Abstract
Lung cancer, a highly malignant disease, greatly affects patients’ quality of life. N6-methyladenosine (m6A) is one of the most common posttranscriptional modifications of various RNAs, including mRNAs and ncRNAs. Emerging studies have demonstrated that m6A participates in normal physiological processes and that its dysregulation is involved in many diseases, especially pulmonary tumorigenesis and progression. Among these, regulators including m6A writers, readers and erasers mediate m6A modification of lung cancer-related molecular RNAs to regulate their expression. Furthermore, the imbalance of this regulatory effect adversely affects signalling pathways related to lung cancer cell proliferation, invasion, metastasis and other biological behaviours. Based on the close association between m6A and lung cancer, various prognostic risk models have been established and novel drugs have been developed. Overall, this review comprehensively elaborates the mechanism of m6A regulation in the development of lung cancer, suggesting its potential for clinical application in the therapy and prognostic assessment of lung cancer.
Subject terms: Epigenetics, Lung cancer
Introduction
According to Global Cancer Statistics 2020, lung cancer deaths are still the leading cause of cancer death, accounting for 18% of total deaths [1]. Histologic classification divides lung cancer into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), the former being subclassified into lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC) and large-cell carcinoma (LCC) [2]. NSCLC accounts for more than 80% of diagnosed lung cancers [3], among which LUAD has surpassed LUSC to become the most common NSCLC. Although targeted therapy and immunotherapy decreased lung cancer incidence (by 2.2–2.3% from 2014 to 2018) [4], inevitable drug resistance remains the biggest obstacle to treatment efficacy. The transdifferentiation of NSCLC with lineage plasticity into SCLC is one of the reasons for its drug resistance, which involves genetic and epigenetic mechanism [5]. Thus, it is necessary to further study the molecular mechanism of lung cancer progression in order to ameliorate drug resistance and find more effective therapeutic targets.
With the progress of research, the understanding of cancer progression has been deepened continuously, from the initial belief that cancer is only driven by genetic alterations, to the discovery that epigenetics is also involved [6]. Epigenetic modification includes DNA methylation, histone modification and RNA modification, among which RNA modification has gradually taken centre stage in the field of cancer research [7]. There are over 160 known posttranscriptional RNA modifications that can affect RNA function by influencing its structure, such as N6-methyladenosine (m6A), N1-methyladenosine (m1A) and 5-methylcytosine [8], among which m6A is the most common. The mainstream m6A site detection methods include m6A sequencing (m6A-seq) and m6A-specific methylated RNA immunoprecipitation. The principle is to enrich m6A-modified RNA fragments through immunocoprecipitation and conduct high-throughput sequencing of these fragments [9, 10]. But because these methods can only capture 100–200 nt RNA fragments and with inadequate resolution, researchers have refined the techniques to improve resolution, such as photo-crosslinking-assisted m6A sequencing [11], m6A individual-nucleotide-resolution cross-linking and immunoprecipitation [12] and m6A-CLIP [13]. Moreover, SCARLET was developed to verify the accuracy of high-throughput detection, which can accurately identify single m6A site on mRNA and lncRNA [14]. And m6A-LAIC-seq can quantitatively analyse the m6A modification level of genes with introduced internal reference RNA [15]. In recent years, some new technologies have also emerged, such as antibodies-independent MAZTER-seq [16], and deep learning-based FunDMDeep-m6A [17]. Liu et al. found that direct RNA sequencing detected m6A with high accuracy, in the form of systematic errors and reduced base-calling qualities [18], and Zhang et al. formed m6A-REF-seq, which can explicitly clarify m6A enrichment patterns near RNA stop codons [19]. Thanks to these techniques, researchers can identify and localise m6A sites at a transcriptome-wide level [9]. It is found that m6A modifies the consensus motif RRm6ACH ([A/G/U] [A/G]m6AC[A/C/U]) in eukaryotic cells, which is highly conservative in both human and mouse and is enriched within long internal exons, near stop codons and in 3′ untranslated regions (3′ UTRs) [9, 10]. The m6A regulation system is composed of ‘writers’, ‘erasers’ and ‘readers’. ‘Writer’ refers to the methyltransferase complex, which accomplishes the methylation of m6A [20]. M6A modification is demethylated by ‘erasers’, fat mass and obesity-associated protein (FTO) and alkB homologue 5 (ALKBH5) [20]. ‘Readers’ include the YT521-B homology (YTH) domain-containing protein, insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) and the heterogeneous nuclear ribonucleoprotein (HNRNP) family, which recognise the m6A-modified transcript and regulate the downstream target RNA [21].
M6A methylation plays a major role in the mRNA life cycle and participates in the transcription, export, translation and degradation of mRNA [22]. It is reported that the m6A reader YTHDC1 activates the SRSF3 pathway to modulate alternative splicing of pre-mRNA in the nucleus [23]. After processing, the mature mRNA is then exported to the cytoplasm with the assistance of m6A regulators METTL3, ALKBH5 and YTHDC1 [24, 25]. Then, YTHDF1 interacts with eukaryotic initiation factor 3 (eIF3) to promote mRNA translation efficiency in an m6A-dependent manner [26]. Finally, m6A regulators promote m6A-modified mRNA degradation via targeting distinct pathways and transcripts, such as YTHDF2 activates deadenylating and endoribonucleolytic cleavage pathways, YTHDC2 recruits 5′-3′ exoribonuclease XRN1 [27]. M6A has also been found in many types of noncoding RNAs (ncRNAs), including long ncRNAs (lncRNAs), microRNAs (miRNAs), circular RNAs (circRNAs), and ribosomal RNAs (rRNAs) [8, 28, 29]. It regulates the stability, transport and decay of ncRNAs; further studies report that ncRNAs can affect m6A modification via distinct signalling pathways [30, 31] and play a powerful role in regulating different cancer types. For instance, miRNA-96 regulates AMPKα2-FTO-m6A/MYC axis to promote the progression of colorectal cancer [32], circVMP1/miR-524-5p-METTL3-m6A/SOX2 axis affects the malignancy of NSCLC [33] and high HNRNPC promotes breast cancer cell proliferation via circBACH2/has-miR-944/HNRNPC axis [34].
M6A modification is associated with many biological processes and diseases. For instance, it contributes to the normal development of haematopoietic system and central nervous system, and abnormal m6A modification regulates target genes to act as promoter or suppressor in the progression of different types of cancer (such as bladder cancer, acute myelocytic leukaemia and lung cancer [20, 35, 36]. Besides, m6A is implicated in the transdifferentiation of tumour cells, as the remodelling of m6A methylation was found in the transformation of pre-B cells into macrophages in acute lymphoblastic leukaemia [37], which encourages in-depth exploration on the role of the shifts in m6A RNA patterns in lung cancer transdifferentiation. The existing studies have shown that LUAD retains its original epigenetic features in the transformation process to LUSC and SCLC, suggesting that epigenetic therapy may reverse TKIs resistance during phenotypic transformation in EGFR-mutant NSCLC [5]. In short, this review focuses on the biological function of m6A methylation in the progression of lung cancer, and its potential in diagnosing and predicting the prognosis of patients, aiming to summarise the new progress of epigenetic modification to guide improved therapy for lung cancer.
The composition and function of the m6A system
Writers
The highly conserved mRNA consensus site is recognised by the methyltransferase complex, which is also called the ‘writer’ [38]. The catalytic core METTL3-METTL14 heterodimer plays a major role in m6A methylation, catalysing the transfer of a methyl group from donor S-adenosyl methionine to adenine nucleobases in acceptor RNA substrates [39]. Although METTL14 is catalytically inactive, it acts as a stabiliser to increase METTL3 activity [40]. Further studies show that Wilms’ tumour 1-associating protein (WTAP) is also noncatalytic but is crucial in regulating and localising METTL3 and METTL14 into target RNA [41]. In brief, METTL3, METTL14 and WTAP combine structurally to form the core component of methyltransferase. Other methyltransferase components form the auxiliary structure of catalysis. Vir-like m6A methyltransferase associated recruits the METTL3/METTL14/WTAP complex and is linked with polyadenylation cleavage factors to mediate site-specific mRNA methylation [42]. RNA binding motif protein 15 (RBM15) can facilitate methylation of the m6A-methyltransferase complex [43]. Zinc finger CCCH domain-containing protein 13 interacts with WTAP to affect METTL3/METTL14/WTAP subcellular localisation [44]. The synergy between all the components of m6A writer ensures normal m6A methylation.
In addition to the m6A epigenetic modification in mRNA, there are other writer-mediated m6A modifications in ncRNA [45]. METTL16, a conserved protein found from bacteria to vertebrates, has been verified to be the methyltransferase for the U6 snRNA. Moreover, METTL16 was found to bind to lncRNA MALAT1, but its consequent role remains unclear [46]. There are also abundant m6A modifications in rRNA. For example, zinc finger CCHC domain-containing protein 4 is the methyltransferase of 28S rRNA and METTL5 acts as a ribosomal RNA writer by forming a heterodimer with tRNA methyltransferase activator subunit 11-2 [47]; studies have found that they are all related to cancer progression.
Readers
M6A readers are a class of RNA-binding proteins (RBPs) that recognise RNA-specific m6A sites to affect RNA metabolism and relevant biological processes [48]; different readers function at different cellular locations. In the nucleus, YTHDC1 exports methylated mRNA and recognises m6A residues to facilitate RNA biological function [43, 49]. In the cytoplasm, YTHDF proteins affect different stages of the mRNA life cycle. YTHDF2 destabilises m6A-containing RNAs [50], while YTHDF1 increases their translation efficiency by interacting with eIF3 [26]. YTHDF3 cooperates with YTHDF1 and YTHDF2 to regulate methylated mRNA decay and translation [51, 52]. In the nucleus, HNRNPC preferentially binds to the m6A-modified site and then affects mRNA maturation [53]. M6A also affects microRNA processing and mRNA alternative splicing mediated by HNRNPA2B1 and HNRNPG, respectively [54, 55].
There are some studies on other m6A readers, but deeper explorations are needed to elucidate their functions. IGF2BPs promote the stability and translation of target mRNAs in an m6A-dependent manner, while fragile X mental retardation protein 1 has the opposite effect [56, 57]. However, whether FMRP and IGF2BPs are direct RBPs remains to be explored.
Erasers
M6A erasers are demethylases that remove the methyl group from m6A, making methylation a reversible process [58]. Studies identified FTO as the first m6A eraser, but the substrates for FTO are still being explored. Initially, experiments revealed that FTO colocalised with nuclear speckles and negatively regulated m6A mRNA levels [59]. However, recent studies also found that FTO preferentially demethylates N6,2′-O-dimethyladenosine (m6Am) in the 5′ cap of mRNA instead of m6A [60]. Thus, a specific site needs to be found to further identify the bona fide FTO substrate.
Another important RNA demethylase, ALKBH5, has similar demethylation activity to that of FTO and with a preference for m6A [24]. The modification ALKBH5 exerts on mRNA subsequently influences mRNA metabolism and has a crucial role in essential biological processes. One of the interesting examples is that ALKBH5 is indispensable in normal spermatogenesis in mouse testes [61]. There are also other members of the ALKBH family that act as demethylases of RNA, and ALKBH3 and ALKBH1 mediate demethylation of m6A tRNA to regulate protein synthesis and affect tumour cells’ malignant progression [62, 63]. Researchers are still exploring the identification of more erasers, their preferential substrates, and their demethylation mechanisms.
The important regulatory function of m6A methylation in lung cancer progression
The role of m6A methylation in lung tumorigenesis and proliferation
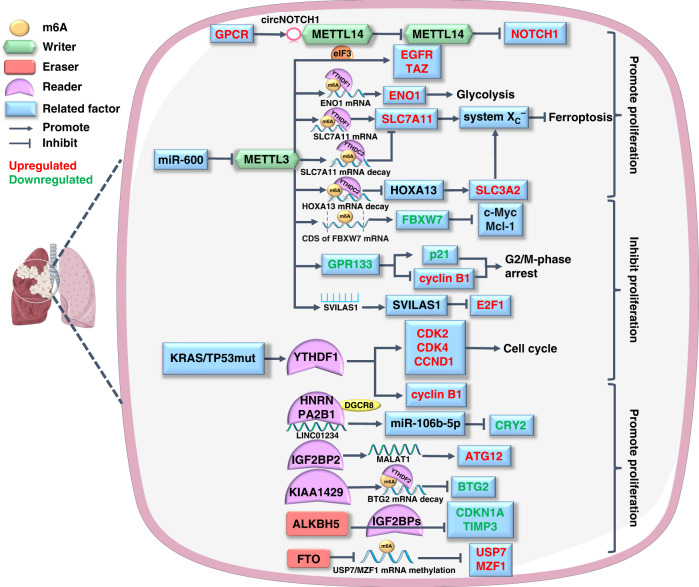
The imbalance of proto-oncogenes and tumour suppressor genes plays an important role in cell carcinogenesis [64] and the effect of m6A methylation on cancer-related genes is mediated by mRNA. The interaction between m6A and ncRNA is also involved in the proliferation of lung cancer cells. The dual regulatory role of m6A modification on tumour cell proliferation will be presented in the following section (Fig. 1).
Fig. 1. The m6A modification regulates the proliferation of lung cancer.
The m6A regulators (including writers, readers and erasers) dually regulate lung cancer proliferation by affecting the expression of proteins in signalling pathways related to glycolysis, ferroptosis and cell cycle. In particular, the effective cooperation between m6A writer and reader is significant for this process. The molecules affected by m6A modification in lung cancer are colour-coded, with red for upregulated and green for downregulated.
The reprogramming of energy metabolism patterns is an important character of cancer, which provides cancer cells with sufficient materials for sustaining proliferation [65]. Recent studies have explored the role of m6A methylation in cancer energy metabolism, especially glucose (Warburg effect), lipid and iron metabolism. Since different m6A regulators act as pivotal hubs in metabolic pathways such as mTOR, PTEN, MAPK and AMPK to regulate tumoral energy homoeostasis, the dysfunction of m6A modification has a great impact on tumorigenesis [66]. One of the typical metabolic characteristics of cancer cells, glycolysis, can be catalysed by enolase1 (ENO1) [67]. Ma et al. found that ENO1 mRNA is m6A methylated by an increase in the writer METTL3 and a decrease in the eraser ALKBH5, which synergise with the reader YTHDF1 to reinforce glycolysis and promote the growth of LUAD by upregulating ENO1 [68]. Moreover, m6A methylation also acts as a ferroptosis inducer to cause lipid peroxidation in LUAD. The antioxidant system XC− plays an important role in ferroptosis, and the solute carriers 7A11 (SLC7A11) and 3A2 are two main members in it [69]. In 2021, another study by Ma et al. reported that the m6A reader YTHDC2 inhibited SLC7A11 via its m6A-reading domain, which impaired XC− system antioxidant activity and induced ferroptosis [70]. Similarly, YTHDC2 also destabilised homeobox A13 mRNA, a transcription factor of SLC3A2, to suppress its transcription in an m6A-dependent manner, acting as a ferroptosis inducer to suppress LUAD growth [71]. In contrast to YTHDC2, the m6A writer METTL3, as a ferroptosis inhibitor, increases SLC7A11 expression by recognising its mRNA m6A modification through YTHDF1 and ultimately inhibits lung cancer cell ferroptosis to promote its proliferation [72]. Another important event in tumorigenesis, hypoxia, can change the metabolic pattern of tumour cells [73]; studies have pointed out the role of YTH family as an m6A recogniser in hypoxic lung cancer progression. Xu et al. revealed that hypoxia-induced YTHDF2 overexpression activated the mTOR/AKT axis via m6A modification, which contributed to LUSC cell proliferation [74]. Additionally, Shi et al. found that under normoxic conditions, YTHDF1 depletion reduced the m6A modification levels of several cell cycle checkpoint regulators, such as cyclin-dependent kinase 2/4 and cyclin D1, and downregulated their expression to retard NSCLC cell proliferation. Further research revealed that YTHDF1 knockdown mediated the hypoxic adaptation of NSCLC cancerous cells by decreasing Kelch-like ECH-associated protein 1 (Keap1) and activating the Nrf2-AKR1C1 axis in an m6A-dependent manner [75].
While affecting the metabolism of cancer cells, m6A regulators including writers, readers and erasers also exhibit regulatory effects on the mRNA of oncogenes and tumour suppressor genes via m6A modification. These kinds of regulations alter the expression of proteins or other tumorigenesis-associated factors, affecting the proliferation of different types of lung tumour cells. First, studies have shed light on how m6A writers are involved in the proliferative behaviour of pulmonary neoplasms by regulating the transcription and translation of different genes. One study reported that the m6A writer METTL3 recruited eIF3 to independently initiate the translation of several oncogenes, such as epidermal growth factor receptor (EGFR) and the Hippo pathway effector TAZ, in lung neoplasms [76]. However, some studies have revealed the role of METTL3 as a tumour inhibitor. For instance, METTL3 can upregulate G protein-coupled receptor 133 in an m6A-dependent manner, which induces G2/M-phase cell cycle arrest of tumour cells to inhibit LUAD growth [77]. In addition, METTL3 installs m6A modification of F-box and WD repeat domain-containing 7 mRNA to upregulate its expression and facilitate its LUAD inhibitory effect [78]. Another m6A methyltransferase, METTL14, degrades Notch homologue 1 mRNA to inhibit the proliferation of NSCLC cells; this tumour-suppressing effect can be competitively reversed by the G protein-coupled oestrogen receptor [79]. Zhang et al. revealed that overexpressed KIAA1429, another core protein of m6A methyltransferase, interacted with the reader YTHDF2 to modulate m6A modification on BTG anti-proliferation factor 2 (BTG2) mRNA to downregulate BTG2, and ultimately promoted LUAD cell proliferation [80]. In addition to m6A writer methylation, the performance of m6A-modified RNAs biological functions also requires readers and specific RBPs. Therefore, abnormal expression of readers will also cause dysregulation of related RNA to affect lung cancer cell proliferative behaviour. Lou et al. found that markedly increased YTHDF1 in KRAS and TP53 co-mutation LUAD patients promoted the proliferation of cancer cells by upregulating cyclin B1 in an m6A-dependent manner [81]. M6A erasers also play important roles in modulating lung tumour growth. Microarray analysis showed that the eraser ALKBH5 demethylated m6A modification of many IGF2BP target genes (such as E2F1, p21 and TIMP3) to promote proliferation of NSCLC cells [82]. Additionally, an experiment carried out by Zhu et al. confirmed that ALKBH5 targeted tissue inhibitors of metalloproteinase 3 (TIMP3) mRNA to decrease its translation by m6A demethylation, which attenuated its tumour suppressive function in NSCLC [83]. Another m6A eraser, FTO, also plays an oncogenic role in the growth of NSCLC. FTO demethylates m6A modification on ubiquitin-specific protease-7 to elevate its expression, which then promotes the proliferation of NSCLC cells through p53-dependent pathways [84]. Moreover, abnormally upregulated FTO demethylates m6A in myeloid zinc finger protein 1 mRNA, which upregulates this proliferation-related transcription factor to contribute to LUSC growth [85].
In addition to mRNA, m6A also interacts with ncRNA to affect lung cancer cell proliferation. M6A regulators modulate ncRNAs in an m6A-dependent manner, and the epigenetic modification of ncRNAs is critical for their subsequent effects on downstream targets [86]. Hu et al. reported that the m6A writer METTL3 was involved in the m6A-methylated stabilisation of the tumour suppressor SVIL antisense RNA 1, which promoted E2 promoter-binding Factor 1 (E2F1) degradation via ubiquitination to inhibit LUAD cell proliferation [87]. Additionally, IGF2BP2 was found to target and upregulate lncRNA MALAT1 in an m6A-dependent manner; the latter then elevated downstream autophagy-related 12 expression to promote NSCLC proliferation both in vitro and in vivo [88]. A comprehensive analysis verified that the m6A-related lncRNA ABALON sponged miR-139-3p to indirectly downregulate NIN1/RPN12 binding protein 1 homologue and inhibit lung cancer growth [89]. The ncRNA can also interact with m6A regulators to influence lung cancer cell proliferation. The m6A reader HNRNPA2B1 combined with biotinylated lncRNA LINC01234 was shown to recruit DiGeorge syndrome critical region 8. The latter promoted miR-106b-5p maturation to exert its inhibitory role on cryptochrome 2 (CRY2), weakening the CRY2 tumour suppressor effect and promoting the proliferation of NSCLC cells [90].
The role of m6A methylation in lung cancer invasion and metastasis
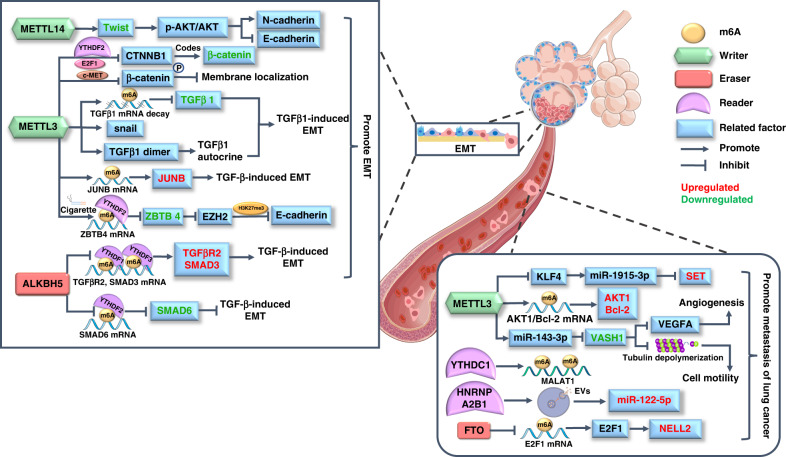
Cell invasion and metastasis are two other features of tumours and are often associated with poor prognosis. In recent years, many studies have uncovered the role of m6A modification in these two processes in lung cancer cells, which provides a new direction for improving the prognosis and survival rate of lung cancer patients (Fig. 2).
Fig. 2. The m6A modification regulates the invasion and migration of lung cancer.
The epithelial-mesenchymal transition (EMT) is a prominent mechanism of lung cancer metastasis, and m6A modification is involved in the regulation of EMT-related signalling pathways on epithelial or mesenchymal markers. Besides, the m6A-modified mRNA and ncRNA also directly modulate the expression of proteins in the pathological process of tumoral invasion and migration, and the aberrant molecules in lung cancer are marked in different colours.
Epithelial-mesenchymal transition (EMT) facilitates the invasion and metastasis of epithelial-derived tumours [91]; several signalling pathways have been implicated in this biological process, such as the TGF-β and Wnt/β-catenin signalling pathways [92]. TGF-β activates classic SMAD signalling or non-SMAD signalling pathways, which target downstream transcription factors such as Snail, Twist and zinc finger E-box binding homeobox, to mediate mesenchymal protein expression. Additionally, the activated β-catenin in Wnt pathway binds to the TCF/LEF family members to initiate transcription of downstream genes (such as c-Myc and cyclin D), and Wnt signalling elicits the expression of Snail1/2 and Twist, which finally decrease epithelial markers like E-cadherin and increase mesenchymal markers like N-cadherin and fibronectin [92]. Both of them contribute to the well-orchestrated programme of EMT. Some recent studies revealed the m6A modification on distinct EMT pathways and molecules, which provides a new perspective to understand the invasive and metastatic mechanisms of lung cancer cells. Recently, Yang et al. revealed that the knockdown of m6A reader METTL14 suppresses the p-AKT/AKT signalling pathway by destabilising Twist, which attenuates EMT and the malignancy of NSCLC [93]. In addition, researchers have found that METTL3 negatively regulates β-catenin by mediating m6A deposition on its coding gene and downregulates c-Met-mediated phosphorylation, thereby inhibiting β-catenin membrane localisation by modulating cell-cell adhesion. Both effects promote β-catenin-related EMT and dissemination of cancer cells [94]. There are also studies focusing on TGF-β, the most predominant molecule in inducing EMT. METTL3 was found to upregulate JUNB proto-oncogene, a critical transcriptional regulator of EMT, by modifying its mRNA in m6A modification, which contributed to TGF-β-induced EMT of lung cancer cells [95]. In addition, another study revealed multiple effects of METTL3 on TGFβ1. The METTL3 knockdown attenuated m6A modification on TGFβ1 mRNA, which upregulated TGFβ1 and increased its protein half-life, secretion and activation. Furthermore, the downregulated Snail caused the inability of TGFβ1 to induce EMT in METTL3-null cells. Therefore, the m6A-related METTL3/TGFβ1/Snail axis appears to be important in lung cancer cell metastasis [96]. In contrast to METTL3, ALKBH5 works as an m6A demethylase to decrease TGFβR2 and SMAD3, and increase SMAD6, impairing TGF-β/SMAD signalling pathway that associated with EMT and tumour metastasis [97]. A study also investigated cigarette smoke (CS)-induced EMT. High METTL3 levels were detected in CS-exposed human bronchial epithelial cells, and they induced m6A modification of zinc finger and BTB domain-containing 4 (ZBTB4) under the targeting of m6A reader YTHDF2. Decreased ZBTB4 then upregulated enhancer of zeste homologue 2 to silence E-cadherin and ultimately promoted EMT [98].
Researchers have also uncovered m6A modification in coding and noncoding RNAs, which are closely associated with the invasion and metastasis of various types of lung cancer. The m6A reader METTL3 methylates AKT serine/threonine kinase 1 (AKT1) mRNA to potentiate its translation; increased AKT1 contributes to advanced TNM stage and lymph node metastasis of NSCLC [99]. METTL3 also increases B-cell lymphoma-2 expression in an m6A-dependent manner and enhances the viability and migration of NSCLC cells [100]. Wang et al. found that the eraser FTO demethylated m6A on E2F1 to upregulate it, which then elevated expression of its downstream target neural epidermal growth factor-like 2 to further enhance cell invasion and metastasis of NSCLC [101]. In regard to the function of m6A regulators on ncRNAs, researchers have found that METTL3 assists miR-143-3p maturation in an m6A-dependent manner and miR-143-3p downregulates vasohibin-1, which weakens the inhibitory effect of the latter on vascular endothelial growth factor A and angiogenesis, promoting lung cancer cells crossing the blood-brain barrier to invade brain tissue [102]. Pan et al. found that METTL3-mediated m6A modification attenuated the transcriptional activity of the miR-1915-3p promoter in NSCLC, and low miR-1915-3p enhanced the migration and EMT of cancer cells through the upregulation of SET nuclear proto-oncogene (SET) [103]. Extracellular vesicles (EVs) are important for the transfer of molecules between different cells, which can cause cancer metastasis [104]. A recent study confirmed that the m6A reader HNRNPA2B1 participates in the selective sorting of miR-122-5p by interacting with its m6A site through the EXO motif and upregulates miR-122-5p in EVs of lung cancer cells to promote hepatic metastasis [105]. In addition, the m6A reader YTHDC1 was discovered to be recruited to nuclear speckles by lncRNA MALAT1, and it recognised the m6A site on MALAT1 to promote lung cancer metastasis [106].
The role of m6A methylation in lung cancer progression
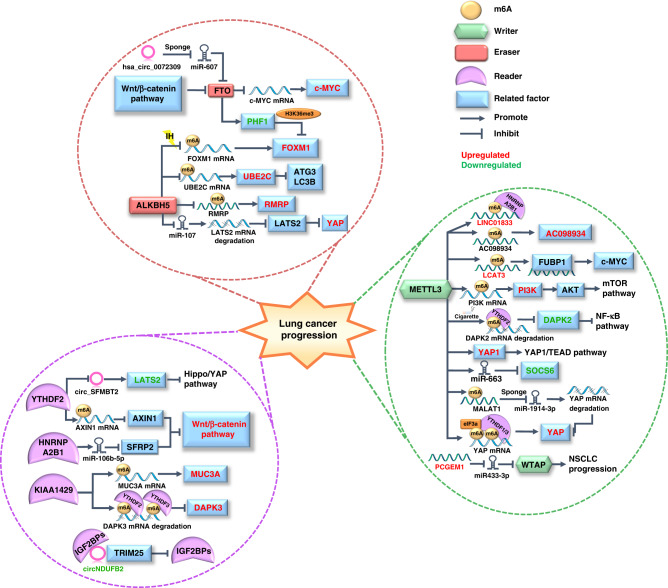
As mentioned above, m6A modification is essential in every biological behaviour of lung cancer cells, such as proliferation or metastasis. In addition to revealing the role of m6A enzymes in every stage of lung cancer progression, the holistic exploration of the role of m6A enzymes in the continuous process of lung cancer cell proliferation, invasion, and metastasis will enable us to better comprehend how the vast m6A regulatory network is finely orchestrated to regulate lung cancer progression (Fig. 3).
Fig. 3. The role of m6A modification in the progression of lung cancer.
The m6A modification contributes to the whole process of biogenesis of lung cancer via its epigenetic modification on various RNAs. The different signalling pathways involved in lung cancer progression are categorised according to the types of m6A regulators and the specific regulated molecules are marked in different colour codes to fully demonstrate the molecular mechanism by which m6A modification regulates lung tumour progress.
First, m6A modification exerts its major role in lung cancer progression by modifying various cancer-related mRNAs. Researchers found that phosphatidylinositol 3-kinase (PI3K) mRNA was upregulated by METTL3-mediated m6A modification and activated the downstream mTOR/AKT signalling pathway, which ultimately promoted lung cancer proliferation and metastasis [107]. The Hippo signalling pathway is another important pathway involved in the progression of the malignant phenotype of cancer, and Yes-associated protein (YAP) is a key effector of it. Jin et al. discovered that the m6A writer METTL3 and eraser ALKBH5 had opposite effects on YAP expression and activity. METTL3 not only recruits eIF3b to enhance YAP translation via interacting with YTHDF1/3 in an m6A-dependent manner but also increases YAP activity by promoting m6A-methylated MALAT1 to sponge miR-1914-3p, which promotes the invasion and metastasis of NSCLC. ALKBH5 suppresses the expression of YAP in YTHDF1/2/3-mediated m6A modification and decreases YAP activity through the miR-107/LATS2 axis to inhibit NSCLC progression. [108, 109]. YAP is also implicated in inflammatory microenvironment-related tumour progression. For example, high expression of IL-6 in the liver microenvironment may be involved in liver metastasis from lung cancer. Ni et al. carried out the in vitro experiment in A549 and H1975 LUAD cells to reveal that METTL3 and m6A methylation were remarkably increased and facilitated the proliferation and metastasis of LUAD cells in hepatic inflammatory microenvironment. The mechanism is that METTL3 methylated YAP1 mRNA in an m6A-dependent manner to enhance its translation, and then YAP1 activated the downstream YAP1/TEAD signalling pathway. Further research is required to investigate how this mechanism affects lung cancer progression in vivo. [110]. Moreover, a study on the association between cigarette smoking and lung cancer also showed a METTL3-mediated mechanism. Cigarette smoke exposure upregulates METTL3 mRNA, which cooperates with the m6A reader YTHDF2 to decrease death-associated protein kinase 2 (DAPK2) via m6A methylation. As a serine/threonine kinase, downregulated DAPK2 phosphorylates nuclear factor kappa-B (NF-κB) to activate the downstream oncogenic pathway, which promotes carcinogenesis of NSCLC [111]. Additionally, one study revealed that the m6A writer KIAA1429 works with the m6A readers YTHDF2/3 to mediate m6A-dependent degeneration of DAPK3 mRNA, reversing its role in inhibiting NSCLC malignancy [112]. Similarly, other studies also report the role of KIAA1429 as a lung tumour promotor that increases m6A levels of mucin 3A to promote the expression of this potential oncogene, facilitating the proliferation and migration of LUAD [113, 114]. There are also investigations of how readers and erasers recognise and demethylate m6A modification on mRNA to regulate key factors in proliferative and metastatic signalling pathways. Elevated YTHDF2 was found to recognise the m6A site on Axin1 mRNA to decrease this negative regulator of the Wnt/β-catenin signalling pathway and promote LUAD progression [115]. In regard to erasers, Guo et al. have discovered that ALKBH5 can demethylate m6A modification on ubiquitin-conjugating enzyme E2C (UBE2C) transcripts and stabilise them, then upregulate UBE2C-activated autophagy repression, which finally promotes NSCLC progression [116]. In addition, Yang et al. showed that low m6A eraser FTO mediated by the Wnt/β-catenin pathway elevated the proto-oncogene c-Myc via m6A demethylation and c-Myc regulated cancer cell aerobic glycolysis, promoting pulmonary tumorigenesis [117]. A body of evidence has confirmed the role of Forkhead-box (FOX) family proteins in carcinogenesis [118–120]; interestingly, both FTO and ALKBH5 can regulate the expression of FOXM1, a typical stemness-related oncogene in this family. Ning et al. revealed that FTO stabilised plant homeodomain finger protein 1 (PHF1) via m6A demethylation and inhibited the FTO/PHF1 axis in LUAD, promoting FOXM1 gene expression by reducing H3K36me3 deposition [121]. For LUAD patients with obstructive sleep apnoea, intermittent hypoxia (IH) is a key link in the malignant progression of cancer. Chao et al. reported elevated FOXM1 after m6A demethylation by ALKBH5 under IH [122]. The above regulation of FOXM1 contributes to LUAD cell growth, invasion and migration [121, 122]. Cancer stem cells (CSCs) have similar stem cell self-renewal capabilities, which are closely associated with the progression of lung tumours [123]. Studies have revealed that the m6A eraser ALKBH5 is transcriptionally regulated by p53 and contributes to the stemness of CSCs and malignancy of lung cancer [124].
Next, m6A modification also regulates the level of ncRNAs, such as miRNAs, lncRNAs and circRNAs, which function in promoting or inhibiting the proliferation, invasion and metastasis of lung cancer cells via different molecular axes [125]. Rong et al. found that the m6A reader HNRNPA2B1 recognised the m6A site of pri-miR-106b-5p to promote its maturation, and miR-106b-5p then degraded SFRP2, a regulator of Wnt/β-catenin signalling, to activate this pathway and enhance LUAD stemness and progression [126]. The m6A writer METTL3 promotes the maturation of miR-663 through m6A modification and then inhibits the expression of downstream target suppressor of cytokine signalling 6 to promote malignant progression of lung cancer [127]. In addition to miRNA, METTL3 also modified lncRNAs in an m6A-dependent manner to stabilise lncRNAs such as LCAT3 and AC098934. LCAT3 interacts with far upstream element binding protein 1 to activate c-Myc, and both LCAT3 and AC098934 promote the proliferation and migration of lung cancer cells [128, 129]. METTL3 upregulates long intergenic non-protein coding RNA 1833 (LINC01833) via its m6A-methyltransferase activity, and LINC01833 is recognised by the m6A reader HNRNPA2B1 to enhance the proliferative and invasive abilities of NSCLC [130]. Yu et al. determined that the m6A eraser ALKBH5 upregulated lncRNA RMRP via m6A demethylation, and high RMRP was positively associated with tumorigenesis of LUAD [131]. A new lncRNA FEZF1-AS1 is also revealed to be upregulated by m6A regulators, which plays an oncogenic role in NSCLC cell proliferation and invasion via the ITGA11/miR-516b-5p axis [132]. In addition, the m6A reader YTHDF2 is confirmed to expedite circ_SFMBT2 decay by recognising the enriched m6A modification on it, which otherwise promotes large tumour suppressor kinase 2 (LATS2) and regulates Hippo/YAP pathway activity to inhibit the proliferation and metastasis of NSCLC cells [133]. The above introduces the molecular mechanism of m6A regulators regulating lung tumour-related ncRNA, then the modulatory role of ncRNAs on m6A regulators will be presented. It has been reported that miR-600 can suppress METTL3 to indirectly affect its m6A methylating effect on downstream pathways, such as the PI3K/AKT signalling pathway, and can ultimately inhibit lung cancer development [134]. In addition, Weng et al. found that lncRNA PCGEM1 could sponge miR-433-3p to reduce the degradation of m6A writer WTAP mRNA, and upregulated WTAP accelerated the proliferation of NSCLC by exerting its m6A catalytic activity [135]. The elevated hsa_circ_0072309 sponges miR-607 to upregulate the m6A eraser FTO, promoting the malignant progression of LUAD [136]. In addition, Li et al. depicted circNDUFB2 as an NSCLC suppressor; one of its functions is acting as a scaffold in an m6A-dependent manner to assist Tripartite motif 25 in degrading m6A reader IGF2BPs and further affect related oncogenic factor expression [137].
The prognostic value of m6A modification in lung cancer
The analysis about the heterogeneity of NSCLC comes out that cancer cells have higher heterogeneity than other cells in tumour microenvironment (TME), especially LUSC. And other sophisticated components in TME contain a variety of subtypes, such as stromal cells and immune cells (including B cells, plasma cells, macrophages, dendritic cells, neutrophil) [138]. It is revealed in another study that heterogeneity of RNA modification-related proteins expression pattern leads to different regulation of oncogenes in different cancer types. For instance, NSUN5 is significantly high in glioblastoma, HEN1 methyltransferase homologue 1 and L antigen family member 3 are frequently upregulated in multiple cancers [139]. Thus, it is important to detect epitranscriptomic heterogeneity in each tumour type for better diagnosis and therapy. Traditional bulk RNA-seq is performed on cell population and researchers end up with an average of gene expression profiles of tumour cells. However, the specific single-cell RNA sequencing (scRNA-seq) opens avenues to analyse tumoral heterogeneity. For example, Hydro-seq, an improved scRNA-seq technology, can be used for non-invasive diagnosis of circulating tumour cells via selecting suitable epitranscriptomic biomarkers. The scRNA-seq can be extended horizontally to the analysis of individual cell genomics, epigenomics, proteomics and metabolomics. Besides, vertically integrated single cell multimodal omics by transcriptomics and other cytotomics also provides new ideas for characterising cells [140].
Tumour heterogeneity would reflect the predictive value of epitranscriptomic prognostic models to some extent, since the higher the tumoral heterogeneity, the greater the burden of tumour somatic mutations, and the more likely tumour to relapse after treatment [141]. But on the other hand, epitranscriptomic biomarkers can exhibit fairly uniform perturbation even in the face of redundant tumour heterogeneity, since translation is a downstream convergence of various oncogenic pathways after RNA epigenetic modification [142]. Recently, many epitranscriptomic models of m6A methylation have focused on lung cancer clustering subgroups that are formed based on heterogeneous expression profile of m6A regulators. These subgroups were divided into high-risk and low-risk groups based on different clinical characteristics of different clusters (such as 5-year survival rate, lymph node metastasis, distant metastasis and prognosis), making these m6A-based prognostic risk models be able to evaluate the clinicopathological characteristics of patients and guide precise therapy in the early stage of lung cancer.
Many studies have demonstrated the prognostic value of m6A regulators as independent factors in predicting various types of lung cancer, whether single or in combination. The prognostic value of overexpressed m6A readers HNRNPC and IGF2BP3 in LUAD was revealed by Guo et al. Multivariate Cox regression analysis referred to HNRNPC and IGF2BP3 as independent prognostic factors, and the Kaplan‒Meier curve showed that their high expression predicted poor prognosis of patients [143, 144]. Another m6A reader, IGF2BP1, was associated with poor prognosis and immune infiltration based on the LUAD TCGA database, suggesting that it can predict the efficacy of immunotherapy in LUAD patients [145]. In addition to single regulators, researchers have also investigated the value of multiple regulator combinations in predicting prognosis. The YTH domain family, an important member of m6A readers, was investigated for its expression profile and correlation with clinical outcomes in LUAD patients. Researchers found that the members of this family are involved in immune cell infiltration; LUAD patients with low expression of YTHDC1/2 and YTHDF1/2/3 were linked to better prognosis [146]. Additionally, different prognostic risk scores based on several m6A regulator expression profiles were constructed, which act as independent prognostic factors to predict the OS of NSCLC patients [147–149]. To predict the prognosis of SCLC patients, a score based on five m6A regulators (G3BP1, METTL5, ALKBH5, IGF2BP3 and RBM15B) was constructed; this prognostic model shows that low-risk cohorts exhibited a better response to adjuvant chemotherapy and PD-1 immunotherapy [150].
In addition to m6A regulators, researchers have also investigated the prognostic value of risk scores constructed from m6A regulatory genes. Zhang et al. established a two-gene risk score based on the interplay between the m6A eraser FTO and the m6A writer METTL3, which is correlated with OS and could be an independent prognostic factor in early diagnosis [151]. Gu et al. constructed a three-m6A-regulatory-gene (WTAP-YTHDC1-YTHDF1) prognostic score for LUSC patients [152], and several studies have also reported different three-gene prognostic risk scores for LUAD patients, such as KIAA1429-RBM15-HNRNPC [153], IGF2BP3-HNRNPA2B1-HNRNPC [154], IGF2BP1-HNRNPC-HNRNPA2B1 [155], ZCRB1-ADH1C-YTHDC2 [156] and HNRNPC-IGFBP3-IGF2BP1 [157]. These aberrant regulators have been validated to be independent prognostic factors for predicting poor prognosis in high-risk patients. Furthermore, researchers systematically analysed DEGs between LUAD tissues and adjacent normal tissues to establish risk scores based on more m6A genes. For instance, Wang et al. built a four-m6A-gene risk score to predict the potential drugs and survival risk of LUAD patients [158], and Wu et al. constructed a five-m6A-gene prognostic signature that can also be used in immune checkpoint blockade therapy of LUAD patients [159]. Researchers also utilised LASSO Cox regression to form m6A-associated 6-gene, 7-gene and 11-gene independent prognostic scores, which are related to clinical features and immune characteristics [49, 160, 161]. Additionally, Liu et al. built an 8‐gene signature for LUAD and a 3‐gene signature for LUSC, which exhibited better prognostic predictive ability in late‐stage NSCLC [162]. In addition to constructing prognostic risk models with abnormally expressed m6A-related genes, Li et al. focused on the prognostic value of m6A regulator gene mutations. They found that the m6A readers YTHDF3, YTHDF1 and YTHDC2 have the most frequent copy number variations. Furthermore, they determined that the copy number loss of FTO and YTHDC2 in NSCLC patients is related to poor prognosis [163].
Since m6A-modified RNAs are implicated in many cellular pathways related to lung cancer progression, some studies have also focused on the role of m6A-regulated RNAs as predictive markers of lung cancer. Sun et al. identified 84 potential m6A-regulated mRNAs related to the OS of LUAD patients, of which 10 were selected to construct an m6A-regulated mRNA signature. Then, they validated its prognostic value and confirmed that high score was associated with poor survival outcomes [164]. Gene alternative splicing can lead to proteomic diversity, and its dysregulation is associated with the progression of lung cancer [165]. After analysis, researchers confirmed the m6A writers METTL3 and RBM15 and reader HNRNPC as splicing factors, and they further discovered seven m6A-related AS events in LUAD and fourteen in LUSC to establish a risk signature, with the high-risk group having worse OS [166]. Researchers have also established an m6A-related lncRNA model to predict lung cancer prognosis. Hou et al. constructed an 11 m6A-related lncRNA risk model, and LUAD patients in different groups had different survival advantages and drug sensitivities, indicating that this could act as an independent prognostic indicator [167]. Additionally, different researchers constructed 13 and 11 m6A-related lncRNA risk models and further proved that these risk scores were correlated with immune cells and TME, which affected malignant tumour progression and prognosis [168, 169]. Then, the investigators further explored mRNA-ncRNA models co-expressed with m6A regulators. They divided NSCLC patients with similar co-expression patterns into one cluster and found that patients in different clusters exhibited different immune responses, PD-L1 expression, and OS, indicating that the prognostic value of this model was better than that of separate RNA models [170]. The above application of m6A methylation in the lung cancer prognostic risk model is summarised in Table 1.
Table 1.
The application of m6A modification in prognostic risk models of different kinds of lung cancer.
| Type of lung cancer | Risk signature | Prognostic factors and biomarkers | Expression in lung tumour | Prognostic | Enriched pathways or biological processes | Ref. |
|---|---|---|---|---|---|---|
| LUAD | regulator | HNRNPC | ↑ | low OS | pyrimidine metabolism, purine metabolism, spliceosome, cell cycle, RNA degradation, RNA polymerase, basal transcription factors, DNA replication, mismatch repair, nucleotide excision repair | [143] |
| IGF2BP3 | ↑ | low OS | G2M checkpoint, E2F targets, mitotic spindle, spermatogenesis, MTORC1 signalling, MYC targets V1, MYC targets V2, glycolysis, epithelial-mesenchymal transition, DNA repair | [144] | ||
| IGF2BP1 | ↑ | poor prognosis | cell cycle-related pathway (positive correlation): organelle fission, spindle organisation, DNA recombination immune regulation (negative correlation): leucocyte activation involved in inflammatory response, mast cell-mediated immunity, cilium or flagellum-dependent cell motility | [145] | ||
|
YTHDC1, YTHDC2 YTHDF1, YTHDF2, YTHDF3 |
↓ ↑ |
low YTHDC2, YTHDF1, YTHDF2 better OS | antigen processing and presentation, hemidesmosome assembly, formation of primary germ layer, appendage development | [146] | ||
| HNRNPA2B1, HNRNPC, KIAA1429, RBM15, METTL3 | ↑ | poor prognosis | cell cycle, DNA replication, RNA degradation, RNA polymerase, nucleotide excision repair, basal transcription factors | [147] | ||
| METTL14, METTL3, ZC3H13, YTHDF1, HNRNPC, RBM15 | ↑ | poor prognosis | cell cycle, RNA degradation, p53 signalling pathway, homologous recombination, mismatch repair | [149] | ||
|
LUAD LUSC |
regulator |
LUAD: IGF2BP1, IGF2BP2, HNRNPC, KIAA1429, HNRNPA2B1 LUSC: CBLL1, HNRNPC, METTL3 |
poor prognosis | [148] | ||
| SCLC | regulator |
G3BP1, METTL5 ALKBH5, IGF2BP3, RBM15B |
↑ | high score: low OS | [150] | |
| LUSC | regulator gene | FTO, METTL3 | ↑ | high‐risk group: low OS | [151] | |
| WTAP, YTHDC1, YTHDF1 | ↑ | high‐risk group: low OS | [152] | |||
| LUAD | regulator gene | KIAA1429, RBM15, HNRNPC | ↑ | poor prognosis | extracellular matrix structural constituent conferring tensile strength, receptor regulator activity, receptor-ligand activity, protein digestion and absorption, cell cycle, neuroactive ligand-receptor interaction signalling pathways | [153] |
| IGF2BP3, HNRNPA2B1, HNRNPC | ↑ | high-risk group: low OS | G2M checkpoint, E2F targets, MTORC1 signalling, MYC targets, DNA repair | [154] | ||
| IGF2BP1, HNRNPC, HNRNPA2B1 | ↑ | poor prognosis | cell cycle, genetic material (purine and pyrimidine) metabolism, DNA replication, translation process (RNA polymerase and RNA degradation) | [155] | ||
|
ZCRB1 ADH1C, YTHDC2 |
↑ ↓ |
poor prognosis | nuclear division, organelle fission, regulation of mRNA metabolic process, regulation of chromosome organisation | [156] | ||
| HNRNPC, IGFBP3, IGF2BP1 | ↑ | high-risk group: low OS | humoral immune response, defence response to bacterium, hormone metabolic process, apical part of the cell, apical plasma membrane, secretory granule lumen, receptor-ligand activity, enzyme inhibitor activity | [157] | ||
| METTL3, IGF2BP2, HNRNPC, HNRNPA2B1 | ↑ | high-risk score: poor prognosis | [158] | |||
| IGF2BP1, IGF2BP2, HNRNPA2B1, METTL3, HNRNPC | ↑ | high-risk group: low OS | cell division, cell proliferation, cell cycle, p53 signalling pathway | [159] | ||
| CLEC3B, TENM3, IGF2BP1, E2F7, ANLN, ANKRD18B, FBN2 | high-risk score: poor prognosis | immune-related biological processes (humoral immune response and regulation of adaptive immune response), cell cycle | [49] | |||
| HNRNPC, METTL3, KIAA1429, YTHDC2, ALKBH5, YTHDF1 | ↑ | high-risk score: poor prognosis | RNA modification, regulation of mRNA metabolic process, mRNA methylation, mRNA modification, regulation of mRNA stability, regulation of RNA stability, RNA methylation, regulation of mRNA catabolic process, mRNA processing, positive regulation of translational initiation | [160] | ||
|
LUSC; LUAD |
regulator gene |
METTL3, YTHDC1, HNRNPC; YTHDC2, METTL3, RBM15, HNRNPC, YTHDF2, YTHDF1, ZC3H13, KIAA1429 |
ZC3H13: ↓ Others: ↑ |
high‐risk score: low OS |
SMAD protein signal transduction, regulation of mitogen-activated protein kinase cascade, positive regulation of cell growth/division. viral carcinogenesis, regulation of actin cytoskeleton, PPAR signalling pathway, PI3K-Akt signalling pathway |
[162] |
| NSCLC | regulator gene | FTO, YTHDC2 | ↓ | poor prognosis | oxidative phosphorylation, ribosome, translation, 3-UTR-mediated translational regulation, metabolism of mRNA, influenza life cycle | [163] |
| LUAD | m6A-related RNA | RFXAP, KHDRBS2, MAPRE3, TXN, EGFR, MGST3, USP1, ARHGEF4, CDHR5, ADAMTS6 |
EGFR: ↑ Others: ↓ |
high-risk score: low OS | RNA biosynthetic processes and developmental process | [164] |
| NSCLC | m6A-related-AS |
LUAD: DGKZ|15540|AP, PMP22|39340|AP (risky) ABCC6|34219|AT, KIAA0586|27718|ES, LDB1|12935|AP, RPS25|19054|ES, S100A14|7729|AP (protective) LUSC: AKR1E2|10639|ES, SSH1|24258|ES (risky) ALPK1|70369|ES, FAM63A|7531|AP, CHMP1A|38102|ES, TSTD2|87013|AT, KIAA1598|13239|AP, ASXL3|45046|AT, VPS37A|82796|ES, TOX2|59455|ES, ZNF544|52429|ES, NOL8|86863|ES, FAM124B|57772|AT, PTCHD4|76446|AT (protective) |
high-risk group: low OS |
LUAD: the GPCR signalling pathway, DNA metabolic process, DNA repair, cellular response to DNA damage stimulus, carbon oxygen lyase activity, cell adhesion molecule binding pathways LUSC: the TGF-beta signalling pathway, peptidyl-serine modification, regulation of actin cytoskeleton, intracellular signalling by second messengers, early endosome pathway |
[166] | |
| LUAD | m6A-related lncRNA | AP001178.1, AL121772.2, AL360270.2, AL358115.1, AF131215.5, AC010999.2, TRAF3IP2-AS1, AC026355.2, ADPGK-AS1, LINC02656, AC012409.4 | high-risk group: low OS | Spliceosomal tri-snRNP complex assembly, formation of quadruple SL/U4/U5/U6 snRNP, mRNA trans-splicing via spliceosome, Golgi lumen, endopeptidase inhibitor activity, peptidase inhibitor activity, etc. | [167] | |
| ADPGK-AS1, AC103591.3, AC018529.1, AC010175.1, AC016747.2, AC007613.1, AC026355.2, ABALON, AC034102.8, AC073316.3, AL031667.3, AC005884.1, TSPOAP1-AS1 |
ABALON: ↑ Others: ↓ |
high-risk group: low OS | the spliceosome, mRNA surveillance, ribosome biogenesis in eukaryotes, viral carcinogenesis, nucleocytoplasmic transport pathway | [168] | ||
|
DLGAP1-AS2, COLCA1, LINC00968, MGC32805, TRG-AS1, FENDRR, RPARP-AS1, TBX5-AS1, CRNDE, TMPO-AS1, ARHGEF26-AS1 |
Upregulated TMPO-AS1 and DLGAP1-AS2: poor OS | high-risk group: low OS | cilium movement, neuronal activity ligand-receptor interaction, NABA matrisome associated | [169] | ||
| NSCLC | Co-expressed mRNAs and ncRNAs |
mRNAs: CPED1, CD302, SCN1A, NEIL3, PTPRM, CCDC68, MTURN, ANLN, LIFR, GPRIN1, DDX11, RNASE1, IGF2BP1, HMGA2, MT-ND6, and LRP8 ncRNAs: SH3BP5-AS1, AL122010.1, AL021368.3, AL162586.1, AC020978.2, AC135050.3, and AC138028.6 |
high-risk group: low OS | G2M checkpoint, mTORC1 signalling, MYC targets V1, E2F targets, PI3K-AKT-mTOR signalling, hypoxia, glycolysis, DNA repair | [170] |
The TME is composed of various immune cells, inflammatory cells and noncellular components and plays an indispensable role in tumorigenesis. In recent studies on lung cancer, researchers have elucidated the prognostic value of m6A modification patterns in TME. Zhu et al. constructed an m6A score of individual tumours by quantifying the m6A modification pattern, in which a low score indicates lower PD-L1 expression and is conducive to immune checkpoint suppression therapy [171]. Another study on NSCLC revealed three m6A modification patterns that corresponded to three tumour immunophenotypes. Researchers then established an m6A score according to the co-DEGs of the three patterns and found that the high-score group with low tumour mutation burden (TMB) was more immune-activated and responded better to immunotherapy [172]. Research specifically on LUAD reported that the scoring system used to quantify m6A patterns was positively correlated with TMB, and the low-score group in the early stage of LUAD was accompanied by a better prognosis [173]. Similarly, based on m6A modification, Jiang et al. created an m6A score to predict the immune response in TME, the efficacy of immunotherapy and the prognosis of LUAD patients [174]. Zhu et al. built another m6A score to predict the response to radiotherapy and immunotherapy in LUAD patients [175].
Application of m6A modification in lung cancer therapy
At present, the clinical treatment of lung cancer includes surgery, radiotherapy, chemotherapy, immunotherapy and targeted therapy. Traditional chemotherapy regimens are toxic to the body, and drug resistance limits their efficacy. In view of these conditions, the development of new treatments appears increasingly critical to prolong the survival time of lung cancer patients and improve their quality of life. Since the indispensable effect m6A modification exerts on lung tumour malignancy, diverse pieces of research have shown that m6A regulators, which are being identified as novel anticancer drug targets, have great potential for the treatment of lung cancer [176, 177].
Several studies have revealed that m6A regulators are implicated in the resistance mechanisms of various lung cancer drugs, which indicates that inhibiting or mimicking these regulators may pave the way for restoring the sensitivity of cancerous cells to existing drugs. Cisplatin (DDP) is the first metal complex found to have anticancer activity and used in lung cancer treatment. As mentioned above, upregulation of the m6A writer METTL3 causes DDP resistance in NSCLC patients by increasing AKT1 expression [99], and deficiency of the m6A reader YTHDF1 also causes DDP resistance via the Keap1/Nrf2 axis [75]. NcRNAs are extensively involved in the development of tumour drug resistance and interact with different m6A regulators to coregulate lung cancer resistance [178–180]. For instance, miRNA that regulates ferroptosis is closely related to drug resistance in lung cancer [181]. Song et al. reported that enriched exosomal miR-4443 upregulated ferroptosis suppressor protein 1 in METTL3-mediated m6A modification, which increased the proliferative marker Ki-67 to exert DDP resistance [182]. In addition to miRNA, Xie et al. demonstrated that circVMP1 can disseminate chemo-resistant characteristics by sponging miR-524-5p to increase the writer METTL3; the latter stabilised SRY-box transcription factor 2 (SOX2) via m6A modification to exert its function in inducing drug resistance [33]. In addition to DDP, tyrosine kinase inhibitors (TKIs), including first-generation gefitinib (GE), erlotinib, second-generation afatinib, and third-generation osimertinib, are novel small-molecule targeted drugs for the treatment of lung cancer. Researchers found that in GE-resistant LUAD cells, upregulated METTL3 and YTHDF2 suppressed circASK1 in an m6A-dependent manner, which blocked the activation of circASK1-mediated ASK1/JNK/p38 pathway [183]. Zhang et al. also revealed that elevated lncRNA SNHG17 through METTL3-mediated m6A modification impaired LATS2 tumour suppressor effect, which facilitated LUAD cell GE resistance [184]. Researchers have also reported the mechanism of LUAD drug resistance to erlotinib, another first-generation EGFR-TKI drug. It was found that the degradation of lncRNA TUSC7 mediated by YTHDF2 and the activation of Notch signalling mediated by METTL3 both contributed to erlotinib resistance [185].
To alleviate drug resistance of lung neoplasms, researchers have tested the combination of TKIs and other types of drugs, and found that m6A modification participated in the specific mechanism during the process of reversing drug resistance. The first-generation TKI crizotinib has been used as a first-line medicine for NSCLC therapy, but drug resistance greatly limits its efficacy [186]. Encouragingly, researchers found that chidamide could cotreat with crizotinib to reduce drug resistance and inhibit lung cancer growth. The mechanism is that chidamide reduces m6A modification of c-MET mRNA by inhibiting the writer METTL3 and WTAP, and NSCLC patients with low c-MET restores sensitivity to crizotinib [187]. In addition, the synergy between β-elemene and GE inhibits METTL3-mediated m6A methylation in a dose-dependent manner, which suppresses the protective effect of METTL3-related autophagy on drug-resistant cells, enhancing GE sensitivity [188]. Meclofenamic acid (MA), a kind of NSAID, is also reported to restore GE sensitivity. GE-MA coadministration inactivates EGFR downstream pathways and downregulates c-Myc expression via the m6A demethylase FTO, promoting NSCLC cell apoptosis [189]. In addition, Li et al. found that metformin cotreatment with TKIs remarkably reversed drug resistance by facilitating m6A writer METTL3 expression, and METTL3-assisted Let-7b-5p maturation by m6A methylation to restore drug sensitivity [190]. Inhibitors of the m6A writer METTL7B may reverse TKI resistance by suppressing METTL7B-mediated m6A modification of antioxidant enzymes such as glutathione peroxidase 4 and superoxide dismutase [191]. Researchers also revealed that high-dose propofol upregulated miR-486-5p in an m6A-dependent manner to suppress the downstream gene RAP1, inactivating the cancer-promoting NF-κB pathway in NSCLC cells to restore DDP effectiveness [192].
In addition to known chemotherapies and targeted therapies, researchers are also working to develop novel treatments for lung cancer and bring better prognosis to patients. Chen et al. found that simvastatin suppressed the m6A writer METTL3 to inhibit the effects of downstream EMT-related genes, exerting the pharmacological effects of ameliorating tumour progression in a dose-dependent manner [193]. In addition, β-elemene can inhibit METTL3 and attenuate its m6A methylation effect on the tumour suppressor gene phosphatase and tension homologue (PTEN), thereby preventing the occurrence of malignant lung tumours and providing better treatment for patients [194]. Some Chinese herbal medicines have also been confirmed to have certain antitumour activities. Isoliquiritigenin, derived from the Chinese herb licorice, inhibits m6A reader IGF2BP3 mediated m6A modification to indirectly inhibit the EMT-related factor TWIST1, performing effective dose-dependent inhibition of cancerous biological behaviour [195]. Another Chinese herbal extract, delicaflavone, was reported to activate antitumour immunity by m6A modification and increase the expression of IFN-γ-responsive genes (such as Stat1 and Irf1) by inhibiting the m6A writer METTL3/14, thereby recruiting CD8+ T cells to initiate the immune response [196]. Researchers found that sevoflurane suppressed FTO-induced abnormal metabolism and proliferation of cancer cells by reducing lncRNA HOTAIR recruitment to FTO, and then suppressing hexokinase 2 expression by demethylating m6A in its mRNA [197]. Additionally, researchers have discovered that after interleukin 37 treatment, the total m6A levels are elevated, and Wnt5a/5b pathways are also impacted, which suggests a cancer suppressive effect [198].
Conclusion and perspective
As one of the most abundant modifications in eukaryotes, m6A modification is involved in almost the entire RNA life cycle, which works by the synergy of methyltransferases, m6A-binding proteins, and demethylases (writers, readers and erasers) [22]. The modification effect of m6A is closely related to many diseases, and this review focuses on the role of m6A in lung cancer. We discussed how the enzymes of m6A modification regulated lung tumour malignant progression, which may act as robust biomarkers to improve the early diagnosis rate for lung cancer patients and present great value for their prognosis and treatment.
Thanks to advances in technology, researchers can comprehensively analyse the abnormal expression of m6A regulators in lung cancer tissues and target genes of m6A. By analysing the m6A targets, researchers found that the m6A present in mRNAs and ncRNAs could regulate their expression, while ncRNAs can also act on m6A regulators, which in turn affect lung cancer progression through various signalling pathways. The m6A modification is also involved in the metabolism and interaction of tumour cells within the TME. Based on the indispensable role of m6A modification in the development of cancerous cells, researchers have constructed many independent m6A-related prognostic models to guide early clinical diagnosis and treatment. In addition, studying the molecular mechanism of m6A modification can also improve the efficacy of treatments in patients with lung cancer.
Although we have obtained deeper insight into lung cancer based on m6A-related mechanisms, the current studies still have some limitations. First, the complex role of each molecule implicated in m6A modification makes it difficult to clearly elucidate how they cooperate with each other to modulate the pathologic process of lung cancer, which requires us to understand their integral role from a higher perspective. Next, some m6A prognostic models use patient data extracted from public databases, which need further validation in other prospective cohorts. Since the small number of samples may limit the universality of their application to the population. Additionally, although there are some studies on m6A-related combination drugs to improve TKI resistance, most of them are still stay at the mechanistic level; further investigations are needed for clinical application.
Here are some new directions and suggestions for further research on m6A and lung cancer. First, research on the mechanism by which m6A affects lung cancer should be expanded to in vivo experiments on the basis of cell experiments, which will help m6A modification use in clinical practice in the future. Second, different types of lung cancer have different m6A-related genes, so the specific exploration of these genes will guide precise targeted therapy. Third, there are a number of prognostic models based on the m6A-related gene signature; the downstream targets of these genes need to be further explored, which will help us determine the potential mechanisms between the DEGs in different risk groups. Finally, as a posttranscriptional modification, the synergistic effect of m6A with posttranslational modification should be further investigated. This would be meaningful for improving the effect of drug combinations and attenuating TKI resistance to bring more progress in this field.
Author contributions
MND, XJZ searched the literature, YFZ provided inspiration and guidance for writing, MND wrote the manuscript and prepared all the figures and tables. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (22006084); Hubei Key Laboratory of Environmental and Health Effects of Persistent Toxic Substances (PTS2019-05; PTS2019-06); and graphical abstract was modified from Servier Medical Art (http://smart.servier.com/).
Competing interests
The authors declare no competing interests.
Consent for publication
All authors consent to publication.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Nicholson AG, Tsao MS, Beasley MB, Borczuk AC, Brambilla E, Cooper WA, et al. The 2021 WHO classification of lung tumors: impact of advances since 2015. J Thorac Oncol. 2022;17:362–87. doi: 10.1016/j.jtho.2021.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 5.Shaurova T, Zhang L, Goodrich DW, Hershberger PA. Understanding lineage plasticity as a path to targeted therapy failure in EGFR-mutant non-small cell lung cancer. Front Genet. 2020;11:281. doi: 10.3389/fgene.2020.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orsolic I, Carrier A, Esteller M. Genetic and epigenetic defects of the RNA modification machinery in cancer. Trends Genet. 2023;39:74–88. doi: 10.1016/j.tig.2022.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Teng PC, Liang Y, Yarmishyn AA, Hsiao YJ, Lin TY, Lin TW, et al. RNA modifications and epigenetics in modulation of lung cancer and pulmonary diseases. Int J Mol Sci. 2021;22:10592. doi: 10.3390/ijms221910592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–d7. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 10.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen K, Lu Z, Wang X, Fu Y, Luo GZ, Liu N, et al. High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew Chem (Int ed Engl) 2015;54:1587–90. doi: 10.1002/anie.201410647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–72. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, et al. A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 2015;29:2037–53. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA. 2013;19:1848–56. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molinie B, Wang J, Lim KS, Hillebrand R, Lu ZX, Van Wittenberghe N, et al. m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat Methods. 2016;13:692–8. doi: 10.1038/nmeth.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Campos MA, Edelheit S, Toth U, Safra M, Shachar R, Viukov S, et al. Deciphering the ‘m(6)A Code’ via antibody-independent quantitative profiling. Cell. 2019;178:731–47.e16. doi: 10.1016/j.cell.2019.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Zhang SY, Zhang SW, Fan XN, Zhang T, Meng J, Huang Y. FunDMDeep-m6A: identification and prioritization of functional differential m6A methylation genes. Bioinforma. 2019;35:i90–i8. doi: 10.1093/bioinformatics/btz316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu H, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, et al. Accurate detection of m(6)A RNA modifications in native RNA sequences. Nat Commun. 2019;10:4079. doi: 10.1038/s41467-019-11713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Chen LQ, Zhao YL, Yang CG, Roundtree IA, Zhang Z, et al. Single-base mapping of m(6)A by an antibody-independent method. Sci Adv. 2019;5:eaax0250. doi: 10.1126/sciadv.aax0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther. 2021;6:74. doi: 10.1038/s41392-020-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608–24. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 23.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A Reader YTHDC1 regulates mRNA Splicing. Mol Cell. 2016;61:507–19. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen ZH, Chen TQ, Zeng ZC, Wang D, Han C, Sun YM, et al. Nuclear export of chimeric mRNAs depends on an lncRNA-triggered autoregulatory loop in blood malignancies. Cell Death Dis. 2020;11:566. doi: 10.1038/s41419-020-02795-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–99. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 29.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue Q, et al. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun. 2017;482:582–9. doi: 10.1016/j.bbrc.2016.11.077. [DOI] [PubMed] [Google Scholar]
- 31.Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Yue C, Chen J, Li Z, Li L, Chen J, Guo Y. microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKα2-FTO-m6A/MYC axis. J Exp Clin Cancer Res. 2020;39:240. doi: 10.1186/s13046-020-01731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie H, Yao J, Wang Y, Ni B. Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Deliv. 2022;29:1257–71. doi: 10.1080/10717544.2022.2057617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lv W, Tan Y, Xiong M, Zhao C, Wang Y, Wu M, et al. Analysis and validation of m6A regulatory network: a novel circBACH2/has-miR-944/HNRNPC axis in breast cancer progression. J Transl Med. 2021;19:527. doi: 10.1186/s12967-021-03196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X, Ye W, Gong Y. The Role of RNA Methyltransferase METTL3 in Normal and Malignant Hematopoiesis. Front Oncol. 2022;12:873903. doi: 10.3389/fonc.2022.873903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q. Current advances in N6-methyladenosine methylation modification during bladder cancer. Front Genet. 2021;12:825109. doi: 10.3389/fgene.2021.825109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bueno-Costa A, Piñeyro D, García-Prieto CA, Ortiz-Barahona V, Martinez-Verbo L, Webster NA, et al. Remodeling of the m(6)A RNA landscape in the conversion of acute lymphoblastic leukemia cells to macrophages. Leukemia. 2022;36:2121–4. doi: 10.1038/s41375-022-01621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269:17697–704. doi: 10.1016/S0021-9258(17)32497-3. [DOI] [PubMed] [Google Scholar]
- 39.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–47. [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Huang J, Zou T, Yin P. Human m(6)A writers: two subunits, 2 roles. RNA Biol. 2017;14:300–4. doi: 10.1080/15476286.2017.1282025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–73. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol Cell. 2018;69:1028–38.e6. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oerum S, Meynier V, Catala M, Tisné C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021;49:7239–55. doi: 10.1093/nar/gkab378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA. Methyltransferase-like protein 16 binds the 3’-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci USA. 2016;113:14013–8. doi: 10.1073/pnas.1614759113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719–33. doi: 10.1093/nar/gkz619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Y, Shi Y, Shen H, Xie W. m(6)A-binding proteins: the emerging crucial performers in epigenetics. J Hematol Oncol. 2020;13:35. doi: 10.1186/s13045-020-00872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou H, Zheng M, Shi M, Wang J, Huang Z, Zhang H, et al. Characteristic of molecular subtypes in lung adenocarcinoma based on m6A RNA methylation modification and immune microenvironment. BMC Cancer. 2021;21:938. doi: 10.1186/s12885-021-08655-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–28. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, et al. Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–7. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan TN. 6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–4. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, et al. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun. 2018;9:420. doi: 10.1038/s41467-017-02770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051–63. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, et al. N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol. 2017;24:870–8. doi: 10.1038/nsmb.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, et al. Reversible methylation of m(6)A(m) in the 5’ cap controls mRNA stability. Nature. 2017;541:371–5. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y, et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc Natl Acad Sci USA. 2018;115:E325–e33. doi: 10.1073/pnas.1717794115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K, et al. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep. 2017;7:42271. doi: 10.1038/srep42271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, Zhang Y, Guo Y, Liu R, Yu Q, Gong L, et al. ALKBH1 promotes lung cancer by regulating m6A RNA demethylation. Biochem Pharm. 2021;189:114284. doi: 10.1016/j.bcp.2020.114284. [DOI] [PubMed] [Google Scholar]
- 64.Scott RE, Wille JJ, Jr., Wier ML. Mechanisms for the initiation and promotion of carcinogenesis: a review and a new concept. Mayo Clin Proc. 1984;59:107–17. doi: 10.1016/S0025-6196(12)60244-4. [DOI] [PubMed] [Google Scholar]
- 65.Kant R, Manne RK, Anas M, Penugurti V, Chen T, Pan BS, et al. Deregulated transcription factors in cancer cell metabolisms and reprogramming. Semin Cancer Biol. 2022;86:1158–74. doi: 10.1016/j.semcancer.2022.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.An Y, Duan H. The role of m6A RNA methylation in cancer metabolism. Mol Cancer. 2022;21:14. doi: 10.1186/s12943-022-01500-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L, Bi R, Yin H, Liu H, Li L. ENO1 silencing impaires hypoxia-induced gemcitabine chemoresistance associated with redox modulation in pancreatic cancer cells. Am J Transl Res. 2019;11:4470–80. [PMC free article] [PubMed] [Google Scholar]
- 68.Ma L, Xue X, Zhang X, Yu K, Xu X, Tian X, et al. The essential roles of m(6)A RNA modification to stimulate ENO1-dependent glycolysis and tumorigenesis in lung adenocarcinoma. J Exp Clin Cancer Res. 2022;41:36. doi: 10.1186/s13046-021-02200-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma L, Chen T, Zhang X, Miao Y, Tian X, Yu K, et al. The m(6)A reader YTHDC2 inhibits lung adenocarcinoma tumorigenesis by suppressing SLC7A11-dependent antioxidant function. Redox Biol. 2021;38:101801. doi: 10.1016/j.redox.2020.101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma L, Zhang X, Yu K, Xu X, Chen T, Shi Y, et al. Targeting SLC3A2 subunit of system X(C)(-) is essential for m(6)A reader YTHDC2 to be an endogenous ferroptosis inducer in lung adenocarcinoma. Free Radic Biol Med. 2021;168:25–43. doi: 10.1016/j.freeradbiomed.2021.03.023. [DOI] [PubMed] [Google Scholar]
- 72.Xu Y, Lv D, Yan C, Su H, Zhang X, Shi Y, et al. METTL3 promotes lung adenocarcinoma tumor growth and inhibits ferroptosis by stabilizing SLC7A11 m(6)A modification. Cancer Cell Int. 2022;22:11. doi: 10.1186/s12935-021-02433-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karakashev SV, Reginato MJ. Progress toward overcoming hypoxia-induced resistance to solid tumor therapy. Cancer Manag Res. 2015;7:253–64. doi: 10.2147/CMAR.S58285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu P, Hu K, Zhang P, Sun ZG, Zhang N. Hypoxia-mediated YTHDF2 overexpression promotes lung squamous cell carcinoma progression by activation of the mTOR/AKT axis. Cancer Cell Int. 2022;22:13. doi: 10.1186/s12935-021-02368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun. 2019;10:4892. doi: 10.1038/s41467-019-12801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–45. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu G, Zhai D, Xie J, Zhu S, Liang Z, Liu X, et al. N(6) -methyladenosine (m(6) A) RNA modification of G protein-coupled receptor 133 increases proliferation of lung adenocarcinoma. FEBS Open Bio. 2022;12:571–81. doi: 10.1002/2211-5463.13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu Y, Chang N, Zhang Y, Zhang X, Xu L, Che Y, et al. METTL3-mediated m(6)A mRNA modification of FBXW7 suppresses lung adenocarcinoma. J Exp Clin Cancer Res. 2021;40:90. doi: 10.1186/s13046-021-01880-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shen Y, Li C, Zhou L, Huang JA. G protein-coupled oestrogen receptor promotes cell growth of non-small cell lung cancer cells via YAP1/QKI/circNOTCH1/m6A methylated NOTCH1 signalling. J Cell Mol Med. 2021;25:284–96. doi: 10.1111/jcmm.15997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang C, Sun Q, Zhang X, Qin N, Pu Z, Gu Y, et al. Gene amplification-driven RNA methyltransferase KIAA1429 promotes tumorigenesis by regulating BTG2 via m6A-YTHDF2-dependent in lung adenocarcinoma. Cancer Commun. 2022;42:609–26. doi: 10.1002/cac2.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lou X, Ning J, Liu W, Li K, Qian B, Xu D, et al. YTHDF1 promotes cyclin b1 translation through m(6)A modulation and contributes to the poor prognosis of lung adenocarcinoma with KRAS/TP53 co-mutation. Cells. 2021;10:1669. doi: 10.3390/cells10071669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsuchiya K, Yoshimura K, Iwashita Y, Inoue Y, Ohta T, Watanabe H, et al. m(6)A demethylase ALKBH5 promotes tumor cell proliferation by destabilizing IGF2BPs target genes and worsens the prognosis of patients with non-small-cell lung cancer. Cancer Gene Ther. 2022;29:1355–1372. doi: 10.1038/s41417-022-00451-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu Z, Qian Q, Zhao X, Ma L, Chen P. N(6)-methyladenosine ALKBH5 promotes non-small cell lung cancer progress by regulating TIMP3 stability. Gene. 2020;731:144348. doi: 10.1016/j.gene.2020.144348. [DOI] [PubMed] [Google Scholar]
- 84.Li J, Han Y, Zhang H, Qian Z, Jia W, Gao Y, et al. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun. 2019;512:479–85. doi: 10.1016/j.bbrc.2019.03.093. [DOI] [PubMed] [Google Scholar]
- 85.Liu J, Ren D, Du Z, Wang H, Zhang H, Jin Y. m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018;502:456–64. doi: 10.1016/j.bbrc.2018.05.175. [DOI] [PubMed] [Google Scholar]
- 86.Coker H, Wei G, Brockdorff N. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim Biophys Acta Gene Regul Mech. 2019;1862:310–8. doi: 10.1016/j.bbagrm.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 87.Hu Z, Zhu L, Zhang Y, Chen B. N6-methyladenosine-induced SVIL antisense RNA 1 restrains lung adenocarcinoma cell proliferation by destabilizing E2F1. Bioengineered. 2022;13:3093–107. doi: 10.1080/21655979.2022.2025697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han L, Lei G, Chen Z, Zhang Y, Huang C, Chen W. IGF2BP2 regulates MALAT1 by serving as an N6-methyladenosine reader to promote NSCLC proliferation. Front Mol Biosci. 2021;8:780089. doi: 10.3389/fmolb.2021.780089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu X, Zuo X, Ma L, Wang Q, Zhu L, Li L, et al. Integrated analysis of the m6A-related lncRNA identified lncRNA ABALON/miR-139-3p/NOB1 axis was involved in the occurrence of lung cancer. Cancer Manag Res. 2021;13:8707–22. doi: 10.2147/CMAR.S339032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen Z, Chen X, Lei T, Gu Y, Gu J, Huang J, et al. Integrative analysis of NSCLC identifies LINC01234 as an oncogenic lncRNA that interacts with HNRNPA2B1 and regulates miR-106b biogenesis. Mol Ther. 2020;28:1479–93. doi: 10.1016/j.ymthe.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Suarez-Carmona M, Lesage J, Cataldo D, Gilles C. EMT and inflammation: inseparable actors of cancer progression. Mol Oncol. 2017;11:805–23. doi: 10.1002/1878-0261.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang F, Yuan WQ, Li J, Luo YQ. Knockdown of METTL14 suppresses the malignant progression of non-small cell lung cancer by reducing Twist expression. Oncol Lett. 2021;22:847. doi: 10.3892/ol.2021.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li J, Xie G, Tian Y, Li W, Wu Y, Chen F, et al. RNA m(6)A methylation regulates dissemination of cancer cells by modulating expression and membrane localization of β-catenin. Mol Ther. 2022;30:1578–96. doi: 10.1016/j.ymthe.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wanna-Udom S, Terashima M, Lyu H, Ishimura A, Takino T, Sakari M, et al. The m6A methyltransferase METTL3 contributes to transforming growth factor-beta-induced epithelial-mesenchymal transition of lung cancer cells through the regulation of JUNB. Biochem Biophys Res Commun. 2020;524:150–5. doi: 10.1016/j.bbrc.2020.01.042. [DOI] [PubMed] [Google Scholar]
- 96.Li J, Chen F, Peng Y, Lv Z, Lin X, Chen Z, et al. N6-Methyladenosine regulates the expression and secretion of TGFβ1 to affect the epithelial-mesenchymal transition of cancer cells. Cells. 2020;9:296. doi: 10.3390/cells9020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun Z, Su Z, Zhou Z, Wang S, Wang Z, Tong X, et al. RNA demethylase ALKBH5 inhibits TGF-β-induced EMT by regulating TGF-β/SMAD signaling in non-small cell lung cancer. FASEB J. 2022;36:e22283. doi: 10.1096/fj.202200005RR. [DOI] [PubMed] [Google Scholar]
- 98.Cheng C, Wu Y, Xiao T, Xue J, Sun J, Xia H, et al. METTL3-mediated m(6)A modification of ZBTB4 mRNA is involved in the smoking-induced EMT in cancer of the lung. Mol Ther Nucleic Acids. 2021;23:487–500. doi: 10.1016/j.omtn.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 99.Shi L, Gong Y, Zhuo L, Wang S, Chen S, Ke B. Methyltransferase-like 3 upregulation is involved in the chemoresistance of non-small cell lung cancer. Ann Transl Med. 2022;10:139. doi: 10.21037/atm-21-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y, Liu S, Zhao T, Dang C. METTL3‑mediated m6A modification of Bcl‑2 mRNA promotes non‑small cell lung cancer progression. Oncol Rep. 2021;46:163. doi: 10.3892/or.2021.8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang Y, Li M, Zhang L, Chen Y, Zhang S. m6A demethylase FTO induces NELL2 expression by inhibiting E2F1 m6A modification leading to metastasis of non-small cell lung cancer. Mol Ther Oncolytics. 2021;21:367–76. doi: 10.1016/j.omto.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen Z, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019;18:181. doi: 10.1186/s12943-019-1108-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 103.Pan H, Pan Z, Guo F, Meng F, Zu L, Fan Y, et al. MicroRNA-1915-3p inhibits cell migration and invasion by targeting SET in non-small-cell lung cancer. BMC Cancer. 2021;21:1218. doi: 10.1186/s12885-021-08961-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tkach M, Théry C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164:1226–32. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 105.Li C, Qin F, Wang W, Ni Y, Gao M, Guo M, et al. hnRNPA2B1-mediated extracellular vesicles sorting of miR-122-5p potentially promotes lung cancer progression. Int J Mol Sci. 2021;22:12866. doi: 10.3390/ijms222312866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang X, Liu C, Zhang S, Yan H, Zhang L, Jiang A, et al. N(6)-methyladenosine modification of MALAT1 promotes metastasis via reshaping nuclear speckles. Dev Cell. 2021;56:702–15.e8. doi: 10.1016/j.devcel.2021.01.015. [DOI] [PubMed] [Google Scholar]
- 107.Cheng FW, Peng LM, Luo D. Methyltransferase-like 3 promotes the progression of lung cancer via activating PI3K/AKT/mTOR pathway. Clin Exp Pharm Physiol. 2022;49:748–58. doi: 10.1111/1440-1681.13647. [DOI] [PubMed] [Google Scholar]
- 108.Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12:135. doi: 10.1186/s13045-019-0830-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 109.Jin D, Guo J, Wu Y, Yang L, Wang X, Du J, et al. m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol Cancer. 2020;19:40. doi: 10.1186/s12943-020-01161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ni XF, Xie QQ, Zhao JM, Xu YJ, Ji M, Hu WW, et al. The hepatic microenvironment promotes lung adenocarcinoma cell proliferation, metastasis, and epithelial-mesenchymal transition via METTL3-mediated N6-methyladenosine modification of YAP1. Aging. 2021;13:4357–69. doi: 10.18632/aging.202397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jin M, Li G, Liu W, Wu X, Zhu J, Zhao D, et al. Cigarette smoking induces aberrant N(6)-methyladenosine of DAPK2 to promote non-small cell lung cancer progression by activating NF-κB pathway. Cancer Lett. 2021;518:214–29. doi: 10.1016/j.canlet.2021.07.022. [DOI] [PubMed] [Google Scholar]
- 112.Xu Y, Chen Y, Yao Y, Xie H, Lu G, Du C, et al. VIRMA contributes to non-small cell lung cancer progression via N(6)-methyladenosine-dependent DAPK3 post-transcriptional modification. Cancer Lett. 2021;522:142–54. doi: 10.1016/j.canlet.2021.08.027. [DOI] [PubMed] [Google Scholar]
- 113.Zhao W, Xie Y. KIAA1429 promotes the progression of lung adenocarcinoma by regulating the m6A level of MUC3A. Pathol Res Pract. 2021;217:153284. doi: 10.1016/j.prp.2020.153284. [DOI] [PubMed] [Google Scholar]
- 114.Sun Y, Sun X, You C, Ma S, Luo Y, Peng S, et al. MUC3A promotes non-small cell lung cancer progression via activating the NFκB pathway and attenuates radiosensitivity. Int J Biol Sci. 2021;17:2523–36. doi: 10.7150/ijbs.59430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Y, Sheng H, Ma F, Wu Q, Huang J, Chen Q, et al. RNA m(6)A reader YTHDF2 facilitates lung adenocarcinoma cell proliferation and metastasis by targeting the AXIN1/Wnt/β-catenin signaling. Cell Death Dis. 2021;12:479. doi: 10.1038/s41419-021-03763-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guo J, Wu Y, Du J, Yang L, Chen W, Gong K, et al. Deregulation of UBE2C-mediated autophagy repression aggravates NSCLC progression. Oncogenesis. 2018;7:49. doi: 10.1038/s41389-018-0054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang X, Shao F, Guo D, Wang W, Wang J, Zhu R, et al. WNT/β-catenin-suppressed FTO expression increases m(6)A of c-Myc mRNA to promote tumor cell glycolysis and tumorigenesis. Cell Death Dis. 2021;12:462. doi: 10.1038/s41419-021-03739-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328:198–206. doi: 10.1016/j.canlet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 119.Liu Y, Ao X, Jia Y, Li X, Wang Y, Wang J. The FOXO family of transcription factors: key molecular players in gastric cancer. J Mol Med. 2022;100:997–1015. doi: 10.1007/s00109-022-02219-x. [DOI] [PubMed] [Google Scholar]
- 120.Liu Y, Wang Y, Li X, Jia Y, Wang J, Ao X. FOXO3a in cancer drug resistance. Cancer Lett. 2022;540:215724. doi: 10.1016/j.canlet.2022.215724. [DOI] [PubMed] [Google Scholar]
- 121.Ning J, Wang F, Bu J, Zhu K, Liu W. Down-regulated m6A reader FTO destabilizes PHF1 that triggers enhanced stemness capacity and tumor progression in lung adenocarcinoma. Cell Death Discov. 2022;8:354. doi: 10.1038/s41420-022-01125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chao Y, Shang J, Ji W. ALKBH5-m(6)A-FOXM1 signaling axis promotes proliferation and invasion of lung adenocarcinoma cells under intermittent hypoxia. Biochem Biophys Res Commun. 2020;521:499–506. doi: 10.1016/j.bbrc.2019.10.145. [DOI] [PubMed] [Google Scholar]
- 123.Sato R, Semba T, Saya H, Arima Y. Concise review: stem cells and epithelial-mesenchymal transition in cancer: biological implications and therapeutic targets. Stem Cells. 2016;34:1997–2007. doi: 10.1002/stem.2406. [DOI] [PubMed] [Google Scholar]
- 124.Liu X, Wang Z, Yang Q, Hu X, Fu Q, Zhang X, et al. RNA demethylase ALKBH5 prevents lung cancer progression by regulating EMT and stemness via regulating p53. Front Oncol. 2022;12:858694. doi: 10.3389/fonc.2022.858694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu Y, Ao X, Yu W, Zhang Y, Wang J. Biogenesis, functions, and clinical implications of circular RNAs in non-small cell lung cancer. Mol Ther Nucleic Acids. 2022;27:50–72. doi: 10.1016/j.omtn.2021.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rong L, Xu Y, Zhang K, Jin L, Liu X. HNRNPA2B1 inhibited SFRP2 and activated Wnt-β/catenin via m6A-mediated miR-106b-5p processing to aggravate stemness in lung adenocarcinoma. Pathol Res Pract. 2022;233:153794. doi: 10.1016/j.prp.2022.153794. [DOI] [PubMed] [Google Scholar]
- 127.Li S, Lu X, Zheng D, Chen W, Li Y, Li F, et al. Methyltransferase-like 3 facilitates lung cancer progression by accelerating m6A methylation-mediated primary miR-663 processing and impeding SOCS6 expression. J Cancer Res Clin Oncol. 2021;148:3485–3499. doi: 10.1007/s00432-022-04128-5. [DOI] [PubMed] [Google Scholar]
- 128.Qian X, Yang J, Qiu Q, Li X, Jiang C, Li J, et al. LCAT3, a novel m6A-regulated long non-coding RNA, plays an oncogenic role in lung cancer via binding with FUBP1 to activate c-MYC. J Hematol Oncol. 2021;14:112. doi: 10.1186/s13045-021-01123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang S, Jin M, Lan X, Wu JL, Zhang Z, Zhao J, et al. LncRNA AC098934 promotes proliferation and invasion in lung adenocarcinoma cells by combining METTL3 and m6A modifications. J Cancer. 2022;13:2662–72. doi: 10.7150/jca.69406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li D, Fu Z, Dong C, Song Y. Methyltransferase 3, N6-adenosine-methyltransferase complex catalytic subunit-induced long intergenic non-protein coding RNA 1833 N6-methyladenosine methylation promotes the non-small cell lung cancer progression via regulating heterogeneous nuclear ribonucleoprotein A2/B1 expression. Bioengineered. 2022;13:10493–503. doi: 10.1080/21655979.2022.2061305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yu H, Zhang Z. ALKBH5-mediated m6A demethylation of lncRNA RMRP plays an oncogenic role in lung adenocarcinoma. Mamm Genome. 2021;32:195–203. doi: 10.1007/s00335-021-09872-6. [DOI] [PubMed] [Google Scholar]
- 132.Song H, Li H, Ding X, Li M, Shen H, Li Y, et al. Long non‑coding RNA FEZF1‑AS1 facilitates non‑small cell lung cancer progression via the ITGA11/miR‑516b‑5p axis. Int J Oncol. 2020;57:1333–47. doi: 10.3892/ijo.2020.5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xu J, Shang Y, Qin X, Gai Y, Cai F, Xiao H, et al. N6-Methyladenosine reader YTHDF2 enhances non-small-cell lung cancer cell proliferation and metastasis through mediating circ_SFMBT2 degradation. Contrast Media Mol Imaging. 2022;2022:1087622. doi: 10.1155/2022/1087622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wei W, Huo B, Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag Res. 2019;11:1177–87. doi: 10.2147/CMAR.S181058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Weng L, Qiu K, Gao W, Shi C, Shu F. LncRNA PCGEM1 accelerates non-small cell lung cancer progression via sponging miR-433-3p to upregulate WTAP. BMC Pulm Med. 2020;20:213. doi: 10.1186/s12890-020-01240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mo WL, Deng LJ, Cheng Y, Yu WJ, Yang YH, Gu WD. Circular RNA hsa_circ_0072309 promotes tumorigenesis and invasion by regulating the miR-607/FTO axis in non-small cell lung carcinoma. Aging. 2021;13:11629–45. doi: 10.18632/aging.202856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021;12:295. doi: 10.1038/s41467-020-20527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wu F, Fan J, He Y, Xiong A, Yu J, Li Y, et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat Commun. 2021;12:2540. doi: 10.1038/s41467-021-22801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Begik O, Lucas MC, Liu H, Ramirez JM, Mattick JS, Novoa EM. Integrative analyses of the RNA modification machinery reveal tissue- and cancer-specific signatures. Genome Biol. 2020;21:97. doi: 10.1186/s13059-020-02009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li X, Wang CY. From bulk, single-cell to spatial RNA sequencing. Int J oral Sci. 2021;13:36. doi: 10.1038/s41368-021-00146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Goldman SL, Hassan C, Khunte M, Soldatenko A, Jong Y, Afshinnekoo E, et al. Epigenetic modifications in acute myeloid leukemia: prognosis, treatment, and heterogeneity. Front Genet. 2019;10:133. doi: 10.3389/fgene.2019.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vaklavas C, Blume SW, Grizzle WE. Translational dysregulation in cancer: molecular insights and potential clinical applications in biomarker development. Front Oncol. 2017;7:158. doi: 10.3389/fonc.2017.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guo W, Huai Q, Zhang G, Guo L, Song P, Xue X, et al. Elevated heterogeneous nuclear ribonucleoprotein C expression correlates with poor prognosis in patients with surgically resected lung adenocarcinoma. Front Oncol. 2020;10:598437. doi: 10.3389/fonc.2020.598437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Guo W, Huai Q, Wan H, Guo L, Song P, Gao S, et al. Prognostic impact of IGF2BP3 expression in patients with surgically resected lung adenocarcinoma. DNA Cell Biol. 2021;40:316–31. doi: 10.1089/dna.2020.6136. [DOI] [PubMed] [Google Scholar]
- 145.Liu J, Li Z, Cheang I, Li J, Zhou C. RNA-binding protein IGF2BP1 associated with prognosis and immunotherapy response in lung adenocarcinoma. Front Genet. 2022;13:777399. doi: 10.3389/fgene.2022.777399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hu K, Yao L, Yan Y, Zhou L, Li J. Comprehensive analysis of YTH domain family in lung adenocarcinoma: expression profile, association with prognostic value, and immune infiltration. Dis Markers. 2021;2021:2789481. doi: 10.1155/2021/2789481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang H, Zhao X, Lu Z. m(6)A RNA methylation regulators act as potential prognostic biomarkers in lung adenocarcinoma. Front Genet. 2021;12:622233. doi: 10.3389/fgene.2021.622233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tan S, Li Z, Li K, Li Y, Liang G, Tang Z, et al. The regulators associated with N6-methyladenosine in lung adenocarcinoma and lung squamous cell carcinoma reveal new clinical and prognostic markers. Front Cell Dev Biol. 2021;9:741521. doi: 10.3389/fcell.2021.741521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sun L, Liu WK, Du XW, Liu XL, Li G, Yao Y, et al. Large-scale transcriptome analysis identified RNA methylation regulators as novel prognostic signatures for lung adenocarcinoma. Ann Transl Med. 2020;8:751. doi: 10.21037/atm-20-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang Z, Zhang C, Luo Y, Wu P, Zhang G, Zeng Q, et al. m(6)A regulator expression profile predicts the prognosis, benefit of adjuvant chemotherapy, and response to anti-PD-1 immunotherapy in patients with small-cell lung cancer. BMC Med. 2021;19:284. doi: 10.1186/s12916-021-02148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang K, Han Z, Zhao H, Liu S, Zeng F. An integrated model of FTO and METTL3 expression that predicts prognosis in lung squamous cell carcinoma patients. Ann Transl Med. 2021;9:1523. doi: 10.21037/atm-21-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gu C, Shi X, Qiu W, Huang Z, Yu Y, Shen F, et al. Comprehensive analysis of the prognostic role and mutational characteristics of m6a-related genes in lung squamous cell carcinoma. Front Cell Dev Biol. 2021;9:661792. doi: 10.3389/fcell.2021.661792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li F, Wang H, Huang H, Zhang L, Wang D, Wan Y. m6A RNA Methylation regulators participate in the malignant progression and have clinical prognostic value in lung adenocarcinoma. Front Genet. 2020;11:994. doi: 10.3389/fgene.2020.00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Guo W, Huai QL, Sun SJ, Guo L, Xue XM, Song P, et al. Development and validation of m6A RNA methylation regulators-based signature in lung adenocarcinoma. Chin Med J. 2021;134:2128–30. doi: 10.1097/CM9.0000000000001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li X, Feng Z, Wang R, Hu J, He X, Shen Z. Expression status and prognostic value of m(6)A RNA methylation regulators in lung adenocarcinoma. Life. 2021;11:619. doi: 10.3390/life11070619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Guo B, Zhang H, Wang J, Wu R, Zhang J, Zhang Q, et al. Identification of the signature associated with m(6)A RNA methylation regulators and m(6)A-related genes and construction of the risk score for prognostication in early-stage lung adenocarcinoma. Front Genet. 2021;12:656114. doi: 10.3389/fgene.2021.656114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang X, Zhao C, Huang D, Liu Z, Liu M, Lin F, et al. A novel M6A-related genes signature can impact the immune status and predict the prognosis and drug sensitivity of lung adenocarcinoma. Front Immunol. 2022;13:923533. doi: 10.3389/fimmu.2022.923533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang Y, Zhao X, Li J, Wang X, Hu W, Zhang X. Four m6A RNA Methylation Gene Signatures And Their Prognostic Values In Lung Adenocarcinoma. Technol Cancer Res Treat. 2022;21:15330338221085373. doi: 10.1177/15330338221085373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wu X, Sheng H, Wang L, Xia P, Wang Y, Yu L, et al. A five-m6A regulatory gene signature is a prognostic biomarker in lung adenocarcinoma patients. Aging. 2021;13:10034–57. doi: 10.18632/aging.202761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhu J, Wang M, Hu D. Deciphering N(6)-Methyladenosine-related Genes Signature To Predict Survival In Lung Adenocarcinoma. Biomed Res Int. 2020;2020:2514230. doi: 10.1155/2020/2514230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhuang Z, Chen L, Mao Y, Zheng Q, Li H, Huang Y, et al. Diagnostic, progressive and prognostic performance of m(6)A methylation RNA regulators in lung adenocarcinoma. Int J Biol Sci. 2020;16:1785–97. doi: 10.7150/ijbs.39046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu Y, Guo X, Zhao M, Ao H, Leng X, Liu M, et al. Contributions and prognostic values of m(6) A RNA methylation regulators in non-small-cell lung cancer. J Cell Physiol. 2020;235:6043–57. doi: 10.1002/jcp.29531. [DOI] [PubMed] [Google Scholar]
- 163.Li N, Chen X, Liu Y, Zhou T, Li W. GenE characteristics and prognostic values of m(6)A RNA methylation regulators in nonsmall cell lung cancer. J Health Eng. 2021;2021:2257066. doi: 10.1155/2021/2257066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sun J, Ping Y, Huang J, Zeng B, Ji P, Li D. N6-Methyladenosine-regulated mRNAs: potential prognostic biomarkers for patients with lung adenocarcinoma. Front Cell Dev Biol. 2021;9:705962. doi: 10.3389/fcell.2021.705962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.David CJ, Manley JL. The search for alternative splicing regulators: new approaches offer a path to a splicing code. Genes Dev. 2008;22:279–85. doi: 10.1101/gad.1643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhao Z, Cai Q, Zhang P, He B, Peng X, Tu G, et al. N6-methyladenosine RNA methylation regulator-related alternative splicing (AS) gene signature predicts non-small cell lung cancer prognosis. Front Mol Biosci. 2021;8:657087. doi: 10.3389/fmolb.2021.657087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hou Q, Zhong Y, Liu L, Wu L, Liu J. Construction of a lung adenocarcinoma prognostic model based on N6-methyl-adenosine-related long noncoding RNA and screening of potential drugs based on this model. Anticancer Drugs. 2022;33:371–83. doi: 10.1097/CAD.0000000000001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhao J, Lin X, Zhuang J, He F. Relationships of N6-methyladenosine-related long non-coding RNAs with tumor immune microenvironment and clinical prognosis in lung adenocarcinoma. Front Genet. 2021;12:714697. doi: 10.3389/fgene.2021.714697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zheng J, Zhao Z, Wan J, Guo M, Wang Y, Yang Z, et al. N-6 methylation-related lncRNA is potential signature in lung adenocarcinoma and influences tumor microenvironment. J Clin Lab Anal. 2021;35:e23951. doi: 10.1002/jcla.23951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dong B, Wu C, Li SH, Huang L, Zhang C, Wu B, et al. Correlation of m6A methylation with immune infiltrates and poor prognosis in non-small cell lung cancer via a comprehensive analysis of RNA expression profiles. Ann Transl Med. 2021;9:1465. doi: 10.21037/atm-21-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhu J, Jiang Y, Wang T, Wu A, Zhou T, Zhang A, et al. Integrative analysis of m6A RNA methylation regulators and the tumor immune microenvironment in non-small-cell lung cancer. Dis Markers. 2022;2022:2989200. doi: 10.1155/2022/2989200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fan Y, Zhou Y, Lou M, Li X, Zhu X, Yuan K. m(6)A Regulator-mediated methylation modification patterns and characterisation of tumour microenvironment infiltration in non-small cell lung cancer. J Inflamm Res. 2022;15:1969–89. doi: 10.2147/JIR.S356841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ye W, Huang T. Correlation analysis of m6A-modified regulators with immune microenvironment infiltrating cells in lung adenocarcinoma. PLoS One. 2022;17:e0264384. doi: 10.1371/journal.pone.0264384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jiang F, Hu Y, Liu X, Wang M, Wu C. Methylation pattern mediated by m(6)A regulator and tumor microenvironment invasion in lung adenocarcinoma. Oxid Med Cell Longev. 2022;2022:2930310. doi: 10.1155/2022/2930310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhu M, Cui Y, Mo Q, Zhang J, Zhao T, Xu Y, et al. Characterization of m(6)A RNA methylation regulators predicts survival and immunotherapy in lung adenocarcinoma. Front Immunol. 2021;12:782551. doi: 10.3389/fimmu.2021.782551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Deng LJ, Deng WQ, Fan SR, Chen MF, Qi M, Lyu WY, et al. m6A modification: recent advances, anticancer targeted drug discovery and beyond. Mol Cancer. 2022;21:52. doi: 10.1186/s12943-022-01510-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xu P, Ge R. Roles and drug development of METTL3 (methyltransferase-like 3) in anti-tumor therapy. Eur J Med Chem. 2022;230:114118. doi: 10.1016/j.ejmech.2022.114118. [DOI] [PubMed] [Google Scholar]
- 178.Zhou X, Ao X, Jia Z, Li Y, Kuang S, Du C, et al. Non-coding RNA in cancer drug resistance: underlying mechanisms and clinical applications. Front Oncol. 2022;12:951864. doi: 10.3389/fonc.2022.951864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Liu Y, Ao X, Wang Y, Li X, Wang J. Long non-coding RNA in gastric cancer: mechanisms and clinical implications for drug resistance. Front Oncol. 2022;12:841411. doi: 10.3389/fonc.2022.841411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Liu Y, Ao X, Ji G, Zhang Y, Yu W, Wang J. Mechanisms of action and clinical implications of MicroRNAs in the drug resistance of gastric cancer. Front Oncol. 2021;11:768918. doi: 10.3389/fonc.2021.768918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zuo YB, Zhang YF, Zhang R, Tian JW, Lv XB, Li R, et al. Ferroptosis in cancer progression: role of noncoding RNAs. Int J Biol Sci. 2022;18:1829–43. doi: 10.7150/ijbs.66917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Song Z, Jia G, Ma P, Cang S. Exosomal miR-4443 promotes cisplatin resistance in non-small cell lung carcinoma by regulating FSP1 m6A modification-mediated ferroptosis. Life Sci. 2021;276:119399. doi: 10.1016/j.lfs.2021.119399. [DOI] [PubMed] [Google Scholar]
- 183.Wang T, Liu Z, She Y, Deng J, Zhong Y, Zhao M, et al. A novel protein encoded by circASK1 ameliorates gefitinib resistance in lung adenocarcinoma by competitively activating ASK1-dependent apoptosis. Cancer Lett. 2021;520:321–31. doi: 10.1016/j.canlet.2021.08.007. [DOI] [PubMed] [Google Scholar]
- 184.Zhang H, Wang SQ, Wang L, Lin H, Zhu JB, Chen R, et al. m6A methyltransferase METTL3-induced lncRNA SNHG17 promotes lung adenocarcinoma gefitinib resistance by epigenetically repressing LATS2 expression. Cell Death Dis. 2022;13:657. doi: 10.1038/s41419-022-05050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Li K, Peng ZY, Gao S, Wang QS, Wang R, Li X, et al. M6A associated TSUC7 inhibition contributed to Erlotinib resistance in lung adenocarcinoma through a notch signaling activation dependent way. J Exp Clin Cancer Res. 2021;40:325. doi: 10.1186/s13046-021-02137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dagogo-Jack I, Shaw AT. Crizotinib resistance: implications for therapeutic strategies. Ann Oncol. 2016;27:iii42–iii50. doi: 10.1093/annonc/mdw305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ding N, You A, Tian W, Gu L, Deng D. Chidamide increases the sensitivity of non-small cell lung cancer to crizotinib by decreasing c-MET mRNA methylation. Int J Biol Sci. 2020;16:2595–611. doi: 10.7150/ijbs.45886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu S, Li Q, Li G, Zhang Q, Zhuo L, Han X, et al. The mechanism of m(6)A methyltransferase METTL3-mediated autophagy in reversing gefitinib resistance in NSCLC cells by β-elemene. Cell Death Dis. 2020;11:969. doi: 10.1038/s41419-020-03148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Chen H, Jia B, Zhang Q, Zhang Y. Meclofenamic acid restores gefinitib sensitivity by downregulating breast cancer resistance protein and multidrug resistance protein 7 via FTO/m6A-Demethylation/c-Myc in non-small cell lung cancer. Front Oncol. 2022;12:870636. doi: 10.3389/fonc.2022.870636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Li K, Gao S, Ma L, Sun Y, Peng ZY, Wu J, et al. Stimulation of Let-7 maturation by metformin improved the response to tyrosine kinase inhibitor therapy in an m6A dependent manner. Front Oncol. 2021;11:731561. doi: 10.3389/fonc.2021.731561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Song H, Liu D, Wang L, Liu K, Chen C, Wang L, et al. Methyltransferase like 7B is a potential therapeutic target for reversing EGFR-TKIs resistance in lung adenocarcinoma. Mol Cancer. 2022;21:43. doi: 10.1186/s12943-022-01519-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ling Q, Wu S, Liao X, Liu C, Chen Y. Anesthetic propofol enhances cisplatin-sensitivity of non-small cell lung cancer cells through N6-methyladenosine-dependently regulating the miR-486-5p/RAP1-NF-κB axis. BMC Cancer. 2022;22:765. doi: 10.1186/s12885-022-09848-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chen WW, Qi JW, Hang Y, Wu JX, Zhou XX, Chen JZ, et al. Simvastatin is beneficial to lung cancer progression by inducing METTL3-induced m6A modification on EZH2 mRNA. Eur Rev Med Pharm Sci. 2020;24:4263–70. doi: 10.26355/eurrev_202004_21006. [DOI] [PubMed] [Google Scholar]
- 194.Feng Y, Li C, Liu S, Yan F, Teng Y, Li X, et al. β-elemene restrains PTEN mRNa degradation to restrain the growth of lung cancer cells via METTL3-mediated N(6) methyladenosine modification. J Oncol. 2022;2022:3472745. doi: 10.1155/2022/3472745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Cui Y, Wu Y, Wang C, Wang Z, Li Y, Jiang Z, et al. Isoliquiritigenin inhibits non-small cell lung cancer progression via m(6)A/IGF2BP3-dependent TWIST1 mRNA stabilization. Phytomedicine. 2022;104:154299. doi: 10.1016/j.phymed.2022.154299. [DOI] [PubMed] [Google Scholar]
- 196.Wang X, Xu D, Chen B, Huang D, Li Z, Sui Y, et al. Delicaflavone represses lung cancer growth by activating antitumor immune response through n6-methyladenosine transferases and oxidative stress. Oxid Med Cell Longev. 2022;2022:8619275. doi: 10.1155/2022/8619275. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 197.Sun X, Li Q, Yang L. Sevoflurane inhibits lncRNA HOTAIR-modulated stability of HK2 mRNA in a m6A-dependent manner to dampen aerobic glycolysis and proliferation in lung cancer. Biomed Res Int. 2022;2022:4668774. doi: 10.1155/2022/4668774. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 198.Mu X, Zhao Q, Chen W, Zhao Y, Yan Q, Peng R, et al. IL-37 confers anti-tumor activity by regulation of m6A methylation. Front Oncol. 2020;10:526866. doi: 10.3389/fonc.2020.526866. [DOI] [PMC free article] [PubMed] [Google Scholar]