Although the role of copper (Cu) in promoting KRas- or BRaf-mutation driven cancers via activating MEK1/2 kinases is known, the mechanism by which the copper transporter SLC31A1 (CTR1) is upregulated in pancreatic cancer (PDAC, KRas mutation) is not defined. In this study, we provide evidence that MEK signal maintains a high level of SLC31A1 through silencing the expression of miR-124-3P (miR-124) via a novel MEK-DNMT1-miR-124 feedback loop in PDAC cells. Further, we reveal that miR-124 directly targets suppression of SLC31A1, and miR-124 introduction together with tetrathiomolybdate (TM) treatment hampered pancreatic cancer growth in vitro and in vivo. Our results demonstrate that a SLC31A1-MEK-DNMT1-miR-124 feedback loop is an important pathway to maintain copper absorption and promote pancreatic cancer progression, and we hope to provide a Cu-chelation as an adjuvant treatment strategy, to block the progression in Kras mutant PDAC patients.
Oncogenes, such as KRas, is frequently activated during the malignant progression of PDAC. Almost 90% patients harbor mutations of the KRas, and activated KRas triggered its downstream BRaf and then MEK-ERK signal pathway.1 Recently, several inhibitors to MEK or ERK have been developed and assayed in various cancers, including PDAC. Among these inhibitors, Cu chelators attract our attention due to its practicality and novelty. Cu enhances phosphorylation of MEK1 in a dose-dependent way, and Cu chelators could reduce the ability of MEK1 to phosphorylate ERK1/2.2
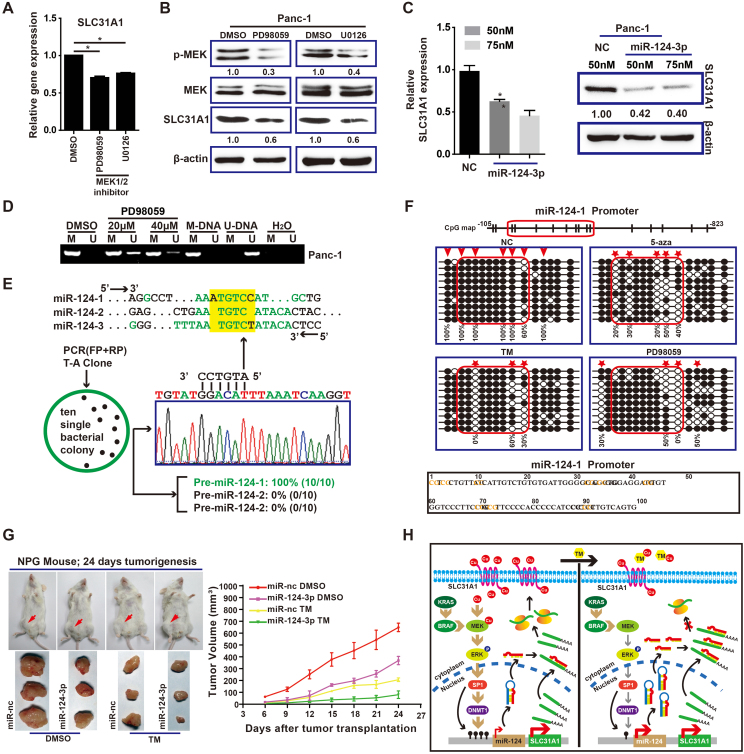
Our previous researches revealed that copper content in pancreatic cancer tissues was significantly higher than that in corresponding adjacent tissues, as well as the copper transporter SLC31A1 was markedly upregulated in PDAC tissues and cells.3 To reveal the proposed link between upregulated levels of SLC31A1 and Kras-MEK pathway, we first found the abnormal upregulation of SLC31A1 in COAD, DLBC, GBM, LGG, PAAD, READ, STAD, and UCEC through the public database GEPIA (Fig. S1A, B). Intriguingly, these cancers mostly occur missense Kras mutations (activated Kras) (Fig. S1C). Furthermore, prognostic survival analysis indicated the lower expression of SLC31A1 was associated with longer survival in PAAD (Fig. S1D). As shown in Figure 1A and B, the suppressive effect of MEK inhibitors on SLC31A1 mRNA level was consistent with that of SLC31A1 protein detection. These data suggest SLC31A1 is closely related to the Kras signal pathway.
Figure 1.
The SLC31A1-MEK-DNMT1-miR-124 feedback loop was revealed by molecular experiments and clinical database analyses. (A, B) qRT-PCR and Western blot were used to measure the expression of SLC31A1 in Panc-1 cells treated with MEK1/2 inhibitors. (C) Effects of miR-124 overexpression on endogenous SLC31A1 expression as measured by qPCR and Western blot. (D) Methylation-specific PCR analyses of the effect of PD98059 on the methylation status in Panc-1 cells. M-DNA methylated human control DNA (bisulfite converted), U-DNA unmethylated human control DNA (bisulfite converted). (E) T-A clone and sequencing analysis are used to distinguish between pre-miR-124-1, 2 and 3. (F) TM and PD98059 induce demethylation of miR-124 promoter. Nonmethylated and methylated CpGs are depicted as open and solid circles, respectively. The red arrows indicate the reported methylation sites and the red asterisk suggests the prominent sites of demethylation. (G) Effects of miR-124 and TM on subcutaneous xenograft tumor formation in NPG mice. (H) Schematic representation of the feedback loop that involves copper, MEK signal, DNMT1 and miR-124 in the regulation of SLC31A1 expression. Each bar in the figure represents the mean ± SEM of triplicates. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
According to our previous studies, SLC31A1 is highly expressed in PADC,3 which suggests the post-transcriptional or transcriptional regulation of SLC31A1 is disordered. Using miRDB, TargetScan, miRanda and DIANA to screen predicted candidate miRNAs that might regulate SLC31A1 expression. We focused on miR-124 since it was the only miRNA by intersection of above independent lists of miRNA-SLC31A1 prediction (Fig. S2A, B). Data from 299 PDAC samples displayed a significant negative correlation between the expression of miR-124 and SLC31A1 (Fig. S2C). Furthermore, miR-124 was significantly reduced in PDAC and pancreatitis patients, compared with the control group (healthy individual) in GEO datasets (GSE24279), suggesting that SLC31A1 is the potential target of miR-124 (Fig. S2D). Q-PCR and Western blot analysis confirmed the expression of SLC31A1 was decreased in Panc-1 cells transfected with miR-124 (Fig. 1C). Finally, dual-luciferase reporter results showed that miR-124 mimic decreased the luciferase activity in the 293 T cells transfected with SLC31A1-Wt plasmid, but not in the cells transfected with SLC31A1-Mut plasmid (Fig. S2E).
Evidence demonstrates that epigenetic silencing of miR-124 is an early event in pancreatic carcinogenesis due to promoter methylation.4 DNMT1 was validated to be elevated in PDAC (Fig. S3A, B). Therefore, we examined the effect of MEK inhibitors on the levels of miR-124 and DNMT1. Results showed that treatment with PD98059 in Panc-1 cells resulted in increases in the level of miR-124, and decrease in that of DNMT1, at both mRNA and protein levels (Fig. S3C, D). Moreover, as shown, the CpG island of miR-124 was methylated in Panc-1 cells, but was demethylated after PD98059 treatment, as indicated by the increases in unmethylated DNA (U) (Fig. 1D). To our knowledge, miR-124 is represented in three different genomic loci on 8p23.1 (miR-124-1), 8q12.13 (miR-124-2) and 20q13.3 (miR-124-3) (Fig. S3E). Based on the difference between miR-124-1, miR-124-2 and miR-124-3 precursors on the RNA loop sequences (Fig. S3F), we performed RT-PCR amplification and sequencing for their RNA loop regions, and calculated the ratio of each type of pre-miR-124 in Panc-1 cells. As shown in Figure 1E, we were surprised to find that all clones were pre-miR-124-1. These results suggest that pre-miR-124-1 is dominant in Panc-1. Next, we observed that the inhibitor of DNA methylation, 5-Aza (3 μM), could increase the level of pre-miR-124 (Fig. S3G), among which most are pre-miR-124-1 (Fig. S3H). This mechanistic link was further confirmed by the effect of 5-Aza on the expression of SLC31A1 (Fig. S3I), which confirmed the intermediary role of DNMT1 on the relation between miR-124 and SLC31A1 expressions.
Recent researches have suggested that DNA methylation could be regulated by MEK-ERK activity in some circumstances.5 As expected, MEK inhibitors increased the expression level of pre-miR-124 in a dose-dependent manner (Fig. S4A). Copper chelator TM could also significantly inhibit DNMT1 and upregulated pre-miR-124 level (Fig. S4B). Moreover, clinical data of PDAC patients from TCGA ENCORI showed significant correlations between NDMT1 vs. SLC31A1, NDMT1 vs. miR-124, and miR-124 vs. SLC31A1, respectively (Fig. S4C). Based on the above data, we proposed that Cu-MEK upregulated the expression of SLC31A1 via the MEK-DNMT1-miR124 signaling axis, as depicted in Figure S4D. Next, we utilized bisulfite sequencing to analyze the methylation status on the promoter regions of miR-124 in Panc-1 cells treated with TM or PD98059, 5-Aza was using as the positive control. We found that TM or PD98059 treatment decreased methylation levels of CpG islands in the promoter of miR-124 at CpG sites 2, 3, 4, and CpG sites 4, 5, respectively (Fig. 1F). Finally, we found that overexpression of miR-124 together with TM significantly decreased cancer cell proliferation, colony formation, and tumor growth in vivo (Fig. S5A, B; Fig. 1G).
In summary, we demonstrate that constitutively activated Kras is highly related with the expression of SLC31A1 in PDAC cells. We show that miR-124 can target decreasing the level of copper transporter SLC31A1 (CTR1), while SLC31A1 modulates MEK signal transduction and DNA Methyltransferase 1 (NDMT1) expression by regulating copper influx. Moreover, DNMT1 acts as a regulator of methylation mediating silence of miR-124, hence, downregulation of DNMT1 increases the level of miR-124. Taken together, these results reveal a feedback loop between miR-124 and MEK signal (Fig. 1H). This feedback loop comprises miR-124, SLC31A1, MEK and DNMT1, which are essential for PDAC growth. What's more, we reveal an intricate signaling network, at least partially, contributing the absorption of copper under physiological conditions in PDAC patients.
Conflict of interests
No potential conflicts of interest are disclosed.
Funding
This study was supported by The National Natural Science Foundation of China (No. 32072801) and Fundamental Research Funds for the Central Universities of China (No. 2572020DY12).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.05.033.
Contributor Information
Ze Yu, Email: Zeyunfu@163.com.
Chun-Bo Teng, Email: chunboteng@nefu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kinsey C.G., Camolotto S.A., Boespflug A.M., et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 2019;25(4):620–627. doi: 10.1038/s41591-019-0367-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brady D.C., Crowe M.S., Turski M.L., et al. Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature. 2014;509(7501):492–496. doi: 10.1038/nature13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu Z., Zhou R., Zhao Y., et al. Blockage of SLC31A1-dependent copper absorption increases pancreatic cancer cell autophagy to resist cell death. Cell Prolif. 2019;52(2) doi: 10.1111/cpr.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang P., Chen L., Zhang J., et al. Methylation-mediated silencing of the miR-124 genes facilitates pancreatic cancer progression and metastasis by targeting Rac1. Oncogene. 2014;33(4):514–524. doi: 10.1038/onc.2012.598. [DOI] [PubMed] [Google Scholar]
- 5.Chu P.C., Lin P.C., Wu H.Y., et al. Mutant KRAS promotes liver metastasis of colorectal cancer, in part, by upregulating the MEK-Sp1-DNMT1-miR-137-YB-1-IGF-IR signaling pathway. Oncogene. 2018;37(25):3440–3455. doi: 10.1038/s41388-018-0222-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.