Abstract
BACKGROUND
The molecular mechanisms of heart failure (HF) are still poorly understood. Circular RNA (circRNA) has been discovered in the heart in increasing numbers of studies. The goal of this research is to learn more about the potential roles of circRNAs in HF.
METHODS & RESULTS
We used RNA sequencing data to identify the characteristics of circRNAs expressed in the heart and discovered that the majority of circRNAs screened were less than 2000 nt. Additionally, chromosomes One and Y had the most and least number of circRNAs, respectively. After excluding duplicate host genes and intergenic circRNAs, a total of 238 differentially expressed circRNAs (DECs) and 203 host genes were discovered. However, only four of the 203 host genes of DECs were examined in HF differentially expressed genes. Another study used Gene Oncology analysis of DECs host genes to elucidate the underlying pathogenesis of HF, and it found that binding and catalytic activity accounted for a large portion of DECs. Immune system, metabolism, and signal transduction pathways were significantly enriched. Furthermore, 1052 potentially regulated miRNAs from the top 40 DECs were collected to build a circRNA-miRNA network, and it was discovered that 470 miRNAs can be regulated by multiple circRNAs, while others are regulated by a single circRNA. In addition, a comparison of the top 10 mRNAs in HF and their targeted miRNAs revealed that DDX3Y and UTY were regulated by the most and least circRNA, respectively.
CONCLUSION
These findings demonstrated circRNAs have species and tissue specific expression patterns; while circRNA expression is independent on host genes, the same types of genes in DECs and DEGs worked in HF. Our findings would contribute to a better understanding of the critical roles of circRNAs and lay the groundwork for future studies of HF molecular functions.
Heart failure (HF), the leading cause of death in the world, is a clinical syndrome characterized by dysfunction of myocardial pump functions to maintain tissue perfusion.[1] Pathophysiologically, cardiac output is low and/or has a pathological distribution. HF is associated with some remodeling processes including adverse cellular, structural, and functional changes in the myocardium,[2] which lead to related clinical manifestations such as pulmonary congestion, dyspnea, and fatigue.[3,4] Despite significant therapeutic advances, current therapeutic approaches do not address the fundamental problem, and the treatments are suboptimal for heart disease. So, the process of molecular regulation in HF needs to be well elucidated.
Hitherto, with the development of whole-genome sequencing studies, non-coding RNAs have emerged as critical molecules of post-transcriptional regulation in HF. CircRNA is a type of rediscovered endogenous RNA with a covalently closed continuous loop.[5] CircRNAs act as molecular sponges to interact with miRNAs or proteins; in addition, circRNAs regulate the transcription process via Pol II.[6-9] Recently, a study demonstrated that circRNAs could directly synthesize proteins as mRNA.[10] Increasing evidence reveals that circRNAs are involved in the progression of cardiovascular diseases. For example, cdr1as and circNCX1 are reported to promote myocardial infarction (MI);[11-12] however, MFCAR inhibits MI by increasing MTP18 expression.[13] Another circRNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/Fam65b pathway.[14] Moreover, circFoxo3 accelerates cardiac senescence by modulating the anti-senescent protein ID-1 and the transcription factor E2F1, as well as the anti-stress proteins FAK and HIF1a.[15] CircRNA HRCR mitigates cardiac hypertrophy by targeting miR-223.[16] CircRNA_000203 and circRNA_010567 facilitate myocardial fibrosis via suppressing miR-26b-5p and miR-141, respectively.[17,18] Given that circRNAs are stable in circulation and their dynamic changes can reflect different stages of cardiovascular diseases, they are considered potential biomarkers. For instance, it is reported that the combination of hsa_circ_0124644 and hsa_circ_0098964 may serve as promising diagnostic biomarker for coronary artery disease.[19] Another circRNA MICRA improves risk classification after MI.[20] Plasma circ_0005870 is significantly decreased in hypertension patients, which may be a potential biomarker for hypertension.[21]
With advances in RNA sequencing technology, extensively expressed circRNAs in heart development and diseases have been found by several researches.[22-27] Nevertheless, these results still remain elusive in terms of systematically understanding the function of circRNAs in heart diseases.
To investigate the potential roles of circRNAs in heart failure, we first used RNA sequencing data to identify the characteristics of circRNAs expressed in the heart. Following that, GO and pathway analyses of DEC host genes were performed to shed light on the potential pathogenesis of HF. Furthermore, the levels of circRNAs and mRNA expression in HF patients were compared. We gathered the top 40 DECs’ interacting miRNAs to build a circRNA-miRNA network, and then compared the top 10 mRNA and their targeted miRNA in HF. As a result, these findings would improve our understanding of the critical roles of circRNAs and lay the groundwork for future studies of HF molecular functions.
METHODS
Data Source
The circRNA expression profiles were collected from five articles about RNA sequencing in heart failure (PMID: 27132142; PMID: 27476877; PMID:27531932; PMID: 28082450 and PMID: 29523209 ).[22-26] Because of datasets from different research centers, group variation had occurred. It was unable to perform the data analysis on interdatasets. Considering these limitations, the average values from each dataset were obtained to represent the expression levels. In these analysis, significantly changed circRNAs were defined by the absolute value of logarithmic-transformed fold change 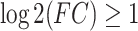 and a P-value less than 0.05.
and a P-value less than 0.05.
GO and Signal Pathways Analysis
The host genes of differentially expressed circRNAs were used for GO and signal pathway analysis. GO enrichment analysis was performed by the PANTHER classifications database;[28] and Reactome database was employed to analyze signal pathway.[29] Significant enrichment thresholds for GO and pathways analysis were adjusted P-value less than 0.05.
Interaction Network Construction Analysis
Based on the degree of differential expression, we have chose the top 40 DECs. The miRNA Binding of circAtlas 2.0 database (http://circatlas.biols.ac.cn) was used predicting downstream miRNAs of DECs. The miRNAs interacted with DEGs were predicted by TargetScan database.[30] Cytoscape was used to establish networks.[31]
Quantitative Real-Time PCR Validation
AC16 cell line were maintained in DMEM/F-12 supplemented with 10% fetal bovine serum (FBS) and 0.6% penicillin and 1% streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. The cells were seeded into 6-well plates and stimulated with 10-6 mol/L angiotensin II (Ang-II) 24 h when they grown about 80% confluency. Total RNA was isolated from cultured cells using TRIzol (Invitrogen, Carlsbad, CA, USA) method and then reversely transcribed by using the TransScript One-step gDNA Removal and cDNA Synthesis SuperMix kit according to the manufacturer’s protocol (Beijing Transgen Biotech, Co., Ltd., Beijing, China). The expression of circRNAs was analyzed on the Applied Biosystems® QuantStudio™ 6 Flex Real-Time PCR System (Thermo Fisher Scientific) with Power SYBR Green PCR Master Mix (TaKaRa, Shiga, Japan). Expression levels of the circRNAs were normalized to GAPDH using the 2-ΔΔ Cq method. The primers of each assayed circRNA/gene were given in Table 1.
Table 1. Primer sequences.
| CircRNA/gene | Forward primer | Reverse primer |
| GAPDH | GGGAAACTGTGGCGTGAT | GAGTGGGTGTCGCTGTTGA |
| NPPA | AGTGAGCCGAATGAAGAAG | GCAGATCGATCAGAGGAGT |
| hsa-intergenic_004876 | CACCAGCCTTCTGAAGGGAAG | CCACAGAGTTATATTGACAAG |
| hsa-ZFY_000001 | GCCAGCTGTGAGGACTACCTAAT | GGCTCTTGTGGCTGCAATTC |
| hsa-KLHL24_000001 | GGCAATATACTGTTACGATCC | CCATTGTTACATCAGTTGACTAT |
| hsa-SDHA_000018 | CGTCTAGAGATGTGGTGTCT | GTGCGATGACACCACGGCACT |
Statistical Analysis
The real-time PCR data were displayed as the mean ± SD and evaluated by the two-tailed Student’s t test. A P-value less than 0.05 was considered to indicate a statistically significant difference. All experiments were repeated at least three times.
RESULTS
Landscape of circRNA Length and Distribution on Chromosomes
Five articles on RNA sequencing data were enrolled in our study.[22-26] A total of 13,094, 9,953 and 16,429 circRNAs were obtained and integrated in rat, mouse and human hearts, respectively. Subsequently, we ranked them according to their length and found that most circRNAs in the heart were < 2000 nt. CircRNAs containing < 500 nt were at the top level (48%, 52%, 47% in rat, mouse and human, respectively) and 500-2000 nt were the second (38%, 39% and 42% in rat, mouse and human, respectively); whereas other lengths only accounted for 14%, 9% and 11% in rat, mouse and human, respectively (Figure 1A; supplemental Figure 1). Strikingly, compared with the analysis in intervertebral disc tissue,[32] we found the distribution of circRNAs length was quite different in various tissues. Furthermore, we counted up the number of circRNAs on each chromosome and discovered the distribution of circRNAs on different chromosomes was not uniform. Chromosome 1 contained the most circRNAs (10.59%) in both human and rat (Figure 1B; supplemental Figure 2A), however, Chromosome 2 had the most circRNAs (8.91%) only in mouse (supplemental Figure 2B). Chromosome Y contained the least circRNAs in all three species: 0.20% in human, 0.10% in mouse and no circRNAs were found in rat (Figure 1B; supplemental Figure 2A & 2B). In addition, we further analyzed the distribution of circRNAs on the unit base of each chromosome. Except for sex chromosomes, the distribution of circRNAs on each chromosome was relatively consistent: 0.0257%-0.0463% in human, 0.0272%-0.0574% in mouse and 0.0286%-0.0608% in rat (Figure 1C; supplemental Figure 2C & 2D). However, only a small amount of circRNAs was distributed on sex chromosomes: 0.0118%, 0.0102% and 0.0137% in Chromosome X of human, mouse and rat, respectively (Figure 1C; supplemental Figure 2C & 2D); and 0.0035%, 0.0011% in human and mouse Chromosome Y, respectively (Figure 1C; supplemental Figure 2D).
Figure 1.
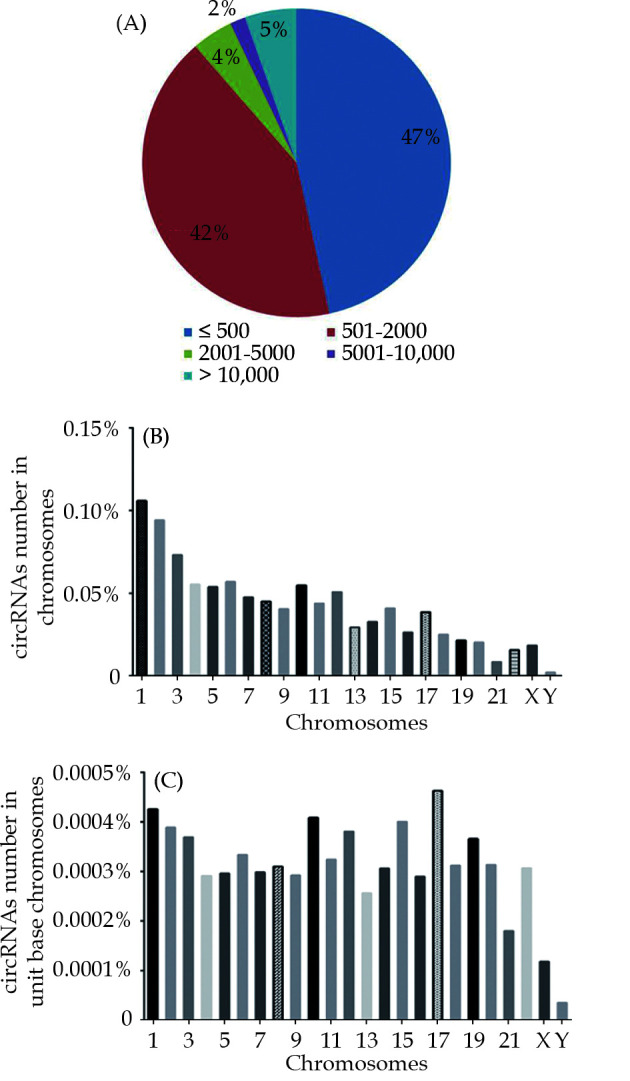
Landscape of circRNA length and distribution on chromosomes in human heart.
(A): Schematic diagram illustrating the distribution of circRNA lengths in human heart; (B): the percentage of total circRNA number on each chromosomes; (C): the percentage of circRNA number on each unit of chromosomes.
Correlation Analysis of DECs and DEGs in Human Heart Failure
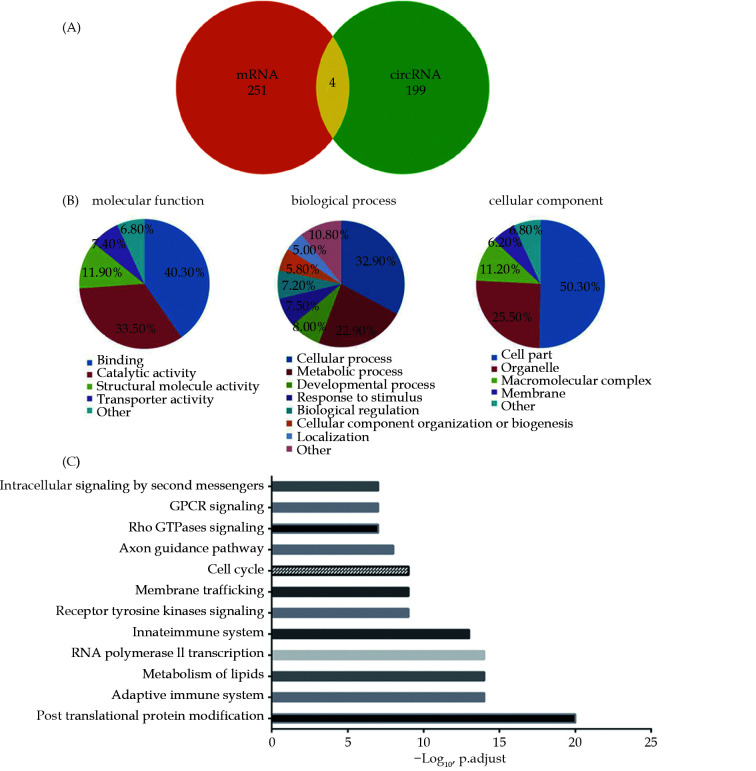
Only one article on circRNAs involvement in human HF was included in our collected data. A total of 238 DECs with 224 up-regulated and 14 down-regulated circRNAs were found and 203 host genes were obtained by excluding duplicate host genes and intergenic circRNAs (supplemental Table 1). Previously, we analyzed mRNA in human HF and found a total of 255 DEGs.[33] In order to figure out the correlation between DECs and DEGs in HF, the corresponding host genes were compared. Surprisingly, only 4 of the 203 host genes of DECs were confirmed in DEGs of HF (Figure 2A). This result suggested DECs and DEGs in HF had different expression patterns.
Figure 2.
The characteristics of DECs in human heart failure.
(A): Venn diagram for the intersections between the host genes of DECs and DEGs; (B): schematic diagram of the gene ontology enrichment analysis of DECs in human heart failure; (C): significantly enriched signaling pathways of DECs in human heart failure.
GO Annotation and Pathway Analyses of DECs Host Genes
In order to precisely clarify the potential roles of DECs in heart failure, the GO of DECs host genes was analyzed with PANTHER database. In the molecular function (MF) category, the two most enriched terms were binding (40.3%) and catalytic activity (33.5%); in biological process (BP) category, the two most enriched terms were cellular process (32.9%) and metabolic process (22.9%); in the cellular component (CC) category, cell part (50.3%) and organelle (25.5%) were the top two clustered terms (Figure 2B). Intriguingly, the results showed that enriched terms of DECs were similar to DEGs’ which we previously analyzed in HF: the top two enriched terms were catalytic activity (39.4%) and protein binding (38.0%) in MF; processes (27.0%) and metabolic processes (25.2%) in BP; cell part (35.8%) and organelle (23.6%) in CC (Figure 2B). Comprehensive analysis of these results demonstrated that the same types of genes in DECs and DEGs worked in HF, although they are not the same.
To further gain a comprehensive insight into the potential roles of DECs, the pathways of 203 host genes obtained from DECs were analyzed with the Reactome database. Pathways related to the immune system, metabolism and signal transduction were significantly enriched. Among these top pathways, four were involved in signal transduction, including receptor tyrosine kinase signaling, Rho GTPase signaling, GPCR signaling and intracellular signaling by second messengers; two participated in the immune system, including the adaptive immune system and the innate immune system. In addition, post-translational protein modification, metabolism of lipids, RNA polymerase II transcription, membrane trafficking, the cell cycle and axon guidance pathways were also significantly enriched (Figure 2C). This result indicated that the immune system and signal transduction pathways had prominent roles in HF, which was consistent with the DEGs analysis.[33]
Construction of the circRNA–miRNA Regulated Network
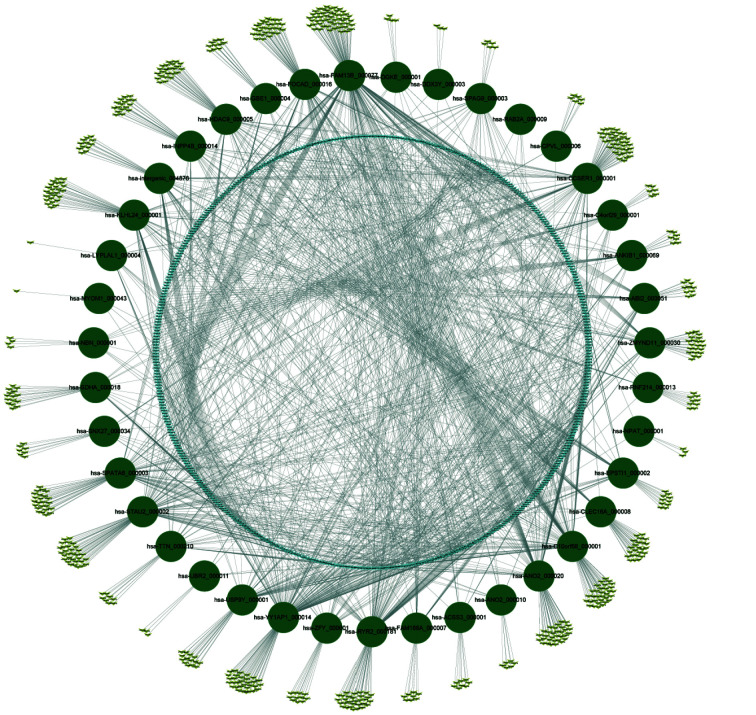
Considering that one role of circRNAs was performed as the sponge of miRNAs to further influence downstream mRNA expression, 1052 downstream miRNAs of the top 40 DECs were predicted by the circAtlas 2.0 database to construct a circRNA–miRNA network. Among these miRNAs, 470 were regulated by more than one circRNA; and the most regulated miRNAs were miR-6756-5p and miR-1226-5p, which could be regulated by 8 circRNAs. Another 582 miRNAs were regulated by a single circRNA (Figure 3). For example, miR-328, which is differentially expressed in HFpEF and HFrEF and in HF and no-HF,[34] could be regulated by hsa-ANO2_000020, hsa-CLEC16A_000008, hsa-FAM13B_000077, hsa-YY1AP1_000014 and hsa-RAB2A_000009; another potential miRNA biomarker for HF, miR-638,[35] would be regulated by only one DEC--hsa-STAU2_000002. Similarly, each circRNA could regulate several miRNAs: from the least 4 miRNAs (regulated by has-MYOM1_000043) to the most 181 miRNAs (regulated by hsa-FAM13B_000077). RyR2, one of the most important genes in cardiac hypertrophy and HF,[36-38] expresses over 100 circRNA isoforms.[23] Hsa-RYR2_000181, one of the top 40 circRNAs derived from RyR2, could regulate 108 miRNAs.
Figure 3.
CircRNA-miRNA network in human heart failure.
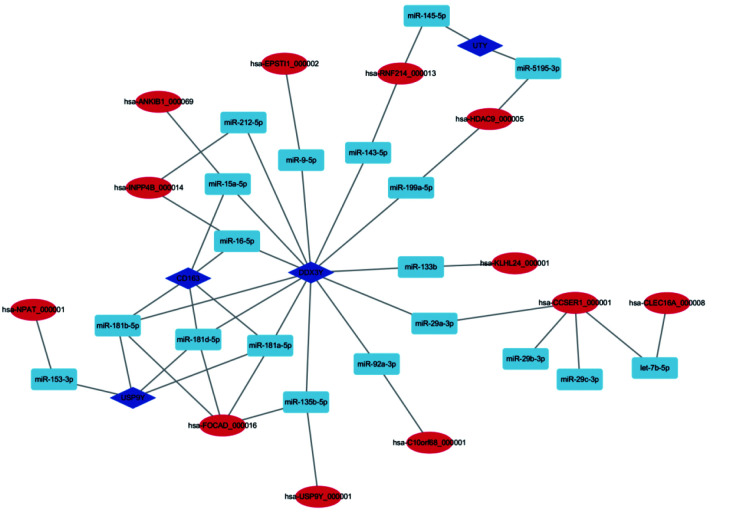
Previously, we analyzed and obtained the top 10 DEGs in HF.[33] In order to further explore the relationship between mRNA and ncRNA in HF, the top 10 DEGs were analyzed based on the circRNA–miRNA network. Finally, 4 mRNAs, 19 miRNAs and 12 circRNAs were found to establish a circRNA–miRNA-mRNA regulation network (Figure 4). DDX3Y, the most regulated gene, could be regulated by 10 circRNAs through 13 miRNAs; and UTY, the least regulated gene, would be regulated by 2 circRNAs through 2 miRNAs. The most regulated circRNA is Has-FOCAD_000016, which could regulate three genes through 4 miRNAs. However, most circRNAs only regulate one miRNA.
Figure 4.
CircRNA-miRNA-mRNA network in human heart failure.
The network consisting of 4 mRNAs, 19 miRNAs and 12 circRNAs was generated by cytoscape.
Real-Time PCR validation
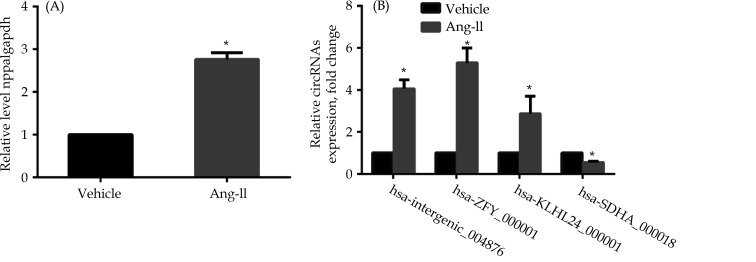
Four DEGs were chosen at random for further evaluation to validate the reliability of the RNA-Seq results analysis. Human heart AC16 cells were stimulated with 10-6 mol/L Ang-II for 24 h. First, nppa, the marker of HF, was detected to verify the model of HF in vitro. Nppa was significantly upregulated in the Ang-II treated group compared to the control group (Figure 5A). Then, Real-Time PCR demonstrated that hsa-intergenic_004876, hsa-ZFY_000001 and hsa-KLHL24_000001 were significantly upregulated (P < 0.05) in the Ang-II treated group compared with the control group; however, hsa-SDHA_000018 was reduced (P < 0.05) in the Ang-II treated group compared with the normal ones (Figure 5B). These results agreed with circRNA RNA-Seq data and suggested the RNA-Seq data were credible.
Figure 5.
Relative expression of circRNAs in AC16 cells with Ang-II.
(A): HF marker nppa expression; (B): hsa-intergenic_004876, hsa-ZFY_000001 and hsa-KLHL24_000001 were significantly upregulated, while hsa-SDHA_000018 was significantly downregulated in HF models compared with normal ones. *P < 0.05.
DISCUSSION
Heart failure, whose pathogenesis has been extensively studied, is the leading cause of global mortality and morbidity. However, the mechanisms are still largely unknown. CircRNAs have been shown in recent studies to act as molecular sponges, splicing regulators, and transcriptional regulators in a variety of cardiovascular diseases. As a result, it is critical to conduct a thorough investigation into the expression profiling of circRNAs in HF. In this study, we first integrated RNA sequencing data and discovered that the majority of circRNAs in the heart were < 2000 nt long. In the human heart, Chromosome One and Chromosome Y have the most and least circRNAs, respectively. The distribution of circRNAs on autosomes was much more uniform across species than that of sex chromosomes. DECs, GO, and pathway analyses all indicated a close relationship between DECs and DEGs in HF. Following that, the top 40 DECs’ potential regulatory miRNAs were gathered to build a circRNA-miRNA network, with 582 miRNAs found to be regulated by a single circRNA and 470 miRNAs found to be regulated by multiple circRNAs. Furthermore, miRNAs can regulate 12 of the top 40 DECs and 4 of the top 10 DEGs.
Recent research has revealed that circRNAs have tissue or developmental stage-specific expression patterns. Xia, et al.[39] discovered over 300,000 circRNAs in humans and mice, with 10.4% of human circRNAs and 34.3% of mouse circRNAs being tissue-specific. The proportion of circRNAs with different length distributions derived from different tissues varies. CircRNAs from the HeLa cell line ranged in length from 150 to 99,934 nucleotides (nt). Approximately 67% of circRNAs have a predicted splicing length of < 10,000 nt, while 46.4% and 20.6% have lengths of < 5,000 and 5000-9999 nt, respectively. Furthermore, 32.0% of circRNAs have a length of 10,000–50,000 nt.[40] CircRNAs are evenly distributed in the human intervertebral disc, with > 10,000 nt being the most abundant (29.34%), whereas containing 5,001-10,000 nt were at the lowest (14.79%); other lengths accounting for about 20%.[32] However, the length of the heart-derived circRNAs differs from that of other organs: most circRNAs in the heart are < 2000 nt and about half of circRNAs have a length of < 500 nt long. According to one study, the lengths of mature circRNAs are measured to determine the pattern of nuclear export.[41] As a result, differences in the length distribution of circRNAs in different tissues may be related to their function.
CircRNAs may be expressed independently of their host genes, according to mounting evidence.[42,43] Our findings also confirmed that most DECs can be expressed independently of their host genes in HF. However, further analysis with GO revealed that the enriched terms of DECs were similar to DEGs' in HF in all three categories previously examined. Furthermore, the pathways analysis shows that DECs in HF are linked to the immune system, metabolism, and signal transduction. It has been reported that sustained immune system activation and lipid metabolism are potential contributors to the progression of heart failure.[44,45] Integration of GO analysis and pathway analysis results in HF suggest that, while circRNA expression is independent of host genes, the same types of genes in DECs and DEGs work in HF. Our findings may shed light on the mechanism of circRNAs in HF.
Numerous studies have investigated the roles of miRNAs in HF, and the circRNA-miRNA network can be a powerful tool for understanding miRNA regulatory mechanisms. Duan, et al.[46] reported that miR-214 inhibited cardiac angiogenesis in HF by regulating the expression of the transcription factor XBP1. Our findings indicated that hsa-ANO2 000020 and hsa-STAU2 000002 could interact with miR-214, implying that they may play roles in cardiac angiogenesis. Furthermore, inhibiting miR-208a improves cardiac function and survival during heart failure.[47] Our research discovered that hsa-EPSTI1 000002 and hsa-ZMYND11 000030 can act as sponges for miR-208a, and that increasing the expression of either of the two circRNAs can improve cardiac remodeling. Previously, we discovered that miR-378 inhibits myocardial fibrosis by inhibiting p38 MAP kinase phosphorylation.[48] Hsa-FAM13B 000077 and hsa-INPP4B 000014 were predicted to regulate miR-378 in the circRNA-miRNA network, and further research into the relationship between the two circRNAs and miR-378 in cardiac fibrosis is warranted. USP9Y and DDX3Y are two of the top five upregulated DEGs in HF, as previously reported.[33] CircRNAs derived from USP9Y and DDX3Y are also upregulated in HF, which is intriguing. The circRNA-miRNA-mRNA network, on the other hand, revealed that there is no miRNA acting as a bridge to connect the circRNAs and mRNAs of USP9Y and DDX3Y, respectively. More research is needed to determine whether USP9Y/DDX3Y and their circRNAs are dependent on HF regulation in the future. Our findings could lead to a better understanding of the molecular mechanisms by which circRNAs regulate HF.
Despite the fact that our analysis is comprehensive, with a high throughput and a large sample size, it has some limitations. It is preferable to detect circRNA in the myocardium of HF patients and healthy controls to validate the reliability of the RNA-Seq results analysis. However, due to donor limitations, we only used AC16 cell lines with Ang-II treatment for the HF model. Another limitation of this study is that we do not focus on the role of circRNAs in translation. Several groups have recently reported that circRNAs are translated.[10,49,50] These studies, however, do not shed much light on the functions of the circRNA-encoded protein products. In the coming years, we expect studies to show the HF impact of circRNA translation and the resulting peptides.
CONCLUSION
In conclusion, a novel and comprehensive analysis of circRNA expression profiles in HF was obtained using RNA seq datasets. The majority of circRNAs in the heart were < 2000 nt in length (89%); additionally, the distribution of heart circRNAs on different chromosomes was not uniform. DECs in HF play roles in the immune system, metabolism, and signal transduction pathways. CircRNAs in the heart may control miRNAs and mRNAs. The findings will be useful for future research into the molecular mechanisms of HF.
Contributor Information
Zhi-Wen DING, Email: zhiwen_d@fudan.edu.cn.
Yun-Zeng ZOU, Email: zou.yunzeng@zs-hospital.sh.cn.
References
- 1.Tanai E, Frantz S Pathophysiology of Heart Failure. Compr Physiol. 2015;6:187–214. doi: 10.1002/cphy.c140055. [DOI] [PubMed] [Google Scholar]
- 2.Birks EJ Molecular changes after left ventricular assist device support for heart failure. Circ Res. 2013;113:777–791. doi: 10.1161/CIRCRESAHA.113.301413. [DOI] [PubMed] [Google Scholar]
- 3.Carlisle MA, Fudim M, DeVore AD, Piccini JP Heart failure and atrial fibrillation, like fire and fury. JACC Heart Fail. 2019;7:447–456. doi: 10.1016/j.jchf.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Tschöpe C, Kherad B, Klein O, et al Cardiac contractility modulation: mechanisms of action in heart failure with reduced ejection fraction and beyond. Eur J Heart Fail. 2019;21:14–22. doi: 10.1002/ejhf.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Li C, Tan C, Liu X Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53:359–365. doi: 10.1136/jmedgenet-2016-103758. [DOI] [PubMed] [Google Scholar]
- 6.Hansen TB, Jensen TI, Clausen BH, et al Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 7.Memczak S, Jens M, Elefsinioti A, et al Circular RNAs are a large classof animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 8.Xing L, Shi Q, Zheng K, et al Ultrasound-mediated microbubble destruction (ummd) facilitates the deliveryof ca19-9 targeted and paclitaxel loaded mpeg-plga-pll nanoparticles inpancreatic cancer. Theranostics. 2016;6:1573–1587. doi: 10.7150/thno.15164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Huang C, Bao C, et al Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Fan X, Mao M, et al Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng HH, Li R, Su YM, et al The Circular RNA Cdr1 as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li MY, Ding W, Tariq MA, et al A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics. 2018;8:5855–5869. doi: 10.7150/thno.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Gan TY, Li N, et al Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou LY, Zhai M, Huang Y, et al The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy viamodulation of the Pink1/ FAM65B pathway. Cell Death Differ. 2019;26:1299–1315. doi: 10.1038/s41418-018-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du WW, Yang W, Chen Y, et al Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 16.Wang K, Long B, Liu F, et al A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 17.Tang CM, Zhang M, Huang L, et al CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep. 2017;7:40342. doi: 10.1038/srep40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou B, Yu JW A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res Commun. 2017;487:769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Z, Li X, Gao C, et al Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep. 2017;7:39918. doi: 10.1038/srep39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salgado-Somoza A, Zhang L, Vausort M, Devaux Y The circular RNA MICRA for risk stratification after myocardial infarction. Int J Cardiol Heart Vasc. 2017;17:33–36. doi: 10.1016/j.ijcha.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu N, Jin L, Cai J Profiling and bioinformatics analyses reveal differential circular RNA expression in hypertensive patients. Clin Exp Hypertens. 2017;39:454–459. doi: 10.1080/10641963.2016.1273944. [DOI] [PubMed] [Google Scholar]
- 22.Jakobi T, Czaja-Hasse LF, Reinhardt R, Dieterich C Profiling and validation of the circular RNA repertoire in adult murine hearts. Genomics Proteomics Bioinformatics. 2016;14:216–223. doi: 10.1016/j.gpb.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Werfel S, Nothjunge S, Schwarzmayr T, et al Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol. 2016;98:103–107. doi: 10.1016/j.yjmcc.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Khan MA, Reckman YJ, Aufiero S, et al RBM20 regulates circular RNA production from the titin gene. Circ Res. 2016;119:996–1003. doi: 10.1161/CIRCRESAHA.116.309568. [DOI] [PubMed] [Google Scholar]
- 25.Tan WL, Lim BT, Anene-Nzelu CG, et al A landscape of circular RNA expression in the human heart. Cardiovasc Res. 2017;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- 26.Lei W, Feng T, Fang X, et al Signature of circular RNAs in human induced pluripotent stem cells and derived cardiomyocytes. Stem Cell Res Ther. 2018;9:56. doi: 10.1186/s13287-018-0793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu M, Wei X, Li M, et al Circular RNA expression profiles of persistent atrial fibrillation in patients with rheumatic heart disease. Anatol J Cardiol. 2019;21:2–10. doi: 10.14744/AnatolJCardiol.2018.35902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mi H, Muruganujan A, Huang X, et al Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v. 14. 0) Nat Protoc. 2019;14:703–721. doi: 10.1038/s41596-019-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jupe S, Fabregat A, Hermjakob H Expression data analysis with Reactome. Curr Protoc Bioinformatics. 2015;49:8.20.1–9. doi: 10.1002/0471250953.bi0820s49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015; 4.
- 31.Doncheva NT, Assenov Y, Domingues FS, Albrecht M Topological analysis and interactive visualization of biological networks and protein structures. Nat Protoc. 2012;7:670–85. doi: 10.1038/nprot.2012.004. [DOI] [PubMed] [Google Scholar]
- 32.Zou F, Ding Z, Jiang J, et al Confirmation and preliminary analysis of circRNAs potentially involved in human intervertebral disc degeneration. Mol Med Rep. 2017;16:9173–9180. doi: 10.3892/mmr.2017.7718. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Jiang Q, Ding Z, et al Identification of a common different gene expression signature in ischemic cardiomyopathy. Genes (Basel) 2018;9:56. doi: 10.3390/genes9010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watson CJ, Gupta SK, O’Connell E, et al MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur J Heart Fail. 2015;17:405–415. doi: 10.1002/ejhf.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong LL, Armugam A, Sepramaniam S, et al Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur J Heart Fail. 2015;17:393–404. doi: 10.1002/ejhf.223. [DOI] [PubMed] [Google Scholar]
- 36.Ding Z, Peng J, Liang Y, et al Evolution of vertebrate ryanodine receptors family in relation to functional divergence and conservation. Int Heart J. 2017;58:969–977. doi: 10.1536/ihj.16-558. [DOI] [PubMed] [Google Scholar]
- 37.Zou Y, Liang Y, Gong H, et al Ryanodine receptor type 2 is required for the development of pressure overload-induced cardiac hypertrophy. Hypertension. 2011;58:1099–1110. doi: 10.1161/HYPERTENSIONAHA.111.173500. [DOI] [PubMed] [Google Scholar]
- 38.Ding Z, Yuan J, Liang Y, et al Ryanodine receptor type 2 plays a role in the development of cardiac fibrosis under mechanical stretch through TGFβ-1. Int Heart J. 2017;58:957–961. doi: 10.1536/ihj.16-572. [DOI] [PubMed] [Google Scholar]
- 39.Xia S, Feng J, Lei L, et al Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984–992. doi: 10.1093/bib/bbw081. [DOI] [PubMed] [Google Scholar]
- 40.Yu D, Li Y, Ming Z, et al Comprehensive circular RNA expression profile in radiation-treated HeLa cells and analysis of radioresistance-related circRNAs. Peer J. 2018;6:e5011. doi: 10.7717/peerj.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang C, Liang D, Tatomer DC, Wilusz JE A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639–644. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gruner H, Cortés-López M, Cooper DA, et al CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6:38907. doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siede D, Rapti K, Gorska AA, et al Identification of circular RNAs with host gene-independent expression in human model systems for cardiac differentiation and disease. J Mol Cell Cardiol. 2017;109:48–56. doi: 10.1016/j.yjmcc.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 44.Carrillo-Salinas FJ, Ngwenyama N, Anastasiou M, et al Heart inflammation: immune cells roles and roads to the heart. Am J Pathol. 2019;189:1482–1494. doi: 10.1016/j.ajpath.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sánchez-Trujillo L, Vázquez-Garza E, Castillo EC, et al Role of adaptive immunity in the development and progression of heart failure: new evidence. Arch Med Res. 2017;48:1–11. doi: 10.1016/j.arcmed.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 46.Duan Q, Yang L, Gong W, et al MicroRNA-214 Is Upregulated in Heart Failure Patients and Suppresses XBP1-Mediated Endothelial Cells Angiogenesis. J Cell Physiol. 2015;230:1964–1973. doi: 10.1002/jcp.24942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montgomery RL, Hullinger TG, Semus HM, et al Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan J, Liu H, Gao W, et al MicroRNA-378 suppresses myocardial fibrosis through a paracrine mechanism at the early stage of cardiac hypertrophy following mechanical stress. Theranostics. 2018;8:2565–2582. doi: 10.7150/thno.22878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Legnini I, Di Timoteo G, Rossi F, et al Circ-ZNF609 Is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pamudurti NR, Bartok O, Jens M, et al Translation of CircRNAs. Mol Cell. 2017;66:9–21. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]