There are numerous online databases dedicated to genes associated with human diseases. The redundancy and lack of consistency of disease terms in these databases pose a significant impediment in high throughput data analysis. To address this, we developed GeDiPNet by compiling data available on gene-disease associations from six popular open access sources namely DisGeNET,1 ClinGen, ClinVar,2 HPO,3 OrphaNet, and PsyGeNET; and curated through several stages including manual curation and merging of disease names to ensure standard vocabulary (Supplementary file 1). GeDiPNet currently includes information on 12,270 genes that have disease association curated from literature records and 20,013 genes have disease association based on text mining.
Comorbid disease prediction using GeDiPNet
The non-redundant, curated data of genes and associated diseases in GeDiPNet can be effectively mined for unraveling shared disease etiologies and novel therapies using algorithms such as enrichment analysis, comorbidity risk prediction and polypharmacological target prediction that are integrated in GeDiPNet.
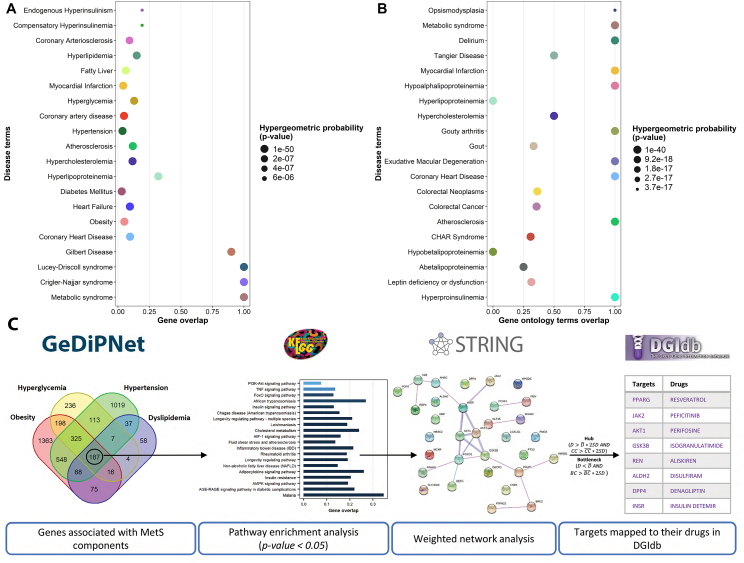
The utility of GeDiPNet for comorbid disease prediction is illustrated using metabolic syndrome (MetS) as a case study. Disease enrichment algorithm allows hypothesis-free identification of comorbid diseases based on probability of genetic and biological processes overlap between the diseases not occurring by chance. The top 20 enriched diseases for MetS genes are displayed in Figure 1. Syndromes related to bilirubin metabolism – Crigler-Najjar syndrome, Lucey-Driscoll syndrome and Gilbert disease are amongst the top most enriched diseases based on genetic overlap (Fig. 1A). Comorbid conditions of MetS such as hyperinsulinemia, leptin deficiency syndrome, hypercholesterolemia, obesity, hypertension and diabetes are most significantly enriched based on functional overlap (Fig. 1B).
Figure 1.
Top 20 enriched diseases (in descending order of significance) identified for MetS based on shared (A) genes and (B) gene ontology terms associated with MetS. (C) Workflow for prediction of potential polypharmacological targets of MetS.
The outcome of the disease enrichment analysis is consistent with our current understanding of MetS. Oxidative stress in MetS is known to be caused due to hyperuricemia and hypobilirubinemia. Therefore, MetS individuals are at higher risk of developing syndromes related to bilirubin metabolism. Individuals with MetS are clinically known to suffer from multimorbidities; primarily linked through obesity and insulin resistance. Hence, it is expected that diseases such as cardiovascular disorders, diabetes, and fatty liver disease are identified to have a higher comorbidity risk with MetS.
Polypharmacological target prediction using GeDiPNet
The importance of managing complex diseases with drugs that act against multiple targets is well known. These polypharmacological targets can be identified based on shared genes amongst the comorbid conditions followed by pathway and network analysis of these genes.
The utility of GeDiPNet in prediction of polypharmacological targets was established using our existing knowledge of four popular polypharmacological drugs and targets. We observed that three of the four known polypharmacological targets, could be correctly identified using GeDiPNet (Supplementary file 2).
The algorithm correctly identified the target gene (PDE5A) and the drug Sildenafil for hypertension and erectile dysfunction. The drug methotrexate represses cell division leading to anti-inflammatory action. Therefore, methotrexate is used for the treatment of inflammatory diseases such as rheumatoid arthritis, Crohn's disease, and psoriasis. GeDiPNet correctly predicted the target ADA and drug methotrexate for these three diseases. Metformin is a well-known drug for treatment of insulin resistance observed in individuals with type II diabetes and PCOS. It promotes insulin signaling by activating phosphatidylinositol 3-kinase (PIK3CA). GeDiPNet, when queried for PCOS and diabetes mellitus correctly identified the target PIK3CA and its drug metformin. Dimethyl fumarate is used for treatment of multiple sclerosis and psoriasis. It mediates its anti-oxidant action via decrease in nuclear translocation of NRF2. Since NRF2 is not yet reported as an associated gene for multiple sclerosis or psoriasis in any of the online databases, the algorithm could not predict the polypharmacological drug or the target in this case. Hence, the utility of the tool to predict a polypharmacological target is dependent on the accuracy and completeness of the data captured in gene-disease databases.
Polypharmacological targets for MetS were predicted by querying the tool with MetS components such as obesity, hyperglycemia, hypertension and dyslipidemia (Fig. 1C). The algorithm identified 32 genes (Supplementary file 3) that included INSR, JAK2, REN, GSK3, DPP4, MAPK, ESR1 and MC4R as potential targets. Many of these targets are currently being used in management of components of MetS, for example: i) hyperglycemia is managed using inhibitors of JAK2 and GS3K; as well as agonists of PPARG and INSR, ii) hypertension is managed using inhibitors of REN and JAK2, and iii) hypertriglyceride is managed by PPARG agonist. In addition to these known targets, few targets identified by the algorithm such as DECR1, involved in fatty acid metabolism and inflammation, and HPGDS, involved in renal programming, are worthy of further validation as therapeutic targets for MetS. GeDiPNet also maps the identified targets to their known drugs from DGIdb4 (Supplementary file 3).
The enhanced utility of GeDiPNet as compared to similar online resources has been demonstrated below using few examples.
-
1.
Disease pathway enrichment analysis: Ocular albinism is a genetic condition, characterized by reduced pigmentation in the retina and visual acuity. It is majorly caused due to dysfunctional G protein coupled receptor 143 (GPR143) and tyrosinase enzymes, that are critical for melanin production. The disease is annotated as “ocular albinism” and “albinism, ocular” in HPO and DisGeNET respectively. Pathway enrichment analysis using Enrichr of 22 genes associated with ocular albinism from HPO identified tyrosine metabolism, nicotine addiction and lysine degradation pathways; whereas analysis of four disease-associated genes from DisGeNET identified renin secretion, and GABAergic synapse, as most significant. Enrichment analysis using GeDiPNet identified melanogenesis pathway to be most enriched followed by GABAergic synapse and tyrosine metabolism. Melanin synthesis pathways play a crucial role in ocular albinism, which is identified only by GeDiPNet.
-
2.
Comorbidity risk prediction: Nicolaides Baraitser Syndrome (NBS) is a neuro-developmental disorder with intellectual disability, seizures and short stature. Prediction of comorbid diseases for NBS, using GeDiPNet, identified coffin-siris syndrome and retinoschisis as most significantly enriched. Coffin-siris syndrome is known to be comorbid and phenotypically very similar to NBS. A case report in 2019 documented association of ocular disorders in a girl with NBS. Retinoschisis is predicted to be comorbid with NBS because of SMARCA2, RS1 and CDKL5 genes commonly associated with the two diseases. The disease association of SMARCA2 is documented in HPO, OrphaNet and curated subset of DisGeNET. ClinVar has cataloged the association of RS1 and CDLK5 genes. The comorbid diseases could be identified by GeDiPNet due to the manually merged data from multiple sources.
-
3.
Drug-target prediction: Behcet syndrome (BS) is a multisystemic inflammatory disorder characterized by oral and genital ulcers, retinal vasculitis and deep vein thrombosis. The pathogenesis of BS is complex and the role of inflammatory cytokines such as TNF-α and IFN-γ is well known. Cyclophosphamide is one of the drugs used in the management of arterial aneurysms in BS. GeDiPNet identified tumor necrosis factor receptor 1 (TNFRSF1A) and cyclophosphamide as a potential target and drug respectively for BS. It is to be noted that the association of TNFRSF1A with BS is only documented in ClinVar database.
The comparative statistics of the data available in GeDiPNet and the other popular online resources are provided in Supplementary file 4. GeDiPNet will be maintained and updated yearly, and can be freely accessed at http://gedipnet.bicnirrh.res.in.
Ethics declaration
Exemption from ethics review was approved by the ICMR-NIRRCH Ethics Committee for Clinical Studies (ICEC/Sci-160/165/2020).
Author contributions
S.I-T planned, designed, and supervised the study. M.S and I.K curated and annotated the data. R.S.B, I.K, and K.P developed the interface and algorithms for analysis. I.K., M.S and S.I-T wrote the manuscript. All authors approved the manuscript.
Conflict of interests
The authors declare that they have no conflict of interests.
Funding
This work (No. RA/1170/11–2021) was supported by grants received from the Department of Biotechnology, India (No. BT/PR40165/BTIS/137/12/2021); Science and Engineering Research Board, India (No. STR/2020/000034).
Data availability
Data download is available in browse pages in http://gedipnet.bicnirrh.res.in.
Acknowledgements
We are grateful to Ms. Ulka Gawde for assisting in the disease data curation and data merging; Mr. Aman Vishvakarma for assisting in interface design and Ms. Kaushiki Prabhudesai, Ms. Sameeksha Bhaye for internally reviewing the database.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.05.034.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Detailed materials and methods (MS Word).
List of popular polypharmacological drugs with associated targets and diseases (MS Word).
Summary of targets identified for MetS (MS Excel).
Comparative statistics of the curated data available in disease databases (MS Word).
List of manually curated disease terms (MS Excel).
References
- 1.Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48(D1):D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landrum M.J., Chitipiralla S., Brown G.R., et al. ClinVar: improvements to accessing data. Nucleic Acids Res. 2020;48(D1):D835–D844. doi: 10.1093/nar/gkz972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Köhler S., Gargano M., Matentzoglu N., et al. The human phenotype ontology in 2021. Nucleic Acids Res. 2021;49(D1):D1207–D1217. doi: 10.1093/nar/gkaa1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freshour S.L., Kiwala S., Cotto K.C., et al. Integration of the drug-gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49(D1):D1144–D1151. doi: 10.1093/nar/gkaa1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed materials and methods (MS Word).
List of popular polypharmacological drugs with associated targets and diseases (MS Word).
Summary of targets identified for MetS (MS Excel).
Comparative statistics of the curated data available in disease databases (MS Word).
List of manually curated disease terms (MS Excel).
Data Availability Statement
Data download is available in browse pages in http://gedipnet.bicnirrh.res.in.