Abstract
Temporal lobe epilepsy (TLE) is the most common and severe form of epilepsy in adults; however, its underlying pathomechanisms remain elusive. Dysregulation of ubiquitination is increasingly recognized to contribute to the development and maintenance of epilepsy. Herein, we observed for the first time that potassium channel tetramerization domain containing 13 (KCTD13) protein, a substrate-specific adapter for cullin3-based E3 ubiquitin ligase, was markedly down-regulated in the brain tissue of patients with TLE. In a TLE mouse model, the protein expression of KCTD13 dynamically changed during epileptogenesis. Knockdown of KCTD13 in the mouse hippocampus significantly enhanced seizure susceptibility and severity, whereas overexpression of KCTD13 showed the opposite effect. Mechanistically, GluN1, an obligatory subunit of N-methyl-D-aspartic acid receptors (NMDARs), was identified as a potential substrate protein of KCTD13. Further investigation revealed that KCTD13 facilitates lysine-48-linked polyubiquitination of GluN1 and its degradation through the ubiquitin-proteasome pathway. Besides, the lysine residue 860 of GluN1 is the main ubiquitin site. Importantly, dysregulation of KCTD13 affected membrane expression of glutamate receptors and impaired glutamate synaptic transmission. Systemic administration of the NMDAR inhibitor memantine significantly rescued the epileptic phenotype aggravated by KCTD13 knockdown. In conclusion, our results demonstrated an unrecognized pathway of KCTD13-GluN1 in epilepsy, suggesting KCTD13 as a potential neuroprotective therapeutic target for epilepsy.
Subject terms: Neuroscience, Neurological disorders
Introduction
Epilepsy is one of the most common and disabling neurological disorders and is characterized by spontaneous recurrent seizures (SRSs) [1]. About 70 million people worldwide suffer from epilepsy [2]. TLE is the most common form of drug-resistant epilepsy (DRE) in adults and imposes severe physical, psychological, and economic burdens on patients and their families [3, 4]. Elucidating the mechanisms underlying TLE is critical to improving current clinical therapies. TLE is traditionally considered to be the result of altered neuronal excitability [5]. Studies of TLE in humans and animal models have shown that protein expression within affected structures such as the hippocampus display extensive dysregulation [5–7].
Ubiquitination is one of the most important post-translational modifications for the maintenance of protein homeostasis [8]. Dysfunctional ubiquitination can result in severe diseases, including epilepsy [9–11]. E3 ubiquitin ligase plays a vital role in determining the specificity of substrates [12–15]. The cullin-RING ligases constitute the largest family of E3 ligases and mainly catalyze the ubiquitination of cellular proteins for degradation by the proteasome [16]. Numerous studies have shown that the epilepsy-associated E3 ubiquitin ligases most likely affect targets and/or pathways associated with the regulation of neuronal excitability [17–20]. Therefore, understanding the role and cellular mechanisms of E3 ubiquitin ligase and its substrates in TLE remains necessary to reveal therapeutic targets.
The 25-member potassium channel tetramerization domain (KCTD) family of proteins shares a conserved bric-a-brac, tramtrak, and broad complex/poxvirus zinc finger domain (BTB/POZ) at the N-terminus [21, 22]. Several human KCTD proteins are associated with neurodevelopmental, neuropsychiatric, and neurodegenerative disorders [23]. For example, mutations in KCTD7 can cause progressive myoclonic epilepsy [24–26]. KCTD13, also known as BACURD1 or POLDIP1, interacts with cullin3 (CUL3) through its N-terminal BTB domain, while the C-terminal binds and recruits substrate proteins to the E3 ubiquitin ligase complex for ubiquitination [23, 27]. KCTD13 is involved in the ubiquitination and degradation of RhoA, a small GTPase protein that plays a key role in neuronal development and synaptic function [28, 29]. Recently, KCTD13 has been found to be involved in the pathogenesis of autism by mediating ubiquitination and degradation of adenosine succinate synthase (ADSS) [30]. Additionally, KCTD13 is located in the 16p11.2 locus, and copy-number variations therein have been implicated in developmental brain disorders, epilepsy, autism spectrum disorders, and schizophrenia [31]. KCTD13 is considered one of the strongest candidate genes for neuropsychiatric disorders in the 16p11.2 locus [32, 33]; however, little is known about its function in epilepsy.
Herein, we provide evidence that KCTD13 plays a critical role in TLE. We show that KCTD13 is required for the total and membrane expression of GluN1, which in turn alters glutamatergic synaptic transmission in TLE. Through further molecular mechanism studies, we demonstrate that KCTD13 regulates ubiquitination of GluN1 and its degradation via the proteasome pathway. Our results suggest that KCTD13, which functions as an adapter for CUL3-based E3 ubiquitin ligase complex, is involved in the pathogenesis of TLE and highlight another intervention strategy against epilepsy.
Results
KCTD13 expression and subcellular distribution in brain tissues of TLE patients and epileptic mice
To investigate the potential role of KCTD13 in TLE, we first examined the expression of KCTD13 in kainic acid (KA) epilepsy model mice using western blot (WB) analysis. Compared with the sham control, the expression of KCTD13 protein increased at 1, 3, and 7 days after KA-induced status epilepticus (SE) but significantly decreased at 14, 30, and 60 days (Fig. 1A, B). In the sham control, we did not observe such dynamic changes in KCTD13 protein expression (Fig. S1A, B). We further examined KCTD13 protein in the temporal lobe of human TLE patients and control non-epileptic traumatic brain injury (TBI) patients. There were no significant differences in age or sex between the two groups (Tables S1 and S2). We found that KCTD13 was significantly reduced in the brain tissues of TLE patients compared with those in TBI controls (Fig. 1C, D).
Fig. 1. Expression and localization of KCTD13 in the brain tissues of mice and patients with TLE.
A, B Representative immunoblots (A) and quantification (B) of KCTD13 protein expression in mice hippocampal lysates at different time points after KA-induced SE compared to that in the sham control. n = 6 mice/group/time point, *p < 0.05, **p < 0.01, unpaired two-tailed Student’s t-test. C, D Representative immunoblots (C) and quantification (D) of KCTD13 protein expression in the temporal lobe of TLE patients compared with that of the control TBI group. n = 12/group, ***p < 0.001, unpaired two-tailed Student’s t-test. E Immunofluorescent labeling of KCTD13 and microtubule-associated protein 2 (MAP2) in cultured primary neurons. F Representative confocal images of KCTD13, MAP2, and glial fibrillary acidic protein (GFAP) immunofluorescence in the brain tissue samples from patients with TLE. G, H Representative confocal images of KCTD13, DAPI, NeuN, and GFAP immunofluorescence in hippocampus from KA epilepsy model mice. I Representative confocal images of colocalization of endogenously expressed KCTD13 with PSD95 and gephyrin in primary neurons.
KCTD13 expression is abundant in brain tissue, but KCTD13 distribution in the hippocampus remains undetermined. We detected abundant expression of KCTD13 in the CA1 region, CA3 region, and dentate gyrus of the mouse hippocampus using immunofluorescence (Fig. S1C). Since cultured primary neurons provide a higher resolution visualization of protein localization than immunofluorescence in vivo, we further investigate the subcellular distribution of KCTD13 in cultured primary neurons. Immunocytochemical assays revealed KCTD13-specific immune signals in both neuron cell bodies and dendrites (Fig. 1E). Next, we investigated the localization of KCTD13 in epileptic brain tissue. We found that KCTD13 was primarily co-expressed with neurons in the brain tissue of TLE patients and a mouse model (Figs. 1F–H and S1D, E). These results indicate that KCTD13 may play a role in epilepsy by directly regulating neurons. To determine the synaptic localization of KCTD13, we performed triple colocalization analysis of KCTD13, excitatory postsynaptic marker postsynaptic density protein 95 (PSD95), and inhibitory postsynaptic marker gephyrin in epileptic brain tissues. We found that KCTD13 colocalized with both excitatory and inhibitory synapses in the neurons (Fig. S1F, G). Immunocytochemical assays revealed that both endogenously expressed and exogenously overexpressed KCTD13 colocalized with both PSD95 and gephyrin (Figs. 1I and S1H), suggesting a possible role for KCTD13 in synaptic transmission.
KCTD13 negatively modulates seizure susceptibility and severity
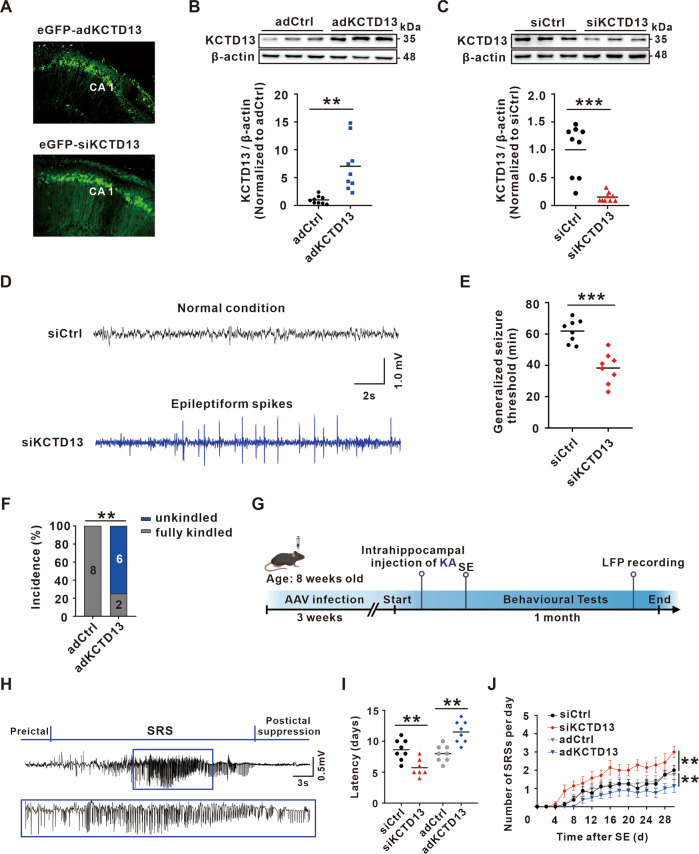
To explore whether the observed dynamic changes of KCTD13 was a concomitant phenomenon or contributed to epileptogenesis, we injected adeno-associated virus (AAV) into mice hippocampal CA1 region to knockdown or overexpression of KCTD13. The expression efficiency of KCTD13 knockdown or overexpression was confirmed after 3 weeks using immunofluorescence (Fig. 2A) and WB (Fig. 2B, C). We then determined if mice with KCTD13 knockdown or overexpression showed any epileptiform responses using intracranial electroencephalographic (EEG) recordings to monitor the local field potential (LFP). Interestingly, epileptiform spikes were recorded in KCTD13 knockdown mice (Fig. 2D) but not in KCTD13 overexpression or control groups (data not shown). After 1 month of behavioral observation, no epileptic seizures were observed in any of the groups. Our results showed that the knockdown of KCTD13 increases the excitability of neural networks.
Fig. 2. KCTD13 regulates seizure susceptibility and severity.
A Immunofluorescent labeling of enhanced green fluorescent protein (eGFP) in hippocampal CA1 region transfected with AAV-eGFP-adKCTD13 or AAV-eGFP-siKCTD13. B, C Immunoblot images and quantification of KCTD13 protein levels in mice hippocampus infected with the indicated AAVs. n = 9/group, **p < 0.01, ***p < 0.001, unpaired two-tailed Student’s t-test. D Typical EEG recordings from mice infected with AAV-siKCTD13. E Effects of KCTD13 knockdown on the threshold of PTZ kindling seizure model. n = 8/group, ***p < 0.001, unpaired two-tailed Student’s t-test. F Incidence rates of fully kindled seizures observed in KCTD13 overexpressing mice following the injection of PTZ. n = 8/group, **p < 0.01, Pearson chi-square test. G Schematic representation showing the experimental design using the intra-hippocampal injection of KA to induce the TLE model. H A typical LFP recording of a spontaneous recurrent seizure in a KA-induced TLE model mouse infected with AAV-siKCTD13. I, J The latency of spontaneous seizure onset (I) and the number of spontaneous seizures per day (J) recorded during a 30-day recording period from the time of SE induction. siKCTD13 group, n = 7; adKCTD13, adCtrl, and siCtrl groups, n = 8/group. **p < 0.01, two-tailed Student’s t-test for (I) and two-way repeated measures ANOVA followed by Bonferroni post hoc test for (J).
Next, we investigated whether altered KCTD13 expression would affect seizure susceptibility on a pentylenetetrazol (PTZ) kindling seizure model, and we found that knockdown of KCTD13 significantly reduced the generalized seizure latency compared with that in the control group (Fig. 2E). However, overexpression of KCTD13 significantly reduced the fully kindled seizure rate (Fig. 2F). We further examined the effect of altered KCTD13 expression on epileptic behavior in a KA-induced TLE model. In this model, SE was induced by intrahippocampal injection of KA, and SRSs appeared after a period of latency, during which behavioral observations and LFP recordings were performed (Fig. 2G). We observed epileptiform discharges (Fig. 2H) in the LFP recordings synchronized with grade IV/V (as defined by Racine’s scale) seizures (Fig. S2A). Behavioral observation revealed that knockdown of KCTD13 significantly reduced the latency (Fig. 2I) and increased the frequency (Fig. 2J) of SRSs. In contrast, overexpression of KCTD13 had the opposite effect (Fig. 2I, J). Together, these results suggested that KCTD13 negatively modulates seizure susceptibility and severity.
KCTD13 negatively regulates glutamatergic synaptic transmission
Epileptic response is often due to alterations in excitatory/inhibitory balance and/or neuronal intrinsic excitability [5]. Thus, we established an in vitro seizure model by removing Mg2+ from the extracellular solution and examined the activities of CA1 pyramidal neurons using whole-cell patch-clamp recordings. We first examined whether KCTD13 had any effect on the intrinsic excitability of pyramidal neurons. We found that the resting membrane potential (Fig. 3A), action potential (AP) firing rate (Figs. 3B–E and S2B), or AP threshold (Figs. 3F and S2C) did not change in the KCTD13 knockdown or overexpression mice, suggesting that KCTD13 does not contribute to the epileptic response by changing intrinsic neuronal excitability. To determine whether KCTD13 regulates synaptic transmission, we examined spontaneous postsynaptic currents. We found that knockdown of KCTD13 increased the frequency and amplitude of spontaneous excitatory postsynaptic currents (sEPSCs) (Fig. 3G, H), while overexpression of KCTD13 decreased the frequency and amplitude of sEPSCs (Fig. 3I, J). However, the frequency and amplitude of spontaneous inhibitory postsynaptic currents (sIPSCs) showed little change in KCTD13 knockdown (Fig. S2D, E) or overexpression (Fig. S2F, G) mice. Our results suggested that altered KCTD13 expression leads to impaired glutamatergic but not GABAergic synaptic transmission.
Fig. 3. KCTD13 regulates glutamatergic synaptic transmission.
A A summary of statistical analyses for resting membrane potential (RMP) recorded from KCTD13 overexpression or knockdown CA1 pyramidal neurons. n = 21 cells, 5 mice, unpaired two-tailed Student’s t-test. B–E Representative traces of recordings in current clamp and average firing rates in response to different depolarizing current injections from KCTD13 knockdown (B, C) or overexpression (D, E) CA1 pyramidal neurons. n = 21 cells, 5 mice, two-way repeated measures ANOVA followed by Bonferroni post hoc test. F Typical spikes (left) and AP threshold (right) obtained from KCTD13 overexpression or knockdown CA1 pyramidal neurons. n = 21 cells, 5 mice, unpaired two-tailed Student’s t-test. G–J Representative traces of sEPSCs (G, I) recorded in siCtrl (n = 15 cells, 4 mice), siKCTD13 (n = 16 cells, 4 mice), adCtrl (n = 15 cells, 4 mice), or adKCTD13 (n = 19 cells, 4 mice) transfected CA1 pyramidal neurons. Cumulative curves of inter-event interval and amplitude of sEPSCs (H, J) and the insets display mean sEPSC frequency and amplitude. **p < 0.01, ***p < 0.001, unpaired two-tailed Student’s t-test.
KCTD13 interacts with GluN1 and negatively regulates GluN1 protein expression in epileptic tissue
To explore the molecular mechanism by which KCTD13 regulates glutamatergic synaptic transmission, hippocampal lysates from KA epileptic mice were subjected to immunoprecipitation combined with electrospray ionization mass spectrometry (ESI-MS). A total of 1535 possible interacting proteins were identified (Table S3). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that the interacting proteins were enriched in the common pathways, such as oxidative phosphorylation, carbon metabolism, and glutamatergic synapse (Fig. 4A), among which the glutamatergic synapse pathway was consistent with our electrophysiological results (Figs. 3 and S2). Therefore, proteins involved in synaptic transmission, i.e., vesicle associated membrane protein 2 (VAMP2), α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR) subunits (GluA1 and GluA2), and NMDAR subunits (GluN1, GluN2A, and GluN2B), were selected for verification. Using co-immunoprecipitation (Co-IP) assays, we found that KCTD13 interacted with GluN1, GluN2A, GluN2B, and VAMP2 (Fig. 4B, C). We next investigated whether KCTD13 regulates the expression of these identified interacting proteins. WB analysis revealed that knockdown of KCTD13 only significantly upregulated GluN1 expression in epileptic brain tissue (Fig. 4D), while overexpression of KCTD13 had the opposite effect (Fig. 4E). We also found endogenous KCTD13 and GluN1 spatially colocalized in cultured primary neurons and epileptic brain tissue (Fig. 4F). We further verified whether KCTD13 could regulate the expression of exogenous GluN1. WB analysis revealed that KCTD13 inhibited the expression of exogenous GluN1 (Fig. 4G). Furthermore, we found that KCTD13 reduced GluN1 in a dose-dependent manner (Fig. 4H). These results suggest that GluN1 may be a potential downstream protein of KCTD13 that is involved in regulating the epileptic phenotype.
Fig. 4. KCTD13 interacts with GluN1 and negatively regulates GluN1 protein expression.
A KEGG pathway analysis of the potential KCTD13-binding proteins identified by ESI-MS of the hippocampal tissue of the KA mice model. B Characterization of interactions between KCTD13 and the indicated proteins by Co-IP. Epileptic mouse brain lysates were precipitated with anti-KCTD13 magnetic beads, and precipitates were subsequently immunoblotted with antibodies to detect GluA1, GluA2, GluN1, GluN2A, GluN2B, and VAMP2. IgG immunoprecipitate was used as a negative control. C Reverse validation of the interaction of GluN1, GluN2A, GluN2B, and VAMP2 with KCTD13 in lysates from the brains of KA-induced epileptic mice by Co-IP assay. D, E WB analysis revealing the total expression of the indicated proteins in KCTD13 knockdown (D) or KCTD13 overexpression (E) KA mice model. n = 6/group, **p < 0.01, ***p < 0.001, unpaired two-tailed Student’s t-test. F Immunofluorescent labeling of KCTD13, GluN1, MAP2, and DAPI in cultured primary neurons and epileptic brain tissues. G, H Immunoblot images of exogenously expressed GluN1 in HEK293T cells transfected with the indicated constructs and analyzed by WB with the indicated antibodies. n = 4 independent experiments.
KCTD13 degrades GluN1 through the ubiquitin-proteasome pathway
KCTD13 may regulate GluN1 protein level by affecting GluN1 transcription or protein stability; therefore, quantitative real-time PCR (qPCR) was performed to investigate whether KCTD13 regulates GluN1 mRNA expression in epileptic brain tissue. Compared with the corresponding control group, knockdown or overexpression of KCTD13 did not significantly affect GluN1 mRNA levels (Fig. 5A). These results suggest that KCTD13 may regulate GluN1 at the post-transcriptional level. To test this hypothesis, we first infected cultured primary neurons with AAVs to regulate the expression of KCTD13 and verified the expression efficiency using WB and immunofluorescence (Fig. S3A–C). In addition, a cycloheximide (CHX) chase assay was used to investigate the effect of KCTD13 on GluN1 stability, and we found that KCTD13 knockdown significantly prolonged the half-life of GluN1 (Fig. 5B, C). Next, we overexpressed KCTD13 in cultured primary neurons and found that KCTD13 down-regulated GluN1 protein levels and that this effect could be rescued by treating neurons with the specific proteasome inhibitor MG132, but not by the lysosomal inhibitor chloroquine (Fig. 5D).
Fig. 5. KCTD13 regulates the ubiquitination of GluN1 and mediates its degradation via the proteasome pathway.
A Quantification of GluN1 expression at the mRNA level detected by qPCR in the hippocampus of epileptic mice. Data were analyzed and normalized to the expression of β-actin. n = 6/group, unpaired two-tailed Student’s t-test. B, C Time course depicting GluN1 degradation with siRNA-mediated KCTD13 knockdown in cultured primary neurons. WB analyses of GluN1 following CHX (500 μM) treatment for the time indicated. GluN1 protein levels were normalized to β-actin and set to 100% at time 0. n = 4, **p < 0.01, two-way ANOVA with Bonferroni’s post hoc correction. D Immunoblot images of GluN1 and KCTD13 protein expression in cultured primary neurons treated with or without the indicated reagents. n = 3 independent experiments. E Co-IP analysis of the interactions between endogenous KCTD13, CUL3, and GluN1 in the hippocampus of KA-induced epileptic mice. IgG (M), mouse IgG, IgG (R), rabbit IgG. F Co-IP analysis of interaction between exogenous KCTD13 BTB domain deletion mutant and endogenous CUL3. G Immunoblot images of exogenously expressed myc-GluN1 protein levels in HEK293T cells transfected with the indicated constructs. n = 4 independent experiments. H Immunoprecipitation analysis to quantify polyubiquitinated GluN1 in KCTD13 knockdown epileptic mice hippocampus. Tissue lysates were immunoprecipitated with anti-GluN1 and analyzed by WB with indicated antibodies. n = 4 independent experiments. I Co-IP analysis of the ubiquitination of exogenously expressed GluN1 in HEK293T cells transfected with indicated constructs. Before cell harvest, MG132 (50 mM) was added into the medium for 6 h. Cell lysates were immunoprecipitated with anti-Myc. n = 3 independent experiments. J Schematic illustration of CUL3, a scaffold protein, which binds with the BTB domain of the substrate recognition adapter KCTD13 and ring finger protein (RBX) to form E3 ubiquitin ligase that regulates GluN1 ubiquitination and proteasomal degradation.
To elucidate whether KCTD13 acts as an adapter for CUL3-based E3 ubiquitin ligase to regulate GluN1, we first identified an interaction between endogenous KCTD13 and CUL3 in epileptic brain tissue using Co-IP assay (Fig. 5E). In addition, we constructed an E3 ligase-deficient KCTD13 mutant by deleting the BTB domain of KCTD13 (denoted as Flag-ΔBTB). We transfected wild-type (WT) KCTD13 or Flag-ΔBTB into HEK293T cells. By Co-IP assay, we found that Flag-ΔBTB could not co-precipitate CUL3 (Fig. 5F). Furthermore, we found that Flag-KCTD13 reduced Myc-GluN1 protein levels, whereas Flag-ΔBTB did not have the same effect (Fig. 5G).
To demonstrate the effect of KCTD13 on the polyubiquitination of GluN1 in vivo, we extracted epileptic hippocampal brain tissue and measured the ubiquitination level of GluN1 using Co-IP assay. We found that knockdown of KCTD13 resulted in a significant decrease in the polyubiquitination level of GluN1 (Fig. 5H), whereas overexpression of KCTD13 had the opposite effect (Fig. S3D). Next, we co-transfected HA-ubiquitin (HA-Ub), Flag-KCTD13, and Myc-GluN1 plasmids into HEK293T cells and found that the polyubiquitination level of exogenous GluN1 was significantly increased after KCTD13 overexpression (Fig. S3E). Notably, the KCTD13 BTB domain deletion mutant lost the ability to mediate GluN1 polyubiquitination (Fig. 5I). Overall, our results suggest that KCTD13 regulates the ubiquitination of GluN1 and mediates its degradation via the proteasome pathway (Fig. 5J).
KCTD13 mediates K48-linked polyubiquitination of GluN1 at lysine residue 860
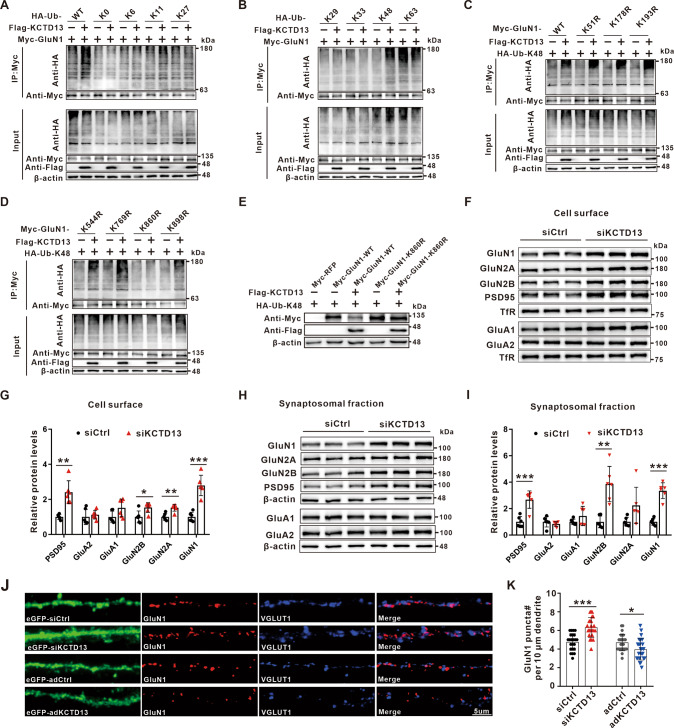
To determine the type of KCTD13-induced GluN1 polyubiquitin chain, expression plasmids for Flag-KCTD13 and Myc-GluN1 were transfected into HEK293T cells together with plasmids encoding WT Ub or Ub mutants. Co-IP and immunoblot analysis showed that KCTD13 promoted the K48-linked polyubiquitination of GluN1 (Fig. 6A, B). Since ubiquitination modification usually occurs in lysine residues of substrate proteins, we further mapped ubiquitination sites on GluN1 that were modified by KCTD13. HEK293T cells were transfected with Flag-KCTD13, Myc-GluN1, and HA-Ub-K48 plasmids. Using ESI-MS, we found that KCTD13-mediated polyubiquitination may occur at lysine (K) residues K51, K178, K193, K544, K769, K860, and K898 of GluN1 (Table S4 and Fig. S4A, B). We then constructed GluN1 mutants in which each of these seven sites had these residues altered to arginine (R). The ubiquitination assay revealed that only the K860R mutant failed to be ubiquitinated by KCTD13, indicating that K860 was a specific modification site of the GluN1 protein (Fig. 6C, D). Furthermore, we found that Myc-GluN1-K860R rescued the downregulation of Myc-GluN1 protein level caused by KCTD13 overexpression (Fig. 6E). These data suggest that KCTD13 acts as an adapter for E3 ubiquitin ligase, binding GluN1 and triggering K48-linked polyubiquitination of GluN1 at K860, which subsequently mediates GluN1 degradation via the proteasome pathway.
Fig. 6. KCTD13 promotes K48-linked polyubiquitination of GluN1 at lysine residue 860 and regulates the surface and synaptosomal expression of glutamate receptors.
A, B Co-IP analysis of the ubiquitination of exogenously expressed GluN1 in HEK293T cells transfected with plasmids of Flag-KCTD13 and Myc-GluN1 together with HA-Ub or its mutants for 48 h. Before cell harvest, MG132 (50 mM) was added into the medium for 6 h. n = 3 independent experiments. C, D Co-IP analysis of the polyubiquitination of GluN1 WT and its mutants, in which the lysine residues at positions 51, 178, 193, 544, 769, 860, and 898 were replaced with arginine individually. The exogenously expressed GluN1 was immunoprecipitated with anti-Myc antibody from HEK293T cells transfected with the indicated plasmids and analyzed by WB with the indicated antibodies. Before cell harvest, MG132 (50 mM) was added into the medium for 6 h. n = 3 independent experiments. E WB analysis of exogenously expressed GluN1 WT and its K860R mutant in HEK293T cells transfected with the indicated plasmids. n = 3 independent experiments. F–I Immunoblot images and quantification of the surface (F, G) and synaptosome (H, I) expression of glutamate receptors in epileptic hippocampal tissues transfected with AAV-siKCTD13 or AAV-siCtrl. n = 6/group, *p < 0.05, **p < 0.01, ***p < 0.001, unpaired two-tailed Student’s t-test. Adjustments were made for multiple comparisons test. J, K Representative confocal images (J) and quantification of GluN1 puncta (K) in cultured primary neurons transfected with siCtrl (n = 23 cells), siKCTD13 (n = 23 cells), adCtrl (n = 25 cells), or adKCTD13 (n = 25 cells) labeled with eGFP, GluN1, and VGLUT1 antibodies. *p < 0.05, ***p < 0.001, unpaired two-tailed Student’s t-test.
KCTD13 negatively regulates the surface expression and synaptosomal localization of GluN1
It remains unclear whether the regulation of GluN1 by KCTD13 is accompanied by changes in surface expression of GluN1. We observed that knockdown of KCTD13 resulted in a significant upregulation of the surface expression of GluN1, as well as GluN2A, GluN2B, and PSD95 in epileptic hippocampal brain tissue (Fig. 6F, G). In contrast, when KCTD13 was overexpressed, the surface expression of GluN1, GluN2A, GluN2B, and GluA1 were significantly down-regulated (Fig. S4C). To further determine the effect of KCTD13 on the expression of NMDARs in synapses, we extracted synaptosomes from epileptic hippocampal tissue. We observed changes in synaptosome receptors consistent with their surface expressions (Figs. 6H, I and S4D). Interestingly, the surface and synaptosome expressions of GluA1 were also significantly changed. We further asked whether the total GluA1 protein level was changed. Surprisingly, we found that KCTD13 also negatively regulated GluA1 but not GluA2 total protein (Fig. S4E, F). In addition, the colocalization of GluN1 and the excitatory presynaptic marker vesicular glutamate transporter 1 (VGLUT1) was detected using immunofluorescence labeling. In KCTD13 knockdown neurons, the number of GluN1 positive puncta were significantly increased, and most of these GluN1 puncta colocalized with VGLUT1 (Fig. 6J, K), further confirming that increased GluN1 was indeed present at synapses. In contrast, when KCTD13 was overexpressed, we found a significant decrease in the number of GluN1 positive puncta (Fig. 6J, K). These results suggest that KCTD13 negatively regulates the total protein expression of GluN1, and this is accompanied by the surface and synaptosome expression changes of glutamate receptors in epileptic brain tissues.
KCTD13 regulates the number of functional synapses and NMDAR-mediated glutamatergic synaptic transmission
Next, we focused on whether the regulation of excitatory receptors by KCTD13 affects synaptic function. We found that KCTD13 regulates the frequency of sEPSCs (Fig. 3G–J), which may be influenced by changes in the probability of presynaptic glutamate release or the number of functional synapses. To test these possibilities, we recorded the EPSC-mediated paired–pulse ratio (PPR) in CA3–CA1 synapses. Neither overexpression nor knockdown of KCTD13 resulted in significant changes in PPR (Fig. 7A, B), suggesting that the probability of presynaptic glutamate release was not altered. Therefore, we used immunocytochemistry to measure the number of functional synapses. The overlap between presynaptic VGLUT1 labeling and postsynaptic PSD95 labeling defines the excitatory synapse [34]. Consistent with the higher sEPSC frequency, the number of excitatory synapses was significantly increased in KCTD13 knockdown neurons, whereas overexpression of KCTD13 showed the opposite change (Fig. 7C, D). Since functional excitatory synapses are mainly localized in dendritic spines [35], we used Golgi staining to investigate the effect of KCTD13 on the morphology of dendritic spines on the apical dendrites of CA1 pyramidal neurons (Fig. 7E). We found a significant increase in the proportion of mature dendritic spines in KCTD13 knockdown brain tissues (Fig. 7F, I), despite no significant difference in the density of dendritic spines (Fig. 7H). In contrast, the density of dendritic spines and the proportion of mature dendritic spines were significantly decreased in KCTD13 overexpressing brain tissues as compared with that in the control group (Fig. 7G, H, J).
Fig. 7. KCTD13 regulates the number of functional glutamatergic synapses.
A, B EPSC-PPR recordings from KCTD13 knockdown (A) or overexpression (B) brain slices. The PPR responses evoked by pairs of stimuli delivered 50 ms apart. siCtrl group, n = 16 cells, 4 mice; siKCTD13 group, n = 17 cells, 5 mice; adCtrl group, n = 16 cells, 4 mice; adKCTD13 group, n = 17 cells, 5 mice; unpaired two-tailed Student’s t-test. C Representative confocal images in cultured primary neurons transfected with siCtrl (n = 19 cells), siKCTD13 (n = 20 cells), adCtrl (n = 20 cells), or adKCTD13 (n = 21 cells) labeled with eGFP, PSD95, and VGLUT1 antibodies. D Analysis of the total number of excitatory synapses identified as VGLUT1/PSD95-positive puncta. **p < 0.01, unpaired two-tailed Student’s t-test. E–J Golgi staining (E–G) and quantitative analysis (H–J) of dendritic spine density and dendritic spine maturity of pyramidal neurons in the CA1 region of the epileptic mice model. The blue arrow indicates apical dendrites of CA1 pyramidal neurons where dendritic spine density was determined. Filopodia dendritic spines (>2 μm) and long thin spines (length >1 μm, head diameter <0.2 μm) were immature spines, while stubby and mushroom spines were mature spines. n = 24/group, *p < 0.05, **p < 0.01, ***p < 0.001, unpaired two-tailed Student’s t-test. Adjustments were made for multiple comparisons test (I, J).
To further verify the excitatory synaptic function, we recorded evoked EPSCs mediated by AMPARs (AMPAR-eEPSCs) and NMDARs (NMDAR-eEPSCs) at the CA3–CA1 synapse. We found that knockdown of KCTD13 resulted in a significant increase in the NMDA/AMPA ratio and NMDAR-eEPSCs (Fig. 8A, B). Conversely, overexpression of KCTD13 significantly reduced NMDAR-eEPSCs and AMPAR-eEPSCs, especially NMDAR-eEPSCs (Fig. 8C, D). By another pharmacological approach to measure NMDAR-eEPSCs, we found the same changes in NMDAR-eEPSC amplitude (Fig. S5A, B, D, E). Interestingly, we also observed that knockdown of KCTD13 significantly prolonged the decay time of NMDAR-eEPSCs, while the overexpression of KCTD13 shortened it (Fig. S5C, F). To further characterize the KCTD13-mediated changes in NMDAR function, we measured changes in NMDAR-eEPSCs at different presynaptic stimulation intensities. We found hyperexcitability of NMDARs in KCTD13 knockdown CA1 pyramidal neurons (Fig. S5G, H). Next, we determined the functional composition of postsynaptic NMDAR subunits by isolating GluN2B-containing NMDARs blocked by Ro 25-6981. We found that the blocking effect of Ro 25-6981 on NMDAR-eEPSCs in KCTD13 knockdown neurons was significantly stronger than that in control neurons (Fig. 8E, F), which indicated that knockdown of KCTD13 leads to a more significant increase in GluN2B-containing NMDARs. Our findings suggest that KCTD13’s regulation of NMDARs may underpin its effect on the epileptic phenotype. For further confirmation, we used low doses of memantine, a noncompetitive inhibitor of the NMDAR that has previously been shown to treat epilepsy or autism caused by enhanced NMDARs function [36, 37]. Daily intraperitoneal injection of memantine (5 mg/kg, for 5 days after KA-induced SE) significantly alleviated the epileptic phenotype caused by KCTD13 knockdown (Fig. 8G, H). Taken together, these results suggest that KCTD13-mediated ubiquitination and degradation of GluN1 is involved in the pathogenesis of TLE.
Fig. 8. KCTD13 regulates NMDAR-mediated glutamatergic synaptic transmission.
A, B Representative traces (A) and quantification (B) of NMDAR-eEPSCs (holding at +40 mV) and AMPAR-eEPSCs (holding at −70 mV) recorded at CA1 pyramidal neurons from KCTD13 knockdown or control brain slices. NMDAR-mediated responses were measured at 50 ms post-stimulus. n = 13 cells, 4 mice, ***p < 0.001, unpaired two-tailed Student’s t-test. C, D Representative traces (C) and quantification (D) of NMDAR-eEPSCs and AMPAR-eEPSCs recorded in CA1 pyramidal neurons from KCTD13 overexpression or control brain slices. n = 13 cells, 4 mice. *p < 0.05, ***p < 0.001, unpaired two-tailed Student’s t-test. E, F Representative traces (E) and quantification (F) of NMDAR-eEPSCs recorded at CA1 pyramidal neurons from KCTD13 knockdown or control slices before and after (20 min) incubation with Ro 25-6981 (1 μM, dissolved in dimethyl sulfoxide (DMSO)). n = 11 cells, 4 mice/group, *p < 0.05, unpaired two-tailed Student’s t-test. G, H Effects of KCTD13 knockdown on the latency (G) and frequency of SRSs (H) in KA mice model treated with or without memantine. n = 6/group, #p > 0.05, *p < 0.05, **p < 0.01, unpaired two-tailed Student’s t-test (G), two-way repeated measures ANOVA (H).
Discussion
Protein homeostasis and the regulatory mechanisms underlying protein synthesis and degradation are essential for the normal functioning of the central nervous system [10, 38]. As an important pathway of endogenous protein degradation, dysfunction of the ubiquitin-proteasome pathway may lead to various neurological diseases, including epilepsy [39–41]. The molecular mechanism of ubiquitination in epilepsy remains to be further elucidated. KCTD13, an adapter for cullin3-based E3 ubiquitin ligase, is abundantly expressed in the brain and is emerging as an important influencing factor in neurological diseases [30, 42, 43]; however, its role in epilepsy remains unclear. Our data suggest that ubiquitination represents an important post-translational mechanism of protein expression regulation during epilepsy in which KCTD13 plays a key role.
Brain insults, including KA-induced SE, can trigger epileptogenesis during which functional and structural reorganization of neuronal networks occurs, resulting in the onset of SRSs. During epileptogenesis, gene expression profiles significantly change [6, 44]. For the first time to date, we investigated the expression of KCTD13 in epileptic brain tissues. We found that KCTD13 protein levels increased progressively, reaching a peak at 3 days after KA-induced SE. Then, the expression of KCTD13 decreased during the chronic phase of post-SE epileptogenesis (Fig. 1A, B). In the behavioral study, we found that KCTD13 knockdown in mice exacerbated the epileptic phenotype, whereas KCTD13 overexpression had a protective effect (Fig. 2). Our results suggest that the altered KCTD13 expression may be involved in the development of epilepsy rather than a simple concomitant phenomenon. We speculate that the upregulated KCTD13 protein in the acute and latent phase of epileptogenesis most likely represents an endogenous anti-epileptogenic adaptive mechanism for self-balance and to protect the brain once pathological processes are initiated. However, in the chronic phase, hyperexcitatory neural networks are formed and the expression level of KCTD13 is significantly reduced, which may contribute to the maintenance of the hyperexcitability of the abnormal neural network and the recurrence of SRSs. The dynamic changes of KCTD13 protein expression during epileptogenesis may be precisely regulated by complex factors, including transcription factors and epigenetic modification. The specific reasons are warrant further exploration. In conclusion, our results suggest that KA-induced SE triggers dynamic changes of KCTD13 protein and is involved in the pathogenesis of epilepsy.
Accumulating evidence suggests that KCTD13 plays an important role in the central nervous system by catalyzing ubiquitylation of various substrate proteins. In neuropsychiatric disorders such as autism, KCTD13 may play a role by ubiquitinating RhoA [27, 43]. A recent study using ubiquitination proteomics analysis showed that ADSS may be a potential substrate of KCTD13 affecting autism [30]. However, it is unclear whether KCTD13 regulates epilepsy through identified substrate proteins such as RhoA and ADSS. To reduce bias, we combined immunoprecipitation and ESI-MS to identify KCTD13-regulated substrate proteins in epilepsy. We found that the potential substrate protein of KCTD13 in epilepsy is GluN1, which is an essential subunit of NMDARs (Fig. 4). The known KCTD13 substrate proteins RhoA and ADSS were not detected in our mass spectrometry identification. This may be due to the low abundance of these two substrate proteins or low affinity between KCTD13 and these two substrates in our animal model.
AMPARs and NMDARs are the two major types of glutamate receptors that transmit presynaptic signals to postsynaptic neurons, and the localization, activation, and abundance of these receptors are highly regulated [45–48]. Their changes in expression and function are closely related to epilepsy [49–51]. In the hippocampus, functional NMDARs are hetero-tetrameric assemblies of two GluN1 subunits and two GluN2A or GluN2B subunits [52, 53]. Here, we identified KCTD13 as a novel ubiquitinated E3 ligase substrate adapter that regulates GluN1 abundance. Our results suggest that KCTD13 binds CUL3 to catalyze ubiquitination of GluN1 and regulate its proteasomal degradation in vitro and in vivo (Fig. 5). Further investigation showed that KCTD13 mainly mediated the K48-linked polyubiquitin of GluN1 (Fig. 6A, B). The K48-linked polyubiquitin chain is the main type of ubiquitin modification that mediates degradation of substrate proteins through the ubiquitin-proteasome pathway [13, 54].
Another important finding was that the surface and synaptosomal expression of GluN1 were consistent with the changes in the total GluN1 protein (Fig. 6F–I). These results suggest that the regulation of GluN1 protein level by KCTD13 may affect synaptic function in epilepsy. We speculate that this is because the knockdown of KCTD13 significantly increases GluN1 stability, while the kinetics of NMDAR endocytosis and exocytosis remain unchanged, thus recycling it to the membrane surface. However, overexpression of KCTD13 resulted in the reduction of GluN1 in synaptosomes, accompanied by the reduction of NMDAR functional subunits GluN2A and GluN2B. We also detected changes in GluA1, GluA2, and PSD95 in synaptosomal fractions. Numerous studies have shown that the localization of AMPARs at mature synapses is primarily associated with NMDAR activation [55–58]. Therefore, we hypothesized that this change may be due to KCTD13 regulating the function of NMDARs, which affects the recruitment of GluA1-containing AMPARs at synapses. In addition, we found that KCTD13 negatively regulated total GluA1 protein (Fig. S4E, F). Although the Co-IP assay did not detect any interaction between KCTD13 and GluA1, a direct regulatory effect of KCTD13 on GluA1 cannot be excluded. It is also possible that KCTD13 indirectly regulates the expression of total GluA1 protein by regulating KCTD13 substrate proteins that are yet unidentified. We observed that the knockdown of KCTD13 increased AMPA- and, to a greater extent, NMDAR-eEPSC amplitude (Fig. 8A, B). This suggests that knockdown of KCTD13 mainly enhances NMDAR-dependent synaptic function. Unexpectedly, we found that knockdown of KCTD13 significantly prolonged the decay time of NMDAR-eEPSCs (Fig. S5C). Previous studies have shown that GluN2B-containing NMDARs have longer decay kinetics [47, 53, 59], suggesting that knockdown of KCTD13 results in assembly of NMDARs at synapses dominated by GluN2B-containing NMDARs. Our study suggests that enhanced NMDAR signaling may underlie the exacerbation of epilepsy caused by decreased KCTD13 expression. Therefore, we performed behavioral rescue experiments and showed that memantine, a noncompetitive NMDA receptor blocker, significantly reduced the susceptibility and severity of epilepsy in KCTD13 knockdown mice (Fig. 8G, H).
As an adapter for E3 ubiquitin ligase, the substrate proteins catalyzed by KCTD13 may be different under different physiological and pathological conditions. Hence, it may not be feasible to discover every single target of KCTD13. However, our data demonstrated that the potential target substrate protein of KCTD13 in epilepsy is GluN1, which is a novel and important discovery. Our findings may offer the possibility to control seizures in DRE patients with reduced KCTD13 expression by targeting the KCTD13-GluN1 pathway, such as by using memantine. Furthermore, the effect of KCTD13 on synaptic proteins and synaptic function also provides a basis for further study of KCTD13 in diseases that are characterized by altered synaptic transmission.
Materials and methods
Antibodies and reagents
The antibodies used in this study are described in Table S5. HiScript II Q RT SuperMix for qPCR (+gDNA wiper) and ChamQ Universal SYBR qPCR Master Mix were purchased from Vazyme Biotech Co., Ltd (Nanjing, China). Protein A/G magnetic beads, chloroquine, and Ro 25-6981 were purchased from MedChemExpress (State of New Jersey, USA). Coomassie Blue Fast Staining and No-decoloring Solution was obtained from Epizyme (Shanghai, China). Synaptosome Isolation Kit and Plasma Membrane Protein Isolation and Cell Fractionation Kit were purchased from Invent Biotechnologies (Minnesota, USA). Tetrodotoxin was obtained from Tocris Bioscience (Bristol, UK). Hito Golgi-Cox OptimStain PreKit (Prepare Solution Kit) was purchased from Hitobiotec Corp (TN, USA). EndoFree Maxi Plasmid Kit V2 was purchased from TIANGEN (Beijing, China). Other reagents not mentioned were purchased from Sigma-Aldrich (St. Louis, USA).
Plasmids
The Flag-tagged WT KCTD13 and KCTD13 BTB domain truncated mutant were constructed by cloning the human cDNA of full-length or truncated KCTD13 into the pcDNA3.1-3xFlag-T2A-eGFP vector. The human cDNAs of GluN1 WT and mutants K51R, K178R, K193R, K544R, K769R, K860R, and K898R were generated by site-directed mutagenesis and cloned in frame with a Myc tag into pcDNA3.1-T2A-TagRFP vectors. HA-tagged WT ubiquitin and HA-tagged ubiquitin mutants K0, K6, K11, K27, K29, K33, K48, and K63 were cloned into pIRES2-TagBFP vectors. All constructs were confirmed by sequencing.
Adeno-associated virus vectors
The AAV9 vectors carrying mouse full-length KCTD13 (adKCTD13), eGFP alone (adCtrl), KCTD13-RNAi (siKCTD13), and scramble-RNAi (siCtrl) were constructed by GeneChem Co., Ltd. (Shanghai, China). For adKCTD13 packaging, a CMV-bGlobin-MCS-eGFP-3Flag-WPRE-hGH polyA vector was used. Empty AAV vectors coding eGFP were used as control. For siKCTD13 packaging, a U6-MCS-CAG-eGFP vector was used. The KCTD13-RNAi sequence was 5′-GCACTTCAGATGACAACTTAC-3′. The scramble-RNAi sequence was 5′-CGCTGAGTACTTCGAAATGTC-3′, which has no target in the mouse.
Human brain tissue
Human brain tissue samples from patients with TLE or TBI who underwent temporal lobe resection were used in this study and collected from the First Affiliated Hospital of Chongqing Medical University. Written informed consent was obtained from patients for the use of brain tissue and access to medical records for research purposes. The collection and use of all specimens were approved by the Ethics Committee of The First Affiliated Hospital of Chongqing Medical University and were conducted in accordance with the Declaration of Helsinki. The brain tissue specimens obtained from the operation were immediately stored in liquid nitrogen until use.
Animals
The experimental animals used were male C57BL/6J mice obtained from the Laboratory Animal Center of Chongqing Medical University (Certificate: SCXK (YU) 2018-0003). Except for neonatal mice used to generate primary neuron cultures, all mice were 6–7 weeks old at the beginning of the experiment. The mice were randomly assigned to each experimental condition. The mice were group-housed in cages and raised on a 12:12-light-dark cycle with free access to food and water. All animal experiments were approved by the Ethics Committee of Chongqing Medical University. All efforts were made to minimize animal suffering and numbers.
Stereotaxic AAV injection
Under anesthesia, AAV9 vectors (siCtrl, siKCTD13, adCtrl or adKCTD13) were injected stereotactically into the mice CA1 hippocampal region (Paxinos Mouse Brain Atlas; coordinates from bregma; Anteroposterior (AP): −2.0 mm; mediolateral (ML): ±1.5 mm; dorsoventral (DV): −1.7 mm) via a micro syringe. All AAV deliveries were performed bilaterally with a volume of 1 μl each at a flow rate of 0.2 μl/min. The needle was kept in place for 5 min after each injection.
KA model
Mice were anesthetized and placed in a stereotaxic apparatus (RWD Life Science Co. Ltd., China). Further, 0.5-μl KA (0.3 μg in 0.5 μl saline) was injected in the right hippocampal CA1 region for 3 min. The mice were allowed to recover from anesthesia at 32 °C and then monitored closely during SE. The behavioral seizure scores were classified according to Racine’s scale [60]. SE usually began 15 min after complete recovery and ended 40 min after SE onset with intraperitoneal diazepam (10 mg/kg). Sham-control mice were treated identically but received the same volume of saline instead of KA. For behavioral tests, trials started 3 weeks post-AAV injection. Mice were video monitored to quantify the latency and the number of SRSs over 30 consecutive days after KA injection. Seizures (Racine’s stages III–V) were quantified during off-line video review by two independent observers, and one was blinded to the treatment.
PTZ-induced seizures
Mice previously transfected with AAVs (siKCTD13, siCtrl, adCtrl, or adKCTD13) were injected with PTZ. Seizures were induced by repetitive intraperitoneal administration of PTZ (10 mg/kg; Sigma-Aldrich) every 10 min until generalized seizures occurred or until the total amount reached 100 mg/kg. Seizures were monitored throughout the experiment and rated on Racine’s scale. The latency to the first generalized seizure (stages IV and V) and PTZ-induced generalized seizure incidence was recorded.
LFP recording
Surgery was performed 1 week prior to recordings. Briefly, to record hippocampal LFP, mice were anesthetized and placed in a stereotaxic apparatus, two stainless steel screws implanted in the anterior cranium served as the ground electrode. For LFP recording, the recording electrode was implanted into the right dorsal hippocampus and connected to the MAP data acquisition system (Plexon; Texas, USA) to collect data. Typical seizure-like event characterized by the disappearance of normal electrophysiological rhythm and the occurrence of a cluster of spontaneous paroxysmal discharges with amplitude greater than 2 times the baseline, frequency greater than 3 Hz, and duration greater than 5 s.
Drug administration
For each behavioral cohort, mice were acclimatized to daily intraperitoneal injections of either the vehicle (saline) or the drug. Memantine was diluted in saline at 10 μl per gram of mouse body weight for administration. Notably, the amount of memantine used was comparable to those from studies in other mouse models of neurodevelopmental disorders, which was expected to result in a reversible blockade of a relatively small fraction of synaptic NMDARs [37].
Western blot
The extraction steps of plasma membrane proteins and synaptosomes were performed according to the manufacturer’s instructions. Total proteins of cells or tissues were lysed with RIPA lysis buffer (Beyotime; Shanghai, China). Lysates containing equal proteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (Millipore, USA). The membranes were incubated with Tris-buffered saline with 0.1% Tween-20 (TBST) containing 5% nonfat milk at room temperature for 60 min and then incubated with primary antibodies diluted in primary antibody dilution buffer (Beyotime) at 4 °C overnight. After 12–16 h, the membranes were washed with TBST and subsequently incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies diluted in TBST at room temperature for 1 h. The signal was detected using an enhanced chemiluminescence reagent (Biosharp, Guangzhou, China) and the Fusion FX7 image analysis system (Vilber Lourmat, Marne-la-Vallée, France).
Immunostaining
For immunofluorescence staining of cryostat sections, mice were anesthetized and perfused with Phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA). Brains were post-fixed in 4% PFA overnight and submerged in 30% sucrose for 3 days. Brain sections (16 μm) were sliced using a Leica frozen microtome. Brain sections were permeabilized with 0.4% Triton X-100 for 20 min, blocked with goat serum working liquid (Boster Biological Technology; Wuhan, China) for 60 min at room temperature, and then incubated with mixed primary antibodies diluted in PBS at 4 °C for overnight. Sections were washed three times with PBS and incubated with appropriate fluorescence-conjugated secondary antibodies for 60 min at room temperature. 4′,6-diamidino-2-phenylindole (DAPI) was used for nucleus staining. For immunocytochemical assays, primary neurons were fixed with PBS solution containing 4% PFA and 4% sucrose (30 min at room temperature). Neurons were permeabilized with 0.1% Triton X-100 for 10 min and blocked with goat serum working liquid for 60 min at room temperature. Then, neurons were incubated with mixed primary antibodies diluted in PBS at 4 °C for overnight. Next, neurons were washed three times with PBS and incubated with fluorescent secondary antibodies at room temperature for 1 h. All images were acquired under a confocal microscope (Leica; Wetzlar, Germany).
Immunoprecipitation
Cells or hippocampal tissue samples were lysed with RIPA lysis buffer (Beyotime), which was supplemented with a protease inhibitor cocktail (MedChemExpress) at 4 °C for 30 min. Lysates containing equal amounts of proteins were incubated with protein A/G magnetic beads (MedChemExpress) bound to anti-Myc, anti-KCTD13, anti-GluN1, anti-GluN2A, anti-GluN2B, anti-VAMP2, anti-Flag, or anti-CUL3 antibodies at 4 °C for 2 h, according to the manufacturer’s instructions. After washing, the proteins were harvested by magnetic separation, and proteins bound on the beads were analyzed by WB.
Mass spectrometry analysis
Total protein was extracted from hippocampal tissue samples 3 days after KA injection. KCTD13-beads were used for immunoprecipitation of proteins interacting with KCTD13. The collected interacting proteins were loaded onto SDS-PAGE. After electrophoresis, the gel was stained with Coomassie blue for 30 min at room temperature. For mass spectrometry analysis, lanes without protein bands on the gel were excised. Mass spectrometric analysis was performed by Applied Protein Technology (Shanghai, China). To identify the ubiquitination modification site of GluN1, HEK293T cells were transfected with Flag-KCTD13, HA-Ub-K48, and Myc-GluN1 plasmids. After 48 h of transfection, cells were lysed with lysis buffer, and the next steps were as described previously. KEGG pathway enrichment analysis was performed using the DAVID bioinformatics web server (https://david.ncifcrf.gov/) by uploading the gene lists from our mass spectrometric analysis.
qPCR
Total RNA was extracted from epileptic hippocampal tissue samples using RNAsimple Total RNA Kit (TIANGEN) according to the manufacturer’s instructions. qPCR analysis was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme). Gene expression levels were normalized to the housekeeping gene β-actin. The sequences of qPCR primers are listed in Supplementary Table 6.
Cell culture and transfection
HEK293T cells were obtained from the American Type Culture Collection. The cells were authenticated and tested for mycoplasma contamination. HEK293T cells were maintained at 37 °C with 5% CO2 and cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco; New York, USA) containing 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (P/S; Gibco). Transient transfection of HEK293T was performed with Lipofectamine 3000 Reagent (Invitrogen) according to the manufacturer’s instructions.
Primary neuronal culture
Cortical and hippocampal tissues from C57BL/6J mice pups at postnatal day 0 were carefully isolated from neonatal mice, transferred to ice-cold D-Hank’s balanced salt solution (Solarbio; Beijing, China), dissected into small fragments, and digested with 0.125% trypsin (Gibco) for 10 min at 37 °C. The cells were seeded on cover slides pretreated with poly-L-lysine (Sigma-Aldrich), and DMEM containing 10% FBS and 1% P/S was added. After 4 h, the DMEM was replaced with neurobasal medium containing 2% B27 supplement, 1% P/S, and 1% L-glutamine (Gibco). Neurons were cultured at 37 °C with 5% CO2. Half of the fresh neurobasal medium was replaced every 3 days.
Ubiquitination assays
HEK293T cells transfected with the expression construct for HA-ubiquitin or its mutant, together with other expression constructs, were treated with MG132 for 6 h and lysed with RIPA lysis buffer. Lysates were subjected to immunoprecipitation followed by WB analysis.
Golgi staining
AAV-transfected mice were sacrificed and decapitated 3 days after KA injection. Brain tissue samples were washed with double-distilled water to remove blood from the surface. Neurons were stained using the Hito Golgi-Cox OptimStain Kit according to the manufacturer’s protocol. Secondary apical dendrites from CA1 pyramidal neurons were imaged for spine analysis. Sections were imaged using a microscope (Leica) at a magnification of ×100 and analyzed with ImageJ software (NIH, Maryland, USA).
Slice preparation
Acute hippocampal slices were prepared from mice that were transfected with AAV-siCtrl, AAV-siKCTD13, AAV-adCtrl, or AAV-adKCTD13 3 weeks ago. Animals were anesthetized with pentobarbital and transcardially perfused with ice-cold oxygenated (95% O2, 5% CO2) slicing solution (2.5 mM KCl, 1.25 mM NaH2PO4, 6 mM MgCl2, 1 mM CaCl2, 220 mM sucrose, 26 mM NaHCO3, 10 mM D-glucose). After decapitation, brains were removed for sectioning in the same ice-cold slice solution using a vibratome (Leica VT1200S, Germany). For whole-cell patch-clamp recordings, 300-μm coronal hippocampal sections were prepared. Slices were recovered in oxygenated artificial CSF (aCSF; 124 mM NaCl, 3 mM KCl, 1.23 mM NaH2PO4, 26 mM NaHCO3, 2 mM MgCl2, 2 mM CaCl2, and 10 mM D-glucose, with pH adjusted to 7.4) at 32 °C for 30 min and at room temperature (25 °C) for an additional 30 min before recording.
Whole-cell patch-clamp recordings
A coronal slice containing hippocampus was placed in a submersion chamber maintained at room temperature and perfused at 3 ml/min with oxygenated Mg2+-free aCSF. All recordings were done with patch pipettes with a resistance between 3–6 MΩ. We performed recordings in the whole-cell configuration using a Multiclamp 700B amplifier (Axon, USA) with 10 kHz digitization and a 2 kHz low-pass Bessel filter. Data were acquired and analyzed using pCLAMP 11.1 software (Molecular Devices Co.; San Jose, CA, USA). Series resistance changes were monitored throughout the experiment and neurons were discarded if the series resistance surpassed 25 MΩ or changed by >20%. Neurons with unstable resting potential or potential >−50 mV were discarded. Transfected hippocampal CA1 pyramidal neurons were identified using eGFP fluorescence and from a pyramidal somatic shape.
For current-clamp recordings, the internal solution contained 60 mM K2SO4, 60 mM NMG, 40 mM HEPES, 4 mM MgCl2, 0.5 mM BAPTA, 12 mM phosphocreatine, 2 mM Na2ATP, and 0.2 mM Na3GTP (pH 7.3, 275-290 mOsm). D-APV (50 μM), DNQX (20 μM), and picrotoxin (100 μM) were added to block synaptic transmission. A current step protocol was used to evoke action potentials by injecting 500 ms long depolarizing current steps of increasing amplitude from –100 pA to 300 pA (△20 pA).
For voltage-clamp recordings, the internal solution used to record EPSCs contained 130 mM Cs-methanesulfonate, 10 mM HEPES, 10 mM CsCl, 4 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 5 mM NMG, 5 mM MgATP, 0.5 mM Na3GTP, and 12 mM phosphocreatine (pH 7.25; 280–300 mOsm). The sIPSCs were recorded by using internal solution that contained 100 mM CsCl, 10 mM HEPES, 1 mM MgCl2, 1 mM EGTA, 30 mM NMG, 5 mM MgATP, 0.5 mM Na3GTP, and 12 mM phosphocreatine (pH 7.25; 280–300 mOsm). Cells were clamped at −70 mV throughout the experiment unless otherwise stated. Recordings were performed in the presence of 100 μM picrotoxin for sEPSCs, 20 μM DNQX and 50 μM D-APV for sIPSCs, 100 μM picrotoxin and 20 μM DNQX for NMDAR-eEPSCs, 100 μM picrotoxin for NMDA/AMPA ratio, and 100 μM picrotoxin for EPSC-PPR. Measurements of the NMDA/AMPA ratio were performed at holding potentials of −70 mV (AMPAR-eEPSCs) or +40 mV (NMDAR-eEPSCs, quantified at 50 ms after the stimulus). The amplitudes and frequencies of sIPSCs and sEPSCs were detected by continuous recording for 300 s by using MiniAnalysis software (Synaptosoft).
Statistics
GraphPad Prism 8 software (San Diego, CA, USA) and IBM SPSS Statistics 23 (New York, USA) were used for statistical analysis. Experimental data are presented as either median with interquartile range or mean ± standard deviation (SD). p < 0.05 was considered statistically significant. The normality of the data was analyzed using Shapiro–Wilk test. When the variance of the data set was significantly different, we used a nonparametric statistical analysis. The two-tailed Student’s t-test was used to compare between two independent groups, and one-way or two-way ANOVA was used for multi-group comparison. Statistical methods for each experiment are detailed in the corresponding figure legend.
Supplementary information
Acknowledgements
We express our gratitude to the patients for the donations of brain tissue and their time and effort devoted to the consent process.
Author contributions
JG, FX, PK and XW designed the study; JG, PK, HG, JL, YL, XT, XX and DX performed the experiments; DX and YM participated in the data and sample collection; JG, PK, HG and ZH analyzed data; JG, FX and ZH wrote the manuscript; XW and FX oversaw the project. All authors read and approved the final paper.
Funding
This work was supported by National Natural Science Foundation of China (Nos. 81922023, 82271496, 81873788, 82001378 and 82171440), Natural Science Foundation of Chongqing (CSTB2022NSCQ-MSX0747 and cstc2021ycjh-bgzxm0035). Future Medical Youth Innovation Team Program of Chongqing Medical University (No. W0043), the Fifth Senior Medical Talents Program of Chongqing for Young and Middle-aged, Middle-aged Medical Excellence Team Program of Chongqing, and Chongqing chief expert studio project, China. Science and Technology Research Program of Chongqing Education Commission (KJQN202200435), Chongqing Talents: Exceptional Young Talents Project (CQYC202005014).
Data availability
Data supporting the conclusions in the paper are present in the paper and the Supplementary Materials. Additional data are available from the corresponding author.
Competing interests
The authors declare no competing interests.
Ethical approval
All animal experiments were approved by the Ethics Committee of Chongqing Medical University. Human brain tissue samples from patients with TLE or TBI were collected from the First Affiliated Hospital of Chongqing Medical University. Written informed consent was obtained from patients for the use of brain tissue and access to medical records for research purposes. The collection and use of all specimens were approved by the Ethics Committee of The First Affiliated Hospital of Chongqing Medical University and in accordance with the Declaration of Helsinki.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xuefeng Wang, Email: xfyp@163.com.
Fei Xiao, Email: feixiao81@126.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01174-5.
References
- 1.Devinsky O, Vezzani A, O’Brien TJ, Jette N, Scheffer IE, de Curtis M, et al. Epilepsy. Nat Rev Dis Primers. 2018;4:18024. doi: 10.1038/nrdp.2018.24. [DOI] [PubMed] [Google Scholar]
- 2.Thijs RD, Surges R, O’Brien TJ, Sander JW. Epilepsy in adults. Lancet. 2019;393:689–701. doi: 10.1016/S0140-6736(18)32596-0. [DOI] [PubMed] [Google Scholar]
- 3.Shlobin NA, Sander JW. Learning from the comorbidities of epilepsy. Curr Opin Neurol. 2022;35:175–80. doi: 10.1097/WCO.0000000000001010. [DOI] [PubMed] [Google Scholar]
- 4.Vinti V, Dell’Isola GB, Tascini G, Mencaroni E, Cara GD, Striano P, et al. Temporal lobe epilepsy and psychiatric comorbidity. Front Neurol. 2021;12:775781. doi: 10.3389/fneur.2021.775781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ren E, Curia G. Synaptic reshaping and neuronal outcomes in the temporal lobe epilepsy. Int J Mol Sci. 2021;22:3860. doi: 10.3390/ijms22083860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfisterer U, Petukhov V, Demharter S, Meichsner J, Thompson JJ, Batiuk MY, et al. Identification of epilepsy-associated neuronal subtypes and gene expression underlying epileptogenesis. Nat Commun. 2020;11:5038. doi: 10.1038/s41467-020-18752-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guelfi S, Botia JA, Thom M, Ramasamy A, Perona M, Stanyer L, et al. Transcriptomic and genetic analyses reveal potential causal drivers for intractable partial epilepsy. Brain. 2019;142:1616–30. doi: 10.1093/brain/awz074. [DOI] [PubMed] [Google Scholar]
- 8.Mabb AM. Historical perspective and progress on protein ubiquitination at glutamatergic synapses. Neuropharmacology. 2021;196:108690. doi: 10.1016/j.neuropharm.2021.108690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poliquin S, Kang JQ. Disruption of the ubiquitin-proteasome system and elevated endoplasmic reticulum stress in epilepsy. Biomedicines. 2022;10:647. doi: 10.3390/biomedicines10030647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J, Tsai NP. Ubiquitination and E3 ubiquitin ligases in rare neurological diseases with comorbid epilepsy. Neuroscience. 2020;428:90–9. doi: 10.1016/j.neuroscience.2019.12.030. [DOI] [PubMed] [Google Scholar]
- 11.Li C, Beauregard-Lacroix E, Kondratev C, Rousseau J, Heo AJ, Neas K, et al. UBR7 functions with UBR5 in the Notch signaling pathway and is involved in a neurodevelopmental syndrome with epilepsy, ptosis, and hypothyroidism. Am J Hum Genet. 2021;108:134–47. doi: 10.1016/j.ajhg.2020.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Htet ZM, Lopez-Alfonzo E, Martin A, Walters KJ. Proteasome interaction with ubiquitinated substrates: from mechanisms to therapies. FEBS J. 2021;288:5231–51. doi: 10.1111/febs.15638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Ferriz A, Ferrando A, Fathinajafabadi A, Farras R. Ubiquitin-mediated mechanisms of translational control. Semin Cell Dev Biol. 2022;132:146–54. [DOI] [PubMed]
- 14.Liu M, Yan M, Lv H, Wang B, Lv X, Zhang H, et al. Macrophage K63-linked ubiquitination of YAP promotes its nuclear localization and exacerbates atherosclerosis. Cell Rep. 2020;32:107990. doi: 10.1016/j.celrep.2020.107990. [DOI] [PubMed] [Google Scholar]
- 15.Wang P, Song J, Ye D. CRL3s: the BTB-CUL3-RING E3 ubiquitin ligases. Adv Exp Med Biol. 2020;1217:211–23. doi: 10.1007/978-981-15-1025-0_13. [DOI] [PubMed] [Google Scholar]
- 16.Baek K, Scott DC, Schulman BA. NEDD8 and ubiquitin ligation by cullin-RING E3 ligases. Curr Opin Struct Biol. 2021;67:101–9. doi: 10.1016/j.sbi.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JE, Lee DS, Kim TH, Park H, Kim MJ, Kang TC. PLPP/CIN-mediated Mdm2 dephosphorylation increases seizure susceptibility via abrogating PSD95 ubiquitination. Exp Neurol. 2020;331:113383. doi: 10.1016/j.expneurol.2020.113383. [DOI] [PubMed] [Google Scholar]
- 18.Zhu J, Lee KY, Jong TT, Tsai NP. C2-lacking isoform of Nedd4-2 regulates excitatory synaptic strength through GluA1 ubiquitination-independent mechanisms. J Neurochem. 2019;151:289–300. doi: 10.1111/jnc.14840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JE, Lee DS, Kim MJ, Kang TC. PLPP/CIN-mediated NEDD4-2 S448 dephosphorylation regulates neuronal excitability via GluA1 ubiquitination. Cell Death Dis. 2019;10:545. doi: 10.1038/s41419-019-1781-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds JP, Jimenez-Mateos EM, Cao L, Bian F, Alves M, Miller-Delaney SF, et al. Proteomic analysis after status epilepticus identifies UCHL1 as protective against hippocampal injury. Neurochem Res. 2017;42:2033–54. doi: 10.1007/s11064-017-2260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smaldone G, Pirone L, Pedone E, Marlovits T, Vitagliano L, Ciccarelli L. The BTB domains of the potassium channel tetramerization domain proteins prevalently assume pentameric states. Febs Lett. 2016;590:1663–71. doi: 10.1002/1873-3468.12203. [DOI] [PubMed] [Google Scholar]
- 22.Pinkas DM, Sanvitale CE, Bufton JC, Sorrell FJ, Solcan N, Chalk R, et al. Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases. Biochem J. 2017;474:3747–61. doi: 10.1042/BCJ20170527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teng X, Aouacheria A, Lionnard L, Metz KA, Soane L, Kamiya A, et al. KCTD: a new gene family involved in neurodevelopmental and neuropsychiatric disorders. CNS Neurosci Ther. 2019;25:887–902. doi: 10.1111/cns.13156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alevy J, Burger CA, Albrecht NE, Jiang D, Samuel MA. Progressive myoclonic epilepsy-associated gene Kctd7 regulates retinal neurovascular patterning and function. Neurochem Int. 2019;129:104486. doi: 10.1016/j.neuint.2019.104486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metz KA, Teng X, Coppens I, Lamb HM, Wagner BE, Rosenfeld JA, et al. KCTD7 deficiency defines a distinct neurodegenerative disorder with a conserved autophagy-lysosome defect. Ann Neurol. 2018;84:766–80. doi: 10.1002/ana.25351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Bogaert P. KCTD7-related progressive myoclonus epilepsy. Epileptic Disord. 2016;18:115–9. doi: 10.1684/epd.2016.0856. [DOI] [PubMed] [Google Scholar]
- 27.Lin GN, Corominas R, Lemmens I, Yang X, Tavernier J, Hill DE, et al. Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron. 2015;85:742–54. doi: 10.1016/j.neuron.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Escamilla CO, Filonova I, Walker AK, Xuan ZX, Holehonnur R, Espinosa F, et al. Kctd13 deletion reduces synaptic transmission via increased RhoA. Nature. 2017;551:227–31. doi: 10.1038/nature24470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H, et al. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell. 2009;35:841–55. doi: 10.1016/j.molcel.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Madison JM, Duong K, Vieux EF, Udeshi ND, Iqbal S, Requadt E, et al. Regulation of purine metabolism connects KCTD13 to a metabolic disorder with autistic features. iScience. 2021;24:101935. doi: 10.1016/j.isci.2020.101935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kusenda M, Vacic V, Malhotra D, Rodgers L, Pavon K, Meth J, et al. The influence of microdeletions and microduplications of 16p11.2 on global transcription profiles. J Child Neurol. 2015;30:1947–53. doi: 10.1177/0883073815602066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golzio C, Willer J, Talkowski ME, Oh EC, Taniguchi Y, Jacquemont S, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–7. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rein B, Yan Z. 16p11.2 copy number variations and neurodevelopmental disorders. Trends Neurosci. 2020;43:886–901. doi: 10.1016/j.tins.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai J, Patzke C, Liakath-Ali K, Seigneur E, Sudhof TC. GluD1 is a signal transduction device disguised as an ionotropic receptor. Nature. 2021;595:261–5. doi: 10.1038/s41586-021-03661-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu C, Liu HJ, Qi L, Tao CL, Wang YJ, Shen Z, et al. Structure and plasticity of silent synapses in developing hippocampal neurons visualized by super-resolution imaging. Cell Discov. 2020;6:8. doi: 10.1038/s41421-019-0139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Song R, Chen W, Strong K, Shrey D, Gedela S, et al. Recurrent seizure-related GRIN1 variant: molecular mechanism and targeted therapy. Ann Clin Transl Neurol. 2021;8:1480–94. doi: 10.1002/acn3.51406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang S, Terzic B, Wang IJ, Sarmiento N, Sizov K, Cui Y, et al. Altered NMDAR signaling underlies autistic-like features in mouse models of CDKL5 deficiency disorder. Nat Commun. 2019;10:2655. doi: 10.1038/s41467-019-10689-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morandell J, Schwarz LA, Basilico B, Tasciyan S, Dimchev G, Nicolas A, et al. Cul3 regulates cytoskeleton protein homeostasis and cell migration during a critical window of brain development. Nat Commun. 2021;12:3058. doi: 10.1038/s41467-021-23123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu J, Lee KY, Jewett KA, Man HY, Chung HJ, Tsai NP. Epilepsy-associated gene Nedd4-2 mediates neuronal activity and seizure susceptibility through AMPA receptors. PLoS Genet. 2017;13:e1006634. doi: 10.1371/journal.pgen.1006634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Judson MC, Wallace ML, Sidorov MS, Burette AC, Gu B, van Woerden GM, et al. GABAergic neuron-specific loss of Ube3a causes angelman syndrome-like EEG abnormalities and enhances seizure susceptibility. Neuron. 2016;90:56–69. doi: 10.1016/j.neuron.2016.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang M, Li Y, Ren J, Hu R, Gao X, Chen L. Epilepsy-associated UBE3A deficiency downregulates retinoic acid signalling pathway. Front Genet. 2021;12:681295. doi: 10.3389/fgene.2021.681295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arbogast T, Razaz P, Ellegood J, McKinstry SU, Erdin S, Currall B, et al. Kctd13-deficient mice display short-term memory impairment and sex-dependent genetic interactions. Hum Mol Genet. 2019;28:1474–86. doi: 10.1093/hmg/ddy436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin Lorenzo S, Nalesso V, Chevalier C, Birling MC, Herault Y. Targeting the RHOA pathway improves learning and memory in adult Kctd13 and 16p11.2 deletion mouse models. Mol Autism. 2021;12:1. doi: 10.1186/s13229-020-00405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conboy K, Henshall DC, Brennan GP. Epigenetic principles underlying epileptogenesis and epilepsy syndromes. Neurobiol Dis. 2020;148:105179. doi: 10.1016/j.nbd.2020.105179. [DOI] [PubMed] [Google Scholar]
- 45.Chen TS, Lai MC, Huang HI, Wu SN, Huang CW. Immunity, ion channels and epilepsy. Int J Mol Sci. 2022;23:6446. doi: 10.3390/ijms23126446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou MH, Chen SR, Wang L, Huang Y, Deng M, Zhang J, et al. Protein kinase C-mediated phosphorylation and alpha2delta-1 interdependently regulate NMDA receptor trafficking and activity. J Neurosci. 2021;41:6415–29. doi: 10.1523/JNEUROSCI.0757-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang YQ, Huang YH, Balakrishnan S, Liu L, Wang YT, Nestler EJ, et al. AMPA and NMDA receptor trafficking at cocaine-generated synapses. J Neurosci. 2021;41:1996–2011. doi: 10.1523/JNEUROSCI.1918-20.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, Bramham CR. Bidirectional dysregulation of AMPA receptor-mediated synaptic transmission and plasticity in brain disorders. Front Synaptic Neurosci. 2020;12:26. doi: 10.3389/fnsyn.2020.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou C, Lippman Bell JJ, Sun H, Jensen FE. Hypoxia-induced neonatal seizures diminish silent synapses and long-term potentiation in hippocampal CA1 neurons. J Neurosci. 2011;31:18211–22. doi: 10.1523/JNEUROSCI.4838-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lippman-Bell JJ, Zhou C, Sun H, Feske JS, Jensen FE. Early-life seizures alter synaptic calcium-permeable AMPA receptor function and plasticity. Mol Cell Neurosci. 2016;76:11–20. doi: 10.1016/j.mcn.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Itoh M, Yamashita M, Kaneko M, Okuno H, Abe M, Yamazaki M, et al. Deficiency of AMPAR-palmitoylation aggravates seizure susceptibility. J Neurosci. 2018;38:10220–35. doi: 10.1523/JNEUROSCI.1590-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marwick KFM, Hansen KB, Skehel PA, Hardingham GE, Wyllie DJA. Functional assessment of triheteromeric NMDA receptors containing a human variant associated with epilepsy. J Physiol. 2019;597:1691–704. doi: 10.1113/JP277292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soares C, Lee KF. A prominent role for triheteromeric GluN1/GluN2A/GluN2B NMDARs at central synapses. J Neurosci. 2013;33:14975–7. doi: 10.1523/JNEUROSCI.3109-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li X, Yang KB, Chen W, Mai J, Wu XQ, Sun T, et al. CUL3 (cullin 3)-mediated ubiquitination and degradation of BECN1 (beclin 1) inhibit autophagy and promote tumor progression. Autophagy. 2021;17:4323–40. doi: 10.1080/15548627.2021.1912270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–6. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 57.Clements L, Harvey J. Activation of oestrogen receptor α induces a novel form of LTP at hippocampal temporoammonic‐CA1 synapses. Br J Pharmacol. 2020;177:642–55. doi: 10.1111/bph.14880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glasgow SD, Labrecque S, Beamish IV, Aufmkolk S, Gibon J, Han D, et al. Activity-dependent Netrin-1 secretion drives synaptic insertion of GluA1-containing AMPA receptors in the hippocampus. Cell Rep. 2018;25:168–82.e6. doi: 10.1016/j.celrep.2018.09.028. [DOI] [PubMed] [Google Scholar]
- 59.Wong JM, Folorunso OO, Barragan EV, Berciu C, Harvey TL, Coyle JT, et al. Postsynaptic serine racemase regulates NMDA receptor function. J Neurosci. 2020;40:9564–75. doi: 10.1523/JNEUROSCI.1525-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Racine RJ. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr Clin Neurophysiol. 1972;32:281–94. doi: 10.1016/0013-4694(72)90177-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the conclusions in the paper are present in the paper and the Supplementary Materials. Additional data are available from the corresponding author.