Abstract
Histidine methylation serves as an intriguing strategy to introduce altered traits of target proteins, including metal ion chelation, histidine-based catalysis, molecular assembly, and translation regulation. As a newly identified histidine methyltransferase, METTL9 catalyzes N1-methylation of protein substrates containing the “His-x-His” motif (HxH, x denotes small side chain residue). Here our structural and biochemical studies revealed that METTL9 specifically methylates the second histidine of the “HxH” motif, while exploiting the first one as a recognition signature. We observed an intimate engagement between METTL9 and a pentapeptide motif, where the small “x” residue is embedded and confined within the substrate pocket. Upon complex formation, the N3 atom of histidine imidazole ring is stabilized by an aspartate residue such that the N1 atom is presented to S-adenosylmethionine for methylation. Moreover, METTL9 displayed a feature in preferred consecutive and “C-to-N” directional methylation of tandem “HxH” repeats that exist in many METTL9 substrates. Collectively, our work illustrates the molecular design of METTL9 in N1-specific methylation of the broadly existing “HxH” motifs, highlighting its importance in histidine methylation biology.
Keywords: Histidine N1-methylation, Methyltransferase, METTL9, His-x-His motif
1. Introduction
Post-translational modifications of proteins play a critical role in life cycle and cell fate decision by introducing altered traits of the modified proteins. As a prevalent type of modification, methylation is usually catalyzed by methyltransferases (MTases), which transfer the methyl group from S-adenosylmethionine (SAM; also known as AdoMet) onto residues including lysine, arginine, histidine, glutamate, glutamine, asparagine, and cysteine, with S-adenosylhomocysteine (SAH; also known as AdoHcy) as the co-product (Jakobsson, 2021). Extensive studies have demonstrated the importance of lysine and arginine methylation for biological regulation and cellular responses (Greer & shi, 2012). Recent studies have shined light on the significant role of histidine methylation involved in various biological pathways. The methylation of histidine side chain brings about new biochemical properties of the modified histidine, such as: increase in molecular volume and hydrophobicity as well as the fixation of histidine's protonation and tautomerization states (Kwiatkowski & drozak, 2020). Methylation of histidine may affect the chelation of divalent metal ions such as Zn2+, Cu2+, and Fe2+ by metal binding proteins (Davydova et al., 2021; Lv et al., 2021). It has been reported that methylation of His73 on β-actin promotes smooth muscle contraction and actin filament stabilization (Wilkinson et al., 2019). Moreover, histidine methylation serve as a key step for converting carnosine (β-alanyl-l-histidine) to anserine (β-alanyl-N-methyl-l-histidine), both are bioactive dipeptides in the brain and muscle (Boldyrev et al., 2013).
Although N-methylated histidine were first discovered in actin (Johnson et al., 1967) and myosin (Huszar & elzinga, 1972) around 50 years ago, only a limited number of proteins are found carrying this modification (Al-Hadid et al., 2014; Carroll et al., 2005; Meyer & mayr, 1987; Raftery et al., 1996; Webb et al., 2010). Recent advancements in high-throughput proteomics have led to the discovery of hundreds of methylated histidine sites in proteins extracted from mammalian tissues and cells (Jakobsson, 2021). Both nitrogen atoms on the imidazole ring of histidine can be methylated, namely N1-methylhistidine (1 MH; also called Nπ-methylhistidine) or N3-methylhistidine (3 MH; also called Nτ-methylhistidine) (Kwiatkowski & drozak, 2020). Three protein histidine methyltransferases (PHMTs) that have been characterized so far, including SETD3 from the SET domain-containing family(Kwiatkowski et al., 2018; Wilkinson et al., 2019), METTL9 and METTL18 from the seven-β-strand (7BS) family (Fig. 1A) (Davydova et al., 2021). Among these three proteins, SETD3 catalyzes the N3-methylation of actin His73 (Kwiatkowski et al., 2018; Wilkinson et al., 2019), while METTL18 catalyzes the N3-methylation of RPL-3 His245 (Malecki et al., 2021). In comparison, METTL9 is the only enzyme that catalyzes the N1-methylation of various substrates containing the histidine-x-histidine motif (HxH, in which ‘x’ represents a small side chain residue), including SLC39A7, S100A9, and NDUFB3, etc. Several studies have recently identified regulatory functions of METTL9 in physiological processes such as tumor growth, antitumor immunity (Lv et al., 2021), innate immune response (Daitoku et al., 2021), and zinc transport between the endoplasmic reticulum (ER) and cytoplasm (Lv et al., 2021).
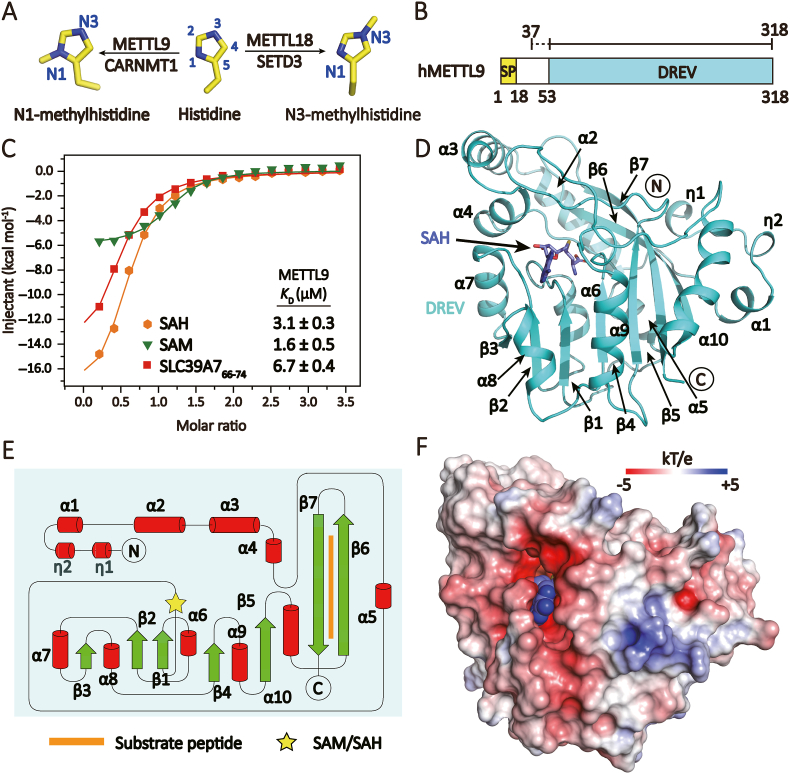
Fig. 1.
Overall structure of METTL9.(A) Position-specific histidine methylation and its writers. (B) Domain architecture of METTL9. Yellow, signal peptide (SP, residue 1–18); cyan, DREV catalytic domain (residue 53–318). The expression frame for crystallization is denoted in the top. Dashed line, invisible region. (C) Isothermal titration calorimetry (ITC) fitting curves of full-length METTL9 titrated with SAM, SAH and unmethylated substrate peptide. (D) Structure of METTL9 (cyan ribbon) bound to SAH (blue sticks). (E) Topology diagram of the METTL9 fold. Yellow star, SAH/SAM cofactors; orange line segment, substrate peptide. (F) Electrostatic surface view of METTL9 bound to SAH (blue spheres). The electrostatic potential is expressed as a spectrum ranging from −5 kT/e (red) to +5 kT/e (blue). (D) and (F) are in the same orientation. (For Interpretation of the references to colour in this figure legend the readers is referred to the web version of this article.)
Although the regulatory function of METTL9 has been reported, the molecular mechanism underlying N1-specific methylation of the “HxH” motif by METTL9 remains blurry. Here we performed structural and biochemical studies of METTL9 with substrate peptides derived from SLC39A7, a critical zinc transporter of the ER and Golgi apparatus (Lv et al., 2021). Our structural and mutagenesis studies demonstrated how METTL9 achieved its enzymatic activity towards N1-specific methylation of the “HxH” motif. Moreover, we revealed preferred consecutive histidine methylation of tandem “HxH” motifs in “C-to-N” direction as a result of unmethylated Hi-2-guided Hi methylation. The precise installation of methyl marks on consensus histidine motifs highlights dedicated molecular mechanisms and suggests the functional importance of histidine methylation biology.
2. Results
2.1. Overall structure of METTL9
Human full-length METTL9 protein contains 318 residues, consisting of a putative N-terminal signal peptide (1–18) and a methyltransferase domain called “DREV” (DORA reverse strand protein) that extends from residue 53 to 318 (Fig. 1B). First, we expressed full length METTL9 and performed binding studies with SAM, SAH, and a substrate peptide SLC39A766-74, respectively. Isothermal titration calorimetry (ITC) results showed that METTL9 binds to SAM, SAH, and peptide SLC39A766-74 with a dissociation constant (KD) of 1.6, 3.1 and 6.7 μM, respectively (Fig. 1C). Next, we performed crystallization screen using different METTL9 constructs, and successfully obtained crystals of human METTL9 (aa 37–318) bound to the co-product SAH. Then we collected diffraction data and solved the binary complex structure at 2.5 Å (Supplementary Table S1).
Based on the electron densities, we were able to build residues 53–318 of METTL9, encompassing the whole DREV domain. The DREV domain contains a canonical SAM-dependent methyltransferase core fold consisting of a seven-stranded β sheet (β1-β7) connected by six flanking α helices (α5-α10) (Fig. 1D and E). The core fold is wrapped around and stabilized by four N-terminal α helices (α1-α4) (Fig. 1D). Of note, N-terminal η2 and α1 buttresses α10 from the top of METTL9, α2 and α3 anchor β6 and β7 edgewise through hydrophobic interactions to stabilize the DREV domain (Supplementary Figure S1). The DREV core domain is conserved among metazoans and sandwiches SAH and substrate at the active center (Fig. 1F and Supplementary Figure S2).
2.2. SAH binding by METTL9
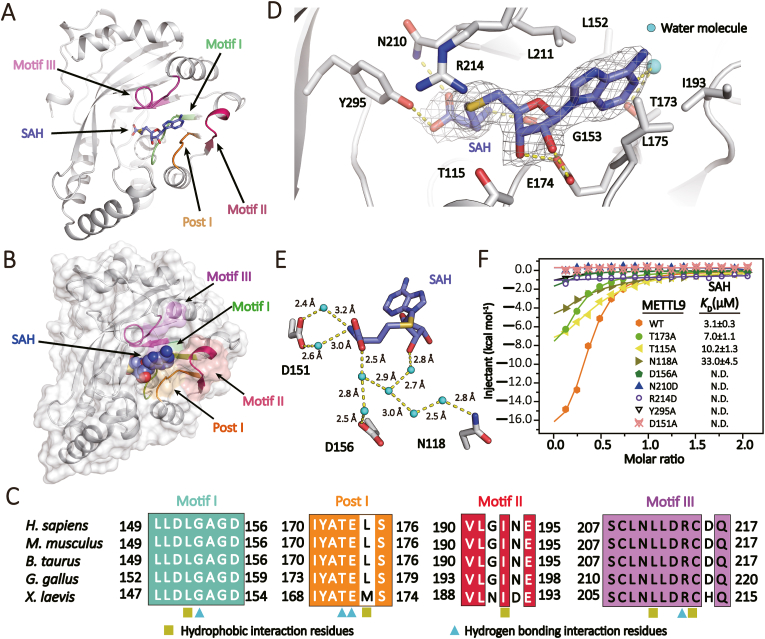
The co-product SAH is bound to a negative-charged pocket formed by four structural motifs: I, post I, II, and III(Martin & mcmillan, 2002) (Fig. 1, Fig. 2C). The adenosine ring of SAH is sandwiched between hydrophobic residues L152, L175, I193, and L211 (Fig. 2D), which distinguishes it from most METTL family members, such as METTL6 and METTL18, that employ an aromatic residue to stabilize the adenosine ring via π-π stacking (Supplementary Figure S3). The ribose ring and methionine backbone are stabilized by hydrogen bonding interactions involving residues N210, G153, E174, and Y295 (Fig. 2D). Besides, the SAH molecule is further stabilized by water-mediated interaction involving residues N118, D151 and D156 (Fig. 2E). To investigate the SAH binding pocket, we conducted mutagenesis studies and all tested mutants retained proper folding similar to the wild-type METTL9, as confirmed by circular dichroism and gel filtration chromatography analysis (Supplementary Figure S4). Our ITC experiments revealed 2–10-fold decrease in SAH binding by T173A, T115A and N118A, and loss of SAH binding by D151A, D156A, N210D, R214D, Y295A (Fig. 2F), stressing their respective contributions in SAH recognition.
Fig. 2.
SAH binding by METTL9. (A) Overall structure of METTL9 (white ribbon) bound to SAH (blue sticks). Four SAH binding motifs are colored coded in pale green (Motif I), orange (Post I), red (motif II), and pink (Motif III), respectively. (B) Surface representation of METTL9 bound to SAH. Orientation and color code are the same as in (A). (C) Sequence alignment of human METTL9 and representative orthologs in vertebrates. Four SAH binding motifs are colored same as in (A). Olive square, hydrophobic interaction residues. Cyan triangles, hydrogen bonding residues. (D) Details of SAH binding pocket. 2Fo-Fc omit maps of SAH is contoured at 2.0 σ level. Sticks, residues directly participate in SAH binding; Cyan spheres, water molecules; Yellow dashes, hydrogen bonding interactions. (E) Water-mediated hydrogen-bonding network in SAH recognition. (F) ITC fitting curves of wild-type and mutant METTL9 titrated with SAH. KD values are listed with standard deviations. N.D., not detected. (For Interpretation of the references to colour in this figure legend the readers is referred to the web version of this article.)
2.3. “HxH” peptide substrate recognition
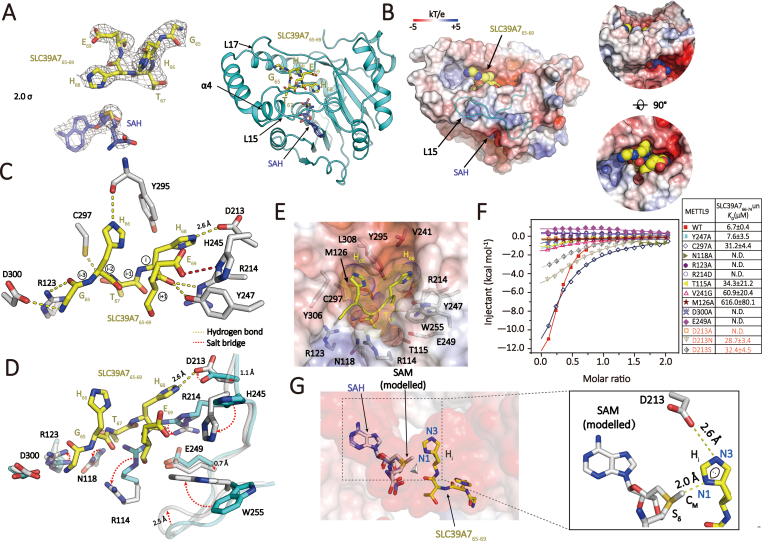
We also determined a ternary complex structure of METTL937-318 bound to SAH and its substrate peptide, SLC39A765-70 (GHTHES), at 3.4 Å resolution (Fig. 3A, left and Supplementary Table S1). In the complex structure, we were able to model residues 65–69 of SLC39A7, which takes on a β-turn like conformation and is inserted into a substrate pocket formed by α4, and loops L15, L17 (Fig. 3A, right). The peptide substrate and SAH are docked into the active center channel from two different directions and largely covered by an extended L15 element (Fig. 3B, left). The SLC39A765-69 is snuggly fitted into a negatively charged pocket (Fig. 3B, right), which corresponds to 534 Å2 decrease in solvent accessible surface area (ASA) accounting for 72% ASA of the pentapeptide 65–69 of SLC39A7.
Fig. 3.
Peptide substrate recognition by METTL9.(A) Overall structure of METTL9 bound to SLC39A7 peptide and SAH. Left, 2Fo-Fc omit maps of SAH and SLC39A765-69 contoured at 2.0 σ level. (B) Surface view of METTL9 with SAH and SLC39A7 peptide. The surface is colored by its electrostatic potential ranging from −5 kT/e (red) to +5 kT/e (blue). SAH and SLC39A7 are depicted as space-filling spheres. The L15 element covering the substrate channel is highlighted as cyan loop. Right, side- and top-views of peptide (yellow spheres) engagement in METTL9 pocket. (C) Interaction network between SLC39A7 peptide and METTL9. Key residues of METTL9 are shown as white sticks. (D) Structural alignment of peptide-free (cyan) and peptide-bound (white) METTL9 substrate pockets, the peptide substrate is depicted as yellow sticks. Red dashed arrows highlight conformational adjustments of key residues. (E) Top view of peptide binding pocket. The pocket is shown as half-transparent electrostatic surface view. “HxH” motif residues are highlighted as “dotted” spheres. Key pocket residues are shown as sticks. (F) ITC fitting curves of wild-type (WT) METTL9 and its pocket residue mutants titrated with substrate SLC39A766-74. Mutant information and binding KD values are listed. (G) Positioning of SAH/SAM and the histidine residue at the center of substrate channel. SAM (white sticks) is modeled based on SAH (blue sticks). Right, close-up view highlighting histidine imidazole ring-centered interactions. Lone pair electron of N1 atom is indicated. (For Interpretation of the references to colour in this figure legend the readers is referred to the web version of this article.)
In the complex structure of METTL9 with substrate peptide, the second histidine (H68) of the “HxH” motif is positioned at the active center for methylation, thereby designated as residue “i”. Multiple hydrogen bonding interactions contribute to the pentapeptide motif (i-3 to i+1) specific recognition by METTL9 (Fig. 3C). These interactions involve residues R123, D300, C297, Y295, D213, R214, Y247 of METTL9 and G65, H66, T67, H68 of the SLC39A7 peptide. Upon peptide substrate engagement, the catalytic center undergoes induced conformational changes, triggering residues R123 and D300 to form hydrogen bonds with the backbone of G65, prompting residues R114 and R214 to flip outwards that enables SLC39A7 peptide insertion (Supplementary Figure S5), as well as residues H245 and W255 that flip inwards to promote E69 recognition at i+1 position (Fig. 3D). Notably, D213 displayed subtle yet critical conformational adjustment to ensure an optimal hydrogen bonding interaction with the N3 atom of H68 (Fig. 3D).
Our structural studies revealed a high degree of shape complementarity between the pentapeptide and METTL9 substrate pocket, in which two histidine residues of the “HxH” motif are anchored in place with the “x” residue buried at the bottom (Fig. 3E). The burial of residue “x” explains the necessity of a small side chain residue for recognition to avoid steric clash. To explore the importance of these pocket residues, we performed mutagenesis and ITC binding studies. Among these mutants, Y247A retained a similar binding KD but displayed largely reduced enthalpy change; T115A, V241G, and C297A reduced binding by ∼6–10 fold; KD of M126A is nearly undetectable, while N118A, R123A, D213A, R214D, D300A, and E249A disrupted peptide binding completely (Fig. 3F). Collectively, these results highlight the importance of the observed polar interactions and van der Waals contacts in METTL9-SLC39A7 recognition (Supplementary Figure S6).
2.4. Histidine N1-specific methylation
Our ternary structure analysis showed that both the methyl donor (SAM) and the methyl acceptor (Hi) are juxtaposed within a substrate channel such that the methyl transfer reaction takes place at the very center (Fig. 3G, left). Based on the SAH structure, we modeled a SAM molecule into the active center, in which the high energy methyl group directly points to the N1 position of the histidine imidazole ring for methyl transfer (Fig. 3G, right). Importantly, residue D213 forms a charge-stabilized hydrogen bond with the N3 atom of histidine side chain, such that N1 is deprotonated and presented to SAM for methylation via an SN2 mechanism (Supplementary Figure S7A) (Guo & guo, 2007; Kwiatkowski & drozak, 2020; Zhang and bruice, 2008b, Zhang and bruice, 2008a). Our ITC results supported the essential role of D213 in substrate binding by METTL9, in which D213A completely disrupted binding, while D213S or D213N weakened the binding by ∼4–6 fold (Fig. 3F).
2.5. Enzymatic and mutagenesis studies
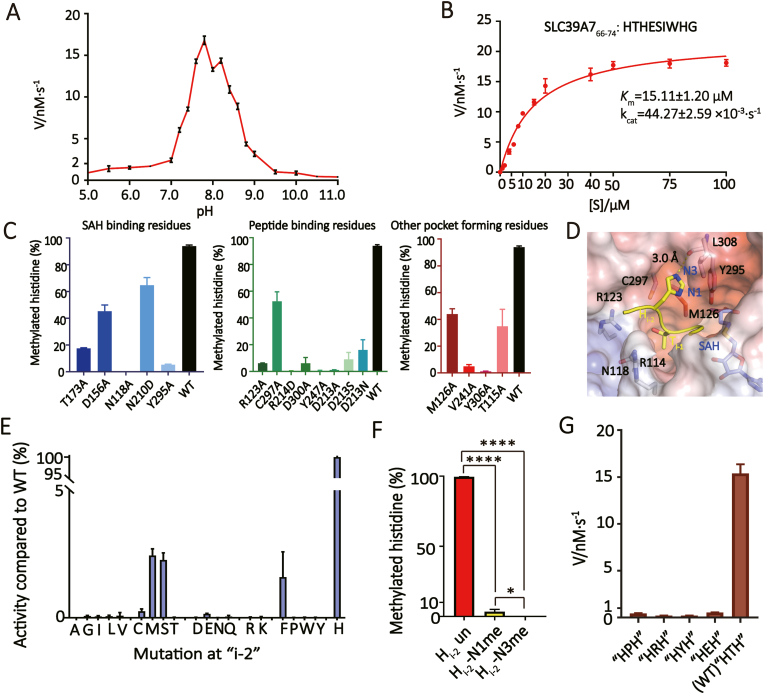
To explore the enzymatic features of METTL9, we performed kinetic studies using a quantitative LC-MS/MS strategy. We used the same peptide for both ITC and kinetic studies (SLC39A766-74, HTHESIWHG), and quantified the enzyme activity of METTL9 at different pH values (pH 5–11). Our results showed that METTL9 maintained relatively high enzymatic activities between pH 7.2–8.4, and we therefore chose pH 7.5 for the subsequent reactions (Fig. 4A). We then measured the kinetic parameters of METTL9 with Km = 15.11 μM and kcat = 44.27 × 10−3 s−1 (Fig. 4B). This level of enzymatic efficiency is comparable to other histidine methyltransferases, such as SETD3 (Kwiatkowski et al., 2018; Wilkinson et al., 2019). Our structural analysis suggested that METTL9 can only methylate the N1 atom of histidine residue Hi (H68). To validate these structural insights, we performed enzymatic assays using unmethylated H68 and N1- or N3-methylated SLC39A766-74 substrates. Mass spectrometry analysis of the products demonstrated that only the unmethylated substrate underwent methylation at the N1 position of H68. No methylation events were detected at the N3 position of H68 or at other histidine residues, such as H66 (i-2) or H73 (i+5) (Supplementary Figure S8). These results highlight the enzymatic specificity of METTL9.
Fig. 4.
Enzymatic analyses of METTL9.(A) The methylation rate of METTL9 at different pH. The pHs were adjusted by mixing different volumes of 50 mM sodium acetate and 50 mM CAPS. The detection was carried out at intervals of 0.2 between pH 7–9 and at intervals of 0.5 for the remaining pH ranges. (B) Michaelis–Menten plots of METTL9-catalyzed histidine N1-methylation of the SLC39A766-74 peptide. The kinetic parameters, Km and kcat were listed. The reaction was carried out under 20 mM Tris-HCl pH 7.5, 50 mM NaCl, and 0.5 mM METTL9. (C)In vitro methylation assays of METTL9 mutants towards the SLC39A766-74 substrate. Three categories of pocket residues are shown as indicated. (D) Close-up view of histidine recognition at i-2 and i-1 position. Key METTL9 residues are shown as sticks. (E) Comparison of catalytic efficiency by METTL9 among wild type and Hi-2-altered peptide substrates. The enzymatic activities are normalized to the wild type peptide substrate. The Y-axis is expressed as the percentage activity, and the X-axis is spread as twenty proteinogenic amino acids. (F) Comparison of enzyme activities of METTL9 towards unmethylated, N1-methylated, and N3-methylated Hi-2 peptides. (G) Comparison of catalytic efficiency by METTL9 among wild type and “xi-1”-altered peptide substrates. Error bars, N = 3, One-way ANOVA with post-hoc analysis.
Next, we expanded the enzymatic assays to METTL9 mutants, which can be divided into three categories: 1) SAM cofactor binding residues; 2) substrate peptide hydrogen bonding residues; 3) other pocket forming residues. For category 1, N210D and Y295A led to nearly complete loss of enzyme activity, while alanine mutations of D156, N118, and T173 decreased enzyme activity by ∼40–80% (Fig. 4C, left). Among category 2, R123A, R214D, D300A, and Y247A displayed enzymatic activity drop by >90%, while C297A reduced enzyme activity by ∼50%, supporting their importance in substrate binding. Consistent with the ITC results (Fig. 3F), D213A not only disrupted peptide binding but also lost enzymatic activity. Although D213S and D213N retained decent substrate binding (Fig. 3F), they lost >80–90% enzymatic activities (Fig. 4C, middle), suggesting a role of an acidic D213 residue in both substrate engagement and enzymatic catalysis. Furthermore, mutations of other pocket-forming residues, such as T115A, M126A, V241A, Y306A, resulted in an enzymatic activity reduction by > 50–90% (Fig, 4C, right), underscoring their contributions to pocket integrity and substate binding.
2.6. Hi-2 is a recognition signature for METTL9
Our structural studies have revealed that the second histidine in the “HxH” motif is the methylation site (Hi), while the first histidine is recognized by a pocket composed of Y295, M126, C297 and L308 (Fig. 4D). Therefore, we speculated that the histidine residue at position “i-2” serves as a recognition signature for efficient methylation. To this end, we conducted enzymatic studies by substituting Hi-2 with other 19 residues. Such an alphabet screening assays showed that while the wild type substrate displayed robust activity, most mutant substrates lost enzymatic activity, except that methionine (M), serine (S) and phenylalanine (F) substitutions retained ∼2%–3% activities (Fig. 4E). These results demonstrate that METTL9 does indeed prefer a histidine residue at the i-2 position.
We also examined the impact of Hi-2 methylation on Hi methylation, and found that pre-existing N1-methylation of Hi-2 retained ∼5% enzymatic activity, while N3-methylation of Hi-2 completely disrupted METTL9 activity towards Hi (Fig. 4F). This demonstrates that METTL9 prefers unmethylated Hi-2 instead of its N1- and N3-methylated forms. The fact that N3-methylation completely disrupted METTL9 activity highlights the importance of the hydrogen bonding interaction between Y295 with unmethylated N3 of Hi-2.
2.7. The “x” residue is restrained by a small x-pocket of METTL9
Previous studies have indicated that the “x" residue in the “HxH” motif of METTL9 substrate must be a small side chain residue, specifically an “ANGST” residue (Davydova et al., 2021). Our structural analysis of METTL9 with substrate peptide confirms this finding, as we observed that the “x" residue at position “i-1″ is anchored at a small x-pocket formed by residues R114, N118, and R123 (Fig. 4D). The shape complementarity of this x-pocket plays a critical role in recognizing the “x" residue (Fig. 3, Fig. 4). To validate this observation, we performed site-directed mutagenesis to replace the threonine (T) residue at position xi-1 with “non-ANGST” residues, such as proline (P), arginine (R), tyrosine (Y), and glutamic acid (E). Our enzymatic assays revealed that these substitutions almost completely abolished the methylation activity (Fig. 4G). Additionally, we observed that these xi-1 substitution peptides lost their binding ability to METTL9 (Supplementary Figure S9). In summary, our findings demonstrate that the “x" residue between two “H" residues is highly restricted by the small x-pocket of METTL9 through shape complementary, which is crucial for optimal substrate recognition and preference.
2.8. Histidine methylation of tandem “HxH” repeats by METTL9
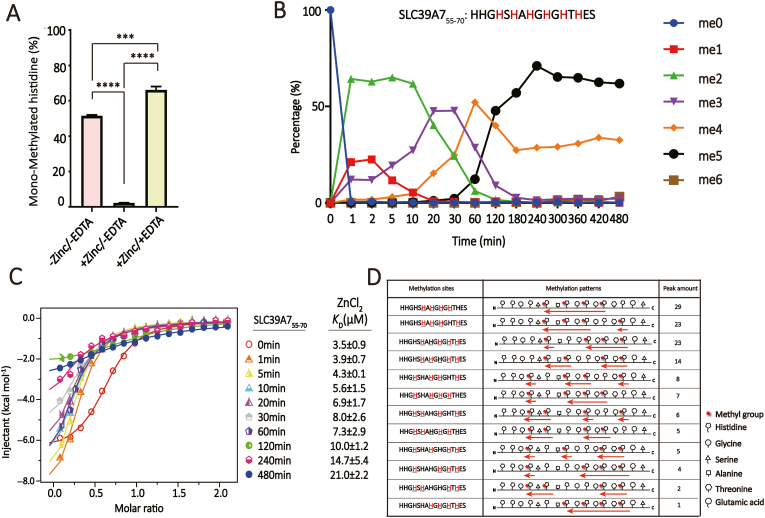
Many METTL9 substrate proteins contain tandem “HxH” motifs, which tend to form zinc finger structures (Davydova et al., 2021). To explore whether zinc coordination by “HxH” motifs could affect METTL9 activity, we synthesized the SLC39A755-70 peptide (55HHGHSHAHGHGHTHES70) rich in “HxH” motifs, and performed enzymatic assays in the presence and absence of zinc ions. We found that histidine methylation was substantially inhibited by the addition of 0.5 mM ZnCl2. Importantly, the enzymatic activity could be completely restored if the zinc ions were removed by EDTA (ethylenediaminetetraacetic acid) (Fig. 5A). These data support that SLC39A755-70 is a zinc binding motif and there exists a competition between zinc coordination by the “HxH” peptide and its efficient methylation by METTL9.
Fig. 5.
Enzymatic assays of tandem “HxH” repeats substrate by METTL9.(A) Impact of zinc coordination on METTL9 activity. Unmethylated “HxH”-rich SLC39A755-70 peptide was used as substrate and mono-methylated product was used to monitor the enzymatic activity. Error bars, N = 3, One-way ANOVA with post-hoc analysis. (B) Time-course Q-TOF-MS based enzymatic assays of METTL9 with unmethylated SLC39A755-70 peptide as substrate. Different methylation states are shown as indicated. The concentration of METTL9 is 0.5 μM, and concentration of substrate peptide is 100 μM. (C) ITC fitting curves of SLC39A755-70 peptide with ZnCl2. KD values are listed with standard deviations. ITC were carried out with three biological replicates. (D) Qualitative detection of methylation patterns of remaining tetra-methylated products after 480 min of reaction. Methylation states and modification sites are highlighted in red with underlines. “C-to-N” consecutive methylation patterns are illustrated in right column with red arrows. Due to technical problems, we were unable to detect the LC-MS/MS data of the penta-methylated product. (For Interpretation of the references to colour in this figure legend the readers is referred to the web version of this article.)
Our structural analysis revealed that, among candidate methylation sites, only the second histidine of the “HxH” motif can be methylated by METTL9. This is supported by our enzymatic assays of the SLC39A755-70 peptide, in which methylation of H55 or H56 was not detected since they do not follow the “Hi-2xHi” rule (Supplementary Figure S10A). By contrast, histidine methylation at other sites could be observed to various extents in a time-course assay (Fig. 5B). After prolonged reaction of >2 h, the SLC39A755-70 peptide could achieve high percentage of penta- or tetra-methylation, yet quite limited higher degree methylations at the steady state (Fig. 5B and Supplementary Figure S10A). Meanwhile, to investigate the impact of METTL9-catalyzed substrate methylation on substrate function, we performed ITC titrations with zinc ions using time-course methylation products. The ITC results indicated that the ability of the SLC39A755-70 peptide to coordinate zinc ions was compromised as the reaction proceeded to reach higher level of methylations (Fig. 5C).
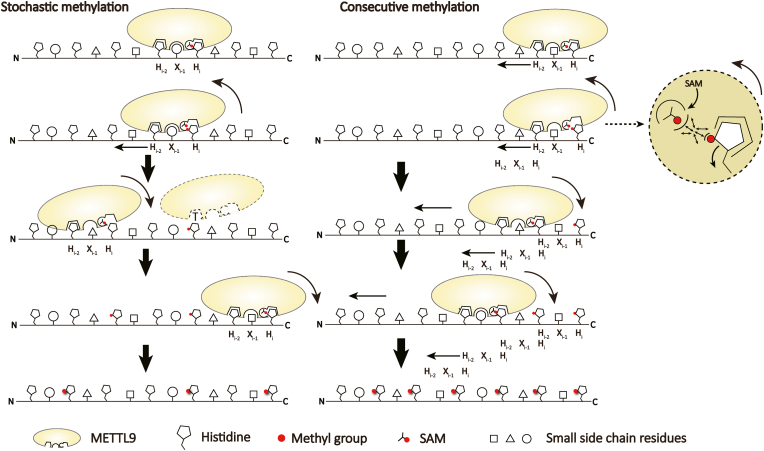
Interestingly, our LC-MS/MS based analysis revealed that the SLC39A755-70 peptide preferred consecutive methylations in “C-to-N” direction (Fig. 5D and Supplementary Table S2). On the one hand, these results reflect an inhibitory effect of methylated Hi-2 on Hi methylation to prevent sequential “N-to-C” methylation; on the other hand, although due to the stochastic site-binding nature of the METTL9 catalytic pocket which causes different methylation initiation sites, the consecutive methylation of tandem “HxH” motifs preferentially takes place in “C-to-N” direction as a result of unmodified Hi-2-guided Hi methylation, which is a unique enzymatic feature of METTL9. Conceivably, Hi methylation shall promote its release from the Hi pocket to avoid steric clash with the incoming methyl donor, which may be coupled to transposition of the adjacent Hi-2 into the Hi pocket for next cycle of methylation. The preference for directional “C-to-N” methylation along with methylated “Hi-2” inhibition of METTL9 may explain the observed low occurrence of hexa- and hepta-methylation of the SLC39A755-70 peptide. Chances are low to achieve these hypermethylation states as it requires not only an initial methylation event occurred to the very C-terminal histidine but also free of interference from unwanted Hi-2 methylation.
2.9. Substrate specificity among different histidine methyltransferases
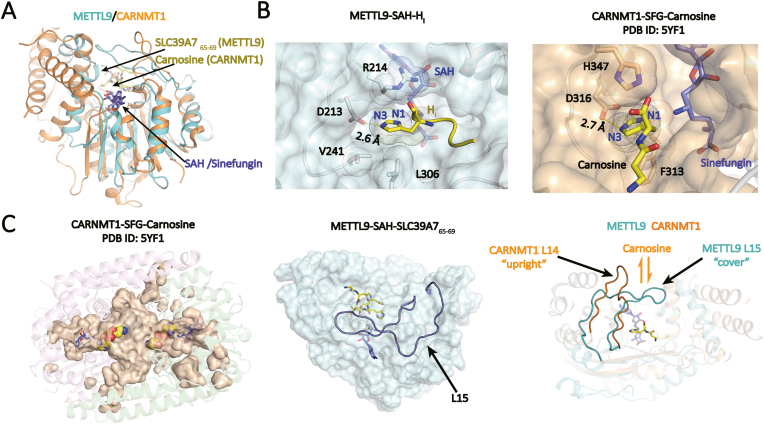
Both METTL9 and CARNMT1 are N1-specific histidine methyltransferases, with physiological substrates of the latter being carnosine or related peptides (Davydova et al., 2021; Drozak et al., 2015). We have previously reported the crystal structure of CARNMT1 bound to carnosine dipeptide (Cao et al., 2018). Structural comparison revealed that the catalytic core domains of METTL9 and CARNMT1 are superimposed with an r.m.s.dCα of 2.64 Å (Fig. 6A); they adopt a similar strategy to achieve N1-position specificity, in which the histidine imidazole ring is anchored in an Asp-lined pocket such that N3 is hydrogen-bonded to the Asp residue (D213 in METTL9 and D316 in CARNMT1) and N1 is presented to SAM for methyl transfer (Fig. 6B). Notably, hydrophobic residues from both sides of substrate histidine residue including R214, V241, and L306 of METTL9 and H347, F313 of CARNMT1 plays a role in confining the imidazole ring from flipping around to further ensure N1-position specificity (Fig. 6B). By contrast, the imidazole ring is less well confined in the case of SETD3, and flipping of the imidazole ring has been observed in structural studies (Dai et al., 2019). Whereas, the histidine N3-position specificity of SETD3 is achieved by a unique “head-to-head” methyl donor-acceptor engagement mode, in which only the N3 atom at the outmost position is available for methylation (Zheng et al., 2020).
Fig. 6.
Structural comparison between METTL9 and CARNMT1.(A) Structural alignment of METTL9 (cyan) and CARNMT1(orange). SAH or Sinefungin is depicted as blue sticks, methyl acceptor substrates are colored yellow. (B) Substrate histidine recognition and catalysis by METTL9 and CARNMT1. Key residues are shown as sticks. (C) Comparison of substrate binding pocket of METTL9 and CARNMT1. Left, CARNMT1 dimer; middle, covering of the SAH and peptide access channel by L15; right, structural alignment of METTL9 and CARNMT1 monomers, highlighting the conformational difference between L14 of CARNMT1 and L15 of METTL9. (For Interpretation of the references to colour in this figure legend the readers is referred to the web version of this article.)
CARNMT1 is a symmetrical dimer with the methyl acceptor substrate pockets confined at the center. Dimer formation blocks protein substrate binding and leads to limited dimension of the center pocket, which explains why CARNMT1 mainly accepts short peptide substrates (Fig. 6C, left). In the cases of METTL9, the interface of the peptide binding pocket is rather exposed, well positioned to allow bulky protein substrate engagement. Intriguingly, in adaption to protein substrate recognition, METTL9 has evolved with an extended L15 element that bends over the substrate channel and participates in the “HxH” motif binding (Fig. 6C, middle and right). Meanwhile, the corresponding loop in CARNMT1 takes on an upright conformation that allows carnosine access from the top through the substrate channel in the context of CARNMT1 dimer (Fig. 6C, right).
3. Discussion
Histidine methylation has aroused great interests with recent advancements in structural and functional characterizations of several histidine methyltransferases, such as CARNMT1, SETD3, METTL18, and METTL9 (Cao et al., 2018; Davydova et al., 2021; Kwiatkowski et al., 2018; Malecki et al., 2021; Wilkinson et al., 2019). Remarkably, these enzymes displayed distinct sequence motif selectivity as well as imidazole ring N1 versus N3 position-specificity, suggesting the regulatory complexity of histidine methylation biology. As an N1-specific protein histidine methylation writer, METTL9 is responsible for ∼80% of histidine N1-methylation in multiple cell lines and tissues (Davydova et al., 2021). Here, our biochemical and structural studies established the molecular basis underlying “HxH” motif and N1-specific methylation by METTL9.
Our structural studies revealed that an “HxH” motif-containing pentapeptide of SLC39A7 is snuggly embedded within the substrate pocket of METTL9, with the latter histidine being the site for methylation and the first histidine being a recognition signature. Unlike SETD3 that involves ∼15 residues for substrate recognition (Dai et al., 2019; Guo et al., 2019; Kwiatkowski et al., 2018; Wilkinson et al., 2019; Zheng et al., 2020), the requirement of ∼5 residues (i-3 to i+1) for engagement explains the observed broader substrate spectrum of METTL9 given the high incidence of short peptide motifs in proteome. Meanwhile, intimate encapsulation of the pentapeptide contributes to micromolar substrate binding by METTL9 for efficient catalysis (Fig. 3B). Remarkably, our mutagenesis studies revealed that many single-residue perturbations within the substrate pocket could completely abolish peptide binding or enzymatic activity of METTL9, even if they did not participate in binding directly (Fig. 3, Fig. 4). These results together with the observed induced fit suggest that substrate engagement by METTL9 is a highly coordinated process that requires an intact native pocket.
Hundreds of histidine methylation sites have been experimentally identified (Kapell & jakobsson, 2021; Ning et al., 2016), and a PROSITE analysis of human proteome revealed about 2237 “HxH” motifs that can be potentially methylated by METTL9 (Sigrist et al., 2013). Using tandem “HxH” repeats motif derived from SLC39A7 as a model substrate, we revealed preferred consecutive methylation events catalyzed by METTL9, notably in “C-to-N” direction (Fig. 7). Depending on its cognate substrate, METTL9-mediated histidine methylation may regulate many cellular functions, including zinc transport (Lv et al., 2021), ER stress (Lv et al., 2021), and innate immunity (Daitoku et al., 2021). Functionally, the consecutive and directional catalysis feature of METTL9 may help to determine its substrate preference and regulate corresponding activities (Fig. 5A and C), such as zinc sensing in cells.
Fig. 7.
Schematic model of tandem “HxH” methylation by METTL9. Methylation of tandem “HxH” repeats by METTL9 can occur in two modes. In the stochastic mode, METTL9 recognizes the “HxH” motif and methylate the Hi residue in a random manner since methylated Hi impairs successive Hi+2 methylation, which leads to products with scattered methylation pattern. In the consecutive mode, recognition of an initial “HxH” motif and subsequent Hi methylation shall promote sequential “C-to-N” methylation, as release of methylated Hi product can be well coupled with relocation of the adjacent Hi-2 residue into the Hi pocket for next round of methylation. Our enzymatic assays suggested that consecutive “C-to-N” methylation exists as the major mode as indicated by big red arrows.
In sum, our structural and biochemical dissections of METTL9 highlight the exquisite molecular design of a histidine methyltransferase in substrate recognition and catalysis. These findings pave the way for future investigations into the functional importance of METTL9 in health and disease. Given the regulatory complexity of histidine methylation and its writers, it remains an intriguing question to characterize corresponding readers or erasers, if they exist, in the years to come.
4. Methods
4.1. Materials
All SLC39A7 peptides (>95% purity) were synthesized at SciLight Biotechnology (Supplementary Table S2). The full-length METTL9 (1–318) and truncated METTL9 (37–318) were cloned into pGood6p vector (modified based on pGEX-6P-1 vector) using Seamless Cloning Kit (Beyotime). All point mutations were generated using the QuikChange (Stratagene) site-directed mutagenesis strategy.
4.2. Protein purification
The full-length (1–318) and the truncated (37–318) METTL9 were expressed in Escherichia coli strain BL21(DE3) and induced overnight by 0.2 mM isopropyl β-D-thiogalactoside (IPTG) at 16 °C in LB medium. The harvested cells were resuspended in 20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 5% glycerol, 2 mM dithiothreitol (DTT), 1% Triton-X100, 1 mM phenylmethylsulfonyl fluoride (PMSF). The resuspended cells were then lysed using UH12 high-pressure homogenizer (Union-Biotech). After centrifugation, the cleared supernatant was loaded onto GST column followed by washing with high-salt buffer containing 20 mM Tris-HCl, pH 7.5, 1 M NaCl, and low-salt buffer containing 20 mM Tris-HCl, pH 7.5, 50 mM NaCl, and 0.1% DDM. Both full-length and truncated METTL9 samples were eluted by an on-column cleavage strategy using home-made GST-P3C protease. The tag-free METTL9 samples were enriched and purified by a HiTrap Q anion-exchange column, and further polished by a Superdex 75 10/300 gel filtration column using an AKTA Purifier 10 systems (GE Healthcare). All purified proteins were aliquoted and stored in buffer containing 20 mM Tris-HCl, pH 7.5, and 50 mM NaCl at ∼10 mg/ml in −80 °C freezer.
4.3. Isothermal titration calorimetry (ITC)
For ITC measurements, synthetic SLC39A7 peptides, SAH ligand, and recombinant proteins were prepared under the same titration buffer containing 50 mM NaCl, 20 mM Tris-HCl, pH 7.5. Protein concentration was measured based on UV 280 nm absorption. Peptide concentrations were determined by weighing in large quantities. After weighing and quantification, each peptide was dissolved in Milli-Q water, aliquoted and freeze-dried for future use. ITC experiments were performed at 15 °C using a MicroCal PEAQ-ITC instrument (Malvern Panalytical). For the SAH-METTL9 (wild type or mutants) titration experiments, 0.5 mM SAH was titrated into 0.05 mM METTL9. For the SLC39A7 peptides-METTL9 (wild type or mutants) titration experiments, 0.5 mM SLC39A7 peptides were titrated into 0.05 mM METTL9 preincubated with 2 mM SAH. Each ITC titration consisted of 17 successive injections with 0.4 μL for the first and 2.4 μL for the rest. The resultant ITC curves were processed using the Origin 7.0 software (OriginLab) according to the “One Set of Sites” fitting model.
4.4. In vitro methyltransferase assay
Methyltransferase assays were carried out in 20 μL volume containing 2 μM purified wild-type or mutant METTL9 (1–318), 100 μM SLC39A7 peptides, 1 mM SAM, 50 mM Tris-HCl, pH 7.5, and 50 mM NaCl. Reactions were performed at 37 °C for 1 h and terminated by adding 0.1% trifluoroacetic acid (TFA) and then subjected to LC-MS detection. For enzyme kinetic analysis, unmethylated substrate at various concentration were incubated with 1 μM of METTL9. Products were analyzed by LC-MS. Less than 20% of the substrate was consumed in all reactions. The kinetic parameters, Km and kcat were calculated by the GraphPad software according to the Michaelis–Menten equation.
4.5. LC-MS/MS detection
For LC-MS analysis, the analytes were separated by a 5 min gradient elution at a flow rate of 0.5 ml/min with an ACQUITY UPLC system, which was directly interfaced with an SYNAPT-G2-Si mass spectrometer produced by Waters company. The analytical column was a Protein BEH C4 silica capillary column (2.1 mm ID, 50 mm length; Made in Ireland) packed with C-4 resin (300 Å, 1.7 μm) purchased from Waters company. Mobile phase A consisted of 0.1% formic acid aqueous solution, and mobile phase B consisted of 100% acetonitrile and 0.1% formic acid.
Aliquots of 10 μL analytes were loaded into an autosampler for electrospray ionization. Samples were analyzed on a Q-TOF mass spectrometer (SYNAPT G2-Si, Waters company) instrument optimized for high-mass protein analysis. The measurements were performed with capillary 3000–3500 V and data were collected over the m/z range of 350–1350. Once having acquired raw electrospray mass spectra, it was extracted and integrated by the MassHunter Qualitative Analysis software (Agilent), and exported for further analysis.
The nanoLC−MS/MS experiments were performed on UltiMate 3000RSLCnano systems (Thermo Scientific, USA) coupled to a Q-Exactive HF mass spectrometer (Thermo Scientific, USA). The Q-Exactive HF mass spectrometer was operated in data-dependent acquisition mode. The 15 most intense multiply charged ions were isolated and fragmented by higher-energy collisional dissociation. MaxQuant (version 1.5.2.8) was used to analyze the raw files. Targeted peptide (HHGHSHAHGHGHTHES) was included in a compiled fasta file with Uniprot human database (downloaded on 20210315) as background. In the database search, N-terminal acetylation, C-terminal NH3 modification as well as methyl (H) was set as variable modifications. The FDR at the PSM, protein and site level were both set to be below 1%. Other paraments were default in MaxQuant.
4.6. Crystallization, data collection and structural determination
Crystallization was performed via the sitting or hanging-drop vapor diffusion method under 16 °C by mixing equal volumes (0.2–1.0 μL) of the protein sample and the reservoir solution. For METTL937-318-SAH crystals, the protein was pre-mixed with 2 mM SAH at a concentration of 8 mg/ml and incubated at 4 °C for 1 h. METTL9-SAH crystals were eventually grown in 100 mM Tris-HCl, pH 8.0, 4% v/v glycerol, 1.8 M NaCl. For METTL937-318-SAH-SLC39A7 peptide crystals, the protein was mixed with SAH, peptide at a molar ratio of 1:5:5 at a concentration of 8 mg/ml and incubated at 4 °C for 1 h. METTL9-SAH-SLC39A7 peptide crystals were eventually grown in 100 mM Hepes-Na, pH 7.0, 20% w/v PEG 4000, 500 mM KCl.
For data collection, crystals were briefly soaked in a cryo-protectant composed of reservoir solution supplemented with 20% glycerol, and then flash-frozen in liquid nitrogen for data collection at 100 K. The data of METTL937-318-SAH was collected at a wavelength of 0.9792 Å at Shanghai Synchrotron Radiation Facility beamline BL02U1, and the data of METTL937-318-SAH-SLC39A7 peptide complex was collected at a wavelength of 0.9792 Å at Shanghai Synchrotron Radiation Facility beamline BL18U2. Data were indexed, integrated, and merged using the HKL2000 software package (Otwinowski & minor, 1997).
The phase problem of METTL937-318-SAH and METTL937-318-SAH-peptide crystals were solved by molecular replacement with the MOLREP software (Vagin & teplyakov, 2010) using the AlphaFold-predicted structure as the search model (Heo & feig, 2020; Varadi et al., 2022). The structures were refined using PHENIX with iterative manual model building using COOT (Liebschner et al., 2019). Detailed structural collection and refinement statistics are shown in Supplementary Table S1.
Data access
The atomic coordinates and structure factors for human METTL9-SAH and human METTL9-SAH-SLC39A7(65–70) have been deposited into Protein Data Bank under accession codes 8GZF and 8GZE, respectively. The Protein Data bank accession number of CARNMT1-SFG-Carnosine is 5YF1.
Funding
This research was supported by grants from the National Natural Science Foundation of China (92153302 to H.L. and M.Y.) and National Key R&D Program of China (2020YFA0803303 and 2021YFA1300103 to H.L.).
Author contributions
H. Li conceived the study. W. Zhao performed structural and biochemical studies. Y. Zhou performed biochemical experiments. K. Wang and M. Ye performed LC-MS analysis. C. Li and Y. Bi assisted with research. H. Li and W. Zhao wrote the manuscript.
Declaration of competing interest
The authors declare no competing interests.
Materials & correspondence
Correspondence and requests for materials should be addressed to Haitao Li.
Acknowledgement
We thank Y. Ou and other members in the Li laboratory for scientific input. We thank the staff at beamline BL18U1 and BL02U1 of the Shanghai Synchrotron Radiation Facility and Dr. S. Fan at Tsinghua Center for Structural Biology for assistance in data collection. We also thank Dr. J. Li and D. Yan at Protein Chemistry and proteomics facility, the Center of Protein Research and Technology in Tsinghua University for their help in mass spectrum.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.cellin.2023.100090.
Appendix ASupplementary data
The following is the supplementary data related to this article:
References
- Al-hadid q., roy k., munroe w., dzialo m.C., chanfreau g.F., clarke s.G. Histidine methylation of yeast ribosomal protein rpl3p is required for proper 60s subunit assembly. Molecular and Cellular Biology. 2014;34:2903–2916. doi: 10.1128/MCB.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldyrev a.A., aldini g., derave w. Physiology and pathophysiology of carnosine. Physiological Reviews. 2013;93:1803–1845. doi: 10.1152/physrev.00039.2012. [DOI] [PubMed] [Google Scholar]
- Cao r., zhang x., liu x., li y., li h. Molecular basis for histidine n1 position-specific methylation by carnmt1. Cell Research. 2018;28:494–496. doi: 10.1038/s41422-018-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll j., fearnley i.M., skehel j.M., runswick m.J., shannon r.J., hirst j., walker j.E. The post-translational modifications of the nuclear encoded subunits of complex i from bovine heart mitochondria. Molecular and Cellular Proteomics. 2005;4:693–699. doi: 10.1074/mcp.M500014-MCP200. [DOI] [PubMed] [Google Scholar]
- Dai s., horton j.R., woodcock c.B., wilkinson a.W., zhang x., gozani o., cheng x. Structural basis for the target specificity of actin histidine methyltransferase setd3. Nature Communications. 2019;10:3541. doi: 10.1038/s41467-019-11554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitoku h., someya m., kako k., hayashi t., tajima t., haruki h., sekiguchi n., uetake t., akimoto y., fukamizu a. Sirna screening identifies mettl9 as a histidine npi-methyltransferase that targets the proinflammatory protein s100a9. Journal of Biological Chemistry. 2021;297 doi: 10.1016/j.jbc.2021.101230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydova e., shimazu t., schuhmacher m.K., jakobsson m.E., willemen h., liu t., moen a., ho a.Y.Y., malecki j., schroer l., pinto r., suzuki t., gronsberg i.A., sohtome y., akakabe m., weirich s., kikuchi m., olsen j.V., dohmae n.…falnes p.O. The methyltransferase mettl9 mediates pervasive 1-methylhistidine modification in mammalian proteomes. Nature Communications. 2021;12:891. doi: 10.1038/s41467-020-20670-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozak j., piecuch m., poleszak o., kozlowski p., chrobok l., baelde h.J., de heer e. Upf0586 protein c9orf41 homolog is anserine-producing methyltransferase. Journal of Biological Chemistry. 2015;290:17190–17205. doi: 10.1074/jbc.M115.640037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer e.L., shi y. Histone methylation: A dynamic mark in health, disease and inheritance. Nature Reviews Genetics. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo h.B., guo h. Mechanism of histone methylation catalyzed by protein lysine methyltransferase set7/9 and origin of product specificity. Proceedings of the National Academy of Sciences of the U S A. 2007;104:8797–8802. doi: 10.1073/pnas.0702981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo q., liao s., kwiatkowski s., tomaka w., yu h., wu g., tu x., min j., drozak j., xu c. Structural insights into setd3-mediated histidine methylation on beta-actin. Elife. 2019;8 doi: 10.7554/eLife.43676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo l., feig m. High-accuracy protein structures by combining machine-learning with physics-based refinement. Proteins. 2020;88:637–642. doi: 10.1002/prot.25847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huszar g., elzinga m. Homologous methylated and nonmethylated histidine peptides in skeletal and cardiac myosins. Journal of Biological Chemistry. 1972;247:745–753. [PubMed] [Google Scholar]
- Jakobsson m.E. Enzymology and significance of protein histidine methylation. Journal of Biological Chemistry. 2021;297 doi: 10.1016/j.jbc.2021.101130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson p., harris c.I., perry s.V. 3-methylhistidine in actin and other muscle proteins. Biochemical Journal. 1967;103:79p. [PMC free article] [PubMed] [Google Scholar]
- Kapell s., jakobsson m.E. Large-scale identification of protein histidine methylation in human cells. Nar genom bioinform. 2021;3 doi: 10.1093/nargab/lqab045. lqab045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski s., drozak j. Protein histidine methylation. Current Protein and Peptide Science. 2020;21:675–689. doi: 10.2174/1389203721666200318161330. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski s., seliga a.K., vertommen d., terreri m., ishikawa t., grabowska i., tiebe m., teleman a.A., jagielski a.K., veiga-da-cunha m., drozak j. Setd3 protein is the actin-specific histidine n-methyltransferase. Elife. 2018;7 doi: 10.7554/eLife.37921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner d., afonine p.V., baker m.L., bunkoczi g., chen v.B., croll t.I., hintze b., hung l.W., jain s., mccoy a.J., moriarty n.W., oeffner r.D., poon b.K., prisant m.G., read r.J., richardson j.S., richardson d.C., sammito m.D., sobolev o.V.…adams p.D. Macromolecular structure determination using x-rays, neutrons and electrons: Recent developments in phenix. Acta crystallography d structure biology. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv m., cao d., zhang l., hu c., li s., zhang p., zhu l., yi x., li c., yang a., yang z., zhu y., zhang k., pan w. Mettl9 mediated n1-histidine methylation of zinc transporters is required for tumor growth. Protein cell. 2021;12:965–970. doi: 10.1007/s13238-021-00857-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki j.M., odonohue m.F., kim y., jakobsson m.E., gessa l., pinto r., wu j., davydova e., moen a., olsen j.V., thiede b., gleizes p.E., leidel s.A., falnes p.O. Human mettl18 is a histidine-specific methyltransferase that targets rpl3 and affects ribosome biogenesis and function. Nucleic Acids Research. 2021;49:3185–3203. doi: 10.1093/nar/gkab088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin j.L., mcmillan f.M. Sam (dependent) i am: The s-adenosylmethionine-dependent methyltransferase fold. Current Opinion in Structural Biology. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- Meyer h.E., mayr g.W. N pi-methylhistidine in myosin-light-chain kinase. Biological Chemistry Hoppe-Seyler. 1987;368:1607–1611. doi: 10.1515/bchm3.1987.368.2.1607. [DOI] [PubMed] [Google Scholar]
- Ning z., star a.T., mierzwa a., lanouette s., mayne j., couture j.F., figeys d. A charge-suppressing strategy for probing protein methylation. Chemical Communications. 2016;52:5474–5477. doi: 10.1039/c6cc00814c. [DOI] [PubMed] [Google Scholar]
- Otwinowski z., minor w. Processing of x-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Raftery m.J., harrison c.A., alewood p., jones a., geczy c.L. Isolation of the murine s100 protein mrp14 (14 kda migration-inhibitory-factor-related protein) from activated spleen cells: Characterization of post-translational modifications and zinc binding. Biochemical Journal. 1996;316(pt 1):285–293. doi: 10.1042/bj3160285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist c.J., de castro e., cerutti l., cuche b.A., hulo n., bridge a., bougueleret l., xenarios i. New and continuing developments at prosite. Nucleic Acids Research. 2013;41:d344–d347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin a., teplyakov a. Molecular replacement with molrep. Acta crystallogr d biol crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- Varadi m., anyango s., deshpande m., nair s., natassia c., yordanova g., yuan d., stroe o., wood g., laydon a., zidek a., green t., tunyasuvunakool k., petersen s., jumper j., clancy e., green r., vora a., lutfi m.…velankar s. Alphafold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Research. 2022;50:d439–d444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb k.J., zurita-lopez c.I., al-hadid q., laganowsky a., young b.D., lipson r.S., souda p., faull k.F., whitelegge j.P., clarke s.G. A novel 3-methylhistidine modification of yeast ribosomal protein rpl3 is dependent upon the yil110w methyltransferase. Journal of Biological Chemistry. 2010;285:37598–37606. doi: 10.1074/jbc.M110.170787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson a.W., diep j., dai s., liu s., ooi y.S., song d., li t.M., horton j.R., zhang x., liu c., trivedi d.V., ruppel k.M., vilches-moure j.G., casey k.M., mak j., cowan t., elias j.E., nagamine c.M., spudich j.A.…gozani o. Setd3 is an actin histidine methyltransferase that prevents primary dystocia. Nature. 2019;565:372–376. doi: 10.1038/s41586-018-0821-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang x., bruice t.C. Mechanism of product specificity of adomet methylation catalyzed by lysine methyltransferases: Transcriptional factor p53 methylation by histone lysine methyltransferase set7/9. Biochemistry. 2008;47:2743–2748. doi: 10.1021/bi702370p. [DOI] [PubMed] [Google Scholar]
- Zhang x., bruice t.C. Product specificity and mechanism of protein lysine methyltransferases: Insights from the histone lysine methyltransferase set8. Biochemistry. 2008;47:6671–6677. doi: 10.1021/bi800244s. [DOI] [PubMed] [Google Scholar]
- Zheng y., zhang x., li h. Molecular basis for histidine n3-specific methylation of actin h73 by setd3. Cell discover. 2020;6:3. doi: 10.1038/s41421-019-0135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.