Abstract
Persistent R-loop accumulation can cause DNA damage and lead to genome instability, which contributes to various human diseases. Identification of molecules and signaling pathways in controlling R-loop homeostasis provide important clues about their physiological and pathological roles in cells. Here, we show that NKAP (NF-κB activating protein) is essential for preventing R-loop accumulation and maintaining genome integrity through forming a protein complex with HDAC3. NKAP depletion causes DNA damage and genome instability. Aberrant accumulation of R-loops is present in NKAP-deficient cells and leads to DNA damage and DNA replication fork progression defects. Moreover, NKAP depletion induced R-loops and DNA damage are dependent on transcription. Consistently, the NKAP interacting protein HDAC3 exhibits a similar role in suppressing R-loop associated DNA damage and replication stress. Further analysis uncovers that HDAC3 functions to stabilize NKAP protein, independent of its deacetylase activity. In addition, NKAP prevents R-loop formation by maintaining RNA polymerase II pausing. Importantly, R-loops induced by NKAP or HDAC3 depletion are processed into DNA double-strand breaks by XPF and XPG endonucleases. These findings indicate that both NKAP and HDAC3 are novel key regulators of R-loop homeostasis, and their dysregulation might drive tumorigenesis by causing R-loop associated genome instability.
Subject terms: Gene regulation, RNA, Cell biology
Introduction
R-loops are three-stranded nucleic acid structures that consist of an RNA:DNA hybrid and a displaced single-stranded DNA (ssDNA), forming by the reannealing of the nascent RNA to the template DNA strand during transcription [1]. R-loops have been identified in many organisms, from bacteria to mammals, where they play regulatory roles in a variety of biological processes, including DNA replication, gene expression, immunoglobulin class switch recombination, DNA methylation and histone modifications, and DNA repair [1–9]. Although these natural occurring R-loops have beneficial roles in physiological contexts, abnormal R-loop accumulation can cause DNA damage and genomic instability, presumably by blocking the progression of replication forks and promoting double-strand breaks (DSBs) [8, 10, 11]. Increased R-loops are often found in many human diseases, such as cancer and neurological disorder [3, 12–14]. Thus, the levels and distribution of R-loops in cells need to be tightly regulated. Previous studies have identified multiple pathways and factors in controlling the formation and resolution of R-loops [3, 5, 8]. For instance, a number of proteins, including topoisomerases, splicing factors and chromatin regulators, function to prevent R-loop formation during transcription [15–26]. Several other proteins are reported to resolve R-loop structures. These proteins include RNA nucleases RNaseH1 and DNA/RNA helicases, which resolve R-loops by degrading or unwinding RNA:DNA hybrid [27–39]. In addition, DNA repair factors, such as BRCA1, BRCA2 and members of the Fanconi Anemia pathway, are also important for suppressing R-loop formation [40–42]. Despite much progress made, identification of novel players in controlling R-loop homeostasis will advance our understanding on the underlying molecular mechanisms as well as how this might relate to disease.
NKAP (NF κB–activating protein) is a highly conserved nucleoplasmic protein across different species [43]. Initially identified to be a mediator for NF κB activation, NKAP has been subsequently shown as a transcriptional repressor via its interaction with the Notch corepressor complex and HDAC3 [44, 45]. Analysis of conditional knockout mice has demonstrated that both NKAP and HDAC3 are involved in T cell development and maturation as well as the maintenance and survival of adult hematopoietic stem cells [46–48]. Mutations in NKAP have been linked to a neurodevelopmental disorder with musculoskeletal features in human patients [49]. Moreover, NKAP regulates RNA splicing and processing through direct interaction with RNA and RNA binding proteins [43]. Recent structure studies have shown that NKAP is a part of post-catalytic P-complex, which is responsible for exon ligation and mRNA release [50]. Notably, NKAP has also been reported to be critical for chromosome alignment during mitosis, indicating its potential role as a tumor suppressor [51]. A study in pancreatic cells reveals that NKAP acts as a m6A reader at the pri-miR-25 splicing site [52]. Thus, NKAP appears to be a multifunctional protein that acts through different molecular mechanisms.
Here, we provide the first evidence that NKAP regulates R-loop accumulation and this novel function of NKAP is essential in preventing DNA damage, genome instability and replication stress. Moreover, we show that aberrant R-loop accumulation associated with NKAP knockdown is transcription dependent. We also find that the NKAP interacting protein HDAC3 is required to prevent R-loop accumulation and promote the progression of replication forks. Detailed analysis indicates that NKAP is stabilized by HDAC3 in a deacetylase activity independent manner. This stabilization is partially mediated by the lysosome/autophagy pathway. We show that NKAP prevents R-loop formation by maintaining the duration of RNA polymerase II (Pol II) pausing. We further demonstrate that R-loops induced by NKAP or HDAC3 depletion are processed by XPF and XPG leading to DNA damage. Collectively, our work uncovers a previously unappreciated function of NKAP and HDAC3 in the suppression of R-loop formation and maintenance of genome integrity.
Material and methods
Cell culture and drug treatment
HeLa, U2OS and HB-8730 hybridoma cell lines were purchased from the American Type Culture Collection (ATCC). hTERT-RPE-1 cell line was a gift from Dr. Chan Kuan Yoow and originally from ATCC. HEK293T cell line was a gift from Dr. Liang Hongqing and originally from ATCC. U2OS, HeLa, HEK293T and HB-8730 cells were cultured in high glucose DMEM medium (Thermo Scientific), supplemented with 10% fetal bovine serum (Thermo Scientific) and 1% penicillin/streptomycin (Thermo Scientific, 100 U/ml penicillin, 100 µg/ml streptomycin). hTERT-RPE-1 cells were cultured in DMEM/F12 medium (Thermo Scientific), supplemented with 10% fetal bovine serum (Thermo Scientific) and 1% penicillin/streptomycin (Thermo Scientific, 100 U/ml penicillin, 100 µg/ml streptomycin). Cells were maintained at 37 °C in a humidified incubator containing 5% CO2 and routinely tested for Mycoplasma contamination according to the manufacturer’s instructions.
5,6-Dichlorobenzimidazole 1-b-D-ribofuranoside (DRB, Sigma-Aldrich) was added for 2 h at a final concentration of 100 μM. HDAC inhibitors were added for 24 h and final concentrations were: Trichostatin A (TSA, 1 μM, MCE), Valproic acid (VPA, 0.5 mM, MCE), or RGFP966 (10 μM, MCE). MG132 (MCE)was added for 6 h at a final concentration of 5 μM. Chloroquine (CQ, MCE) was added for 24 h at a final concentration of 20 μM. 3-Methyladenine (3-MA, MCE) was added for 24 h at a final concentration of 5 mM.
RNH1 or NKAP overexpression was induced by treatment of the cell culture with 1 µg/ml of doxycycline (DOX, MCE) for 24 h.
Plasmids
To create pCW57.1-RNH1-Flag-mRFP, open reading frame of RNASEH1 (accession number NM_001286834.3) was amplified from human cDNA, fused to SV40 NLS and Flag-mRFP tag, and then cloned into the pCW57.1 vector with Nhe I and Bam I restriction sites. Open reading frame of HDAC3 (accession number NM_001355039.2) was amplified from human cDNA, fused to Flag tag, and then cloned into the pcDNA3.1 vector with Nhe I and EcoR I restriction sites to generate pcDNA3.1-Flag-HDAC3. siRNA-resistant HDAC3 construct was then produced by introducing silent mutations in the targeting region for HDAC3 siRNA by site-directed mutagenesis (Q5 Site-Directed Mutagenesis Kit, NEB). K25A (Lysine to Alanine) and Y298F (Tyrosine to Phenylalanine) mutations in HDAC3 sequence were introduced by site-directed mutagenesis (Q5 Site-Directed Mutagenesis Kit, NEB). Open reading frame of NKAP (XM_017029842.1) was amplified from human cDNA, fused to Myc tag, and then cloned into the pCMV-GFP vector with Nhe I and Xho I restriction sites to generate pCMV-Myc-NKAP-GFP. The truncated construct of NKAP (pCMV-Myc-NKAP-C) was generated by cloning C-terminal fragment of NKAP with the Myc tag into the pCMV-GFP vector, and then GFP sequence was deleted by inverse PCR. A fragment of Myc-NKAP-GFP was amplified from pCMV-Myc-NKAP-GFP and cloned into the pCW57.1 vector with Nhe I and Sal I restriction sites to generate pCW57.1-Myc-NKAP-GFP. siRNA-resistant NKAP construct (pCW57.1-Myc-NKAPRes-GFP) was then produced by introducing silent mutations in the targeting region for NKAP siRNA by site-directed mutagenesis (Q5 Site-Directed Mutagenesis Kit, NEB). RS-domain deleted NKAP construct (pCW57.1-Myc-NKAPRes+∆RS-GFP) was generated from pCW57.1-Myc-NKAPRes-GFP using an inverse PCR technique. The primers used for amplification are listed in Table S1.
siRNA and plasmid transfection
Transfections of siRNAs were performed using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. siRNAs were synthesized by Genepharma company (Shanghai) and used at a final concentration of 50 nM. The sequences of siRNA oligonucleotides are listed in Key Resources Table. For transient ectopic gene expression, transfection of various plasmids were done using EndoFectin (GeneCopoeia) or Lipofectamine 3000 (Thermo Fisher Scientific) following the manufacturer’s instructions.
Lentivirus production
Lentiviral particles were generated by transfecting HEK293T cells with the lentiviral plasmid (pCW57.1-RNH1-Flag-mRFP, pCW57.1- Myc-NKAPRes-GFP or pCW57.1- Myc-NKAP∆RS-GFP), psAX2 and pMD2.G at a ratio of 3:2:1. Transfection was performed using EndoFectin Max (GeneCopoeia) according to the manufacturer’s protocol. The medium was replaced with normal medium 6 h after transfection. Virus-containing supernatant was collected and filtered (0.45 µm) 48 h and 72 h after transfection. Cells were transduced by exposure to the viral supernatant in the presence of polybrene (8 µg/ml) for 24 h and selected using puromycin (2 µg/ml) for 72 h. Finally, three stable cell lines (U2OS-TetOn-RNaseH1, U2OS-TetOn-NKAP and U2OS-TetOn- NKAP∆RS) were obtained.
Western blot and Co-immunoprecipitation (Co-IP)
For Western blot, cells were harvested and lysed with RIPA buffer (150 mM NaCl, 0.5% Triton X-100, 50 mM Tris-HCl pH 7.5, 1 mM EDTA, 0.1% SDS, Protease and Phosphatase Inhibitor Cocktail (Sigma-Aldrich)) on ice for 30 min. Samples were run on SDS–PAGE protein gels and transferred to PVDF membranes (Millipore) via wet transfer. Membranes were then blocked with 5% non-fat milk in TBST (0.1% Tween) for a minimum of 1 h before incubating with the appropriate primary antibodies overnight at 4 °C. After washing three times (10 min each) in TBST, membranes were then incubated with secondary horseradish peroxidase (HRP)-conjugated antibodies with 5% non-fat milk in TBST for 1 h. Protein bands were detected using Immobilon Western HRP mix Reagent (Millipore). Primary antibodies used were: rabbit anti-NKAP (Abcam, ab121121,1:1000), rabbit anti-RPA32 (Proteintech, 10412-1-AP, 1:1000), rabbit anti-RPA32 S33 (Bethyl Laboratories, A300-246A, 1:1000), mouse anti-γH2AX (Abcam, ab26350, 1:1000), rabbit anti-H2AX (Cell Signaling Technology, 2595 S, 1:1000), rabbit anti-Histone H3K9ac (Active Motif, 39917, 1:1000), mouse anti-Histone H3(Active Motif, 61976, 1:4000),rabbit anti-53BP1 (Novus Bio, NB100-304, 1:1000), rabbit anti-LC3 (Proteintech, 14600-1-AP, 1:1000), rabbit anti-HDAC3 (Abcam, ab32369, 1:1000), rabbit anti-XPF (Cell Signaling Technology, 13465, 1:1000), rabbit anti-XPG (Proteintech, 11331-1-AP, 1:1000), rabbit anti-DHX9 (Proteintech, 17721-1-AP, 1:4000), mouse anti-RNA polymerase II (Active Motif, 91151,1:2000), mouse anti-RNA polymerase II phospho S5 (Abcam, ab5131,1:2000), mouse anti-RNA polymerase II phospho S2 (Active Motif, 91116,1:2000), rabbit anti-GAPDH (Proteintech, 10494-1-AP, 1:8000), mouse anti-αTubulin (Sigma-Aldrich, T9026, 1:8000), mouse anti-Myc (Abcam, ab18185, 1:8000) and mouse anti-Flag (Sigma-Aldrich, F1804, 1:8000).
For co-immunoprecipitations, cells were washed with cold 1xPBS and lysed in ice-cold lysis buffer with NP40 (150 mM NaCl, 50 mM Tris-HCl pH 7.5, 1 mM EDTA and 1% Nonidet P40, Protease Inhibitor Cocktail) for 30 min at 4 °C under rotation. Lysates were centrifuged at 12,000 g for 15 min at 4 °C. While 50 μl of clear supernatant was saved as input, the rest was incubated with added antibodies (mouse anti-Myc (Abcam, ab18185), mouse anti-Flag (Sigma-Aldrich, F1804), mouse anti-HDAC3 (Cell Signaling Technology, 3949), mouse anti-S9.6 antibody or IgG (30000-0-AP, Proteintech)) overnight or 3 h at 4 °C under rotation. Samples were then incubated with Protein A/G beads (Sigma-Aldrich) or Protein A/G Magnetic beads (MCE) for 2 h at 4 °C under rotation. After centrifuging at 1100 g for 5 min at 4 °C, the beads were collected and washed three times in PBS. Proteins were eluted from beads by adding loading buffer and heating at 95 °C for 5 min. All Samples were then analyzed by Western blot.
Immunofluorescence analysis
For γH2AX, 53BP1 and p-RPA2 staining, cells were fixed with 4% paraformaldehyde (PFA) for 15 min at room temperature and then permeabilized with 0.5% Triton X-100 buffer for 15 min on ice. Next, samples were blocked with 5% BSA in PBS for 30 min at room temperature before incubating with specific primary antibodies overnight at 4 °C. After washing three times with PBS (10 min each), samples were incubated with appropriate secondary antibodies for 2 h at room temperature. Nuclei were counterstained with 0.5 µg/ml 4′,6-diamidino-2-phenylindole (DAPI). For S9.6 staining, cells were fixed with ice-cold methanol for 15 min and stained as described above. Primary antibodies used were: mouse anti-γ-H2AX (Abcam, ab26350, 1:400), rabbit anti-53BP1 (Novus Bio NB100-304, 1:400), rabbit p-RPA2 S33 (A300-246A, Bethyl Laboratories, 1:400), rabbit anti-Flag (Proteintech, 20543-1-AP, 1:1000), rabbit anti-Myc (Proteintech, 16286-1-AP, 1:1000), rabbit anti-Nucleolin (ABclonal, A20910, 1:400), mouse anti-S9.6 (Kerafast, ENH001, 1:200) or mouse anti-S9.6 antibody (1:200) purified from the HB-8730 hydridoma cell line. Secondary antibodies used were: Goat anti-Rabbit Alexa Fluor 488 (Thermo Fisher Scientific, A-11008, 1:1000), Goat anti-Mouse Alexa Cy3 (Thermo Fisher Scientific, M30010, 1:1000), Goat anti-Mouse Alexa Fluor 488 (Thermo Fisher Scientific, A-11001, 1:1000), Goat anti- Rabbit Cy3 (Thermo Fisher Scientific, A10520, 1:1000) and Goat anti-Rabbit Cy5 (Thermo Fisher Scientific, A-10523, 1:1000). Samples were mounted in Vectashield and visualized with the Olympus FV1000 confocal microscope. In each independent experiment, at least 200 cells were measured per condition.
EdU and EU staining
For EdU or EU incorporation experiments, cells were pulse-labeled with 10 µM EdU or EU for 60 min at 37 °C before being fixed and blocked as described above. The click reaction was carried out using the Click-iT EdU Alexa Fluor 555 kit (Thermo Fisher Scientific, C10338) or Click-iT™ RNA Alexa Fluor™ 488 Imaging Kit (Thermo Fisher Scientific, C10329) according to the manufacturer’s protocol. Briefly, the reaction mixture was added to cells for 30 min at room temperature, followed by three washes in PBS (10 min each). Nuclei were counterstained with DAPI. Samples were mounted in Vectashield and visualized with the Olympus FV1000 confocal microscope.
Alkaline and neutral comet assay
Comet assay was performed as previously described [53]. Briefly, cells were collected and mixed with 0.7% low-melting point agarose at 37 °C. The mixture was placed on the precoated slides. Cells were lysed at 4 °C in lysis buffer for 1 h under dark condition. Next, the slides were rinsed with alkaline (2.5 M NaCl, 100 mM EDTA, 10 mM Tris, 1% sarkosyl, 1% Triton X-100, pH 13.0) or neutral (100 mM Tris, 300 mM Na Acetate, pH 9.0) electrophoresis solution and incubated for 1 h in the dark for DNA unwinding. Electrophoresis was performed at 18 V for 1 h in the dark. Slides were washed and dried with ethanol, and transferred to 37 °C for complete dry in the dark. DNA was stained with SYBR green (Solarbio, 1:10,000) for 10 min at room temperature. Images were taken with a Nikon ECLIPSE 80i microscope. Tail moments of the comets were quantified using OpenComet plugin in ImageJ software.
DNA fiber analysis
DNA fiber analysis was performed as previously described [54]. Briefly, cells were pulse-labeled with 25 mM 5-Iodo-20-deoxyuridine (IdU, MCE, HY-B0307) for 20 min, followed by 250 mM 5-Chloro-20-deoxyuridine (CldU, Sigma-Aldrich, C6891) for 20 min. Next, cells were harvested and lysed on glass slides in spreading buffer (200 mM Tris-HCl pH 7.4, 50 mM EDTA and 0.5% SDS). After 10 min, the slides were tilted at 45 degree and the resulting DNA spreads were air-dried and fixed in methanol/acetic acid (3:1) for 2 min. The DNA fibers on slides were denatured with 2.5 M HCl for 80 min, washed three times with PBS (5 min each) and blocked with 5% BSA in PBS for 30 min. Standard immunofluorescence staining was then performed with rat anti-BrdU/CldU (detects CldU, ab6326, Abcam, 1:500) and mouse anti-IdU/BrdU (detects IdU, 347580, Becton Dickinson, 1:200) antibodies. Secondary antibodies used were Goat anti-Rat Cy3 (Thermo Fisher Scientific, PA128714, 1:1000) and Goat anti-Mouse Alexa Fluor 488 (Thermo Fisher Scientific, A-11001, 1:1000). Images were acquired on a Leica Axio Observe microscope. IdU and CldU tract lengths were measured using ImageJ. In each independent experiment, at least 100 fibers were measured per condition.
Dot blot analysis
Dot blot analysis was performed as previously described [55]. Genomic DNA was extracted with phenol/chloroform extraction and ethanol precipitation and digested overnight at 37 °C using restriction enzyme mix (HindIII, EcoRI, XhoI, BamHI and AgeI, NEB). Digested genomic DNA was incubated with or without 50U/ml RNase H (NEB) for 3 h at 37 °C. gDNA was diluted in 6× saline sodium citrate (SSC) buffer and spotted onto a Nylon membrane (Millipore) using the Bio-Dot® microfiltration apparatus (Bio-Rad). After UV-crosslinking (0.12 J/m2) exposure for 10 min, the membrane was blocked with 5% non-fat milk in TBST for 1 h at room temperature before incubating with the mouse S9.6 antibody (Kerafast, ENH001, 1:1000) or mouse anti-dsDNA antibody (Abcam, ab27156, 1:4000) in 1% BSA in TBST overnight at 4 °C. After washing three times (10 min each) in TBST, the membrane was then incubated with secondary horseradish peroxidase (HRP)-conjugated antibodies in 5% milk for 1 h at room temperature. Signals were developed using Immobilon Western HRP mix reagent (Millipore).
Metaphase spreads
Metaphase spreads were performed according to the optimized protocol [56, 57]. Briefly, cells were treated with 100 μL Colchicine (10 μg/ml) for 3 h, and then trypsinized and collected by centrifuging at 1000 rpm for 5 min. The supernatant was discarded and the cell pellet was resuspended in PBS. After adding ice-cold KCl solution (0.075 M), samples were incubated at 37 °C for 30 min before centrifuging at 1000 rpm for 5 min. The pellet was then resuspended in ice-cold methanol: glacial acetic acid (3:1) for fixation. After removing fixation reagents, 10 μl of the cell suspension was dropped onto an acetic-acid-humidified slide for the spreading of metaphase chromosomes. The spreads were air-dried and stained with Giemsa solution (Solarbio) for 20 min before mounting with neutral resins (Solarbio). Images were acquired on a Nikon ECLIPSE 80i microscope.
Cell cycle analysis by flow cytometry
For cell cycle assay, cells transfected with siRNAs for 48 h were harvested, washed with PBS, and fixed with cold 70% ethyl alcohol in the dark for 2 h. After being washed twice with cold PBS, cells were treated with 200 mg/mL RNase A for 30 min at 37 °C. Next, cells were stained with 100 mg/mL propidium iodide for 30 min at room temperature. The samples were immediately analyzed by flow cytometry (ACEA NovoCyte Flow Cytometer). Cell cycle phase distribution was determined using FlowJo.
Quantitative real-time PCR
Total RNA was extracted from cells using TRIZOL reagent (Thermo Fisher Scientific) followed by DNase I treatment (Thermo Fisher Scientific). cDNA was synthesized using SuperScript Reverse Transcriptase III with random hexamers (Thermo Fisher Scientific). Real time quantitative PCR (qPCR) was performed on an ABI Prism 7900 system (Applied Biosystems) with SYBR qPCR Mix (Vazyme). All primers used for qPCR are listed in Table S1. Quantification for target gene expression was performed using the 2−ΔΔCt method and GAPDH was used as normalization control.
DNA:RNA immunoprecipitation (DRIP) and qPCR analysis
DRIP analysis was performed as previously described [58]. Briefly, genomic DNA was isolated with phenol/chloroform extraction and ethanol precipitation. Extracted genomic DNA was digested overnight at 37 °C using restriction enzyme mix (HindIII, EcoRI, XhoI, BamHI and AgeI, NEB), followed by treatment or not with RNase H. For each DRIP, 8 µg of digested DNAs were subject to immunoprecipitation with 5 µg of S9.6 antibody using DRIP binding buffer (10 mM NaPO4 pH 7.0, 140 mM NaCl, and 0.05% Triton X-100) overnight at 4 °C. Following that, Protein A/G beads (Sigma-Aldrich) were added to the samples for an additional 2 h incubation at 4 °C. The beads were then collected by centrifugation. DNAs were subsequently eluted from the beads with elution buffer (50 mM Tris pH 8.0, 10 mM EDTA, 0.5% SDS) and treated with Proteinase K (Sigma-Aldrich) for 45 min at 55 °C. DNAs were then purified with phenol/chloroform extraction and ethanol precipitation and resuspended in TE buffer (10 mM Tris-HCl pH 8.0 and 10 mM EDTA). Quantitative PCR was performed at the indicated gene region using the primers listed in Table S1. The relative R-loop level in each gene loci was determined as percentage of input.
Chromatin immunoprecipitation sequencing (ChIP-seq)
U2OS cells were transfected with siRNAs for 48 h and fixed with 1% formaldehyde at room temperature for 10 min. After quenching with 125 mM glycine, cells were washed two times with ice-cold PBS and scraped in 1 ml PBS by centrifuging at 2000 × g for 5 min. Cell pellets were resuspended in 500 µL lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) and spun at 2000 × g for 10 min at 4 °C. The supernatant was discarded. cOmplete Protease Inhibitor Cocktail (Roche) and phosphatase inhibitors (Roche) were added to all buffers. The pellet was washed two times in lysis buffer and prepared for sonication using a Bioruptor (30 s on, 30 s off, 10 rounds of 10 min) at 4 °C. Five percent of the solution was reserved as input. Protein A/G Magnetic Beads (MCE) was prepared according to manufacturer’s specifications and either anti-RNA polymerase II (Active Motif, 91151) was conjugated to the magnetic beads. Equal amounts (5–10 μg) of sheared chromatin were subjected to chromatin immunoprecipitations for 4 h at 4 °C on a rotating wheel. Chromatin-bound beads were washed once with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.0, 150 mM NaCl), once with high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH 8.0, 300 mM NaCl), once with lithium chloride wash buffer (0.25 M LiCl, 1% NP-40, 1% Sodium Deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8.0) and twice in TE buffer. Cross-linked chromatin immunoprecipitates were eluted in elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS) containing 40 μg/ml RNase A at 37 °C for 30 min, followed by incubation at 65 °C overnight to reverse crosslinks. After reversal of the crosslink, DNA was purified by MinElute PCR Purification Kit (Qiagen). The pulled down material and input DNA were size-selected, and ligated to Illumina barcoded adaptors, using NEBNext® Ultra™ II DNA Library Prep Kit (NEB, USA, Catalog #: E7645L) on Illumina HiSeq 6000 platform. ChIP-seq data were aligned to Human genome reference (hg38 assembly) with Bowtie. Mapping quality was assessed with SAMtools and Python scripts. Peak-calling for ChIP-seq data was done using MACS2 with a q value of 0.01. DeepTools or ChIPseeker was used to compute and draw profiles on positions of interest (peaks, TSS, TES). Further analyses were done in R (http://www.R-project.org), with Bioconductor packages for graphic representation. The ChIP-seq datasets are available in the Gene Expression Omnibus (GEO) repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE229412.
Statistical analysis
Prism v6 (GraphPad Software) was used to perform Mann–Whitney U-test or student’s t test for statistical differences between groups, where noted. Error bars represent standard error of the mean (SEM). In general, a P value < 0.05 was considered as statistically significant (****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05).
Results
NKAP deficiency causes DNA damage and genome instability
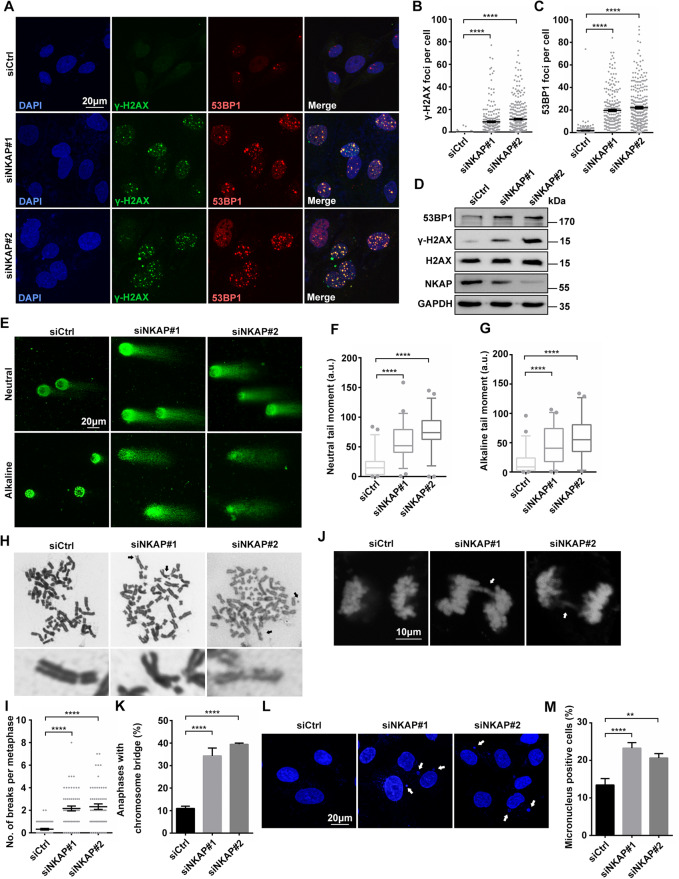
SUMOylated NKAP in mitosis was shown to be required for correct alignment of metaphase chromosomes [51]. Consistent with this, it was demonstrated that depletion of NKAP resulted in chromosomal instability and possibly contributed to tumorigenesis [51]. However, it remains unknown whether loss of NKAP triggers other events leading to genomic instability and tumorigenesis, independently of mitosis. In order to further illustrate the biological function of NKAP in human cells, we first asked whether it has a role in preventing DNA damage. For this purpose, we transiently reduced NKAP levels in U2OS cells by transfection with siRNAs and performed immunofluorescent staining to determine the levels of DNA damage, as detected by the formation of γH2AX and 53BP1 foci, two markers of DNA damage. Two NKAP siRNAs targeting different regions of the NKAP transcript were utilized, and the knockdown efficiency of these two siRNAs was confirmed by qRT-PCR and Western blot (Supplementary Fig. S1). Compared to the controls, depletion of NKAP by siRNA resulted in a significant increase in γH2AX and 53BP1 foci, indicating that NKAP depletion leads to increased DNA damage (Fig. 1A–C). Increased H2AX phosphorylation and 53BP1 expression were also detected by Western blot of the total lysates of NKAP knockdown cells (Fig. 1D). Importantly, the increased levels of γH2AX and 53BP1 seen with NKAP siRNA was rescued by expressing a siRNA-resistant NKAP, indicating reduced NKAP is responsible for the DNA damage phenotype (Supplementary Fig. S2). On the contrary, expression of a RS-domain mutated form of NKAP did not rescue the RNAi effect (Supplementary Fig. S3). Further, NKAP knockdown induced DNA damage was not specific to U2OS cells, as it was also observed in both HeLa cells and non-transformed hTert-RPE-1 cells (Supplementary Fig. S4). Consistently, NKAP depletion cells showed an accumulation of DNA breaks, as determined by the increased comet tail moment observed in both neutral and alkaline comet assays (Fig. 1E–G). Concomitant with intense DNA damage, NKAP depletion caused a significantly increase of chromosome breaks as compared with the control (Fig. 1H and I). Furthermore, anaphase bridges and micronuclei were increased in NKAP knockdown cells (Fig. 1J–M). We observed that a higher percentage of micronuclei associated with γH2AX signals upon NKAP knockdown (Supplementary Fig. S5). Altogether, these results suggest that loss of NAKP leads to accumulation of DNA damage and genome instability.
Fig. 1. Knockdown of NKAP leads to DNA damage and genome instability.
A–C Representative images (A) and quantification (B, C) of γH2AX and 53BP1 foci in U2OS cells following transfection with control or NKAP siRNAs for 48 h. The graph shows the number of foci per cell. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). D Western blot analysis using indicated antibodies. Lysates were prepared from cells following transfection with control or NKAP siRNAs for 48 h. GAPDH serves as a loading control. E–G Representative images (E) and quantification (F, G) of DNA breaks measured by neutral and alkaline comet assay in cells following transfection with control or NKAP siRNAs for 48 h. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). H and I Representative images (H) and quantification (I) of metaphase chromosomes in cells following transfection with control or NKAP siRNAs. Arrows indicate examples of chromosomes with breaks, and zoom in images are shown below. The graph shows the number of chromosome breaks per metaphase. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). J and K Representative images (J) and quantification (K) of chromosomes with anaphase bridges in cells following transfection with control or NKAP siRNAs for 48 h. The graph shows the percentage of anaphase cells with chromosome bridges. ****P < 0.0001 (paired Student’s t test). L and M Representative images (L) and quantification (M) of nuclei with micronuclei in cells following transfection with control or NKAP siRNAs for 48 h. The graph shows the percentage of cells containing micronuclei. ****P < 0.0001; **P < 0.01 (paired Student’s t test). Scale bar, 20 μm in (A, E and L); 10 μm in (J). a.u. indicates arbitrary units. Data are presented as Mean ± standard error of the mean (SEM).
R-loops drives DNA damage in NKAP depleted cells
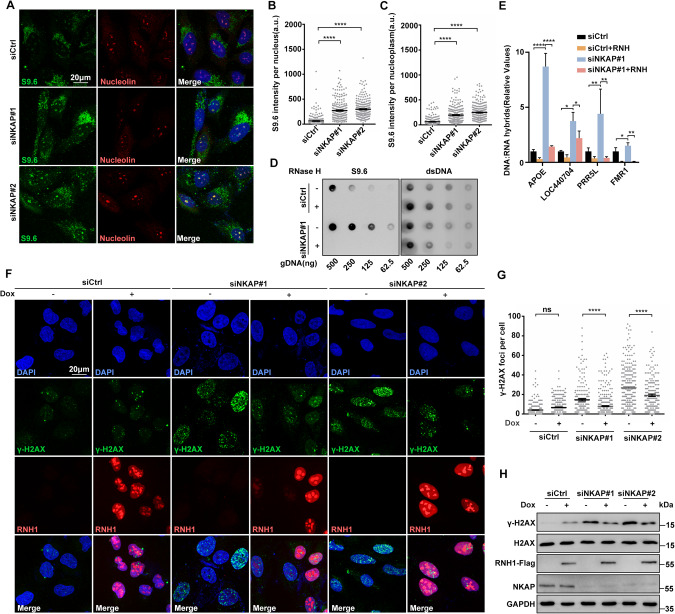
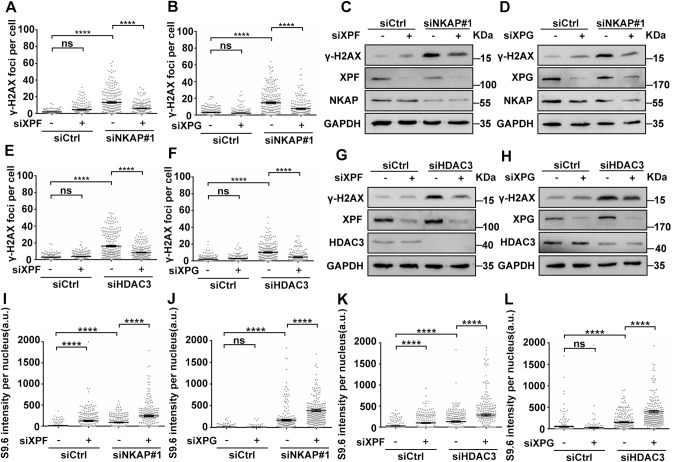
NKAP has been shown to be a part of the post-catalytic spliceosome (P-complex) that is crucial for pre-mRNA splicing [50]. Defects in RNA splicing factors, such as SLU7, lead to increased genomic instability attributed to the accumulation of R-loops [59]. Therefore, we hypothesized that the DNA damage and genome instability we observed in NKAP depleted cells was a result of increased R-loop formation. To test this, we performed immunofluorescent staining of RNA:DNA hybrids using the S9.6 monoclonal antibody, which recognizes RNA:DNA hybrids [60]. As compared to the control, NKAP knockdown in U2OS cells led to higher levels of RNA:DNA hybrids, indicating abnormal R-loop accumulation (Fig. 2A–C). Increased S9.6 staining was also observed in HeLa and hTERT-RPE-1 cells treated with siRNA targeting NKAP (Supplementary Fig. S6). Importantly, the S9.6 signals were significantly reduced upon the overexpression of RNaseH1 under the control of a doxycycline-inducible promoter, which degrades RNA:DNA hybrids (Supplementary Fig. S7A–C). We noticed that the cytoplasmic S9.6 signals were increased upon NKAP depletion, and this was alleviated under RNaseH1 overexpression (Supplementary Fig. S7D). Moreover, R-loop accumulation was observed in NKAP knockdown cells using a Dot blot assay (Fig. 2D). Given recent concerns about the specificity of imaging with S9.6, we also performed DNA:RNA immunoprecipitation experiments followed by quantitative PCR (DRIP-qPCR) at a set of R-loop prone loci [61, 62]. These includes APOE, LOC440704, PRP5L, and FMR1 genes. We found that depletion of NKAP by siRNA resulted in a significant increase in signals at these loci (Fig. 2E). In vitro RNaseH treatment abolished these changes in DRIP-qPCR signal observed upon NKAP knockdown, further confirming the signals were induced by RNA:DNA hybrids (Fig. 2E).
Fig. 2. NKAP depletion induced R-loop accumulation contributes to DNA damage.
A–C Representative images (A) and quantification (B and C) of S9.6 fluorescence staining in cells following transfection with control or NKAP siRNAs for 48 h. anti-Nucleolin antibody was used to detect Nucleolin and label the nucleolus. The graph shows the S9.6 fluorescence intensity per nucleus (B) and nucleoplasm (C). The value for the nucleoplasm was determined by the signal intensity per nucleus with subtraction of the nucleolar signal. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). D Dot blot analysis of R-loops using the S9.6 antibody. Genomic DNA was extracted from cells following transfection with control or NKAP siRNAs for 48 h. RNase H treatment was conducted alongside and used as a negative control. dsDNA was used as an internal sample loading control. E DRIP-qPCR analysis of the indicated loci in cells following transfection with control or NKAP siRNAs for 48 h. In vitro RNase H treatment was conducted prior to pulldown and served as a control. The graph shows the relative value of R-loop level in each gene loci which was normalized with respect control. ****P < 0.0001; **P < 0.01; *P < 0.05 (paired Student’s t test). F and G Representative images (F) and quantification (G) of γH2AX foci in cells following transfection with control or NKAP siRNAs for 48 h and subsequent treatment with (+) or without (−) doxycycline (DOX) to induce RNH1 overexpression. The graph shows the number of foci per cell. ***P < 0.001 (Mann– Whitney U-test, two-tailed). H Western blot analysis using indicated antibodies. Lysates were prepared from cells following transfection with control or NKAP siRNAs for 48 h and subsequent treatment with (+) or without (−) doxycycline (DOX) to induce RNH1 overexpression. Each value shows the quantification of γH2AX in relative to H2AX. GAPDH serves as a loading control. Scale bar, 20 μm. a.u. indicates arbitrary units. Data are presented as Mean ± standard error of the mean (SEM).
To further assess R-loop accumulation as a potential mechanism for driving DNA damage, we overexpressed RNaseH1 to resolve R-loops in NKAP knockdown cells and then examined DNA damage. RNaseH1 overexpression significantly reduced the number of γH2AX foci in NKAP depletion cells as compared to the control (Fig. 2F and G). Consistently, Western blot analysis of γH2AX levels in protein extracts from NKAP depletion cells with RNaseH1 overexpression also showed decreased levels of γH2AX (Fig. 2H). Moreover, our neutral and alkaline comet assays showed excess DNA damage in NKAP depletion cells was suppressed by overexpression of RNaseH1 (Supplementary Fig. S8). These data demonstrate that the increased level of R-loops is a driver of DNA damage in NKAP deficient cells.
NKAP depletion induced R-loops impair replication fork progression
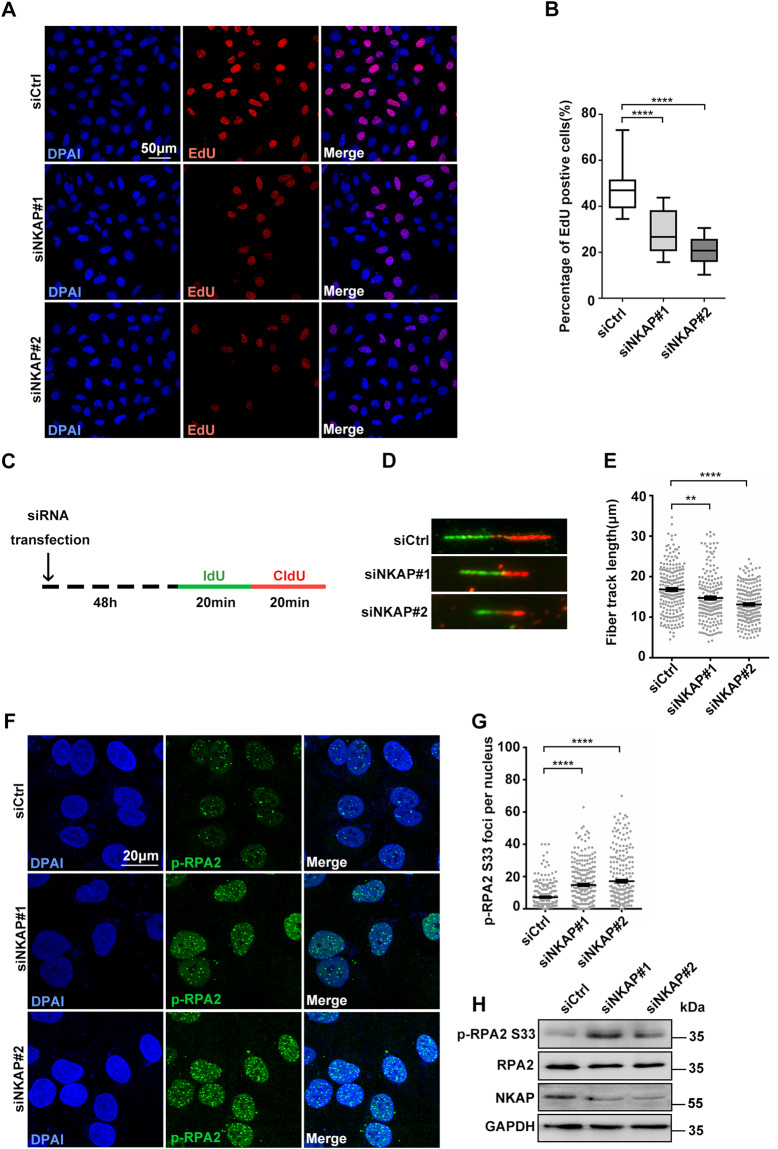
Given that accumulation of R-loops can lead to impairment of replication fork progression, we next examined the impact of NKAP depletion on DNA replication in U2OS cells. For this purpose, we first performed a 5-ethynyl-2′-deoxyuridine (EdU) incorporation experiment to measure overall DNA synthesis in NKAP depleted cells. The percentage of cells incorporating EdU was decreased in NKAP depleted cells compared to the control cells (Fig. 3A and B), indicating a reduction in DNA synthesis. Next, we used DNA fiber assays to determine the progression of DNA replication forks. Cells were double pulse-labeled with the thymidine analogs (5-iodo-2′-deoxyuridine (IdU) and 5-chloro-2′-deoxyuridine (CldU)) sequentially for 20 min each to allow the examination of replication fork movement (Fig. 3C). Measuring the lengths of both IdU and CldU tracks, we found that NKAP depleted cells had a significant reduction of average track length compared to the control cells (Fig. 3D and E), indicating that the replication forks moved slower upon NKAP knockdown. Furthermore, we examined the phosphorylation of RPA2 on serine 33, a marker of DNA replication stress, and found the number of pRPA2 S33 foci was elevated in NKAP depleted cells (Fig. 3F and G). Consistently, the level of pRPA2 S33 by Western Blot was higher in NKAP depleted cells (Fig. 3H). Importantly, overexpression of RNaseH1 suppressed all these effects (Supplementary Fig. S9), strongly suggesting that R-loops accumulated in NKAP depleted cells impair replication fork progression.
Fig. 3. Defective DNA replication in NKAP depletion cells.
A and B Representative images (A) and quantification (B) of EdU incorporation in cells following transfection with control or NKAP siRNAs for 48 h. The graph shows the percentage of EdU positive cells. ****P < 0.0001 (paired Student’s t test). C Schematic representation of the DNA fiber assay. Cells were transfected with control or NKAP siRNAs for 48 h and then incubated with 5′-iodo-2′-deoxyuridine (25 μM IdU, 20 min) followed by 5′-chloro-2′-deoxyuridine (250 μM CldU, 20 min) as indicated. D and E Representative images (D) and quantification (E) of DNA fibers in cells following transfection with control or NKAP siRNAs for 48 h. DNA fibers were labeled by IdU (Green) and CldU (Red). The graph shows the track length of DNA fibers. ****P < 0.0001; **P < 0.01 (Mann–Whitney U-test, two-tailed). F and G Representative images (F) and quantification (G) of p-RPA2 foci in cells following transfection with control or NKAP siRNAs for 48 h. The graph shows the number of p-RPA2 foci per nucleus. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). H Western blot analysis using indicated antibodies. Lysates were prepared from cells following transfection with control or NKAP siRNAs for 48 h. GAPDH serves as a loading control. Scale bar, 50 μm in (A); 20 μm in (F). a.u. indicates arbitrary units. Data are presented as Mean ± standard error of the mean (SEM).
R-loop accumulation and DNA damage in NKAP deficient cells are dependent on transcription
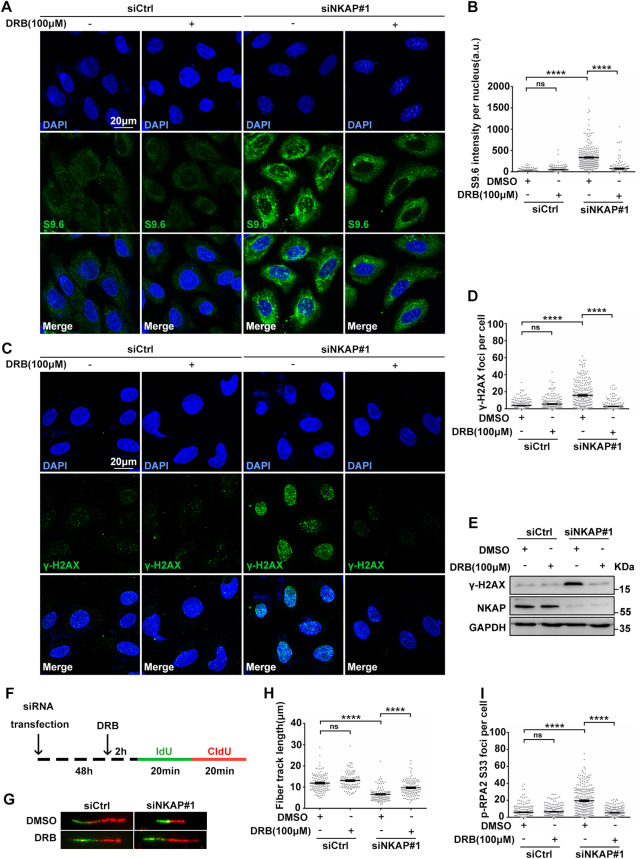
Previous studies have shown that R-loop accumulation is dependent on transcription, we next asked whether active transcription is involved NKAP depletion induced R-loop accumulation and DNA damage. For this purpose, we treated cells with 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB), a RNA polymerase II inhibitor, to block transcription and examined the effects. Inhibition of transcription by DRB reduced R-loop formation in NKAP depleted cells (Fig. 4A and B), indicating that R-loop accumulation is dependent on transcription. Moreover, DRB treatment suppressed the levels of DNA damage in NKAP depleted cells, as determined by the level of γH2AX in both immunofluorescence staining and Western blot experiments (Fig. 4C–E). In addition, we also performed DNA fiber assays to examine replication fork progression upon DRB treatment (Fig. 4F). Consistently, DRB partially reversed the effect of NKAP depletion on the fiber track length (Fig. 4G and H), supporting the notion that R-loop accumulation proceeds defective replication fork progression. Lastly, the increased number of p-RPA2 S33 loci in NKAP depleted cells was suppressed by DRB as well (Fig. 4I), confirming that the observed DNA stress is dependent on transcription. Taken together, these data demonstrate that NKAP depletion induced R-loop accumulation and DNA damage are dependent on transcription.
Fig. 4. NKAP depletion induced R-Loops accumulation and DNA damage are dependent on transcription.
A and B Representative images (A) and quantification (B) of S9.6 fluorescence staining in cells following transfection with control or NKAP siRNAs for 46 h and subsequent treatment with or without DRB (100 μM) for 2 h. The graph shows the S9.6 intensity per nucleus. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). C and D Representative images (C) and quantification (D) of γH2AX foci in cells following transfection with control or NKAP siRNAs for 46 h and subsequent treatment with or without DRB (100 μM) for 2 h. The graph shows the number of γH2AX foci per cell. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). E Western blot analysis using indicated antibodies. Lysates were prepared from cells following transfection with control or NKAP siRNAs for 46 h and subsequent treatment with or without DRB (100 μM) for 2 h. GAPDH serves as a loading control. F Schematic representation of the DNA fiber assay. Cells were transfected with control or NKAP siRNAs for 46 h and then treated with or without DRB (100 μM) for 2 h before incubating with IdU (25 μM, 20 min) followed by CldU (250 μM, 20 min) as indicated. G and H Representative images (G) and quantification (H) of DNA fibers in cells following transfection with control or NKAP siRNAs for 46 h and subsequent treatment with or without DRB (100 μM) for 2 h. DNA fibers were labeled by IdU (Green) and CldU (Red). The graph shows the track length of DNA fibers. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). I Quantification of p-RPA2 foci in cells following transfection with control or NKAP siRNAs for 46 h and subsequent treatment with or without DRB (100 μM) for 2 h. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). Scale bar, 20 μm. a.u. indicates arbitrary units. Data are presented as Mean ± standard error of the mean (SEM).
The NKAP-interacting protein HDAC3 regulates R-loop homeostasis
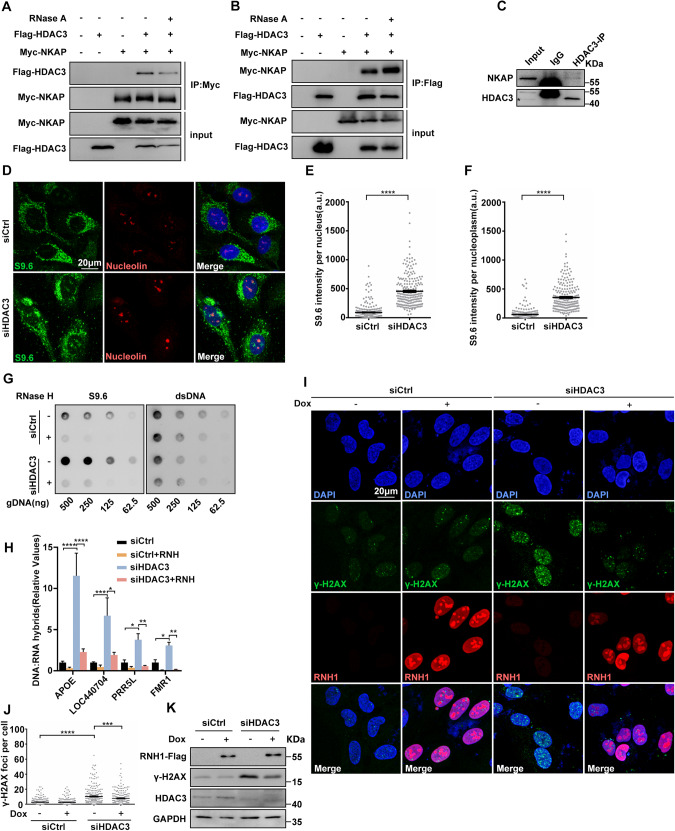
To further explore the function of NKAP in controlling R-loop homeostasis, we searched for NKAP-interacting proteins from the literatures. The histone deacetylase 3 (HDAC3) causes our attention, as it forms a complex with NKAP and regulates transcription [44]. Previous studies have identified the interaction between NKAP and HDAC3 as well as their roles in transcription regulation [44, 46]. Interestingly, HDAC3 is known to prevent DNA damage, while the underlying mechanism was unknown [63, 64]. Thus, we suspected that NKAP functions in R-loop homeostasis and maintenance of genome stability are involved in HDAC3. We first confirmed the physical association between NKAP and HDAC3 in U2OS cells. Co-immunoprecipitation (Co-IP) assays revealed that NKAP interacted with HDAC3 when ectopically expressed (Fig. 5A). In a reverse Co-IP experiment, the interaction between NKAP and HDAC3 was also observed (Fig. 5B). An additional Co-IP experiment further verified the interaction between endogenous NKAP and HDAC3 (Fig. 5C). Moreover, treatment with RNase A did not abolish these interactions (Fig. 5A and B). These results confirm the interaction of NKAP and HDAC3, and further suggest that the interaction is not dependent on RNA.
Fig. 5. Knockdown of HDAC3 results in R-loop accumulation that triggers DNA damage.
A and B Western blot analysis of co-IP between Myc-NKAP and Flag-HDAC3 proteins expressed in HEK293T cells. The IP was performed using anti-Myc (A) or anti-Flag (B) antibodies with cell lysates treated with (+) or without (−) RNase A. C Western blot analysis of co-IP between endogenous HDAC3 and NKAP proteins in U2OS cells. The IP was performed using anti-HDAC3 antibody with cell lysates. D–F Representative images (D) and quantification (E and F) of S9.6 fluorescence staining in cells following transfection with control or HDAC3 siRNAs for 48 h. anti-Nucleolin antibody was used to detect Nucleolin and label the nucleolus. The graph shows the S9.6 fluorescence intensity per nucleus (E) and nucleoplasm (F). The value for the nucleoplasm was determined by the signal intensity per nucleus with subtraction of the nucleolar signal. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). G Dot blot analysis of R-loops using the S9.6 antibody. Genomic DNA was extracted from cells following transfection with control or HDAC3 siRNAs for 48 h. RNase H treatment was conducted alongside and used as a negative control. dsDNA was used as an internal sample loading control. H DRIP-qPCR analysis of the indicated loci in cells following transfection with control or HDAC3 siRNAs for 48 h. In vitro RNase H treatment was conducted prior to pulldown and served as a control. The graph shows the relative value of R-loop level in each gene loci which was normalized with respect control. ****P < 0.0001; **P < 0.01; *P < 0.05 (paired Student’s t test). I and J Representative images (I) and quantification (J) of γH2AX foci in cells following transfection with control or HDAC3 siRNAs for 48 h and subsequent treatment with (+) or without (−) doxycycline (DOX) to induce RNH1 overexpression. The graph shows the number of foci per cell. ****P < 0.0001; ***P < 0.001 (Mann–Whitney U-test, two-tailed). K Western blot analysis using indicated antibodies. Lysates were prepared from cells following transfection with control or HDAC3 siRNAs for 48 h and subsequent treatment with (+) or without (−) doxycycline (DOX) to induce RNH1 overexpression. GAPDH serves as a loading control. Scale bar, 20 μm. a.u. indicates arbitrary units. Data are presented as Mean ± standard error of the mean (SEM).
Next, we determined whether HDAC3 functions to regulate R-loop homeostasis as NKAP. Knockdown of HDAC3 by siRNA led to a significant increase of S9.6 signals compared to the control (Fig. 5D–F). Importantly, this increase was suppressed by RNaseH1 overexpression (Supplementary Figs. S10A and B). A Dot blot analysis showed that similar R-loop accumulation was observed in HDAC3 knockdown cells (Fig. 5G). We also used DRIP-qPCR assay to detect R-loop accumulation in APOE, LOC440704, PRP5L, and FMR1 genes, and found that higher levels of R-loops in HDAC3 depleted cells compared to the control cells (Fig. 5H). RNaseH treatment also suppressed these effects (Fig. 5H). Together, these data support that HDAC3 is a novel regulator of R-loop homeostasis. As it has been shown that loss of HDAC3 causes DNA damage, we then want to examine the potential association between R-loop accumulation and DNA damage in HDAC3 depleted cells. Similar to previously studies, elevated levels of DNA damage were observed in HDAC3 depleted cells, as characterized by the higher number of γ-H2Ax loci and increased level of γ-H2Ax (Fig. 5I–K). Interestingly, overexpression of RNaseH1 suppressed these effects, suggesting that R-loop accumulation contributes to DNA damage in HDAC3 deficient cells (Fig. 5I–K). Moreover, the average track length of DNA fibers was reduced in HDAC3 depleted cells, which was rescued by overexpression of RNaseH1 (Supplementary Fig. S10C and D). Both the number of p-RPA2-S33 foci and level of p-RPA2-S33 were increased upon HDAC3 knockdown and this was suppressed by RNaseH1 overexpression (Supplementary Fig. S10E and F). Therefore, these results demonstrate that HDAC3 displays a similar function with NAKP in preventing R-loop accumulation and DNA damage.
Disruption of NKAP/HDAC3 complex triggers R-loop accumulation and DNA damage
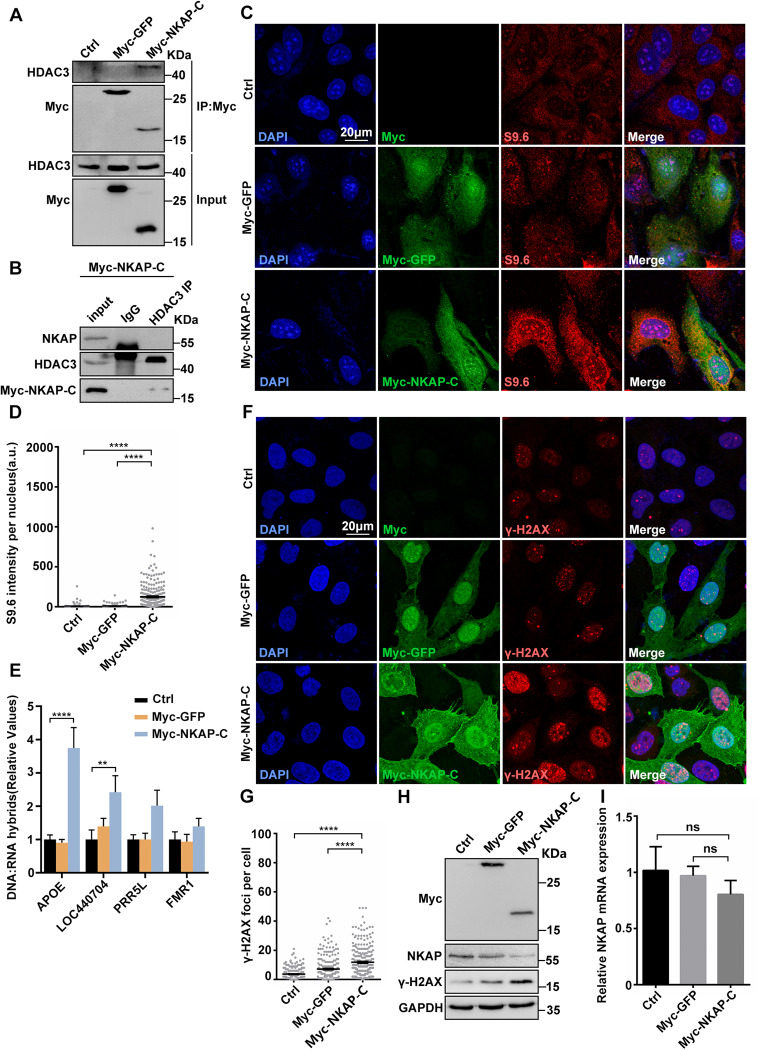
It has been previously shown that the C-terminal DUF926 domain of NKAP specifically binds to HDAC3 [44]. To illustrate the importance of NKAP-HDAC3 interaction in preventing R-loop accumulation and genome instability, we next overexpressed this C-terminal DUF926 domain of NKAP (Myc-NKAP-C), which was supposed to disrupt the NKAP/HDAC3 complex. To verify this, we first performed the co-IP experiment to assess the interaction between the Myc-NKAP-C fragment and HDAC3. Indeed, the Myc-NKAP-C fragment interacted with HDAC3 in the extracts from Myc-NKAP-C overexpressing cells, but not from control cells (Fig. 6A). Consistently, native NKAP and HDAC3 interaction was not detected in the presence of overexpressed Myc-NKAP-C (Fig. 6B). Importantly, overexpression of Myc-NKAP-C resulted in a significant increase of S9.6 signals as compared to the control (Fig. 6C and D). DRIP-qPCR analysis further revealed that higher signals were detected at those previously examined R-loop prone loci when the C-terminal fragment of NKAP was overexpressed and the value for two loci were statistically significant (Fig. 6E). Thus, the disruption of HDAC3/NKAP complex leads to the accumulation of R-loops. Furthermore, the higher number of γ-H2Ax foci and increased levels of H2AX were observed when overexpressing C-terminal fragment of NKAP, indicating that the disruption of HDAC3/NKAP complex also causes DNA damage (Fig. 6F–H). We noticed that overexpression of the C-terminal fragment of NKAP led to a reduction of NKAP protein level (Fig. 6H). This NKAP downregulation was not due to reduction of NKAP mRNA transcription, as determined by the RT-qPCR analysis (Fig. 6I). Together, these findings support the notion that the intact HDAC3/NKAP complex is crucial for preventing R-loop accumulation and DNA damage.
Fig. 6. Overexpression of NKAP C-terminal DUF926 domain causes R-loop accumulation and DNA damage.
A Western blot analysis of co-IP between HDAC3 and Myc-GFP or Myc-NKAP-C expressed in HEK293T cells. Lysates were prepared from cells transfected with control, pCMV-Myc-GFP or pCMV-Myc-NKAP-DUF926 plasmids. The IP was performed using anti-Myc antibody with cell lysates. B Western blot analysis of co-IP between endogenous HDAC3 and NKAP proteins in pCMV-Myc-NKAP-DUF926 overexpression U2OS cells. The IP was performed using anti-HDAC3 antibody with cell lysates. C and D Representative images (C) and quantification (D) of S9.6 fluorescence staining in cells transfected with control, pCMV-Myc-GFP or pCMV-Myc-NKAP-DUF926 plasmids. The graph shows the S9.6 fluorescence intensity per nucleus. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). E DRIP-qPCR analysis of the indicated loci in cells following transfection with control, pCMV-Myc-GFP or pCMV-Myc-NKAP-DUF926 plasmids. The graph shows the relative value of R-loop level in each gene loci which was normalized with respect control. ****P < 0.0001; **P < 0.01; *P < 0.05 (paired Student’s t test). F and G Representative images (F) and quantification (G) of γH2AX foci in cells transfected with control, pCMV-Myc-GFP or pCMV-Myc-NKAP-DUF926 plasmids. The graph shows the number of foci per cell. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). H Western blot analysis using indicated antibodies. Lysates were prepared from cells transfected with control, pCMV-Myc-GFP or pCMV-Myc-NKAP-DUF926 plasmids. GAPDH serves as a loading control. I RT-qPCR analysis of NKAP and HDAC3 mRNA levels. Total RNAs were isolated from cells following transfection with control, pCMV-Myc-GFP or pCMV-Myc-NKAP-DUF926 plasmids. GAPDH serves as a control. ***P < 0.001(paired Student’s t test). Scale bar, 20 μm. a.u. indicates arbitrary units. Data are presented as Mean ± standard error of the mean (SEM).
NKAP stability is regulated by HDAC3
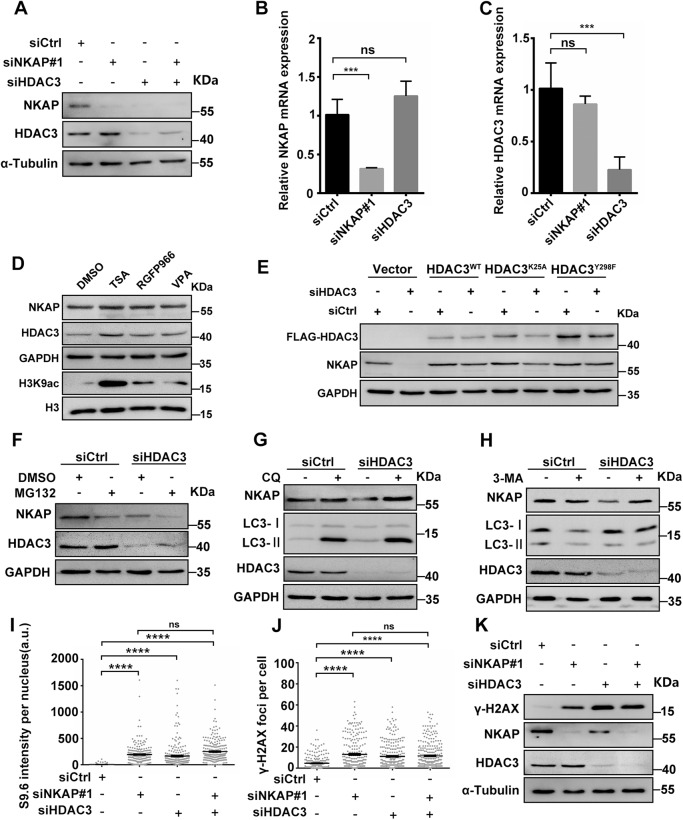
Given that HDAC3 and NKAP exist in a stable protein complex to regulate R-loop homeostasis and DNA damage, we next wanted to assess whether their respective protein stabilities might be affected by each other. Western blot analysis revealed that NKAP protein levels were dramatically decreased in cells depleted of HDAC3, while HDAC3 protein levels were not obviously altered when NKAP was depleted (Fig. 7A). This is consistent with our previous results in cells overexpressing C-terminal fragment of NKAP (Fig. 6H). We further performed RT-qPCR analysis to measure mRNA levels, and found that the mRNA level of NKAP was not reduced in HDAC3 depleted cells (Fig. 7B). In addition, NKAP depletion did not affect HDAC3 mRNA levels (Fig. 7C). Thus, these data suggest that HDAC3 facilitates NKAP protein stability by forming a protein complex.
Fig. 7. NKAP protein is stabilized by HDAC3 independent of its deacetylase activity.
A Western blot analysis using indicated antibodies. Lysates were prepared from cells following transfection with control, NKAP, HDAC3 or a combination of both siRNAs for 48 h. α-Tubulin serves as a loading control. B and C RT-qPCR analysis of NKAP and HDAC3 mRNA levels. Total RNAs were isolated from cells following transfection with control, NKAP or HDAC3 siRNAs for 48 h. GAPDH serves as a control. ***P < 0.001(paired Student’s t test). D Western blot analysis using indicated antibodies. Lysates were prepared from control (DMSO) cells or cells treated with Trichostatin A (TSA, 5 μM), Valproic acid (VPA, 0.5 mM), or RGFP966 (10 μM) for 24 h. GAPDH serves as a loading control. E Western blot analysis using indicated antibodies. Lysates were prepared from siCtrl or siHDAC3 cells transfected with empty vector, siRNA-resistant form of Flag-HDAC3WT, Flag-HDAC3K25A or Flag-HDAC3Y298F plasmids. GAPDH serves as a loading control. F Western blot analysis using indicated antibodies. Lysates were prepared from cells following transfection with control or HDAC3 siRNAs for 42 h and subsequent treatment with DMSO or MG132 (5 μM) for 6 h. GAPDH serves as a loading control. G Western blot analysis using indicated antibodies. Lysates were prepared from cells following transfection with control or HDAC3 siRNAs for 24 h and subsequent treatment with or without Chloroquine (CQ, 20 μM) for 24 h. GAPDH serves as a loading control. H Western blot analysis using indicated antibodies. Lysates were prepared from cells following transfection with control or HDAC3 siRNAs for 24 h and subsequent treatment with or without 3-Methyladenine (3-MA, 5 mM) for 24 h. GAPDH serves as a loading control. I Quantification of S9.6 fluorescence staining in cells following transfection with control, NKAP, HDAC3 or a combination of both siRNAs for 48 h. The graph shows the S9.6 intensity per nucleus. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). J Quantification of γH2AX foci in cells following transfection with control, NKAP, HDAC3 or a combination of both siRNAs for 48 h. The graph shows the number of γH2AX foci per cell. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). K Western blot analysis using indicated antibodies. Lysates were prepared from cells following transfection with control, NKAP, HDAC3 or a combination of both siRNAs for 48 h. α-Tubulin serves as a loading control. a.u. indicates arbitrary units. Data are presented as Mean ± standard error of the mean (SEM).
As HDAC3 functions majorly as lysine deacetylases to target histones as well as nonhistone proteins, it is conceivable that the stabilization of NKAP by HDAC3 is mediated by lysine deacetylation. To test this idea, we first asked whether the enzyme activity of HDAC3 is responsible for the protein stability of NKAP. Treatment of wild type U2OS cells with the pan-HDAC inhibitor trichostatin A (TSA) or a selective class I HDAC inhibitor Valproic acid (VPA) did not lead to reduction of NKAP protein levels (Fig. 7D). Using a highly selective HDAC3 inhibitor RGFP966 also did not cause changes in NKAP protein levels (Fig. 7D). Western blot analysis showed higher levels of H3K9ac upon treatment of the inhibitors, confirming the inhibition of HDAC3 activity by these inhibitors (Fig. 7D). These data suggest that the enzyme activity of HDAC3 is not required for controlling NKAP protein stability. To further verify this, we constructed HDAC3 wild type and deacetylase deficient mutant plasmid and performed the rescue experiments in HDAC3 depleted cells. While overexpression of HDAC3 wild type plasmid rescued the effect of HDAC3 depletion on NKAP protein level, similar rescue was also observed when overexpressing HDAC3 with K25A or Y298F mutations, which have been reported to abolish HDAC3 deacetylase enzymatic activity (Fig. 7E) [65].
To further explore the possible mechanism underlying the maintenance of NKAP protein stability, we examined the proteasomal and lysosomal protein degradation pathways. Inhibition of the proteasome by MG132 did not restore NKAP levels in HDAC3 depleted cells (Fig. 7F). We noticed that a further reduction of NKAP levels upon MG132 treatment, although the reason for this was unclear. Interestingly, inhibition of the lysosome by chloroquine (CQ) restored the levels of NKAP in HDAC3 depleted cells (Fig. 7G). Similarly, treatment with another autophagy inhibitor 3-Methyladenine (3-MA) also brought up NKAP levels in HDAC3 depleted cells (Fig. 7H). Therefore, these findings suggest that the degradation of NKAP acts through the lysosomal pathway in HDAC3 deficient cells.
The above data suggest that HDAC3 may act upstream of NKAP within the same pathway to regulate R-loop homeostasis and DNA damage. To confirm this, HDAC3 and NKAP were depleted separately or together to allow us to examine the resulting effects. Compared with either HDAC3 or NKAP depletion alone, combined depletion resulted in a similar accumulation of R-loops (Fig. 7I). Furthermore, a similar increase of DNA damage was also observed in cells depleted of HDAC3, NKAP alone or together (Fig. 7J and K). To further support the role of HDAC3 in controlling R-loops through stabilizing NKAP, we performed the NKAP overexpression experiment. We found that overexpression of NKAP was able to reduce R-loops and DNA damage in HDAC3 knockdown cells, as revealed by the staining of S9.6 and γH2AX (Supplementary Fig. S11A–D). Elevated γH2AX levels was confirmed by western blot analysis (Supplementary Fig. S11E). Together, these data show that HDAC3 controls NKAP protein stability, which is mediated by the lysosomal pathway.
NKAP prevents R-loop formation by regulating RNA polymerase II pausing
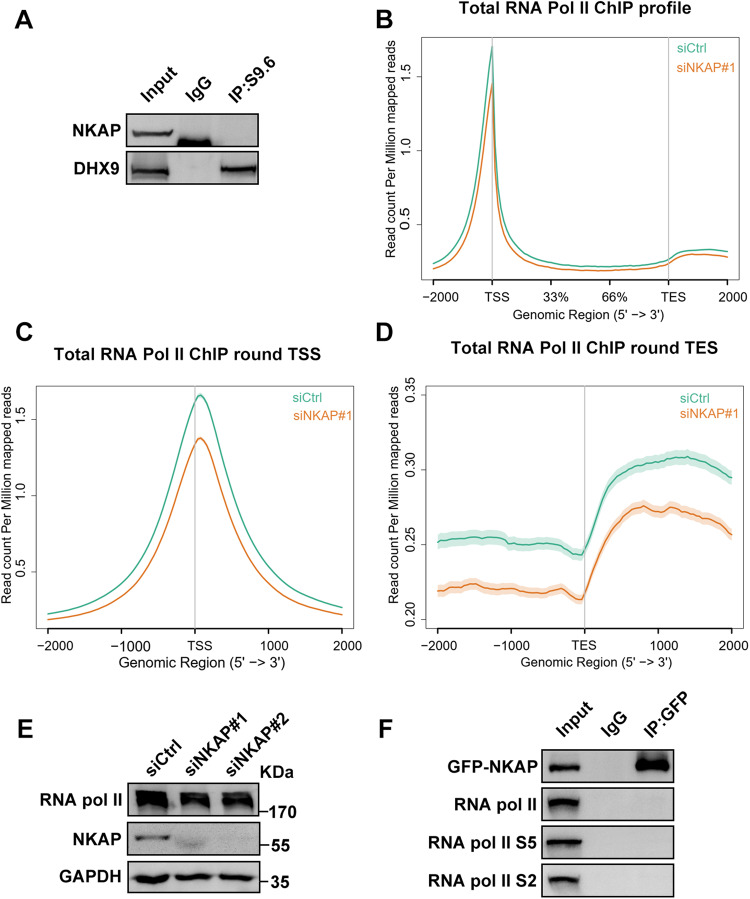
As the impact of HDAC3 on R-loops is mediated through the stability of NKAP, we then focused on NKAP and explored the potential role of NKAP in regulating R-loop homeostasis. In order to do this, we performed a series of experiments. First, we examined the interaction between NKAP and R-loops using the coimmunorecipitation (Co-IP) assay, and the result showed that there was no detectable interaction between them (Fig. 8A). As a control, DHX9 interacted with R-loops as reported previously [66] (Fig. 8A). This led us to conclude that NKAP likely functions in preventing R-loop formation but not the resolution of the R-loops. As NKAP has been previously shown to be part of the splicing complex, we consider that R-loop accumulation in NKAP depletion cells might be due to the defects in coupling between RNA splicing and transcription [50]. It has been proposed that R-loop accumulation often occurs following RNA polymerase II pausing at transcription start sites [21, 67–69]. To test this possibility, we performed ChIP-Seq analysis with an antibody against RNA polymerase II. We found that the distribution of Pol II signals was decreased at the transcription start sites (TSSs) and transcription end sites (TESs) upon depletion of NKAP (Fig. 8B–D). Moreover, knockdown of NKAP did not reduce the protein level of Pol II, ruling out the possible effect of reduced expression of Pol II (Fig. 8E). The reduction in Pol II occupancy suggests that a potential mechanism by which NKAP prevents R-loop formation by maintaining the duration of Pol II pausing. To further characterize the relationship between NKAP and Pol II, we tested the possible interaction between NKAP and Pol II as well as the Ser2 and Ser5 phosphorylated forms of Pol II. Our Co-IP analysis showed that there were no detectable interactions between NKAP and various forms of Pol II, indicating that the connection between NKAP and Pol II might be indirect (Fig. 8F). Taken together, we suggest that stimulation of Pol II elongation promotes R-loop accumulation in NKAP deficient cells.
Fig. 8. Depletion of NKAP reduces Pol II occupancy at TSS and TES sites.
A Western blot analysis of RNA:DNA hybrid IP using anti-NKAP or anti-DHX9 antibodies. The IP was performed using S9.6 antibodies with cell lysates. B Average distribution of total RNA Pol II from transcription start site (TSS) to transcription end site (TES) of all RefSeq transcripts in control and NKAP knockdown cells. C Average distribution of total RNA Pol II around TSS in control and NKAP knockdown cells. D Average distribution of total RNA Pol II around TES in control and NKAP knockdown cells. E Western blot analysis using indicated antibodies. Lysates were prepared from cells following transfection with control or NKAP siRNAs for 48 h. GAPDH serves as a loading control. F Western blot analysis of GFP-NKAP IPs using antibodies against total RNA Pol II, RNA Pol II S5 or RNA Pol II S2. The IP was performed using anti-GFP antibodies with cell lysates.
R-loops accumulated in NKAP or HDAC3 deficient cells are processed by XPF and XPG
It has been shown that R-loops can be actively processed into DNA double-strand breaks by the transcription coupled nucleotide excision repair endonucleases XPF and XPG [70]. To explore the roles of XPF and XPG in the processing of R-loops, we tested whether these two endonucleases are responsible for R-loop associated DNA damage observed in NKAP or HDAC3-deficient cells. If XPF or XPG indeed has a role in processing NKAP or HDAC3-deficiency-induced R-loop into double-strand DNA break, then we would expect to see a reduction in DNA damage and an increase in R-loop levels in NKAP or HDAC3 knockdown cells with concomitant XPF or XPG knockdown. Depletion of XPF or XPG by siRNA clearly reduced the number of γH2AX foci observed upon knockdown of NKAP (Fig. 9A and B, and Supplementary Fig. S12A and B). Consistently, Western blot analysis confirmed that XPF or XPG knockdown decreased the induction of γH2AX in NKAP deficient cells (Fig. 9C and D). Similar results were obtained upon concurrent knockdown of XPF or XPG with HDAC3 (Fig. 9E–H, and Supplementary Fig. S13A and B). It is possible that the effects of XPF and XPG knockdown were indirect due to reduced DNA replication and overall transcription. To rule out this possibility, we first used flow cytometry-based DNA content analysis to examine cell cycle progression in XPF and XPG knockdown cells. As compared to the control, knockdown of XPF did not cause alterations in cell cycle regulation (Supplementary Fig. S14A and B). A slight increase in cell population in the S phase was observed in XPG knockdown cells with a concomitant decrease in the G1-phase population (Supplementary Fig. S14A and B). In addition, we used 5-ethynyluridine (EU) incorporation assay to visualize overall transcription. The results showed that the EU signal was comparable between control and XPF knockdown cells (Supplementary Fig. S14C and D). We also noticed that the EU signal intensity was slightly higher in XPG knockdown cells as compared to the control (Supplementary Fig. S14C and D). These data together indicate that knockdown of XPF or XPG does not reduce DNA replication or transcription, further supporting their direct roles in NKAP and HDAC3 deficient cells.
Fig. 9. R-loops are processed into DNA double-strand breaks by XPF and XPG in NKAP or HDAC3 depletion cells.
A Quantification of γH2AX foci in cells following transfection with control, XPF, NKAP or a combination of both siRNAs for 48 h. The graph shows the number of foci per cell. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). B Quantification of γH2AX foci in cells following transfection with control, XPG, NKAP or a combination of both siRNAs for 48 h. The graph shows the number of foci per cell. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). C and D Western blot analysis using indicated antibodies. Lysates were prepared from cells as in (A and B), respectively. GAPDH serves as a loading control. E Quantification of γH2AX foci in cells following transfection with control, XPF, HDAC3 or a combination of both siRNAs for 48 h. The graph shows the number of foci per cell. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). F Quantification of γH2AX foci in cells following transfection with control, XPG, HDAC3 or a combination of both siRNAs for 48 h. The graph shows the number of foci per cell. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). G and H Western blot analysis using indicated antibodies. Lysates were prepared from cells as in E and F, respectively. GAPDH serves as a loading control. I–L Quantification of S9.6 fluorescence staining in cells following transfection with indicated siRNAs for 48 h. The graph shows the S9.6 intensity per nucleus. ****P < 0.0001 (Mann–Whitney U-test, two-tailed). a.u. indicates arbitrary units. Data are presented as Mean ± standard error of the mean (SEM).
Next, we examined the levels of R-loops after codepletion of either XPF or XPG with NKAP. More R-loops were accumulated when either XPF or XPG was knocked down with NKAP compared to knockdown of NKAP alone (Fig. 9I and J, and Supplementary Fig. S15A and B). We noticed that R-loops were slightly increased in XPF knockdown cells (Fig. 9I and J, and Supplementary Fig. S15A). Similar effects on R-loop accumulation were observed when XPF or XPG was codepleted with HDAC3 (Fig. 9K and L, and Supplementary Fig. S16A and B). Together, these results indicate that XPF and XPG are responsible for NKAP/HDAC3 depletion induced DNA damage through the processing of R-loops.
Discussion
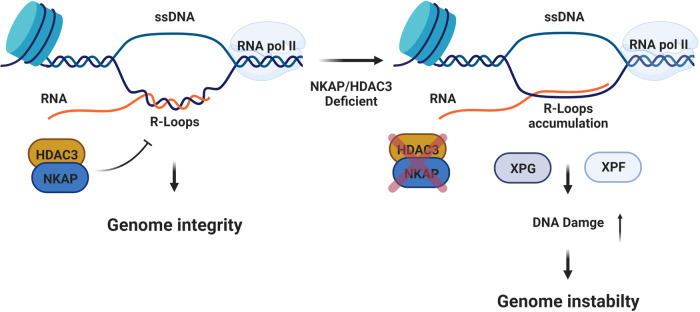
In this study, we show that NKAP has a function in preventing R-loop associated genome instability. Moreover, loss of NKAP results in DNA replication stress mediated by the formation of R-loops. Consistent with the association between NKAP and HDAC3, depletion of HDAC3 leads to R-loop mediated genome instability and DNA replication stress. Interestingly, we show that HDAC3 regulates NKAP stability in a deacetylase independent manner. Furthermore, we provide evidence that NKAP functions to prevent R-loop formation through regulating RNA polymerase II pausing. Last, we find that R-loops induced by NKAP or HDAC3 depletion are processed by the endonuclease XPF and XPG. These results not only provide new insights into the function of NKAP and HDAC3 in R-loop metabolism, but also support a model in which the NKAP/HDAC3 complex prevent DNA replication stress and open the possibility of using R-loop regulators to develop anticancer agents (Fig. 10).
Fig. 10. Working model showing the role of NKAP and HDAC3 in preventing R-loop formation and genome instability.
In wild type cells, NKAP and HDAC3 form a complex to block unscheduled R-loop formation. This will help to maintain genome integrity. However, this mechanism is defective in NKAP or HDAC3 deficient cells. Thus, accumulated R-loops results in increased DNA damage and genome instability. Both XPG and XPF are crucial for processing aberrant R-loops. The figure was created with Biorender.com.
NKAP has an RS domain at the N-terminus and plays important roles in splicing [43]. Previous studies have linked RNA splicing factors with R-loop associated genome instability [21, 25]. Consistent with this, NKAP depletion cells accumulate DNA breaks monitored as H2AX foci and R-loops as stained with S9.6 antibody, and also show chromosome bridges and micronuclei. Importantly, accumulation of DNA breaks can be partially suppressed by the overexpression of RNase H1, indicating that R-loops are responsible for the genomic instability generated in NKAP knockdown cells. In addition, accumulation of R-loops and DNA damage in NKAP deficient cells also depends on transcription, indicating that active transcription is a feature of R-loop occurring regions in the genome. NKAP is one component of the human spliceosome and interacts with other splicing factors [43, 50]. We have shown that the RNA polymerase II occupancy is reduced in NKAP depletion cells, and this indicates that stimulation of Pol II elongation might mediate the effect on aberrant R-loop formation. This is consistent with the idea that the coordination between RNA splicing and transcriptional elongation is essential for preventing R-loop formation [71, 72]. However, the interaction between NKAP and Pol II is not detected, indicating that other factors are involved in the connection between NKAP and Pol II. The regulation of R-loop homeostasis by splicing factors is also involved in other mechanisms. For instance, a recent study reports that the splicing factor XAB2 interacts with XPF and XPG and the protein complex is recruited on R-loops under conditions that favor R-loop formation [73]. It remains unknown whether XAB2 also regulates Pol II pausing. In NKAP depletion cells, we observe the increase of nuclear R-loops as well as cytoplasmic R-loops. This is consistent with recent findings that cytoplasmic R-loops and ssDNA moieties are triggered by the formation of nuclear R-loops [74, 75]. Importantly, it has been shown that these cytoplasmic R-loops and ssDNA moieties can active an immune response [74, 75]. Further studies are needed to examine whether cytoplasmic R-loops in NKAP depletion cells also elicit such an immune response.
The NKAP/HDAC3 complex has been extensively studied in the immune system through the conditional knockout mice [44, 46, 76, 77]. Similar phenotypes are observed when either NKAP or HDAC3 are deleted, supporting the importance of their interactions [44, 46, 76, 77]. It has previously been known that depletion of HDAC3 leads to DNA damage and genome instability [63, 64]. Consistent with this, we show that loss of HDAC3 causes DNA damage in human U2OS cells. Importantly, we also find that HDAC3 depletion causes R-loop accumulation and overexpression of RNase H1 could partially suppress DNA damage in HDAC3 deficient cells. These findings provide novel molecular mechanism underlying the role of HDAC3 in preventing DNA damage and maintaining genome integrity. Disrupting the interaction between NKAP and HDAC3 could give rise to similar phenotypes as in NKAP or HDAC3 depletion cells, further supporting the integrity of the NKAP/HDAC3 complex is essential for preventing R-loop associated genome instability. Interestingly, we find that NKAP protein levels are reduced in the absence of HDAC3, indicating a potential regulation of NKAP by HDAC3. Our detailed analysis reveals that the regulation of NKAP by HDAC3 does not dependent on the deacetylase activity and key residues involved in deacetylation function of HDAC3. These findings support the notion that HDAC3 could function to stabilize NKAP through a deacetylase independent mechanism. We provide evidence that overexpression of NKAP was able to reduce R-loops and DNA damage in HDAC3 deficient cells, further supporting the role of HDAC3 in controlling R-loop homeostasis through stabilizing NKAP. We show that the degradation of NKAP in HDAC3 deficient cells could be mediated by the lysosome/autophagy pathway. However, the detailed molecular mechanism requires further investigation.
We have shown that DNA damage defects are partially rescued in cells depleted for NKAP or HDAC3 when XPF or XPG is knocked down. Furthermore, the levels of R-loops are further increased in these cells. These findings support that endonucleases XPF and XPG function to process unscheduled R-loops into DNA breaks in NKAP or HDAC3 deficient cells. Notably, while knockdown of XPG does not alter R-loop levels, XPF knockdown alone causes a slightly increase in R-loop accumulation. One possible explanation could be that XPF also plays a role in processing naturally occurring R-loops.
Consequently, we propose a model in which NKAP acts with HDAC3 in preventing R-loop associated genome instability (Fig. 10). NKAP and HDAC3 form a complex and associate with the nascent RNA directly or indirectly during transcription, such association protects the genome from accumulation of aberrant R-loops. It would be interesting to explore the genomic distribution of these R-loops structures in NKAP or HDAC3 deficient cells in the future. Complete understanding of the deregulation of NKAP and HDAC3 leading to the accumulation of R-loops in human cells could open new direction for designing new therapeutic approaches in cancer treatment.
Supplementary information
Acknowledgements
We thank Prof Jun Huang for providing S9.6 antibodies during initial studies. We are grateful to Profs Jun Ma, Chih-Hung Hsu, Feng He, Pengfei Xu, Hongqing Liang and Tinggang Chew for helpful discussions.
Author contributions
WG and WL contributed to the study design. XZ performed all the experiments, prepared the figures and analyzed data with the help from JD, YL, XJ and CW. WG, WL, and XY provided resource and supervision. XZ and WG wrote the paper with input from all authors.
Funding
This study was supported by the National Natural Science Foundation of China (grant 31970668) and the National Key Research and Development Program of China (2018YFC1003200).
Data availability
The authors declare that [the/all other] data supporting the findings of this study are available within the paper [and its supplementary information files].
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Weiguo Lu, Email: lbwg@zju.edu.cn.
Wanzhong Ge, Email: wanzhongge@zju.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01182-5.
References
- 1.Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–24. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet. 2015;16:583–97. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 3.García-Muse T, Aguilera A. R Loops: From Physiological to Pathological Roles. Cell. 2019;179:604–18. doi: 10.1016/j.cell.2019.08.055. [DOI] [PubMed] [Google Scholar]
- 4.Grunseich C, Wang IX, Watts JA, Burdick JT, Guber RD, Zhu Z, et al. Senataxin Mutation Reveals How R-Loops Promote Transcription by Blocking DNA Methylation at Gene Promoters. Mol Cell. 2018;69:426–37. doi: 10.1016/j.molcel.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crossley MP, Bocek M, Cimprich KA. R-Loops as Cellular Regulators and Genomic Threats. Mol Cell. 2019;73:398–411. doi: 10.1016/j.molcel.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chédin F. Nascent Connections: R-Loops and Chromatin Patterning. Trends Genet: TIG. 2016;32:828–38. doi: 10.1016/j.tig.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sollier J, Cimprich KA. Breaking bad: R-loops and genome integrity. Trends Cell Biol. 2015;25:514–22. doi: 10.1016/j.tcb.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niehrs C, Luke B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat Rev Mol Cell Biol. 2020;21:167–78. doi: 10.1038/s41580-019-0206-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–96. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gómez-González B, García-Rubio M, Bermejo R, Gaillard H, Shirahige K, Marín A, et al. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 2011;30:3106–19. doi: 10.1038/emboj.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011;25:2041–56. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wells JP, White J, Stirling PC. R Loops and Their Composite Cancer Connections. Trends Cancer. 2019;5:619–31. doi: 10.1016/j.trecan.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Richard P, Manley JL. R Loops and Links to Human Disease. J Mol Biol. 2017;429:3168–80. doi: 10.1016/j.jmb.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groh M, Gromak N. Out of balance: R-loops in human disease. PLoS Genet. 2014;10:e1004630. doi: 10.1371/journal.pgen.1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayona-Feliu A, Barroso S, Munoz S, Aguilera A. The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription-replication conflicts. Nat Genet. 2021;53:1050–63. doi: 10.1038/s41588-021-00867-2. [DOI] [PubMed] [Google Scholar]
- 16.Prendergast L, McClurg UL, Hristova R, Berlinguer-Palmini R, Greener S, Veitch K, et al. Resolution of R-loops by INO80 promotes DNA replication and maintains cancer cell proliferation and viability. Nat Commun. 2020;11:4534. doi: 10.1038/s41467-020-18306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam FC, Kong YW, Huang Q, Vu Han T-L, Maffa AD, Kasper EM, et al. BRD4 prevents the accumulation of R-loops and protects against transcription-replication collision events and DNA damage. Nat Commun. 2020;11:4083. doi: 10.1038/s41467-020-17503-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards DS, Maganti R, Tanksley JP, Luo J, Park JJH, Balkanska-Sinclair E, et al. BRD4 Prevents R-Loop Formation and Transcription-Replication Conflicts by Ensuring Efficient Transcription Elongation. Cell Rep. 2020;32:108166. doi: 10.1016/j.celrep.2020.108166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T, Wallis M, Petrovic V, Challis J, Kalitsis P, Hudson DF. Loss of TOP3B leads to increased R-loop formation and genome instability. Open Biol. 2019;9:190222. doi: 10.1098/rsob.190222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JJ, Lee SY, Gong F, Battenhouse AM, Boutz DR, Bashyal A, et al. Systematic bromodomain protein screens identify homologous recombination and R-loop suppression pathways involved in genome integrity. Genes Dev. 2019;33:1751–74. doi: 10.1101/gad.331231.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, Chen J-Y, Huang Y-J, Gu Y, Qiu J, Qian H, et al. The Augmented R-Loop Is a Unifying Mechanism for Myelodysplastic Syndromes Induced by High-Risk Splicing Factor Mutations. Mol Cell. 2018;69:412–25. doi: 10.1016/j.molcel.2017.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, Bedford MT. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol Cell. 2014;53:484–97. doi: 10.1016/j.molcel.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–58. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, et al. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat Cell Biol. 2009;11:1315–24. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–78. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Salas-Armenteros I, Pérez-Calero C, Bayona-Feliu A, Tumini E, Luna R, Aguilera A. Human THO-Sin3A interaction reveals new mechanisms to prevent R-loops that cause genome instability. EMBO J. 2017;36:3532–47. doi: 10.15252/embj.201797208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosler T, Conte F, Longo GMC, Mikicic I, Kreim N, Mockel MM, et al. R-loop proximity proteomics identifies a role of DDX41 in transcription-associated genomic instability. Nat Commun. 2021;12:7314. doi: 10.1038/s41467-021-27530-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frame JM, North TE. Ddx41 loss R-loops in cGAS to fuel inflammatory HSPC production. Dev Cell. 2021;56:571–2. doi: 10.1016/j.devcel.2021.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Yu Z, Mersaoui SY, Guitton-Sert L, Coulombe Y, Song J, Masson J-Y, et al. DDX5 resolves R-loops at DNA double-strand breaks to promote DNA repair and avoid chromosomal deletions. NAR Cancer. 2020;2:zcaa028. doi: 10.1093/narcan/zcaa028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérez-Calero C, Bayona-Feliu A, Xue X, Barroso SI, Muñoz S, González-Basallote VM, et al. UAP56/DDX39B is a major cotranscriptional RNA-DNA helicase that unwinds harmful R loops genome-wide. Genes Dev. 2020;34:898–912. doi: 10.1101/gad.336024.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Björkman A, Johansen SL, Lin L, Schertzer M, Kanellis DC, Katsori A-M, et al. Human RTEL1 associates with Poldip3 to facilitate responses to replication stress and R-loop resolution. Genes Dev. 2020;34:1065–74. doi: 10.1101/gad.330050.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mersaoui SY, Yu Z, Coulombe Y, Karam M, Busatto FF, Masson J-Y, et al. Arginine methylation of the DDX5 helicase RGG/RG motif by PRMT5 regulates resolution of RNA:DNA hybrids. EMBO J. 2019;38:e100986. doi: 10.15252/embj.2018100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lockhart A, Pires VB, Bento F, Kellner V, Luke-Glaser S, Yakoub G, et al. RNase H1 and H2 Are Differentially Regulated to Process RNA-DNA Hybrids. Cell Rep. 2019;29:2890–900. doi: 10.1016/j.celrep.2019.10.108. [DOI] [PubMed] [Google Scholar]
- 34.Zhao H, Zhu M, Limbo O, Russell P. RNase H eliminates R-loops that disrupt DNA replication but is nonessential for efficient DSB repair. EMBO Rep. 2018;19:e45335. doi: 10.15252/embr.201745335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ribeiro de Almeida C, Dhir S, Dhir A, Moghaddam AE, Sattentau Q, Meinhart A, et al. RNA Helicase DDX1 Converts RNA G-Quadruplex Structures into R-Loops to Promote IgH Class Switch Recombination. Mol Cell. 2018;70:650–62. doi: 10.1016/j.molcel.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song C, Hotz-Wagenblatt A, Voit R, Grummt I. SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev. 2017;31:1370–81. doi: 10.1101/gad.300624.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amon JD, Koshland D. RNase H enables efficient repair of R-loop induced DNA damage. eLife. 2016;5:e20533. doi: 10.7554/eLife.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wahba L, Amon JD, Koshland D, Vuica-Ross M. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell. 2011;44:978–88. doi: 10.1016/j.molcel.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell. 2015;57:636–47. doi: 10.1016/j.molcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.García-Rubio ML, Pérez-Calero C, Barroso SI, Tumini E, Herrera-Moyano E. Rosado IV, et al. The Fanconi Anemia Pathway Protects Genome Integrity from R-loops. PLoS Genet. 2015;11:e1005674. doi: 10.1371/journal.pgen.1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhatia V, Barroso SI, García-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–5. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- 43.Burgute BD, Peche VS, Steckelberg A-L, Glöckner G, Gaßen B, Gehring NH, et al. NKAP is a novel RS-related protein that interacts with RNA and RNA binding proteins. Nucleic Acids Res. 2014;42:3177–93. doi: 10.1093/nar/gkt1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pajerowski AG, Nguyen C, Aghajanian H, Shapiro MJ, Shapiro VS. NKAP is a transcriptional repressor of notch signaling and is required for T cell development. Immunity. 2009;30:696–707. doi: 10.1016/j.immuni.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen DY, Li ZQ, Yang Q, Zhang JB, Zhai ZH, Shu HB. Identification of a nuclear protein that promotes NF-kappa B activation. Biochem Bioph Res Co. 2003;310:720–4. doi: 10.1016/j.bbrc.2003.09.074. [DOI] [PubMed] [Google Scholar]
- 46.Shapiro MJ, Lehrke MJ, Chung JY, Romero Arocha S, Shapiro VS. NKAP Must Associate with HDAC3 to Regulate Hematopoietic Stem Cell Maintenance and Survival. J Immunol (Baltim, Md: 1950) 2019;202:2287–95. doi: 10.4049/jimmunol.1800862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu FC, Pajerowski AG, Nelson-Holte M, Sundsbak R, Shapiro VS. NKAP is required for T cell maturation and acquisition of functional competency. J Exp Med. 2011;208:1291–304. doi: 10.1084/jem.20101874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pajerowski AG, Shapiro MJ, Gwin K, Sundsbak R, Nelson-Holte M, Medina K, et al. Adult hematopoietic stem cells require NKAP for maintenance and survival. Blood. 2010;116:2684–93. doi: 10.1182/blood-2010-02-268391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fiordaliso SK, Iwata-Otsubo A, Ritter AL, Quesnel-Vallières M, Fujiki K, Nishi E, et al. Missense Mutations in NKAP Cause a Disorder of Transcriptional Regulation Characterized by Marfanoid Habitus and Cognitive Impairment. Am J Hum Genet. 2019;105:987–95. doi: 10.1016/j.ajhg.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai K. A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation. Science. 2019;363:710–4. doi: 10.1126/science.aaw5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li T, Chen L, Cheng J, Dai J, Huang Y, Zhang J, et al. SUMOylated NKAP is essential for chromosome alignment by anchoring CENP-E to kinetochores. Nat Commun. 2016;7:12969. doi: 10.1038/ncomms12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, et al. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10:1858. doi: 10.1038/s41467-019-09712-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu Y, Liu Y, Yang C. Evaluating In Vitro DNA Damage Using Comet Assay. J Visualized Exp. 2017;128:56450. [DOI] [PMC free article] [PubMed]
- 54.Chang EY-C, Tsai S, Aristizabal MJ, Wells JP, Coulombe Y, Busatto FF, et al. MRE11-RAD50-NBS1 promotes Fanconi Anemia R-loop suppression at transcription-replication conflicts. Nat Commun. 2019;10:4265. doi: 10.1038/s41467-019-12271-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shiromoto Y, Sakurai M, Minakuchi M, Ariyoshi K, Nishikura K. ADAR1 RNA editing enzyme regulates R-loop formation and genome stability at telomeres in cancer cells. Nat Commun. 2021;12:1654. doi: 10.1038/s41467-021-21921-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao B, Lin J, Rong L, Wu S, Deng Z, Fatkhutdinov N, et al. ARID1A promotes genomic stability through protecting telomere cohesion. Nat Commun. 2019;10:4067. doi: 10.1038/s41467-019-12037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukherjee C, Tripathi V, Manolika EM, Heijink AM, Ricci G, Merzouk S, et al. RIF1 promotes replication fork protection and efficient restart to maintain genome stability. Nat Commun. 2019;10:3287. doi: 10.1038/s41467-019-11246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanz LA, Chédin F. High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat Protoc. 2019;14:1734–55. doi: 10.1038/s41596-019-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jimenez M, Urtasun R, Elizalde M, Azkona M, Latasa MU, Uriarte I, et al. Splicing events in the control of genome integrity: role of SLU7 and truncated SRSF3 proteins. Nucleic Acids Res. 2019;47:3450–66. doi: 10.1093/nar/gkz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, et al. Characterization of Monoclonal-Antibody to DNA.Rna and Its Application to Immunodetection of Hybrids. J Immunol Methods. 1986;89:123–30. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- 61.Smolka JA, Sanz LA, Hartono SR, Chedin F. Recognition of RNA by the S9.6 antibody creates pervasive artifacts when imaging RNA:DNA hybrids. J Cell Biol. 2021;220:e202004079. doi: 10.1083/jcb.202004079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chédin F, Hartono SR, Sanz LA, Vanoosthuyse V. Best practices for the visualization, mapping, and manipulation of R-loops. EMBO J. 2021;40:e106394. doi: 10.15252/embj.2020106394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18:436–47. doi: 10.1016/j.ccr.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bhaskara S, Chyla BJ, Amann JM, Knutson SK, Cortez D, Sun ZW, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun Z, Feng D, Fang B, Mullican SE, You SH, Lim HW, et al. Deacetylase-Independent Function of HDAC3 in Transcription and Metabolism Requires Nuclear Receptor Corepressor. Mol Cell. 2013;52:769–82. doi: 10.1016/j.molcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cristini A, Groh M, Kristiansen MS, Gromak N. RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage. Cell Rep. 2018;23:1891–905. doi: 10.1016/j.celrep.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zatreanu D, Han Z, Mitter R, Tumini E, Williams H, Gregersen L, et al. Elongation Factor TFIIS Prevents Transcription Stress and R-Loop Accumulation to Maintain Genome Stability. Mol Cell. 2019;76:57–69. doi: 10.1016/j.molcel.2019.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shivji MKK, Renaudin X, Williams CH, Venkitaraman AR. BRCA2 Regulates Transcription Elongation by RNA Polymerase II to Prevent R-Loop Accumulation. Cell Rep. 2018;22:1031–9. doi: 10.1016/j.celrep.2017.12.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X, Chiang HC, Wang Y, Zhang C, Smith S, Zhao X, et al. Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat Commun. 2017;8:15908. doi: 10.1038/ncomms15908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sollier J, Stork CT, García-Rubio ML, Paulsen RD, Aguilera A, Cimprich KA. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–85. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shinriki S, Hirayama M, Nagamachi A, Yokoyama A, Kawamura T, Kanai A, et al. DDX41 coordinates RNA splicing and transcriptional elongation to prevent DNA replication stress in hematopoietic cells. Leukemia. 2022;36:2605–20. doi: 10.1038/s41375-022-01708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zardoni L, Nardini E, Brambati A, Lucca C, Choudhary R, Loperfido F, et al. Elongating RNA polymerase II and RNA:DNA hybrids hinder fork progression and gene expression at sites of head-on replication-transcription collisions. Nucleic Acids Res. 2021;49:12769–84. doi: 10.1093/nar/gkab1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goulielmaki E, Tsekrekou M, Batsiotos N, Ascensao-Ferreira M, Ledaki E, Stratigi K, et al. The splicing factor XAB2 interacts with ERCC1-XPF and XPG for R-loop processing. Nat Commun. 2021;12:3153. doi: 10.1038/s41467-021-23505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crossley MP, Song C, Bocek MJ, Choi JH, Kousorous J, Sathirachinda A, et al. R-loop-derived cytoplasmic RNA-DNA hybrids activate an immune response. Nature. 2023;613:187–94. doi: 10.1038/s41586-022-05545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chatzidoukaki O, Stratigi K, Goulielmaki E, Niotis G, Akalestou-Clocher A, Gkirtzimanaki K, et al. R-loops trigger the release of cytoplasmic ssDNAs leading to chronic inflammation upon DNA damage. Sci Adv. 2021;7:eabj5769. doi: 10.1126/sciadv.abj5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dash B, Shapiro MJ, Chung JY, Romero Arocha S, Shapiro VS. Treg-specific deletion of NKAP results in severe, systemic autoimmunity due to peripheral loss of Tregs. J Autoimmun. 2018;89:139–48. doi: 10.1016/j.jaut.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hsu FC, Belmonte PJ, Constans MM, Chen MBW, McWilliams DC, Hiebert SW, et al. Histone Deacetylase 3 Is Required for T Cell Maturation. J Immunol. 2015;195:1578–90. doi: 10.4049/jimmunol.1500435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that [the/all other] data supporting the findings of this study are available within the paper [and its supplementary information files].