Abstract
Sterile α motif and histidine/aspartic acid domain containing protein 1 (SAMHD1) is a deoxynucleoside triphosphate (dNTPs) triphosphohydrolase that can hydrolyze dNTPs into deoxynucleosides and triphosphates to keep the balance of the intracellular dNTPs pool. Moreover, it has been reported that SAMHD1 plays a role in regulating cell proliferation and the cell cycle, maintaining genome stability and inhibiting innate immune responses. SAMHD1 activity is regulated by phosphorylation, oxidation, SUMOylation, and O-GlcNAcylation. SAMHD1 mutations have been reported to cause diseases, including chronic lymphocytic leukemia and mantle cell lymphoma. SAMHD1 expression in acute myeloid leukemia predicts inferior prognosis. Recently, it has been revealed that SAMHD1 mediates the resistance to anti-cancer drugs. This review will focus on SAMHD1 function and regulation, the association between SAMHD1 and hematological malignancies and will provide updated information on SAMHD1 mediating resistance to nucleoside analogue antimetabolites, topoisomerase inhibitors, platinum-derived agents and DNA hypomethylating agents. Furthermore, histone deacetylase inhibitors and tyrosine kinase inhibitors indirectly increase anti-cancer drug resistance by increasing SAMDH1 activity. We herein highlight the importance of the development of novel agents targeting SAMHD1 to overcome treatment resistance of hematological malignancies, which would be an opportunity to improve the outcome of patients with refractory hematological malignancies.
Keywords: Anti-cancer drugs, Hematological malignancies, Resistance mechanism, SAMHD1, Regulation
Introduction
In 2000, sterile α motif and histidine/aspartic acid domain-containing protein 1 (SAMHD1) was firstly identified in the cDNA library of dendritic cells by Xuetao Cao and his colleagues and was named dendritic cell-derived interferon γ (IFN-γ)-induced protein (DCIP), as it is a homologue of mouse IFN-γ induced protein MG11 with 72% protein sequence identity.1 SAMHD1 contains 626 amino acid residues with two domains arranged in tandem, an N-terminal sterile α motif for protein or nucleic acid binding, and a histidine/aspartic residue domain with enzyme activity core site.2, 3, 4 In 2009, SAMHD1 mutation was identified as a cause of Aicardi-Goutières syndrome (AGS), a congenital encephalopathy with homeostasis disturbance of IFN-α.5 SAMHD1 was reported as a deoxynucleoside triphosphate (dNTPs) triphosphohydrolase, which could catalyze the hydrolysis of dNTPs into deoxynucleoside and triphosphate.3 In 2011, Laguette et al demonstrated that SAMHD1 could alleviate human immunodeficiency virus 1 (HIV-1) infection.6 Since then, SAMHD1 has been regarded an effective antiretroviral protein.6, 7, 8
Recently, SAMHD1 mutations have been reported in many hematological malignancies, including chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL), T-cell prolymphocytic leukemia (T-PLL), and classical Hodgkin lymphoma (cHL).9, 10, 11, 12, 13, 14, 15, 16, 17, 18 Moreover, SAMHD1 mediates the resistance to several anti-cancer drugs, and higher expression of SAMHD1 predicts a worse prognosis in several types of cancer.9,19,20 In this review, we will focus on the function and regulation of SAMHD1, its expression and mutations in hematological malignancies, and its role in inducing therapeutic resistance in hematological malignancies, to enable a better understanding of SAMHD1 targeting to overcome treatment resistance in hematological malignancies.
SAMHD1 functions
Regulating cell proliferation, restricting virus infection, and controlling cell cycle
SAMHD1 maintains the balance of a cellular dNTP pool by hydrolyzing dNTPs into deoxynucleosides and triphosphates, and conversely SAMHD1 activity is regulated by dNTPs.3,21,22 A SAMHD1 single unit has little dNTPase activity, and SAMHD1 function is optimal in the homologous tetramer. Guanosine triphosphate/deoxyguanosine triphosphate (GTP/dGTP) binding to guanine-specific allosteric sites (A1) and dNTPs binding to other allosteric sites (A2) are important activators for SAMHD1 homologous tetramer formation.22, 23, 24 Furthermore, dNTPs are the elementary materials for DNA synthesis, and DNA synthesis is the premise for cell proliferation. Thus, SAMHD1 regulates cell proliferation by controlling the efficiency of DNA synthesis.19,25,26 Moreover, SAMHD1 restricts HIV and retrovirus at the early stage of infection by depleting the major dNTPs for reverse transcription and complementary DNA replication.3 SAMHD1 plays a role in cell cycle regulation by controlling cells transforming from S phase to G2 phase.27 Phosphorylation of SAMHD1 at Thr592 by cyclinA2/cyclin-dependent kinase (cyclinA2/CDK) complex in S phase decreases its dNTPase activity.28 Phosphorylated SAMHD1 with ribonucleotide reductase, a key enzyme of de novo synthesis of dNTPs, maintains sufficient dNTPs for DNA replication and regulates the cell cycle S/G2 transition.27,29 Thus, SAMHD1 regulates cell proliferation, restricts HIV and retrovirus infection, and controls the cell cycle.
Maintaining genome stability
Felip et al reported that loss or downregulation of SAMHD1 induces cell genome instability, thereby making the cell susceptible to DNA damage agents.19 SAMHD1 maintains genome stability by facilitating cell homologous recombination when cells undergo DNA double-strand breaks (DSBs) by harmful agents like chemo-agents and irradiation. In this process, SAMHD1 conserved C-terminal combines with CtBP-interacting protein (CtIP), and the complex locates at the DNA breakpoint promoting the DNA end resection and initiating the homologous recombination.30 During DSB conditions, aberrant end joining and abnormal insertion of dNTPs causing genomic instability often happen in cancer cells, while SAMHD1 can shorten repair joints and decrease dNTPs level to avoid abnormal insertion and maintain genome integrity.31
In the development process of mature B-cells, cells somatic hypermutation (SHM) generates different high affinity variable regions of antibodies, and class switch recombination (CSR) changes constant regions. CSR occurs in with the generation of DNA double strands breaking followed by non-homologous end joining. In the translocation process of mature B-cells CSR and IgH/c-Myc, SAMHD1 maintains a low cellular dNTPs level to decrease the insertion of DSB point nucleotides.32,33 Thus, SAMHD1 can maintain genome integrity during DNA damage or under normal physiological conditions.
Inhibiting innate immune response
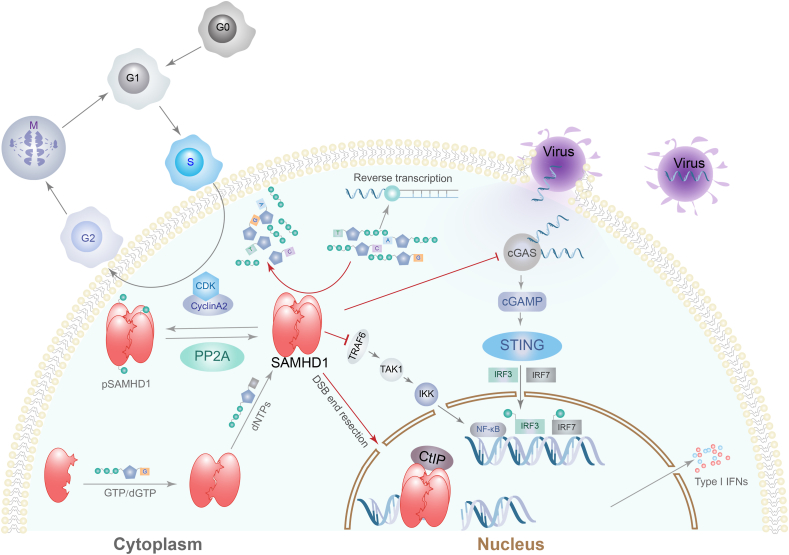
SAMHD1 plays a role in innate immune responses through direct actions on cytokines, viral DNA replication, and RNA reverse transcription. In previous studies, it was shown that SAMHD1 mutations lead to AGS. AGS is characterized by spontaneous IFN-α secretion, which suggests that SAMHD1 inhibits IFN-α production.5 During virus infection, viral DNA replication in the cytoplasm is sensed by cyclic guanosine monophosphate-adenosine monophosphate (cGAMP) synthase (cGAS), which catalyzes the second messenger cGAMP synthesis to activate downstream stimulator of interferon genes (STING) molecule, leading to interferon regulatory factor 3 (IRF3) and IRF7 phosphorylation and inducing type Ι IFN production.34 Coquel et al reported that SAMHD1 inhibits single-strand DNA production by activating MRE11 exonuclease to resect nascent DNA at replication folk, and activates checkpoint of ATR-CHK1 to stop DNA replication. Thus, SAMHD1 leads to a low level of cytosolic DNA by blocking viral DNA replication or RNA reverse transcription and also by decreasing dNTPs level, which is insufficient to be sensed by cGAS and activates downstream immune responses and type Ι IFN production.35 The NF-κB pathway is essential for inflammatory responses and innate immunity.36,37 Espada et al reported that SAMHD1 directly suppresses TNF receptor-associated factor 6 (TRAF6) and the transforming growth factor β-activated kinase 1 (TAK1) axis to inhibit activation of the NF-κB signaling pathway.38 Thus, SAMHD1 is a negative regulator of the innate immune response. SAMHD1 biological functions have been summarized and displayed in Figure 1.
Figure 1.
SAMHD1 hydrolyzes deoxynucleoside triphosphate (dNTPs), regulates the cell cycle, and inhibits innate immunity. SAMHD1 subunit forms tetramer mediates by GTP/dGTP binding to SAMHD1 guanine-specific allosteric sites (A1) and dNTPs binding to allosteric sites (A2). SAMHD1 tetramers are phosphorylated by the cyclinA2/CDK complex in S/G2 transition to inhibit SAMHD1 dNTPase activity and are dephosphorylated by protein phosphatase 2A (PP2A). SAMHD1 hydrolyzes dNTPs into deoxynucleoside and triphosphate to keep the intracellular balance of the dNTPs pool. SAMHD1 restricts retrovirus infections at reverse transcription stage by depleting intracellular dNTPs. SAMHD1 inhibits innate immunity by suppressing virus DNA replication in the cytoplasm and by keeping cytoplasm single-strand DNA at a low level insufficient to initiate cGAS-STING pathway to induce type Ι interferon secretion. SAMHD1 inhibits the TRAF6-TAK1axis to inhibit activation of the NF-κB signaling pathway. SAMHD1 binds the CtBP-interacting protein (CtIP) to mediate DNA double-strand break (DSB) end resection to initiate DNA homologous recombination repair. pSAMHD1, phosphorylated SAMHD1; TNF receptor-associated factor 6 (TRAF6); transforming growth factor β-activated kinase 1 (TAK1); IKK, inhibitor of kappa B kinase; cGAS, cyclic GMP-AMP synthase; cGAMP, cyclic guanosine monophosphate-adenosine monophosphate; STING, stimulator of interferon genes; IRF, interferon regulatory factor.
SAMHD1 expression and activity regulation
Expression regulation
The mechanism of SAMHD1 expression regulation remains unknown. However, some studies have revealed that microRNA may play a role in SAMHD1 expression. MicroRNA is a type of small non-coding RNA that can negatively regulate gene expression by a post-transcriptional mechanism. MicroRNA complementarily binds to target messenger RNA (mRNA) and leads to mRNA cleavage.39 Specifically, microRNA-181 (miR-181) and miR-30a could bind to the 3′ untranslated region (3′-UTR) of SMAHD1 mRNA and down-regulate SAMHD1 expression.40, 41, 42, 43 It has been reported that miR-181 and miR-30a are down-regulated by type Ι and II IFNs in human monocytes but not in macrophages and dendritic cells. This suggests that SAMHD1 regulation by IFNs is cell-type dependent.43
Phosphorylation down-regulates SAMHD1 dNTPase activity
In proliferating cells, SAMHD1 is a substrate for cyclinA2/CDK complex and is phosphorylated at Thr592 in the S phase.7,28,44 Phosphorylation at Thr592 in S phase down-regulates SAMHD1 activity to reduce dNTPs hydrolysis, and keep sufficient dNTPs concentration for DNA replication. SAMHD1 is down-regulated but not inactive in phosphorylated state, and phosphorylated SAMHD1 cooperates with the ribonucleotide reductase in S/G2 transition in a fine-tuning manner to keep the balance of the dNTPs pool for DNA replication.27 In the cell cycle of M/G1 transition, phosphatase protein phosphatase 2A (PP2A) mediates SAMHD1 dephosphorylation and reactivation.45 Disturbance of SAMHD1 phosphorylation at Thr592 would arrest cells in the S phase.28
SAMHD1 phosphorylation relies on the cell cycle and on the cyclinA2/CDK complex. Additionally, the cyclinA2/CDK complex is negatively regulated by cyclin-dependent kinase inhibitors (CKIs), including p21, p27, and p57, which bind the cyclinA2/CDK complex and inhibit its kinase activity.46 In DNA damaged cells, phosphorylated p53 tetramers act as a transcription factor upregulating p21 and p27 expression.47,48 Moreover, activation of Toll-like receptor 4 (TLR4) in infected cells up-regulates p21 and inhibits CDK1.49 Thus, when cells undergo DNA damage or get infected, p21 and p27 mediate inhibition of the cyclinA2/CDK complex, which decreases SAMHD1 phosphorylation and leads to cell cycle arrest. Conversely, phosphorylation at Thr592 keeps SAMHD1, thereby localizing in the nucleus where it is protected from oxidation by cytoplasmic reactive oxygen species (ROS).29
Oxidation inhibits SAMHD1 oligomerization
SAMHD1 has been reported to be regulated by the redox switch.50,51 Oxidation by cellular ROS occurs on SAMHD1 amino acid residues Cys341, Cys350 and Cys522 50, 51. SAMHD1 antiviral activity is dependent on redox transformation to form a stable tetramer and to induce catalysis activity.50 Cys341 and Cys350 oxidation are indispensable for SAMHD1 oligomerization and activity. Previous studies have reported that Cys522 oxidation does not influence SAMHD1 oligomerization and dNTPase activity, but Cys522 mutation makes SAMHD1 resistant to redox switch regulation.50 Thus, Cys522 oxidation may be promising for SAMHD1 oxidation on Cys341 and Cys350 residues. This research reveals that oxidation of Cys341 and Cys522 restricts retrovirus infection, but does not influence dNTPs pools and Thr592 phosphorylation, suggesting that SAMHD1 restriction of retrovirus infection has different mechanisms other than depleting dNTPs.50 However, the full mechanism remains to be elucidated. Moreover, the oxidation process is reversible and only cytoplasmic SAMHD1 is oxidized by ROS in the cytoplasm.51
SUMOylation and O-GlcNAcylation enhance SAMHD1 virus restriction
SUMOylation is a mechanism of post-translational modifications, similar to ubiquitinoylation, in which a single sentrin/small ubiquitin-like modifier (SUMO) conjugates to a protein substrate on lysine residues and the catalytic reaction occurs through activation of E1, conjugation of E2, and ligation of E3 enzymes. Unlike ubiquitinoylation, SUMOylation does not mediate target protein degradation, but changes cellular localization, activity, or stability of proteins.52 Martinat et al reported that SAMHD1 SUMOylation occurs mainly at lysine residues of 469, 595 and 622. SUMOylation at Lys595 and phosphorylation at Thr592 are two independent events, but SUMOylation at Lys595 endows dephosphorylated SAMHD1 antiviral activities.4,53 Hu et al reported another mechanism of the regulation of SAMHD1 antiviral activity in chronic hepatitis B virus (HBV) infection conditions. An HBV infection increases SAMHD1 O-linked-N-acetylglucosaminylation (O-GlcNAcylation) at Ser93 through the hexosamine biosynthesis pathway (HBP). The O-GlcNAcylation at Ser93 enhances the stability of SAMHD1 tetrameter and enhances the activity of SAMHD1 virus restriction.54
SAMHD1 expression and mutation in hematological malignancies
SAMHD1 expression
SAMHD1 is expressed in several hematological malignancies, including chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL), T-cell prolymphocytic leukemia (T-PLL), acute myeloid leukemia (AML), and classical Hodgkin lymphoma (cHL).9,10,12,13,18,20,55 In our previous study, we found that MCL patients with higher SAMHD1 expression were associated with an inferior chemotherapy response rate and a shorter overall survival.9 Merrien et al reported that blastoid/pleomorphic MCL had a higher SAMHD1 expression, which positively correlated with MCL ki-67 proliferation index.25 Moreover, Roider et al reported that SAMHD1 expression in B-cell lymphoma cell lines inversely correlates with the effect of cytarabine single agent treatment in vitro. However, as MCL chemotherapy regime contains many agents other than cytarabine, no significant difference in complete remission rate and survival between SAMHD1 positive and negative patients were found.13,25 In AML, high SAMHD1 expression predicts short progression-free survival, relapse-free survival and overall survival (OS).20 In cHL, SAMHD1 is positive in Hodgkin and Reed-Sternberg (HRS) cells of 31% of cHL patients, and SAMHD1 positive cHL patients have a higher disease progression rate, a shorter disease-free survival, and an inferior OS. Therefore, SAMHD1 expression is an adverse factor for cHL prognosis.18
SAMHD1 mutation
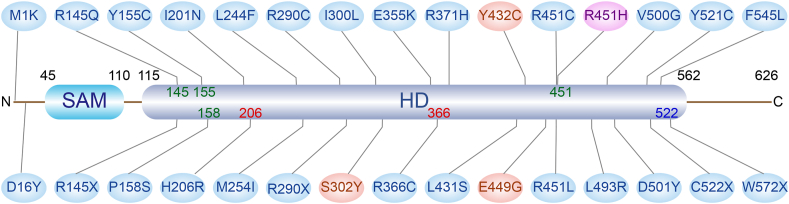
SAMHD1 mutation has been reported in several hematological malignancies. Mutations of SAMHD1 exist in 7.1%–10% of MCL,9,13,14 9.6%–11% of CLL,12,55 and 18% of T-PLL.10 It has been proposed that SAMHD1 mutations could be a driver event of CLL11,16 and MCL tumorigenesis.14 Mutation sites and functions are displayed in Figure 2. Most reported SAMHD1 mutations are single nucleotide substitutions, causing variations in amino acids. Mutation sites on SAMHD1 involve allosteric sites, catalysis sites, and regulatory sites. The GTP/dGTP mediates SAMHD1 tetramer formation and maintains its stability by binding to amino acid residues, including Arg145, Tyr155, Pro158 and Arg451.56 His206 and Arg366 are in active sites and Cys522 is the residue for regulation of oxidation. SAMHD1 mutations lead to the increase of cellular dNTPs, and cell cycle disturbance may lead to tumorigenesis.21 Furthermore, cells with SAMHD1 mutations are vulnerable to agents that cause DNA damage, and thus they are at an increased risk of tumor formation.30,31 For CLL and MCL, SAMHD1 mutations affect the process of IgH/c-Myc translocation, increase insertion mutations and aberrant end joining during CSR, and induce B-cell lymphoma.32 However, whether SAMHD1 mutation affects tumor prognosis is not yet known.
Figure 2.
SAMHD1 mutations in CLL and MCL. SAMHD1 has 626 amino acid residues, including a sterile α motif domain SAM (45–110) and a histidine/aspartic acid (HD) domain (115–562). In the schematic diagram, mutations of CLL are marked in blue and mutations of MCL are marked in red. R451H in purple is reported in both CLL and MCL. Among these mutation sites, Arg145, Tyr155, Pro158, and Arg451 are the key residues of allosteric sites; His206 and Arg366 are in composition of the catalytic active center; and Cys355 is the cysteine for oxidation regulation. Other functions of mutation sites displayed have not yet been elucidated.
SAMHD1 and anti-cancer drug resistance
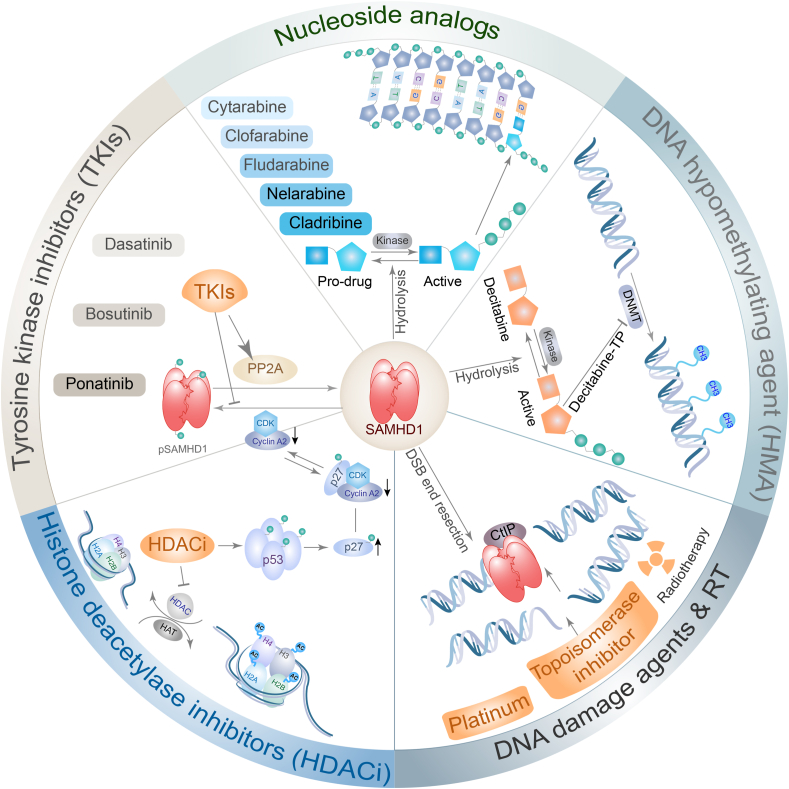
SAMHD1 has been reported to be associated with resistance to several types of anti-cancer drugs used in hematological malignancies, including nucleotide analogue antimetabolites, topoisomerase inhibitors, platinum-derived agents and DNA hypomethylating agents (HMAs). Histone deacetylase inhibitors (HDACi) and tyrosine kinase inhibitors (TKIs) mediate SAMHD1 dephosphorylation and indirectly increase resistance to other anti-cancer drugs. The underlying mechanisms of SAMHD1 mediating resistance of anti-cancer drugs are diverse (Fig. 3).
Figure 3.
SAMHD1 mediates resistance of multiple anti-cancer drugs. Nucleoside analogues are prodrug forms and are phosphorylated to active forms by cellular kinases. SAMHD1 catalyzes the hydrolysis of phosphorylated nucleoside analogues into inactive prodrug forms and triphosphate. DNA hypomethylating agent (HMA), decitabine, is phosphorylated to decitabine-TP and inhibits DNA methyltransferase (DNMT). SAMHD1 hydrolyzes decitabine-TP to decitabine and triphosphate. Topoisomerase inhibitor, platinum-derived agents, and radiotherapy mediates DNA double-strand breaks (DSBs) in cancer cells and kill cancer cells. SAMHD1 binds CtBP-interacting protein (CtIP) and facilitates DSB end resection to initiate DNA homologous recombination and increase resistance to DNA damage agents. Histone deacetylase inhibitors (HDACi) up-regulate p53/p27phosphorylation. Phosphorylated p27 binds the cyclinA2/CDK complex to inhibit its function of mediating SAMHD1 phosphorylation. Tyrosine kinase inhibitors (TKIs) inhibit intracellular kinase to phosphorylate SAMHD1 and increase PP2A activity to dephosphorylated SAMHD1. HDACi and TKIs decrease phosphorylated SAMHD1 and indirectly increase SAMHD1 mediating the resistance of other agents. HAT, histoneacetyltransferase.
Nucleoside analogue antimetabolites
Nucleoside analogue antimetabolites are a large class of agents that mimic dNTPs. During cancer cell replication, they incorporate into DNA inhibiting DNA polymerase activity, thereby resulting in the termination of DNA synthesis. Thus, cell proliferation is stopped and the cells undergo apoptosis.57 Nucleoside analogue antimetabolites are wildly used in AML, myelodysplastic syndromes (MDS), acute lymphoblastic leukemia (ALL), lymphoma, and myeloma. Nucleoside analogue antimetabolites are prodrugs that are converted to active metabolites by a group of kinases that phosphorylate substrates of nucleosides and nucleotides. The nucleoside analogue antimetabolites are phosphorylated to monophosphate analogues, diphosphate and triphosphate forms by various cellular enzymes.57 These phosphorylated nucleoside analogues can bind the SAMHD1 catalytic pocket and are dephosphorylated by SAMHD1, thus leading to intracellular low concentration of effective nucleoside analogues.58 Using this mechanism, SAMHD1 promotes resistance to a variety of nucleoside analogue antimetabolites, including cytarabine, clofarabine, fludarabine, nelarabine, and cladribine.20,58, 59, 60, 61 For example, cytarabine is phosphorylated by kinases into cytarabine-TP, while SAMHD1 hydrolyzes cytarabine-TP to cytarabine and triphosphate. However, gemcitabine is not influenced by SAMHD1, because gemcitabine is not only a nucleoside analogue but also an inhibitor of ribonucleotide reductase. Gemcitabine inhibition of ribonucleotide reductase leads to a decrease in de novo synthesis of dNTPs and in the intracellular concentration of dNTPs. Low dNTPs concentration influences SAMHD1 allosteric transformation into a tetramer and decrease SAMHD1 dNTPs activity. Thus, gemcitabine indirectly leads to inhibition of SAMHD1 activity and reduced gemcitabine-TP hydrolysis.62
DNA damage agents
DNA damage agents include topoisomerase inhibitors and platinum-derived agents.19,30,63 Topoisomerase inhibitors, including etoposide, and platinum-derived agents, such as cisplatin and carboplatin, are the backbone of the treatment of hematological malignancies. Radiotherapy is generally applied in lymphoma.64, 65, 66 The common mechanism of topoisomerase inhibitors, platinum-derived agents and even radiotherapy to kill cancer cells is by induction of DNA damage.67, 68, 69, 70 These DNA damage agents and radiotherapy lead to cancer cell DSB, while SAMHD1 could mediate DNA damage repair by facilitating DSB end resection, which is the first step of homologous recombination repair.30 DNA damage induces phosphorylation and tetramerization of p53, leading to the phosphorylation of p21, which binds CDK1. CDK1 is an essential kinase that promotes SAMHD1 phosphorylation. Thus, DNA damage agents decrease SAMHD1 phosphorylation and increase SAMHD1 activity through the p53/p21/CDK1 pathway. SAMHD1 inversely repairs DNA damage and increases DNA stability, resulting in DNA damage agents and chemotherapy resistance.63
HMAs
HMAs, including decitabine and azacytidine, are cytosine nucleoside analogues, which suppress DNA methyltransferases (DNMTs), deplete DNA methylation, and induce DNA damage and cell apoptosis.71 Decitabine and azacytidine are phosphorylated to the active form of decitabine-TP and azacytidine-TP by the action of cellular enzymes. Decitabine-TP and azacytidine-TP can incorporate into nucleic acid, while decitabine-TP mainly incorporates into DNA and azacytidine-TP incorporates into both DNA and RNA.72 Decitabine and azacytidine are mainly applied in AML and MDS.73, 74, 75, 76 Oellerich et al reported that SAMHD1 compromises decitabine, but not azacytidine, efficacy in AML and MDS. The underlying mechanism relies on the fact that decitabine-TP is a substrate of SAMHD1, while azacytidine-TP is not. SAMHD1 hydrolysis of decitabine-TP leads to low intracellular levels and limited efficacy of decitabine. High SAMHD1 expression is associated with cellular insensitivity to decitabine and a poor response to decitabine therapy in AML patients. Thus, SAMHD1 expression is a biomarker of decitabine response.77
HDACi
Histones are the coil of chromatin, which could regulate DNA replication and transcription through posttranslational modification mechanisms, such as methylation, acetylation, phosphorylation and ubiquitination.78 Histone acetylation leads to chromatin in a loose and active status for DNA replication and gene expression, while deacetylation in reverse status. Histone deacetylases (HDACs), also called eraser, is the key enzyme removing histone acetylation.79 The abnormal activation of HDACs resulting in histone hypoacetylation and leading to aberrant gene expression is a major mechanism of tumorigenesis. HDACi including vorinostat, romidepsin, belinostat, panobinostat and chidamide were approved by FDA and are mainly applied in T cell lymphoma, MDS and multiple myeloma.80, 81, 82, 83, 84, 85 HDACi could mediate histone hyperacetylation and arrest cells at G1 phase by inhibiting of cyclinA2/CDK complex through the pathway of p53/p27/CDK1, which could lead to SAMHD1 dephosphorylation.86,87 Mlcochova et al have revealed that HDACi (vorinostat and panobinostat) could mediate SAMHD1 dephosphorylation, increase SAMHD1 activity and indirectly increase SAMHD1 mediating anti-cancer drug resistance.87
TKI
Tyrosine kinases (TKs) are a group of enzymes that mediate substrate phosphorylation of signaling proteins and modulate their activities. Cell receptors such as the epidermal growth factor receptor (EGFR), vascular endothelial growth factor receptor (VEGFR) and fibroblast growth factor receptor (FGFR) belong to the TK family.88 The most famous TKI agent is imatinib, the first generation BCR-ABL inhibitor, and is mainly used in chronic myeloid leukemia and BCR-ABL-positive ALL. Dasatinib is a second generation TKI, with potent inhibitory effects and targeting multiple kinases other than BCR-ABL, such as SRC kinase and c-Kit. TKIs, including dasatinib, bosutinib, and ponatinib induce SAMHD1 activation by decreasing SAMHD1 phosphorylation, among which dasatinib provides the most potent effect.89, 90, 91 BCR-ABL inhibits PP2A expression. Therefore, TKIs targeting BCR-ABL induce the activation of PP2A, and mediate dephosphorylation and activation of SAMHD1.91,92 In conclusion, TKIs indirectly induce SAMHD1 activation resulting in SAMHD1 mediating resistance of multiple anti-cancer drugs.
Future perspectives
SAMHD1 plays a vital role in the therapeutic resistance of hematological malignancies. Herold et al reported that they used virus-like particles to deliver viral accessory protein x (Vpx) to the AML cell line, THP-1. Vpx could target the SAMHD1 C-terminal region and conjugate to the DNA damage-binding protein (DDB1) and comprising cullin4A (CUL4)-associated factors 1 (DCAF1), an E3 ligase substrate adaptor, and promote SAMHD1 degradation by the proteasome.93,94 Depleting SAMHD1 significantly improved cytarabine sensitivity in hematological malignancies.95 This study demonstrated that targeting SAMHD1 could reverse resistance to cancer cytarabine. It is possible that chemoresistance may be overcome by depleting SAMHD1.
However, there is a lack of SAMHD1-specific inhibitors. To overcome that, other inhibitors can be used. For example, the CDK4/6 selective inhibitor, palbociclib, can modulate SAMHD1 phosphorylation, and synergistically enhance antimetabolite-based drug effects.96 However, the underlying mechanism remains to be further elucidated and whether palbociclib can enhance other types of anti-cancer drugs, such as topoisomerase inhibitors, platinum-derived agents and HMAs is not yet known. Mauney et al used high-throughput screening to identify several agents that inhibit the activities of SAMHD1 dNTPase. Eight promising agents were found to be potent and specific SAMHD1 dNTPase inhibitors. Among them, retinoic acid, l-thyroxine, amrinone, and montelukast are clinically used, while lomofungin, ergotamine, hexestrol, and sulindac are not used in the clinic.97 However, the inhibitory efficacy to SAMHD1 of these agents has not been identified in vivo.
Conclusions
In conclusion, SAMHD1 plays a vital role in the development, therapeutic resistance, and prognosis of hematological malignancies. To guide clinical treatment, in the future, SAMHD1 may be used as a novel risk stratification biomarker for hematological malignancies, such as MCL, AML, and cHL. In addition, the development of SAMHD1-specific inhibitors may overcome therapeutic resistance and improve patients’ prognosis.
Author contributions
Tao Wang was responsible for writing the manuscript, and Ping Liu contributed to graphs. Jianming Yang conducted writing the review and revised the manuscript. We would like to thank MogoEdit (https://www.mogoedit.com) for its English editing during the preparation of this manuscript.
Conflict of interests
The authors declare no competing financial interests.
Funding
This work was supported by the funding of National Natural Science Foundation of China (No. 81770209).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.06.001.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Li N., Zhang W., Cao X. Identification of human homologue of mouse IFN-gamma induced protein from human dendritic cells. Immunol Lett. 2000;74(3):221–224. doi: 10.1016/s0165-2478(00)00276-5. [DOI] [PubMed] [Google Scholar]
- 2.Morris E.R., Caswell S.J., Kunzelmann S., et al. Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis. Nat Commun. 2020;11(1):3165. doi: 10.1038/s41467-020-16983-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstone D.C., Ennis-Adeniran V., Hedden J.J., et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 4.Martinat C., Cormier A., Tobaly-Tapiero J., et al. SUMOylation of SAMHD1 at Lysine 595 is required for HIV-1 restriction in non-cycling cells. Nat Commun. 2021;12(1):4582. doi: 10.1038/s41467-021-24802-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice G.I., Bond J., Asipu A., et al. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41(7):829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laguette N., Sobhian B., Casartelli N., et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.St Gelais C., de Silva S., Hach J.C., et al. Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J Virol. 2014;88(10):5834–5844. doi: 10.1128/JVI.00155-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonucci J.M., St Gelais C., de Silva S., et al. SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity. Nat Med. 2016;22(10):1072–1074. doi: 10.1038/nm.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T., Yue W., Tang G., et al. SAMHD1 mutations and expression in mantle cell lymphoma patients. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.763151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansson P., Klein-Hitpass L., Choidas A., et al. SAMHD1 is recurrently mutated in T-cell prolymphocytic leukemia. Blood Cancer J. 2018;8(1):11. doi: 10.1038/s41408-017-0036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns A., Alsolami R., Becq J., et al. Whole-genome sequencing of chronic lymphocytic leukaemia reveals distinct differences in the mutational landscape between IgHVmut and IgHVunmut subgroups. Leukemia. 2018;32(2):332–342. doi: 10.1038/leu.2017.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guièze R., Robbe P., Clifford R., et al. Presence of multiple recurrent mutations confers poor trial outcome of relapsed/refractory CLL. Blood. 2015;126(18):2110–2117. doi: 10.1182/blood-2015-05-647578. [DOI] [PubMed] [Google Scholar]
- 13.Roider T., Wang X., Hüttl K., et al. The impact of SAMHD1 expression and mutation status in mantle cell lymphoma: an analysis of the MCL Younger and Elderly trial. Int J Cancer. 2021;148(1):150–160. doi: 10.1002/ijc.33202. [DOI] [PubMed] [Google Scholar]
- 14.Nadeu F., Martin-Garcia D., Clot G., et al. Genomic and epigenomic insights into the origin, pathogenesis, and clinical behavior of mantle cell lymphoma subtypes. Blood. 2020;136(12):1419–1432. doi: 10.1182/blood.2020005289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amin N.A., Seymour E., Saiya-Cork K., et al. A quantitative analysis of subclonal and clonal gene mutations before and after therapy in chronic lymphocytic leukemia. Clin Cancer Res. 2016;22(17):4525–4535. doi: 10.1158/1078-0432.CCR-15-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuh A., Becq J., Humphray S., et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood. 2012;120(20):4191–4196. doi: 10.1182/blood-2012-05-433540. [DOI] [PubMed] [Google Scholar]
- 17.Rentoft M., Lindell K., Tran P., et al. Heterozygous colon cancer-associated mutations of SAMHD1 have functional significance. Proc Natl Acad Sci U S A. 2016;113(17):4723–4728. doi: 10.1073/pnas.1519128113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xagoraris I., Vassilakopoulos T.P., Drakos E., et al. Expression of the novel tumour suppressor sterile alpha motif and HD domain-containing protein 1 is an independent adverse prognostic factor in classical Hodgkin lymphoma. Br J Haematol. 2021;193(3):488–496. doi: 10.1111/bjh.17352. [DOI] [PubMed] [Google Scholar]
- 19.Felip E., Gutiérrez-Chamorro L., Gómez M., et al. Modulation of DNA damage response by SAM and HD domain containing deoxynucleoside triphosphate triphosphohydrolase (SAMHD1) determines prognosis and treatment efficacy in different solid tumor types. Cancers. 2022;14(3):641. doi: 10.3390/cancers14030641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider C., Oellerich T., Baldauf H.M., et al. SAMHD1 is a biomarker for cytarabine response and a therapeutic target in acute myeloid leukemia. Nat Med. 2017;23(2):250–255. doi: 10.1038/nm.4255. [DOI] [PubMed] [Google Scholar]
- 21.Franzolin E., Pontarin G., Rampazzo C., et al. The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc Natl Acad Sci U S A. 2013;110(35):14272–14277. doi: 10.1073/pnas.1312033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu C., Gao W., Zhao K., et al. Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat Commun. 2013;4:2722. doi: 10.1038/ncomms3722. [DOI] [PubMed] [Google Scholar]
- 23.Li Y., Kong J., Peng X., et al. Structural insights into the high-efficiency catalytic mechanism of the sterile α-motif/histidine-aspartate domain-containing protein. J Biol Chem. 2015;290(49):29428–29437. doi: 10.1074/jbc.M115.663658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji X., Wu Y., Yan J., et al. Mechanism of allosteric activation of SAMHD1 by dGTP. Nat Struct Mol Biol. 2013;20(11):1304–1309. doi: 10.1038/nsmb.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merrien M., Wasik A.M., Ljung E., et al. Clinical and biological impact of SAMHD1 expression in mantle cell lymphoma. Virchows Arch. 2022;480(3):655–666. doi: 10.1007/s00428-021-03228-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ballana E., Esté J.A. SAMHD1: at the crossroads of cell proliferation, immune responses, and virus restriction. Trends Microbiol. 2015;23(11):680–692. doi: 10.1016/j.tim.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Tramentozzi E., Ferraro P., Hossain M., et al. The dNTP triphosphohydrolase activity of SAMHD1 persists during S-phase when the enzyme is phosphorylated at T592. Cell Cycle. 2018;17(9):1102–1114. doi: 10.1080/15384101.2018.1480216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan J., Hao C., DeLucia M., et al. CyclinA2-cyclin-dependent kinase regulates SAMHD1 protein phosphohydrolase domain. J Biol Chem. 2015;290(21):13279–13292. doi: 10.1074/jbc.M115.646588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batalis S., Rogers L.C., Hemphill W.O., et al. SAMHD1 phosphorylation at T592 regulates cellular localization and S-phase progression. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.724870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daddacha W., Koyen A.E., Bastien A.J., et al. SAMHD1 promotes DNA end resection to facilitate DNA repair by homologous recombination. Cell Rep. 2017;20(8):1921–1935. doi: 10.1016/j.celrep.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akimova E., Gassner F.J., Schubert M., et al. SAMHD1 restrains aberrant nucleotide insertions at repair junctions generated by DNA end joining. Nucleic Acids Res. 2021;49(5):2598–2608. doi: 10.1093/nar/gkab051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Husain A., Xu J., Fujii H., et al. SAMHD1-mediated dNTP degradation is required for efficient DNA repair during antibody class switch recombination. EMBO J. 2020;39(15) doi: 10.15252/embj.2019102931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thientosapol E.S., Bosnjak D., Durack T., et al. SAMHD1 enhances immunoglobulin hypermutation by promoting transversion mutation. Proc Natl Acad Sci U S A. 2018;115(19):4921–4926. doi: 10.1073/pnas.1719771115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glück S., Guey B., Gulen M.F., et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol. 2017;19(9):1061–1070. doi: 10.1038/ncb3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coquel F., Silva M.J., Técher H., et al. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature. 2018;557(7703):57–61. doi: 10.1038/s41586-018-0050-1. [DOI] [PubMed] [Google Scholar]
- 36.Taniguchi K., Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 37.Kim E.T., Roche K.L., Kulej K., et al. SAMHD1 modulates early steps during human cytomegalovirus infection by limiting NF-κB activation. Cell Rep. 2019;28(2):434–448. doi: 10.1016/j.celrep.2019.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Espada C.E., St Gelais C., Bonifati S., et al. TRAF6 and TAK1 contribute to SAMHD1-mediated negative regulation of NF-κB signaling. J Virol. 2021;95(3):e01970. doi: 10.1128/JVI.01970-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dragomir M.P., Knutsen E., Calin G.A. Classical and noncanonical functions of miRNAs in cancers. Trends Genet. 2022;38(4):379–394. doi: 10.1016/j.tig.2021.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Pilakka-Kanthikeel S., Raymond A., Atluri V.S., et al. Sterile alpha motif and histidine/aspartic acid domain-containing protein 1 (SAMHD1)-facilitated HIV restriction in astrocytes is regulated by miRNA-181a. J Neuroinflammation. 2015;12:66. doi: 10.1186/s12974-015-0285-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohnken R., Kodigepalli K.M., Mishra A., et al. MicroRNA-181 contributes to downregulation of SAMHD1 expression in CD4+ T-cells derived from Sèzary syndrome patients. Leuk Res. 2017;52:58–66. doi: 10.1016/j.leukres.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin C., Peng X., Liu F., et al. MicroRNA-181 expression regulates specific post-transcriptional level of SAMHD1 expression in vitro. Biochem Biophys Res Commun. 2014;452(3):760–767. doi: 10.1016/j.bbrc.2014.08.151. [DOI] [PubMed] [Google Scholar]
- 43.Riess M., Fuchs N.V., Idica A., et al. Interferons induce expression of SAMHD1 in monocytes through down-regulation of miR-181a and miR-30a. J Biol Chem. 2017;292(1):264–277. doi: 10.1074/jbc.M116.752584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cribier A., Descours B., Valadão A.L.C., et al. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 2013;3(4):1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 45.Schott K., Fuchs N.V., Derua R., et al. Dephosphorylation of the HIV-1 restriction factor SAMHD1 is mediated by PP2A-B55α holoenzymes during mitotic exit. Nat Commun. 2018;9(1):2227. doi: 10.1038/s41467-018-04671-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bury M., Le Calvé B., Ferbeyre G., et al. New insights into CDK regulators: novel opportunities for cancer therapy. Trends Cell Biol. 2021;31(5):331–344. doi: 10.1016/j.tcb.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 47.Kciuk M., Gielecińska A., Mujwar S., et al. Cyclin-dependent kinases in DNA damage response. Biochim Biophys Acta Rev Cancer. 2022;1877(3) doi: 10.1016/j.bbcan.2022.188716. [DOI] [PubMed] [Google Scholar]
- 48.Hume S., Grou C.P., Lascaux P., et al. The NUCKS1-SKP2-p21/p27 axis controls S phase entry. Nat Commun. 2021;12(1):6959. doi: 10.1038/s41467-021-27124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mlcochova P., Winstone H., Zuliani-Alvarez L., et al. TLR4-mediated pathway triggers interferon-independent G0 arrest and antiviral SAMHD1 activity in macrophages. Cell Rep. 2020;30(12):3972–3980. doi: 10.1016/j.celrep.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z., Bhattacharya A., White T., et al. Functionality of redox-active cysteines is required for restriction of retroviral replication by SAMHD1. Cell Rep. 2018;24(4):815–823. doi: 10.1016/j.celrep.2018.06.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mauney C.H., Rogers L.C., Harris R.S., et al. The SAMHD1 dNTP triphosphohydrolase is controlled by a redox switch. Antioxidants Redox Signal. 2017;27(16):1317–1331. doi: 10.1089/ars.2016.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang H.M., Yeh E.T.H. SUMO: from bench to bedside. Physiol Rev. 2020;100(4):1599–1619. doi: 10.1152/physrev.00025.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saiada F., Zhang K., Li R. PIAS1 potentiates the anti-EBV activity of SAMHD1 through SUMOylation. Cell Biosci. 2021;11(1):127. doi: 10.1186/s13578-021-00636-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu J., Gao Q., Yang Y., et al. Hexosamine biosynthetic pathway promotes the antiviral activity of SAMHD1 by enhancing O-GlcNAc transferase-mediated protein O-GlcNAcylation. Theranostics. 2021;11(2):805–823. doi: 10.7150/thno.50230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clifford R., Louis T., Robbe P., et al. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood. 2014;123(7):1021–1031. doi: 10.1182/blood-2013-04-490847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu C.H., Bhattacharya A., Persaud M., et al. Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification. Nat Commun. 2021;12(1):731. doi: 10.1038/s41467-021-21023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shelton J., Lu X., Hollenbaugh J.A., et al. Metabolism, biochemical actions, and chemical synthesis of anticancer nucleosides, nucleotides, and base analogs. Chem Rev. 2016;116(23):14379–14455. doi: 10.1021/acs.chemrev.6b00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knecht K.M., Buzovetsky O., Schneider C., et al. The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1. Proc Natl Acad Sci U S A. 2018;115(43):E10022–E10031. doi: 10.1073/pnas.1805593115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herold N., Rudd S.G., Sanjiv K., et al. SAMHD1 protects cancer cells from various nucleoside-based antimetabolites. Cell Cycle. 2017;16(11):1029–1038. doi: 10.1080/15384101.2017.1314407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rothenburger T., Thomas D., Schreiber Y., et al. Differences between intrinsic and acquired nucleoside analogue resistance in acute myeloid leukaemia cells. J Exp Clin Cancer Res. 2021;40(1):317. doi: 10.1186/s13046-021-02093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rassidakis G.Z., Herold N., Myrberg I.H., et al. Low-level expression of SAMHD1 in acute myeloid leukemia (AML) blasts correlates with improved outcome upon consolidation chemotherapy with high-dose cytarabine-based regimens. Blood Cancer J. 2018;8(11):98. doi: 10.1038/s41408-018-0134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rudd S.G., Tsesmetzis N., Sanjiv K., et al. Ribonucleotide reductase inhibitors suppress SAMHD1 ara-CTPase activity enhancing cytarabine efficacy. EMBO Mol Med. 2020;12(3) doi: 10.15252/emmm.201910419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mlcochova P., Caswell S.J., Taylor I.A., et al. DNA damage induced by topoisomerase inhibitors activates SAMHD1 and blocks HIV-1 infection of macrophages. EMBO J. 2018;37(1):50–62. doi: 10.15252/embj.201796880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hawkes E.A., Barraclough A., Sehn L.H. Limited-stage diffuse large B-cell lymphoma. Blood. 2022;139(6):822–834. doi: 10.1182/blood.2021013998. [DOI] [PubMed] [Google Scholar]
- 65.Eichenauer D.A., Engert A. How I treat nodular lymphocyte-predominant Hodgkin lymphoma. Blood. 2020;136(26):2987–2993. doi: 10.1182/blood.2019004044. [DOI] [PubMed] [Google Scholar]
- 66.Yamaguchi M., Suzuki R., Oguchi M. Advances in the treatment of extranodal NK/T-cell lymphoma, nasal type. Blood. 2018;131(23):2528–2540. doi: 10.1182/blood-2017-12-791418. [DOI] [PubMed] [Google Scholar]
- 67.Zhang C., Xu C., Gao X., et al. Platinum-based drugs for cancer therapy and anti-tumor strategies. Theranostics. 2022;12(5):2115–2132. doi: 10.7150/thno.69424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaswal S., Nehra B., Kumar S., et al. Recent advancements in the medicinal chemistry of bacterial type II topoisomerase inhibitors. Bioorg Chem. 2020;104 doi: 10.1016/j.bioorg.2020.104266. [DOI] [PubMed] [Google Scholar]
- 69.Wong K.C.W., Johnson D., Hui E.P., et al. Opportunities and challenges in combining immunotherapy and radiotherapy in head and neck cancers. Cancer Treat Rev. 2022;105 doi: 10.1016/j.ctrv.2022.102361. [DOI] [PubMed] [Google Scholar]
- 70.Ghanbari-Movahed M., Kaceli T., Mondal A., et al. Recent advances in improved anticancer efficacies of camptothecin nano-formulations: a systematic review. Biomedicines. 2021;9(5):480. doi: 10.3390/biomedicines9050480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zavras P.D., Shastri A., Goldfinger M., et al. Clinical trials assessing hypomethylating agents combined with other therapies: causes for failure and potential solutions. Clin Cancer Res. 2021;27(24):6653–6661. doi: 10.1158/1078-0432.CCR-21-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Šimoničová K., Janotka Ľ., Kavcová H., et al. Different mechanisms of drug resistance to hypomethylating agents in the treatment of myelodysplastic syndromes and acute myeloid leukemia. Drug Resist Updates. 2022;61 doi: 10.1016/j.drup.2022.100805. [DOI] [PubMed] [Google Scholar]
- 73.Welch J.S., Petti A.A., Miller C.A., et al. TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N Engl J Med. 2016;375(21):2023–2036. doi: 10.1056/NEJMoa1605949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lübbert M., Grishina O., Schmoor C., et al. Valproate and retinoic acid in combination with decitabine in elderly nonfit patients with acute myeloid leukemia: results of a multicenter, randomized, 2 × 2, phase II trial. J Clin Oncol. 2020;38(3):257–270. doi: 10.1200/JCO.19.01053. [DOI] [PubMed] [Google Scholar]
- 75.Garcia-Manero G., Santini V., Almeida A., et al. Phase III, randomized, placebo-controlled trial of CC-486 (oral azacitidine) in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2021;39(13):1426–1436. doi: 10.1200/JCO.20.02619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pratz K.W., Jonas B.A., Pullarkat V., et al. Measurable residual disease response and prognosis in treatment-naïve acute myeloid leukemia with venetoclax and azacitidine. J Clin Oncol. 2022;40(8):855–865. doi: 10.1200/JCO.21.01546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oellerich T., Schneider C., Thomas D., et al. Selective inactivation of hypomethylating agents by SAMHD1 provides a rationale for therapeutic stratification in AML. Nat Commun. 2019;10(1):3475. doi: 10.1038/s41467-019-11413-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Millán-Zambrano G., Burton A., Bannister A.J., et al. Histone post-translational modifications - cause and consequence of genome function. Nat Rev Genet. 2022 doi: 10.1038/s41576-022-00468-7. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 79.Nitsch S., Zorro Shahidian L., Schneider R. Histone acylations and chromatin dynamics: concepts, challenges, and links to metabolism. EMBO Rep. 2021;22(7) doi: 10.15252/embr.202152774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim Y.H., Bagot M., Pinter-Brown L., et al. Mogamulizumab versus vorinostat in previously treated cutaneous T-cell lymphoma (MAVORIC): an international, open-label, randomised, controlled phase 3 trial. Lancet Oncol. 2018;19(9):1192–1204. doi: 10.1016/S1470-2045(18)30379-6. [DOI] [PubMed] [Google Scholar]
- 81.Sekeres M.A., Othus M., List A.F., et al. Randomized phase II study of azacitidine alone or in combination with lenalidomide or with vorinostat in higher-risk myelodysplastic syndromes and chronic myelomonocytic leukemia: north American Intergroup Study SWOG S1117. J Clin Oncol. 2017;35(24):2745–2753. doi: 10.1200/JCO.2015.66.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bachy E., Camus V., Thieblemont C., et al. Romidepsin plus CHOP versus CHOP in patients with previously untreated peripheral T-cell lymphoma: results of the Ro-CHOP phase III study (conducted by LYSA) J Clin Oncol. 2022;40(3):242–251. doi: 10.1200/JCO.21.01815. [DOI] [PubMed] [Google Scholar]
- 83.O'Connor O.A., Horwitz S., Masszi T., et al. Belinostat in patients with relapsed or refractory peripheral T-cell lymphoma: results of the pivotal phase II BELIEF (CLN-19) study. J Clin Oncol. 2015;33(23):2492–2499. doi: 10.1200/JCO.2014.59.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laubach J.P., Schjesvold F., Mariz M., et al. Efficacy and safety of oral panobinostat plus subcutaneous bortezomib and oral dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma (PANORAMA 3): an open-label, randomised, phase 2 study. Lancet Oncol. 2021;22(1):142–154. doi: 10.1016/S1470-2045(20)30680-X. [DOI] [PubMed] [Google Scholar]
- 85.Shi Y., Dong M., Hong X., et al. Results from a multicenter, open-label, pivotal phase II study of chidamide in relapsed or refractory peripheral T-cell lymphoma. Ann Oncol. 2015;26(8):1766–1771. doi: 10.1093/annonc/mdv237. [DOI] [PubMed] [Google Scholar]
- 86.Chun P. Histone deacetylase inhibitors in hematological malignancies and solid tumors. Arch Pharm Res (Seoul) 2015;38(6):933–949. doi: 10.1007/s12272-015-0571-1. [DOI] [PubMed] [Google Scholar]
- 87.Mlcochova P., Sutherland K.A., Watters S.A., et al. A G1-like state allows HIV-1 to bypass SAMHD1 restriction in macrophages. EMBO J. 2017;36(5):604–616. doi: 10.15252/embj.201696025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Novellis D., Cacace F., Caprioli V., et al. The TKI era in chronic leukemias. Pharmaceutics. 2021;13(12):2201. doi: 10.3390/pharmaceutics13122201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Neviani P., Harb J.G., Oaks J.J., et al. PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. J Clin Invest. 2013;123(10):4144–4157. doi: 10.1172/JCI68951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bermejo M., López-Huertas M.R., García-Pérez J., et al. Dasatinib inhibits HIV-1 replication through the interference of SAMHD1 phosphorylation in CD4+ T cells. Biochem Pharmacol. 2016;106:30–45. doi: 10.1016/j.bcp.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 91.Szaniawski M.A., Spivak A.M., Cox J.E., et al. SAMHD1 phosphorylation coordinates the anti-HIV-1 response by diverse interferons and tyrosine kinase inhibition. mBio. 2018;9(3):e00819. doi: 10.1128/mBio.00819-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Amarante-Mendes G.P., Rana A., Datoguia T.S., et al. BCR-ABL1 tyrosine kinase complex signaling transduction: challenges to overcome resistance in chronic myeloid leukemia. Pharmaceutics. 2022;14(1):215. doi: 10.3390/pharmaceutics14010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schwefel D., Groom H.C.T., Boucherit V.C., et al. Structural basis of lentiviral subversion of a cellular protein degradation pathway. Nature. 2014;505(7482):234–238. doi: 10.1038/nature12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hrecka K., Hao C., Gierszewska M., et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Herold N., Rudd S.G., Ljungblad L., et al. Targeting SAMHD1 with the Vpx protein to improve cytarabine therapy for hematological malignancies. Nat Med. 2017;23(2):256–263. doi: 10.1038/nm.4265. [DOI] [PubMed] [Google Scholar]
- 96.Castellví M., Felip E., Ezeonwumelu I.J., et al. Pharmacological modulation of SAMHD1 activity by CDK4/6 inhibitors improves anticancer therapy. Cancers. 2020;12(3):713. doi: 10.3390/cancers12030713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mauney C.H., Perrino F.W., Hollis T. Identification of inhibitors of the dNTP triphosphohydrolase SAMHD1 using a novel and direct high-throughput assay. Biochemistry. 2018;57(47):6624–6636. doi: 10.1021/acs.biochem.8b01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.