Abstract
Irritable bowel syndrome (IBS) is a complex multifactorial condition including alterations of the gut–brain axis, intestinal permeability, mucosal neuro-immune interactions, and microbiota imbalance. Recent advances proposed epigenetic factors as possible regulators of several mechanisms involved in IBS pathophysiology. These epigenetic factors include biomolecular mechanisms inducing chromosome-related and heritable changes in gene expression regardless of DNA coding sequence. Accordingly, altered gut microbiota may increase the production of metabolites such as sodium butyrate, a prominent inhibitor of histone deacetylases. Patients with IBS showed an increased amount of butyrate-producing microbial phila as well as an altered profile of methylated genes and micro-RNAs (miRNAs). Importantly, gene acetylation as well as specific miRNA profiles are involved in different IBS mechanisms and may be applied for future diagnostic purposes, especially to detect increased gut permeability and visceromotor dysfunctions. In this review, we summarize current knowledge of the role of epigenetics in IBS pathophysiology.
Keywords: Irritable bowel syndrome, Epigenetic, DNA methylation, miRNA
Introduction
Irritable bowel syndrome (IBS) is a disorder of gut–brain interaction (DGBI) characterized by recurrent abdominal pain associated with defecation or change in bowel habits [1]. According to ROME IV criteria, four IBS subgroups are identified: IBS with diarrhoea (IBS-D), IBS with constipation (IBS-C), IBS with mixed bowel habits (IBS-M), and unclassified IBS (IBS-U) [2]. A large subgroup of IBS patients experiences extra-intestinal symptoms, including psychiatric and mood disorders [3–5]. Early life adverse events (EAEs), comprising psychological and physical stress as well as traumatic experiences during childhood have been identified as a predisposing factor for IBS development [6, 7]. Several factors are involved in the pathophysiology of IBS including unbalanced gut microbiota [8], low-grade immune activation [9], overactive serotonergic system [10], and intestinal barrier dysfunction [11]. Regarding immune system involvement, the observation of mucosal infiltration with immune cells, in particular mast cells, in IBS patients has been backed up by mechanistic evidence. This demonstrates an effect on the epithelial permeability, and enteric nervous system function [12–16]. More recently, epigenetic modifications such as chromatin remodelling, DNA methylation, and non-coding RNAs have been indicated among the players involved in IBS development (Tables 1 and 2) [17–21]. Additionally, gut microbiota can modulate the intestinal host’s gene expression by its metabolites and in turn can be epigenetically regulated by the host. Additionally, gut microbiota can modulate the intestinal host’s gene expression by its metabolites [22] and in turn can be epigenetically regulated by the host [23].
Table 1.
Main ncRNAs involved in IBS pathophysiology and identified as regulators of IBS mechanisms
| miRNA [ref] | miRNA expression (↓ or ↑) in IBS vs HC | Target gene or pathway (identified in human samples and/or cells)# | Observed correlation with GI function | Clinical condition and samples analysed (n); (human, cells, and animal model) |
|---|---|---|---|---|
| Human studies | ||||
|
miR16 [112] |
↓ | 5HT-4 | Motility and stool form | IBS-D (n 14) & HC (n 17); Jejunum biopsies |
|
miR-16a [19] |
↓ | CLDN2 | Increased IP | IBS-D (n 43) & HC (n 23); Proximal jejunum biopsies (Watson capsule) |
|
miR-24 [114] |
↑ | SERT | Increased VHS | IBS (n 10) & HC (n 10); intestinal mucosa biopsies |
| miR-29a | ||||
| [74] | ↑ | GLUL | Increased IP | IBS-D (n 19) & HC (n 10); duodenum and colon biopsies and blood microvesicles |
| [77] | ↑ | ZO-1 & CLDN1 | Increased IP | IBS-D (n 21) & HC (n 16); colon biopsies |
| [118] | ↑ | HTR7 | Increased VHA | IBS (n 10) & HC (n 10); colon biopsies |
|
miR-29a/b [75] |
↑ | CLDN1 & NKRF | Increased IP | IBS-C (n 74), IBS-D (n 109), & HC (n 36); Colon and jejunal biopsies |
|
miR-103 [112] |
↓ | 5HT-4 | Motility and stool form | IBS-D (n 14) & HC (n 17); Jejunum biopsies |
|
miR-125b [19] |
↓ | CGN | Increased IP | IBS-D (n 43) & HC (n 23); Proximal jejunum biopsies (Watson capsule) |
| miR-148b-5p | ||||
| [82] | ↑ | RGS-2 | Increased IP | IBS-D (n 20) & HC (n 20); HT-29 cells cultured with serum exosomes (IBS-exo & HC-exo) |
| [95] | ↑ | § | IBS-C (n 14), IBS-D (n 17), & HC (n 30); | |
|
miR-199a [18] |
↓ | TRPV1 | Increased VHS | IBS-D (n 45) & HC (n 40); colon biopsies |
|
miR-199b [66] |
↓ | / | Increased coliform count | IBS-C (n 20), IBS-D (n 18), IBS-M (n 32) & HC (n 20); serum |
|
miR-219a-5p [81] |
↓ | ITGB1BP1, ABC transports C1 & C5, CAMK1D | Impaired barrier function | IBS-C (n 15), IBS-D (n 14), & HC (n 15); Colon biopsies |
|
miR-338-3p [81] |
↓ | MAPK signaling | Increased VHS | IBS-C (n 15), IBS-D (n 14), & HC (n 15); Colon biopsies |
|
miR-510 [111] |
δ; functional variant 5HT3 | Increased 5HT3E expression | Symptoms associated with female IBS-D |
IBS-C (n 100), IBS-D (n 200), & HC (n 100)—discovery study; IBS-D (n 119), & HC (n 195) – a replication study |
|
lncRNA H19 [86] |
↑ | §z | Increased IP | IBS-D (n 10), & HC (n 10); |
| Animal studies | ||||
|
miR-21-5p [65] |
↑ | PTEN & PDCD4 | Increased IP | GF mice & Conventional mice |
|
miR-24 [114] |
↑ | SERT | Increased VHS and inflammation | TNBS-induced IBS mouse model & Control |
| miR-29a | ||||
| [76] | ↑ | CLDN1, Collagen I, collagen IV, FBN & ITGB1, | Increased IP | IUGR PN (n 6) & NPN (n 12) |
| [77] | ↑ | ZO-1 & CLDN1 | Increased IP | TNBS-induced IBS-D model & Control |
| [118] | ↑ | HTR7 | Increased VHA | WAS-induced IBS models: miR-29a knockout & wild-type mice |
|
miR-29/a/b [75] |
↑ | CLDN1 & NKRF | Increased IP | TNBS & WAS-induced IBS models: miR-29a/b knockout & wild-type mice |
|
miR-144 [78] |
↑ | Occludin & ZO-1 | Increased IP | Acid acetic-induced IBS-D rat model (n 20) & Control (n 20) |
|
miR-181c-5p [101] |
↑ | IL1A, TNFα, IL2, IL6 | Decreased low-grade inflammation | AWR-induced IBS rat model (n 56) & Control (n 15) |
|
miR-200a [79] |
↑ | CNR1 & SERT | Increased VHA | Unpredictable chronic stress-induced IBS-D rat model (n 20) & Control (n 20) |
|
miR-325-5p [141] |
↓ | CCL2 | Increased VHS | Acid acetic rat model & CRD used to induce pain behavior |
|
miR-495 [140] |
↓ | PI3K/AKT path via PKIB | Increased VHS | Acid acetic-induced IBS-D mouse model (n 10) & Control (n 10) |
| In vitro studies | ||||
|
miR200b [80] |
↑ | TNFα | Decreased TEER & paracellular permeability | Caco2 cell line |
|
miR-490-5p [91] |
↑ | Tryptase & PAR2 | Promotes cell proliferation and resistance to apoptosis | p815 mast cell line |
#Target gene expression inversely correlated with miRNAs expression; §Target not investigated; δNo change in miRNA expression but disruption of an miRNA-binding site;
AWR abdominal withdraw reflection, CAMK1D calcium/calmodulin dependent protein kinase ID, CCL2 C–C motif chemokine ligand 2, CLDN1 Claudin 1, CLDN2 Claudin 2, CGN Cingulin, CRD colorectal distention, FBN fibronectin, IP intestinal permeability, ITGB1 Integrin-β1, ITGB1BP1 integrin subunit beta 1 binding protein 1, IUGR PN intrauterine growth restricted porcine neonates, NPN normal porcine neonates, RGS-2 regulator of G protein signaling-2, TEER transepithelial electrical resistance TNBS 2,4,6- trinitrobenzenesulfonic, VHA visceral hyperalgesia, VHS visceral hypersensitivity, WAS water avoidance stress
Table 2.
Gene methylations involved in IBS mechanisms
| Methylated Genes | Expression (↓ or ↑) | Regulation &/or observed function | Biomarkers analysed | Clinical condition and samples analysed (n); (human, cells and animal model); biological sources | Reference |
|---|---|---|---|---|---|
| Human studies | |||||
| SSPO, GSTM1, GSTM5, TPPP | ↑ | Association of SSPO methylation with high HADs and PSS scores | HAD and PSS scores | IBS-D (n 10), IBS-C (n 8), IBS-M (n 9), HC (n 23); whole blood (PBMCs) | [20] |
| ADCYAP1 | ↓ | ||||
| AKAP12; PRKAR1B | ↑ | ||||
| Animal studies | |||||
| GR, CRF | GR↑, CRF↓ | Increase visceral pain | VMR to CRD; GR, CFR expression | WAS rat model & HC; amygdala tissue | [137] |
| H3K9 methylation in the promoter region of claudin-1, ZOs, and occludin | ↑ | IL-6 promotes H3K9me2/me3, preventing GR transcriptional factors binding on the TJs genes promoter region; with the consequent increase in permeability and visceral pain | IL-6, GR, claudin-1, ZOs, occludin | WAS rat model & HC | [71] |
| GR (NC3R1), CNR1 | ↑ | WAS increased DNMT1-mediated CNR1 methylation | VMR to CRD, GR, TRPV1, CNR1 | WAS rat model & HC | [21] |
CNR1 cannabinoid receptor 1, COX2 cyclooxygenase-2, CRD colorectal distension, CRF corticotropin-releasing factor, DNMT1 DNA methyltransferases 1, GR glucocorticoid receptor, HC healthy control, IFNγ interferon γ, IL-1β interleukin-1β IL-6 interleukin-6, IL-8 interleukin-8, IkB inhibitor of kB, NF-κB nuclear factor kappa-light-chain-enhancer of activated B cells, PBMCs peripheral blood mononuclear cells, PSS perceived stress scale, VMR visceromotor response, WAS water-avoidance stress
In this review, we will focus on current findings about epigenetic mechanisms and non-coding RNA identified in IBS patients and in early life stress animal models, thus revealing possible novel targets for future diagnostic and therapeutic tools.
Epigenetic and non-coding RNAs
The term “epigenetics” refers to the whole biomolecular mechanism inducing chromosome-related, heritable changes of gene expression, regardless of DNA coding sequence [24]. Hence, the current definition comprises upstream modifications of chromatin and DNA structure, occurring at the gene promoter site but not the downstream modulation of transcripts provided by non-coding RNA molecules. However, for convenience, in this review the locution “epigenetic modification/regulation/factors” will also include non-coding RNA functions.
Nucleosomes represent chromatin’s functional units and determine its relaxed or condensed form, namely euchromatin and heterochromatin, respectively. Nucleosomes are octamers composed of 4 couples of histones with the genomic DNA wrapped around each one. Histone acetylation induces histone distancing and a loosened chromatin structure, favouring the access of transcription complexes to the promoter sequence. Conversely, the loss of acetylic groups condenses chromatin structure and inhibits gene transcription (Fig. 1) [25]. Histone acetyl transferase (HATs) and histone deacetylases (HDACs) are the enzymes responsible for the association and detachment of acetylic groups on histones. In addition, histones can undergo several other enzyme-mediated modifications such as methylation, biotinylation, phosphorylation, sumoylation, ubiquitination, and serotonylation (see following paragraph), which is more recently discovered. These processes may either promote or repress gene transcription [26]. Conversely, methylation of genomic DNA on the 5’ terminal in correspondence to cytosine–guanine dinucleotides inhibits the attachment of transcription complexes. DNA methyl transferase (DNMTs) and ten-eleven translocation enzymes mediate cytosine methylation and demethylation, respectively [27].
Fig. 1.
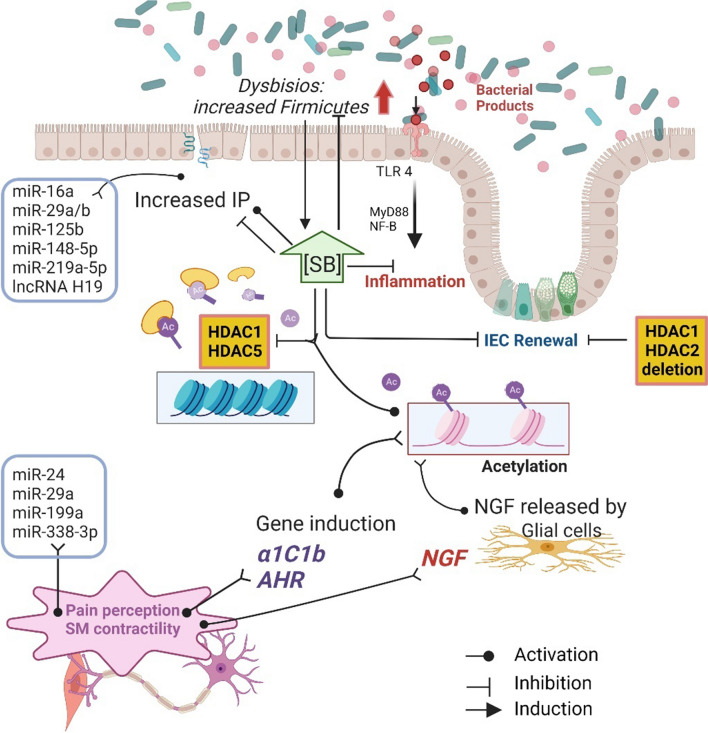
Epigenetic factors involved in gut–brain axis dysregulation – Histone acetylation and miRNA expression triggering biomolecular pathways involved in IBS. The underlying hypothesis envisages miRNA such as miR-24 and miR-29a having a direct involvement in pain perception and increased IP. An akin effect is induced by SB, a great HDAC inhibitor, produced at high levels by Firmicutes. In contrast, high SB levels were shown to restore IP and dysbiosis. Increased histone acetylation elicits NGF production by enteric glial cells resulting in higher SM contractility and pain perception. High SB levels reduce TLR4-mediated inflammation and, allegedly by a dose-dependent mechanism, both impair and restore intestinal permeability. AHR aromatic hydrocarbon receptor, IEC intestinal epithelial cells, NGF nerve growth factor, SB sodium butyrate, VIP vasoactive intestinal protein
Around 90% of the total RNA in a normal cell do not translate genetic information into any protein product. A dramatic step-forward of last decade clarified that this remarkable component of the transcriptome owes regulatory functions. Nowadays, research is focused on the causal links between non-coding RNAs and a vast number of biological functions in health and disease [28, 29]. Among the various categories [30], much attention has been shed on microRNA (miRNA) and long non-coding RNA (lncRNA) and their involvement in inflammatory and functional GI disorders [31, 32]. miRNAs are 20–22-base-long RNA oligomers modulating gene expression by binding complementary mRNAs on specific sequences and inducing the inhibition of the translation or the degradation of mRNA [33, 34]. Differently, lncRNAs are oligo sequences spanning 200–100.000 nucleotides in length, stemming from DNA regions flanking gene promoter sequences. lncRNA can transcribe adjunctive miRNA to form RNA macromolecules with the capacity to absorb target miRNA through a sponge-like mechanism. The function of lncRNAs has not yet been elucidated, but they likely have several regulatory roles, including the modulation of gene expression by chromatin remodelling [35]. Moreover, non-coding RNAs can signal outside the cellular space complexed with RNA-binding proteins or included in extracellular vesicles [36, 37].
Epigenetic and non-coding RNAs in IBS
Host–microbiome interaction
Perturbations of the complex homeostasis between host and intestinal microbial species in IBS have been largely studied in the last decade. Luminal bacteria participate in digestive functions including the digestion of Fermentable, oligo-, di-, monosaccharides, and polyols (FODMAPs). These are the primary source of short-chain fatty acids (SCFAs) which are the key factors in the interplay with the host [17, 38–40] and are likely involved in IBS pathophysiology. Available evidence suggests a role for SCFAs in intestinal inflammation, intestinal barrier integrity, motility, and gut–brain axis regulation [41]. In particular, among the most abundant SCFAs in the colon, Tana and colleagues demonstrated that acetate and propionate are increased, together with total organic acids, in IBS patients compared to controls, and positively correlated with symptom severity [42]. Another SCFA, Sodium butyrate (SB), a renowned HDAC inhibitor [43, 44] which has gained much attention for its debated role as an immune modulator [38, 45–47], it is currently studied predominantly in IBS pathophysiology [40]. SB exerts its activity by either receptor coupling or by the activation of specific genes involved in intestinal motility and immune activation [45, 48, 49]. Recent studies unravelled a pleiotropic effect of SB with more than 2400 genes regulated by histone acetylation as a result of SB-induced HDAC inhibition (Fig. 1, Table 3). Although there are still no data on the involvement of the SB-HDAC mechanism in IBS, it is reasonable to assume it on the basis of the following evidence. Additionally, SB exposure strongly affects the expression of ligand-activated aromatic hydrocarbon receptor (AHR) in cell lines [50], which in turn mediates stem cell proliferation as well several other processes including xenobiotic metabolism, adaptive immunity and, as more recently assessed, intestinal motility [51–53]. Indeed, Obata and colleagues elegantly detailed a causal link between microbiota colonization and colonic motor function by the activity of AHR [53]. Microbiota colonization of germ-free (GF) mice induced AHR expression in colonic neurons, which in turn activated different genes, including the kcnj12, coding for the Kir2.2 inward rectifier potassium channel, involved in neuronal depolarization. In addition, AHR knock-out mice showed a faster intestinal transit compared to wild-type mice as well as a lower amplitude of colonic migrating motor complex of isolated colonic muscular strips. This indicates that AHR increases colon peristalsis by an autonomous fashion regardless of central circuits [53]. Furthermore, AHR promoter activity was increased by 5 to 7 folds in different cell lines upon HDAC inhibition and in particular, because of n-butyrate treatment [50]. In addition, Marinelli and colleagues indicated SB as a possible AHR ligand [49]. In line with these studies, colonization of GF mice with microbiota derived by conventionally fed mice increased histone 3 (H3) and histone 4 (H4) acetylation which correlated with SCFA amount [54]. Analogously, Gills and colleagues described an increased acetylation of H3 and H4 localized at the promoter region of the serotonin transporter (SERT). A lower SERT level was detected in response to SB treatment in vitro as well as in vivo in the distal ileum and colon tract of mice fed with pectin, a soluble dietary fibre producing elevated levels of SCFA (see paragraph on serotonin signalling) [55]. Finally, a further alteration of luminal content could be exerted by high SB by altering the homeostasis of sodium and calcium transport [56]. Accordingly, a recent meta-analysis suggests an ameliorative effect of a low-FODMAP diet on IBS symptoms [57]. Moreover, luminal Firmicutes, major contributors of SB production in the human gut, were increased in patients with IBS [58], as well as in animal models, [59]; their intestinal load also positively correlated with IBS symptom scores [60]. Although it is clear that SB is involved in key pathways of IBS pathophysiology, further studies are necessary to clarify its role, particularly in relation to other factors influencing its concentration. Indeed, experimental data demonstrated that the effect of SB is dose-dependent, therefore, it is fundamental to consider factors including diet, antibiotics, and probiotics, which could influence its final concentration [61].
Table 3.
Histone acetylation associated with IBS mechanisms
| Histone ID acetylation site | Acetylation (↑ or↓) | Regulation &/or observed function | Clinical condition and samples analysed (n); (human, cells, and animal model); biological sources | Reference |
|---|---|---|---|---|
| Animal studies | ||||
| H4K12 | ↓ | HDAC inhibition ameliorate VHS | Stress-induced VHS rat model (maternal separation stress) (n 20) & HC (n 10); spinal and lumbosacral DRGs | [128] |
| H3K9—CACNA1 | ↑ | increased intestinal motility associated with VIP and α1C1b increased expression | TNBS rat model (n 6) & HC (n 6); colon resection | [124] |
| Overall acetylation | ↑ | SB induces NGF released by EGCs | SB supplemented CRL2690 cells; Colonic hypersensitivity induced by butyrate enemas in rats (n 8 treated & n 8 control); Colon resections | [125] |
| Overall acetylation | ↓ | HDAC inhibition ameliorates VHS | WAS rat model & HC; amygdala tissue | [137] |
| H3 | ↑ | BDNF-mediated VHS | CPS and chronic stress in adult offspring (WAS or FSS); spinal and lumbosacral DRGs | [133] |
| HDAC1 | ↓ | |||
| H3K9, H3K18 | ↓ | HDAC inhibition ameliorate VHS | FSS rat model & HC | [138] |
| H3K9 | ↓ | HDAC inhibition ameliorates estradiol-induced VHS | ovariectomized rats, 17β-estradiol treated & untreated | [139] |
| ↑ | Increased VHS; HDAC inhibition ameliorate VHS via SERT expression | Unpredictable early life stress rat model; central nucleus of the amygdala | [136] | |
| H3 | ↑ | Increased VHS via TRPV1 overexpression | WAS rat model; spinal and lumbosacral DRGs | [21] |
| In vitro studies | ||||
| Overall acetylation | ↓ | SCFAs reduce LPS-induced permeability, inhibiting NLPR3 and autophagy | SCFAs treated Caco-2 cell line | [38] |
| Overall acetylation | ↓ | SB induces IL-10RA by STAT3 activation resulting in enhanced permeability | SB treated T84 cell line | [17] |
| H3, H4 | ↑ | HDAC inhibitions decreased SERT expression | SB or TSA-treated Caco-2 cell line | [55] |
Specific sites of acetylation are labelled with a number of acetylated histones followed by the number of the acetylated lysine on the histone tail where stated (e.g. H3K9)
BDNF brain-derived neuronal growth factor, CPS chronic prenatal stress, FSS forced swim stress, TNBS tri-nitro benzene sulphonic acid, TSA trichostatin, WAS water avoidance stress, HDAC histone deacetylases, DRGs dorsal root ganglia, MS maternal separation, SAHA suberoylanilide hydroxamic acid, VIP vasoactive intestinal peptide, SB sodium butyrate, NGF neuronal growth factor, EGCs enteric glial cells, VMR visceromotor response, CRD colorectal distension, SCFA short chain fatty acids, ZO-1 zonula occludens-1, IL-1β interleukin-1β, IL-18 interleukin 18, TSA trichostatin A, IL-10 RA interleukin 10 receptor α, WAS water -avoidance-induced stress, veh. vehicle, FS forced swim, DMSO dimethyl sulphoxide, mGluR metabotropic glutamate receptor, CRH corticotropin-releasing hormone, GR glucocorticoid receptor, CeA central nucleus of the amygdala, HAT histone acetyl-transferases, TRPV1 transient receptor potential cation channel subfamily V member 1
A recent study investigated the effects of SB on visceral sensitivity, highlighting the role of Interleukin-1 Receptor-Associated Kinase 1 (IRAK1). Despite the reduced sample size involved in the study, results showed the SB ability to reduce IRAK1 expression both in vivo and in vitro and, in turn, to smooth the visceral hypersensitivity in the IBS mice model [62].
Intestinal miRNAs act as modulators at host-microbial interface. Exosomes containing specific miRNA sequences derived by intestinal epithelial cells (IECs) were detected in mice feces. In addition, transgenic mice unable to produce IECs-derived miRNA showed an increased luminal content of Firmicutes and Proteobacteria phila, as well as variations of specific bacterial families [23]. Moreover, chemical-induced inflammation altered the miRNA profile in mouse models, further confirming the importance of IEC as a gut primary source of miRNA [63].
Several studies showed a distinct miRNA pattern in GF mice compared to mice with conventional gut microbiota [59, 64] (Fig. 1 and Table 1). In addition, miRNA profiling comparing GF and antibiotic-treated mice indicated specific downregulated miRNA, strengthening the hypothesis of an eventual application of miRNA patterns as biomarkers for microbiota impairment and dysbiosis [59, 65]. In line with this, a lower level of miR-199b was detected in blood samples of patients with IBS compared to healthy controls (HC), which inversely correlated with a higher concentration of coliform bacteria [66]. miR-199b is encoded within the introns of the DNM1 gene, a member of the dynamin GTPase family proteins, crucial in the formation of endocytic vesicles, a fundamental step in endocytosis [67]. A study by Zhou and co-workers on IBs patients correlated a decreased miR-199b expression with augmented visceral hypersensitivity and abdominal pain through TRPV1 upregulation [18]. Additionally, considering the role of miR-199a/b in endocytosis, its involvement in intestinal barrier permeability cannot be excluded. A further study on GF mice colonized with faecal content, derived from naïve mice, showed reduced expression of miR-665, targeting Abcc3, which is a transport protein involved in biliary and intestinal excretion of organic anions [68]. Overall, in this study, miRNA concentration was found to increase in the colon compared to ileum tissue. This finding could reflect a positive correlation of miRNA content with increasing bacterial load [68]. Interestingly, there was a higher level of miRNA in the lower intestinal tract of both GF and conventional mice compared to the small intestine. The authors ascribed this difference to different structures and functions of the small and large intestines. In this regard, it would be interesting to address whether the intestinal microbiota might have contributed phylogenetically to the inherited genetic traits also through miRNA induction. Finally, other microbial species such as virus and fungi showed the capacity to influence host immune defences through the inter-kingdom transfer of miRNA or miRNA-like molecules [69]. In this context, it is worth mentioning that several viral species could influence bacterial homeostasis through miRNA expression [70]. However, the impact of this interaction in shaping the host’s gene expression is still undetermined.
Intestinal barrier permeability
The intestinal barrier is the first interface between the gastrointestinal tract and the environment. It is essential to avoid the passage of harmful antigens and bacteria into the deeper layers of the gut wall, but on the other side, it must allow the passage of nutrients, ions, and molecules useful for the body [11].
A recent study by Wiley and coworkers demonstrated the IL-6 ability to promote the methylation of lysine 9 on histone 3 (H3K9me2/me3) and prevent the binding of transcriptional factors, like glucocorticoid receptor (GR), in the promoter region of tight junction (TJ) genes in young adult male rats subjected to water avoidance and human cells [71]. In addition, intestinal epithelial HDAC1/2 deficiency decreases the expression of claudin-3 (CLDN3), a component of epithelial thigh junctions [72]. In line with this, local ablation of HDAC1 and HDAC2 in the intestine of mouse models and in cultured intestinal organoids induced a marked loss of enteric stem cells, which could accordingly impair epithelial renewal and barrier integrity [73]. Furthermore, lysine 9 on histone 3 (H3K9) methylation decreases claudin-1 (CLDN1), ZOs, and occludin expression in epithelial cells, resulting in paracellular permeability increase [71].
An altered miRNA profile has been related to intestinal barrier dysfunctions in IBS-D patients [19, 74–77]. In particular, a study reported increased miR-29a expression in gut tissues and blood microvesicles derived from patients with IBS, showing higher lactulose/mannitol urine fraction [i.e., a measure of intestinal permeability (IP)] compared to HC [74]. In the same study, the authors highlighted significantly enhanced epithelial permeability following miR-29a overexpression, whereas an opposite effect was observed after miR-29a inhibition in human colonic epithelial cells (FHC) and human IECs of the small intestine (FHs74Int) [74]. A further study identified the miRNA family members miR-29a and miR-29b associated with increased IP in a subset of patients with IBS-D characterized by increased IP, but not in IBS-D patients, showing normal permeability, in IBS-C patients, or in HC [75]. In particular, in this study higher miR-29a/b levels were associated with a lower expression of tight junction components and with the disruption of CLDN1 and nuclear factor repressing -κb- factor (NKRF) mRNAs. Furthermore, the authors showed a milder increase of IP in miR-29a/b−/− mice subjected to either water avoidance stress (WAS) or trinitrobenzene sulfonic acid (TNBS) treatment, compared to wild-type mice [75]. In line with these findings, increased miR-29a level was associated with higher IP in a model of intrauterine growth restriction foetus in pigs [76]. Interestingly, the increase in miR-29a was associated with a decreased expression of TJs and extracellular matrix proteins, which impair cell growth and intestinal epithelial integrity [76] (Fig. 1). These findings are supported by an in vitro experiment showing an improvement in the monolayer integrity by increasing cell proliferation and transepithelial electric resistance following miR-29a inhibition [76]. More recently, a study reported that miRNA-29a was upregulated in the colonic epithelium of patients with IBS-D, while ZO-1 and CLDN1 were downregulated and the apical junctional complex was discontinuous [77]. In the same study, a TNBS-induced IBS-D mouse model, the treatment with miRNa-29a inhibitor induced an increase of ZO-1 and CLDN1 expression, confirming their role in the intestinal mucosal barrier [77]. Other miRNAs potentially involved in IP impairments are miR-21-5p, miR-144, and miR-200a—identified in animal models [65, 78, 79]—and miR-16 and miR-125b-5p—identified in humans [19]. Regarding animal models, Nakata and colleagues described that miR-21-5p induces ADP ribosylation factor 4 (ARF4), a member of the Ras GTPase superfamily which impairs intestinal barrier functions, via silencing ARF4-negative regulators PTEN and PDCD4 [65]. Another study highlighted that higher expression of miR-144, targeting ZO-1 and occludin mRNAs was associated with increased IP in a mouse model of IBS-D [78]. Regarding the miR-200 family, an in vitro study demonstrated that miR-200b expression inhibits paracellular permeability hindering TNFα-induced disruption of TJs [80]. Interestingly, upregulation of a member of the same family, miR-200a, increased visceromotor response in vivo by targeting cannabinoid receptor 1 (CNR1) and serotonin transporter (SERT) transcripts [79]. On the contrary, decreased miR-16 and miR-125b-5p levels correlated with increased IP in colonic biopsies of IBS-D patients compared to HC [19]. In particular, authors demonstrated these two miRNAs regulate the expression of TJ proteins encoded by CLDN2 (target of miR-16) and CNG (target of miR-125b) which, in turn, modulate intestinal epithelial barrier function and correlate with major clinical symptoms [19]. A recent work evaluated miRNA levels in the sigmoid biopsies of IBS patients and identified two miRNA, miR-219a-5p and miR-338-3p, significantly decreased in IBS compared to controls [81]. In addition, miR-219a-5p inhibition induced a permeability increase in colonic epithelial cell line NCM460 while the inhibition of miR-338-3p caused alterations in the mitogen-activated protein kinase (MAPK) signalling [81]. A further study performed on human colonic epithelium HT-29 cells highlighted the involvement of miR-148b-5p in epithelial barrier regulation. In particular, HT-29 cells, incubated with serum-derived exosomes from patients with IBS, showed increased permeability by up-regulating miR-148b-5p to suppress RGS2 expression. Interestingly, the effect on permeability was abrogated by interfering with miR-148b-5p expression [82].
The impairment of intestinal permeability in IBS has also been associated with a decreased expression of aquaporins (AQP) particularly AQP 1,3 [83]. Interestingly, a further work showed a decreased expression of lncRNA H19, which positively correlated with AQP1 and AQP3, in IBS-D patients [84].
Neuro-immune interactions
An increased number of mucosal immune cells, particularly mast cells, have been reported in IBS patients, and low-grade inflammation is considered as one of the most important underlying pathophysiological mechanisms of this syndrome [85–87]. In addition, mast cells lie in proximity to nerve endings in the gut wall, a key position for the crosstalk between the immune and nervous systems of the gut [88]. Moreover, activated mast cells in proximity of nerve endings correlate significantly with the severity and frequency of abdominal pain in IBS patients [85, 88]. Interestingly, miRNAs regulate both immune and nervous systems along with control signal exchange in neuro-immune interactions involved in pain pathways [89]. For instance, miR-490-5p expression promotes mast cell proliferation and resistance to apoptosis, probably via multiple targets, including the tryptase/PAR-2 signal path [90]. Moreover, the downregulation of miR-125b and miR-16 correlates with increased mast-cell counts in the jejunum biopsies of IBS-D patients [19].
A further study carried out on an established IBS animal model provided evidence that miR-181c-5p overexpression determined IL1A downregulation exerting anti-inflammatory effects in IBS [91].
An altered interaction between sexual hormone production and immune response was reported in IBS patients [92–94]. Recently, three different miRNAs, namely miR-145, miR-148-5p, and miR-592, involved in these interactions were found dysregulated in IBS patients. In particular, miR-145 and miR-592 expressions were decreased in IBS-C and IBS-D patients, respectively, while miR-148-5p levels were higher in IBS-D patients compared to control subjects [95]. A recent study highlighted the involvement of eosinophils in IBS-D, via peripheral CRF. The same study also reported the corticotropin-releasing hormone receptor 1 gene (CRHR1) downregulation and the up-regulation of the membrane protein SNAP23 related to vascular transport in IBS-D [96]. Numerous lines of evidence suggest a positive effect of HDAC inhibition at the central level and an immune modulation mediated by microglial cells [97–100]. In particular, Jaworska and colleagues showed that these effects are induced by SB, which also promoted central neurogenesis in rat pups subjected to central ischemic insult [100]. Furthermore, rats perfused with SB into the cecum showed a modified enteric neuronal plasticity favouring the reinforcement of the cholinergic system associated with an increased visceral hypersensitivity as described in the following paragraphs [48]. Since SB can cross blood–brain-barrier due to its low-molecular weight, its effect at a central level has been thoroughly investigated. A recent study showed how dietary intervention could suppress microglia activation in the CNS after an excessive alcohol intake mainly through suppressing neuroinflammation [101]. The emerging clue is that SB could restore gut dysbiosis, upgrading beneficial bacteria and their metabolites. This improves the intestinal microenvironment, thereby exerting a protective effect on the intestinal mucosal barrier and nervous system. However, further studies are warranted to clarify SB-induced gut-brain neuroplastic changes and to define the underlying cellular and molecular mechanisms, particularly those related to HDAC-mediated gene expression. Lately, several studies delved more into these mechanisms showing a protective role of SB on different in vivo models of ischemic stroke inhibiting the expression of proinflammatory cytokines TNF-α and nitric oxide synthase-1, while upregulating IL-10 [102, 103]. Still at a central level, akin results showed improved neuromotor capabilities of mice models of Parkinson’s Disease treated with SB, linking this effect to an inhibition of the TLR4/MyD88/NF-B inflammatory pathway, as well as to a beneficial effect on intestinal barrier elicited by a reduction of dysbiosis [102]. Although related to a different experimental model, these results are in contrast with a possible detrimental activity of SB described previously in different experimental models. Yet, a clear causal link clarifying SB-induced epigenetic changes in regulating neuro-immune interactions at a central level and IBS is still missing.
Serotonin signalling
The intestinal tract represents the major site of serotonin (5-hydroxytryptamine, 5-HT) production. The enterochromaffin cells produce up to 95% of the total 5-HT in the human body. 5-HT regulates intestinal motility, peristalsis, and contributes to several physiological functions such as neurogenesis and bone mass accrual [104]. The serotoninergic system comprises 15 different receptors of which 5HT3 and 5HT4 are the main targets for therapeutic strategies currently applied in IBS-D and IBS-C treatment, respectively. Indeed, experimental evidence showed a dysregulation of 5HT system in at least a subgroup of IBS, and more recently the influence of the gut microbiota to host serotoninergic system has become the target of research [105, 106]. This also includes a novel scenario where resident bacteria produce 5HT to activate host’s enterochromaffin cells [107].
From a biochemical standpoint, 5HT is also responsible for the covalent binding of proteins mediated by transglutaminases (TG), a process defined as serotonylation [108] which occurs at trimethylated histone 4 (H4K3me3). A recent milestone study described a causal link between serotonylation of H4K3me3 via TG2 and euchromatin levels, setting off this process as an additional epigenetic modification mostly occurring in the brain and colon [109]. Interestingly, H4K3me3 levels were associated with intestinal permeability and a decrease of Lactobacillus plantarum L168 in the intestinal tract of a Drosophila Melanogaster model [110].
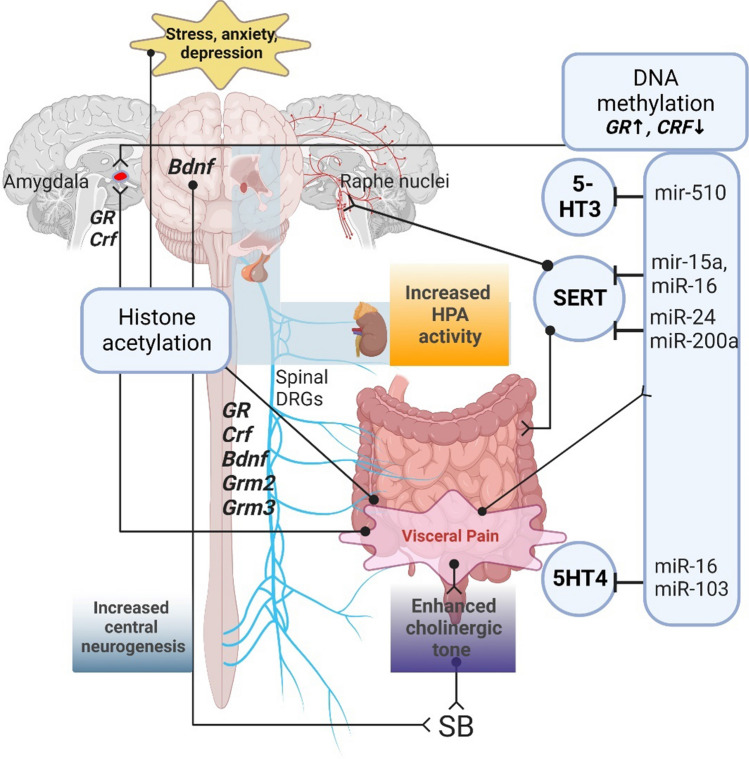
Coming to non-coding RNA dysregulation mediated by genetic variants, Kapeller and colleagues showed that the 5HT3 variant c.*76G>A, located in the 3’UTR regulatory region of the E subunit (5HT3E), was associated with female IBS-D. The authors demonstrated that this variant causes the loss of the binding site of miR-510 inducing an increased expression of 5HT3 [111]. Another study reported a significant downregulation of miR16 and miR-103 in jejunum biopsies of IBS-D patients compared to controls [112]. Decreased levels of these miRNAs correlated with IBS symptom severity through 5-HT4 mRNA targeting [112]. 5-HT recaption and metabolism by enterocytes are mediated by SERT and monoamine oxidase activity, respectively [109, 113]. A growing consensus indicates an altered SERT expression in IBS patients [114, 115]. Interestingly, numerous studies described the involvement of different miRNAs in the regulation of SERT gene expression. miR-16, previously mentioned as a 5HT3 inhibitor and associated with high mast cell count, was associated with reduced SERT expression at a central level in mice raphe nuclei [116]. These results were further confirmed by Moya and colleagues which, additionally, observed a repressive activity on SERT exerted by miR-15a [117]. Furthermore, miR-200a upregulation was associated with a downregulation of both CNR1 and SERT in a rat model of IBS-D [79]. Finally, miR-24 was increased in the colonic mucosa of IBS patients compared to healthy subjects as well as in mice models induced by TNBS, together with a decrease of SERT levels [114]. In addition, the above-mentioned miR-29a can modulate the translation of the HTR7 protein, a G-protein-coupling receptor of 5-HT. Specifically, the overexpression of miR-29a, observed in the colon tissue of IBS patients and in WAS model, reduces the HTR7 expression, enhancing visceral hyperalgesia [118].
Dysregulation of HPA axis and visceral hypersensitivity
Intestinal over-responsiveness to noxious stimuli is a crucial feature in IBS, resulting from several molecular mechanisms promoting peripheral and central sensitization [119]. Recent findings showed a higher colorectal contraction in response to corticotropin-releasing factor (CRF), and an altered activity at a central level upon colorectal distension (CRD), confirming the contribution of the HPA axis in IBS visceral pain [120]. In addition, it has been observed an increased amount of CRF in the cytoplasmic granules of jejunal eosinophils in IBS-D patients positively correlated with IBS clinical severity, chronic stress, and depression [96]. Importantly, the evidence demonstrated that subjects reporting EAEs and/or psychiatric disorders showing an increased activity of HPA-axis [3, 7] are prone to develop visceral hypersensitivity and IBS in adulthood [6, 121]. Notably, HDAC inhibitor MS-275 showed a beneficial effect on the anxiety-prone mice model, proposing H3 acetylation as a possible biomarker for the development of anxiety therapies [122].
From the scientific databases consulted in the present work we were not able to retrieve any data on epigenetic variation in visceral hypersensitivity associated with IBS with no central comorbidities, which has been extensively described by Mahurkar-Joshi and Chang [123]. Notably, two independent animal studies detected an increased visceromotor response (VMR) to CRD associated with HDAC inhibition induced by SB treatment, further suggesting SB as a contributor to IBS symptoms [124, 125].
Translational models demonstrated that stress or painful events in early life predisposed to chronic pain [126] and higher visceral hypersensitivity [127] via epigenetic regulation. Moloney and colleagues showed that in a rat model of maternal separation, visceral hypersensitivity was associated with a reduction of histone 4 lysine 12 (H4K12) acetylation in tissue samples from the spinal cord. Furthermore, inhibition of HDAC by treating animals with suberoylanilide hydroxamic acid (SAHA) reduced the evacuation frequency and pain sensation triggered by CRD [128]. In line with this, histone acetylation as well as DNA methylation was increased in male rats subjected to WAS and corticosterone treatment [21]. Moreover, increased signalling of brain-derived neurotrophic factor (BDNF) and glucocorticoid gene expression detected in animal models of EAEs are mediated by epigenetic factors [129, 130]. Notably, BDNF was increased in the colonic biopsies of patients with IBS [131, 132] and its concentration correlated with IBS symptom severity [131] and visceral hypersensitivity [132]. BDNF upregulation derived by H3 acetylation and HDAC1 inhibition was associated with enhanced VMR only in female rats subjected to heterotypic intermittent chronic prenatal stress [133]. This is also consistent with the altered regulation of BDNF observed in HPA-related disorders associated with early life stress [129, 134]. Interestingly, Lambert and colleagues extensively described a stress-dependent induction of glucocorticoid receptor (GR) mediated by BDNF, showing an exclusive, synergic activity of dexamethasone and BDNF in inducing an altered transcriptomic trait of central neurons [135]. As none of the functions of the detected mRNA profile was ascribable to BDNF nor GR activity taken alone, the authors hypothesized an epigenetic regulation exerted by BDNF on GR as a possible underlying mechanism. In line with this evidence, increased acetylation of lysine 9 on histone 3, (H3K9) was displayed at both GR and CRF promoter sites in a rat model of odour-induced stress showing enhanced VMR [136] (Fig. 2). In addition, in the amygdala of rats, a higher VMR induced by WAS was associated with an increased promoter methylation and reduced expression of GR. On the other side, CRF promoter methylation was decreased with a concomitant increase in CRF expression. In addition, visceral hypersensitivity was decreased upon treatment with the HDAC inhibitor TSA [137]. Hong and colleagues showed that rats subjected to WAS displayed an increase in DNA methylation of the glucocorticoid receptor (NR3C1) and CNR1 promoters and increased histone acetylation of transient receptor potential vanilloid type 1 (TRPV1) promoter [21]. As previously mentioned, IL-6 promotes H3K9 methylation in a GR-mediated way in colon epithelial cells and prevents TJs proteins expression, resulting in paracellular permeability increment and visceral hyperalgesia increase [71].
Fig. 2.
Epigenetic factors involved in brain-gut axis dysregulation. The scheme reports gene acetylation and miRNAs evoking visceral pain by interacting with key players of serotonergic (i.e., 5-HT3, 5-HT4, SERT), cholinergic, HPA response, and neurogenesis. SERT serotonin transporter, SB sodium butyrate, GR glucocorticoid receptor, CRF corticotropin-releasing factor, BDNF brain-derived neurotrophic factor, GRM glutamate receptor genes
Experimental models of post-infectious IBS showed that increased vasoactive intestinal peptide (VIP) levels in the muscolaris externa mediated an upregulation of the voltage-gated calcium receptor 1.2b subunit α1C1b through H3K9. This could allegedly increase intracellular calcium flux resulting in enhanced smooth muscle contractions [124].
Rats with increased VMR induced by forced swim showed a decreased acetylation (H3K9, H3K18) and expression levels of glutamate receptor genes Grm 2 and Grm3 [138]. Furthermore, the same authors detected a lower H3K9 acetylation of Grm 2 associated with an increased VMR derived by estradiol substitutive supplementation in ovariectomized female rats [139]. These data indicate gonadal hormones as possible contributors to the development of visceral hypersensitivity induced by early-life stress events. Notably, the VMR of both ovariectomized and forced-swim models was reduced by restored levels of GR induced by HDAC inhibition [138, 139]. In line with these findings, the glutamatergic system might also be dampened by a reduction of glutamate decarboxylase described in pain disorders as well as by the inhibition of the GLUL gene by miR-29a described in IBS patients [74].
In a rat model of IBS-D and in human colonic biopsies of patients with IBS-D, elevated levels of miR-200a have been shown. MiR-200a seems to be involved in increasing visceral hypersensitivity by acting on CNR1 and SERT [79]. Moreover, the miR-495 was poorly expressed in a mice IBS-D model, whereas PKIB, the miR-495 target gene was upregulated. On the other hand, overexpression of miR-495 and the consequent suppression of PKIB can inhibit the PI3K/AKT signalling pathway and decreases visceral sensitivity [140]. An additional study reported that a decreased miR-325-5p expression correlates with CCL2 upregulation in a rat model of chronic visceral pain [141]. The involvement of CCL2 in inflammatory and neurodegenerative diseases and functional dyspepsia [141–143] point to the involvement of the miR-325-5p/CCL2 signalling in chronic visceral pain in IBS patients and other patients with the gastrointestinal disorder as well.
Conclusions
Increasing experimental evidence suggests the involvement of epigenetic regulation in IBS pathophysiology. Residing intestinal microbiome could represent a hub between the different mechanisms involved in IBS, whilst epigenetic factors should be reckoned as contributors for implementing these mechanisms in the host. Moreover, recent findings describe crypt enterocytes as key players in host-microbiome interaction mediated by both histone modifications [49, 53] and miRNA signalling [23]. The indirect activity of microbial metabolites such as that exerted by SB-induced HDAC inhibition represents a complex multifactorial regulation of adaptive immunity, intestinal motility, the permeability of gut barrier, and abdominal pain. At the host interface, extra-cellular miRNA produced by intestinal epithelial and stem cells contribute to the equilibrium between residing microbial species [65]. With regard to ncRNA, many miRNAs have been identified as implicated in the pathophysiology of IBS. Among these, the miR-19 family implicated in both visceral hypersensitivity and augmented intestinal permeability could represent key biomarkers in IBS diagnosis and treatment, particularly for IBS-D patients. However, additional studies are needed to confirm the role of this miRNA family and identify enzymes and signalling paths involved in IBS complexity.
An important limitation in IBS preclinical research pertains to the complexity and heterogeneity of this disease, which pose a hurdle to the development of a reliable and valid experimental model. For this reason, there is no animal model of IBS mimicking all the symptoms typical of this condition. Nowadays, there are only models imitating some aspects of IBS. Neonatal maternal separation, water avoidance, and wrap restraint stress are useful to study the mechanisms underlying psychological stress, but they cannot be considered a model of IBS. Several studies on epigenetics in IBS belong from animal models, representing a major limitation in the evaluation of the results.
Future studies should include a large cohort of well-characterized patients to decipher the molecular mechanisms, including epigenetic ones, underlying IBS pathophysiology and to identify new therapeutic options.
Acknowledgements
G. Dothel was supported by the Ministry of Education, University and Research of Italy [MIUR, Grant number 2015Y3C5KP_002 to SA]. S. Monesmith is supported by a Fulbright Research Scholarship (year 2022)
Abbreviations
- AHR
Aromatic hydrocarbon receptor
- ARF4
ADP ribosylation factor 4
- BDNF
Brain-derived neurotrophic factor
- CNR1
Cannabinoid receptor 1
- CRD
Colorectal distension
- CRF
Corticotropin-releasing factor
- DNMTs
DNA methyl transferase
- DRG
Dorsal root ganglia
- EAEs
Early-life adverse events
- EGC
Enteric glial cells
- EV
Extracellular vesicles
- FSS
Forced swim stress
- FODMAPs
Fermentable oligo- mono- di-saccharides and polyoils
- GF
Germ free
- GMR
Glutamate metabotropic receptor
- GR
Glucocorticoid receptor
- HATs
Histone acetyl transferase
- HDACs
Histone deacetylases
- HPA
Hypothalamic–pituitary–adrenal
- IECs
Intestinal epithelial cells
- IP
Intestinal permeability
- lncRNA
Long non-coding RNA
- miRNA
MicroRNA
- MS
Maternal separation
- NGF
Neuronal growth factor
- RGS2
Regulator of G protein signalling 2
- SAHA
Suberoylanilide hydroxamic acid
- SB
Sodium butyrate
- SCFAs
Short chain fatty acids
- SERT
Serotonin transporter
- TNBS
Trinitrobenzene sulfonic acid
- TSA
Trichostatin
- VIP
Vasoactive intestinal protein
- VMR
Visceromotor response
- WAS
Water avoidance stress
- 5HT
5-Hydroxytryptamine serotonin
- 5HT3
Serotonin receptor 3
- 5HT4
Serotonin receptor 4
Funding
Open access funding provided by Alma Mater Studiorum - Università di Bologna within the CRUI-CARE Agreement.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Giovanni Dothel and Maria Raffaella Barbaro share co-first authorship.
References
- 1.Lacy BE, Mearin F, Chang L, Chey WD, Lembo AJ, Simren M, et al. Bowel disorders. Gastroenterology. 2016;150:1393–1407. doi: 10.1053/j.gastro.2016.02.031. [DOI] [PubMed] [Google Scholar]
- 2.Drossman DA, Hasler WL. Rome IV-functional GI disorders: disorders of gut–brain interaction. Gastroenterology. 2016;150:1257–1261. doi: 10.1053/j.gastro.2016.03.035. [DOI] [PubMed] [Google Scholar]
- 3.Ng QX, Sen SAY, Loke W, Venkatanarayanan N, Lim DY, Yeo WS. Systematic review with meta-analysis: the association between post-traumatic stress disorder and irritable bowel syndrome. J Gastroenterol Hepatol. 2019;34:68–73. doi: 10.1111/jgh.14446. [DOI] [PubMed] [Google Scholar]
- 4.Fond G, Loundou A, Hamdani N, Boukouaci W, Dargel A, Oliveira J, et al. Anxiety and depression comorbidities in irritable bowel syndrome (IBS): a systematic review and meta-analysis. Eur Arch Psychiatry Clin Neurosci. 2014;264:651–660. doi: 10.1007/s00406-014-0502-z. [DOI] [PubMed] [Google Scholar]
- 5.Sibelli A, Chalder T, Everitt H, Workman P, Windgassen S, Moss-Morris R. A systematic review with meta-analysis of the role of anxiety and depression in irritable bowel syndrome onset. Psychol Med. 2016;46:3065–3080. doi: 10.1017/S0033291716001987. [DOI] [PubMed] [Google Scholar]
- 6.Chitkara DK, Van Tilburg MAL, Blois-Martin N, Whitehead WE. Early life risk factors that contribute to irritable bowel syndrome in adults: a systematic review. Am J Gastroenterol. 2008;103:765–774. doi: 10.1111/j.1572-0241.2007.01722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Videlock EJ, Adeyemo M, Licudine A, Hirano M, Ohning G, Mayer M, et al. Childhood trauma is associated with hypothalamic-pituitary-adrenal axis responsiveness in irritable bowel syndrome. Gastroenterology. 2009;137:1954–1962. doi: 10.1053/j.gastro.2009.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chong PP, Chin VK, Looi CY, Wong WF, Madhavan P, Yong VC. The Microbiome and irritable bowel syndrome—a review on the pathophysiology, current research and future therapy. Front Microbiol. 2019;10:42. doi: 10.3389/fmicb.2019.01136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simrén M, Öhman L. Pathogenesis of IBS: role of inflammation, immunity and neuroimmune interactions. Nat Rev Gastroenterol Hepatol. 2010;7:163–173. doi: 10.1038/nrgastro.2010.4. [DOI] [PubMed] [Google Scholar]
- 10.Stasi C, Bellini M, Bassotti G, Blandizzi C, Milani S. Serotonin receptors and their role in the pathophysiology and therapy of irritable bowel syndrome. Tech Coloproctol. 2014;18:613–621. doi: 10.1007/s10151-013-1106-8. [DOI] [PubMed] [Google Scholar]
- 11.Barbara G, Barbaro MR, Fuschi D, Palombo M, Falangone F, Cremon C, et al. Inflammatory and microbiota-related regulation of the intestinal epithelial barrier. Front Nutr. 2021;8:22. doi: 10.3389/fnut.2021.718356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbara G, Feinle-Bisset C, Ghoshal UC, Santos J, Vanner SJ, Vergnolle N, et al. The intestinal microenvironment and functional gastrointestinal disorders. Gastroenterology. 2016;150:1305–1318. doi: 10.1053/j.gastro.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 13.Bashashati M, Moossavi S, Cremon C, Barbaro MR, Moraveji S, Talmon G, et al. Colonic immune cells in irritable bowel syndrome: a systematic review and meta-analysis. Neurogastroenterol Motil. 2018;30:13192. doi: 10.1111/nmo.13192. [DOI] [PubMed] [Google Scholar]
- 14.Hasler WL, Grabauskas G, Singh P, Owyang C. Mast cell mediation of visceral sensation and permeability in irritable bowel syndrome. Neurogastroenterol Motil. 2022;34:25. doi: 10.1111/nmo.14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguilera-Lizarraga J, Hussein H, Boeckxstaens GE. Immune activation in irritable bowel syndrome: what is the evidence? Nat Rev Immunol. 2022;22:674–686. doi: 10.1038/s41577-022-00700-9. [DOI] [PubMed] [Google Scholar]
- 16.Vanuytsel T, Bercik P, Boeckxstaens G. Understanding neuroimmune interactions in disorders of gut-brain interaction: from functional to immune-mediated disorders. Gut. 2023;72:787–798. doi: 10.1136/gutjnl-2020-320633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng L, Kelly CJ, Battista KD, Schaefer R, Lanis JM, Alexeev EE, et al. Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor-dependent repression of claudin-2. J Immunol. 2017;199:2976–2984. doi: 10.4049/jimmunol.1700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Q, Yang L, Larson S, Basra S, Merwat S, Tan A, et al. Decreased miR-199 augments visceral pain in patients with IBS through translational upregulation of TRPV1. Gut. 2016;65:797–805. doi: 10.1136/gutjnl-2013-306464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez C, Rodinõ-Janeiro BK, Lobo B, Stanifer ML, Klaus B, Granzow M, et al. miR-16 and miR-125b are involved in barrier function dysregulation through the modulation of claudin-2 and cingulin expression in the jejunum in IBS with diarrhoea. Gut. 2017;66:1597–1610. doi: 10.1136/gutjnl-2016-311477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahurkar S, Polytarchou C, Iliopoulos D, Pothoulakis C, Mayer EA, Chang L. Genome-wide DNA methylation profiling of peripheral blood mononuclear cells in irritable bowel syndrome. Neurogastroenterol Motil. 2016;28:410–422. doi: 10.1111/nmo.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong S, Zheng G, Wiley JW. Epigenetic regulation of genes that modulate chronic stress-induced visceral pain in the peripheral nervous system. Gastroenterology. 2015;148:148–157.e7. doi: 10.1053/j.gastro.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul B, Barnes S, Demark-Wahnefried W, Morrow C, Salvador C, Skibola C, et al. Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin Epigenet. 2015;7:112. doi: 10.1186/s13148-015-0144-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S, da Cunha AP, Rezende RMM, Cialic R, Wei Z, Bry L, et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe. 2016;19:32–43. doi: 10.1016/j.chom.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deans C, Maggert KA. What do you mean, “epigenetic”? Genetics Genetics. 2015;199:887–896. doi: 10.1534/genetics.114.173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allfrey VG, Faulkner R, Mirsky AE. Acetylatiom and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32:42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y, Rao A. Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet. 2014;30:464. doi: 10.1016/j.tig.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deveson IW, Hardwick SA, Mercer TR, Mattick JS. The dimensions, dynamics, and relevance of the mammalian noncoding transcriptome. Trends Genet. 2017;33:464–478. doi: 10.1016/j.tig.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179:1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halawi H, Camilleri M. Pharmacogenetics and the treatment of functional gastrointestinal disorders. Pharmacogenomics. 2017;18:1085–1094. doi: 10.2217/pgs-2017-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 34.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yarani R, Mirza AH, Kaur S, Pociot F. The emerging role of lncRNAs in inflammatory bowel disease. Exp Mol Med. 2018;50:54. doi: 10.1038/s12276-018-0188-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 38.Feng Y, Wang Y, Wang P, Huang Y, Wang F. Short-chain fatty acids manifest stimulative and protective effects on intestinal barrier function through the inhibition of NLRP3 inflammasome and autophagy. Cell Physiol Biochem. 2018;49:190–205. doi: 10.1159/000492853. [DOI] [PubMed] [Google Scholar]
- 39.Jia Q, Zhang L, Zhang J, Pei F, Zhu S, Sun Q, et al. Fecal microbiota of diarrhea-predominant irritable bowel syndrome patients causes hepatic inflammation of germ-free rats and berberine reverses it partially. Biomed Res Int. 2019;20:19. doi: 10.1155/2019/4530203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Q, Jia Q, Song L, Duan L. Alterations in fecal short-chain fatty acids in patients with irritable bowel syndrome: a systematic review and meta-analysis. Medicine. 2019;98:22. doi: 10.1097/MD.0000000000014513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat Rev Gastroenterol Hepatol. 2019;16:461–478. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- 42.Tana C, Umesaki Y, Imaoka A, Handa T, Kanazawa M, Fukudo S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:512. doi: 10.1111/j.1365-2982.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- 43.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133:52. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 44.Yuille S, Reichardt N, Panda S, Dunbar H, Mulder IE. Human gut bacteria as potent class I histone deacetylase inhibitors in vitro through production of butyric acid and valeric acid. PLoS ONE. 2018;13:e5014. doi: 10.1371/journal.pone.0201073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee C, Kim BG, Kim JH, Chun J, Im JP, Kim JS. Sodium butyrate inhibits the NF-kappa B signaling pathway and histone deacetylation, and attenuates experimental colitis in an IL-10 independent manner. Int Immunopharmacol. 2017;51:47–56. doi: 10.1016/j.intimp.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 46.Martin-Gallausiaux C, Béguet-Crespel F, Marinelli L, Jamet A, Ledue F, Blottière HM, et al. Butyrate produced by gut commensal bacteria activates TGF-beta1 expression through the transcription factor SP1 in human intestinal epithelial cells. Sci Rep. 2018;8:1–13. doi: 10.1038/s41598-018-28048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vancamelbeke M, Laeremans T, Vanhove W, Arnauts K, Ramalho AS, Farré R, et al. Butyrate does not protect against inflammation-induced loss of epithelial barrier function and cytokine production in primary cell monolayers from patients with ulcerative colitis. J Crohns Colitis. 2019;13:1351–1361. doi: 10.1093/ecco-jcc/jjz064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soret R, Chevalier J, De Coppet P, Poupeau G, Derkinderen P, Segain JP, et al. Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology. 2010;138:22. doi: 10.1053/j.gastro.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 49.Marinelli L, Martin-Gallausiaux C, Bourhis JM, Béguet-Crespel F, Blottière HM, Lapaque N. Identification of the novel role of butyrate as AhR ligand in human intestinal epithelial cells. Sci Rep. 2019;9:52. doi: 10.1038/s41598-018-37019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garrison PM, Rogers JM, Brackney WR, Denison MS. Effects of histone deacetylase inhibitors on the Ah receptor gene promoter. Arch Biochem Biophys. 2000;374:161–171. doi: 10.1006/abbi.1999.1620. [DOI] [PubMed] [Google Scholar]
- 51.Nebert DW, Roe AL, Dieter MZ, Solis WA, Yang Y, Dalton TP. Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem Pharmacol. 2000;59:65–85. doi: 10.1016/S0006-2952(99)00310-X. [DOI] [PubMed] [Google Scholar]
- 52.Quintana FJ, Sherr DH. Aryl hydrocarbon receptor control of adaptive immunity. Pharmacol Rev. 2013;65:1148–1161. doi: 10.1124/pr.113.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Obata Y, Castaño Á, Boeing S, Bon-Frauches AC, Fung C, Fallesen T, et al. Neuronal programming by microbiota regulates intestinal physiology. Nature. 2020;578:284–289. doi: 10.1038/s41586-020-1975-8. [DOI] [PubMed] [Google Scholar]
- 54.Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, et al. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell. 2016;64:982–992. doi: 10.1016/j.molcel.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gill RK, Kumar A, Malhotra P, Maher D, Singh V, Dudeja PK, et al. Regulation of intestinal serotonin transporter expression via epigenetic mechanisms: role of HDAC2. Am J Physiol Cell Physiol. 2013;304:C334–C341. doi: 10.1152/ajpcell.00361.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Binder HJ, Mehta P. Short-chain fatty acids stimulate active sodium and chloride absorption in vitro in the rat distal colon. Gastroenterology. 1989;96:989–996. doi: 10.1016/0016-5085(89)91614-4. [DOI] [PubMed] [Google Scholar]
- 57.van Lanen AS, de Bree A, Greyling A. Correction to: Efficacy of a low-FODMAP diet in adult irritable bowel syndrome: a systematic review and meta-analysis. Eur J Nutr. 2021;60:3523. doi: 10.1007/s00394-021-02620-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 59.Moloney GM, Viola MF, Hoban AE, Dinan TG, Cryan JF. Faecal microRNAs: indicators of imbalance at the host-microbe interface? Benef Microbes. 2018;9:175–183. doi: 10.3920/BM2017.0013. [DOI] [PubMed] [Google Scholar]
- 60.Tap J, Derrien M, Törnblom H, Brazeilles R, Cools-Portier S, Doré J, et al. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology. 2017;152:111–123.e8. doi: 10.1053/j.gastro.2016.09.049. [DOI] [PubMed] [Google Scholar]
- 61.Kannampalli P, Shaker R, Sengupta JN. Colonic butyrate-algesic or analgesic? Neurogastroenterol Motil. 2011;23:975–979. doi: 10.1111/j.1365-2982.2011.01775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He Y, Tan Y, Zhu J, Wu X, Huang Z, Chen H, et al. Effect of sodium butyrate regulating IRAK1 (interleukin-1 receptor-associated kinase 1) on visceral hypersensitivity in irritable bowel syndrome and its mechanism. Bioengineered. 2021;12:1436–1444. doi: 10.1080/21655979.2021.1920324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anzola A, González R, Gámez-Belmonte R, Ocón B, Aranda CJ, Martínez-Moya P, et al. miR-146a regulates the crosstalk between intestinal epithelial cells, microbial components and inflammatory stimuli. Sci Rep. 2018;8:62. doi: 10.1038/s41598-018-35338-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoban AE, Stilling RM, Moloney M, G, Moloney RD, Shanahan F, Dinan TG,, et al. Microbial regulation of microRNA expression in the amygdala and prefrontal cortex. Microbiome. 2017;5:102. doi: 10.1186/s40168-017-0321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakata K, Sugi Y, Narabayashi H, Kobayakawa T, Nakanishi Y, Tsuda M, et al. Commensal Microbiota-induced microRNA modulates intestinal epithelial permeability through the small GTPase ARF4. J Biol Chem. 2017;292:15426–15433. doi: 10.1074/jbc.M117.788596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mansour MA, Sabbah NA, Mansour SA, Ibrahim AM. MicroRNA-199b expression level and coliform count in irritable bowel syndrome. IUBMB Life. 2016;68:335–342. doi: 10.1002/iub.1495. [DOI] [PubMed] [Google Scholar]
- 67.Aranda JF, Canfran-Duque A, Goedeke L, Suarez Y, Fernandez-Hernando C. The miR-199-dynamin regulatory axis controls receptor-mediated endocytosis. J Cell Sci. 2015;128:3197–3209. doi: 10.1242/jcs.165233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dalmasso G, Nguyen HTT, Yan Y, Laroui H, Charania MA, Ayyadurai S, et al. Microbiota modulate host gene expression via microRNAs. PLoS ONE. 2011;6:e5014. doi: 10.1371/journal.pone.0019293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Libertucci J, Young VB. The role of the microbiota in infectious diseases. Nat Microbiol. 2019;2:435–445. doi: 10.1038/s41564-018-0278-4. [DOI] [PubMed] [Google Scholar]
- 70.Cardin SE, Borchert GM. Viral MIcroRNAs, host microRNAs regulating viruses, and bacterial microRNA-like RNAs. Methods Mol Biol. 2017;1617:39–56. doi: 10.1007/978-1-4939-7046-9_3. [DOI] [PubMed] [Google Scholar]
- 71.Wiley JW, Zong Y, Zheng G, Zhu S, Hong S. Histone H3K9 methylation regulates chronic stress and IL-6-induced colon epithelial permeability and visceral pain. Neurogastroenterol Motil. 2020;32:e13941. doi: 10.1111/nmo.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Turgeon N, Blais M, Gagnè J-M, Tardif V, Boudreau F, Perreault N, et al. HDAC1 and HDAC2 restarin the intestinal inflammatory response bt regulating intestinal eoithelial cell differentiation. PLoS ONE. 2013;8:e73785. doi: 10.1371/journal.pone.0073785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zimberlin CD, Lancini C, Sno R, Rosekrans SL, McLean CM, Vlaming H, et al. HDAC1 and HDAC2 collectively regulate intestinal stem cell homeostasis. FASEB J. 2015;29:2070–2080. doi: 10.1096/fj.14-257931. [DOI] [PubMed] [Google Scholar]
- 74.Zhou QQ, Souba WW, Croce CM, Verne GN. MicroRNA-29a regulates intestinal membrane permeability in patients with irritable bowel syndrome. Gut. 2010;59:775–784. doi: 10.1136/gut.2009.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou Q, Costinean S, Croce CM, Brasier AR, Merwat S, Larson SA, et al. MicroRNA 29 targets nuclear factor-κB-repressing factor and Claudin 1 to increase intestinal permeability. Gastroenterology. 2015;148:158–169.e8. doi: 10.1053/j.gastro.2014.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu Y, Wang W, Yuan T, Fu L, Zhou L, Lin G, et al. MicroRNA-29a mediates the impairment of intestinal epithelial integrity induced by intrauterine growth restriction in pig. Am J Physiol Gastrointest Liver Physiol. 2017;312:G434–G442. doi: 10.1152/ajpgi.00020.2017. [DOI] [PubMed] [Google Scholar]
- 77.Zhu H, Xiao X, Shi Y, Wu Y, Huang Y, Li D, et al. Inhibition of miRNA-29a regulates intestinal barrier function in diarrhea-predominant irritable bowel syndrome by upregulating ZO-1 and CLDN1. Exp Ther Med. 2020;20:2. doi: 10.3892/etm.2020.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hou Q, Huang Y, Zhu S, Li P, Chen X, Hou Z, et al. MiR-144 increases intestinal permeability in IBS-D rats by targeting OCLN and ZO1. Cell Physiol Biochem. 2023;44:2256–2268. doi: 10.1159/000486059. [DOI] [PubMed] [Google Scholar]
- 79.Hou Q, Huang Y, Zhang C, Zhu S, Li P, Chen X, et al. MicroRNA-200a targets cannabinoid receptor 1 and serotonin transporter to increase visceral hyperalgesia in diarrhea-predominant irritable bowel syndrome rats. J Neurogastroenterol Motil. 2018;24:656. doi: 10.5056/jnm18037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shen Y, Zhou M, Yan J, Gong Z, Xiao Y, Zhang C, et al. miR-200b inhibits TNF-α-induced IL-8 secretion and tight junction disruption of intestinal epithelial cells in vitro. Am J Physiol Gastrointest Liver Physiol. 2017;312:G123–G132. doi: 10.1152/ajpgi.00316.2016. [DOI] [PubMed] [Google Scholar]
- 81.Mahurkar-Joshi S, Rankin CR, Videlock EJ, Soroosh A, Verma A, Khandadash A, et al. The colonic mucosal microRNAs, microRNA-219a-5p, and microRNA-338-3p are downregulated in irritable bowel syndrome and are associated with barrier function and MAPK signaling. Gastroenterology. 2021;160:2409–2422.e19. doi: 10.1053/j.gastro.2021.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xing Y, Xue S, Wu J, Zhou J, Xing F, Li T, et al. Serum exosomes derived from irritable bowel syndrome patient increase cell permeability via regulating miR-148b-5p/RGS2 signaling in human colonic epithelium cells. Gastroenterol Res Pract. 2021;2021:6655. doi: 10.1155/2021/6655900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chao G, Zhang S. Aquaporins 1, 3 and 8 expression and cytokines in irritable bowel syndrome rats’ colon via cAMP-PKA pathway. Int J Clin Exp Pathol. 2018;11:4117. [PMC free article] [PubMed] [Google Scholar]
- 84.Chao G, Wang Z, Yang Y, Zhang S. LncRNA H19 as a competing endogenous RNA to regulate AQP expression in the intestinal barrier of IBS-D patients. Front Physiol. Front Physiol. 2021;11:607026. doi: 10.3389/fphys.2020.602076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barbara G, Stanghellini V, De Giorgio R, Cremon C, Cottrell GS, Santini D, et al. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology. 2004;126:693–702. doi: 10.1053/j.gastro.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 86.Matricon J, Meleine M, Gelot A, Piche T, Dapoigny M, Muller E, et al. Review article: associations between immune activation, intestinal permeability and the irritable bowel syndrome. Aliment Pharmacol Ther. 2012;36:1009–1031. doi: 10.1111/apt.12080. [DOI] [PubMed] [Google Scholar]
- 87.Vara EJ, Brokstad KA, Hausken T, Lied GA. Altered levels of cytokines in patients with irritable bowel syndrome are not correlated with fatigue. Int J Gen Med. 2018;11:285–291. doi: 10.2147/IJGM.S166600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barbara G, Wang B, Stanghellini V, de Giorgio R, Cremon C, Di Nardo G, et al. Mast cell-dependent excitation of visceral-nociceptive sensory neurons in irritable bowel syndrome. Gastroenterology. 2007;132:26–37. doi: 10.1053/j.gastro.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 89.Kress M, Hüttenhofer A, Landry M, Kuner R, Favereaux A, Greenberg D, et al. microRNAs in nociceptive circuits as predictors of future clinical applications. Front Mol Neurosci. 2013;6:33. doi: 10.3389/fnmol.2013.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ren HX, Zhang FC, Luo HS, Zhang G, Liang LX. Role of mast cell-miR-490-5p in irritable bowel syndrome. World J Gastroenterol. 2017;23:93–102. doi: 10.3748/wjg.v23.i1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ji LJ, Li F, Zhao P, Weng LP, Wei J, Yan J, et al. Silencing interleukin 1α underlies a novel inhibitory role of miR-181c-5p in alleviating low-grade inflammation of rats with irritable bowel syndrome. J Cell Biochem. 2019;120:15268–15279. doi: 10.1002/jcb.28794. [DOI] [PubMed] [Google Scholar]
- 92.Heitkemper MM, Chang L. Do fluctuations in ovarian hormones affect gastrointestinal symptoms in women with irritable bowel syndrome? Gend Med. 2009;6(Suppl 2):152–167. doi: 10.1016/j.genm.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meleine M, Matricon J. Gender-related differences in irritable bowel syndrome: potential mechanisms of sex hormones. World J Gastroenterol. 2014;20:6725–6743. doi: 10.3748/wjg.v20.i22.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Weaver KR, Melkus GD, Fletcher J, Henderson WA. Relevance of sex and subtype in patients with IBS: an exploratory study of gene expression. Biol Res Nurs. 2020;22:13–23. doi: 10.1177/1099800419889189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jacenik D, Cygankiewicz AI, Fichna J, Mokrowiecka A, Małecka-Panas E, Krajewska WM. Estrogen signaling deregulation related with local immune response modulation in irritable bowel syndrome. Mol Cell Endocrinol. 2018;471:89–96. doi: 10.1016/j.mce.2017.07.036. [DOI] [PubMed] [Google Scholar]
- 96.Salvo-Romero E, Martínez C, Lobo B, Rodiño-Janeiro BK, Pigrau M, Sánchez-Chardi AD, et al. Overexpression of corticotropin-releasing factor in intestinal mucosal eosinophils is associated with clinical severity in diarrhea-predominant irritable bowel syndrome. Sci Rep. 2020;10:20706. doi: 10.1038/s41598-020-77176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang S, Fujita Y, Matsuzaki R, Yamashita T. Class I histone deacetylase (HDAC) inhibitor CI-994 promotes functional recovery following spinal cord injury. Cell Death Dis. 2018;9:460. doi: 10.1038/s41419-018-0543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim T, Song S, Park Y, Kang S, Seo H. HDAC inhibition by valproic acid induces neuroprotection and improvement of PD-like behaviors in LRRK2 R1441G transgenic mice. Exp Neurobiol Exp Neurobiol. 2019;28:504–515. doi: 10.5607/en.2019.28.4.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu Y, Dou J, Wan X, Leng Y, Liu X, Chen L, et al. Histone Deacetylase inhibitor MS-275 alleviates postoperative cognitive dysfunction in rats by inhibiting hippocampal neuroinflammation. Neuroscience. 2019;417:70–80. doi: 10.1016/j.neuroscience.2019.08.020. [DOI] [PubMed] [Google Scholar]
- 100.Jaworska J, Zalewska T, Sypecka J, Ziemka-Nalecz M. Effect of the HDAC inhibitor, sodium butyrate, on neurogenesis in a rat model of neonatal hypoxia-ischemia: potential mechanism of action. Mol Neurobiol Mol Neurobiol. 2019;56:6341–6370. doi: 10.1007/s12035-019-1518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wei H, Yu C, Zhang C, Ren Y, Guo L, Wang T, et al. Butyrate ameliorates chronic alcoholic central nervous damage by suppressing microglia-mediated neuroinflammatio and modulating themicrobiome-gut–brain axis. Biomed Pharmacother. 2023;160:114308. doi: 10.1016/j.biopha.2023.114308. [DOI] [PubMed] [Google Scholar]
- 102.Guo TT, Zhang Z, Sun Y, Zhu R-Y, Wang F-X, Ma L-J, et al. Neuroprotective effects of sodium butyrate by restoring gut microbiota and inhiiting TLR4 signaling in mice with MPTP-induced Parkinson's disease. Nutrients. 2023;15:930. doi: 10.3390/nu15040930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Patnala R, Arumugam TV, Gupta N, Dheen ST. HDAC inhibitor sodium butyrte-mediated epigenetic regulation enhances neuroprotective function of microglia during ischemic stroke. Mol Neurobiol. 2017;54:6391–6411. doi: 10.1007/s12035-016-0149-z. [DOI] [PubMed] [Google Scholar]
- 104.Terry N, Margolis KG. Serotonergic mechanisms regulating the GI tract: experimental evidence and therapeutic relevance. Handb Exp Pharmacol. 2017;239:319–342. doi: 10.1007/164_2016_103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mars RAT, Yang Y, Ward T, Houtti M, Priya S, Lekatz HR, et al. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell. 2020;182:1460–1473.e17. doi: 10.1016/j.cell.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mishima Y, Ishihara S. Enteric microbiota-mediated serotonergic signaling in pathogenesis of irritable bowel syndrome. Int J Mol Sci. 2021;22:10235. doi: 10.3390/ijms221910235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bhattarai Y, Williams BB, Battaglioli EJ, Whitaker WR, Till L, Grover M, et al. Gut Microbiota-produced tryptamine activates an epithelial G-protein-coupled receptor to increase colonic secretion. Cell Host Microbe. 2018;23:775–785.e5. doi: 10.1016/j.chom.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Walther DJ, Stahlberg S, Vowinckel J. Novel roles for biogenic monoamines: from monoamines in transglutaminase-mediated post-translational protein modification to monoaminylation deregulation diseases. FEBS J. 2011;278:4740–4755. doi: 10.1111/j.1742-4658.2011.08347.x. [DOI] [PubMed] [Google Scholar]
- 109.Farrelly LA, Thompson RE, Zhao S, Lepack AE, Lyu Y, Bhanu NV, et al. Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature. 2019;567:535–539. doi: 10.1038/s41586-019-1024-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen K, Luan X, Liu Q, Wang J, Chang X, Snijders AM, et al. Drosophila histone demethylase KDM5 regulates social behavior through immune control and gut microbiota maintenance. Cell Host Microbe. 2019;25:537–552.e8. doi: 10.1016/j.chom.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kapeller J, Houghton LA, Mönnikes H, Walstab J, Möller D, Bönisch H, et al. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17:2967–2977. doi: 10.1093/hmg/ddn195. [DOI] [PubMed] [Google Scholar]
- 112.Wohlfarth C, Schmitteckert S, Härtle JD, Houghton LA, Dweep H, Fortea M, et al. miR-16 and miR-103 impact 5-HT4 receptor signalling and correlate with symptom profile in irritable bowel syndrome. Sci Rep. 2017;7:14680. doi: 10.1038/s41598-017-13982-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wade PR, Chen J, Jaffe B, Kassem IS, Blakely RD, Gershon MD. Localization and function of a 5-HT transporter in crypt epithelia of the gastrointestinal tract. J Neurosci. 1996;16:2352–2364. doi: 10.1523/JNEUROSCI.16-07-02352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liao XJ, Mao WM, Wang Q, Yang GG, Wu WJ, Shao SX. MicroRNA-24 inhibits serotonin reuptake transporter expression and aggravates irritable bowel syndrome. Biochem Biophys Res Commun. 2016;469:288–293. doi: 10.1016/j.bbrc.2015.11.102. [DOI] [PubMed] [Google Scholar]
- 115.Barbaro MR, Di Sabatino A, Cremon C, Giuffrida P, Fiorentino M, Altimari A, et al. Interferon-γ is increased in the gut of patients with irritable bowel syndrome and modulates serotonin metabolism. Am J Physiol Gastrointest Liver Physiol. 2016;310:G439–G447. doi: 10.1152/ajpgi.00368.2015. [DOI] [PubMed] [Google Scholar]
- 116.Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science. 2010;329:1537–1541. doi: 10.1126/science.1193692. [DOI] [PubMed] [Google Scholar]
- 117.Moya PR, Wendland JR, Salemme J, Fried RL, Murphy DL. miR-15a and miR-16 regulate serotonin transporter expression in human placental and rat brain raphe cells. Int J Neuropsychopharmacol. 2013;16:621–629. doi: 10.1017/S1461145712000454. [DOI] [PubMed] [Google Scholar]
- 118.Zhu H, Xiao X, Chai Y, Li D, Yan X, Tang H. MiRNA-29a modulates visceral hyperalgesia in irritable bowel syndrome by targeting HTR7. Biochem Biophys Res Commun. 2019;511:671–678. doi: 10.1016/j.bbrc.2019.02.126. [DOI] [PubMed] [Google Scholar]
- 119.Farmer AD, Aziz Q. Gut pain & visceral hypersensitivity. Br J pain. 2013;7:39–47. doi: 10.1177/2049463713479229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kano M, Muratsubaki T, Van Oudenhove L, Morishita J, Yoshizawa M, Kohno K, et al. Altered brain and gut responses to corticotropin-releasing hormone (CRH) in patients with irritable bowel syndrome. Sci Rep. 2017;7:12425. doi: 10.1038/s41598-017-09635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bradford K, Shih W, Videlock EJ, Presson AP, Naliboff BD, Mayer EA, et al. Association between early adverse life events and irritable bowel syndrome. Clin Gastroenterol Hepatol. 2012;10:385–390.e3. doi: 10.1016/j.cgh.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sah A, Sotnikov S, Kharitonova M, Schmuckermair C, Diepold RP, Landgraf R, et al. Epigenetic mechanisms within the cingulate cortex regulate innate anxiety-like behavior. Int J Neuropsychopharmacol. 2019;22:317–328. doi: 10.1093/ijnp/pyz004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mahurkar-Joshi S, Chang L. Epigenetic mechanisms in irritable bowel syndrome. Front psychiatry. 2020;11:805. doi: 10.3389/fpsyt.2020.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li Q, Winston JH, Sarna SK. Developmental origins of colon smooth muscle dysfunction in IBS-like rats. Am J Physiol Gastrointest Liver Physiol. 2013;305:503–512. doi: 10.1152/ajpgi.00160.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Long X, Li M, Li LX, Sun YY, Zhang WX, Zhao DY, et al. Butyrate promotes visceral hypersensitivity in an IBS-like model via enteric glial cell-derived nerve growth factor. Neurogastroenterol Motil. 2018;30:e13227. doi: 10.1111/nmo.13227. [DOI] [PubMed] [Google Scholar]
- 126.Beggs S, Currie G, Salter MW, Fitzgerald M, Walker SM. Priming of adult pain responses by neonatal pain experience: maintenance by central neuroimmune activity. Brain. 2012;135:404–417. doi: 10.1093/brain/awr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Prusator DK, Greenwood-Van MB. Sex-related differences in pain behaviors following three early life stress paradigms. Biol Sex Differ. 2016;7:29. doi: 10.1186/s13293-016-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moloney RD, Stilling RM, Dinan TG, Cryan JF. Early-life stress-induced visceral hypersensitivity and anxiety behavior is reversed by histone deacetylase inhibition. Neurogastroenterol Motil. 2015;27:1831–1836. doi: 10.1111/nmo.12675. [DOI] [PubMed] [Google Scholar]
- 129.Daskalakis NP, De KER, Yehuda R, Malaspina D, Kranz TM. Early life stress effects on glucocorticoid-BDNF interplay in the hippocampus. Front Mol Neurosci. 2015;8:1–13. doi: 10.3389/fnmol.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vaiserman AM, Koliada AK. Early-life adversity and long-term neurobehavioral outcomes: epigenome as a bridge? Hum Genomics. 2017;11:34. doi: 10.1186/s40246-017-0129-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yu YB, Zuo XL, Zhao QJ, Chen FX, Yang J, Dong YY, et al. Brain-derived neurotrophic factor contributes to abdominal pain in irritable bowel syndrome. Gut. 2012;61:685–694. doi: 10.1136/gutjnl-2011-300265. [DOI] [PubMed] [Google Scholar]
- 132.Zhang Y, Qin G, Liu DR, Wang Y, Yao SK. Increased expression of brain-derived neurotrophic factor is correlated with visceral hypersensitivity in patients with diarrhea-predominant irritable bowel syndrome. World J Gastroenterol. 2019;25:269–281. doi: 10.3748/wjg.v25.i2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Winston JH, Li Q, Sarna SK. Chronic prenatal stress epigenetically modifies spinal cord BDNF expression to induce sex-specific visceral hypersensitivity in offspring. Neurogastroenterol Motil. 2014;26:715–730. doi: 10.1111/nmo.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Silberman DM, Acosta GB, Zorrilla Zubilete MA. Long-term effects of early life stress exposure: role of epigenetic mechanisms. Pharmacol Res. 2016;109:64–73. doi: 10.1016/j.phrs.2015.12.033. [DOI] [PubMed] [Google Scholar]
- 135.Lambert WM, Xu C-F, Neubert TA, Chao MV, Garabedian MJ, Jeanneteau FD. Brain-derived neurotrophic factor signaling rewrites the glucocorticoid transcriptome via glucocorticoid receptor phosphorylation. Mol Cell Biol. 2013;33:3700–3714. doi: 10.1128/MCB.00150-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Louwies T, Greenwood-Van MB. Sex differences in the epigenetic regulation of chronic visceral pain following unpredictable early life stress. Neurogastroenterol Motil. 2020;32:4. doi: 10.1111/nmo.13751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tran L, Chaloner A, Sawalha AH, Greenwood V-M. Importance of epigenetic mechanisms in visceral pain induced by chronic water avoidance stress. Psychoneuroendocrinology. 2013;38:898–906. doi: 10.1016/j.psyneuen.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 138.Cao DY, Bai G, Ji Y, Karpowicz J, Traub RJ. EXPRESS: Histone hyperacetylation modulates spinal type II metabotropic glutamate receptor alleviating stress-induced visceral hypersensitivity in female rats. Mol Pain. 2016;12:2. doi: 10.1177/1744806916660722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cao DY, Bai G, Ji Y, Traub RJ. Epigenetic upregulation of metabotropic glutamate receptor 2 in the spinal cord attenuates oestrogen-induced visceral hypersensitivity. Gut. 2015;64:1913–1920. doi: 10.1136/gutjnl-2014-307748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fei L, Wang Y. microRNA-495 reduces visceral sensitivity in mice with diarrhea-predominant irritable bowel syndrome through suppression of the PI3K/AKT signaling pathway via PKIB. IUBMB Life. 2020;72:1468–1480. doi: 10.1002/iub.2270. [DOI] [PubMed] [Google Scholar]
- 141.Wu R, Zhang P-A, Liu X, Zhou X, Xu M, Jiang X, et al. Decreased miR-325-5p contributes to visceral hypersensitivity through post-trasncriptional upregulation of CCL2 in rat dorsal root ganglia. Neurosci Bull. 2019;35:791–801. doi: 10.1007/s12264-019-00372-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Van Steenwinckel J, Auvynet C, Sapienza A, Reaux-Le Goazigo A, Combadiere C, Melik PS. Stromal cell-derived ccl2 drives neuropathic pain states through myeloid cell infiltration in injured nerve. Brain Behav Immun. 2015;45:198–210. doi: 10.1016/j.bbi.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 143.Xiromerisiou G, Marogianni C, Lampropoulos IC, Dardiotis E, Speletas M, Ntavaroukas P, et al. Peripheral inflamamtory markers TNF-α and CCL2 revisited: association with Parkinson's disease severity. Int J Mol Sci. 2022;24:264. doi: 10.3390/ijms24010264. [DOI] [PMC free article] [PubMed] [Google Scholar]