Abstract
Small non-coding RNAs are potential diagnostic biomarkers for lung cancer. Mitochondria-derived small RNA (mtRNA) is a novel regulatory small non-coding RNA that only recently has been identified and cataloged. Currently, there are no reports of studies of mtRNA in human lung cancer. Currently, normalization methods are unstable, and they often fail to identify differentially expressed small non-coding RNAs (sncRNAs). In order to identify reliable biomarkers for lung cancer screening, we used a ratio-based method using mtRNAs newly discovered in human peripheral blood mononuclear cells. In the discovery cohort (AUC = 0.981) and independent validation cohort (AUC = 0.916) the prediction model of eight mtRNA ratios distinguished lung cancer patients from controls. The prediction model will provide reliable biomarkers that will allow blood-based screening to become more feasible and will help make lung cancer diagnosis more accurate in clinical practice.
Keywords: Lung cancer, Mitochondria-derived small RNAs, Plasma, Ratio, Small non-coding RNAs
Introduction
Lung cancer remains the leading cause of cancer-related death worldwide.1 Recently, significant efforts have been focused on the detection of lung cancer through low-dose computed tomography (LDCT) scanning. Although lung cancer screening with low-dose CT benefits the patients, screening costs are expensive.2 Moreover, annual LDCT screening with conventional chest radiography and reveal that LDCT screening reached a 20% decrease in lung cancer mortality after only three rounds of screening.3 Although studies on biomarker for the detection of lung cancer are numerous, their diagnostic accuracy was low.4, 5, 6, 7 Hence, it is crucial to find inexpensive and reliable methods to detect lung cancer.
In the process of developing and progressing lung cancer, mitochondria play an essential role in maintaining cell energy metabolism.8 A total of 37 genes are found in the human mitochondrial DNA (mtDNA). These include 2 rRNAs, 22 tRNAs, and 13 protein-coding genes.9 In addition, only about 12% of the individual small RNAs determinate were encoded in mitochondria. Pathophysiological functions and cancer development may be influenced by mtRNAs. While various sncRNAs, including miRNA, snoRNA, and piRNA, have been extensively studied in the diagnosis of lung cancer, little is known about small RNAs derived from mitochondria (mtRNAs).4,10, 11, 12 A majority of sncRNA normalization studies use synthetic external controls or published endogenous miRNAs as controls. sncRNA studies cannot directly use these references due to their labile nature. The ratio transformation method can be used for the difficult task of normalizing sncRNA data for the purpose of identifying reliable biomarkers.13, 14, 15, 16 The internal or external control normalization-based method requires two assumptions. A first assumption is that the sncRNAs and the internal control are both affected by the same systematic influences; the second assumption is the true internal control values are the same for all samples. In contrast, the ratio-based method supposes that all sncRNAs in a sample share the identical systematic factors. We propose a new method for lung cancer diagnosis by using peripheral blood mtRNAs and ratio transformation.
Methods
Datasets
Gene Expression Omnibus (GEO) repository (GSE148861, GSE148862) was used for discovery and independent validation. SPORTS1.1 software was used to align each small RNA-seq to extract mtRNA expression levels.17 Initially, all miRNA-seq FASTQ files were cleaned of adapters using NF-core/smrnaseq software.18 STAR was used to align the trimmed sequence reads to the mitotRNAdb database.19 Counts were obtained using the htseq-count script from the HTSeq tools.20 MetImp 1.2 imputed missing values.21
Ratio transformation and statistical analysis
Using a ratio transformation method, we were able to stabilize the mtRNA expression profile.13 mtRNA1-mtRNA2 transformation were performed using the following equation: mtRNA1 to mtRNA2 = mtRNA1/mtRNA2. We analyzed differentially expressed (DE) mtRNAs in the discovery group using unpaired Student's t-test after log transformation. Feature Selection and modeling were conducted by Random Forest. ROC curves were graphed using the “precrec” package. Random forest regression was performed using “randomForest” package. Principal Component Analysis (PCA) was performed using “ggfortify” package. A p-value of 0.05 has been assigned to statistical significance.
Diagnostic model
Differentially expressed (DE) (FDR <0.01) and fold change (| Log2(fold change) | > 1) ratio-mtRNAs were involved in the dianostic model. Based on the random Forest model, we calculated the mean decrease in accuracy and mean decrease Gini of every transformed mtRNA. Choosing features according to the overlapping of the top ten mean decreases in accuracy and mean decreases in Gini (Table S1). A final prediction model was built based on selected features.
Bioinformatic prediction and GO enrichment analysis
The minimum free-energy hybridization (energy < −20 kcal/mol) of eleven significantly differentially expressed mtRNAs and target mRNA were predicted by RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid). Gene Ontology (GO) analysis was utilized to profound examine the potential molecular function (MF) and cellular component (CC) of predicted target genes.
Results
Patient cohorts
Table 1 summarizes the clinical features of two lung cancer cohorts. We analyzed 76 cases, comprising 51 lung cancer patients and 25 healthy controls. The lung cancer group in the discovery cohort had an average age of 62.5 ± 0.28, while the control group had an average age of 52.7 ± 1.0. The lung cancer group had 23 (63.8%) adenocarcinoma, 6 (16.7%) squamous cell carcinoma, and 5 (13.9%) small cell lung carcinoma, respectively. A total of 13 (36.1%) patients were smokers and 18 (50%) patients had distant metastasis. A comparison of lung cancer and control groups showed an average age of 45 ± 1.85 versus 60 ± 0.76 in the validation dataset. The lung cancer group had 9 (60.0%) adenocarcinoma, 3 (20.0%) squamous cell carcinoma, and 3 (20.0%) small cell lung carcinoma, respectively. There are 3 (20%) had distant metastasis and 4 (26.7%) patients were smokers.
Table 1.
Clinical characteristics of discovery and independent validation lung cancer cohort.
| Cohort | Discovery | Validation |
|---|---|---|
| Num. of patients | 49 | 27 |
| Age in years, mean (SD) | ||
| Control | 52.7(1.0) | 45(1.85) |
| Cancer | 62.5(0.28) | 60(0.76) |
| Males, count (%) | ||
| Control | 7/13(53.8) | 6/12(50.0) |
| Cancer | 21/36(58 0.3) | 10/15(66.6) |
| Cancer type (%) | ||
| ADC | 23/36(63.8) | 9/15(60) |
| SCC | 6/36(16.7) | 3/15(20) |
| SCLC | 5/36(13.9) | 3/15(20) |
| Other | 2/36(5.6) | |
| Smoking history (%) | ||
| YES | 13/36(36.1) | 4/15(26.7) |
| NO | 23/36(63.9) | 11/15(73.3) |
| Distant metastasis (%) | ||
| YES | 18/36(50) | 3/15(20) |
| NO | 18/36(50) | 12/15(80) |
| Lymph node involvement (%) | ||
| YES | 15/36(41.7) | 4/15(26.7) |
| NO | 21/36(58.3) | 11/15(73.3) |
| Tumor stages (%) | ||
| Stage I | 10/36(27.8) | 4/15(26.7) |
| Stage II | 2/36(5.5) | 3/15(20) |
| Stage III | 6/36(16.7) | 5/15(33.3) |
| Stage IV | 18/36(50) | 3/15(20) |
The molecular signature composed of mtRNAs
A total of 15 mtRNA species were identified in peripheral blood samples of human subjects (Table S2). By classifying mtRNAs according to their parent tRNA types, there are thirteen types of mtRNAs (i.e., mt-tRNA-His, mt-tRNA-Ser). Sequences of mtRNA ranged in length from 16 to 38 nucleotides, with an average of 25.5 nucleotides.
Eight dysregulated mtRNAs diagnostic model were constructed in training dataset
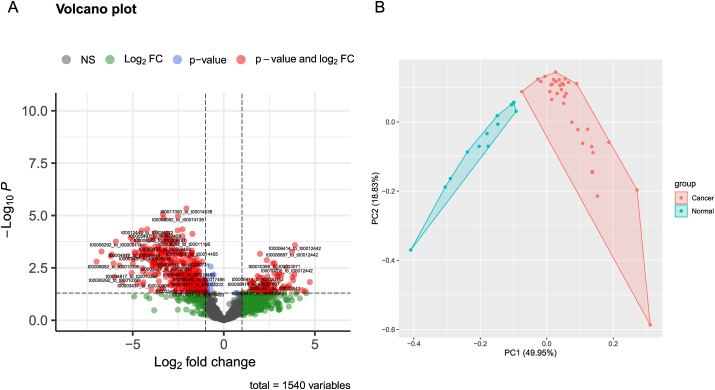
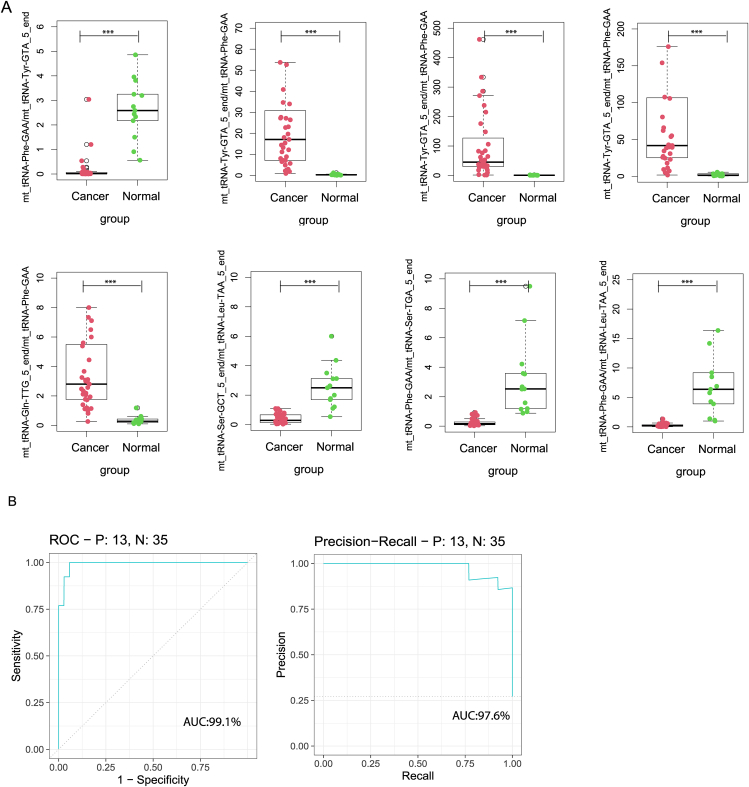
Following the criteria described in the methods section, 106 mtRNA pairs were significantly different by student t-test (Fig. 1A and Table S3). Unsupervised hierarchical clustering PCA analysis was performed based on the DE mtRNA pairs. The PCA plot demonstrated a totally distinct expression pattern of the mtRNA pairs signature between the lung cancer and control group (Fig. 1B). RF was used to select the most effective variables from 106 mtRNA-pairs to build prediction models. As a result of the RF mean accuracy decrease, eight mtRNA pairs were selected, including up-regulated mt_tRNA-Tyr-GTA_5_end/mt_tRNA-Phe-GAA, mt_tRNA-Tyr-GTA_5_end/mt_tRNA-Phe-GAA, mt_tRNA-Tyr-GTA_5_end/mt_tRNA-Phe-GAA, mt_tRNA-Gln-TTG_5_end/mt_tRNA-Phe-GAA and down-regulated mt_tRNA-Phe-GAA/mt_tRNA-Tyr-GTA_5_end, mt_tRNA-Ser-GCT_5_end/mt_tRNA-Leu-TAA_5_end, mt_tRNA-Phe-GAA/mt_tRNA-Ser-TGA_5_end, mt_tRNA-Phe-GAA/mt_tRNA-Leu-TAA_5_end in cancer samples (Fig. 2A). The model's performance was evaluated using the receiver operating characteristic curve (ROC curve) and the Precision-Recall curve (PR curve). The area under the receiver operating characteristic (ROC) curve (AUC) for cancer subjects was 0.991 (AUC), while the AUC for the PR curve was 0.976 (Fig. 2B).
Figure 1.
mtRNA diagnostic panel. (A) Volcano plot of differentially expressed mtRNAs between lung cancer and control group. (B) Principal Component Analysis (PCA) plot.
Figure 2.
The performance of the prediction model in the discovery cohort. (A) Eight model selected mtRNA expression levels in discovery cohort. (B) ROC curve and PR curve of the diagnostic prediction model with mtRNA markers in discovery cohort. ∗∗∗P < 0.001.
The expression of eight dysregulated mtRNAs in external validation set were highly consistent with those of the training set
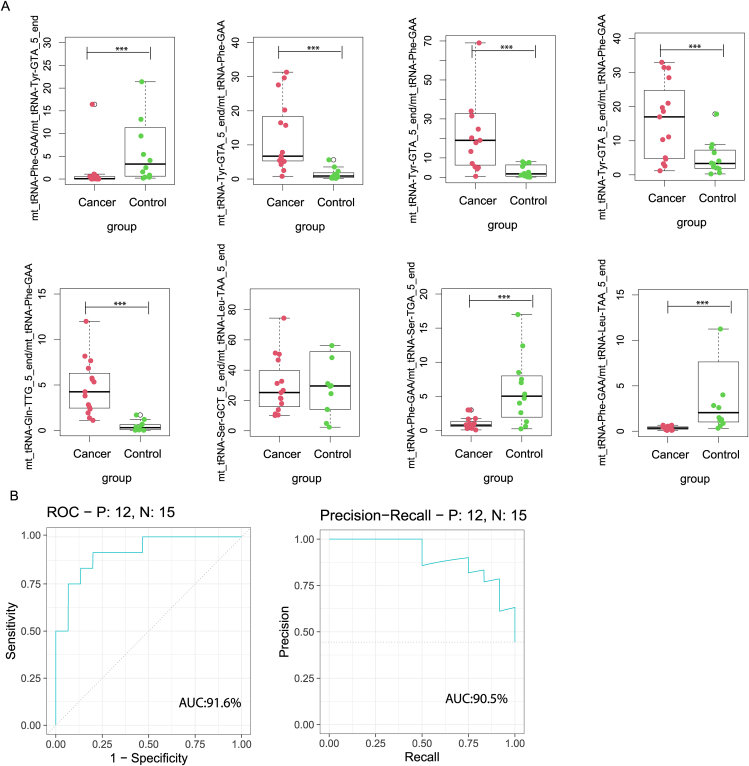
The prediction model was further evaluated in an independent validation cohort. Within the validation dataset, the boxplot shows the levels of expression of eight mtRNAs (Fig. 3A). The remaining variation is significant, except for t00016493_to_t00024522, which is marginal for the validation cohort. The AUC in the prediction model was 0.916, and the AUC in the PR curve was 90.5 (Fig. 3B), which indicates the high classification power for lung cancer screening.
Figure 3.
The performance of the prediction model in the independent validation cohort. (A) Eight model selected mtRNA expression levels in independent validation data set. (B) ROC curve and PR curve of the diagnostic prediction model with mtRNA markers in the independent validation cohort. ∗∗∗P < 0.001.
Bioinformatic function prediction
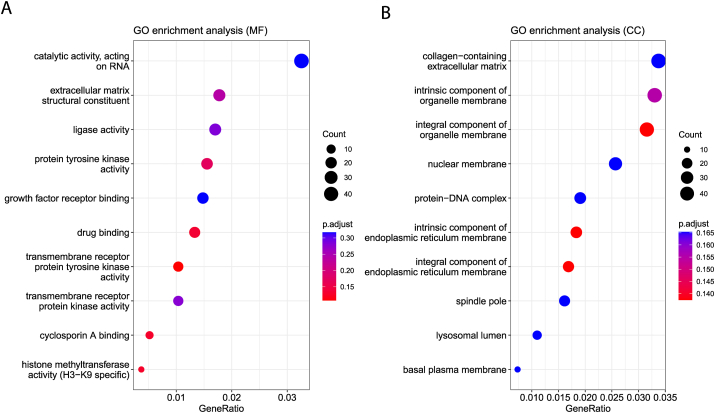
To investigate the regulatory function of the ten mtRNA signatures, we utilized the RNAhybrid, bioinformatic functional prediction tool. Molecular function (MF) and cellular component of GO analysis show that cellular component is catalytic activity acting on RNA and collagen−containing extracellular matrix terms over-represented significantly (Fig. 4A, B).
Figure 4.
Bioinformatic prediction and GO enrichment analysis. (A) Gene Ontology analysis of molecular function. (B) Gene Ontology analysis of cellular component.
Discussion
Although substantial improvements have been made over the past several decades, lung cancer remains the leading cause of cancer mortality worldwide. The early and accurate diagnosis is vital to enhancing the survival rate of patients with lung cancer. Due to the significant expense and high false-discovery rate, the achievement of CT screening is inadequate.22 Hence, the availability of blood-based screening could improve lung cancer patient uptake. Our present study aimed to identify the new peripheral blood small non-coding RNAs as predictive biomarkers for lung cancer diagnosis. In this article, we use the peripheral blood mtRNA, which is first applied to lung cancer, to detect lung cancer diagnostic biomarkers by ratio-based method and machine learning method. The panel of the peripheral blood mtRNA biomarkers can discriminate lung cancer patients from control subjects with ROC curve AUC = 0.991 and PR curve AUC = 0.976. The diagnostic performance of the biomarkers was further validated in an independent validation set (ROC AUC = 0.916, PR AUC = 0.905). ROC analysis has shown that the set of eight mtRNAs have the ability to better distinguish the lung cancer from normal than any other subset of mtRNAs. It is known from the literature that the evidence for mtRNA levels decreasing in several cancers is mutually consistent.23,24 These mtRNAs may act as tumour suppressors. Hence, the discovery might be a significant characteristic if they are applied for the diagnosis of lung cancer.
Mitochondria are complex eukaryotic organelles for maintaining cell energy metabolism. Mitochondrial dysfunction has been involved in an overabundance of human diseases, most distinctly in cancer and aging.25,26 The respiratory chain of mitochondrial metabolism produces considerable levels of reactive oxygen species (ROS) which can alter the structure of the respiratory chain and cause mitochondrial DNA damage.27 Furthermore, due to the lack of protective histones, introns, and efficient DNA repair systems, mitochondrial DNA (mtDNA) obtains 10-fold more mutations than nuclear genomic DNA. In particular, damages to mitochondrial DNA are susceptible to smoking. In lung cancer, the frequency of mtDNA mutation was significantly higher in non-smokers when compared to smokers.8 The functional analysis of the mtRNA-regulated genes showed a clear bias towards catalytic activity acting on RNA and collagen-containing extracellular matrix. The interaction between mtRNAs and mRNA targets may function as a miRNA-like active structure.
In summary, we have confirmed that a differential mtRNA expression profile exists in the peripheral blood of lung cancer patients. Moreover, the prediction model has been established that may have diagnostic potential to improve the non-invasive detection of Lung cancer.
Limitations
While models have been established for the diagnosis of lung cancer, the functions of these mtRNAs remain unclear. Further research is needed on this issue. Moreover, the prediction model may be not easy to use by those with little familiarity with the coding.
Author contributions
Youping Deng, Gang Huang, Wei Chen and Zhougui Ling had the concept and established the investigation. Shaoqiu Chen, Zongtao Yu, Ying Tang, and Zhenming Tang collected and processed data. Shaoqiu Chen and Ting Gong drafted the manuscript, and all authors checked the manuscript and approved the version for publication.
Conflict of interests
The authors report no conflict of interests.
Funding
Dr.Deng is partially supported by the National Institutes of Health (NIH) grants 1R01CA223490, 5P30GM114737, 5P20GM103466, 5U54MD007601, 5P30CA071789, 1R01CA230514, U54CA143727 and P20GM139753.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.07.013.
Contributor Information
Zhougui Ling, Email: lzg228@163.com.
Gang Huang, Email: huangg@sumhs.edu.cn.
Wei Chen, Email: chenwei6311@163.com.
Youping Deng, Email: dengy@hawaii.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., et al. Cancer statistics, 2021. CA A Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.The National Lung Screening Trial Research Team Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seijo L.M., Peled N., Ajona D., et al. Biomarkers in lung cancer screening: achievements, promises, and challenges. J Thorac Oncol. 2019;14(3):343–357. doi: 10.1016/j.jtho.2018.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y., Zitello E., Guo R., et al. The function of LncRNAs and their role in the prediction, diagnosis, and prognosis of lung cancer. Clin Transl Med. 2021;11(4):e367. doi: 10.1002/ctm2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu L., Ai J., Long H., et al. Integrative microRNA and gene profiling data analysis reveals novel biomarkers and mechanisms for lung cancer. Oncotarget. 2016;7(8):8441–8454. doi: 10.18632/oncotarget.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Z., Chen H., Zhu Y., et al. Global lipidomics reveals two plasma lipids as novel biomarkers for the detection of squamous cell lung cancer: a pilot study. Oncol Lett. 2018;16(1):761–768. doi: 10.3892/ol.2018.8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H., Liu H., Zou H., et al. Evaluation of plasma miR-21 and miR-152 as diagnostic biomarkers for common types of human cancers. J Cancer. 2016;7(5):490–499. doi: 10.7150/jca.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts E.R., Thomas K.J. The role of mitochondria in the development and progression of lung cancer. Comput Struct Biotechnol J. 2013;6:e201303019. doi: 10.5936/csbj.201303019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larriba E., Rial E., Del Mazo J. The landscape of mitochondrial small non-coding RNAs in the PGCs of male mice, spermatogonia, gametes and in zygotes. BMC Genom. 2018;19(1):634. doi: 10.1186/s12864-018-5020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uddin A., Chakraborty S. Role of miRNAs in lung cancer. J Cell Physiol. 2018:1–10. doi: 10.1002/jcp.26607. [DOI] [PubMed] [Google Scholar]
- 11.Mourksi N.E., Morin C., Fenouil T., et al. snoRNAs offer novel insight and promising perspectives for lung cancer understanding and management. Cells. 2020;9(3):541. doi: 10.3390/cells9030541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fathizadeh H., Asemi Z. Epigenetic roles of PIWI proteins and piRNAs in lung cancer. Cell Biosci. 2019;9:102. doi: 10.1186/s13578-019-0368-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng Y., Zhu Y., Wang H., et al. Ratio-based method to identify true biomarkers by normalizing circulating ncRNA sequencing and quantitative PCR data. Anal Chem. 2019;91(10):6746–6753. doi: 10.1021/acs.analchem.9b00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen J., Chang S.I., Lee E.S., et al. Determination of cluster number in clustering microarray data. Appl Math Comput. 2005;169(2):1172–1185. [Google Scholar]
- 15.Dou Y., Zhu Y., Ai J., et al. Plasma small ncRNA pair panels as novel biomarkers for early-stage lung adenocarcinoma screening. BMC Genom. 2018;19(1):545. doi: 10.1186/s12864-018-4862-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuo Y., Chen S., Yan L., et al. Development of a tRNA-derived small RNA diagnostic and prognostic signature in liver cancer. Genes Dis. 2021;9(2):393–400. doi: 10.1016/j.gendis.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi J., Ko E.A., Sanders K.M., et al. SPORTS1.0: a tool for annotating and profiling non-coding RNAs optimized for rRNA-and tRNA-derived small RNAs. Dev Reprod Biol. 2018;16(2):144–151. doi: 10.1016/j.gpb.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ewels P.A., Peltzer A., Fillinger S., et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38(3):276–278. doi: 10.1038/s41587-020-0439-x. [DOI] [PubMed] [Google Scholar]
- 19.Jühling F., Mörl M., Hartmann R.K., et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37(Database issue):D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei R., Wang J., Su M., et al. Missing value imputation approach for mass spectrometry-based metabolomics data. Sci Rep. 2018;8(1):663. doi: 10.1038/s41598-017-19120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinsky P.F., Gierada D.S., Black W., et al. Performance of lung-RADS in the National lung screening trial: a retrospective assessment. Ann Intern Med. 2015;162(7):485–491. doi: 10.7326/M14-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reznik E., Wang Q., La K., et al. Mitochondrial respiratory gene expression is suppressed in many cancers. Elife. 2017;6:e21592. doi: 10.7554/eLife.21592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reznik E., Miller M.L., Şenbabaoğlu Y., et al. Mitochondrial DNA copy number variation across human cancers. Elife. 2016;5:e10769. doi: 10.7554/eLife.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nam H.S., Izumchenko E., Dasgupta S., et al. Mitochondria in chronic obstructive pulmonary disease and lung cancer: where are we now? Biomarkers Med. 2017;11(6):475–489. doi: 10.2217/bmm-2016-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bratic A., Larsson N.G. The role of mitochondria in aging. J Clin Invest. 2013;123(3):951–957. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Servais S., Couturier K., Koubi H., et al. Effect of voluntary exercise on H2O2 release by subsarcolemmal and intermyofibrillar mitochondria. Free Radic Biol Med. 2003;35(1):24–32. doi: 10.1016/s0891-5849(03)00177-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.