Abstract
Objective
Biases within probabilistic electronic phenotyping algorithms are largely unexplored. In this work, we characterize differences in subgroup performance of phenotyping algorithms for Alzheimer’s disease and related dementias (ADRD) in older adults.
Materials and methods
We created an experimental framework to characterize the performance of probabilistic phenotyping algorithms under different racial distributions allowing us to identify which algorithms may have differential performance, by how much, and under what conditions. We relied on rule-based phenotype definitions as reference to evaluate probabilistic phenotype algorithms created using the Automated PHenotype Routine for Observational Definition, Identification, Training and Evaluation framework.
Results
We demonstrate that some algorithms have performance variations anywhere from 3% to 30% for different populations, even when not using race as an input variable. We show that while performance differences in subgroups are not present for all phenotypes, they do affect some phenotypes and groups more disproportionately than others.
Discussion
Our analysis establishes the need for a robust evaluation framework for subgroup differences. The underlying patient populations for the algorithms showing subgroup performance differences have great variance between model features when compared with the phenotypes with little to no differences.
Conclusion
We have created a framework to identify systematic differences in the performance of probabilistic phenotyping algorithms specifically in the context of ADRD as a use case. Differences in subgroup performance of probabilistic phenotyping algorithms are not widespread nor do they occur consistently. This highlights the great need for careful ongoing monitoring to evaluate, measure, and try to mitigate such differences.
Keywords: machine learning, phenotyping, subgroup performance
INTRODUCTION
Machine learning (ML) algorithms are computational tools that enable computers to learn patterns and make predictions or decisions without being explicitly programmed, but rather learned from the underlying data. The widespread adoption of ML algorithms for risk stratification has unearthed plenty of cases of racial/ethnic biases within algorithms—from x-ray images to electronic health records (EHRs) and clinical notes.1–5 When built without careful weightage, calibration, and bias-proofing, ML algorithms can give wrong recommendations, thereby worsening health disparities faced by communities of color. Medical researchers in fields like dermatology,6 pharmacovigilance,7 and clinical-decision support,8 to name a few, have started to examine biases inherently embedded within ML algorithms via the features used, quality of datasets, types of ML algorithms, and design decisions. Until the beginning of 2023, there have been over 600 published papers in PubMed, that address the evaluation and mitigation of racial bias in clinical ML models,9 with some pieces providing very insightful ideas (eg, dividing bias in statistical and social),10 listing challenges (eg, adaptive learning, clinical implementation, and evaluating outcomes),11 as well as strategies (eg, reporting clarity, using denoising strategies, explainability, among others12 on how to think about bias, where can it be present,13 and how it can be mitigated. On the implementation/deployment side, researchers have proposed how to introduce/represent these models to end-users14 and some best practices within the field.15–18
In the broader ML community, Kleinberg et al19 showed that a probabilistic classification to be “fair” to different groups should satisfy 3 inherent conditions: (1) calibration within groups, (2) balance for the negative class, and (3) balance for the positive class. However, these conditions cannot be satisfied all at once, which has led to the development of numerous other “fairness measures”20–22 that overlap and create confusion.23 While most of these metrics apply to algorithms directly, they have not been analyzed in the context of medicine24–26 until late 2019, with mixed and at times contradictory findings. When applied to medicine, other factors need to be considered, such as the clinical utility and benefit of the model.27
Rule-based phenotyping has been the de facto method for identifying cohorts of patients belonging to any given condition/phenotype.28 This method requires clinicians to agree on a set of clinical elements organized in logical rules that best represent the targeted phenotype. Two of the biggest disadvantages of this approach are that: (1) the rules are rigid, meaning that they do not allow patients that have missing key data points to be included and (2) these definitions are expert-driven and very time-consuming/expensive to construct. Most recently, different ML-based approaches have gained traction, because they are data driven and allow more flexibility for patient inclusion.29 Specifically, patients are assigned probability scores, rather than a binary label. In this work, we used a probabilistic score approach to examine racial bias in EHR data of disease phenotypes that impact older adult patients. Other analytical approaches for phenotyping like latent class analysis30,31 and unsupervised clustering algorithms have been used in this context, but out of scope of our evaluation.
The National Institute on Aging has defined Alzheimer’s disease and related dementias (ADRD) as a series of complex brain disorders that affect millions of Americans, more specifically an estimate of 5 million in 2014, which is projected to grow to 13.9 million by 2060.32 This growth will have a deleterious impact on individuals, their families, long-term care facilities, health care providers, and health care systems. The negative impact of ADRD on minority older adults cannot be overstated. In this study, we selected ADRD phenotypes as they impact all older adults, and especially in communities of color.33–36 We utilize electronic phenotyping to characterize subgroup performance, which could lead to algorithmic biases (if any) in this context. A review of existing literature identified only one study by Straw and Wu37 presenting a sex-stratified analysis of ML models for liver disease prediction. In this work, instead of a sex-stratified analysis, we build a more detailed and robust gender-stratified analysis to identify bias from a broader perspective, nicely providing an additional view of the problem than Straw and Wu. Additional prior research has examined racial bias in the context of dementia.38 To our knowledge, this is the first study to evaluate the impact of racial subgroup performance, within probabilistic phenotyping models, on older adults in the context of mild cognitive impairment (MCI), Alzheimer’s disease, and Parkinson’s disease. Moreover, our study, which uses a larger EHR dataset, found very different conclusions on racial subgroup performance in dementia, thereby demonstrating the usefulness of our evaluation framework that was built for EHR data in the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) format. This CDM has over 700 million patient records, across the world, converted to it.
OBJECTIVE
In this study, we characterized the racial subgroup in performance of probabilistic electronic phenotyping algorithms developed from EHR datasets. Without using race as a modeling variable, we hypothesized that (1) probabilistic algorithms perform differently for different racial groups, (2) the difference in performance is tied to data availability for different racial groups, and (3) not all algorithms show the same level of racial subgroup performance differences.
MATERIALS AND METHODS
Dataset
The dataset utilized for this work is from deidentified data from the Stanford Medicine Research data Repository, consisting of over 3 million patients with clinical data from 2008 until the end of 2018. This dataset has been converted to the OMOP CDM version 5.3 and used in multiple Observational Health Data Sciences and Informatics (OHDSI) studies over the years. Table 1 shows the overall demographics of the dataset. Of particular interest is the underlying racial distribution, which will be highlighted throughout the rest of this study.
Table 1.
Demographics of patients from 2008 to 2018
| Overall |
|||
|---|---|---|---|
| n = 3 113 080 | |||
| Gender (%) | |||
| Missing | 5806 | (0.2) | |
| Female | 1 673 410 | (53.8) | |
| Male | 1 433 864 | (46.1) | |
| Race (%) | |||
| Asian | 303 800 | (9.8) | |
| Black | 97 399 | (3.1) | |
| Native American | 6819 | (0.2) | |
| Other | 393 466 | (12.6) | |
| Pacific Islander | 23 142 | (0.7) | |
| Unknown | 1 071 563 | (34.4) | |
| White | 1 216 891 | (39.1) | |
| Ethnicity (%) | |||
| Hispanic/Latino | 338 820 | (10.9) | |
| Non-Hispanic | 1 602 753 | (51.5) | |
| Unknown | 1 171 507 | (37.6) | |
| Age (mean [sd]) | 46.7 | (25.11) | |
Rule-based algorithms as our gold standard
We used the rule-based phenotype algorithms validated and available on the Health Data Research UK (HDR UK) Phenotype library39 for the following conditions: dementia40 and Parkinson’s disease.41 We adapted them into OHDSI ATLAS cohort definitions for their application on our OMOP-converted data. The rule-based phenotype definitions for MCI were adapted from Jongsiriyanyong and Limpawattana,42 and for Alzheimer’s disease we used the clinical definition by Holmes.43
Manually curated and clinically validated sets of patients are more robust but also significantly more time and resource intensive. Thus, community-approved rule-based definitions from organizations like the Phenotype Knowledgebase44 and the HDR UK Phenotype library45 have become the community approved methodology to computationally identify phenotypes.28,46 These rule-based definitions, while clinically validated, are quite rigid and show less flexibility than other approaches,28 this leads to many potential patients being excluded if certain codes or conditions are just not presented in their health record because of lack of coding or errors. One major limitation of these rule-based definitions is that they could have any hard-coded biases due to lack of representative subgroup data when they were built or due to different coding practices of subgroup members. However, our approach will not encode these as the probabilistic definitions are data driven as explained in the following section.
Probabilistic phenotypes with APHRODITE
Instead of relying on rigid rules to define medical condition phenotypes, newer data-driven approaches leverage ML to build statistical models to classify patients. These approaches have gained traction in the last few years,28 because they allow subjects to have a degree of probability of belonging to a phenotype, making them more flexible and able to catch people that have clinical codes missing or incomplete data. The methodology used for this work, Automated PHenotype Routine for Observational Definition, Identification, Training and Evaluation (APHRODITE), was designed with this flexibility in mind. Specifically, it relies on building statistical models for phenotypes based on an initial cohort of patients selected using high-precision keywords/clinical codes. The models were built using weak supervision where the patient’s entire clinical record up until the first appearance of the selected keyword/clinical code. This approach is semisupervised as only an initial keyword/clinical code is needed and everything else is data driven. This approach was introduced by Agarwal et al47 and was made into an R package48 which works on standardized data to the OMOP CDM format. In this work, we will use all data from: Conditions, Procedures, Medications, Diagnosis, and Observation codes as variables for modeling.
Experimental framework
In order to identify racial subgroup performance variations within the probabilistic models, we built the following steps into a framework that we can later reuse for a wider variety of phenotypes. We started by selecting 3 of the most popular classical ML models to evaluate: LASSO,49 since regression-based classifiers are widely used for statistical learning purposes with EHR/medical data,50 Random Forest (RF),51 and support vector machines (SVMs).52 Note that the 3 models listed above are the ones supported by default by APHRODITE; however, any model supported by caret R package can also be included.53 Next, we selected matched cases and controls to build our probabilistic models. The patients were matched by age, race, gender, and length of clinical record. We then stratified by the patient’s race for our multipronged evaluation, which consists of: traditional model (all races merged together), balanced model (we balance based on equal distribution of patients for each race), single race only model, and the leave-one-out combinations, which take one race out of the model building process in a systematic way. Following our usual practice, we used 75% of the data to train the model and a 25% unseen set to test the model, in addition to 5-fold cross-validation.
For evaluation, we used the traditional metrics: accuracy, which is the fraction of assignments the model identified correctly; sensitivity, which is the proportion of positives that are correctly identified; and specificity, which measures the proportion of negatives that are correctly identified. In addition, we used variation in order to measure difference between models in the following way:
This measurement allowed us to evaluate the first 3 metrics in a similar context and show how different models are compared with each other. Note that while phenotyping algorithm performance is important, this is not the key point of this work, we present general model classification accuracy in order to put performance variance between phenotypes in context.
RESULTS
Phenotyping algorithms
First, we checked that the probabilistic phenotyping algorithms performed well when compared against their rule-based definitions. Table 2 shows APHRODITE’s performance to select and identify the “gold-standard” patients identified by the rule-based phenotype definitions.
Table 2.
Rule-based and APHRODITE phenotype selection overlap
| Phenotype | Cases identified by rule-based definition | Cases identified by APHRODITE keywords | Initial overlap (%) | Classified by APHRODITE model (prob over 90%) | Total overlap (%) |
|---|---|---|---|---|---|
| Dementia | 13 213 | 16 998 | 95.95 | 471 | 99.52 |
| MCI | 7915 | 8292 | 94.14 | 399 | 99.18 |
| Alzheimer’s disease | 11 401 | 12 828 | 89.04 | 1137 | 99.02 |
| Parkinson’s disease | 5989 | 6644 | 79.50 | 896 | 94.46 |
Our results show that APHRODITE and its probabilistic models are successful at identifying almost the same patients for each phenotype when compared with the rule-based definition, which served as our gold standard.
Table 3 shows the demographics of the patients identified for each phenotype. While our gender classes are almost balanced, due to the large imbalance in the race categories, the performance of ML models will vary because of the smaller training sample sizes.
Table 3.
Patient demographics for the evaluated phenotypes
| Phenotype | Cases | Controls | Gender |
Race |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Female (%) | Male (%) | Asian (%) | Black (%) | Native Amer. (%) | Pacific Island (%) | White (%) | Unknown (%) | |||
| Dementia | 16 998 | 16 998 | 56.22 | 43.78 | 11.07 | 4.96 | 0.27 | 0.80 | 60.33 | 22.57 |
| MCI | 8292 | 8292 | 49.82 | 50.18 | 11.73 | 3.88 | 0.22 | 0.92 | 60.06 | 23.19 |
| Alzheimer’s disease | 12 828 | 12 828 | 60.05 | 39.95 | 12.08 | 4.85 | 0.25 | 0.69 | 63.03 | 19.11 |
| Parkinson’s disease | 6644 | 6644 | 39.30 | 60.70 | 12.06 | 1.34 | 0.23 | 0.32 | 63.11 | 22.95 |
A few things to note are that: (1) the racial distributions for some of the categories like Native American and Pacific Islanders are very low and (2) there were many patients listed as unknown race, which were removed from our evaluation.
Our evaluation framework produced over 1200 plots and charts evaluating the performance of probabilistic models built under multiple conditions. Figure 1 presents model classification accuracies for the 4 phenotypes and the 3 ML algorithms we utilized (ie, 15 parameters) for the 12 different data subsets evaluated. This figure sets the precedent of the importance of the model variance evaluation and how it will change models from being potentially useful, to highly unreliable.
Figure 1.
Model classification accuracy result (y-axis) for all ML algorithms and all phenotypes. In this context, accuracy is defined as the fraction of assignments the model identified correctly.
These results demonstrate that most full models (ie, the classical ones), which are the purple bars, have 70%–90% classification accuracy for the given phenotype. These results are only for illustration purposes and to put the following figures of model variance in perspective. We are not trying to find the best performing models in general, but rather show their subgroup variation, when races are stratified.
Evaluation Scenario 1: Building models with individual racial subgroups
In Figures 2 and 3, we show the classification and sensitivity variance for the RF models between our models built for individual races and compared across all races evaluation. These figures are designed as heatmaps to visually highlight the severity of the variance and place the attention of the researcher on the more relevant sections. For example, we built models using only White patients, and compared their performance when classifying patients from all other races. We used RF as our choice algorithm to illustrate our results due to its solid average performance during our experimentation, and due to the more explainable nature of its models.
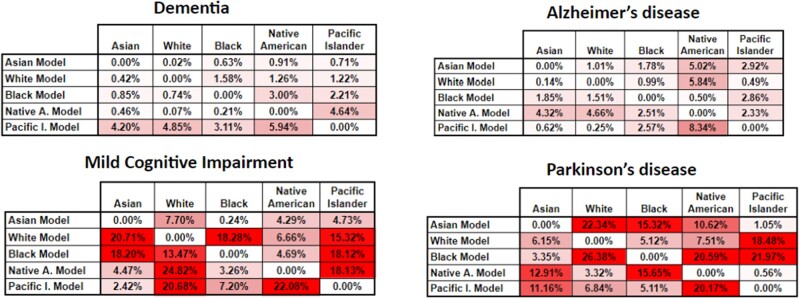
Figure 2.
Classification accuracy variance for the RF algorithm for our 4 phenotypes. In this heatmap, the higher tone of red a cell has, represents bigger variance from the base comparison, or in other words more bias.
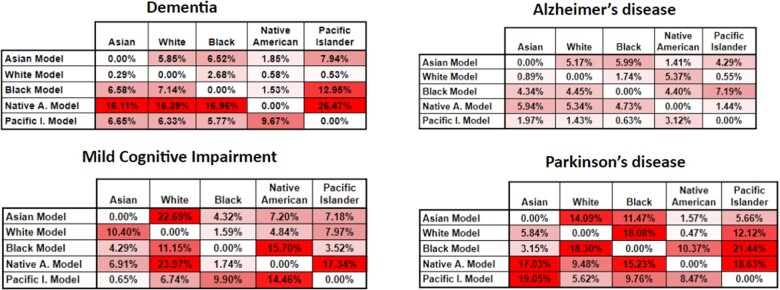
Figure 3.
Classification sensitivity variance for the RF algorithm for our 4 phenotypes. In this heatmap, the higher tone of red a cell has, represents bigger variance from the base comparison, or in other words more bias.
Figure 2 shows some very interesting results for the dementia and Alzheimer’s disease phenotypes, illustrating that the variance between models is not that pronounced (<8% at the worst case). This shows that models built with the individual races are generalizable enough across races, at least for these 2 phenotypes. For the remaining 3 phenotypes the variance increases up to 27%, rendering a classification model with an accuracy of less than 80%, almost equal to random picking. These results showcase the need to evaluate these combinations carefully, particularly before deployment of any phenotyping model.
The sensitivity variance for the models in Figure 3 shows a similar trend as Figure 2—that is, less variance for the dementia and Alzheimer’s phenotypes. However, there is a considerable increase in the variance of the Native American model, most likely due to the fact that this racial group is highly underrepresented in the dataset used; thus, this model is unable to properly generalize across races, particularly when measuring sensitivity.
Evaluation Scenario 2: Full models, balanced models, and leave-one-out
We now switch to evaluate the models in a more traditional sense of using a model with all data available—one that balances the classes but limits the number of samples to the minimum available in any given class. Note that we did not use SMOTE54 or any sampling techniques in this work, as this is not ideal when using clinical data,55,56 since depending on the method used, it adds nonrepresentative extra data. The other scenarios we evaluated include leaving one class out in the model building process. We then applied the built models to the individual classes of patients in the testing set (fully unseen patients). Figures 4 and 5 report these results for classification accuracy and sensitivity.
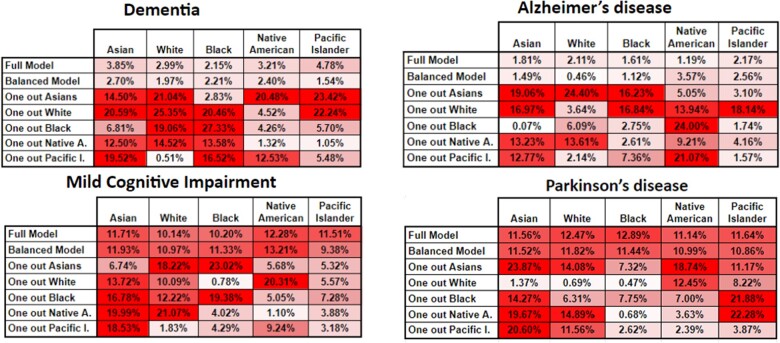
Figure 4.
Classification accuracy variance for the RF algorithm for our 4 phenotypes. In this heatmap, the higher tone of red a cell has, represents bigger variance from the base comparison, or in other words more bias.
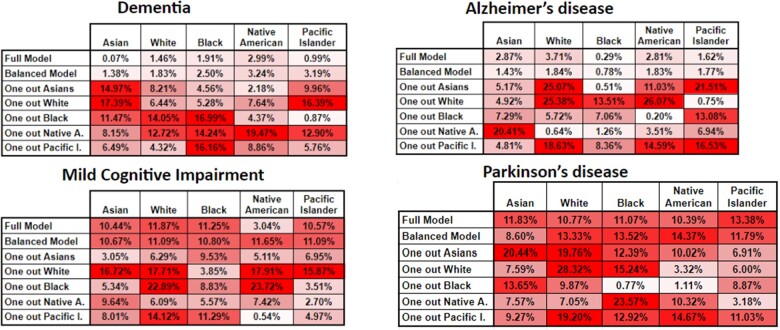
Figure 5.
Classification sensitivity variance for the RF algorithm for our 4 phenotypes. In this heatmap, the higher tone of red a cell has, represents bigger variance from the base comparison, or in other words more bias.
We again see very similar patterns for the dementia and Alzheimer’s disease phenotypes, where the variance is quite low. One thing to note is that there is always variance here as the calculation is performed on the base comparison model performance only classifying the given class (on the unseen test set) versus the base comparison model performance of all classes on the unseen test set. One very interesting result here is that for the phenotype models with less subgroup variance, taking out certain classes brings their overall performance down by considerable amounts (up to 26%) sometimes, which could be mostly due to removing the data-dominant race. In other scenarios, the accuracy variance is not that high, particularly when classifying the races with very small representation in the original models.
Figure 5 shows that classification sensitivity variance is also affected in a similar way as for the classification accuracy in almost the exact same way with the variance trends being very similar. This also reinforces the original finding, namely that 2 of the phenotypes show small variance, across our experimental evaluation and the other 3 have different, but increasing, degrees of variance. These results demonstrate that probabilistic phenotype models need to be carefully examined and improved before they can be used in a clinical setting.
DISCUSSION
We designed our experimental framework to provide a fully automated and standardized (on top of the OMOP CDM and using APHRODITE R package) way to demonstrate if any given probabilistic phenotyping model has racial subgroup variance and estimate how much. As shown in the “Results” sections, we have 2 different evaluation scenarios, which build probabilistic models in different stratified ways to provide flexibility and insight into how the differently built models will vary given popular ML metrics. Note that any identified anomaly in subgroup performance does not automatically translate into the algorithm leading “harming” subgroups, as addressing or “fixing” those anomalies might actually produce worse performing algorithms for all subgroups as found by Pfohl et al.57 In some cases having different algorithms for subgroups or a human in the loop might58 be a better approach to not affect any group of patients.
For the phenotypes of our case study, the first scenario clearly demonstrates that 2 phenotypes: dementia and Alzheimer’s disease present the least amount of subgroup variance (Figures 2 and 3), both in terms of classification accuracy and sensitivity. This is highlighted by their 2 variance figures showing the least amount of red cells. Racial representation in those cohorts could be one possible reason these phenotypes show less subgroup variance, but while they have some of the highest numbers of cases used for training (Table 2), their racial representation is nearly the same as in the whole dataset. An interesting observation is that accuracy shows very little variance whereas the sensitivity results show a higher amount of it. This might indicate that while the stratified models do well as a whole, there might be additional small variance when trying to detect the positive class. However, this can also be explained as an artifact of building the model with the Native American patients, which had the least representation in the full dataset (>0.25%). A common solution to address underrepresentation has been to oversample this class or undersample some of the others. However, our second scenario shows that this does not significantly improve the level of subgroup variance, as other researchers have shown, at least for predictive tasks.55,56 Rather, we recommend using federated learning, as this is starting to be more accepted in large research networks,59 or use ensembles using multiple datasets, when available within a single site or research facility.60 The second evaluation scenario shows that the full model and the balanced model perform very consistently for the phenotypes of dementia and Alzheimer’s disease, which have less subgroup variance. However, it also shows very striking results on the leave-one-out models, particularly when removing the racial groups with the larger representations and when removing a particular racial group and then trying to classify only patients of that group (Figures 4 and 5). These results show the need to consider such detailed analyses before trying to use any of these models in clinical practice.
Regarding the phenotypes—MCI and Parkinson’s disease—with more subgroup variance, we observed some very dramatic variance changes (average of 10%) between most of the experimental models. This indicates that those phenotypes are quite sensitive to any of the racial groups being removed, particularly shown by the leave-one-out models from the second scenario (Figures 4 and 5). These figures also show that even for the full model and balanced scenarios, predicting on individual classes brings considerable accuracy and sensitivity variance. These findings translate to how sensitive these models are to any type of shift in the underlying dataset that is used to train the model and how to evaluate them. We theorize that this sensitivity is due to higher variance in coding practices between racial groups for these 2 phenotypes. These results strongly demonstrate the need for rigorous experimental evaluation before any kind of deployment or testing in production environments (eg, hospitals). While there are plenty of experimental evaluations to analyze, our framework automates the work for researchers, and it only needs human interpretation of its findings.
The limitations of this work are the following: we evaluated 3 ML algorithms based on their popularity and level of use within the field. However, with new algorithms constantly being introduced, the results could vary dramatically when other algorithms are introduced. We decided to keep the same algorithms as in our previous works48,61 since we know how those perform to build probabilistic models using APHRODITE. The flexibility behind APHRODITE allows for other models to be evaluated within the presented framework as long as they have an R package available. We decided to only evaluate the variance metric as it gives a stronger and more interpretable signal on how the model differs from each other. For this study, we needed to keep the number of evaluations and experiments to a reasonable amount to be able to explain this work and its merits. However, any other metric can be configured into our framework. Lastly, our case study phenotypes were evaluated on a single dataset, and, in future work, we plan to fully leverage the OHDSI community to conduct a network study examining racial bias of phenotypic algorithms.62,63 One major item to note is that self-reported race is usually error prone and very often incomplete (missing in up to 23% of the patients selected for the phenotypes evaluated), these factors could lead to some of the results being artifacts of this phenomenon.
CONCLUSION
As we have demonstrated in this work, subgroup performance variance can certainly be found in probabilistic phenotype algorithms that categorize older adults, particularly for phenotypes like MCI, and Parkinson’s disease. As a result of this subgroup variance, models perform up to 30% worse under our multiple model building scenarios. Thus, it is critical for institutions to extensively test and rigorously evaluate their phenotyping models. We found that some phenotypes like dementia and Alzheimer’s disease were more resistant to this subgroup variance as indicated by their very small variance under all of our testing scenarios, meaning that these models could be potentially used safely. Rigorous testing allows researchers to be more confident of the performance of these models under different racial distributions. In terms of clinical relevance, our contribution will allow researchers to build mode robust ML models to identify patients with ADRDs. This could lead to less biased clinical trial eligibility selection, and to less biased disease identification and progression detection algorithms, which are data driven and not only rule-based. Our main contribution is the framework to fully automate this process when the institution has data in the OMOP CDM and can run our extension to the APHRODITE package. This work is particularly important as biomedical scientists and medical professionals strive to make informed conclusions and diagnoses of older adult patients.
ACKNOWLEDGMENTS
The authors would like to thank Jessica Moon, PhD, PMP for her comments and proofreading of this article.
Contributor Information
Juan M Banda, Department of Computer Science, College of Arts and Sciences, Georgia State University, Atlanta, Georgia, USA.
Nigam H Shah, Stanford Center for Biomedical Informatics Research, Stanford University School of Medicine, Stanford, California, USA.
Vyjeyanthi S Periyakoil, Stanford Department of Medicine, Palo Alto, California, USA; VA Palo Alto Health Care System, Palo Alto, California, USA.
FUNDING
This work was supported by the National Institute on Aging of the National Institutes of Health (3P30 AG059307-02S1).
AUTHOR CONTRIBUTIONS
All authors: conception and design, drafting the manuscript, revising the manuscript, approval of submitted version, and accountability of own contributions. JMB: data analysis and data interpretation.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
DATA AVAILABILITY
The EHR data used in this study cannot be shared for ethical/privacy reasons. All codes are available in the following location: https://github.com/OHDSI/Aphrodite.
REFERENCES
- 1. Chen IY, Szolovits P, Ghassemi M.. Can AI help reduce disparities in general medical and mental health care? AMA J Ethics 2019; 21: E167–79. [DOI] [PubMed] [Google Scholar]
- 2. Obermeyer Z, Powers B, Vogeli C, Mullainathan S.. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019; 366 (6464): 447–53. [DOI] [PubMed] [Google Scholar]
- 3. Seyyed-Kalantari L, Liu G, McDermott M, Chen IY, Ghassemi M. CheXclusion: Fairness gaps in deep chest X-ray classifiers. In: Altman RB, Dunker AK, Hunter L, Ritchie MD, Murray T, Klein TE, eds. Biocomputing 2021. Kohala Coast, Hawaii: World Scientific; 2021: 232–43. [PubMed]
- 4. Burlina P, Joshi N, Paul W, Pacheco KD, Bressler NM.. Addressing artificial intelligence bias in retinal diagnostics. Transl Vis Sci Technol 2021; 10 (2): 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thompson HM, Sharma B, Bhalla S, et al. Bias and fairness assessment of a natural language processing opioid misuse classifier: detection and mitigation of electronic health record data disadvantages across racial subgroups. J Am Med Inform Assoc 2021; 28 (11): 2393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Daneshjou R, Smith MP, Sun MD, Rotemberg V, Zou J.. Lack of transparency and potential bias in artificial intelligence data sets and algorithms: A scoping review. JAMA Dermatol 2021; 157 (11): 1362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kompa B, Hakim JB, Palepu A, et al. Artificial intelligence based on machine learning in pharmacovigilance: A scoping review. Drug Saf 2022; 45 (5): 477–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Čartolovni A, Tomičić A, Lazić Mosler E.. Ethical, legal, and social considerations of AI-based medical decision-support tools: A scoping review. Int J Med Inform 2022; 161: 104738. [DOI] [PubMed] [Google Scholar]
- 9. Huang J, Galal G, Etemadi M, Vaidyanathan M.. Evaluation and mitigation of racial bias in clinical machine learning models: Scoping review. JMIR Med Inform 2022; 10 (5): e36388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parikh RB, Teeple S, Navathe AS.. Addressing bias in artificial intelligence in health care. JAMA 2019; 322 (24): 2377. 10.1001/jama.2019.18058 [DOI] [PubMed] [Google Scholar]
- 11. DeCamp M, Lindvall C.. Latent bias and the implementation of artificial intelligence in medicine. J Am Med Inform Assoc 2020; 27 (12): 2020–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vokinger KN, Feuerriegel S, Kesselheim AS.. Mitigating bias in machine learning for medicine. Commun Med (Lond) 2021; 1: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gianfrancesco MA, Tamang S, Yazdany J, Schmajuk G.. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern Med 2018; 178 (11): 1544–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sendak MP, Gao M, Brajer N, Balu S.. Presenting machine learning model information to clinical end users with model facts labels. NPJ Digit Med 2020; 3: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de Hond AAH, Leeuwenberg AM, Hooft L, et al. Guidelines and quality criteria for artificial intelligence-based prediction models in healthcare: A scoping review. NPJ Digit Med 2022; 5 (1): 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rajpurkar P, Chen E, Banerjee O, Topol EJ.. AI in health and medicine. Nat Med 2022; 28 (1): 31–8. [DOI] [PubMed] [Google Scholar]
- 17. Luo W, Phung D, Tran T, et al. Guidelines for developing and reporting machine learning predictive models in biomedical research: A multidisciplinary view. J Med Internet Res 2016; 18 (12): e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu X, Glocker B, McCradden MM, Ghassemi M, Denniston AK, Oakden-Rayner L.. The medical algorithmic audit. Lancet Digit Health 2022; 4 (5): e384–97. [DOI] [PubMed] [Google Scholar]
- 19. Kleinberg J, Mullainathan S, Raghavan M.. Inherent trade-offs in the fair determination of risk scores. In: 8th Innovations in Theoretical Computer Science Conference (ITCS 2017), Schloss Dagstuhl-Leibniz-Zentrum Fuer Informatik GmbH, Wadern/Saarbruecken, Germany; 2017. 10.4230/LIPIcs.ITCS.2017.43. [DOI] [Google Scholar]
- 20. Chouldechova A, Roth A. The frontiers of fairness in machine learning, arXiv [cs.LG]; 2018. http://arxiv.org/abs/1810.08810.
- 21. Beutel A, Chen J, Doshi T, et al. Putting fairness principles into practice: Challenges, metrics, and improvements. In: Proceedings of the 2019 AAAI/ACM Conference on AI, Ethics, and Society, Association for Computing Machinery, New York, NY, USA; 2019: 453–59.
- 22. Chouldechova A, Roth A.. A snapshot of the frontiers of fairness in machine learning. Commun ACM 2020; 63 (5): 82–9. [Google Scholar]
- 23. Castelnovo A, Crupi R, Greco G, Regoli D, Penco IG, Cosentini AC. A clarification of the nuances in the fairness metrics landscape, arXiv [cs.LG]; 2021. http://arxiv.org/abs/2106.00467. [DOI] [PMC free article] [PubMed]
- 24. Xu J, Xiao Y, Wang WH, et al. Algorithmic fairness in computational medicine, bioRxiv; 2022. 10.1101/2022.01.16.21267299. [DOI] [PMC free article] [PubMed]
- 25. McCradden MD, Joshi S, Mazwi M, Anderson JA.. Ethical limitations of algorithmic fairness solutions in health care machine learning. Lancet Digit Health 2020; 2 (5): e221–3. [DOI] [PubMed] [Google Scholar]
- 26. Chen RJ, Chen TY, Lipkova J, et al. Algorithm fairness in AI for medicine and healthcare, arXiv [cs.CV]; 2021. http://arxiv.org/abs/2110.00603.
- 27. Char DS, Shah NH, Magnus D.. Implementing machine learning in health care—addressing ethical challenges. N Engl J Med 2018; 378 (11): 981–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Banda JM, Seneviratne M, Hernandez-Boussard T, Shah NH.. Advances in electronic phenotyping: From rule-based definitions to machine learning models. Annu Rev Biomed Data Sci 2018; 1: 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hripcsak G, Albers DJ.. High-fidelity phenotyping: Richness and freedom from bias. J Am Med Inform Assoc 2018; 25 (3): 289–94. 10.1093/jamia/ocx110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sinha P, Calfee CS, Delucchi KL.. Practitioner’s guide to latent class analysis: Methodological considerations and common pitfalls. Crit Care Med 2021; 49 (1): e63–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodríguez A, Ruiz-Botella M, Martín-Loeches I, et al. ; COVID-19 SEMICYUC Working Group. Deploying unsupervised clustering analysis to derive clinical phenotypes and risk factors associated with mortality risk in 2022 critically ill patients with COVID-19 in Spain. Crit Care 2021; 25 (1): 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matthews KA, Xu W, Gaglioti AH, et al. Racial and ethnic estimates of Alzheimer’s disease and related dementias in the United States (2015–2060) in adults aged ≥65 years. Alzheimers Dement 2019; 15 (1): 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fredriksen-Goldsen KI, Kim H-J, Barkan SE, Muraco A, Hoy-Ellis CP.. Health disparities among lesbian, gay, and bisexual older adults: Results from a population-based study. Am J Public Health 2013; 103 (10): 1802–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dunlop DD, Manheim LM, Song J, Chang RW.. Gender and ethnic/racial disparities in health care utilization among older adults. J Gerontol B Psychol Sci Soc Sci 2002; 57 (4): S221–33. [DOI] [PubMed] [Google Scholar]
- 35. Ward JB, Gartner DR, Keyes KM, Fliss MD, McClure ES, Robinson WR.. How do we assess a racial disparity in health? Distribution, interaction, and interpretation in epidemiological studies. Ann Epidemiol 2019; 29: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson KS. Racial and ethnic disparities in palliative care. J Palliat Med 2013; 16 (11): 1329–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Straw I, Wu H.. Investigating for bias in healthcare algorithms: A sex-stratified analysis of supervised machine learning models in liver disease prediction. BMJ Health Care Inform 2022; 29 (1): e100457. 10.1136/bmjhci-2021-100457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gianattasio KZ, Ciarleglio A, Power MC.. Development of algorithmic dementia ascertainment for racial/ethnic disparities research in the US Health and Retirement Study. Epidemiology 2020; 31 (1): 126–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kuan V, Denaxas S, Gonzalez-Izquierdo A, et al. A chronological map of 308 physical and mental health conditions from 4 million individuals in the English National Health Service. Lancet Digit Health 2019; 1 (2): e63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Phenotype Library. (n.d.). https://phenotypes.healthdatagateway.org/phenotypes/PH148/version/296/detail/ (accessed June 15, 2022).
- 41. Phenotype Library. (n.d.). https://phenotypes.healthdatagateway.org/phenotypes/PH77/version/154/detail/ (accessed June 15, 2022).
- 42. Jongsiriyanyong S, Limpawattana P.. Mild cognitive impairment in clinical practice: A review article. Am J Alzheimers Dis Other Demen 2018; 33 (8): 500–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Holmes C. Genotype and phenotype in Alzheimer’s disease. Br J Psychiatry 2002; 180: 131–4. [DOI] [PubMed] [Google Scholar]
- 44. Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: A catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016; 23 (6): 1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Denaxas S, Gonzalez-Izquierdo A, Direk K, et al. UK phenomics platform for developing and validating electronic health record phenotypes: CALIBER. J Am Med Inform Assoc 2019; 26 (12): 1545–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hripcsak G, Albers DJ.. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc 2013; 20 (1): 117–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Agarwal V, Podchiyska T, Banda JM, et al. Learning statistical models of phenotypes using noisy labeled training data. J Am Med Inform Assoc 2016; 23 (6): 1166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Banda JM, Halpern Y, Sontag D, Shah NH.. Electronic phenotyping with APHRODITE and the Observational Health Sciences and Informatics (OHDSI) data network. AMIA Jt Summits Transl Sci Proc 2017; 2017: 48–57. [PMC free article] [PubMed] [Google Scholar]
- 49. Tibshirani R. Regression shrinkage and selection via the LASSO. J Roy Stat Soc Ser B 1996; 58 (1): 267–88. [Google Scholar]
- 50. Jones AM. Models for health care. In: Clements MP, Hendry DF, eds. The Oxford Handbook of Economic Forecasting. Oxford University Press; 2011. [Google Scholar]
- 51. Breiman L. Random forests. Mach Learn 2001; 45 (1): 5–32. [Google Scholar]
- 52. Cortes C, Vapnik V.. Support-vector networks. Mach Learn 1995; 20 (3): 273–97. [Google Scholar]
- 53. Kuhn M. Building predictive models in R using the caret package. J Stat Soft 2008; 28 (5): 1–26. [Google Scholar]
- 54. Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP.. SMOTE: Synthetic minority over-sampling technique. J Artif Intell Res 2002; 16: 321–57. [Google Scholar]
- 55. van den Goorbergh R, van Smeden M, Timmerman D, Van Calster B.. The harm of class imbalance corrections for risk prediction models: Illustration and simulation using logistic regression. J Am Med Inform Assoc 2022; 29 (9): 1525–34. 10.1093/jamia/ocac093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pfohl SR, Zhang H, Xu Y, Foryciarz A, Ghassemi M, Shah NH.. A comparison of approaches to improve worst-case predictive model performance over patient subpopulations. Sci Rep 2022; 12 (1): 3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pfohl SR, Foryciarz A, Shah NH.. An empirical characterization of fair machine learning for clinical risk prediction. J Biomed Inform 2021; 113: 103621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Verghese A, Shah NH, Harrington RA.. What this computer needs is a physician: Humanism and artificial intelligence. JAMA 2018; 319 (1): 19–20. [DOI] [PubMed] [Google Scholar]
- 59. Xu J, Glicksberg BS, Su C, Walker P, Bian J, Wang F.. Federated learning for healthcare informatics. J Healthc Inform Res 2021; 5 (1): 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reps JM, Williams RD, Schuemie MJ, Ryan PB, Rijnbeek PR.. Learning patient-level prediction models across multiple healthcare databases: Evaluation of ensembles for increasing model transportability. BMC Med Inform Decis Mak 2022; 22: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kashyap M, Seneviratne M, Banda JM, et al. Development and validation of phenotype classifiers across multiple sites in the observational health data sciences and informatics network. J Am Med Inform Assoc 2020; 27 (6): 877–83. 10.1093/jamia/ocaa032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hripcsak G, Ryan PB, Duke JD, et al. Characterizing treatment pathways at scale using the OHDSI network. Proc Natl Acad Sci USA 2016; 113 (27): 7329–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hripcsak G, Duke JD, Shah NH, et al. Observational health data sciences and informatics (OHDSI): Opportunities for observational researchers. Stud Health Technol Inform 2015; 216: 574–8. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The EHR data used in this study cannot be shared for ethical/privacy reasons. All codes are available in the following location: https://github.com/OHDSI/Aphrodite.