDP. The overall phenomenon of transcriptional repression at the promoter and operator sites involves the C-terminal domain of CytRN, other proteins like CAP (catabolite activator protein) and the inducer cytidine, all of which modulate the binding of DNA to CytRN (25). Nevertheless, our conclusions are concerned with the elementary functional binding reaction of CytRN with DNA and affirm that only the excited state and not the disordered form is capable of forming the specific DNA-CytRN complex. Second, our classification of the mechanism as CS pertains only to global overall changes in structure between the free and DNA-bound forms of CytRN; they do not rule out small IF-like adjustments of side chains in response to DNA binding. The orientation of side chains proximal to the DNA binding site, such as L30 and V36, is different between DNA-bound and excited CytRN, suggesting that the HTH motif reorganizes itself in the complex to accommodate the DNA molecule (Fig. 3J). Additional evidence for IF-like behavior is seen near the end of the recognition helix H2, where the terminal N25 forms hydrogen bonds with the bound DNA in the structure of the LacR/operator DNA complex (64, 65), but the corresponding residue N32 is oriented differently in the CytRN excited state (fig. S28A). Unlike for most of CytRN, CEST-derived 15N chemical shifts of N32 (fig. S28B) are measurably different in the DNA-bound and excited conformations, reflecting the IF-like alterations in CytRN structure occurring after the DNA binding event. Third, our observation of a CS mechanism in the case of molecular recognition by CytRN raises the intriguing question of what determines whether an IDP will adopt a folding-before-binding or a folding-upon-binding pathway. We hypothesize that a folding-before-binding route is likely for IDPs where the bound form exhibits a globally folded conformation with substantial secondary structure and tertiary contacts. On the other hand, IDPs which form short helical or β-strand elements [referred to in literature as molecular recognition features (MoRFs) (66)] connected by linear unstructured polypeptide segments in the bound state may require more assistance from the partner protein for stabilizing these MoRFs and therefore adopt a folding-upon-binding pathway. While there are several reports documenting folding-upon-binding of these MoRFs (8, 16, 66), mechanistic data demonstrating folding-before-binding are scarce. However, some support for the above hypothesis comes from the work of Kataoka and coworkers (67), who showed using unstructured or partially structured mutants of SNase that folding can occur before or after binding depending on the context of the mutation. The Δ140-149 deletion mutant that was incapable of forming key long-range interactions necessary for folding required inhibitor binding to occur first and stabilize the folded conformation. In contrast, the 33Ala34 insertion mutation destabilized the folded conformation, but the inhibitor bound to the sparsely populated folded state, resulting in folding occurring before binding. On the other hand, a number of studies have shown the preexistence of local secondary and tertiary structure in disordered ensembles of proteins such as Sendai virus nucleoprotein (68, 69), PDEγ (70), and WASP (71), but it is unclear whether this structure is required for binding or if it has to dissolve before the recognition event (17). We also speculate that a number of instances of folding-before-binding may have been missed in literature, simply because the excited folded conformations are transiently and sparsely populated and therefore invisible to most biophysical methods. Fourth, the CEST data clearly indicate that there is no measurable kinetic coupling between the disordered ensemble and the specific DNA-CytRN complex and that virtually all the flux to this bound state traverses through the excited conformation. This rules out models that involve a rapid conformational search by the disordered state and subsequent folding on the DNA once the specific operator sequence is located. However, models where the unstructured state scans the DNA by 1D diffusion, dissociates transiently to adopt the folded conformation and then binds to the specific operator site cannot be excluded by our data, since binding in such models eventually occurs via the excited state. Last, we note that the CS and IF pathways are limiting mechanisms, and conformational changes accompanying molecular recognition events can switch from a CS to an IF mechanism at high ligand concentrations (62). However, flux calculations (fig. S29) confirm that CytRN recognizes DNA via a CS mechanism over the entire range of physiologically relevant DNA concentrations (1 nM to 1 μM), and sizeable flux along the IF pathway is seen only at unrealistic DNA concentrations above 10 mM.
The functional regulation of a bacterial IDP by a conformationally excited state is pertinent because bacteria, unlike eukaryotes, are unable to substantially expand the structural and functional repertoire of their proteomes through posttranslational modifications. Bacteria also appear to be limited in their ability to use membraneless condensates for spatiotemporal control of cellular processes (72). It is, therefore, appropriate that a bacterial IDP relies on a more primitive disorder-to-order regulatory switch that operates simply by fine-tuning the balance between hydrophobicity and intrinsic net charge to eventually modulate the conformational free energy surface; in turn, this suggests that dynamic regulatory pathways may be more prevalent in bacterial IDPs as a way to expand the functionality of their limited genetic material. In addition, HTH motifs are ubiquitous nucleic acid recognition modules that have diversified enormously in the architectural context in which they are found, as well in the function they perform in the cell (73). The appearance of a folding-before-binding mode of DNA recognition in a HTH-containing bacterial protein suggests that this mechanism was available early on in evolution and might have become incorporated into intrinsically disordered transcriptional repressors in higher organisms as well.
The disorder-to-order transition in CytRN can be viewed as a regulatory protein switch (Fig. 5B), where stochastic transitions take each molecule from the native disordered state into a binding-competent folded conformation. It is only in this folded state that CytRN is able to transduce an input signal, the presence of operator DNA, into a functional output (transcriptional repression). CytRN falls under the fold-switching category of naturally occurring switches (74), albeit with one of the folds being entirely disordered. Although the exact role of disorder in the DBD of CytR is yet to be deciphered, we adapt a model proposed in literature (75, 76), in which the disordered state serves to reset the CytRN regulatory switch (Fig. 5B). Since natively disordered CytRN has a large chain entropy, folding and subsequent complex formation with DNA will incur a free energy cost, which has been estimated in literature to be of the order of 2.5 kcal/mol (77). Studies on the c-Myb/CBP-KIX system show that the extent of disorder is correlated with koff values (78), implying that this 2.5 kcal/mol free energy penalty associated with disorder in CytRN could decrease the lifetime of the CytRN-DNA complex and offer an attractive route to turn off the regulatory switch. Thus, while the disorder-to-order transition “turns on” the regulatory switch and primes it to bind DNA, the disorder in the native state drives the switch toward the “off” position.
The loss in chain entropy in going from a disordered to the folded DNA-bound state of CytRN is opposed by the gain in entropy of water and counter-ions that are released from the interface upon complex formation (79, 80). In particular, statistical-mechanical methods incorporating molecular models for hydration water have shown that water entropy contributes appreciably to the binding free energy in cases where there is shape complementarity at the interface (81). Since DNA molecules selectively bind to folded CytRN, where the recognition helices are preorganized into a suitable geometric arrangement and do not incur a severe loss in conformational entropy upon binding, the entropy of mixing from liberated ions and water is likely to substantially augment the free energy of interaction.
Last, the dynamic regulatory switch in CytRN is another example of molecular recognition occurring via a thermally accessible excited state. It emphasizes the point that dynamics is pivotal for function and that a comprehensive understanding of the relationship between structure and function cannot be obtained merely by studying the native state alone. In the context of IDPs, a thorough characterization of the free energy surface is particularly vital, not only because IDPs lack the stable structure traditionally deemed to be necessary for function but also because they have recently gained importance as targets for pharmacological intervention (82).
MATERIALS AND METHODS
Overexpression and purification of isotope-labeled proteins for NMR spectroscopy
U-15N labeled CytR
Full-length Escherichia coli CytR (UniProt ID: P0ACN7) was cloned between the Nde I and Hind III sites in the pET-29b(+) vector along with an N-terminal 6× His-tag, followed by a linker and a Tobacco Etch Virus (TEV) cleavage site. E. coli BL21(DE3) cells were used for protein expression, and 15N isotope labeling was carried out by growing cells in M9 minimal media containing 15NH4Cl (1 g/liter). Cells were grown at 37°C until an optical density of 0.8, induced with 1 mM isopropyl β-d-1-thiogalactopyranoside, and then shaken at 20°C for ~20 hours. Cells were harvested by centrifugation (at 4°C and 13,000 rpm), and the cell pellet were resuspended in buffer A [20 mM tris (pH 7.5), 1 mM β-mercaptoethanol, 1 mM EDTA, and 300 mM NaCl] containing 2 mM MgCl2 and a Roche protease inhibitor tablet. Cells were lysed with lysozyme (7.5 mg/g of cell pellet) and sonication, following which the suspension was incubated with phenylmethylsulfonyl fluoride (1 mM final concentration), deoxyribonuclease I (50 mg/g of cell pellet), and polyethyleneimine (0.04% of the total volume) at room temperature for 1 hour. The suspension was then centrifuged for 30 min at 13,000 rpm and 4°C, and the clarified lysate was loaded on a Ni-NTA column. His-tagged protein was eluted from the column using buffer A containing 250 mM imidazole, and the tag was subsequently cleaved using TEV protease while simultaneously dialyzing against buffer A at room temperature. Tagless CytR was then recovered by passing the solution again through the Ni-NTA column. Flow-through and wash fractions containing CytR were concentrated and purified using size-exclusion chromatography on a Superdex 200 column equilibrated in 150 mM ammonium acetate. Pure fractions were checked with SDS–polyacrylamide gel electrophoresis (SDS-PAGE), pooled, dialyzed into 22 mM sodium phosphate buffer (pH 7), flash-frozen and stored at −80°C.
U-15N and U-15N,13C labeled CytRN
Untagged CytRN (E. coli CytR1–66) was expressed either from the pTXB1 vector as a fusion protein with a chitin binding domain (samples 1 and 3; table S1) or from a pET-29b(+) vector (all other samples of WT CytRN). Overexpression and purification details for expression from the pTXB1 vector are reported by Munshi et al. (83). With the pET-29b(+) vector, CytRN was cloned between the Nde I and Hind III sites and overexpressed as described above for full-length CytR. U-15N and U-15N,13C isotope–labeled CytRN were obtained from cells grown in M9 minimal media supplemented with 15NH4Cl (1 g/liter) without (U-15N) or with 13C6-glucose (3 g/liter; U-15N,13C). CytRN was purified using a previously described protocol with small modifications (33). Briefly, the clarified cell lysate was subjected to 60% ammonium sulfate precipitation. The precipitate was removed by centrifugation, and the supernatant containing CytRN was loaded on an SP-sepharose cation exchange column. CytRN elutes at an ionic strength of 540 to 640 mM. Fractions containing CytRN were pooled, concentrated, and purified on a Superdex 75 size exclusion column (120 ml bed volume), from which CytRN elutes at a volume of ~70 ml. Fractions were checked for purity using SDS-PAGE, and pure CytRN was dialyzed against 150 mM ammonium acetate, lyophilized, and stored at −20°C. Excited state populations (pE) vary slightly depending on whether CytRN is overexpressed and purified as a chitin binding domain or as a tagless construct, but exchange rate constants (kex,DE) (table S2) and excited state chemical shifts are the same between samples within measurement error.
U-15N,13C A29V/A48M, U-15N A29V, and U-15N V44A CytRN
A29V and V44A mutations were generated in the above construct of CytRN by site-directed mutagenesis using the method of overlapping primers. For A29V/A48M CytRN, the A29V single point mutation was introduced using site directed mutagenesis in CytRN cloned in the pTXB1 vector, and the A48M mutation was subsequently introduced in the A29V CytRN mutant. Isotope-labeled CytRN variants were overexpressed and purified using the same protocol as for WT CytRN. U-15N,13C A29V/A48M CytRN was overexpressed in minimal media as described above for WT CytRN/pTXB1, and the purification was carried out as reported previously (83).
Oligonucleotide preparation
Single-stranded udp half-site DNA (ssDNA) sequences were purchased from Sigma-Aldrich (5′-ATTTATGCAACGCA-3′). Forward and reverse ssDNA were dissolved in 22 mM sodium phosphate buffer (pH 7), and their concentrations were estimated using ultraviolet absorbance at 260 nm. A 1.1-x buffer was used for dissolving the DNA to account for the 10% D2O added as NMR lock solvent. Equimolar concentrations of forward and reverse DNA strands were mixed, and the DNA was annealed by first incubating at 95°C for 15 min on a dry bath and then lowering the temperature slowly by turning off the dry bath. The double stranded DNA was stored at −20°C for further use.
Sample preparation
All NMR experiments were carried out on samples in 20 mM sodium phosphate buffer (pH 7.0) containing 10% D2O for the field-frequency lock. Sample concentrations ranged from 60 μM (full-length CytR) to 2 mM. The details of the various CytRN samples used are summarized in table S1.
Sample preparation for RDC measurements
Polyacrylamide stretched gels
Six percent PAGs were cast using a 30% acrylamide:bisacrylamide (29:1) mixture made in tris buffer (pH 8.8) and polymerized in a 6-mm gel casting chamber (www.newera-spectro.com). After the gel solidified (~ 15 min), the gel was dialyzed multiple times against water to remove the buffer components and air-dried. Before making RDC measurements, the gel was soaked in 550 μl of protein solution containing 10% D2O for about 10 to 12 hours using the same casting chamber. This gel was then transferred to a 4.2-mm (inner diameter) RDC NMR tube with the help of a piston driver. Twenty microliters of buffer was added to both ends to prevent drying of the gel, and the bottom of the sample tube was sealed with a gel end plug. At the top, the gel was capped with a support rod carrying a top-gel plug at the sample end. The support rod was held in place with a support cap that attaches to the NMR tube.
C8E5/n-octanol
Before preparing the protein sample, a 10% C8E5/n-octanol mixture was prepared in the following manner (51): 36 μl of C8E5 (Sigma-Aldrich) was dissolved in 222 μl of sodium phosphate buffer (pH 7) and 30 μl of D2O. The C8E5 solution was kept on ice throughout and mixed thoroughly. Twelve microliters of n-octanol was added to the above C8E5 solution in aliquots of 4 μl, and the mixture was vortexed after each addition. The solution went from yellow to turbid, and lastly, a clear viscous lyotropic phase was obtained. A 2H 1D spectrum was recorded on this sample, and a residual quadrupolar splitting from the 2H nucleus of HDO molecules of 43 Hz was observed. Subsequently, 300 μl of the same buffer lacking the protein was added to dilute the mixture to 5% C8E5.
A similar 10% C8E5/octanol mixture was then prepared and diluted to 5% with buffer containing 1.6 mM 15N-labeled CytRN (sample 10). RDC measurements were collected on this sample as described below. The 2H residual quadrupolar splitting for the final sample was 30.2 Hz.
NMR data collection and analysis
NMR data collection and processing
All NMR experiments were carried out at 287 K using a 14.1-T (1H Larmor frequency of 600 MHz) Agilent DD2 spectrometer equipped with cryogenically cooled or room temperature single-axis gradient triple resonance probes or a 16.4-T (1H Larmor frequency of 700 MHz) Bruker Avance Neo spectrometer equipped with a room temperature TXI single-axis gradient triple resonance probe. NMR spectra were processed using NMRPipe (84) and visualized using the NMRDraw (84) and NMRFAM-Sparky software packages (85).
Backbone resonance assignments of CytRN
The backbone chemical shifts of WT (sample 1) and A29V/A48M CytRN (sample 13) were assigned using a 2D 1H-15N HSQC and standard 3D triple resonance datasets HNCACB, CBCA(CO)NH, HN(CA)CO, and HNCO (86). Assignments of WT CytRN were directly transferable to the 1H-15N HSQC spectra of A29V (fig. S21) and V44A CytRN (fig. S23), as the CSPs to the disordered state are very small (0.078 and 0.247 ppm in 15N and 0.004 and 0.07 ppm in 1H averaged over 55 residues for A29V and 51 residues for V44A CytRN, respectively).
CEST data acquisition
15N, 1HN, 13Cα, and 13C′ CEST data were acquired on U-15N,13C samples of CytRN (samples 1 and 3) using pulse sequences reported in literature (40–43). The details of the experiments are summarized in table S4. The B1 field was calibrated using the method reported by Guenneugues et al. (87). CEST profiles were obtained as a plot of the offset frequency versus the ratio of the peak intensity (I/I0) where I and I0 are the intensities obtained in the presence and absence of an exchange duration in the CEST pulse sequence. 15N CEST profiles of A29V and V44A CytRN were acquired using samples 11 and 12, respectively. Variable temperature 15N CEST data were acquired on sample 2 using the DANTE-CEST (D-CEST) pulse sequence (60).
Excited state RDC measurements were made using separate isotropic and aligned samples for each alignment medium (samples 7 to 10). 15N CEST data for RDC measurements were acquired by setting the power level of the 1H decoupling module during Tex to 0 W. 1HN CEST profiles were collected without any modification to the previously reported pulse sequence (41). Details of data acquisition are reported in table S6.
15N CEST data on DNA-bound CytRN (sample 5) were acquired using the D-CEST pulse sequence (60). D-CEST and DRD-CEST data were collected on a Bruker 700-MHz spectrometer. The DANTE train consisted of pulses applied at a radiofrequency field of 3.57 kHz. DRD-CEST measurements were carried out by keeping DANTE sweep window (swDANTE) fixed to a value of 254 Hz, which corresponds to the separation in 15N frequency between the excited state and the DNA-bound state resonances of D34 (table S8).
CPMG data acquisition
CPMG experiments probing the temperature dependence of the excited state population and lifetime were acquired on sample 2 at 287 K (700 MHz, 800 MHz), 295 K (700 MHz), and 302 K (700 MHz), while CPMG profiles in the presence and absence of DNA were acquired on sample 6 (600 MHz). All CPMG data were acquired using a constant-time CPMG pulse sequence where 15N coherence is in-phase at the start of the 30-ms exchange period (88). CPMG pulsing frequencies ranged from 33 to 1000 Hz. CPMG profiles are plotted as R2,eff versus νCPMG, where
where I and I0 are peak intensities in the presence or absence of the exchange period.
Analysis of CEST profiles
15N CEST of WT CytRN without DNA
15N CEST data acquired at two B1 fields for WT CytRN were fit to the two-state Bloch-McConnell equations (89) using the software package ChemEx (https://github.com/gbouvignies/ChemEx). Twelve residues, A16, A19, S22, A24, T25, N32, T40, R49, G52, Y53, L54, and Q56, were selected since they had isolated resonances in the 1H-15N HSQC that showed well-separated major and minor dips in CEST profiles, and these residues were used to get estimates of pE and kex. Monte Carlo error distributions of pE and kex were obtained by fitting 1000 replica datasets to the two-state Bloch-McConnell equations using the same fitting protocol described above. These replicas were constructed from the fit parameters by adding random noise with zero mean and the same SD as the error in I/I0, assuming that the noise originates from an underlying Gaussian distribution. plots for pE (kex) were constructed by keeping pE (kex) fixed at various values during the fitting routine and repeating the fit.
For some of the overlapped resonances, the overlap was resolved at 298 K, and CEST profiles acquired at 298 K were used to assign excited state chemical shifts to each of the overlapped residues. In a subset of these cases, CEST profiles showed two minor dips, one originating from each of the two overlapped peaks. In such cases, CEST profiles were fit to a sum of Lorentzians to extract residue-specific chemical shifts of the excited state. Six residues were eliminated from the analysis because they were severely overlapped in the HSQC spectra and the overlap could not be resolved at higher temperature.
15N CEST of A29V and V44A CytRN
15N CEST profiles of A29V CytRN were fit to a two-state model of conformational exchange using ChemEx as described for WT CytRN. Eight residues (A16, A19, S22, T25, T40, Y53, L54 and Q56) were chosen for the global fitting routine to determine pE and kex. Since only one intensity dip was present in all CEST profiles of V44A CytRN, these CEST profiles could not be fit to the two-state Bloch-McConnell equations. Instead, an upper limit to the population of the excited state was estimated as described in the legend to fig. S23.
13Cα, 13C′, and 1HN data
13Cα, 13C′, and 1HN chemical shifts of the CytRN excited state were extracted from the corresponding CEST profiles, either by fitting the data to a two-state exchange model within ChemEx or by fitting the data to a sum of Lorentzians. In case of 1HN CEST, each dip is a difference of two Lorentzians separated by 1JNH, which was kept fixed at a value of 93 Hz during the fitting procedure.
15N and 1HN CEST data for measuring RDCs
15N and 1HN CEST profiles acquired on isotropic and aligned samples were modeled as linear combinations of Lorentzians, where each dip in 15N (1HN) CEST is a sum (difference) of two Lorentzians. The frequency difference between the two Lorentzians for a dip gives the 1JNH value for the isotropic sample and the 1JNH + RDC value for the aligned sample.
D- and DRD-CEST data
D- and DRD-CEST data acquired on DNA-bound 15N CytRN were modeled using three-state Bloch-McConnell equations within the ChemEx software package. D-CEST data collected at five B1 fields (8.8, 15.9, 24.8, 28.7, and 34.7 Hz) and DRD-CEST data collected at three B1 fields (25.0, 28.9, and 34.9 Hz) for D34 were globally fit to either linear (D↔E↔B and E↔D↔B) or triangular models of chemical exchange. In each modeling protocol, all parameters except kex,DE were allowed to float while kex,DE was fixed to the value obtained in the absence of DNA. The excited state population (pE) was not fixed to the value in the absence of DNA because there are indications in literature that the folded conformation of CytRN is stabilized by the electrostatic field of the DNA molecule (90). surfaces for kex,EB and kex,DB were constructed by fixing the respective parameters to specific values and determining the from the fit.
Analysis of CPMG profiles
CPMG profiles were fit to the two-state model of conformational exchange using the ChemEx. At each temperature, nonoverlapped peaks with acceptable signal-to-noise having Rex > 5 s−1 were chosen for analysis. Profiles at one (295 K, 302 K) or two B0 fields (287 K) were fit globally either with (295 K, 302 K) or without CEST data (287 K) to extract populations and kex,DE values.
Chemical shift perturbations
CSPs between the excited state and the disordered conformation were calculated from CEST-derived 15N, 1HN, 13Cα, and 13C′ chemical shifts for the excited state and assigned backbone shifts of the native state using the formula
Here, i is the residue index and CSPi are the residue-specific CSPs. Ni is the number of nuclei for the ith residue for which excited state chemical shifts are available, and j is the index that runs from 1 to Ni. Δωij are the chemical shift differences between the excited and ground states for the ith residue and the jth nucleus, while Δωj,RMS is 1 SD of the distribution in protein chemical shifts for the 15N (6.10 ppm), 1HN (0.93 ppm), 13Cα (2.94 ppm), or 13C′ (4.07 ppm) nuclei based on data deposited in the Biological Magnetic Resonance Databank (BMRB).
Secondary structural propensity
SSP calculations were performed using the SSP software package. While 13Cα, 13Cβ, 1HN, 15N, and 1Hα chemical shifts obtained from BMRB (accession number 17419) were used to determine SSP scores for the DNA-bound state, only 13Cα, 13Cβ, and 1Hα chemical shifts were used for the disordered form (32).
Acknowledgments
We thank L. E. Kay (University of Toronto) for providing the pulse sequence codes used in this work; N. Sgourakis, S. Nerli, J. Nagesh, and S. Sankaran for help with CS-Rosetta; and L. E. Kay, P. Vallurupalli, S. Sarma, B. Sathyamoorthy, and R. Rosenzweig for stimulating discussions. D. K. Chaurasiya is gratefully acknowledged for carrying out Monte Carlo sampling. Funding: This work was supported by DBT/Wellcome Trust India Alliance Fellowship IA/I/18/1/503614 (A.S.); DST/SERB Core Research Grant CRG/2019/003457 (A.S.); Start-up grant from the Indian Institute of Science Bangalore (A.S.); infrastructural support from the following programs of the Government of India: DST-FIST, UGC-CAS, and the DBT-IISc partnership program (A.S.); fellowship support from CSIR (K.M.); and fellowship support from the Indian Institute of Science Bangalore (B.N.).
Author contributions: Conceptualization: A.N.N. and A.S. Methodology: K.M. and A.S. Investigation: K.M., B.N., S.M., A.N.N., and A.S. Visualization: K.M., B.N., and A.S. Funding acquisition: A.N.N. and A.S. Project administration: A.N.N. and A.S. Supervision: A.S. Writing—original draft: A.S. Writing—review and editing: K.M., B.N., S.M., A.N.N., and A.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The backbone resonance assignments of WT CytRN are deposited in the BMRB (accession number: 51449).
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S29
Tables S1 to S8
References
REFERENCES AND NOTES
- 1.Uversky V. N., Intrinsically disordered proteins and their “mysterious”(Meta) physics. Front. Phys. 7, 10 (2019). [Google Scholar]
- 2.Wright P. E., Dyson H. J., Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 16, 18–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babu M. M., The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem. Soc. Trans. 44, 1185–1200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.N. Bolik-Coulon, G. Bouvignies, L. Carlier, F. Ferrage, in Intrinsically Disordered Proteins. (Elsevier, 2019), pp. 65–92. [Google Scholar]
- 5.Gianni S., Dogan J., Jemth P., Coupled binding and folding of intrinsically disordered proteins: What can we learn from kinetics? Curr. Opin. Struct. Biol. 36, 18–24 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Fisher C. K., Stultz C. M., Constructing ensembles for intrinsically disordered proteins. Curr. Opin. Struct. Biol. 21, 426–431 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiefhaber T., Bachmann A., Jensen K. S., Dynamics and mechanisms of coupled protein folding and binding reactions. Curr. Opin. Struct. Biol. 22, 21–29 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Wright P. E., Dyson H. J., Linking folding and binding. Curr. Opin. Struct. Biol. 19, 31–38 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bah A., Vernon R. M., Siddiqui Z., Krzeminski M., Muhandiram R., Zhao C., Sonenberg N., Kay L. E., Forman-Kay J. D., Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 519, 106–109 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Vernon R. M., Forman-Kay J. D., First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol. 58, 88–96 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Riback J. A., Zhu L., Ferrolino M. C., Tolbert M., Mitrea D. M., Sanders D. W., Wei M. T., Kriwacki R. W., Brangwynne C. P., Composition-dependent thermodynamics of intracellular phase separation. Nature 581, 209–214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi J.-M., Pappu R. V., The stickers and spacers framework for describing phase behavior of multivalent intrinsically disordered proteins. Biophys. J. 118, 492a (2020).31839263 [Google Scholar]
- 13.C. J. Oldfield, V. N. Uversky, A. K. Dunker, L. Kurgan, in Intrinsically Disordered Proteins. (Elsevier, 2019), pp. 1–34. [Google Scholar]
- 14.Smith N. C., Kuravsky M., Shammas S. L., Matthews J. M., Binding and folding in transcriptional complexes. Curr. Opin. Struct. Biol. 66, 156–162 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Shammas S. L., Crabtree M. D., Dahal L., Wicky B. I., Clarke J., Insights into coupled folding and binding mechanisms from kinetic studies. J. Biol. Chem. 291, 6689–6695 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugase K., Dyson H. J., Wright P. E., Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447, 1021–1025 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Robustelli P., Piana S., Shaw D. E., Mechanism of coupled folding-upon-binding of an intrinsically disordered protein. J. Am. Chem. Soc. 142, 11092–11101 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Giri R., Morrone A., Toto A., Brunori M., Gianni S., Structure of the transition state for the binding of c-Myb and KIX highlights an unexpected order for a disordered system. Proc. Natl. Acad. Sci. U.S.A. 110, 14942–14947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogers J. M., Oleinikovas V., Shammas S. L., Wong C. T., de Sancho D., Baker C. M., Clarke J., Interplay between partner and ligand facilitates the folding and binding of an intrinsically disordered protein. Proc. Natl. Acad. Sci. U.S.A. 111, 15420–15425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider R., Blackledge M., Jensen M. R., Elucidating binding mechanisms and dynamics of intrinsically disordered protein complexes using NMR spectroscopy. Curr. Opin. Struct. Biol. 54, 10–18 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Rogers J. M., Wong C. T., Clarke J., Coupled folding and binding of the disordered protein PUMA does not require particular residual structure. J. Am. Chem. Soc. 136, 5197–5200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tretyachenko-Ladokhina V., Cocco M. J., Senear D. F., Flexibility and adaptability in binding of E. coli cytidine repressor to different operators suggests a role in differential gene regulation. J. Mol. Biol. 362, 271–286 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Lewis M., The lac repressor. C. R. Biol. 328, 521–548 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Weickert M. J., Adhya S., A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 267, 15869–15874 (1992). [PubMed] [Google Scholar]
- 25.Barbier C. S., Short S. A., Senear D. F., Allosteric mechanism of induction of CytR-regulated gene expression: CytR REPRESSOR-CYTIDINE INTERACTION. J. Biol. Chem. 272, 16962–16971 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Slijper M., Bonvin A., Boelens R., Kaptein R., Refined structure of lac repressor headpiece (1-56) determined by relaxation matrix calculations from 2D and 3D NOE data: Change of tertiary structure upon binding to the lac operator. J. Mol. Biol. 259, 761–773 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Nagadoi A., Morikawa S., Nakamura H., Enari M., Kobayashi K., Yamamoto H., Sampei G., Mizobuchi K., Schumacher M. A., Brennan R. G., Nishimura Y., Structural comparison of the free and DNA-bound forms of the purine repressor DNA-binding domain. Structure 3, 1217–1224 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Penin F., Geourjon C., Montserret R., Böckmann A., Lesage A., Yang Y. S., Bonod-Bidaud C., Cortay J. C., Nègre D., Cozzone A. J., Deléage G., Three-dimensional structure of the DNA-binding domain of the fructose repressor from Escherichia coli by 1H and 15N NMR. J. Mol. Biol. 270, 496–510 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Romero P., Obradovic Z., Dunker A. K., Sequence data analysis for long disordered regions prediction in the calcineurin family. Genome Inform. 8, 110–124 (1997). [PubMed] [Google Scholar]
- 30.Li X., Romero P., Rani M., Dunker A. K., Obradovic Z., Predicting protein disorder for N-, C-and internal regions. Genome Inform. 10, 30–40 (1999). [PubMed] [Google Scholar]
- 31.Dosztányi Z., Prediction of protein disorder based on IUPred. Protein Sci. 27, 331–340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsh J. A., Singh V. K., Jia Z., Forman-Kay J. D., Sensitivity of secondary structure propensities to sequence differences between α-and γ-synuclein: Implications for fibrillation. Protein Sci. 15, 2795–2804 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moody C. L., Tretyachenko-Ladokhina V., Laue T. M., Senear D. F., Cocco M. J., Multiple conformations of the cytidine repressor DNA-binding domain coalesce to one upon recognition of a specific DNA surface. Biochemistry 50, 6622–6632 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Sekhar A., Kay L. E., An NMR view of protein dynamics in health and disease. Annu. Rev. Biophys. 48, 297–319 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Munshi S., Rajendran D., Naganathan A. N., Entropic control of an excited folded-like conformation in a disordered protein ensemble. J. Mol. Biol. 430, 2688–2694 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vallurupalli P., Sekhar A., Yuwen T., Kay L. E., Probing conformational dynamics in biomolecules via chemical exchange saturation transfer: A primer. J. Biomol. NMR 67, 243–271 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Sekhar A., Kay L. E., NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc. Natl. Acad. Sci. U.S.A. 110, 12867–12874 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitra S., Oikawa H., Rajendran D., Kowada T., Mizukami S., Naganathan A. N., Takahashi S., Flexible target recognition of the intrinsically disordered DNA-binding domain of CytR monitored by single-molecule fluorescence spectroscopy. J. Phys. Chem. B 126, 6136–6147 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naganathan A. N., Orozco M., The conformational landscape of an intrinsically disordered DNA-binding domain of a transcription regulator. J. Phys. Chem. B 117, 13842–13850 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Vallurupalli P., Bouvignies G., Kay L. E., Studying “invisible” excited protein states in slow exchange with a major state conformation. J. Am. Chem. Soc. 134, 8148–8161 [DOI] [PubMed] [Google Scholar]
- 41.Yuwen T., Sekhar A., Kay L. E., Separating dipolar and chemical exchange magnetization transfer processes in 1H‐CEST. Angew. Chem. Int. Ed. 56, 6122–6125 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Long D., Sekhar A., Kay L. E., Triple resonance-based 13Cα and 13Cβ CEST experiments for studies of ms timescale dynamics in proteins. J. Biomol. NMR 60, 203–208 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Vallurupalli P., Kay L. E., Probing slow chemical exchange at carbonyl sites in proteins by chemical exchange saturation transfer NMR spectroscopy. Angew. Chem. Int. Ed. 52, 4156–4159 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Wishart D. S., Interpreting protein chemical shift data. Prog. Nucl. Magn. Reson. Spectrosc. 58, 62–87 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Kalodimos C. G., Boelens R., Kaptein R., Toward an integrated model of protein−DNA Recognition as inferred from NMR studies on the lac repressor system. Chem. Rev. 104, 3567–3586 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Berjanskii M. V., Wishart D. S., A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc. 127, 14970–14971 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Prestegard J. H., Al-Hashimi H., Tolman J. R., NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q. Rev. Biophys. 33, 371–424 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Chen K., Tjandra N., The use of residual dipolar coupling in studying proteins by NMR. Top Curr. Chem. 326, 47–67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao B., Zhang Q., Measuring residual dipolar couplings in excited conformational states of nucleic acids by CEST NMR spectroscopy. J. Am. Chem. Soc. 137, 13480–13483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chou J. J., Gaemers S., Howder B., Louis J. M., Bax A., A simple apparatus for generating stretched polyacrylamide gels, yielding uniform alignment of proteins and detergent micelles. J. Biomol. NMR 21, 377–382 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Rückert M., Otting G., Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 122, 7793–7797 (2000). [Google Scholar]
- 52.Shen Y., Lange O., Delaglio F., Rossi P., Aramini J. M., Liu G., Eletsky A., Wu Y., Singarapu K. K., Lemak A., Ignatchenko A., Arrowsmith C. H., Szyperski T., Montelione G. T., Baker D., Bax A., Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. U.S.A. 105, 4685–4690 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nerli S., Sgourakis N. G., CS-ROSETTA. Methods Enzymol. 614, 321–362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Y., Bax A., SPARTA+: A modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR 48, 13–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zweckstetter M., NMR: Prediction of molecular alignment from structure using the PALES software. Nat. Protoc. 3, 679–690 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Munshi S., Subramanian S., Ramesh S., Golla H., Kalivarathan D., Kulkarni M., Campos L. A., Sekhar A., Naganathan A. N., Engineering order and cooperativity in a disordered protein. Biochemistry 58, 2389–2397 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L., The FoldX web server: An online force field. Nucleic Acids Res. 33, W382–W388 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Changeux J.-P., Edelstein S., Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol. Rep. 3, 19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vallurupalli P., Tiwari V. P., Ghosh S., A double resonance CEST experiment to study multistate protein conformational exchange: An application to protein folding. J. Phys. Chem. Lett. 10, 3051–3056 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Yuwen T., Kay L. E., Bouvignies G., Dramatic decrease in CEST measurement times using multi-site excitation. ChemPhysChem 19, 1707–1710 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Freeman R., Morris G. A., The ‘DANTE’ experiment. J. Magn. Reson. 213, 244–246 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Hammes G. G., Chang Y.-C., Oas T. G., Conformational selection or induced fit: A flux description of reaction mechanism. Proc. Natl. Acad. Sci. U.S.A. 106, 13737–13741 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cordier F., Hartmann B., Rogowski M., Affolter M., Grzesiek S., DNA recognition by the brinker repressor – An extreme case of coupling between binding and folding. J. Mol. Biol. 361, 659–672 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Bell C. E., Lewis M., A closer view of the conformation of the Lac repressor bound to operator. Nat. Struct. Biol. 7, 209–214 (2000). [DOI] [PubMed] [Google Scholar]
- 65.Kalodimos C. G., Bonvin A. M., Salinas R. K., Wechselberger R., Boelens R., Kaptein R., Plasticity in protein–DNA recognition: lac repressor interacts with its natural operator O1 through alternative conformations of its DNA-binding domain. EMBO J. 21, 2866–2876 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mohan A., Oldfield C. J., Radivojac P., Vacic V., Cortese M. S., Dunker A. K., Uversky V. N., Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 362, 1043–1059 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Onitsuka M., Kamikubo H., Yamazaki Y., Kataoka M., Mechanism of induced folding: Both folding before binding and binding before folding can be realized in staphylococcal nuclease mutants. Proteins 72, 837–847 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Schneider R., Maurin D., Communie G., Kragelj J., Hansen D. F., Ruigrok R. W. H., Jensen M. R., Blackledge M., Visualizing the molecular recognition trajectory of an intrinsically disordered protein using multinuclear relaxation dispersion NMR. J. Am. Chem. Soc. 137, 1220–1229 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Jensen M. R., Houben K., Lescop E., Blanchard L., Ruigrok R. W. H., Blackledge M., Quantitative conformational analysis of partially folded proteins from residual dipolar couplings: Application to the molecular recognition element of Sendai virus nucleoprotein. J. Am. Chem. Soc. 130, 8055–8061 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Song J., Guo L. W., Muradov H., Artemyev N. O., Ruoho A. E., Markley J. L., Intrinsically disordered γ-subunit of cGMP phosphodiesterase encodes functionally relevant transient secondary and tertiary structure. Proc. Natl. Acad. Sci. U.S.A. 105, 1505–1510 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim A. S., Kakalis L. T., Abdul-Manan N., Liu G. A., Rosen M. K., Autoinhibition and activation mechanisms of the Wiskott–Aldrich syndrome protein. Nature 404, 151–158 (2000). [DOI] [PubMed] [Google Scholar]
- 72.Cohan M. C., Pappu R. V., Making the case for disordered proteins and biomolecular condensates in bacteria. Trends Biochem. Sci. 45, 668–680 (2020). [DOI] [PubMed] [Google Scholar]
- 73.Aravind L., Anantharaman V., Balaji S., Babu M. M., Iyer L. M., The many faces of the helix-turn-helix domain: Transcription regulation and beyond. FEMS Microbiol. Rev. 29, 231–262 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Alberstein R. G., Guo A. B., Kortemme T., Design principles of protein switches. Curr. Opin. Struct. Biol. 72, 71–78 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou H.-X., Intrinsic disorder: Signaling via highly specific but short-lived association. Trends Biochem. Sci. 37, 43–48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shammas S. L., Mechanistic roles of protein disorder within transcription. Curr. Opin. Struct. Biol. 42, 155–161 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Teilum K., Olsen J. G., Kragelund B. B., Globular and disordered–The non-identical twins in protein-protein interactions. Front. Mol. Biosci. 2, 40 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shammas S. L., Travis A. J., Clarke J., Allostery within a transcription coactivator is predominantly mediated through dissociation rate constants. Proc. Natl. Acad. Sci. U.S.A. 111, 12055–12060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jayaram B., Jain T., The role of water in protein-DNA recognition. Annu. Rev. Biophys. Biomol. Struct. 33, 343–361 (2004). [DOI] [PubMed] [Google Scholar]
- 80.Dragan A. I., Li Z., Makeyeva E. N., Milgotina E. I., Liu Y., Crane-Robinson C., Privalov P. L., Forces driving the binding of homeodomains to DNA. Biochemistry 45, 141–151 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Hayashi T., Oshima H., Mashima T., Nagata T., Katahira M., Kinoshita M., Binding of an RNA aptamer and a partial peptide of a prion protein: Crucial importance of water entropy in molecular recognition. Nucleic Acids Res. 42, 6861–6875 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen J., Kriwacki R. W., Intrinsically disordered proteins: Structure, function and therapeutics. J. Mol. Biol. 430, 2275–2277 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Munshi S., Gopi S., Subramanian S., Campos L. A., Naganathan A. N., Protein plasticity driven by disorder and collapse governs the heterogeneous binding of CytR to DNA. Nucleic Acids Res. 46, 4044–4053 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A., NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 85.Lee W., Tonelli M., Markley J. L., NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.J. Cavanagh, W. J. Fairbrother, A. G. Palmer III, N. J. Skelton, Protein NMR spectroscopy: Principles and Practice, (Elsevier, Academic Press, 1995), pp. 587. [Google Scholar]
- 87.Guenneugues M., Berthault P., Desvaux H., A method for determining B1 field inhomogeneity. Are the biases assumed in heteronuclear relaxation experiments usually underestimated? J. Magn. Reson. 136, 118–126 (1999). [DOI] [PubMed] [Google Scholar]
- 88.Hansen D. F., Vallurupalli P., Kay L. E., An improved 15N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. J. Phys. Chem. B 112, 5898–5904 (2008). [DOI] [PubMed] [Google Scholar]
- 89.McConnell H. M., Reaction rates by nuclear magnetic resonance. J. Chem. Phys. 28, 430 (1958). [Google Scholar]
- 90.Munshi S., Gopi S., Asampille G., Subramanian S., Campos L. A., Atreya H. S., Naganathan A. N., Tunable order–Disorder continuum in protein–DNA interactions. Nucleic Acids Res. 46, 8700–8709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yuwen T., Kay L. E., A new class of CEST experiment based on selecting different magnetization components at the start and end of the CEST relaxation element: An application to 1H CEST. J. Biomol. NMR 70, 93–102 (2018). [DOI] [PubMed] [Google Scholar]
- 92.Y. Shen, A. Bax, in Artificial neural networks. (Springer, 2015), pp. 17–32. [Google Scholar]
- 93.Korzhnev D. M., Religa T. L., Banachewicz W., Fersht A. R., Kay L. E., A transient and low-populated protein-folding intermediate at atomic resolution. Science 329, 1312–1316 (2010). [DOI] [PubMed] [Google Scholar]
- 94.Bhattacharya A., Tejero R., Montelione G. T., Evaluating protein structures determined by structural genomics consortia. Proteins: Struct. Funct. Bioinform. 66, 778–795 (2007). [DOI] [PubMed] [Google Scholar]
- 95.Cornilescu G., Marquardt J. L., Ottiger M., Bax A., Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J. Am. Chem. Soc. 120, 6836–6837 (1998). [Google Scholar]
- 96.Sass H.-J., Musco G., Stahl S. J., Wingfield P. T., Grzesiek S., Solution NMR of proteins within polyacrylamide gels: Diffusional properties and residual alignment by mechanical stress or embedding of oriented purple membranes. J. Biomol. NMR 18, 303–309 (2000). [DOI] [PubMed] [Google Scholar]
- 97.Cayley S., Lewis B. A., Guttman H. J., Record M. T. Jr., Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity: Implications for protein-DNA interactions in vivo. J. Mol. Biol. 222, 281–300 (1991). [DOI] [PubMed] [Google Scholar]
- 98.Stickle D. F., Vossen K. M., Riley D. A., Fried M. G., Free DNA concentration in E. coli estimated by an analysis of competition for DNA binding proteins. J. Theor. Biol. 168, 1–12 (1994). [DOI] [PubMed] [Google Scholar]
- 99.Vitalis A., Pappu R. V., ABSINTH: A new continuum solvation model for simulations of polypeptides in aqueous solutions. J. Comput. Chem. 30, 673–699 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sickmeier M., Hamilton J. A., LeGall T., Vacic V., Cortese M. S., Tantos A., Szabo B., Tompa P., Chen J., Uversky V. N., Obradovic Z., Dunker A. K., DisProt: The database of disordered proteins. Nucleic Acids Res. 35, D786–D793 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tamiola K., Mulder F. A., Using NMR chemical shifts to calculate the propensity for structural order and disorder in proteins. Biochem. Soc. Trans. 40, 1014–1020 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S29
Tables S1 to S8
References