Abstract
Cancer stem cells (CSCs) are a minority population of cancer cells with stemness and multiple differentiation potentials, leading to cancer progression and therapeutic resistance. However, the concrete mechanism of CSCs in hepatocellular carcinoma (HCC) remains obscure. We found that in advanced HCC tissues, collagen I was upregulated, which is consistent with the expression of its receptor DDR1. Accordingly, high collagen I levels accompanied by high DDR1 expression are associated with poor prognoses in patients with HCC. Collagen I-induced DDR1 activation enhanced HCC cell stemness in vitro and in vivo. Mechanistically, DDR1 interacts with CD44, which acts as a co-receptor that amplifies collagen I-induced DDR1 signaling, and collagen I-DDR1 signaling antagonized Hippo signaling by facilitating the recruitment of PP2AA to MST1, leading to exaggerated YAP activation. The combined inhibition of DDR1 and YAP synergistically abrogated HCC cell stemness in vitro and tumorigenesis in vivo. A radiomic model based on T2 weighted images can noninvasively predict collagen I expression. These findings reveal the molecular basis of collagen I-DDR1 signaling inhibiting Hippo signaling and highlight the role of CD44/DDR1/YAP axis in promoting cancer cell stemness, suggesting that DDR1 and YAP may serve as novel prognostic biomarkers and therapeutic targets in HCC.
Subject terms: Cancer stem cells, Protein-protein interaction networks
Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent causes of cancer-related mortality worldwide [1]. High tumor recurrence and metastasis rates are the primary reasons for the high mortality rate associated with HCC [2]. Chronic hepatitis, fibrosis, and cirrhosis increase the risk of HCC. The intratumor of HCC is constantly accompanied by desmoplasia and cirrhosis [3, 4]. Inflammation and extracellular matrix (ECM) deposition are characteristics of cirrhosis and the tumor microenvironment (TME). ECM serves as a physical scaffold that binds cells and tissues together and is pivotally involved in biochemical and biophysical signal transduction. Matrix deposition and remodeling cause tumor stiffness. Increased stiffness activates biomechanical signaling pathways that promote proliferation, invasiveness, and metastasis of cancer cells [5]. Recent studies have demonstrated that the ECM, primarily collagen, contributes to immune exclusion in tumors with collagen-rich stroma [6–8]. Collagens are the primary and essential components of the ECM, an integral element in mechanics, and are hot off the press in cancer microenvironment research [9]. Collagens resist tensile stress because they stiffen as they are stretched in response to store growth-induced anxiety. Intratumor stress significantly decreases after collagen digestion [10].
High matrix stiffness induces and enhances the stemness of HCC cells [11]. Cancer stem cells (CSCs) are a group of cancer cells with stemness properties that possess self-renewal and differentiation capacities, leading to tumor progression, therapy resistance, metastasis, and recurrence [12, 13]. Increased ECM stiffness can be a physical barrier that protects CSCs from chemotherapeutic agents and CD8+ T cell killing [14]. Changes in biomechanical properties, such as intrinsic softness, may be unique markers of CSCs [15].
In addition to external mechanics’ effects, collagen’s maintenance effect on tumor stem cells remains unclear. Collagen regulates cell function through its corresponding receptor discoidin domain receptor 1 (DDR1) [16]. DDR1 belongs to the receptor tyrosine kinase (RTKs) family and is mainly expressed in epithelial cells and mammalian tissues. Its specific ligands are natural collagen I~VI [17]. Our previous study showed that DDR1 enhanced hepatocellular carcinoma metastasis by recruiting PSD4 to ARF6 [18] and promoted HCC proliferation through the mTORC1 signaling pathway [19]. However, the role and precise mechanism of DDR1 in maintaining hepatocellular carcinoma stemness remain unclear.
Numerous upstream signals induce YAP/TAZ activity, including mechanical forces, cell adhesion, cell polarity, tyrosine kinase receptors, and cellular metabolism [20, 21]. Upon Hippo signaling off, nuclear YAP/TAZ are translocated from the cytoplasm, combined with the TEA domain transcription factor family (TEAD 1–4 [22]), directing gene expression programs [23, 24]. Based on the YAP-on and YAP-off classifications, restraining YAP may be an attractive therapeutic strategy for HCC, defined as the YAP-on stage [25]. YAP/TAZ are essential for cancer cells to reprogram into cancer stem cells and induce tumor initiation, progression, and metastasis [26]. However, the mechanism by which Hippo signaling affects HCC stemness has not yet been elucidated.
As one of the upstream receptors, CD44 is a broadly distributed non-kinase transmembrane glycoprotein that belongs to the cell adhesion molecule (CAMs) family and is involved in cell physiological and pathological processes [27, 28]. Recent studies aim at the new function that CD44 acted as a co-receptor that mediated multiple RTKs inducing intracellular signal transduction [29, 30]. As an RTK family member, we speculated that DDR1 might be modified by CD44.
A preoperative and non-invasive method for evaluating collagen content is vital for surgeons because of the role of collagen I in HCC. Radiomics is an emerging imaging technique that can extract high-throughput imaging features from medical images and is frequently applied to predict the biological behavior of various tumors [31, 32]. Using radiomics signatures, we constructed a model for predicting collagen I expression in HCC.
This study showed that DDR1 stimulated by collagen I, inhibited the Hippo pathway in HCC cells to maintain stemness through YAP nuclear translocation. Specifically, collagen I-induced DDR1 stimulation was augmented by the co-receptor CD44. DDR1 recruits PP2AA for MST1/2 dephosphorylation upon collagen I stimulation, activating YAP translocation. Therefore, we identified a direct association between collagen I-DDR1 signaling and Hippo-YAP signaling in promoting HCC cell stemness. Furthermore, our studies indicate that inhibiting DDR1 signaling combined with disrupting YAP function could be a novel treatment method for HCC. A radiomics model based on T2-weighted images (T2WI) can evaluate relative collagen I expression to predict the prognosis and guide clinical treatment.
Results
High collagen I and DDR1 level predicts poor prognosis in HCC patients
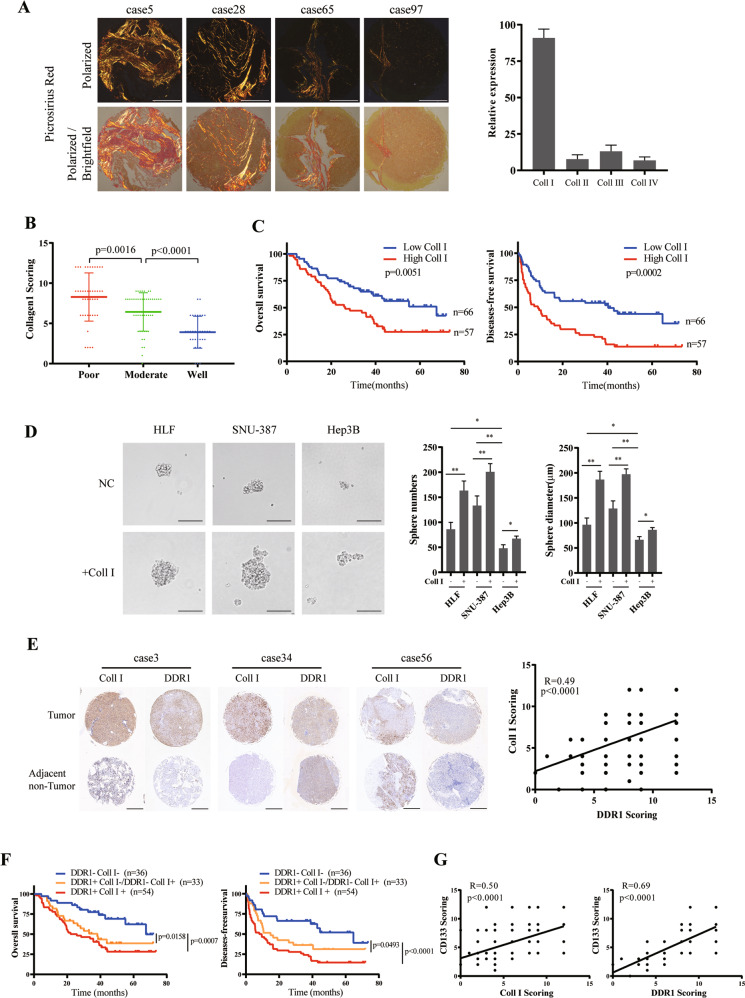
To analyze the types of collagens in the intratumor of HCC, we performed picro-sirius red staining in a tissue microarray, including 123 pair of HCC tissue samples. The results showed that collagen I occupied the highest proportion of type I~IV collagens (Fig. 1A). To explore the clinical significance of collagen I in HCC, we analyzed collagen I expression through IHC staining in the same tissue microarray with corresponding clinicopathological features from Tongji Hospital (Supplementary Table S1). Compared with well-differentiated HCC tissues, collagen I was mainly highly expressed in moderately differentiated HCC tissues and was highest in poorly differentiated tissues (Fig. 1B). Kaplan–Meier analyses demonstrated that patients with high collagen I expression in tumors had a lower overall survival rate and a higher recurrence rate than patients with low collagen I expression (Fig. 1C). Tumor sphere-forming assays showed the ability of collagen I in promoting stemness of various differentiated HCC cell lines (Fig. 1D, Supplementary S1A). HLF and SNU-387 cells represented a higher level of stem cell markers cells and epithelial-mesenchymal transition (EMT) genes among HCC cell lines [33]. Among the collagen I-specific receptors [34], silencing DDR1 significantly reduced the HLF and SNU-387 formed spheres stemness induced by collagen I (Supplementary S1B–D).
Fig. 1. Clinical significance of Collagen I and DDR1 in clinical HCC patients.
A Representative picrosirius red staining for collagen types in specimens of 123 HCC patients (scale bar = 500 μm). B Immunohistochemistry (IHC) staining analyses of collagen I in specimens of HCC patients with tumor differentiation. C Based on IHC staining, cohorts were divided into two groups: Collagen I high (IHC score >6), n = 57; Collagen I low (IHC score ≤6), n = 66. Kaplan–Meier’s analyses of collagen I expression level with the overall survival and disease-free survival rate in HCC patients. D Effects of SNU-387, HLF, or Hep3B with or without collagen I stimulation on sphere-forming capacity, including sphere numbers and diameters (scale bar = 200 μm). E Representative result of IHC staining for collagen I and DDR1 in 123 pair specimens of HCC patients (scale bar = 500 μm) (Left). Correlation of collagen I and DDR1 IHC staining score (Right). F Kaplan–Meier’s analyses of 3 groups of collagen I and DDR1 expression levels with the overall survival and disease-free survival rate in HCC patients. Redline: Collagen I high DDR1 high, n = 54; Orange line: Collagen I low DDR1 high or Collagen I high DDR1 low, n = 33; Blue line: Collagen I low DDR1 low, n = 36. G Correlation of collagen I, DDR1 and CD133 IHC staining score. Pearson’s test was used to calculate the correlation between protein expression levels. The log-rank test was used to calculate the differences between individual groups. Two-tailed unpaired Student’s t-test was performed. Each bar represents the mean ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001.
To examine the clinical significance of DDR1 in HCC, we analyzed the levels of DDR1 in the same tissue microarray and TCGA databases. The results showed that DDR1 positively correlated with collagen I (Fig. 1E, Supplementary S1E). Based on collagen I and DDR1 expression, patients were sorted into three groups: Col1+DDR1+ (n = 54), double-high expression of collagen I/DDR1; Col1+DDR1-or Col1-DDR1+ (n = 33), single-high expression of collagen I or DDR1; Col1-DDR1- (n = 36), double-low expression of collagen I/DDR1. Compared to the other groups, the Col1+DDR1+ group showed the shortest overall survival time and disease-free survival time (Fig. 1F). Also, DDR1 expression was elevated in HCC tissues compared to those in the corresponding adjacent non-tumor tissues. DDR1 was highly expressed in advanced TNM stages, poor tumor differentiation, advanced BCLC stage, and cirrhosis (Supplementary S1F, Supplementary Table S2). Kaplan–Meier analyses demonstrated that patients with high DDR1 expression had a lower overall survival rate and a higher recurrence rate (Supplementary S1G). From the TCGA database, collagen I and DDR1 mRNA expression were positively correlated with CD133 (the marker of HCC stem cells [35]) and consisted of IHC staining results in the tissue microarray (Supplementary S1H, Fig. 1G). In summary, the relationship between collagen I and DDR1 in clinical samples suggested that the synergistic impact of collagen I and DDR1 yielded a poor clinical outcome in HCC patients. They might play an essential role in the occurrence and stemness of HCC.
Collagen I-DDR1 signaling enhances characteristics of HCC stemness
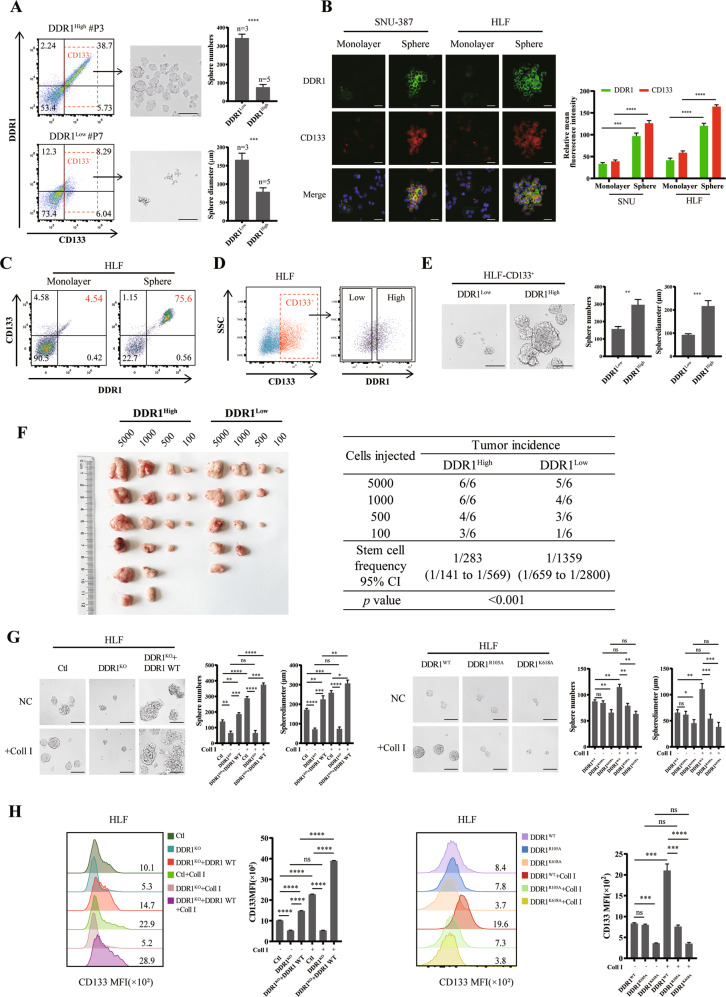
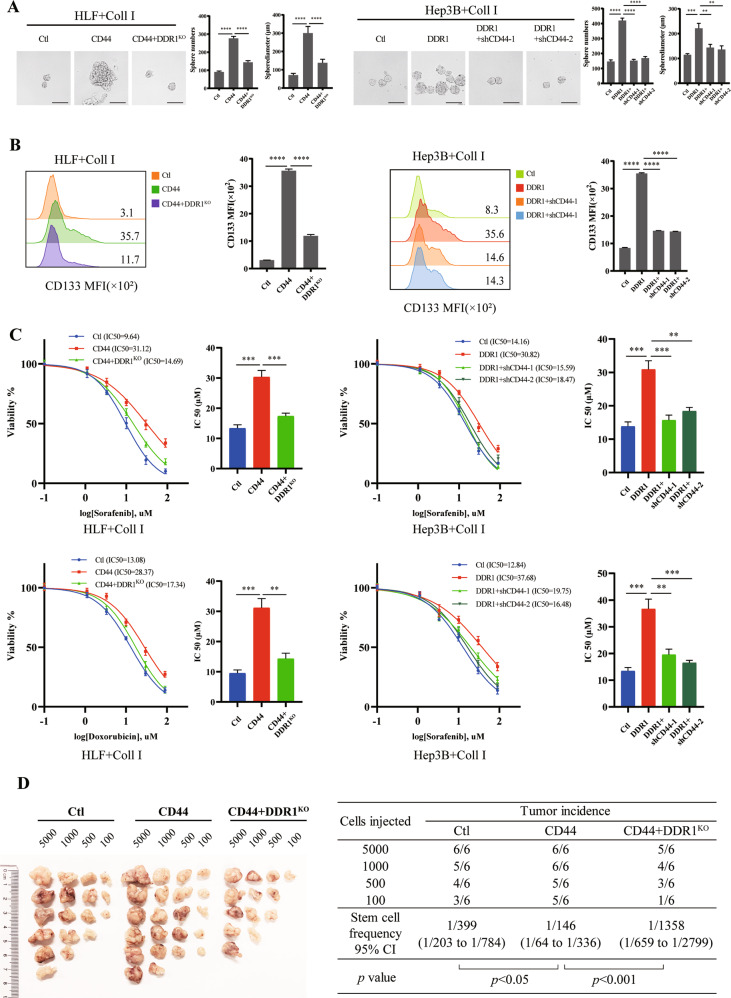
To verify the effect of DDR1 on HCC cell stemness in vitro, we performed sphere-forming assays and flow cytometry cell sorting. We used fluorescence-activated cell sorting (FACS) to sort CD133 high-expressed (CD133+) cells from 8 independent HCC patients. Compared to DDR1 low expressed in CD133+ cells (DDR1+ CD133+ < 30%, n = 5), DDR1 high expressed cells (DDR1+ CD133+ ≥ 30%, n = 3) had increased sphere formation ability (size and number profile) (Fig. 2A, Supplementary S2A). Immunofluorescent and flow cytometry analyses showed that the protein level of DDR1 was strongly upregulated in SNU-387 and HLF spheres (referred to as CSCs) compared to the corresponding adherent cells in monolayers (referred to as non-CSCs) (Fig. 2B–C, Supplementary S2B). We sorted DDR1High and DDR1Low stem cells from CD133+ SNU-387 and HLF (Fig. 2D, Supplementary S2C) and determined their capacities for sphere-forming. CD133+ DDR1High cells formed significantly more abundant and larger spheres than CD133+ DDR1Low cells (Fig. 2E, Supplementary S2D). In vivo, we injected NOD/SCID mice subcutaneously to conduct a limiting dilution assay with the above cells. CD133+ DDR1High cells markedly strengthened the tumor-forming ability and increased stem cell frequency (Fig. 2F).
Fig. 2. Collagen I-DDR1 enhances the stemness of HCC.
A HCC patients derived CD133+ cells were purified by FACS sorting and cultured in sphere forming medium, sphere numbers and diameters was counted (scale bar = 200 μm). B Representative images of immunofluorescence (IF) for DDR1 (green), CD133 (red), and nuclei (blue) in SNU-387 and HLF cells of monolayer or sphere culture (bar = 10 μm). C Flow cytometry analyses of the CD133 and DDR1 fluorescence intensity in HLF of monolayer or sphere culture. D DDR1High and DDR1Low HLF cells were purified form flow-sorted CD133+ cells by FACS sorting. E Representative images of sphere-formation derived from DDR1High and DDR1Low HLF cells. Effects of sphere-forming capacity including sphere numbers and diameters (scale bar = 200 μm). F Extreme limiting dilution analysis of DDR1High and DDR1Low cells of HLF. Cells were serially diluted and then subcutaneously injected into 4-week-old male NOD/SCID mice (n = 12/group) (Left). CSCs frequency was calculated using ELDA software(Right). G Representative images of sphere-formation derived from DDR1 knock-out or DDR1 mutation of HLF cells. Effects of sphere-forming capacity including sphere numbers and diameters (scale bar = 200 μm). H Flow cytometry analyses of the CD133 mean fluorescence intensity in DDR1 knock-out or DDR1 mutation HLF cells. Pearson’s test was used to calculate the correlation between protein expression levels. Two-tailed unpaired Student’s t-test was performed. Each bar represents the mean ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001.
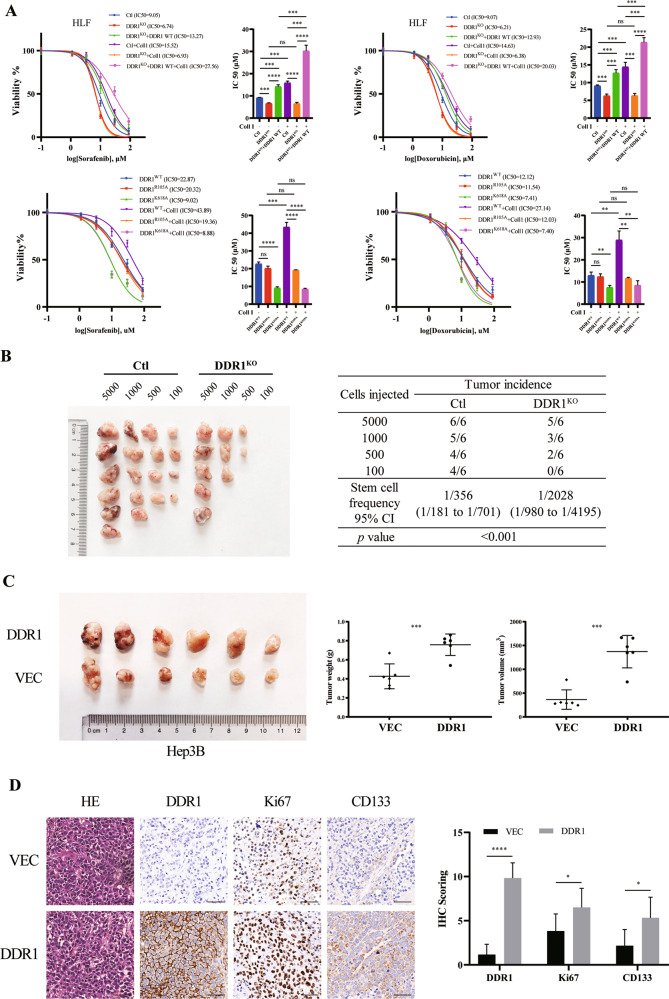
To further examine the role of DDR1 in tumor stemness, we established DDR1 knock-out and DDR1 mutation HLF cell lines using CRISPR/Cas9 gene editing technology, respectively (Supplementary S2E–F). We screened CD133+ stem cells for subsequent experiments of sphere formation, colony formation, and drug resistance assays. As crucial transcription factors and membrane proteins in maintaining the self-renewal potential of stem cells [36], the mRNA expression of SOX2, NANOG and EPCAM were decreased in DDR1 knock-out (DDR1KO) cells, and this effect could be reversed within wild type DDR1 rescued (DDR1KO + DDR1 WT) or collagen I stimulation. The collagen-binding-defective mutation (DDR1R105A [37]) and DDR1 kinase-dead mutation (DDR1K618A [37]) did not up-regulate SOX2, NANOG, and EPCAM mRNA expression by collagen I (Supplementary S2G). Furthermore, DDR1 knock-out reduced tumor-sphere formation efficiency, the proportion of CD133 positive cells, drug resistance to sorafenib or doxorubicin, and single-cell colony formation, and these effects were reversed by DDR1 restored. In contrast, DDR1R105A and DDR1K618A mutants failed to improve the stemness effects by collagen I stimulation (Fig. 2G–H, Fig. 3A, Supplementary S3A). Besides, DDR1 knock-down reduced these CSC-related biological effects in SNU-387 (Supplementary S3B–F). In vivo, we injected NOD/SCID mice with CD133+ DDR1KO and control cells to conduct a limiting dilution assay. DDR1 knock-out markedly weakened the tumor-forming ability and decreased stem cell frequency (Fig. 3B).
Fig. 3. Collagen I-DDR1 enhances the stemness of HCC.
A DDR1 knock-out or DDR1 mutation HLF cells were treated with sorafenib or doxorubicin. Cell viability was determined compared with the untreated cells, and IC50 values were calculated. B Extreme limiting dilution analysis of DDR1KO and control cells of HLF. Cells were serially diluted and then subcutaneously injected into 4-week-old male NOD/SCID mice (n = 12/group) (Left). CSCs frequency was calculated using ELDA software(Right). C 4-week-old male BALB/c nude mice were subcutaneously injected by DDR1 overexpression and control cells of Hep3B (n = 6/group) (Left). Tumor weight and volume were shown (Right). D Representative images of HE and IHC staining of DDR1, Ki67, and CD133 from BALB/c nude mice xenograft (scale bar = 100 μm). Two-tailed unpaired Student’s t-test was performed. Each bar represents the mean ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001.
We used BALB/c nude mice xenograft model to examine the role of DDR1 in HCC development. The subcutaneous tumors formed by DDR1 overexpression cells were larger and heavier than those formed by control cells (Fig. 3C). IHC results showed that the protein levels of DDR1, Ki67, and CD133 were higher in DDR1 overexpression tumors (Fig. 3D). Together, the aforementioned research confirmed that DDR1 promotes cell stemness and tumorigenesis in HCC in vitro and in vivo, depending on its kinase activity and collagen I stimulation.
Collagen I -induced DDR1 activation is synergistically promoted by CD44
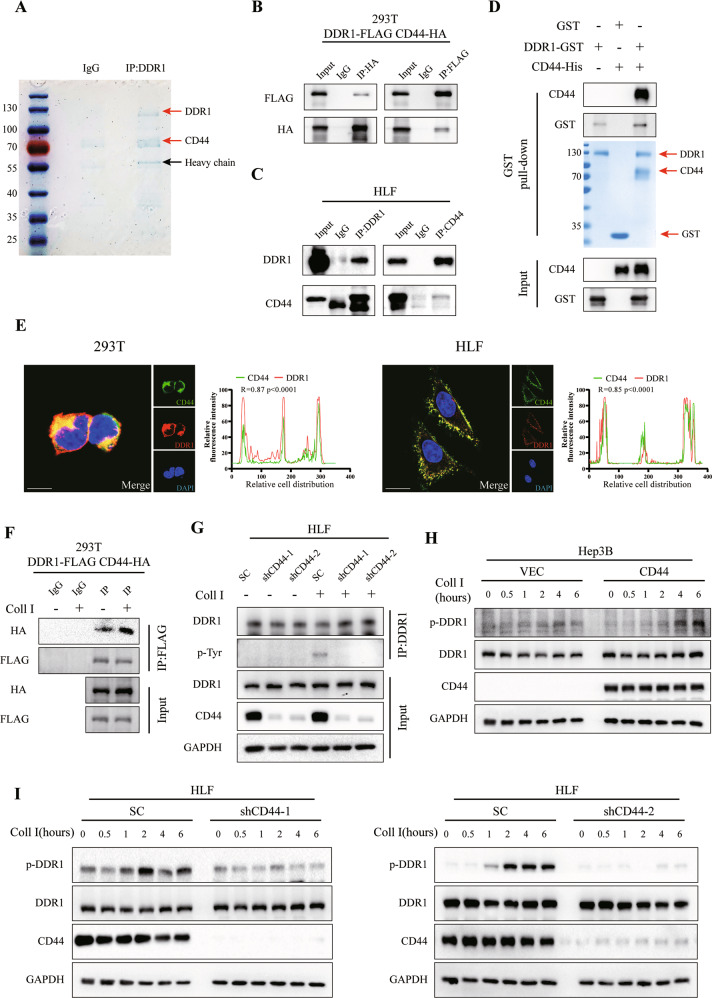
To investigate the molecular mechanism of DDR1 in hepatocellular carcinoma, we previously studied its interacting partners using a combined IP/MS approach [18]. Cells were transfected with FLAG-DDR1 and immunoprecipitated using an anti-FLAG antibody. We found that CD44 (a CSCs marker) was one of the potential interacting partners of DDR1 (Fig. 4A). Co-immunoprecipitation (Co-IP) and glutathione S-transferase (GST) pull-down assays were performed. The results from exogenous (HEK-293T) and endogenous (HLF) assays showed that DDR1 interacted and was combined with CD44 (Fig. 4B–D). In addition, immunofluorescence and laser confocal scanning detected the spatial co-localization of exogenous DDR1-FLAG and CD44-HA in HEK-293T cells. A similar finding was observed for endogenous DDR1 and CD44 in wild-type HLF cells (Fig. 4E). In addition to the mRNA and protein levels. The regulation was not observed between CD44 and DDR1 in q-PCR and western blot assays (Supplementary S4A–D). Next, we performed a co-IP assay with or without collagen I activation. We found that collagen I could increase the interaction between DDR1 and CD44 (Fig. 4F). CD44 can act as a co-receptor to mediate receptor internalization, activation, degradation, or regulation of receptor kinase activity [38–40], and we speculated whether CD44 could regulate collagen I-induced DDR1 activation. Therefore, co-IP assays indicated that CD44 knock-down decreased tyrosine phosphorylation of DDR1 (Fig. 4G). Overexpression of CD44 in Hep3B promoted the phosphorylation of DDR1 induced by collagen I treatment in a time-dependent manner (Fig. 4H). Knock-down of CD44 reduced this effect in the HLF cells (Fig. 4I). Our results suggested that CD44 promotes the collagen I-induced activation of DDR1 in HCC cells.
Fig. 4. CD44 interacts with DDR1 and facilitates DDR1 phosphorylation.
A HLF cell lysates were immunoprecipitated with control rabbit IgG or anti-DDR1, ten percent of the cell extracts were loaded as an input, and western blot and coomassie blue staining were performed. B Co-immunoprecipitation (Co-IP) and western blot assays for exogenously proteins DDR1-FLAG and CD44-HA were performed in HEK-293T cells. Cross-validation by immunoprecipitation with anti-HA, immunoblotting with anti- FLAG, immunoprecipitation with anti-FLAG, and immunoblotting with anti-HA. C Co-IP and immunoblotting assays for endogenously proteins DDR1 and CD44 were performed in HLF cells. Cross-validation by immunoprecipitation with anti-CD44, immunoblotting with anti-DDR1, immunoprecipitation with anti-DDR1, and immunoblotting with anti-CD44. D GST pull-down assay showing direct interaction between purified recombinant-DDR1 and CD44. E Representative confocal images of immunofluorescence (IF) for CD44 (green), DDR1 (red), and nuclei (blue) in HEK-293T and HLF cells (bar = 10 μm). F DDR1-FLAG and CD44-HA were co-transfected into HEK-293T cells and stimulated with or without collagen I. The protein levels were analyzed by immunoprecipitation and western blot. G HLF-shCD44 cells were stimulated with or without collagen I, followed by immunoprecipitation with anti-DDR1 antibody and immunoblotting with anti- phosphorylated tyrosine. H Hep3B-CD44 cells were stimulated with or without collagen I for the indicated time periods. Phospho-DDR1 levels were measured using western blot. I HLF-shCD44 cells were stimulated with or without collagen I for the indicated time periods. Phospho-DDR1 levels were measured using western blot. Pearson’s test was used to calculate the correlation between protein expression levels. Two-tailed unpaired Student’s t-test was performed. Each bar represents the mean ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001.
To investigate whether the function of CD44 depends on DDR1 in HCC, we overexpressed CD44 in DDR1KO HLF cells and knocked down CD44 in DDR1 overexpressed Hep3B cells. DDR1 depletion partially diminished the sphere formation efficiency of HCC cells, CD133 positive stem-like-cell proportion, chemoresistance, and single-cell colony formation driven by CD44 overexpression. Overexpression of DDR1 partially enhanced CSCs characteristics and was reduced by CD44 down-regulation (Fig. 5A–C, Supplementary S4E–F). We performed in vivo limiting dilution tumorigenicity assays. The tumorigenic cells of CD44 overexpression were significantly more abundant than the control group, and this effect could be impaired by DDR1 knocked out (Fig. 5D). In summary, the above evidence suggests that DDR1 is a crucial protein required for CD44-mediated HCC cell stemness signaling.
Fig. 5. CD44 cross-talking with DDR1 enhances HCC stemness in vitro and vivo.
A Effects of DDR1 knock-out with CD44 overexpression or overexpressed DDR1 with CD44 knock-down HCC cells on sphere-forming capacity, including sphere numbers and diameters (scale bar = 200 μm). B Flow cytometry analyses of the CD133 mean fluorescence intensity in DDR1 knock-out with CD44 overexpression or overexpressed DDR1 with CD44 knock-down HCC cells. C DDR1 knock-out with CD44 overexpression or overexpressed DDR1 with CD44 knock-down HCC cells were treated with sorafenib or doxorubicin. Cell viability was determined compared with the untreated cells, and IC50 values were calculated. D Extreme limiting dilution analysis of CD44/DDR1KO and control cells of HLF. Cells were serially diluted and then subcutaneously injected into 4-week-old male NOD/SCID mice (n = 12/group) (Left). CSCs frequency was calculated using ELDA software (Right). Two-tailed unpaired Student’s t-test was performed. Each bar represents the mean ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001.
DDR1 inactivates Hippo signaling by facilitating the recruitment of PP2AA to MST1
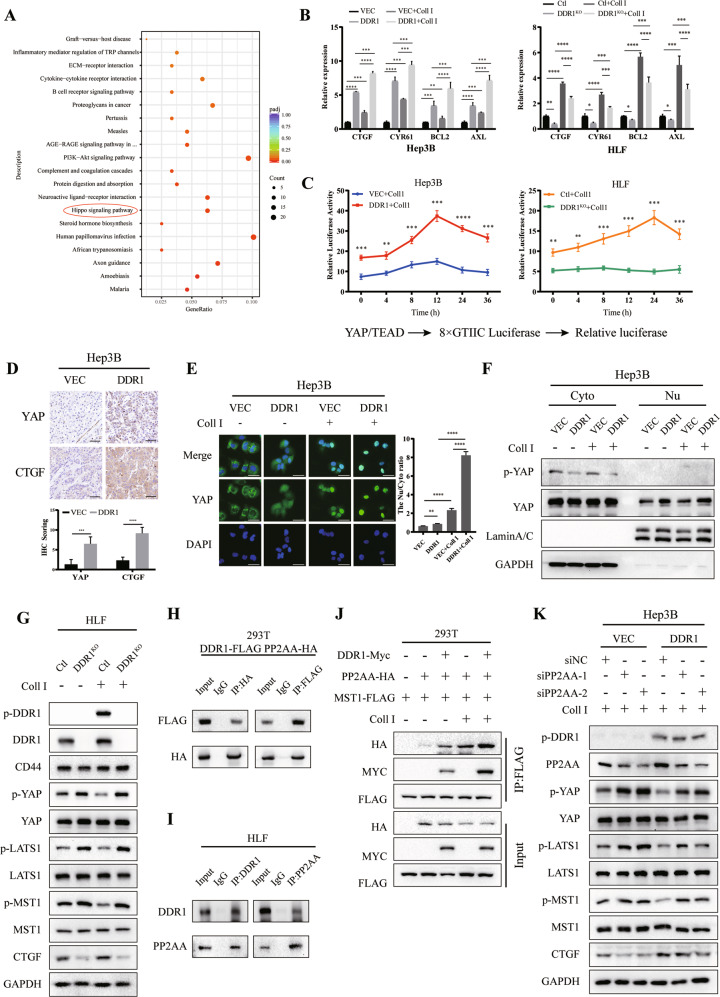
To identify the pathways responsible for the influence of DDR1 on HCC stemness, we performed RNA sequencing of HLF shDDR1 cells and control cells. KEGG pathway enrichment analyses revealed that PI3K-Akt signaling and Hippo signaling were altered by DDR1 loss (Fig. 6A, Supplementary Table S3). The western blot results excluded PI3K-Akt signaling that total PI3K, Akt, PTEN, and their phosphorylated forms were not changed by DDR1 depletion or collagen I treatment (Supplementary S5A). However, based on TCGA database, DDR1 was positively associated with Hippo signaling downstream proteins YAP, CTGF, BCL2, and AXL (Supplementary S5C). Treatment with collagen I inhibited YAP phosphorylation and promoted CTGF expression in the meanwhile that treated with 7rh or verteporfin could reverse the levels of phosphorylated YAP and CTGF in HLF and SNU-387 (Supplementary S5B). It was suggested that collagen I might facilitate YAP downstream genes expression in a DDR1 and YAP-dependent manner.
Fig. 6. DDR1 impedes Hippo signaling through inactivating YAP.
A KEGG pathway enrichment analysis indicates that the Hippo signaling pathway was significantly affected by HLF-shDDR1 cells treated with collagen I. B mRNA expression levels of Hippo pathway downstream-associated genes in DDR1 overexpression or knock-out HCC cell lines. C Luciferase reporter assays in DDR1 overexpression or knock-out HCC cells transfected with YAP/TAZ-responsive synthetic promoter-reporter plasmids and collagen I treatment. D Representative images of IHC staining of YAP and CTGF from BALB/c nude mice xenograft (scale bar = 100 μm). E Representative images of immunofluorescence assays display nucleus/cytoplasm localization of YAP in DDR1 overexpressed Hep3B cells with collagen I (scale bar = 30 μm). F Nucleus/cytoplasm fractionation and western blot analysis of DDR1 overexpressed cells to show YAP nuclear translocation. GAPDH and Lamin A/C were used as cytoplasmic and nuclear markers, respectively. G Effects of protein expression or phosphorylation of Hippo signaling components in DDR1 knock-out HLF cells with collagen I. H Co-IP and western blot assays for exogenously proteins DDR1-FLAG and PP2AA-HA were performed in HEK-293T cells. Cross-validation by immunoprecipitation with anti-HA, immunoblotting with anti- FLAG, immunoprecipitation with anti-FLAG, and immunoblotting with anti-HA. I Co-IP and western blot assays for endogenously proteins DDR1 and PP2AA were performed in HLF cells. Cross-validation by immunoprecipitation with anti-CD44, immunoblotting with anti-DDR1, immunoprecipitation with anti-DDR1, and immunoblotting with anti- PP2AA. J DDR1-Myc, PP2AA-HA, and MST1- FLAG were simultaneously transfected in HEK-293T cells with or without collagen I treatment, and evaluate the collagen I contributed to the indicated protein interaction and stability by western blot. K DDR1 overexpressed cells transfected with siPP2AA or control siRNA in the presence of collagen I treatment. Western blot analyzed the indicated protein from cell lysate. Two-tailed unpaired Student’s t-test was performed. Each bar represents the mean ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001.
Since Hippo signaling is closely associated with tumor stemness, we examined the effects of DDR1 on gene expression downstream of the Hippo signaling pathway. The results showed that the mRNA levels of CTGF, CYR61, BCL2, and AXL were upregulated in the DDR1 overexpression Hep3B, the presence of collagen I enhanced these effects. In contrast, DDR1 knocked out significantly reduced the expression of CTGF, CYR61, BCL2, and AXL (Fig. 6B). The dual-luciferase reporter system suggested that activated DDR1 enhanced the YAP/TEAD-responsive element activity within collagen I (Fig. 6C). IHC staining for BALB/c nude mice xenograft showed that YAP and CTGF were upregulated in DDR1 overexpression xenograft (Fig. 3B, Fig. 6D). Similarly, YAP and CTGF was upregulated in CD44 overexpression xenograft (Fig. 5D, Supplementary S5D). In vitro, immunofluorescence assay, and nucleic-cytoplasmic fractionation indicated that DDR1 overexpression and collagen I promoted the translocation of YAP into the cell nucleus (Fig. 6E–F). Western blot showed that collagen I-stimulated DDR1 phosphorylation reduced MST1 phosphorylation and enhanced YAP activation, whereas DDR1 knock-out showed the opposite effect (Fig. 6G). These results indicated that collagen I-DDR1 signaling could induce YAP activation.
Protein phosphatase two scaffold subunit A alpha (PP2AA; PPP2R1A) as a canonical upstream phosphatase could dephosphorylate MST1/2 and inactivate Hippo signaling [41]. We analyzed DDR1 interacting partners [18]. PP2AA is a potential interacting protein with DDR1 validated by exogenous (HEK-293T) and endogenous (HLF) co-IP assays and glutathione S-transferase (GST) pull-down assay (Fig. 6H–I, Supplementary S5E). Recruiting assays demonstrated that DDR1 promoted the recruitment of PP2AA to MST1 in a collagen-dependent manner (Fig. 6J). PP2AA depletion attenuated collagen I-DDR1 signaling-induced MST1 and LAST1 dephosphorylation, leading to YAP activation (Fig. 6K). These results indicated that PP2AA is necessary for collagen I-DDR1 signaling-induced YAP activation.
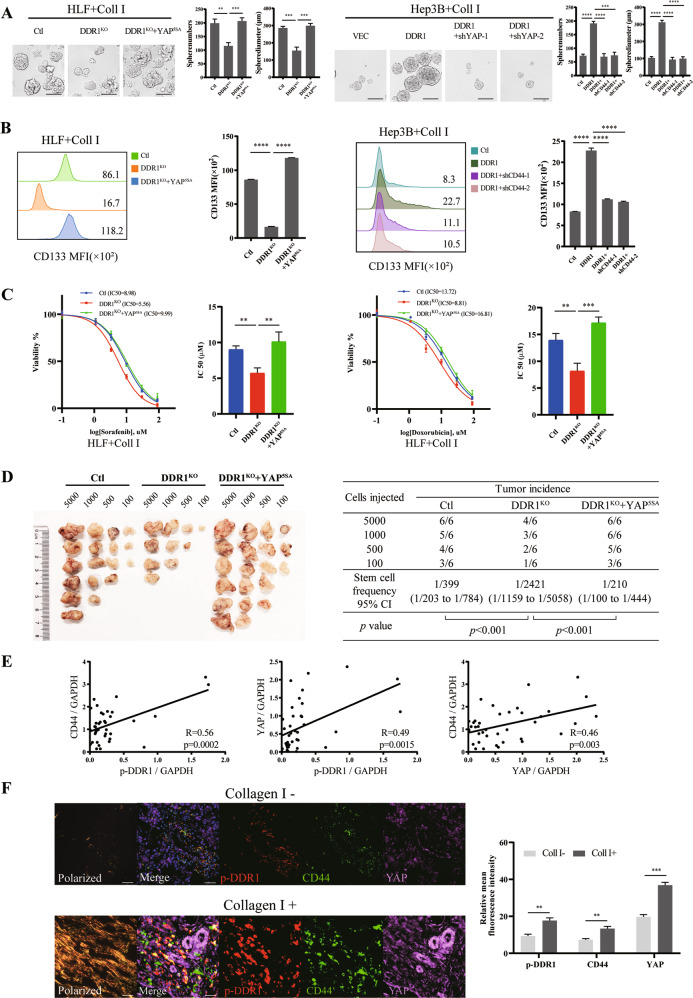
Determine the function of YAP in HCC depending on DDR1. We overexpressed YAP5SA (activated YAP) in DDR1KO HLF cells and knocked down YAP in DDR1 overexpressed Hep3B cells (Supplementary S6A). Knocking out DDR1 diminished the HCC cells in sphere formation efficiency, CD133 positive stem-like-cell proportion, chemoresistance, and single-cell colony formation, and CSCs characteristics could be rescued by YAP5SA overexpression. DDR1 overexpression enhanced CSCs characteristics and was reduced by YAP down-regulation (Fig. 7A–C, Supplementary S6B–C). In vivo limiting dilution tumorigenicity assays showed the tumorigenic cells of DDR1KO were significantly increased by YAP5SA amplification (Fig. 7D). The mRNA levels analyses from the tumors showed that CTGF, CYR61, BCL2, and AXL were downregulated in the DDR1KO cells formed tumor, and the supplement of YAP5SA promoted these mRNA expressions (Supplementary S6D). Western blot from the tumors showed that DDR1 depletion upregulated p-YAP and decreased total YAP and CTGF, and could be reversed by YAP5SA overexpression (Supplementary S6E). IHC results confirmed the protein levels of YAP and CTGF further (Supplementary S6F). In summary, the above evidence suggests that YAP is a crucial protein required for DDR1-mediated HCC cell stemness signaling.
Fig. 7. YAP cross-talking with DDR1 enhances HCC stemness in vitro and vivo.
A Effects of DDR1 knock-out with YAP5SA overexpression or overexpressed DDR1 with YAP knock-down HCC cells on sphere-forming capacity, including sphere numbers and diameters (scale bar = 200 μm). B Flow cytometry analyses of the CD133 mean fluorescence intensity in DDR1 knock-out with YAP5SA overexpression or overexpressed DDR1 with YAP knock-down HCC cells. C DDR1 knock-out with YAP5SA overexpression HLF cells were treated with sorafenib or doxorubicin. Cell viability was determined compared with the untreated cells, and IC50 values were calculated. D Extreme limiting dilution analysis of YAP5SA/DDR1KO and control cells of HLF. Cells were serially diluted and then subcutaneously injected into 4-week-old male NOD/SCID mice (n = 12/group) (Left). CSCs frequency was calculated using ELDA software (Right). E Statistical analyses showed that p-DDR1, CD44, and YAP were pairwise positively upregulated in HCC tissues from western blot. F Representative images of picrosirius red staining polarized field and multiplex fluorescent immunohistochemical staining images of p-DDR1, CD44, and YAP in collagen I poor/abundant HCC sample, respectively (scale bar = 100 μm). Pearson’s test was used to calculate the correlation between protein expression levels. Two-tailed unpaired Student’s t-test was performed. Each bar represents the mean ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001.
In clinical samples, we evaluated p-DDR1, CD44, and YAP protein levels in 39 tissue samples from patients with HCC undergoing surgery at our hospital. The results showed that p-DDR1, CD44, and YAP levels were positively pairwise correlated in HCC tissues (Supplementary S7A, Fig. 7E). Based on the Sirius red staining, the samples were separated into collagen I-positive (collagen I+) and collagen I-negative (collagen I−) expression groups. We then performed multicolor immunofluorescence (IF) analyses to evaluate the expression of p-DDR1, CD44, and YAP (Supplementary S7B, Fig. 7F). As abundant collagen I deposition in HCC, p-DDR1, CD44, and YAP levels were more intense expressed than collagen I negative patients. It was confirmed the positive correlations of p-DDR1, CD44 and YAP in HCC patients.
Combined inhibition of DDR1 and YAP induces synergistic effects in HCC cell stemness
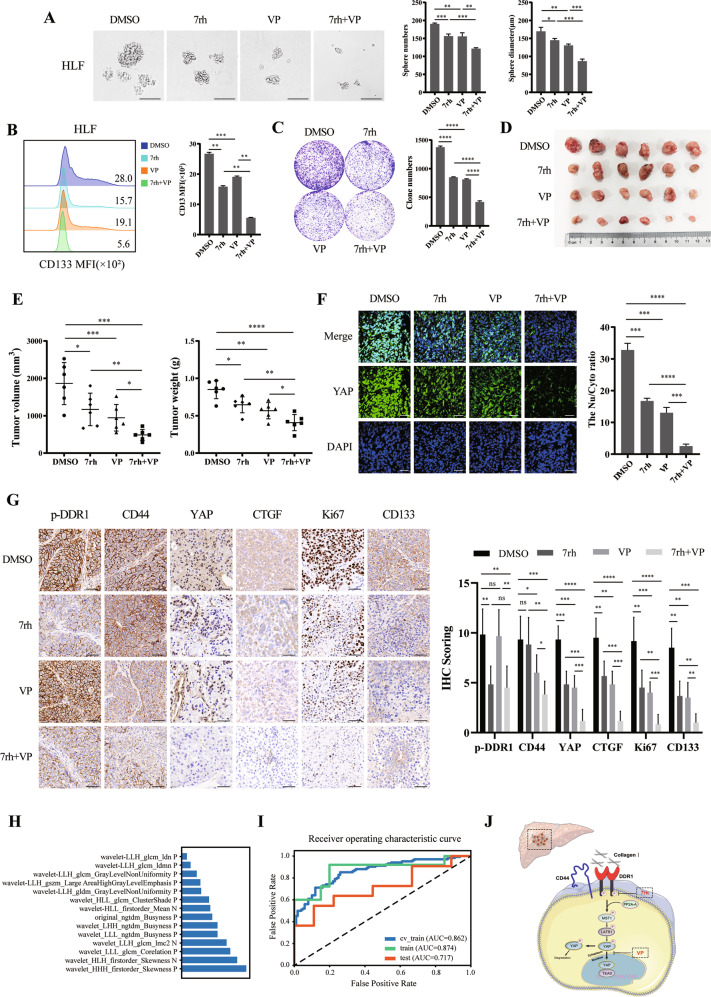
To further verify whether DDR1 exerted its effects via Hippo/YAP signaling, we used in vitro and in vivo treatments with the DDR1 inhibitor (7rh) [42] and the YAP inhibitor (verteporfin; VP) [43]. 7rh is an ATP-competitive oral DDR1-specific small-molecule inhibitor [44]. Verteporfin (VP), an inhibitor of YAP/TAZ, was identified in a small library of FDA-approved compounds and has been reported to sterically hinder the interaction between YAP and TEAD [45]. Treatment with 7rh or VP alone reduced the number of cells with sphere formation efficiency, CD133+ cell proportion, and colony formation and reduced the volume and weight of xenografts, respectively. Combination treatment with 7rh and VP further attenuated these effects compared to single agents alone (Fig. 8A–E).
Fig. 8. Co-inhibition of DDR1 and Hippo/YAP signaling impedes tumor proliferation and radiomics model of prediction on Collagen I expression.
A Effects of treatment with 7rh, VP, or combination therapy in HLF cells on sphere-forming capacity, sphere numbers, and diameters were counted. (Scale bar = 200 μm). B Flow cytometry analyses of the CD133 mean fluorescence intensity in 7rh, VP, or combination therapy treatment in HLF cells. C Colony formation assay of the 7rh, VP, or combination therapy treatment in HLF cell lines. D 4 -week-old male BALB/c nude mice were subcutaneously injected by sphere-formed HLF cells, divided into DMSO, 7rh, VP, and combination groups randomly for 2 weeks of treatment (n = 6/group). E Tumor weight and volume were analyzed from BALB/c nude mice with DMSO, 7rh or VP treatment. F Representative images of immunofluorescence assays display nucleus/cytoplasm localization of YAP in tumor from BALB/c nude mice xenograft (scale bar = 100 μm). G Representative images of IHC staining of p-DDR1, CD44, YAP, CTGF, Ki67, and CD133 form BALB/c nude mice xenograft (scale bar = 100 μm). H The important 14 features of the SVM classifier model in diagnosing HCC collagen I expression. I The ROC curves of models in train and test sets, respectively. J The schematic diagram shows that collagen I/DDR1 enhances HCC stemness through Hippo signaling. Two-tailed unpaired Student’s t-test was performed. Each bar represents the mean ± SD. *p < 0.05, **p < 0.01 and ***p < 0.001.
The mRNA levels analyses from the BALB/c nude mice tumors showed that CTGF, CYR61, BCL2, and AXL were downregulated in 7rh or VP monotherapy group, and the combined therapy decreased these mRNA levels further (Supplementary S7C). Western blot analyses showed that 7rh or VP increased p-YAP level and reduced total YAP and CTGF, 7rh with VP treatment enhanced further (Supplementary S7D). Tumor immunofluorescence assay yielded that 7rh or VP monotherapy inhibited the translocation of YAP into the cell nucleus, combination treatment further reduced nucleic YAP expression (Fig. 8F). IHC results validated that YAP, CTGF, Ki67, and CD133 staining was more intense in control tumors than in 7rh or VP treatment tumors. The combination group yielded the lowest YAP, CTGF, Ki67, and CD133 expression in tumors (Fig. 8G). The combined treatment showed a favorable effect on anti-tumor therapy.
Clinical-radiomics predictive model
Among the patients in the tissue microarray, we acquired 92 T2WI images, and a total of 65 patients with complete imaging data were finally included in this retrospective study. The prediction model depends on the collagen I IHC score. Patients were divided into collagen I low (IHC score ≤6) and high (IHC score >6) groups the same as before. Model performance was evaluated using receiver operating characteristic (ROC) curve analysis. The results showed that the model based on 14 features obtained the highest AUC for the validation data set (Fig. 8H). At this point. The AUC and prediction accuracy of the model were 0.717 and 0.700, respectively, for the testing data-set. The clinical statistics for diagnosis and selected features are shown (Fig. 8I).
Discussion
Dysregulation of the matricellular components of the tumor microenvironment has been linked to the development of metastases in multiple cancer types. The ECM is a non-cellular three-dimensional macromolecular network composed of collagens, elastin, laminins, fibronectin, proteoglycans, and other glycoproteins [46]. The exact mechanism of ECM in tumors has not yet been elucidated. In the present study, we aimed to identify the crucial components—collagen and its receptor DDR1. We found that the phosphorylation of DDR1 by collagen I contributes to maintaining HCC cell stemness. In an attempt to identify collagen I-DDR1 crucial for regulating CSCs, we found collagen I-DDR1 signaling antagonized Hippo signaling with the help of CD44 amplification. The combination of DDR1 and YAP inhibition is a logical target for anti-cancer stem cell-directed therapies. A radiomic predictive model for collagen I can distinguish between chemotherapeutic efficacy and predicted prognosis. In recurrent HCC, mRNA and protein levels of DDR1 are higher than those in non-recurrent HCC [47]. High DDR1 expression is associated with advanced tumor stages [48]. Similar to the observations in our study, patients with high DDR1 expression had worse OS and DFS rates with accompanying collagen I deposition.
As a well-accepted theoretical model, cancer stem cells give rise to chemoresistance, metastasis, and tumor recurrence. The CSCs model has been verified in various solid tumors, including HCC [49]. Extracellular matrix (ECM) rigidity segregates CSCs from chemotherapeutic agents as a physical defender and plays a critical role in tumor progression, especially in cancer cell stemness [14]. ECM stiffness contributes to NANOG activation and cancer stemness [50].
Extensive studies have demonstrated that ECM and collagens can cross-talk with cancer cells, including cancer stem cells, to promote tumor progression [51, 52]. Rich collagens in airway smooth muscle cells contribute to tumor cell colonization in the lung and participate in cancer stemness, progression, and treatment [4, 53–55]. The above studies revealed that collagens could act as ligands that cross-talk with collagen receptors in the surrounding tumor cells [56] and that DDR1-dependent collagen remodeling contributes to immune exclusion in breast cancer [8]. This evidence suggests that CSCs tend to be enriched in stiffer microenvironments, which warrants further in-depth studies. Our study demonstrated that through the corresponding receptor - DDR1, exogenous collagen I enhanced HCC cell stemness. Higher collagen I expression was positively correlated with DDR1 expression in clinical samples with poor prognosis. We demonstrated that collagen I-DDR1 signaling contributes to maintaining HCC stemness and progression.
DDR1, an RTK, is upregulated in multiple cancers [57, 58]. We demonstrated that DDR1 mediates regulatory mechanisms involved in maintaining HCC cell stemness. Functionally, DDR1 is dependent on collagen I stimulation and its kinase activity. It interacts with CD44, which amplifies collagen I-induced DDR1 phosphorylation signaling, CD44/DDR1/YAP signaling axis acts as an inducer of the stemness of HCC cells by DDR1 activation. The results have not determined other components of the ECM that enhance collagen I-induced DDR1 activation, and the role of different types of collagens in HCC stemness deserves further study.
CD44 promotes migration, invasion, and tumor progression by interacting with various proteins. Nevertheless, the regulatory relationship between CD44 and Hippo signaling is not directly precise [59, 60]. Rho A, merlin, and PI3K/Akt have been implicated in CD44-Hippo signaling [61–63]. A previous study demonstrated that CD44 acts as a ligand-binding membrane protein and co-receptor that mediates ligand-receptor-induced intracellular signal transduction [38]. In this study, CD44 was found to function as a co-receptor of DDR1 by negotiating and magnifying collagen I-DDR1 phosphorylation. Our research revealed a new interaction and was complementary to the analyses of the relationship between CD44 and the Hippo signaling pathway.
Hippo signaling is an evolutionarily conserved pathway consisting of a network of signals to modulate tissue growth and organ size, and dysregulation of Hippo signaling contributes to human cancer progression [43, 64, 65]. A recent study indicated that DDR1 could sense the increasing matrix stiffness through control YAP/TAZ activity [66]. Our results confirmed that collagen I - DDR1 signaling maintained HCC cell stemness by inhibiting Hippo signaling. STRIPAK integrates upstream signals to regulate the activities of MST1/2 and MAP4Ks, leading to the initiation of Hippo signaling [41]. We found that DDR1 is an adaptor protein that recruits PP2AA to MST1. Our study is the first to discover that collagen I-DDR1 signaling to suppress Hippo signaling in maintaining HCC stemness.
Moreover, the DDR1 inhibitor–7rh unveiled a significant treatment effect on pancreatic ductal adenocarcinoma and KRAS-mutant lung adenocarcinoma [67–69]. Originally, verteporfin was in clinical use as a photosensitizer in photocoagulation therapy for macular degeneration. It shows potential anti-cancer and anti-antifibrotic treatment in clinical trials and basic experiments [70–73]. In our research, the combination of 7rh and VP therapy had synergistic effects on reducing HCC cell stemness. In addition, VP decreased CD44 levels, and this positive feedback loop may be inactivated by disrupting DDR1 phosphorylation. These findings provide a theoretical basis and present novel therapeutic targets for HCC progression.
Since collagen I content estimates matter in the prognosis of inhibitor treatment, we can rely on radiomics to provide a non-invasive evaluation method for collagen I expression and needle biopsy. Radiomics applied within clinical-decision support systems to improve diagnostic, prognostic, and predictive accuracy are gaining importance in cancer research [74]. In this retrospective study, we constructed and validated T2WI radiomic signatures for the preoperative prediction of collagen I expression status in HCC. The radiomic model could predict collagen I expression and confirm its predictive value, which could be used to stratify patients into groups based on collagen I level to guide treatment. This approach might allow clinicians to select more personalized and effective treatment strategies, as it demands more imaging samples and practice in the future.
In summary, our results illustrate that collagen I-DDR1 signaling magnified by CD44 acts as a tumor motivator to promote HCC cell stemness by recruiting PP2AA to activate YAP. In HCC tissues, increased collagen I and DDR1 expression predicts poor prognosis and thus can serve as promising biomarkers and novel therapeutic strategies for HCC.
Methods and material
Cell culture
SNU-387, Hep3B, HLF, MHCC97H, and HEK-293T cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Gibco, New York, USA), 100 U/ml penicillin and 100 mg/ml streptomycin at 37 °C with 5% CO2. All the HCC cell lines were obtained from the Hepatic Surgery Center, Tongji Hospital, Huazhong University of Science and Technology, China. HEK-293T cells were purchased from the China Center for Type Culture Collection (Wuhan, China).
Primary liver cancer cells isolation
Tumor tissue derived from HCC patients was carefully dissected to remove necrotic tissue, connective tissue, and blood vessels. After tumor tissue was cut into 1 mm3 pieces, about 5 ml collagenase type IV solution was directly added to the tumor for digestion at 37 °C for 10 ~ 20 min. After blowing up-down several times using the pipette, the collagenase solution mix was passed through the sterile 70 μm nylon filters to collect single-cell solution. Cells were centrifuged and washed using a culture medium three times. Freshly isolated tumor cells were cultured in DMEM/F12 medium supplemented with B27 supplement minus vitamin A, 20 ng/mL human recombinant EGF (hEGF), 20 ng/mL bFGF, and 1% ITS supplement. The medium was changed every five days. All procedures were approved by the Ethics Committee of Tongji Hospital and conducted according to the Declaration of Helsinki Principles. Written and informed consent was obtained from each patient.
CRISPR-Cas9 assay, small interfering RNA, plasmids, and lentivirus
To generate DDR1 knock-out HCC cell lines. A pair of sgRNAs were cloned into the empty backbone of lenti-CRISPR v2. The following sgRNA sequences are listed in Supplementary Table S4. Lenti-CRISPR v2 plasmid containing the sgDDR1-1 and sgDDR1-2 sequences were transfected into HEK293T cells with the psPAX2(plasmid #12260) packaging plasmid and pMD2.G(plasmid #12259) envelope-expressing plasmid. The virus was harvested twice at 48 h and 72 h post-transfection and was concentrated using PEG8000. HLF was infected with the concentrated virus and 0.8 μg/ml polybrene, then selected with 5 μg/ml puromycin after 48 h virus transfection for three days. Finally, all cells were diluted into single cells and seeded into 96-well plates. Genomic DNA was extracted from each positive clone, followed by sequencing the PCR products spanning the knock-out sites. Genotyping was performed by sequencing PCR products amplified from the following primers: DDR1 forward: 5′-CACTCCTAGCCTTGACCCTGT-3′; DDR1 reverse: 5′- AGAGATGAAGGAGATTTCGCTDDRG-3′. Transient small interfering RNA (siRNA) assays were described before [75]. Sequences of siRNA are listed in Supplementary Table S4. Briefly, synthesizing siRNA duplexes were produced and validated by Ribobio (Guangzhou, China). Two target siRNA sequences were applied for a gene, and one non-targeting sequence (negative control, scramble). Human DDR1 (NM_001954.4) cDNA, CD44 (NM_001001389.2) cDNA, YAP5SA, and pcDNA3.1 plasmids were gifts from the Hepatic Surgery Center, Tongji Hospital, Huazhong University of Science and Technology, China. pBABE-puro (plasmid #1764), gag/pol (plasmid #14887), pMD2.G, pLKO.1-TRC cloning vector (plasmid #10878), and psPAX2 were purchased from Addgene (Cambridge, MA, USA). Human DDR1 cDNA was cloned into the pBABE-puro retroviral vector and identified via sequencing (TSINGKE, Beijing, China). The CD44-overexpressing lentivirus was purchased from DesignGene Biotechnology (Shanghai, China). To construct pLKO.1-scramble, pLKO.1-shDDR1, pLKO.1- shCD44, pLKO.1-shYAP1 plasmid, two target double-stranded oligonucleotides (shRNA) sequences, and one non-targeting sequence (negative control, scramble) were annealed and cloned into the pLKO.1 vector. The shRNA oligo-pair sequences are listed in Supplementary Table S4. Viral production, infection, and establishment of stable cell clones have been described previously [76]. The pcDNA3.1 plasmid inserted by FlAG- or HA-tagged DDR1, FlAG- or HA-tagged CD44, FlAG-tagged PP2AA were constructed according to the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China) and were identified by sequencing (TSINGKE, Beijing, China).
The generation of mutant cell lines by PE editing
The establishment of DDR1 R105A/K618A mutant in HLF followed as described [77]. Briefly, for PE editing, the epegRNAs and nicking sgRNAs were designed by PrimeDesign [78], then the epegRNAs were cloned into pU6-tevopreq1-GG-acceptor (plasmid #174038), and the nicking sgRNAs was cloned into pU6-pegRNA-GG-acceptor (plasmid #132777). The HLF cell was plated at a density of 1 × 105 per well of a 6-well plate 18 h before transfection. Then, 1.6ug PEmax-BSD, 0.198ug constructed epegRNA vector, 0.088ug constructed Nicking sgRNA vector, 0.8ug pcDNA3.1-MLH1dn were together transfected used FectinMore Transfection Reagent (Chamot, Shanghai, China). After 72 h transfection of plasmids into HLF, the cells were selected with 5 μg/ml blasticidin after 48 h virus transfection for three days. All of these survived cells were seeded into 96-well plates. Genomic DNA was extracted from each positive clone, followed by sequencing of the PCR products spanning the gene editing sites (the design of PCR primers must amplify a region spanning at least from 25 bp upstream of the epegRNA-guided nick to 25 bp downstream of the 3′ flap generated by the RT or any secondary nick). Genotyping was performed by sequencing PCR products amplified from the following primers: DDR1-R105A forward: 5′- GGTGTTTCCCAAGGAG GAGG-3′; reverse: 5′-TAGCTCCGGGAGAACTCCTT-3′; DDR1-K618A forward: 5′-CGACAGCCCTCAAGATCTGG-3′; reverse: 5′-CCATTGTTCTCTCTAGATGGC-3′. The epegRNA and ngRNA sequences of DDR1R105A and DDR1K618A are listed in Supplementary Table S4.
Collagen I-induced DDR1 phosphorylation
Cell lines were serum starved for 10 h, then stimulated with 30 μg/mL collagen I in cell culture medium for 24 h at 37 °C, left unstimulated as a control. The supernatant was removed, and PBS washed cells for three times to remove collagen I then were harvested for lysed or subsequent experiments.
Sphere-formation assay
To calculate sphere-forming efficiency, cells were single-cell sorted into 96-well plates coated with an ultra-low attachment surface (Corning). The cells were grown under anchoring-independent conditions in selective serum-free DMEM/F12 medium supplemented with B27 supplement minus vitamin A, 20 ng/mL hEGF, and 20 ng/mL bFGF. After seven days, the spheres formed were counted. Experiments were repeated at least three times.
Colony formation assay
Cells were suspended and seeded at a density of 2000 cells per well. After three weeks of cultivation, the cells were fixed and stained with 10% formalin and 0.1% crystal violet. The relative number of colonies was counted. Experiments were repeated at least three times.
Flow cytometric analysis
Cells were stained with PE-DDR1(Santa Cruz Biotechnology) or APC-conjugated CD133 (BD Biosciences) antibody in PBS with 2% FBS at 4 °C for 30–60 mins. Isotype-matched mouse immunoglobulins were used as the controls. The samples were sorted with FACSMelody (BD Biosciences) or analyzed using the CytoFLEX flow cytometer (Beckman Coulter). Flow J (version 10) software was used for analyzing the flow cytometry data.
Drug resistance and IC50
Cells in the logarithmic growth phase were uniformly inoculated into 96-well plates (1000/well), and different diluted drug concentrations were added to the medium. Six replicate wells were used for each concentration gradient assay. The cells were cultured in an incubator for 48 h. Subsequently, 10 μL of CCK-8 solution and 90 μL of culture medium was added, followed by incubation in the dark at 37 °C for 1 h. Absorbance at 450 nm was detected using a microplate reader. The half maximal inhibitory concentration (IC50) was analyzed based on GraphPad Prism (Version 8, GraphPad Software, San Diego, CA).
Coomassie blue staining and mass spectrometry
HEK-293 T cells transiently transfected with FLAG-DDR1 or FLAG-vector were lysed in IP lysis buffer (25 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 10% glycerol, and protease inhibitor cocktail), and IP assays were performed as described previously [18]. The eluted proteins were separated by SDS-PAGE followed by Coomassie Blue Fast Staining Solution. Mass spectrometry was performed and analyzed using the ptm-bio lab (PTM BIO, Hangzhou, China).
Reverse transcription PCR and real-time quantitative PCR (q-PCR)
Total cell RNA was extracted using TRIzol. Reverse transcription was carried out using HiScript II Q Select RT SuperMix (+gDNA wiper) according to the manufacturer’s instructions. Real-time fluorescence quantitative PCR was performed using the ChamQ Universal SYBR qPCR Master Mix. Gene expression levels were normalized to those of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in the same samples. Each sample was analyzed independently in triplicate. The primers used are listed in Supplementary Table S5.
Western blot, co-immunoprecipitation (co-IP)
Western blot and co-immunoprecipitation assays were performed as previously described [18]. Briefly, cells were collected and lysed on ice using IP lysis buffer. Lysates were incubated with protein G agarose for 2 h and immunoprecipitated with indicated antibodies overnight at 4 °C. The lysates were incubated with protein G agarose beads for 1 h followed by 1wash using IP lysis buffer and three washes with washing buffer (300 mM NaCl, 1.0 mM EDTA, 25 mM Tris-HCl, pH7.4, 1.0% NP-40). The beads were eluted with 2 × SDS-PAGE loading buffer and subjected to western blot.
Glutathione S transferase (GST) pull-down
The human DDR1 cDNA encoding was subcloned into a pET-42b vector. Following transformation and amplification in BL21(DE3) E. coli, recombinant GST-DDR1 fusion proteins were purified by GST Purification MagBeads. 5 μg GST or 5 μg GST-DDR1 was incubated with recombinant human 5 μg His-CD44 protein (ABclonal Technology, Wuhan, China) in PBS at 4 °C for 4 h under constant mixing. The bound proteins were incubated and immunoprecipitated with GST antibody-protein A beads. After washing away the unbound proteins three times, the bound proteins were analyzed by SDS-PAGE and western blot.
Immunofluorescence and confocal microscopy imaging
Immunofluorescence assays were performed as described previously [75]. Briefly, after the indicated treatments, cells were cultured on coverslips for 12 h, fixed in 4% paraformaldehyde for 15 min at room temperature, and permeabilized with 0.5% Triton X-100 for 5 min. After blocking, slides were incubated with primary antibody overnight at 4 °C in a humidified box. The slides were then washed thrice and incubated with secondary antibody for 4 h at room temperature in a humidified box. Finally, the cell nuclei were stained with 40, 60-diamidino-2-phenylindole (DAPI) for 5 min. The resulting signals were visualized using a confocal laser scanning microscope (Olympus FV1000, Tokyo, Japan). The relative fluorescence intensity was analyzed and quantify using the Image-Pro Plus 6.0 image software.
Dual-luciferase reporter assay
Cells were seeded in a 24-well plate at a density of 10000 cells per well. The next day, cells were co-transfected with 200 ng pGL4.17 and 4 ng pRL-TK plasmids, and transfections were performed using Lipofectamine3000 according to the manufacturer’s instructions. 12 h after transfection, cells were replaced with fresh medium and allowed to grow for 48 h. Luciferase activity was detected with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, United States) using a GloMax 20/20 Luminometer (Promega, Madison, WI, United States). Firefly luciferase activity was normalized to the renilla luciferase activity.
Reagents and antibodies
Puromycin, trypsin-EDTA, Opti-MEM, and polybrene were obtained as previously described [19]. Rattail collagen I (#354236) were purchased from Corning (NY, USA). 7rh, and verteporfin were purchased from MedChemExpress (Shanghai, China). The 50 × B27 supplement (minus vitamin A), 50 × ITS, TRIzol and Lipofectamine 3000 reagent were purchased from Thermo Fisher Scientific (MA, United States). human recombinant EGF (hEGF), basic FGF(bFGF) were purchased from R&D system. ClonExpress II One Step Cloning Kit, Mut Express II Fast Mutagenesis Kit V2, CCK-8 solution, HiScript II Q Select RT SuperMix (+gDNA wiper) and ChamQ Universal SYBR qPCR Master Mix were purchased from Vazyme (Nanjing, China). FectinMore Transfection Reagent was purchased from Chamot (Shanghai, China). Coomassie Blue Fast Staining Solution was purchased from Beyotime Biotechnology (Shanghai, China). GST Purification MagBeads was purchased from Absin Bioscience (Shanghai, China). All the antibodies used in this study are listed in Supplementary Table S6.
Human tissue specimens and immunohistochemical analysis
From February 1, 2014 to December 31, 2015, 123 human tissues were authorized by the Hepatic Surgery Center, Tongji Hospital, Huazhong University of Science and Technology (Wuhan, China). All the participants provided written informed consent. Patients who had received radiotherapy or chemotherapy before surgery were excluded. Survival time was calculated from the date of surgery to the date of death or last follow-up. Tumor staging was performed according to the Seventh edition of the Tumor-Node-Metastasis (TNM) Classification of the International Union Against Cancer [79]. HCC samples (n = 123) were used to generate a tissue microarray (Shanghai Biochip Co., Ltd. Shanghai, China). The fundamental processes of the Immunohistochemistry assay have been previously mentioned [18]. The pathological types of paraffin-embedded slides were rechecked by HE staining before IHC analysis. A DAB substrate kit (Zsbio Commerce Store) was used according to the manufacturer’s instructions. Scores for staining frequency (0 < 10%, 1 = 10–25%, 2 = 26–50%, 3 = 51–75%, 4 > 75%) and intensity (0 = negative, 1 = weak, 2 = moderate, 3 = intense staining) were used. A DAB substrate kit (Zsbio Commerce Store) was used according to the manufacturer’s instructions. The overall staining score (OSS) was calculated by multiplying the staining area percentage score by the intensity score. 0–6 were considered low expression, and 9–12 were considered high expression. The results were scored by two pathologists who were blinded to clinicopathological data. All procedures were approved by the Ethics Committee of Tongji Hospital and conducted according to the Declaration of Helsinki Principles. Written and informed consent was obtained from each patient.
Picrosirius red staining and polarization microscopy
The paraffin-fixed HCC specimens were sliced into 4 μm thick sections. Then, the sections were mounted on glass slides and stained with Picric acid–Sirius red (0.1% Sirius red in saturated aqueous Picric acid). Stained sections were examined with polarizing microscope (Nikon Eclipse Ti2, Tokyo, Japan) to detect collagen types. Five randomly selected images per section were captured and determined the optical density of collagen types. Under polarizing microscope, collagen I present closely packed, thick, non-argyrophilic, strongly birefringent, yellow or red fibers. Collagen II present loose collagenous feltwork displaying a weak birefringence of a varying color. Collagen III present loose framework of thin, argyrophilic, weakly birefringent, greenish fibers. Collagen IV present thin, amorphous, weakly birefringent basal laminae [80]. Collagen I, II, III and IV were analyzed and quantify using the Image-Pro Plus 6.0 image software [81].
Extreme limiting dilution xenograft tumor formation
Four weeks old male NOD/SCID mice were randomly divided into two or three groups (12 mice per group). The sorted CD133+ cells were diluted and bilateral transplanted by subcutaneous injection. Tumors were harvested at the end of the experiment for documentation. Stem cell frequency was calculated using the extreme limiting dilution analysis (ELDA) software [82]. Animal assays were carried out according to Wuhan Medical Experimental Animal Care Guidelines.
Drug treatments in vivo
Sphere-formed HLF cells (2 × 105) were suspended and subcutaneously injected into 4-week-old BALB/c nude mice. After 2 weeks, the animals were orally administered 50 mg/kg verteporfin and 50 mg/kg 7rh twice daily for two weeks. Drugs delivered by oral gavage were dissolved in DMSO and diluted in corn oil. Control mice were treated with the vehicle following an identical procedure. Mice were killed (within 48 h of the last treatment), and samples were obtained for histopathological and immunohistochemical analysis.
RNA-seq
Cells were lysed using TRIzol reagent. RNA extraction, library construction, high-throughput sequencing, and data analyses were conducted by Novogene Technology Co., Ltd. (Beijing, China).
Tumor segmentation and feature extraction
65 patients from IHC analyses with complete imaging data were retrospectively recruited. We randomly selected 45 cases as the training data set (25/20 = positive/negative) and another 20 cases as the independent testing data set (11/9 = positive/negative). Two independent readers manually delineated all MR images as high-resolution T2-weighted images (T2WI) using an open-source software package (ITK-SNAP, version 3.6.0, www.itksnap.org). Tumors were outlined as regions of interest (ROIs). Two types of images, “original images” and “wavelet images,” were used for the analysis in this study. “Original images” were the images without any transformation, and “wavelet images” were used for image denoising and improving image quality. The original images underwent a three-dimensional (i.e., x, y, and z directions) wavelet transformation using the PyWavelet package in Python. Each image was filtered by a high bandpass filter or low band-pass in the three directions, resulting in 8 combinations of different decompositions: LLH, LHL, HLL, LHH, HHL, HLH, HHH, and LLL (H means high, and L means low). The FeAture Explorer Pro (FAE Pro, V 0.4.1) was used to extract radiomic features. The shape features were extracted only from the “original images.” 18 types of first-order statistical features, 14 types of shape features, and 75 types of texture features [24 Gray Level Co-occurrence Matrix (GLCM) + 14 Gray Level Dependence Matrix (GLDM) + 16 Gray Level Run Length Matrix (GLRLM) + 16 Gray Level Size Zone Matrix (GLSZM) + 5 Neighborhood Gray-tone-difference Matrix (NGTDM)], a total of 107 “original images” features were used for analysis in our study. “Wavelet images” contained 144 first-order statistical features [8 wavelet images × 18] and 600 texture features [8 wavelet images × 75]. All 851 characteristics were extracted from the visible primary tumors, recorded, and stored quantitatively (Supplementary Table S7).
Predictive models establishment
To remove the imbalance in the training data set, we used the synthetic minority oversampling technique (SMOTE) to balance the positive/negative samples. Normalization was applied to the feature matrix. Each feature vector was subtracted from the mean value of the vector and divided by its length. Because the dimension of the feature space was high, we compared the similarity of each feature pair. If the PCC value of the feature pair was greater than 0.99, one of them was removed. After this process, the feature space dimension was reduced, and each feature was independent. Before building the model, we used analysis of variance (ANOVA) to select features. The F-value was calculated to evaluate the relationship between the features and labels. We sorted the features according to their corresponding F-values and assigned a specific number of features to build the model. We used a support vector machine (SVM) as the classifier. The kernel function can map the features into a higher dimension to search the hyper-plane to separate cases with different labels. To determine the hyper-parameter of the model (e.g., the number of features), we applied a cross-validation 5-fold on the training data set. The hyper-parameters were selected according to model performance on the validation data-set. The model performance was evaluated using receiver operating characteristic (ROC) curve analysis. The area under the ROC curve (AUC) was calculated for quantification. The above processes were implemented using the FeAture Explorer Pro (FAEPro, V 0.4.1) on Python (3.7.6).
Statistical analysis
All the results were based on at least three replicates. Statistical analyses were performed using GraphPad Prism (Version 8, GraphPad Software, San Diego, CA). Categorical variables were compared using Fisher’s exact test (two-tailed χ2 test) or Pearson’s chi-squared test. Statistical significance for comparing the continuous variables were using the non-parametric two-sided Mann–Whitney U-test. To compare several groups with a control group, one-way ANOVA followed by Dunnett’s multiple comparison test was applied. Two-way ANOVA followed by Bonferroni post-test was applied to compare several groups with a control group over time. Survival curves were generated using the Kaplan–Meier procedure. Underlying assumptions for these tests, including sample independence, variance equality, and normality were assumed to be met although not explicitly examined. Survival curves and hazard ratios were evaluated using log-rank tests. Statistical significance was set at p < 0.05.
Supplementary information
Acknowledgements
We thank all study subjects for their participation in this study. Thanks to the Hubei BIOSSCI Biotech Co., Ltd for the technical support in IHC.
Author contributions
WGZ contributed to the conception. XCZ and WGZ contributed to the study design. HFL and ZGZ form the overarching research goals and aims. YXX and XCZ wrote the main manuscript text. YXX, JHZ, YXZ and YLP contributed to the acquisition, analysis, and interpretation of the data. WGZ supervised the research activity planning. JJL and YXL verified the results/experiments and other research outputs. WGZ support the finance of the project. YW and JPZ critically revised the manuscript. All authors read and approve the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81874149 to Wanguang Zhang; No. 82103606 to Xiaochao Zhang).
Data availability
Research data supporting this publication are available from the StarBase website [83]. All relevant data in this research are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
All procedures were approved by the Ethics Committee of Tongji Hospital and conducted according to the Declaration of Helsinki Principles. Written and informed consent was obtained from each patient.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yi-xiao Xiong, Xiao-chao Zhang, Jing-han Zhu.
Contributor Information
Hui-fang Liang, Email: lianghuifang1997@126.com.
Zhan-guo Zhang, Email: zhanguo_tjh@hust.edu.cn.
Wan-guang Zhang, Email: wgzhang@tjh.tjmu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01166-5.
References
- 1.Villanueva A. Hepatocellular carcinoma. N. Engl J Med. 2019;380:1450–62. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 2.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124–34. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 3.Zamor PJ, DeLemos AS, Russo MW. Viral hepatitis and hepatocellular carcinoma: etiology and management. J Gastrointest Oncol. 2017;8:229–42. doi: 10.21037/jgo.2017.03.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma H, Chang H, Bamodu OA, Yadav VK, Huang T, Wu ATH, et al. Collagen 1A1 (COL1A1) is a reliable biomarker and putative therapeutic target for hepatocellular carcinogenesis and metastasis. Cancers. 2019;11:786. doi: 10.3390/cancers11060786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nia HT, Munn LL, Jain RK. Physical traits of cancer. Sci (Am Assoc Advancement Sci) 2020;370:546. doi: 10.1126/science.aaz0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng DH, Rodriguez BL, Diao L, Chen L, Wang J, Byers LA, et al. Collagen promotes anti-PD-1/PD-L1 resistance in cancer through LAIR1-dependent CD8+ T cell exhaustion. Nat Commun. 2020;11:4520. doi: 10.1038/s41467-020-18298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuczek DE, Larsen AMH, Thorseth M, Carretta M, Kalvisa A, Siersbæk MS, et al. Collagen density regulates the activity of tumor-infiltrating T cells. J Immunother Cancer. 2019;7:68. doi: 10.1186/s40425-019-0556-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X, Wu B, Chiang H, Deng H, Zhang X, Xiong W, et al. Tumour DDR1 promotes collagen fibre alignment to instigate immune exclusion. Nature. 2021;599:673–78. [DOI] [PMC free article] [PubMed]
- 9.Nissen NI, Karsdal M, Willumsen N. Collagens and cancer associated fibroblasts in the reactive stroma and its relation to cancer biology. J Exp Clin Cancer Res. 2019;38:115. doi: 10.1186/s13046-019-1110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stylianopoulos T, Martin JD, Chauhan VP, Jain SR, Diop-Frimpong B, Bardeesy N, et al. Causes, consequences, and remedies for growth-induced solid stress in murine and human tumors. Proc Natl Acad Sci USA. 2012;109:15101–8. doi: 10.1073/pnas.1213353109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.You Y, Zheng Q, Dong Y, Xie X, Wang Y, Wu S, et al. Matrix stiffness-mediated effects on stemness characteristics occurring in HCC cells. Oncotarget. 2016;7:32221–31. doi: 10.18632/oncotarget.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular carcinoma. Nat Rev Dis Prim. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 13.Clarke MF. Clinical and therapeutic implications of cancer stem cells. N. Engl J Med. 2019;380:2237–45. doi: 10.1056/NEJMra1804280. [DOI] [PubMed] [Google Scholar]
- 14.Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225–38. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lv J, Liu Y, Cheng F, Li J, Zhou Y, Zhang T, et al. Cell softness regulates tumorigenicity and stemness of cancer cells. EMBO J. 2021;40:e106123. doi: 10.15252/embj.2020106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh Y. Discoidin domain receptors: microenvironment sensors that promote cellular migration and invasion. Cell Adh Migr. 2018;12:378–85. doi: 10.1080/19336918.2018.1460011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agarwal G, Mihai C, Iscru DF. Interaction of discoidin domain receptor 1 with collagen type 1. J Mol Biol. 2007;367:443–55. doi: 10.1016/j.jmb.2006.12.073. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Hu Y, Pan Y, Xiong Y, Zhang Y, Han M, et al. DDR1 promotes hepatocellular carcinoma metastasis through recruiting PSD4 to ARF6. Oncogene. 2022;41:1821–34. [DOI] [PMC free article] [PubMed]
- 19.Pan Y, Han M, Zhang X, He Y, Yuan C, Xiong Y, et al. Discoidin domain receptor 1 promotes hepatocellular carcinoma progression through modulation of SLC1A5 and the mTORC1 signaling pathway. Cell Oncol. 2022;45:163–78. doi: 10.1007/s13402-022-00659-8. [DOI] [PubMed] [Google Scholar]
- 20.Dey A, Varelas X, Guan K. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat Rev Drug Discov. 2020;19:480–94. [DOI] [PMC free article] [PubMed]
- 21.Meng Z, Moroishi T, Guan K. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamar JM, Stern P, Liu H, Schindler JW, Jiang Z, Hynes RO. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci USA. 2012;109:E2441–50. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Passaniti A, Brusgard JL, Qiao Y, Sudol M, Finch-Edmondson M. Roles of RUNX in Hippo pathway signaling. (Springer Singapore, Singapore). 2017;435-48. [DOI] [PMC free article] [PubMed]
- 24.Pobbati AV, Hong W. A combat with the YAP/TAZ-TEAD oncoproteins for cancer therapy. Theranostics. 2020;10:3622–35. doi: 10.7150/thno.40889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson JD, Huang K, Pacal M, McCurdy SR, Lu S, Aubry A, et al. Binary pan-cancer classes with distinct vulnerabilities defined by pro- or anti-cancer YAP/TEAD activity. Cancer Cell. 2021;39:1115–34. doi: 10.1016/j.ccell.2021.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zöller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254–67. doi: 10.1038/nrc3023. [DOI] [PubMed] [Google Scholar]
- 28.Morath I, Hartmann TN, Orian-Rousseau V. CD44: More than a mere stem cell marker. Int J Biochem Cell Biol. 2016;81:166–73. doi: 10.1016/j.biocel.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Brown RL, Wei Y, Zhao P, Liu S, Liu X, et al. CD44 splice isoform switching determines breast cancer stem cell state. Genes Dev. 2019;33:166–79. doi: 10.1101/gad.319889.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dhar D, Antonucci L, Nakagawa H, Kim JY, Glitzner E, Caruso S, et al. Liver cancer initiation requires p53 inhibition by CD44-enhanced growth factor signaling. Cancer Cell. 2018;33:1061–77. doi: 10.1016/j.ccell.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen M, Cao J, Hu J, Topatana W, Li S, Juengpanich S, et al. Clinical-radiomic analysis for pretreatment prediction of objective response to first transarterial chemoembolization in hepatocellular carcinoma. Liver Cancer. 2021;10:38–51. doi: 10.1159/000512028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X, Zhang HL, Liu QP, Sun SW, Zhang J, Zhu FP, et al. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol. 2019;70:1133–44. doi: 10.1016/j.jhep.2019.02.023. [DOI] [PubMed] [Google Scholar]
- 33.Caruso S, Calatayud AL, Pilet J, La Bella T, Rekik S, Imbeaud S, et al. Analysis of liver cancer cell lines identifies agents with likely efficacy against hepatocellular carcinoma and markers of response. Gastroenterology. 2019;157:760–76. doi: 10.1053/j.gastro.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Cox TR. The matrix in cancer. Nat Rev Cancer. 2021;21:217–38. doi: 10.1038/s41568-020-00329-7. [DOI] [PubMed] [Google Scholar]
- 35.Ma S, Chan KW, Hu L, Lee TKW, Wo JYH, Ng IOL, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–56. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 36.Nio K, Yamashita T, Kaneko S. The evolving concept of liver cancer stem cells. Mol Cancer. 2017;16:4. doi: 10.1186/s12943-016-0572-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hidalgo-Carcedo C, Hooper S, Chaudhry SI, Williamson P, Harrington K, Leitinger B, et al. Collective cell migration requires suppression of actomyosin at cell-cell contacts mediated by DDR1 and the cell polarity regulators Par3 and Par6. Nat Cell Biol. 2011;13:49–58. doi: 10.1038/ncb2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 39.Joosten SPJ, Spaargaren M, Clevers H, Pals ST. Hepatocyte growth factor/MET and CD44 in colorectal cancer: partners in tumorigenesis and therapy resistance. Biochimica et Biophysica Acta Rev Cancer. 2020;1874:188437. doi: 10.1016/j.bbcan.2020.188437. [DOI] [PubMed] [Google Scholar]
- 40.Wang W, Zhang H, Liu S, Kim CK, Xu Y, Hurley LA, et al. Internalized CD44s splice isoform attenuates EGFR degradation by targeting Rab7A. Proc Natl Acad Sci. 2017;114:8366–71. doi: 10.1073/pnas.1701289114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen R, Xie R, Meng Z, Ma S, Guan K. STRIPAK integrates upstream signals to initiate the Hippo kinase cascade. Nat Cell Biol. 2019;21:1565–77. doi: 10.1038/s41556-019-0426-y. [DOI] [PubMed] [Google Scholar]
- 42.Dai W, Liu S, Wang S, Zhao L, Yang X, Zhou J, et al. Activation of transmembrane receptor tyrosine kinase DDR1-STAT3 cascade by extracellular matrix remodeling promotes liver metastatic colonization in uveal melanoma. Signal Transduct Target Ther. 2021;6:176. doi: 10.1038/s41392-021-00563-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Z, Zhou JK, Wang K, Chen H, Qin S, Liu J, et al. PDLIM1 inhibits tumor metastasis through activating hippo signaling in hepatocellular carcinoma. Hepatology. 2020;71:1643–59. doi: 10.1002/hep.30930. [DOI] [PubMed] [Google Scholar]
- 44.Gao M, Duan L, Luo J, Zhang L, Lu X, Zhang Y, et al. Discovery and optimization of 3-(2-(Pyrazolo[1,5-a] pyrimidin-6-yl) ethynyl) benzamides as novel selective and orally bioavailable discoidin domain receptor 1 (DDR1) inhibitors. J Med Chem. 2013;56:3281–95. doi: 10.1021/jm301824k. [DOI] [PubMed] [Google Scholar]
- 45.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee S, Anders RA, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–5. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karamanos NK, Theocharis AD, Neill T, Iozzo RV. Matrix modeling and remodeling: a biological interplay regulating tissue homeostasis and diseases. Matrix Biol: J Int Soc Matrix Biol. 2019;75-76:1–11. doi: 10.1016/j.matbio.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jian Z, Sun J, Chen W, Jin H, Zheng J, Wu Y. Involvement of discoidin domain 1 receptor in recurrence of hepatocellular carcinoma by genome-wide analysis. Med Oncol. 2012;29:3077–82. doi: 10.1007/s12032-012-0277-x. [DOI] [PubMed] [Google Scholar]
- 48.Shen Q, Cicinnati VR, Zhang X, Iacob S, Weber F, Sotiropoulos GC, et al. Role of microRNA-199a-5p and discoidin domain receptor 1 in human hepatocellular carcinoma invasion. Mol Cancer. 2010;9:227. doi: 10.1186/1476-4598-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toh TB, Lim JJ, Hooi L, Rashid MBMA, Chow EK. Targeting Jak/Stat pathway as a therapeutic strategy against SP/CD44+ tumorigenic cells in Akt/β-catenin-driven hepatocellular carcinoma. J Hepatol. 2020;72:104–18. doi: 10.1016/j.jhep.2019.08.035. [DOI] [PubMed] [Google Scholar]
- 50.Pankova D, Jiang Y, Chatzifrangkeskou M, Vendrell I, Buzzelli J, Ryan A, et al. RASSF1A controls tissue stiffness and cancer stem-like cells in lung adenocarcinoma. EMBO J. 2019;38:e100532. doi: 10.15252/embj.2018100532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biondani G, Zeeberg K, Greco MR, Cannone S, Dando I, Dalla PE, et al. Extracellular matrix composition modulates PDAC parenchymal and stem cell plasticity and behavior through the secretome. FEBS J. 2018;285:2104–24. doi: 10.1111/febs.14471. [DOI] [PubMed] [Google Scholar]
- 53.Ezzoukhry Z, Henriet E, Piquet L, Boyé K, Bioulac-Sage P, Balabaud C, et al. TGF-β1 promotes linear invadosome formation in hepatocellular carcinoma cells, through DDR1 up-regulation and collagen I cross-linking. Eur J Cell Biol. 2016;95:503–12. doi: 10.1016/j.ejcb.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 54.Juin A, Di Martino J, Leitinger B, Henriet E, Gary AS, Paysan L, et al. Discoidin domain receptor 1 controls linear invadosome formation via a Cdc42-Tuba pathway. J Cell Biol. 2014;207:517–33. doi: 10.1083/jcb.201404079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong C, Tao B, Tang F, Yang X, Peng T, You J, et al. Remodeling cancer stemness by collagen/fibronectin via the AKT and CDC42 signaling pathway crosstalk in glioma. Theranostics. 2021;11:1991–2005. doi: 10.7150/thno.50613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu S, Xu H, Wang W, Li S, Li H, Li T, et al. The role of collagen in cancer: from bench to bedside. J Transl Med. 2019;17:309. doi: 10.1186/s12967-019-2058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinzelmann-Schwarz VA, Gardiner-Garden M, Henshall SM, Scurry J, Scolyer RA, Davies MJ, et al. Overexpression of the cell adhesion molecules DDR1, Claudin 3, and Ep-CAM in metaplastic ovarian epithelium and ovarian cancer. Clin Cancer Res. 2004;10:4427–36. doi: 10.1158/1078-0432.CCR-04-0073. [DOI] [PubMed] [Google Scholar]
- 58.Valencia K, Ormazábal C, Zandueta C, Luis-Ravelo D, Antón I, Pajares MJ, et al. Inhibition of collagen receptor discoidin domain receptor-1 (DDR1) reduces cell survival, homing, and colonization in lung cancer bone metastasis. Clin Cancer Res. 2012;18:969–80. doi: 10.1158/1078-0432.CCR-11-1686. [DOI] [PubMed] [Google Scholar]
- 59.Lai C, Lin C, Liao W, Hour T, Wang H, Chuu C. CD44 promotes migration and invasion of docetaxel-resistant prostate cancer cells likely via induction of hippo-yap signaling. Cells. 2019;8:295. doi: 10.3390/cells8040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orian-Rousseau V. CD44 acts as a signaling platform controlling tumor progression and metastasis. Front Immunol. 2015;6:154. doi: 10.3389/fimmu.2015.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fan Z, Xia H, Xu H, Ma J, Zhou S, Hou W, et al. Standard CD44 modulates YAP1 through a positive feedback loop in hepatocellular carcinoma. Biomed Pharmacother. 2018;103:147–56. doi: 10.1016/j.biopha.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 62.Li S, Li C, Zhang Y, He X, Chen X, Zeng X, et al. Targeting mechanics-induced fibroblast activation through CD44-RhoA-YAP pathway ameliorates crystalline silica-induced silicosis. Theranostics. 2019;9:4993–5008. doi: 10.7150/thno.35665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu Y, Stamenkovic I, Yu Q. CD44 attenuates activation of the hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 2010;70:2455–64. doi: 10.1158/0008-5472.CAN-09-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han Y. Analysis of the role of the Hippo pathway in cancer. J Transl Med. 2019;17:116. doi: 10.1186/s12967-019-1869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z, Qiu N, Yin J, Zhang J, Liu H, Guo W, et al. SRGN crosstalks with YAP to maintain chemoresistance and stemness in breast cancer cells by modulating HDAC2 expression. Theranostics. 2020;10:4290–307. doi: 10.7150/thno.41008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ngai D, Mohabeer AL, Mao A, Lino M, Bendeck MP. Stiffness-responsive feedback autoregulation of DDR1 expression is mediated by a DDR1-YAP/TAZ axis. Matrix Biol. 2022;110:129–40. doi: 10.1016/j.matbio.2022.05.004. [DOI] [PubMed] [Google Scholar]
- 67.Ambrogio C, Gómez-López G, Falcone M, Vidal A, Nadal E, Crosetto N, et al. Combined inhibition of DDR1 and Notch signaling is a therapeutic strategy for KRAS-driven lung adenocarcinoma. Nat Med. 2016;22:270–7. doi: 10.1038/nm.4041. [DOI] [PubMed] [Google Scholar]
- 68.Nokin M, Darbo E, Travert C, Drogat B, Lacouture A, San José S, et al. Inhibition of DDR1 enhances in vivo chemosensitivity in KRAS-mutant lung adenocarcinoma. JCI Insight. 2020;5:e137869. [DOI] [PMC free article] [PubMed]
- 69.Aguilera KY, Huang H, Du W, Hagopian MM, Wang Z, Hinz S, et al. Inhibition of discoidin domain receptor 1 reduces collagen-mediated tumorigenicity in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2017;16:2473–85. doi: 10.1158/1535-7163.MCT-16-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huggett MT, Jermyn M, Gillams A, Illing R, Mosse S, Novelli M, et al. Phase I/II study of verteporfin photodynamic therapy in locally advanced pancreatic cancer. Br J Cancer. 2014;110:1698–704. doi: 10.1038/bjc.2014.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao X, Wang X, Fang L, Lan C, Zheng X, Wang Y, et al. A combinatorial strategy using YAP and pan-RAF inhibitors for treating KRAS-mutant pancreatic cancer. Cancer Lett. 2017;402:61–70. doi: 10.1016/j.canlet.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 72.Garcia-Rendueles MER, Ricarte-Filho JC, Untch BR, Landa I, Knauf JA, Voza F, et al. NF2 loss promotes oncogenic RAS-induced thyroid cancers via YAP-dependent transactivation of RAS proteins and sensitizes them to MEK inhibition. Cancer Discov. 2015;5:1178–93. doi: 10.1158/2159-8290.CD-15-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Islam MS, Afrin S, Singh B, Jayes FL, Brennan JT, Borahay MA, et al. Extracellular matrix and Hippo signaling as therapeutic targets of antifibrotic compounds for uterine fibroids. Clin Transl Med. 2021;11:e475. doi: 10.1002/ctm2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–62. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 75.Wu Y, Ding Z, Jin G, Xiong Y, Yu B, Sun Y, et al. Autocrine transforming growth factor-β/activin A-Smad signaling induces hepatic progenitor cells undergoing partial epithelial-mesenchymal transition states. Biochimie. 2018;148:87–98. doi: 10.1016/j.biochi.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 76.Wu Y, Wang W, Peng XM, He Y, Xiong YX, Liang HF, et al. Rapamycin upregulates connective tissue growth factor expression in hepatic progenitor cells through TGF-beta-smad2 dependent signaling. Front Pharm. 2018;9:877. doi: 10.3389/fphar.2018.00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Doman JL, Sousa AA, Randolph PB, Chen PJ, Liu DR. Designing and executing prime editing experiments in mammalian cells. Nat Protoc. 2022;17:2431–68. doi: 10.1038/s41596-022-00724-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hsu JY, Grunewald J, Szalay R, Shih J, Anzalone AV, Lam KC, et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat Commun. 2021;12:1034. doi: 10.1038/s41467-021-21337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou L, Rui JA, Ye DX, Wang SB, Chen SG, Qu Q. Edmondson-Steiner grading increases the predictive efficiency of TNM staging for long-term survival of patients with hepatocellular carcinoma after curative resection. World J Surg. 2008;32:1748–56. doi: 10.1007/s00268-008-9615-8. [DOI] [PubMed] [Google Scholar]
- 80.Montes GS. Structural biology of the fibres of the collagenous and elastic systems. Cell Biol Int. 1996;20:15–27. doi: 10.1006/cbir.1996.0004. [DOI] [PubMed] [Google Scholar]
- 81.Qiang G, Zhang L, Yang X, Xuan Q, Shi L, Zhang H, et al. Effect of valsartan on the pathological progression of hepatic fibrosis in rats with type 2 diabetes. Eur J Pharmacol. 2012;685:156–64. doi: 10.1016/j.ejphar.2012.04.028. [DOI] [PubMed] [Google Scholar]
- 82.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–8. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 83.Li J, Liu S, Zhou H, Qu L, Yang J. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic acids Res. 2014;42:D92–7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Research data supporting this publication are available from the StarBase website [83]. All relevant data in this research are available from the corresponding author upon reasonable request.