Abstract
Organic fertilizers can partially replace chemical fertilizers to improve agricultural production and reduce negative environmental impacts. To study the effect of organic fertilizer on soil microbial carbon source utilization and bacterial community composition in the field of rain-fed wheat, we conducted a field experiment from 2016 to 2017 in a completely randomized block design with four treatments: the control with 100% NPK compound fertilizer (N: P2O5: K2O = 20:10:10) of 750 kg/ha (CK), a combination of 60% NPK compound fertilizer with organic fertilizer of 150 kg/ha (FO1), 300 kg/ha (FO2), and 450 kg/ha (FO3), respectively. We investigated the yield, soil property, the utilization of 31 carbon sources by soil microbes, soil bacterial community composition, and function prediction at the maturation stage. The results showed that (1) compared with CK, organic fertilizer substitution treatments improved ear number per hectare (13%−26%), grain numbers per spike (8%−14%), 1000-grain weight (7%−9%), and yield (3%−7%). Organic fertilizer substitution treatments increased the total nitrogen, available nitrogen, available phosphorus, and soil organic matter contents by 26%, 102%, 12%, and 26%, respectively, compared with CK treatments. Organic fertilizer substitution treatments significantly advanced the partial productivity of fertilizers. (2) Carbohydrates and amino acids were found to be the most sensitive carbon sources for soil microorganisms in different treatments. Particularly for FO3 treatment, the utilization of β-Methyl D-Glucoside, L-Asparagine acid, and glycogen by soil microorganisms was higher than other treatments and positively correlated with soil nutrients and wheat yield. (3) Compared with CK, organic fertilizer substitution treatments increased the relative abundance of Proteobacteria, Acidobacteria, and Gemmatimonadetes and decreased the relative abundance of Actinobacteria and Firmicutes. Interestingly, FO3 treatment improved the relative abundance of Nitrosovibrio, Kaistobacter, Balneimonas, Skermanella, Pseudomonas, and Burkholderia belonging to Proteobacteria and significantly boosted the relative abundance of function gene K02433 [the aspartyl-tRNA (Asn)/glutamyl-tRNA (Gln)]. Based on the abovementioned findings, we suggest FO3 as the most appropriate organic substitution method in rain-fed wheat fields.
Keywords: organic manure partial substitution chemical fertilizer, soil microbial carbon sources metabolic characteristics, soil bacteria community, bacteria functional prediction, soil nutrients, wheat yield
1. Introduction
Fertilization is crucial for modern agricultural production. Fertilizer types and application rates greatly affect crop production, soil properties, and soil microorganisms (Geisseler et al., 2017). The application of chemical fertilizers can increase the yield of crops by more than 50% (Tang et al., 2020). However, in the past 20 years, with the substantial amount of chemical fertilizer application, the phenomenon that its returns were diminishing appeared in agricultural production (Peng et al., 2021), especially in the arid areas on the Loess Plateau with insufficient precipitation and poor soil texture and quality. Excessive chemical fertilizer aggravates soil acidification, hardening, and nutrient loss, thereby reducing crop yield (Hui et al., 2018; Li, 2021).
Fertilization has significant effects on the activity of soil microbes (Francioli et al., 2016). Fertilizer application also influences the evolutions in the soil pH, available potassium, SOC, total N, and microbial biomass C and N, which were observed upon fertilizer application (Ji et al., 2018). The application of organic fertilizer with rich nutrients and a slow-release rate can not only alleviate the deterioration of soil physiochemical and biological properties caused by chemical fertilizer inputs but also supply various nutrients for crop growth, promote microbial reproduction, and enhance the ability of soil to retain fertilizer and water (Karuku et al., 2018). Previous studies have revealed that the combined application of organic fertilizer and chemical fertilizers significantly increased the crop yield and reproduction of soil microorganisms (Dominchin et al., 2021; Han et al., 2021a), as well as improved soil fertility and soil bacterial community composition (Wang et al., 2021). Soil microbiota plays key roles in regulating the soil micro-ecological environment, promoting the uptake of nutrient elements in roots, and maintaining soil fertility and sustainability (Li et al., 2022). The higher proportion of organic fertilizer increased soil bacterial diversity (Liu et al., 2021).
Recent studies on high-throughput sequencing have provided new insights into soil microbial diversity and community composition under long-term organic and inorganic fertilization. However, (1) How does soil microbial community composition change as the proportion of organic fertilizer instead of chemical fertilizer increases? (2) What is the soil microbial carbon source utilization under the organic substitution treatments? (3) How is the relationship between the soil microbial community composition and carbon source utilization? and (4) What is the best organic substitution treatment? To answer these questions, we measured the relevant parameters of soil nutrients, investigated the utilization of soil microbial carbon sources using the Biolog-eco microplate method, and analyzed bacterial community structure using 16s rRNA gene Illumina MiSeq high-throughput sequencing under different substitution proportions of organic fertilizer for partial chemical fertilizer in a rain-fed wheat field. The differential analysis and correlation analysis were used to reveal the potential effects of the community composition of specific bacterial taxa on carbon source utilization capacity, functional genes, and their association with soil nutrients. The study results provided useful information for improving the quality and efficiency of agricultural production as well as reducing the negative impact of fertilizers on the environment.
2. Materials and methods
2.1. Study site description
The experiment was conducted from 2016 to 2017 in Yuanqu County (35°14.4′N, 111°43.3′E), Shanxi Province, which is located on the Loess Plateau in northwest China. This region has a sub-humid, warm, and continental monsoon climate with a mean frost-free period of 236 days and an average annual precipitation, temperature, and sunshine time of 631 mm, 13.5°C, and 2 026.2 h, respectively. The soil texture is medium loam and classified as cinnamon red vertical structural loess (Cui et al., 2018; Hui et al., 2018). Before sowing on 30 September 2016, the soil properties in the study field were recorded as follows: pH of 8.0, 10.5 g kg−1 soil organic matter (SOM), 0.71 g kg−1 total nitrogen (TN), 86.3 mg kg−1 alkali-hydrolyzable nitrogen N (AN), 14.5 mg kg−1 available phosphorus (AP), and 112.3 mg kg−1 available potassium (AK) in the 0–20 cm deep soil layer.
2.2. Experimental design and treatments
The field experiment was performed in a completely randomized block design with three biological replicates for each treatment, including the control with 100% chemical fertilizer of 750 kg/ha (CK), a combination of 60% chemical fertilizer with organic fertilizer of 150 kg/ha (FO1), 300 kg/ha (FO2), and 450 kg/ha (FO3), respectively (Table 1). There were 12 plots, and the plot size was 660 m2 (15 m × 44 m). Before sowing winter wheat (cultivar Yannong-21; 112.5 kg/ha) on 30 September 2016, we applied organic fertilizer with a depth of 40–60 cm in the soil. Chemical fertilizer was applied to all plots at the time of sowing using the wide ridge and narrow furrow sowing methods with wide ridges (with a 25-cm wide base and 12 cm height) and narrow furrows (depth 8 cm, sown into the top edges of the furrow, rows spaced 12 cm). Other field management measures, such as irrigation and weeding, were carried out routinely. The organic fertilizer (organic matter ≥45%, N: 2.5%, P2O5: 1.4%, and K2O: 1.6%) and chemical fertilizer (NPK compound fertilizer, N: P2O5: K2O = 20:10:10) were purchased from Guilong Co., Ltd., China.
Table 1.
Experiment treatments and fertilization methods.
| Treatments | Application of chemical fertilizer (chemical NPK) (kg/ha) | Proportion (%) | Reduction of chemical fertilizer (chemical NPK) (kg/ha) | Proportion (%) | Replacement of organic fertilizer (organic matter + organic NPK) (kg/ha) | Proportion (%) |
|---|---|---|---|---|---|---|
| CK | 750 | 100 | 0 | 100 | 0 | 0 |
| FO1 | 450 | 60 | 300 | 40 | 150 | 20 |
| FO2 | 450 | 60 | 300 | 40 | 300 | 40 |
| FO3 | 450 | 60 | 300 | 40 | 450 | 60 |
2.3. Sampling and chemical analysis
The wheat was harvested and yield tested on 8 June 2017. After the harvest, all the wheat in each plot was threshed and weighed. To determine the grain moisture content, we used a fast moisture meter (PM-8188 New, Kett, Japan; three times in each plot). According to the standard moisture content (13%), the actual yield of each plot was obtained.
Soil samples were collected before harvesting. Three sampling sites with 1 m2 were chosen randomly in each plot, and five subsamples from each site were collected at a depth of 0–20 cm using a sterilized 4 cm-diameter soil-drilling sampler, respectively. After sieving through a 2-mm mesh to filter out roots, large rocks, and other impurities, five subsamples were mixed into one sample and divided into three parts for subsequent analyses. One part (fresh soil) was placed in a 50-ml centrifuge tube and then kept at −80°C for microbial sequencing analysis. One part (fresh soil) was stored at 4°C for soil microbial carbon utilization. The last part was air-dried and used to determine the soil's chemical properties.
The SOM, TN, and AN contents were determined using the potassium dichromate volumetric method, the semi-micro Kjeldahl method, and the alkaline hydrolysis diffusion method (He et al., 2022), respectively. The AP content was determined in a 0.5 mol L−1 NaHCO3 extraction using the molybdenum-antimony anti-spectrophotometric method (Sun et al., 2019). The AK content was determined in a 1.0 mol L−1 NH4OAc extraction using leaching-flame photometry (Bell et al., 2005).
2.4. Soil DNA extraction, quantitative real-time PCR (q-PCR), and Illumina sequencing
Microbial DNA was extracted from the frozen soil samples (0.5 g wet weight) using a Fast DNA SPIN Extraction Kit (MP Biomedicals, Santa Ana, CA, USA) following the manufacturer's instructions. DNA quality was determined using a NanoDrop2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). The primer pairs 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) targeting the bacterial 16S rRNA gene V3-V4 hypervariable region were used for polymerase chain reaction (PCR) amplification (Langille et al., 2013). The sample-specific 7-bp barcodes were incorporated into the primers for multiplex sequencing. The PCR mixtures included FastPfu buffer (5 ×), primer (5 μM), dNTP mixture (2.5 mM), template DNA, and H2O for a total volume of 50 μl. PCR assays were performed as follows: 95°C for 2 min, followed by 30 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. PCR products were quantified and pooled in equimolar concentrations, and the paired-end reads (250 nucleotides) were generated using an Illumina HiSeq 2500 platform at Personal Biotechnology Co., Ltd., Shanghai, China.
The raw sequencing data were subjected to quality filtering and double-terminal sequence ligation using QIIME2. The data filtering were described previously (Peng et al., 2021): removal of sequences containing Ns (fuzzy bases or base mismatch number >1 in the 5′-terminal primer matching); removal of sequences containing the same consecutive base number >8; removal of sequences shorter than 150 bp. The UCLHIME in MOTHUR software (version 1.31.2, http://www.mothur.org/) was used to obtain high-quality sequences for subsequent analysis. The UCLUST method in QIIME was used to cluster the high-quality sequences at 97% sequence identity, and the longest sequence in each class was selected as the representative sequence. The Blast method in QIIME was used to align the Green Genes database (release 13.8, http://greengenes.secondgenome.com/) to obtain the taxonomic information of each OTU (operational taxonomic unit). The OTUs with abundances < 0.001% of the total sequences were removed to obtain a simplified OTU list for subsequent analysis. PICRUSt2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) was used for bacterial function prediction (Russo et al., 2023); the closed OTU table was obtained from QIIME and compared to the KEGG (Kyoto Encyclopedia of Genes and Genomes) database to determine different bacterial community functions.
2.5. Carbon source utilization by soil microorganisms
The carbon source utilization ability of soil microorganisms was estimated using the average well-color development (AWCD) and determined using the Biolog-ECO microplate method (Chen et al., 2017). The procedure involved weighing a fresh soil sample equivalent to 5 g of dry soil into a sterile conical flask. Then, 45 ml of 0.85% sterile NaC1 solution was added, followed by sealing and shaking at 2,000 r/min for 15 min. The supernatant was collected after letting the solution stand for 15 min. Moreover, the supernatant was diluted 100-fold with sterile NaCl solution on a clean bench and inoculated into Biolog-eco microplates with an 8-channel sample applicator at 150−1 μl per well. Inoculated plates were incubated in a 25°C biochemical incubator for 10 days, and the good absorbance (OD) at 590 nm was read every 24 h during the incubation period. The absorbance was measured according to the following equation:
where Ci is the OD value of each carbon source well, R is the OD value of the control well, and n is the number of carbon source types in the culture medium (31 in this study).
2.6. Statistical analyses
Origin (2019) was used to plot the line chart of microbial characteristics. SPSS (SPSS 19.0, SPSS Inc., Chicago, USA) was used for variance analysis, principal component analysis (PCA), Spearman correlation analysis (P-value), and cluster analysis. The relationship between soil microbial characteristics and soil properties was determined using Spearman correlation analysis. One-way analysis of variance (ANOVA) was used to analyze the different organic substitution patterns for significant differences (P < 0.05), such as soil physical and chemical properties and microbial characteristics. The difference between the means was represented with different letters by using the least significant difference (LSD; P < 0.05).
3. Results
3.1. Rain-fed wheat yield characteristics, soil chemical properties, and partial productivity of fertilizers
Crop yield and soil fertility depend on the contents of the main nutrients in the soil, which are largely determined by fertilization. The treatments of organic fertilizer substitution for partially chemical fertilizer significantly affected soil nutrient contents, wheat yield, and yield components in rain-fed wheat fields (Table 2). Compared with the control (CK), the organic fertilizer substitution treatments increased the TN, AN, AP, and SOM by 1.33%−41.33%, 17.73%−147.19%, 7.76%−12.30%, and 6.10%−46.15%, respectively. Organic fertilizer substitution treatments also increased ear number, grain numbers per spike, and 1,000-grain weight by 13.21%−26.00%, 8.33%−11.11%, and 8.09%-9.36%, respectively. Therefore, grain yield was increased by 3.45–6.92%.
Table 2.
Soil fertility and the economic characteristics of wheat during wheat maturity in rain-fed wheat field under different treatments.
| Treatments | Ear number (104/ha) | Number of grains per spike | 1,000 grain weight (g) | Yield (kg/ha) |
|---|---|---|---|---|
| CK | 477 ± 3Cc | 36 ± 2Ab | 33.86 ± 1.16Bb | 5337.34 ± 170.31Ab |
| FO1 | 540 ± 2Bb | 39 ± 2Aab | 36.63 ± 0.77Aa | 5521.49 ± 258.34Aab |
| FO2 | 546 ± 9Bb | 41 ± 4Aa | 36.36 ± 0.94ABa | 5503.61 ± 322.35Aab |
| FO3 | 601 ± 7Aa | 40 ± 1Aa | 37.03 ± 0.08Aa | 5706.45 ± 262.97Aa |
| TN (g/kg) | AN (mg/kg) | AP (mg/kg) | SOM (g/kg) | |
| CK | 0.75 ± 0.01Bc | 15.34 ± 0.90Cc | 14.55 ± 0.75Ab | 7.54 ± 0.23Cd |
| FO1 | 0.76 ± 0.02Bc | 18.06 ± 0.01Bb | 15.68 ± 0.38Aab | 8.00 ± 0.01Cc |
| FO2 | 1.01 ± 0.01Ab | 37.02 ± 0.90Aa | 16.91 ± 0.28Aa | 9.51 ± 0.35Bb |
| FO3 | 1.06 ± 0.03Aa | 37.92 ± 0.01Aa | 16.34 ± 1.23Aa | 11.02 ± 0.01Aa |
Different lowercase letters in the same column indicate differ significantly at the 0.05 level (P < 0.05), while different capital letters indicate differ extremely significantly at the 0.01 level (P < 0.01). CK, control; FO1, combination of 60% chemical fertilizer with organic manure of 150 kg/ha; FO2, combination of 60% chemical fertilizer with organic manure of 300 kg/ha; FO3, combination of 60% chemical fertilizer with organic manure of 450 kg/ha. TN, total nitrogen; AN, available nitrogen; AP, available phosphorus; SOM, soil organic matter.
Organic fertilizer treatments for the soil fertility index significantly increased soil AN, TN, SOM, and AP content, respectively, compared to CK (Table 2). This fact indicates that the partial replacement of chemical fertilizer with organic fertilizer is beneficial for the transformation and accumulation of soil N/C/P nutrients.
No significant differences were found between FO2 and FO3 in yield or the values of AN and AP. The results showed that an equal amount of substitution (40%) and an increased substitution (60%) of organic fertilizer may improve the soil chemical properties at a stable wheat yield level in rain-fed.
Compared with CK, the organic substitution treatment significantly increased the partial productivity of N (143%), P (131%), and K (128%) fertilizers, respectively (Table 3).
Table 3.
Inorganic fertilizer application amount and fertilizer partial productivity of different treatments.
| Treatments | Application amount of inorganic fertilizer (kg/ha) | Partial productivity/(kg·kg−1) | ||||
|---|---|---|---|---|---|---|
| N | P2O5 | K2O | N | P | K | |
| CK | 150 | 75 | 75 | 35.58 ± 1.14Cc | 71.16 ± 2.27Cc | 71.16 ± 2.27Cc |
| FO1 | 60 | 30 | 30 | 86.61 ± 4.05Aa | 172.01 ± 8.05Aa | 170.42 ± 7.97Aa |
| FO2 | 60 | 30 | 30 | 81.53 ± 4.78Aab | 160.92 ± 9.43Aa | 158.15 ± 9.26Ab |
| FO3 | 60 | 30 | 30 | 80.09 ± 3.69Bb | 157.20 ± 7.24Bb | 153.40 ± 7.07Bb |
Different lowercase letters in the same column indicate differ significantly at the 0.05 level (P < 0.05), while different capital letters indicate differ extremely significantly at the 0.01 level (P < 0.01). CK, control; FO1, combination of 60% chemical fertilizer with organic manure of 150 kg/ha; FO2, combination of 60% chemical fertilizer with organic manure of 300 kg/ha; FO3, combination of 60% chemical fertilizer with organic manure of 450 kg/ha. TN, total nitrogen; AN, available nitrogen; AP, available phosphorus; SOM, soil organic matter.
3.2. Soil carbon source utilization by soil microorganisms
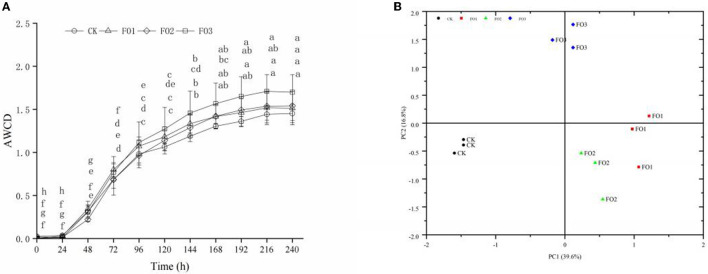
No significant difference was found in AWCD values at 24 h of incubation in different soil treatments, indicating that the use of carbon sources by soil microorganisms was weak. However, the AWCD values increased sharply from 24 to 144 h, reaching their highest value at 144 h (P < 0.05). The AWCD values continued to increase slowly from 144 to 240 h but without significant differences (Figure 1A). This indicated that the soil microbial activity and carbon source utilization ability were strongest at the “inflection point” of 144 h. Of all treatments, CK showed the lowest AWCD values, and the highest values were observed in FO3 (P < 0.05).
Figure 1.
Average well color development (AWCD) (A) and principal component analysis (B) of carbon sources utilization profiles of soil microbes in Biolog Ecoplate of soil samples under different treatments. (A) Different lowercase letters indicate significant differences between different incubation times in the same treatment (P < 0.05). CK, control (circles); FO1, combination of 60% chemical fertilizer with organic manure of 150 kg/ha (triangles); FO2, combination of 60% chemical fertilizer with organic manure of 300 kg/ha; FO3, combination of 60% chemical fertilizer with organic manure of 450 kg/ha (squares). (B) CK, control (filled circles); FO1, FOL combination of 60% chemical fertilizer with organic manure of 150 kg/ha (filled squares); FO2, combination of 60% chemical fertilizer with organic manure of 300 kg/ha (filled triangles); FO3, combination of 60% chemical fertilizer with organic manure of 450 kg/ha (filled diamonds); PC, principal component.
The PCA was used to analyze the utilization of 31 carbon sources as substrates at 144 h by soil microbes (Figure 1B). The two principal component factors (PC1 and PC2), acquired from 31 carbon sources, could explain 39.6 and 16.8% of the variables, respectively. The scores of CK and FO treatments were well separated in the PC1 axis. FO3 was clearly separated from other treatments in the PC2 axis. As shown in Table 4, based on the eigenvector coefficients (loadings) of greater than 0.60 for PC1 and PC2, there were 18 types of carbon sources significantly in PC1, including four carbohydrates, four amino acids, three carboxylic acids, three polymers, two amines, and two phenolic acids, among which carbohydrates and amino acids accounted for 44.4% and contributed the most to PC1. There were five types of carbon sources significantly present in PC2, including carbohydrates (3), carboxylic acids (1), and phenolic acids (1), among which carbohydrates contributed the most, accounting for 60%. In short, carbohydrates and amino acids were the most sensitive carbon sources for soil microorganisms. Additionally, the dominant carbon sources with PC1 and PC2 were D-Mannitol (carbohydrates) and β-Methyl D-Glucoside (carbohydrates), respectively (Table 4). The above results show that organic fertilizer substitution treatments improved the carbohydrate and amino acid utilization abilities of soil microorganisms in rain-fed wheat fields.
Table 4.
Main carbon sources and corresponding PC loadings in different treatments.
| Main carbon sources types | PC1 | Main carbon sources types | PC2 |
|---|---|---|---|
| Carbohydrates | Carbohydrates | ||
| D-Mannitol | 0.892 | β-Methyl-D-glucoside | 0.924 |
| Glucose-1-phosphate | 0.880 | I-erythritol | −0.795 |
| D, L-a-glycerol | −0.855 | D-Galactonic acid y-lactone | 0.647 |
| a-D-Lactose | 0.720 | Carboxylic acids | |
| Amino acids | y-Hydroxybutyric acid | −0.670 | |
| L-Threonine | −0.832 | Phenolic acids | |
| L-Phenylalanine | −0.800 | 2-Hydroxybenzoic acid | 0.602 |
| L-Asparagine | 0.763 | ||
| L-Serine | 0.712 | ||
| Carboxylic acids | |||
| a-Ketobutyric acid | −0.827 | ||
| Pyruvic acid methyl ester | 0.784 | ||
| Itaconic acid | 0.755 | ||
| Polymers | |||
| Glycogen | −0.856 | ||
| 80Tween 80 | 0.769 | ||
| a-Cyclodextrin | 0.663 | ||
| Amines | |||
| Phenylethyl-amine | −0.795 | ||
| Putrescine | 0.696 | ||
| Phenolic acids | |||
| 2-Hydroxybenzoic acid | −0.695 | ||
| 4-Hydroxybenzoic acid | 0.672 |
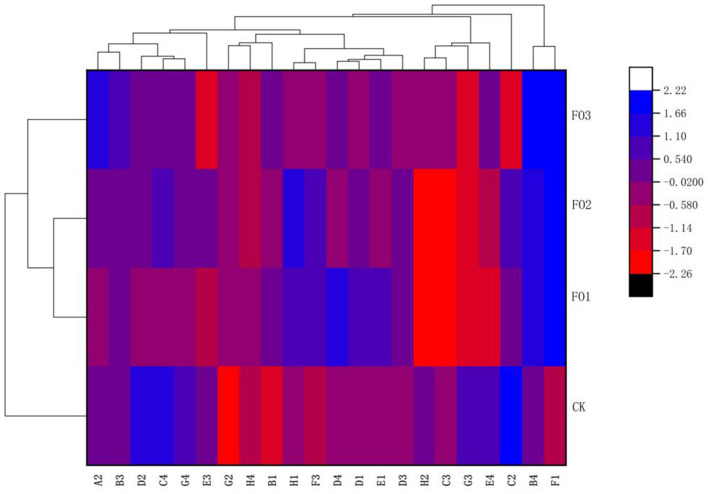
Furthermore, through the cluster analysis of the utilization ability of the 22 types of carbon sources mentioned above on the basis of the AWCD values (Figure 2), three clusters were obtained: one cluster was observed in FO1 and FO2, and two clusters were found in CK and FO3, respectively. The results showed that soil microorganisms' carbon source utilization types for organic fertilizer and chemical fertilizer treatments differed significantly. For example, soil microbes in the organic fertilizer substitution treatments could utilize more F1 (glycogen) than those in CK, but in CK, a larger amount of C2 (I-Erythritol), D2 (D-Mannitol), and C4 (L-Phenylalanine) was used by soil microorganisms. In addition, a larger amount of A2 (β-Methyl D-Glucoside) and B4 (L-Asparagine) was utilized by soil microorganisms in FO3, but more D4 (L-Serine) and H1 (α-D-Lactose) were used by soil microorganisms in FO1 and FO2, respectively (Figure 2).
Figure 2.
Cluster diagram of soil microbial carbon sources utilization under different treatments. The carbon sources are listed on the X-axis and treatments on the Y-axis. For the heat map, the left side of the cluster tree is a treatment cluster tree and the above cluster tree is the carbon sources duster tree. A2, represents β-methyl-D-glucoside; B3, D-galacturonic acid; D2, D-mannitol; C4, L-phenylalanine; G4, phenylethyl-amine; E3, y-hydroxybutyric acid; G2, Glucose-1-Phosphate; H4, Putrescine, B1, pyruvic acid methyl ester; H1, a-D-Lactose; F3, itaconic acid; D4, L-Serine; D1, Tween 80; E1, a-Cyclodextrin; D3, 4-hydroxybenzoic acid; H2, D,L-a-glycerol; C3, 2-hydroxybenzoic acid; G3, a-ketobutyric acid; E4, L-threonine; C2, I-Erythritol; B4, L-asparagine; Fl, represents Glycogen. While CK, shows control; FO1, combination of 60% chemical fertilizer with organic manure of 150 kg/ha; FO2, combination of 60% chemical fertilizer with organic manure of 300 kg/ha; and corresponding FO3, combination of 60% chemical fertilizer with organic manure of 450 kg/ha. The numbers for different colors of the columns represent the abundance of the genera or genes in different samples.
3.3. Soil bacterial community composition and PICRUSt functional predictive analysis
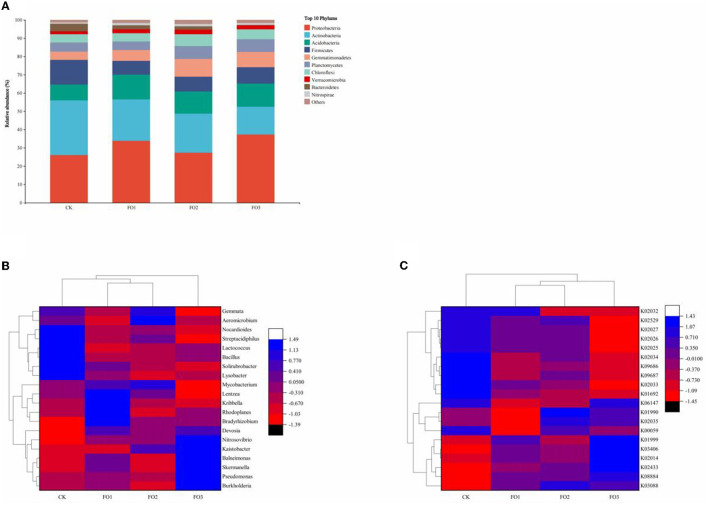
Figure 3A depicts the relative abundance of the top 10 bacteria at the phylum level in all treatments. The most dominant bacterial phyla were Proteobacteria, Actinobacteria, Acidobacteria, Firmicutes, and Gemmatimonadetes. Proteobacteria were abundant in all treatments (>25%). Additionally, its relative abundance was higher in organic fertilizer treatments than in CK. The relative abundance of Actinobacteria was the highest (30%) in the CK. The cluster analysis of relative abundance at the genus level of bacteria (Figure 3B) was divided into two main categories, namely FO3 and other treatments. FO1 and FO2 were further clustered into one category. Among the top 20 abundantly identified soil bacterial genera, 10 genera (Burkholderia, Skermanella, Lysobacter, Kaistobacter, Balneimonas, Bradyrhizobium, Rhodoplanes, Pseudomonas, Nitrosovibrio, and Devosia) belonged to Proteobacteria, seven genera (Kribbella, Solirubrobacter, Aeromicrobium, Mycobacterium, Streptacidiphilus, Lentzea, and Nocardioides) belonged to Actinobacteria, two genera (Bacillus and Lactococcus) belonged to Firmicutes, and one genus (Gemmata) belonged to Planctomycetes. In CK, Nocardioides, Streptacidiphilus, Lactococcus, Bacillus, Solirubrobacter, and Lysobacter were dominant and significantly higher than in other treatments. In FO3, the relative abundance of Nitrosovibrio, Kaistobacter, Balneimonas, Skermanella, Pseudomonas, and Burkholderia was significantly increased. For example, the relative abundance of Nitrosovibrio followed the following order: FO3 > FO2 > FO1 > CK. These results indicated that the organic fertilizer substitution for a partial chemical fertilizer regime significantly affected soil bacterial community composition in rain-fed wheat fields.
Figure 3.
Changes in the relative abundance of soil bacteria at the phylum level (A) among different treatments. The heatmap of relative abundances of the top 20 identified bacteria at the genus level (B) and bacterial community KEGG Orthologs (KOs) for different soil samples (C). CK, shows control; FO1, combination of 60% chemical fertilizer with organic manure of 150 kg/ha; FO2, combination of 60% chemical fertilizer with organic manure of 300 kg/ha; FO3, combination of 60% chemical fertilizer with organic manure of 450 kg/ha. Numbers of different colors of the columns represent the abundance of the genera or genes in different samples.
The relative abundance of KEGG Orthologs (KOs) of the bacterial community exhibited significant variations among treatments, and different soil samples were grouped into three clusters (Figure 3C); CK and FO3 treatments were two separate clusters, and FO1 was clustered with FO2. In CK, K02034 (the peptide/nickel transport system permease protein), K09686 (the antibiotic transport system permease protein), K09687 (the antibiotic transport system ATP-binding protein), and K01692 (the enoyl-CoA hydratase) were dominant compared to other treatments. FO3 significantly increased the relative abundance of K01999 (the branched-chain amino acid transport system substrate-binding protein), K03406 (the methyl-accepting chemotaxis protein), K02014 (the iron complex outer membrane receptor protein), and K02433 [the aspartyl-tRNA (Asn)/glutamyl-tRNA (Gln)]. We found that the relative abundance of K02433 also followed the order FO3 > FO2 > FO1 > CK, which is consistent with the change trends of the relative abundance of Nitrosovibrio. Similarly, the relative abundance changes of K03406 and K02014 were consistent with those of Skermanella and Burkholderia.
3.4. Correlation analysis between soil carbon source utilization, KEGG Orthologs, soil nutrients, and wheat yield under CK and FO3 treatments
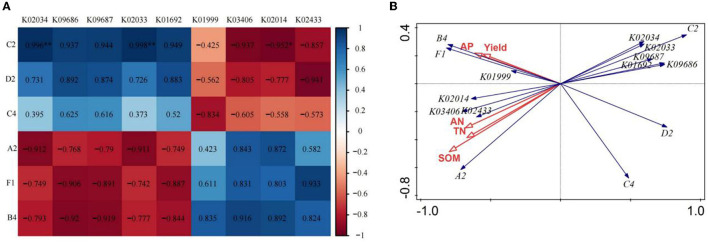
Based on the above analysis, we chose the CK and FO3 treatments to conduct the correlation analysis between soil carbon source utilization, KEGG Orthologs (KOs), soil nutrients, and wheat yield. As shown in Figure 4A, the utilization of C2 (I-Erythritol) by soil microorganisms was significantly negatively correlated with K02014 (iron complex in vitro membrane receptor protein). However, it showed a very significant positive correlation with K02034 (permease protein of the peptide/nickel transport system) and K02033 (peptide/nickel transport system permease protein). The utilization of D2 (D-Mannitol) by soil microorganisms was positively correlated with K09686 (the antibiotic transport system permease protein) and K09687 (the antibiotic transport system ATP-binding protein) and was negatively correlated with K02433 [the aspartyl-tRNA (Asn)/glutamyl-tRNA (Gln)]. The utilization of A2 (β-Methyl-D-Glucoside) by soil microorganisms was negatively correlated with K02034 and K02033 and was positively correlated with K02014. The utilization of F1 (glycogen) and B4 (L-Asparagine) by soil microorganisms was negatively correlated with K09686 and K09687 and positively correlated with K02433.
Figure 4.
Correlation analysis of dominant soil carbon metabolism and dominant KEGG Orthologs (KOs) in rain-fed wheat field (A); redundancy analysis of soil nutrient and yield with dominant carbon source utilization and dominant bacteria KEGG Orthologs (B). A2, represents β-Methyl-D-Glucoside; D2, D-Mannitol; C4, L-phenylalanine; C2, I-Erythritol; B4, L-asparagine; Fl, represents glycogen. TN, total nitrogen; AN, available nitrogen; AP, available phosphorus; SOM, soil organic matter. While CK, shows control; FO1, combination of 60% chemical fertilizer with organic manure of 150 kg/ha; FO2, combination of 60% chemical fertilizer with organic manure of 300 kg/ha; and corresponding FO3, combination of 60% chemical fertilizer with organic manure of 450 kg/ha. The numbers for different colors of the columns represent the abundance of the genera or genes in different samples.
The redundancy analysis of wheat yield, soil nutrients, soil microbial carbon source utilization, and KEGG Orthologs (KOs; Figure 4B) showed that the utilization of C2 (I-Erythritol), D2 (D-Mannitol) and C4 (l-Phenylalanine) by soil microorganisms was negatively correlated with wheat yield, SOM, TN, AN, AP, and the utilization of A2 (β -Methyl-D-Glucoside), B4 (L-Asparagine) and F1 (glycogen) by soil microorganisms. It was also found that wheat yield, SOM, TN, AN, and AP were negatively correlated with K02033, K02034, K09687, and K09686 but were positively correlated with K01999, K02014, K03406, and K02433.
4. Discussion
4.1. Changes of soil nutrients, wheat yield, partial productivity of fertilizer, soil microbial carbon source utilization, and KEGG Orthologs in a rain-fed wheat field
Soil microorganisms had corresponding nutrient preferences, which may alleviate nutrient limitations (Cui et al., 2018). The organic fertilizer application could increase the contents of carbohydrates and amino acids in soil (Xu et al., 2003) to maintain the soil microbial groups metabolizing these two types of carbon sources (Mao et al., 2020; Yang et al., 2020). The organic fertilizer application promoted the metabolism of amino acids (L-Asparagine) by soil microorganisms (Kumar et al., 2017). In this study, we found that the sensitive carbon sources for soil microorganisms that were greatly affected were mainly carbohydrates and amino acids. The carbon sources utilized by soil microorganisms were mainly I-Erythritol (C2), D-mannitol (D2), and L-Phenylalanine (C4) in CK, while they were mainly β-methyl D-glucoside (A2), L-Asparagine (B4), and glycogen (F1) in FO3. This indicated that applying organic fertilizer changed soil microorganisms' ability to utilize different carbon sources (Tang et al., 2019).
Previous studies have discovered that after the addition of exogenous p-hydroxybenzoic acid (pHA) to the soil, the contents of AN, AP, and SOM in the soil reduced (Harper, 1977; Iii and Stuart, 1995; Zavarzina et al., 2019), which promoted the growth of nitrogen-fixing bacteria, ammonifying bacteria, denitrifying bacteria, and nitroso bacteria in the soil and inhibited the growth of nitrifying bacteria (Gu et al., 2020). Another study showed that the Nitrovibrio genus was enriched in organic fertilizer treatments during the wheat season (Shi et al., 2020). This study showed that organic fertilizer substitution treatments could promote the use of phenols (4-hydroxybenzoic acid, the main chemical component of root exudates of wheat and other crops, has strong allelopathy and may lead to lower crop yield) by soil microorganisms and increase the contents of TN, SOM, AN, and AP in soil and the relative abundance of Nitrovibrio. Therefore, organic fertilizer substitution for chemical fertilizer could promote the metabolism of these carbon sources, thereby improving soil nutrients and wheat yield (Han et al., 2021a; Liu et al., 2021). The soil microorganisms' utilization degree of β-Methyl-D-Glucoside, L-asparagine, and glycogen was higher in the FO3 treatment. This might be related to the highest relative abundance of Skermanella, which can utilize D-Glucose, D-Mannitol, lactose, sucrose, aspartate, and pyruvate (Subhash et al., 2017).
This study shows that the relative abundance of K02014 (iron complex outer membrane receptor protein) is consistent with Burkholderia (Lin et al., 2019), which is known for being involved in several iron uptake pathways, including siderophore production and the ability to absorb heme in infected hosts (Pongmala et al., 2022). K02433 represents an aminoacyl tRNA synthetase that can specifically recognize aspartic acid and glutamyl amine and form aminoacyl tRNA with the corresponding tRNA. Ammonium is the main nitrogen source of plants, and it is assimilated into asparagine through the catalytic reaction of asparagine synthetase (GLN), which then forms aspartic amino acid through different amino acid transferases to complete the recycling in ammonia sources of different forms (Cid et al., 2022). In this study, the abundance of related enzyme genes in the above amino acid metabolism process showed clear enrichment in FO3. Therefore, it was speculated that organic fertilizer could accelerate the conversion of ammonium nitrogen into glutamine and further into various amino acids after entering the soil environment, effectively improving the utilization rate of ammonium nitrogen uptake by crops. Indeed, the degree of utilization of B4 (L-Asparagine) by soil microorganisms in FO3 was significantly higher than that of other carbon sources. Organic fertilizer can accelerate the abundance of Pseudomonas and promote the formation of biofilms between specific bacterial groups, which is conducive to plant disease resistance (Chen et al., 2023).
Moreover, the increased abundance of Pseudomonas also promoted soil nitrogen metabolism (Fu et al., 2023). The enrichment of K03406 (the methyl-accepting chemotaxis protein) in FO3 is also important evidence. The results showed that the interaction between microorganisms and soil was greatly affected by organic fertilizer.
The results also showed that the partial productivity of fertilizer was significantly increased under the organic fertilizer substitution treatments, indicating that the utilization efficiency of fertilizer can be increased by optimizing the application of fertilizer (Liu et al., 2018).
4.2. Relationship between soil carbon source utilization, KEGG Orthologs, soil nutrients, and wheat yield under CK and FO3 treatment
Redundancy analysis of soil nutrients, wheat yield, soil microbial carbon sources, and KEGG Orthologs (KOs) showed that the microbial utilization of A2 (β-Methyl-D-Glucoside), B4 (L-Asparagine), and F1 (glycogen) carbon sources was positively correlated with KOs (K01999, K02014, K03406, K02433), soil nutrients (SOM, TN, AN, and AP), and wheat yield.
In particular, soil microbial utilization of B4 and F1 showed a significantly positive correlation with K01999, wheat yield, and AP. K01999 (the branched-chain amino acid transport system is a substrate-binding protein). Branched-chain amino acids are a class of essential amino acids with various physiological and metabolic roles (Virlouvet et al., 2011). The carbon skeleton required for amino acid synthesis comes from glycolysis, photosynthesis, the oxidative pentose phosphate pathway, and the tricarboxylic acid cycle (TCA). Therefore, the synthesis of amino acids is the hub of carbon and nitrogen metabolism (Xin et al., 2019). B4 (L-Asparagine) is one of the metabolites of the TCA cycle and the related amino acid metabolism pathway (Baslam et al., 2020; Han et al., 2021b). Phosphorus-solubilizing bacteria can transform insoluble phosphorus, which is difficult for plants to absorb and utilize, into effective forms and improve the utilization efficiency of soil phosphorus (Kalayu, 2019). Pseudomonas is agricultural soil's main phosphorus-solubilizing bacteria (PSB) (Wang et al., 2022). Soil AP content was closely related to soil bacterial community and wheat yield, indicating that soil phosphorus was an important factor driving soil microbial community and wheat yield changes (Chen et al., 2021).
The utilization of an A2 carbon source by soil microorganisms was significantly positively correlated with K02433, K02014, and SOM. This showed that the application of organic fertilizer could increase SOM content, improve soil microbial diversity, stimulate soil microbial activity (Li et al., 2020), cause organic carbon decomposition, and then cause dramatic changes in soil carbon sources (Haiming et al., 2020). In this way, plants could uptake and utilize a large number of available nutrients (Zhao et al., 2021), thus increasing the plant biomass, and the high plant biomass will secrete more simple organic matter to further improve microbial activity.
5. Conclusion
In the present study, organic fertilizer substitution treatments affected carbon source utilization by soil microorganisms and microbial community composition. Particularly, FO3 treatment improved the utilization of carbohydrates and amino acids by soil microorganisms and enriched bacterial genera (Skermanella, Pseudomonas, and Burkholderia). Furthermore, when subjected to FO3 treatment, the dominant KOs (K01999, K03406, K02014, and K02433) were strongly correlated with soil nutrients, contributing to soil nutrient cycling and crop productivity. In conclusion, adding organic fertilizer was crucial for improving soil quality, promoting agricultural production, regulating soil microbial metabolic capacity, and changing bacterial community composition. In addition, FO3 treatment is recommended as an appropriate fertilization method in this study.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI - PRJNA948916.
Author contributions
ZY and XL conceived and designed the experiments. XL and WL conducted the experiments. WY, JC, and AA analyzed the data. MS and ZG contributed analysis tools. XL, AA, and ZY wrote the article. All authors contributed to the article and approved the submitted version.
Funding Statement
This study was supported by the National Key Technology R&D Program of China (2015BAD23B04-2), the Special Funds for Scientific Research on Public Causes (201503120), the Shanxi Characteristic Wheat Industry Technology Innovative Strategic Alliance ([2020]17), the Ministerial and Provincial Co-Innovation Centre for Endemic Crops Production with High-quality and Efficiency in Loess Plateau (SBGJXTZX-21), and the Modern Agriculture Industry Technology System Construction (CARS-03-01-24).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Baslam M., Mitsui T., Sueyoshi K., Ohyama T. (2020). Recent advances in carbon and nitrogen metabolism in C3 plants. Int. J. Mol. Sci. 22, 318. 10.3390/ijms22010318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell T., Newman J. A., Silverman B. W., Turner S. L., Lilley A. K. (2005). The contribution of species richness and composition to bacterial services. Nature 436, 1157–1160. 10.1038/nature03891 [DOI] [PubMed] [Google Scholar]
- Chen W., Wu Z., Liu C., Zhang Z., Liu X. (2023). Biochar combined with Bacillus subtilis SL-44 as an eco-friendly strategy to improve soil fertility, reduce Fusarium wilt, and promote radish growth. Ecotoxicol. Environ. Saf. 251, 114509. 10.1016/j.ecoenv.2023.114509 [DOI] [PubMed] [Google Scholar]
- Chen X., Jin Y., Ou B., Zhong J., Lin F., Wu H. (2017). Analysis of soil microbial functional diversity of different understory planting dictyophora land based on Biolog-ECO technology. Genomics Appl. Biol. 36, 370–375. 10.13417/j.gab.036.000370 [DOI] [Google Scholar]
- Chen Y., Li S., Liu N., He H., Cao X., Lv C., et al. (2021). Effects of different types of microbial inoculants on available nitrogen and phosphorus, soil microbial community, and wheat growth in high-P soil. Environ. Sci. Pollut. Res. 28, 23036–23047. 10.1007/s11356-020-12203-y [DOI] [PubMed] [Google Scholar]
- Cid G. A., Francioli D., Kolb S., Tandron-Moya Y. A., von Wiren N., Hajirezaei M.-R. (2022). Elucidating the systemic response of wheat plants under waterlogging based on transcriptomic and metabolic approaches. bioRxiv. [preprint]. 10.1101/2022.08.03.502608 [DOI] [PubMed] [Google Scholar]
- Cui X., Zhang Y., Gao J., Peng F., Gao P. (2018). Long-term combined application of manure and chemical fertilizer sustained higher nutrient status and rhizospheric bacterial diversity in reddish paddy soil of Central South China. Sci. Rep. 8, 1–11. 10.1038/s41598-018-34685-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominchin M. F., Verdenelli R. A., Berger M. G., Aoki A., Meriles J. M. (2021). Impact of N-fertilization and peanut shell biochar on soil microbial community structure and enzyme activities in a Typic Haplustoll under different management practices. Eur. J. Soil Biol. 104, 103298. 10.1016/j.ejsobi.2021.103298 [DOI] [Google Scholar]
- Francioli D., Schulz E., Lentendu G., Wubet T., Buscot F., Reitz T. (2016). Mineral vs. organic amendments: microbial community structure, activity and abundance of agriculturally relevant microbes are driven by long-term fertilization strategies. Front. Microbiol. 7, 1446. 10.3389/fmicb.2016.01446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S., Deng Y., Zou K., Zhang S., Duan Z., Wu X., et al. (2023). Dynamic variation of Paris polyphylla root-associated microbiome assembly with planting years. Planta 257, 61. 10.1007/s00425-023-04074-7 [DOI] [PubMed] [Google Scholar]
- Geisseler D., Linquist B. A., Lazicki P. A. (2017). Effect of fertilization on soil microorganisms in paddy rice systems – a meta-analysis. Soil Biol. Biochem. 115, 452–460. 10.1016/j.soilbio.2017.09.018 [DOI] [Google Scholar]
- Gu Y., Wang Y., Wang P., Wang C., Ma J., Yang X., et al. (2020). Study on the diversity of fungal and bacterial communities in continuous cropping fields of chinese chives (Allium tuberosum). BioMed. Res. Int. 2020, 2020. 10.1155/2020/3589758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiming T., Xiaoping X., Chao L., Xiaochen P., Kaikai C., Weiyan L., et al. (2020). Microbial carbon source utilization in rice rhizosphere and nonrhizosphere soils with short-term manure N input rate in paddy field. Sci. Rep. 10, 6487. 10.1038/s41598-020-63639-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Dong Y., Zhang M. (2021a). Chemical fertilizer reduction with organic fertilizer effectively improve soil fertility and microbial community from newly cultivated land in the Loess Plateau of China. Appl. Soil Ecol. 165, 103966. 10.1016/j.apsoil.2021.103966 [DOI] [Google Scholar]
- Han M., Zhang C., Suglo P., Sun S., Wang M., Su T. (2021b). L-Aspartate: an essential metabolite for plant growth and stress acclimation. Molecules 26, 1887. 10.3390/molecules26071887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. L. (1977). Population Biology of Plants. London: Academic press. [Google Scholar]
- He H., Peng M., Lu W., Hou Z., Li J. (2022). Commercial organic fertilizer substitution increases wheat yield by improving soil quality. Sci. Total Environ. 851, 158132. 10.1016/j.scitotenv.2022.158132 [DOI] [PubMed] [Google Scholar]
- Hui L., Xue J.-f., Gao Z.-q., Xue N.-w., Yang Z.-p. (2018). Response of yield increase for dryland winter wheat to tillage practice during summer fallow and sowing method in the Loess Plateau of China. J. Integr. Agric. 17, 817–825. 10.1016/S2095-3119(17)61806-929150801 [DOI] [Google Scholar]
- Iii C., Stuart F. (1995). New cog in the nitrogen cycle. Nature 377, 199–200. 10.1038/377199a0 [DOI] [Google Scholar]
- Ji L., Wu Z., You Z., Yi X., Ni K., Guo S., et al. (2018). Effects of organic substitution for synthetic N fertilizer on soil bacterial diversity and community composition: a 10-year field trial in a tea plantation. Agric. Ecosyst. Environ. 268, 124–132. 10.1016/j.agee.2018.09.008 [DOI] [Google Scholar]
- Kalayu G. (2019). Phosphate solubilizing microorganisms: promising approach as biofertilizers. Int. J. Agron. 2019, 1–7. 10.1155/2019/491725633911103 [DOI] [Google Scholar]
- Karuku G., Onwonga R., Kathumo V. (2018). Effects of tillage practices, cropping systems and organic inputs on soil nutrient content in Machakos County. J. Agric. Sustain. 12. 10.5897/AJAR2018.13444 [DOI] [Google Scholar]
- Kumar U., Shahid M., Tripathi R., Mohanty S., Kumar A., Bhattacharyya P., et al. (2017). Variation of functional diversity of soil microbial community in sub-humid tropical rice-rice cropping system under long-term organic and inorganic fertilization. Ecol. Indic. 73, 536–543. 10.1016/j.ecolind.2016.10.014 [DOI] [Google Scholar]
- Langille M. G., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. 10.1038/nbt.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Zhang D., Song Z., Ren L., Jin X., Fang W., et al. (2022). Organic fertilizer activates soil beneficial microorganisms to promote strawberry growth and soil health after fumigation. Environ. Pollut. 295, 118653. 10.1016/j.envpol.2021.118653 [DOI] [PubMed] [Google Scholar]
- Li S. (2021). Reduced Tillage Increases Grain Yield through Improving Soil Properties and Water Use Efficiency. Liège, Belgium: University of Liège. [Google Scholar]
- Li Y., Liu X., Zhang L., Xie Y., Cai X., Wang S., et al. (2020). Effects of short-term application of chemical and organic fertilizers on bacterial diversity of cornfield soil in a karst area. J. Soil Sci. Plant Nutr. 20, 2048–2058. 10.1007/s42729-020-00274-2 [DOI] [Google Scholar]
- Lin W., Lin M., Zhou H., Wu H., Li Z., Lin W. (2019). The effects of chemical and organic fertilizer usage on rhizosphere soil in tea orchards. PLoS ONE 14, e0217018. 10.1371/journal.pone.0217018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Shu A., Song W., Shi W., Li M., Zhang W., et al. (2021). Long-term organic fertilizer substitution increases rice yield by improving soil properties and regulating soil bacteria. Geoderma 404, 115287. 10.1016/j.geoderma.2021.115287 [DOI] [Google Scholar]
- Liu T., Huang J., Chai K., Cao C., Li C. (2018). Effects of N fertilizer sources and tillage practices on NH3 volatilization, grain yield, and N use efficiency of rice fields in Central China. Front. Plant Sci. 9, 385. 10.3389/fpls.2018.00385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H., Wang K., Wang Z., Peng J., Ren N. (2020). Metabolic function, trophic mode, organics degradation ability and influence factor of bacterial and fungal communities in chicken manure composting. Bioresour. Technol. 302, 122883. 10.1016/j.biortech.2020.122883 [DOI] [PubMed] [Google Scholar]
- Peng J., Han X., Li N., Chen K., Yang J., Zhan X., et al. (2021). Combined application of biochar with fertilizer promotes nitrogen uptake in maize by increasing nitrogen retention in soil. Biochar. 3, 367–379. 10.1007/s42773-021-00090-6 [DOI] [Google Scholar]
- Pongmala K., Pierret A., Oliva P., Pando A., Davong V., Rattanavong S., et al. (2022). Distribution of Burkholderia pseudomallei within a 300-cm deep soil profile: implications for environmental sampling. Sci. Rep. 12, 8674. 10.1038/s41598-022-12795-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo N., Floridia V., D'Alessandro E., Lopreiato V., Pino A., Chiofalo V., et al. (2023). Influence of olive cake dietary supplementation on fecal microbiota of dairy cows. Front. Microbiol. 14, 1406. 10.3389/fmicb.2023.1137452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Liu X., Zhang Q., Gao P., Ren J. (2020). Biochar and organic fertilizer changed the ammonia-oxidizing bacteria and archaea community structure of saline–alkali soil in the North China Plain. J. Soils Sediments 20, 12–23. 10.1007/s11368-019-02364-w [DOI] [Google Scholar]
- Subhash Y., Yoon D.-E., Lee S.-S. (2017). Skermanella mucosa sp. nov., isolated from crude oil contaminated soil. Antonie Van Leeuwenhoek 110, 1053–1060. 10.1007/s10482-017-0878-7 [DOI] [PubMed] [Google Scholar]
- Sun S., Sun H., Zhang D., Zhang J., Cai Z., Qin G., et al. (2019). Response of soil microbes to vegetation restoration in coal mining subsidence areas at Huaibei coal mine, China. Int. J. Environ. Res. Public Health 16, 1757. 10.3390/ijerph16101757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Li C., Xiao X., Shi L., Cheng K., Wen L., et al. (2020). Effects of short-term manure nitrogen input on soil microbial community structure and diversity in a double-cropping paddy field of southern China. Sci. Rep. 10, 1–9. 10.1038/s41598-020-70612-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Xiao X., Xu Y., Li C., Cheng K., Pan X., et al. (2019). Utilization of carbon sources in the rice rhizosphere and nonrhizosphere soils with different long-term fertilization management. J. Basic Microbiol. 59, 621–631. 10.1002/jobm.201800736 [DOI] [PubMed] [Google Scholar]
- Virlouvet L., Jacquemot M.-P., Gerentes D., Corti H., Bouton S., Gilard F., et al. (2011). The ZmASR1 protein influences branched-chain amino acid biosynthesis and maintains kernel yield in maize under water-limited conditions. Plant Physiol. 157, 917–936. 10.1104/pp.111.176818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Li Y., Zhang J., Wang X., Hong J., Qiu C., et al. (2022). Transcriptome profiling analysis of phosphate-solubilizing mechanism of pseudomonas strain W134. Microorganisms 10, 1998. 10.3390/microorganisms10101998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang G., Guo T., Xing Y., Mo F., Wang H., et al. (2021). Effects of plastic mulch and nitrogen fertilizer on the soil microbial community, enzymatic activity and yield performance in a dryland maize cropping system. Eur. J. Soil Sci. 72, 400–412. 10.1111/ejss.12954 [DOI] [Google Scholar]
- Xin W., Zhang L., Zhang W., Gao J., Yi J., Zhen X., et al. (2019). An integrated analysis of the rice transcriptome and metabolome reveals differential regulation of carbon and nitrogen metabolism in response to nitrogen availability. Int. J. Mol. Sci. 20, 2349. 10.3390/ijms20092349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. C., Shen Q. R., Ran W. (2003). Content and distribution of forms of organic N in soil and particle size fractions after long-term fertilization. Chemosphere 50, 739–745. 10.1016/S0045-6535(02)00214-X [DOI] [PubMed] [Google Scholar]
- Yang Y., Awasthi M. K., Bao H., Bie J., Lei S., Lv J. (2020). Exploring the microbial mechanisms of organic matter transformation during pig manure composting amended with bean dregs and biochar. Bioresour. Technol. 313, 123647. 10.1016/j.biortech.2020.123647 [DOI] [PubMed] [Google Scholar]
- Zavarzina A. G., Nikolaeva T. N., Demin V. V., Lapshin P. V., Makarov M. I., Zavarzin A. A., et al. (2019). Water-soluble phenolic metabolites in lichens and their potential role in soil organic matter formation at the pre-vascular stage. Eur. J. Soil Sci. 70, 736–750. 10.1111/ejss.12822 [DOI] [Google Scholar]
- Zhao M., Zhao J., Yuan J., Hale L., Wen T., Huang Q., et al. (2021). Root exudates drive soil-microbe-nutrient feedbacks in response to plant growth. Plant Cell Environ. 44, 613–628. 10.1111/pce.13928 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI - PRJNA948916.