Abstract
Cysteine dioxygenase type 1 (CDO1), belonging to the mammalian non-heme Fe(II) dioxygenases family, is a key enzyme for cysteine catabolism. Its activity and expression is regulated through multiple mechanisms. CDO1 is involved in a spectrum of physiological processes including lipid metabolism, adipogenesis, osteoblastic differentiation, redox homeostasis, fertility, bile acid metabolism, sulfide metabolism, and organismal growth and development. Many of these processes are regulated directly or indirectly by CDO1-mediated metabolism of cysteine. In pathophysiological processes, the degree of CDO1 promoter methylation is closely related to the progression and malignancy of tumors, and overexpression of CDO1 will promote ferroptosis of cancer cells. Moreover, CDO1 may ameliorate metabolic disorders through the taurine-mediated improvement of lipid metabolism and insulin sensitivity and improve neurodegenerative diseases by regulating cysteine level. Therefore, elucidation of the mechanisms underlying the role of CDO1 would provide a clearer view of the therapeutic potential and possible risks of targeting this important enzyme.
Keywords: Anti-tumor, Antioxidation, Cysteine, Promoter methylation, Taurine
Abbreviations
- CcO
Cytochrome c oxidase
- CDO1
Cysteine dioxygenase type 1
- C/EBPα
CCAAT/enhancer-binding protein
- C/EBP
CCAAT/enhancer-binding protein α
- CSA
Cysteine sulfinic acid
- CSAD
CSA decarboxylase
- CSD
Cysteine sulfinate decarboxylase
- CYS-SO32-
Cysteine-S-sulfate
- Fe-S
Iron-sulfur clusters
- GOT1
Cytosolic aspartate aminotransferase
- BMSC
Bone marrow-derived mesenchymal stem cells
- H-bond
Hydrogen bond
- HDH
Hypotaurine dehydrogenase
- PGC-1α
Peroxisome proliferator activated receptor γ coactivator 1-α
- PPARγ
Peroxisome proliferator-activated receptor γ
- SUOX
Sulfite oxidase
- UCP1
Uncoupling Protein 1
- 5′-UTR
5′-untranslated region.
Introduction
CDO1, a mammalian non-heme Fe(II) dioxygenase, belongs to the cupin superfamily. In mammals, CDO1 is expressed mainly in adipose tissue, liver, brain, small intestine, lung, and kidney.1,2 In recent years, CDO1 has been widely studied as a physiological and pathophysiological regulator of the organisms. In physiological conditions, CDO1 can catalyze l-cysteine to produce cysteine sulfinic acid (CSA).1 CSA is further decomposed into taurine and sulfate, which alleviates cysteine accumulation. The accumulation of cysteine in the cells will lead to neurotoxicity and cytotoxicity,3 which will cause motor neuron disease (MND), Parkinson's disease (PD) and Alzheimer's disease (AD).4 Therefore, insufficiency or dysfunction of CDO1 is associated with an increased risk of cysteine accumulation and related diseases. Recently, there has been growing recognition of the important links between CDO1 and lipid metabolism and adipogenesis, redox homeostasis, and osteogenesis. Although mounting evidence suggests that CDO1 is an important regulator in the physiological processes of organisms, the physiological role of CDO1 and its underlying mechanism are not fully understood and need to be further explored. In addition, its enzyme activity has important medical implications. Since it was discovered, the pathophysiological role of CDO1 has been investigated in various disease models in both humans and animals. CDO1 has been extensively studied in cancer, metabolic disorders and neurodegenerative diseases.4,5 As a consequence, the role of CDO1 in disease models has been verified and its potential applications have been explored in a therapeutic setting. However, knowledge of the mechanisms underlying the pathophysiological role of CDO1 is still limited. This review aims at providing an overview of the properties of CDO1 and its functional role in physiological and pathophysiological processes, which could be instructive to further understanding the biological feature of CDO1 and its application in the prevention or treatment of diseases.
Molecular feature of CDO1
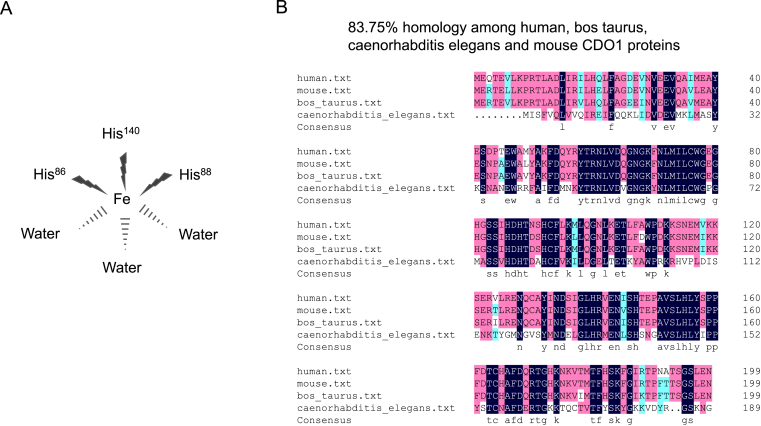
CDO1 is a mammalian non-heme Fe(II) dioxygenase that has several unique characteristics. Normally, the active site of the non-heme Fe enzyme is composed of iron, two histidine, and a carboxylate side chain (2-His-1-carboxylate facial triad). Most of these enzyme catalyze the O2 reaction by using the Fe(II) to activate O2 or using the substrate to activate O2 through a redox process.6 However, in the case of CDO1, its active site does not adopt the above structure and catalytic mode. Firstly, in CDO1, the carboxylate side chain in the conventional triad facial structure is replaced by histidine and forms a 3-His structure, which has a great influence on the electronic structure of the cysteine-bound Fe(II) CDO1 active site, thus affecting the catalysis of oxygen.7 Secondly, the structure of the active center of CDO1, which is the mononuclear iron center, is coordinated with the Nε2 atoms of three histidine ligands (His86, His88, and His140) and water molecules bound to the catalytic iron (Fig. 1A).8,9 Other highly conserved residues directly lining the active site include Tyr58, Arg60, Ser153, and His155, and they can interact with each other to assist the correct positioning of cysteine in the process of enzyme–substrate (ES) complex formation.8 For example, Tyr58 and Arg60 can form hydrogen bond (H-bond) to water in the active site and thus the carboxylate group of the substrate is also recognized. In addition, as shown in a previous study,8 Ser153-Oγ H-bonds to His155-Nε2, and His155-Nδ2 H-bonds to Tyr157-OH, which will form a triad of Ser153·His155·Tyr157. In particular, CDO1 belongs to the typical β-barrel fold of the cupin superfamily, which contains two short but partially conserved sequence motifs—GX5HXHX3–6EX6G and GX5–7PXGX2HX3N,10 and the rare cysteinyl-tyrosine intramolecular cross-link (between Cys93 and Tyr157), but no other member of the cupin dioxygenase family contains the Cys-Tyr cofactor.9,11
Figure 1.
The structure of CDO1 enzyme active center and sequence alignment of CDO1 proteins among species. (A) The enzyme active center structure of CDO1 protein. The mononuclear iron center is coordinated with the Nε2 atoms of three histidine ligands (His,86 His,88 and His140) and water molecules bound to the catalytic iron. (B) Comparison of CDO1 protein sequences of human, bos taurus, caenorhabditis elegans and mouse. Among the 4 sequences, 2 identical residues are represented in blue; 3 identical residues are represented in red; 4 identical residues are represented in black.
Mouse CDO1 gene is located on chromosome 18-NC_000084.7, with a full-length of 15 Kb.1 The full-length of mouse CDO1 consists of five exons and four introns, with all of the intron/exon splice junctions conforming to the AG/TC rule. The splice junction and all intron sizes of mice CDO1 gene are identical to rat CDO1 gene and are 93% similar to the human CDO1 gene.1 Exons 2, 3, and 4 are identical in size among human, mouse and rat species, while exon 1 is slightly larger in the human CDO1 gene.12 Exon 1 contains the 5′-untranslated region (5′-UTR) and the translational start codon. The 5′-UTR of mouse and rat CDO1 contains 213 bp, but the human CDO1 gene has a longer 5′-UTR of 260 bp.13 Furthermore, the nucleotide sequence of the 5′-UTR of CDO1, 120 bp upstream from the ATG start codon, is G + C rich (76%) and contains the TATA-box-like sequence, GC boxes, GRE and CRE-like sequences,1,14 which play an important role in controlling the CDO1 transcription and expression. Furthermore, a variety of consensus cis-acting elements were also identified in the 5′-UTR, including the ones for the hepatic nuclear factor 3β (HNF-3β), HNF-3/forkhead homolog (HFH-1, HFH-2, and HFH-3), and the CCAAT/enhancer-binding protein β (C/EBPβ), all of which are consistent with the tissue-specific expression of CDO1.12 Like the CDO1 gene, the homology among human, bos taurus, caenorhabditis elegans and mouse CDO1 proteins was as high as 83.75% by comparison of protein sequence (Fig. 1B). The high homology of CDO1 protein among species suggests a critical role of CDO1 in biological processes. Interestingly, the protein of rat CDO1 was analyzed by SDS-PAGE and it was found that the protein of CDO1 had a bimolecular band with the molecular mass of 23 kDa and 22.5 kDa and a pI value of 5.5, and the band with a lower molecular mass had higher enzyme activity.15,16
The regulatory mechanism of CDO1 activity and expression in physiological and pathophysiological conditions
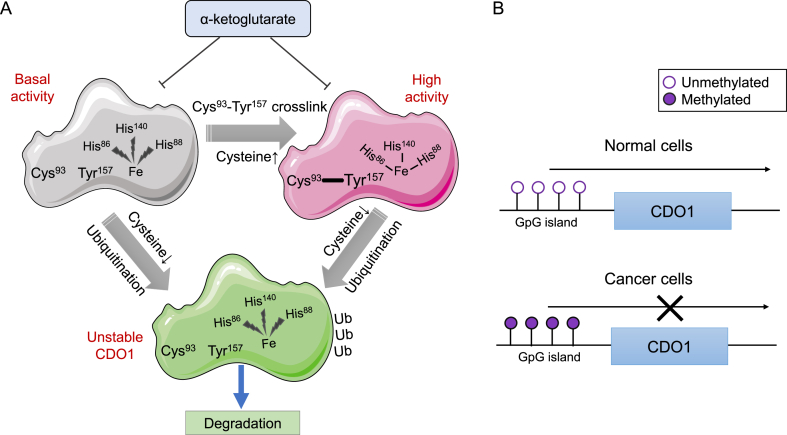
CDO1 has two different levels of activity, and its higher activity is because of the existence of Cys-Tyr cofactor.11 The activity of CDO1 catalytic efficiency was increased by ∼10 fold and its catalytic life time was prolonged when the structure of Cys-Tyr cofactor is formed.11,17 The Cys-Tyr cofactor formation depends on the surrounding cysteine level,11 and once formed, it will substantially increase both the catalytic efficiency and stability of CDO1 (Fig. 2A), thus catabolizing cysteine better.15 In addition, in terms of the catalytic mechanism, Driggers, CM et al18 found that the Cys-Tyr crosslink was important for promoting the Fe-N(Cys) coordination. The reason is that Tyr-OH of the Cys-Tyr crosslink can affect iron coordination, thus promoting the binding of cysteine to iron and improving the catalytic efficiency of CDO1. In addition, the Cys-Tyr cofactor can enhance the binding force of CDO1 active center (iron and 3-His) as shown in Figure 2A.11 However, the latest research shows that the Cys-Tyr cofactor is not essential for the oxidation activity of CDO1, and the formation of cofactors is only a concomitant reaction but not a prerequisite.19 Wang YJ et al19 explained that the process of co-factor formation includes H-abstraction, C–S bond formation, intramolecular migration, etc, and the overall barriers of cofactor formation in CDO1 are always higher than CDO1 oxidation of cysteine. In general, CDO1 can well catalyze the decomposition of cysteine. When the concentration of cysteine is high, the Cys-Tyr cofactor will be formed, and then regulate the concentration of cysteine. Although the formation of co-factors is not necessary for the basic activity of CDO1, it is formed by substrate regulation and may be regarded as a special feed-forward activation of CDO1 enzyme activity (Fig. 2A). In addition, CDO1 is the first known dioxygenase inhibited by α-ketoglutarate (Fig. 2A).11 It may be because the side chain of α-ketoglutarate carboxylate is negatively charged, which forms a competitive inhibition with the thiolate group of cysteine, thus blocking the binding of the thiolate group of cysteine to the active site of CDO1. Together these studies provide important insights into the reciprocal regulation between CDO1 enzyme activity and cysteine.
Figure 2.
The expression and activity of CDO1 is regulated by multiple mechanisms. (A) CDO1 has two different levels of enzyme activities (basal and high), and its higher enzyme activity is because of the existence of Cys-Tyr cofactor. The Cys-Tyr cofactor formation depends on the surrounding cysteine levels. And once formed, it will substantially increase both the catalytic efficiency and stability of CDO1, thus catabolizing cysteine better. In addition, the Cys-Tyr cofactor can enhance the binding force of CDO1 active center (iron and 3-His). The α-ketoglutarate can function as an inhibitor of CDO1 by blocking the binding of the thiolate group of cysteine to the active site of CDO1. Moreover, the ubiquitin-26S system can mediate rapid ubiquitination and degradation of CDO1 under conditions of low cysteine availability. (B) The expression of CDO1 was tightly controlled by promoter methylation. In cancerous cells, aberrant hypermethylation on the CDO1 promoter is a common event, which will lead to transcriptional inactivation and silencing of CDO1 gene. As a tumor suppressor gene, the decreased expression of CDO1 could promote the occurrence and development of cancers.
Based on the above ideas, CDO1 can catalyze cysteine oxidation. Meanwhile, cysteine may also have a unique ability to regulate the expression level of CDO1. Martha HS et al20 found that the amount of cysteine significantly affects the half-life of CDO1 protein in rat primary hepatocyte cells. In that study, the ubiquitin-26S system can mediate rapid ubiquitination and degradation of CDO1 under conditions of low cysteine availability. In addition, when the concentration of cysteine is high, the ubiquitination of CDO1 is blocked. The ubiquitination and proteasome-mediated degradation of CDO1 was confirmed by in vivo experiments.21 In that study, the CDO1 protein level can be promoted by pharmacological inhibition of the 26S proteasome complex in rat liver and kidney. Thus, cysteine, as the substrate of CDO1, can hinder the ubiquitination of CDO1, preventing it from proteasome degradation.
In epigenetics, DNA methylation is closely related to the occurrence and development of cancer. The methylation of genomic DNA can regulate gene expression, contribute to the development and progression of tumor cells and abnormal methylation of genomic DNA is considered to be a hallmark in the early stage of cancer.22 There are CG-rich regions known as CpG Island that can be located within the 5′-UTR regions including promoter, untranslated region and exon 1.22 CpG islands are usually not methylated in normal cells,22 but aberrant hypermethylation leads to transcriptional inactivation and gene silencing which are common events in the carcinogenesis process.23,24 CDO1 is a gene with aberrant promoter methylation in many human cancers and it functions as a tumor suppressor gene.25 The expression of CDO1 was tightly controlled by promoter methylation, suggesting that CDO1 promoter methylation and silencing of CDO1 may be a common event in human carcinogenesis.5 Thus, the promoter methylation-mediated silencing or abnormal expression of CDO1 could contribute to the occurrence and development of diseases, such as cancers (Fig. 2B).
Therefore, the activity and expression level of CDO1 is regulated through multiple mechanisms. Understanding the above regulatory mechanisms may help to better control the functional role of CDO1 in various physiological and pathophysiological processes.
Physiological roles of CDO1
Amino acid metabolism
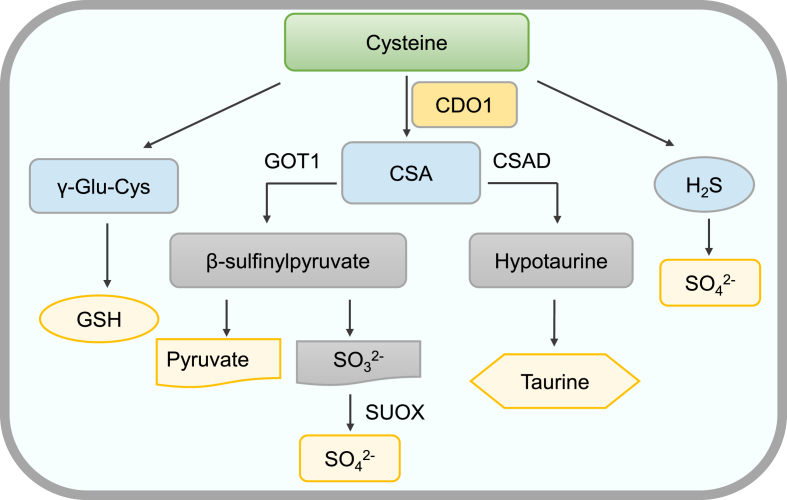
The metabolism of amino acids and related biomolecules are often dependent on mononuclear non-heme iron enzymes that activate O2 for the oxidation of organic substrates.26 CDO1 is a mononuclear non-heme ferritin enzyme that mainly regulates the homeostasis of cysteine in response to changes in sulfur amino acid levels (Fig. 3). CDO1 oxidizes cysteine to CSA, which is further metabolized to taurine or pyruvate plus sulfate.27 The oxidative pathway leads to the production of taurine and sulfate in a ratio of approximately 2:1.28 In mammals, cysteine catabolism depends on CDO1 which oxidizes cysteine to cysteine sulfinate. In biomimetic studies, Fe(III) superoxide species is considered as an efficient oxidant of mononuclear nonheme Fe(II) complexes, which acts as an oxidant in the activation of C−H bond. And it can attack the sulfur atom of the cysteine ligand by the terminal oxygen atom of the superoxide group, which is followed by the formation of a sulfinic acid product.29 This is also the reason why CDO1, as a non-heme Fe(II) dioxygenases enzyme, can transfer two oxygen atoms of oxygen to the sulfur of cysteine, forming CSA. CSA is further metabolized by two pathways.27 In one pathway, CSA is converted to hypotaurine by cysteine sulfinate decarboxylase (CSD); hypotaurine is eventually oxidated to taurine by hypotaurine dehydrogenase (HDH).30 In the other pathway, CSA is transaminated by the cytosolic aspartate aminotransferase (GOT1) to produce β-sulfinylpyruvate, which spontaneously dissociates to give rise to pyruvate and sulfite. Sulfite is also oxidized to sulfate (SO42−) by sulfite oxidase (SUOX).28
Figure 3.
CDO1 catalyzes the metabolic pathway of cysteine. CDO1 can transfer two oxygen atoms of oxygen to the sulfur of cysteine, forming CSA. CSA is converted to hypotaurine by CSAD; hypotaurine is eventually oxidated to taurine by HDH. In addition, CSA is transaminated by GOT1 to produce β-sulfinylpyruvate, which spontaneously dissociates to give rise to pyruvate and sulfite. Sulfite is further oxidized to by SUOX. On the other hand, CDO1 reduces the synthesis of GSH and H2S by promoting cysteine metabolism. CSA, cysteine sulfinic acid CSAD, cysteine sulfinate decarboxylase; HDH, hypotaurine dehydrogenase; GOT1, cytosolic aspartate aminotransferase; , sulfate; SUOX, sulfite oxidase.
Taurine has various biological functions and is involved in the processes of bile acid conjugation, modification of mitochondrial tRNAs, nervous development, antioxidation and immune defense.31 In addition, abnormally elevated cysteine can cause oxidative stress and cytotoxicity in certain cell types.4 CDO1 is involved in synthesizing taurine and regulates the level of cysteine. In order to gain insight into the effects of CDO1 on cysteine and taurine metabolism and the consequence of a loss of CDO1 activity. Ueki I et al27 found that the knockout of CDO1 (CDO1−/−) in mice led to extremely low taurine levels in liver and plasma and somewhat elevated cysteine levels, which was consistent with the CDO1-dependent cysteine catabolic pathways. Besides, the CDO1−/− mice exhibited connective tissue pathology, growth deficit and postnatal mortality. Moreover, supplementation of mice with taurine improved survival of male pups but had little effect on the phenotype of CDO1−/− mice.27 Likewise, this study showed that the higher expression level of CDO1, the higher level of taurine was detected in mice. And the taurine level was highest in CDO1+/+ mice, intermediate in CDO1+/− mice, and lowest in CDO1−/− mice. On the other hand, in mice with liver-specific deletion of CDO1, the cysteine levels in liver and plasma increased, and the abundance of CDO1 and the proportion of more active CDO1 increased in extrahepatic tissues, including kidney, brown fat, gonadal fat and pancreas.32 Additionally, extrahepatic tissues of mice with liver-specific deletion of CDO1 also had higher levels of hypotaurine, while taurine was maintained at normal levels.32 The biosynthesis of taurine is mainly in the liver, while the maintenance of the normal level of taurine and hypotaurine in mice with liver-specific deletion of CDO1 indicate that the extrahepatic tissue can make up for the lack of cysteine metabolism in the liver.
Lipid metabolism and adipogenesis
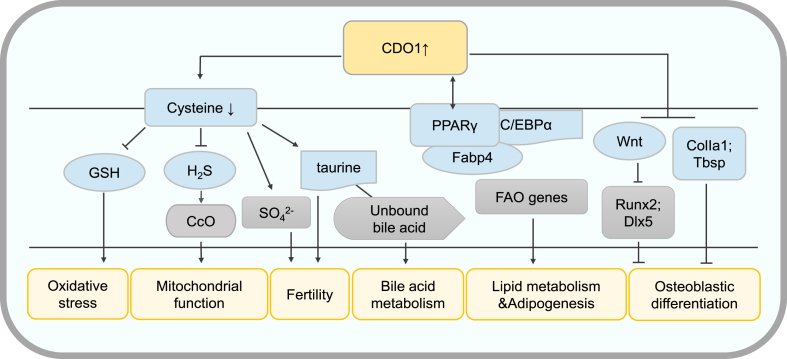
Lipid metabolism is a highly controlled process that involves several positive and negative regulators, while the role of CDO1 in lipid metabolism has been investigated in some studies (Fig. 4). CDO1 is highly expressed in adipose tissue.2 A study showed that the expression of CDO1 was decreased in the adipose tissue of mice with diet-induced or genetically based obesity and insulin resistance.2 And another study demonstrated that hepatic fatty acid oxidation (FAO) was generally impaired and serum leptin levels were low in CDO1−/− mice, which could lead to increased fat storage in liver.33 Furthermore, the expression of CDO1 is up-regulated during adipogenic differentiation of human bone marrow-derived mesenchymal stem cells (hBMSC) and human adipose tissue-derived preadipocytes, suggesting that CDO1 could play a role in the regulation of adipogenesis.34,35 Similarly, in the study of mice bone marrow-derived mesenchymal stem cells (mBMSC) and 3T3-L1 preadipocytes in vitro, it was also found that CDO1 expression was up-regulated during adipogenic differentiation. And siRNA-mediated depletion of CDO1 significantly suppressed adipogenic differentiation of mBMSC and 3T3-L1 cells.36 The peroxisome proliferator-activated receptor γ (PPARγ) is an important adipogenic transcription factor, which can promote fatty acid uptake, triglyceride formation and fat storage in lipid droplets, and also increase insulin sensitivity and glucose metabolism.37 CCAAT/enhancer-binding protein α (C/EBPα) plays a critical role in the late stages of adipogenesis and is important for maintaining the functions of mature adipocytes.38 A study shows that CDO1 knockdown in preadipocytes could significantly suppress the expression of adipogenesis-related genes, including PPARγ, C/EBPα, and fatty acid binding protein-4 (Fabp4).36 Mechanistically, the down-regulation of CDO1 impaired the binding of PPARγ to the promoters of C/EBPα, and Fabp4, indicating that CDO1 may be required for the transcriptional activity of PPARγ.36 On the other hand, previous studies had shown that PPARγ and C/EBPα, in a synergistic way, could be involved in the transactivation of CDO1 expression.39 And the CDO1 gene contains a putative C/EBP-binding site in its 5′-UTR. It was shown in C/EBPα-null mice that CDO1 expression in white adipose tissue (WAT) requires C/EBPα.40 Taken together, CDO1 and PPARγ may be integrated as a positive feedback loop, in which PPARγ initials transcription of CDO1, and CDO1 in turn facilitates PPARγ binding to the promoters of target genes, thereby promoting adipogenesis and maintaining the function of adipocytes. Overall, these studies highlight that CDO1 plays an important role in lipid metabolism and adipocyte differentiation, of which the mechanisms needs further investigation.
Figure 4.
The role and mechanism of CDO1 in physiological processes. CDO1 can promote cysteine metabolism, and then reduce the synthesis of GSH, leading to oxidative stress. Similarly, cysteine decomposition leads to decreased H2S synthesis and promotes mitochondrial function. CDO1 can promote the production of taurine, which can improve fertility and promote bile acid metabolism, and CDO1-mediated sulfate metabolism also plays an important role in fertility. In addition, CDO1 can significantly promote the expression of adipocyte differentiation-related genes, including PPARγ, C/EBPα and Fabp4, thereby facilitating adipogenesis. Cdo1 may regulate hepatic lipid metabolism by regulating FAO. CDO1 is a negative regulator of osteogenesis. It can inhibit the expression of osteogenic related genes, such as ColIa1 and Tbsp, and CDO1 inhibits Wnt signaling and restricts Wnt-induced expression of osteogenic transcriptional factors in BMSC, such as Runx2 and Dlx5. BMSC, bone marrow-derived mesenchymal stem cells; C/EBPα, CCAAT/enhancer-binding protein α; ColIa1, collagen, type I, alpha 1; Dlx5, distal-less homeobox 5; Fabp4, fatty acid binding protein-4; FAO, fatty acid oxidation; PPARγ, peroxisome proliferator-activated receptor γ; Runx2, runt-related transcription factor 2; Tbsp integrin binding sialoprotein.
Regulation of redox homeostasis
Glutathione, as the main cellular thiol and redox buffer of mammals, is an important antioxidant.41 It is a primary cellular antioxidant that maintains redox balance and defends against oxidative stress.42 CDO1, as a rate-limiting enzyme in cysteine catabolism, catalyzes the irreversible conversion of cysteine to CSA, which is the main way to eliminate cellular cysteine.43 And cysteine is an important substrate for the synthesis of glutathione, in which sulfhydryl side chain forms the active moiety of glutathione.44 Therefore, CDO1 is capable of indirectly modulating the total glutathione pool by regulating the intracellular cysteine level (Fig. 4). A study has shown that overexpression of CDO1 can significantly decrease cellular GSH levels by approximately 40%.45 In HepG2/C3A cells, the regulatory effect of CDO1 on glutathione can significantly affect the antioxidant capacity of cells.44 An increased expression of CDO1 in HepG2/C3A cells could lead to oxidative stress by decreasing the amount of cysteine and, in turn, limiting the amount of substrate available for glutathione synthesis.44 Interestingly, CDO1-mediated hepatic cysteine catabolism is inhibited by bile acids. Attenuation of this bile acids effect by cholestyramine, a bile acids sequestrant, induced the hepatic expression of CDO1 to deplete the hepatic free cysteine pool and alter GSH synthesis, which weakens the capacity of antioxidation and results in a decrease in sensitivity to oxidative injury in the liver.45 These results emphasize the potential of bile acids in the regulation of cysteine and glutathione homeostasis. Collectively, the findings above establish a causal relationship between CDO1 and oxidative stress.
Fertility
Some studies have shown that CDO1−/− mice exhibit postnatal mortality, growth deficit, and connective tissue pathology, suggesting a critical role of CDO1 in fertility (Fig. 4).27 It is known that cysteine is the main substrate of sulfate production in the body, and its decomposition is mainly mediated by CDO1. Ueki I et al27 crossed the CDO1+/− mice to generate CDO1−/−, CDO1+/−, and CDO1+/+ mice. Interestingly, severe male infertility was observed when the reproductive ability was tested by natural mating of CDO1−/− males with WT females, suggesting that CDO1 may play an important role in male fertility.27 Furthermore, by using CDO1−/− mice, it was demonstrated that epididymal CDO1 plays a key role in post-testicular sperm maturation and a slight increase in the frequency of sperm head morphological abnormalities was detected in CDO1−/− mice.46 Taurine and hypotaurine are involved in osmotic regulation and antioxidative defense in cells. They are shown to enhance antioxidation capacity, promote motility and increase fertilization ability in spermatozoa.47 CDO1−/− mice had a severe lack of taurine and hypotaurine in their epididymal intraluminal fluid and sperm. Further studies have shown that lack of taurine causes flagellar angulation, and taurine plays a critical role in sperm osmoregulation.46 Moreover, supplementing taurine can rescue the impaired fertility. However, the specific mechanism of how taurine facilitates fertility needs further investigation.
Sulfate plays an important role in the growth and development of fetus.48 Fetus mainly obtains the supply of sulfate through maternal circulation. When the sulfate content in maternal circulation is too low, the fetal sulfate supply is insufficient, which will cause severe fetal abnormalities and death in mice.49 Some studies have shown that the abundance of CDO1 in fetal tissue increases continuously from the second trimester of pregnancy and plays an important role in the fetal sulfate supply in the third trimester of pregnancy.50 Therefore, CDO1-mediated sulfate metabolism could contribute to the sulfate balance of the fetus and plays an important role in fetal growth and development.
Osteoblastic differentiation
Bone marrow-derived mesenchymal stem cells (BMSC) can differentiate into osteoblast and mainly participate in bone synthesis and mineralization in bone formation.51 The process of osteogenic differentiation of BMSC can be categorized into three processes, a commitment to osteoprogenitor cells, differentiation into pre-osteoblasts and osteoblast maturation.51 Zhao XF et al52 demonstrated that CDO1 expression was elevated during osteoblastic differentiation of BMSC in vitro (Fig. 4). And knockdown of CDO1 led to an increased expression of osteogenic related genes, such as Collagen, type I, alpha 1 (ColIa1), and integrin binding sialoprotein (Tbsp). Consistently, knockdown of CDO1 enhanced alkaline phosphatase (early marker of osteoblastic differentiation) activity and promoted mineralization. On the contrary, the signal factors associated with osteogenic differentiation were impaired by overexpression of CDO1 in BMSC.52 The mechanism of CDO1-mediated inhibition of osteogenesis was explored further. Overexpression of CDO1 inhibited Wnt signaling and restricted Wnt-induced expression of osteogenic transcriptional factors in BMSC, such as RUNX family transcription factor 2 (Runx2) and distal-less homeobox 5 (Dlx5).52 Therefore, CDO1 suppressed the differentiation from BMSC to osteoblasts, and dysregulation of CDO1 expression may be correlated with the bone-related diseases.
Metabolism of sulfide
CDO1 is the initial and rate-limiting enzyme involved in the oxidative degradation of cysteine to inorganic sulfate, which is believed to be the main source of sulfate in vivo (Fig. 4).53 The process of generating H2S and sulfate from sulfur-containing amino acid are the main desulfurization methods in vivo. Cysteine can be decomposed into sulfate mediated by CDO1. Cysteine can also produce H2S and be further metabolized to sulfate in an CDO1-independent way. Cellular cysteine concentrations are a major determinant of the flux of cysteine to H2S.28 CDO1 can indirectly regulate this process. Some studies have shown that plasma sulfate levels were slightly higher in CDO1−/− mice than in CDO1+/− or CDO1+/+ mice, and the tissue levels of acid-labile sulfide were elevated in CDO1−/− mice.27 Similarly, CDO1−/− mice exhibited increased levels of urinary thiosulfate.54 Previous studies in CDO1−/− mice showed that the absence of CDO1 promoted cysteine desulfurization, resulting in the overproduction of H2S and thiosulfate.28,54 The content of H2S is determined by the balance between its production rate and its clearance rate. On the one hand, excessive production of H2S can lead to the inhibition of mitochondrial cytochrome c oxidase activity and further damage electronic transmission.54 On the other hand, H2S has been identified as an important gaseous signaling molecule, and an appropriate amount of H2S is beneficial to the body, while excessive H2S will lead to toxicity.33,55 Therefore, CDO1-mediated cysteine desulfurization plays a critical role in maintaining the appropriate levels of H2S and sulfate in the body and ensuring the functional role of H2S as a signal molecule.
Bile acid metabolism
In the enterohepatic system, bile acids play an important role in the regulation of metabolic homeostasis, immune response, and cell proliferation.56,57 Bile acids are efficiently conjugated to glycine or taurine to form N-acyl amidated bile acids in hepatocytes.58 Bile acid amidation increases bile acid aqueous solubility and enhances its digestive function in the gut,59 and the mouse bile acid pool contains primarily taurine-conjugated bile acids.60 Murine bile acids undergo conjugation with taurine in the liver prior to their secretion in the bile, and the production of taurine in the liver requires CDO1. Stipanuk MH et al61 used the CDO1-null mouse model and found that hepatic taurine was depleted to only 3–5% of that in wild-type mice, and there was a dramatic decrease in the hepatic concentration of taurine-conjugated bile acids with a dramatic increase in unconjugated and glycine-conjugated bile acids.61 The effects of taurine deficiency on the expression of proteins involved in sulfur amino acid and bile acid metabolism were examined in CDO1-null mice. It was found that the expression of hepatic genes, including CSA decarboxylase (CSAD), betaine homocysteine methyltransferase (Bhmt), cholesterol 7α-hydroxylase (Cyp7a1), and cytochrome P450 3A11 (Cyp3a11), was strongly affected by taurine depletion in the CDO1-null mice. Dietary taurine supplementation in CDO1-null mice restored hepatic expression levels of the above four genes and their corresponding proteins. Therefore, the deletion of CDO1 will lead to a decrease in taurine production, which in turn reduces the binding of taurine to unbound bile acids, and ultimately affects the pathway of bile acids metabolism and the expression of genes related to bile acids metabolism. In short, taurine promotes the metabolism of bile acid by combining with unbound bile acids and this process is indirectly regulated by CDO1 (Fig. 4). Further studies are needed to examine the functional role of CDO1 in enterohepatic circulation of bile acid.
Pathophysiological roles of CDO1
Cancer
Cancer is caused by aberrant gene regulation, including up-regulation of oncogenes and down-regulation of cancer suppressor genes. Down-regulation of CDO1 mRNA and protein levels were observed in cancer cell lines and tumors.5 Moreover, forced expression of full-length CDO1 gene in human cancer cells markedly decreased the tumor cell growth both in vitro and in vivo, whereas knockdown of CDO1 increased tumor cell growth in vitro.5 In the carcinogenic process of cancer, abnormal CDO1 gene regulation can occur through epigenetic changes in CDO1 gene promoter.23 Brait et al5 performed TaqMan-MSP on the CDO1 promoter in tumor tissues derived from the breast, esophagus, lung, bladder, and stomach, the results showed that CDO1 gene promoter was highly methylated, of which the sensitivity and specificity were over 78% in tumors of all tissue types tested. Furthermore, treatment with 5-aza-2′-deoxycytidine (a drug that promotes demethylation) reactivated the CDO1 expression in most cancer cell lines, indicating that the transcriptional expression of CDO1 is closely correlated with its promoter methylation level.5 And high methylation level of the CDO1 promoter region has been found in intraductal papillary mucinous neoplasm,62 colorectal cancer,5,63, 64, 65, 66, 67 biliary tract cancer,68 non-small cell lung cancer,69, 70, 71 breast cancer,72, 73, 74, 75 clear-cell renal cell cancer,76 esophagus adenocarcinoma,77,78 gastric cancer,79, 80, 81 small bowel cancer,82 hepatocellular carcinoma83 and endometrial cancer.84 The degree of methylation of CDO1 promoter is closely related to the progression and prognosis of cancer.76,85,86 Therefore, the methylation level of CDO1 promoter can reflect the progression and malignancy of cancer (Fig. 5).
Figure 5.
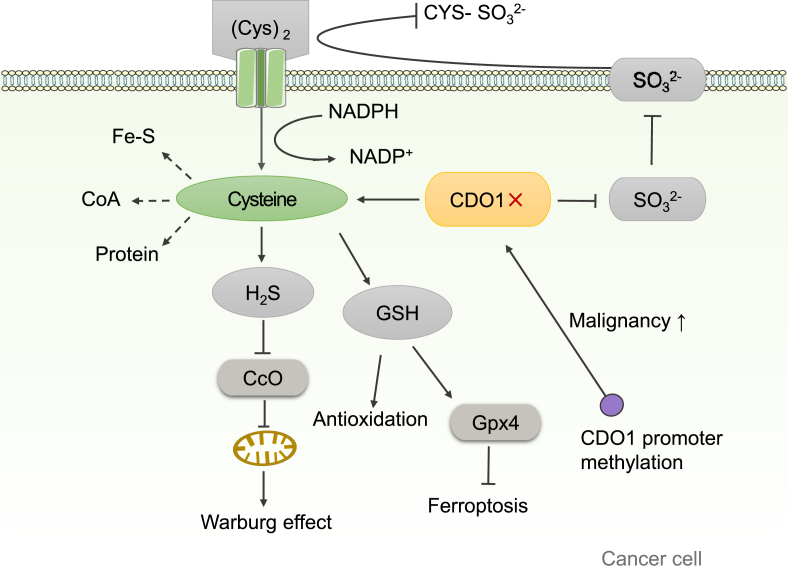
The effect of CDO1 on cancer. CDO1 reduces intracellular level of cysteine by catabolizing cysteine. Down-regulation of CDO1 increases the intracellular level of cysteine by decreasing the catabolism of cysteine. In addition, CDO1 silencing can reduce the production of and inhibit the reaction of cystine with to produce cysteine-S-sulfate . Because cystine is the resource for intracellular cysteine, reduced formation of will increase the level of free cystine, thereby indirectly increasing intracellular cysteine level. Intracellular cysteine is involved in the synthesis of proteins, CoA and Fe-S, which are important for cancer cell proliferation. Furthermore, extracellular cystine can enter the cell through its transporter and NADPH as an electron donor for the production of cysteine from cystine, which consumes a large amount of NADPH. Silencing of CDO1 will increase the intracellular level of cysteine, which would decrease the need for the transportation of cystine from extracellular area, leading to decreased consumption of NADPH and increased NADPH/NADP+ ratio. It is known that increased NADPH/NADP + ratio will promote cancer cell proliferation. Increased promoter methylation of CDO1 results in down-regulation of CDO1 gene expression, thereby increasing the malignancy of cancer. In cancer cells, CDO1 silencing can promote the production of GSH, and then improve the adaption of cancer cells to oxidative stress. In addition, when CDO1 is inhibited, the synthesis of GSH increased, which in turn increases the activity of GPX4 and inhibits the ferroptosis of cancer cells. Furthermore, CDO1 silencing leads to the increase of H2S production, which inhibits mitochondrial function and in turn promotes Warburg effect in cancer cells. Therefore, CDO1 could function as a tumor suppressor gene. Fe-S, iron-sulfur clusters; CcO, cytochrome c oxidase; Gpx4, glutathione peroxidase 4.
In cancer cells, the silencing of CDO1 gene may enhance the ability of cancer cells to adapt to oxidative stress by enhancing the antioxidative capacity of the cells, as shown in Figure 5.75 Cysteine is a source for the biosynthesis of glutathione, a major antioxidant that is decreased in the presence of CDO1.44 In a previous study, glutathione was shown to increase significantly in PCa cancer cells compared to normal control cells.87 Therefore, in cancer cells, CDO1 silencing could promote the production of GSH, and then improve the adaption of cancer cells to oxidative stress. Many cancer cells, in particularly at the advanced stage, exhibit higher basal levels of ROS compared to normal cells as a result of an imbalance between the levels of oxidants and antioxidants. Therefore, cancer cells have higher basal levels of endogenous oxidative stress than normal cells.88 Under persistently increased ROS production, cancer cells adapt to such stress to decrease oxidative damage and ROS-induced apoptosis by developing an enhanced, endogenous detoxification capacity.88 And a related study shows that CDO1-induced reduction in ROS detoxification sensitizes breast cancer cells to anthracycline treatment.75,89 Increased ROS production makes cancer cells more vulnerable to damage by further ROS insults caused by exogenous ROS-generating agents such as anthracyclines.88 Enhanced antioxidative capacity not only allows cancer cells to survive under oxidative stress and to facilitate cancer cell transformation and metastasis,90 but also leads to resistance to ROS-generating agents.91 Therefore, the silencing of CDO1 may enhance the detoxification of ROS and enhance the survival of cancer cells.
Ferroptosis is a recently discovered form of iron-dependent non-apoptotic cell death.92 Recently, ferroptosis is considered to be an effective physiological mechanism for mediating cancer cell death. It is characterized by loss of the activity of the lipid repair enzyme, glutathione peroxidase 4 (GPX4).93 GPX4, as a GSH-dependent enzyme, inhibits ferroptosis of cells by inhibiting the production of ROS.94 In gastric cancer cells, when CDO1 is inhibited, the synthesis of GSH increased, which in turn increases the activity of GPX4 and inhibits the ferroptosis of gastric cancer cells.95 Therefore, overexpression of CDO1 in cancer cells may be a potential method for the treatment of cancer, which may lead to ferroptosis of cancer cells through the GSH–GPX4 signal pathway, and then promotes the death of cancer cells.
In terms of cancer cells proliferation, a study shows that the up-regulation of CDO1 significantly weakens the proliferation of lung cancer cells.71 However, CDO1 silencing can promote cysteine accumulation and cancer cell proliferation. Generally, cysteine is involved in the synthesis of proteins, CoA and iron-sulfur clusters (Fe-S), which are necessary for cancer cell proliferation. Down-regulation of CDO1 increases the intracellular level of cysteine by decreasing the catabolism of cysteine. Besides, CDO1 silencing can reduce the production of SO32− and inhibit the reaction of cystine with to produce cysteine-S-sulfate (CYS-). Because cystine is the resource for intracellular cysteine, reduced formation of CYS-SO32- will increase the level of free cystine, thereby indirectly increasing intracellular cysteine level. Furthermore, extracellular cystine can enter the cell through its transporter and NADPH as an electron donor for the production of cysteine from cystine, which consumes a large amount of NADPH. Silencing of CDO1 will increase the intracellular level of cysteine, which would decrease the need for the transportation of cystine from extracellular area, leading to decreased consumption of NADPH and increased NADPH/NADP+ ratio. It is known that NADPH is critical for both antioxidation and cellular biosynthetic processes, and increased NADPH/NADP+ ratio will promote cancer cell proliferation. Therefore, by increasing intracellular cysteine and NADPH levels, CDO1 silencing facilitates tumorigenesis (Fig. 5).
Aberrant up-regulation of H2S-producing enzymes is frequently observed in different cancer types.96 At excessive concentration of H2S, it will seriously inhibit cytochrome c oxidase (CcO) and cause mitochondrial respiratory dysfunction.97 In fact, the destruction of the function of CcO will lead to the increase of Warburg effect and carcinogenicity.98,99 In tumor cells, CDO1 silencing can lead to mitochondrial respiratory dysfunction and inhibit mitochondrial oxidative phosphorylation.100 Meanwhile, CDO1 deficiency will increase the rate of cysteine metabolism to H2S.54 Therefore, H2S contributes to enhanced Warburg effect and mitochondrial dysfunction in human cancer, and silencing of the CDO1 gene may be a key signal that increases H2S level to activate the Warburg effect pathway and accelerate mitochondrial dysfunction (Fig. 5). In view of all that has been mentioned so far, the potential interest for future studies will be the role of CDO1 in regulating mitochondrial function and glycolysis and the potential mechanism of H2S and sulfide in tumorigenesis.
Obesity and metabolic disorders
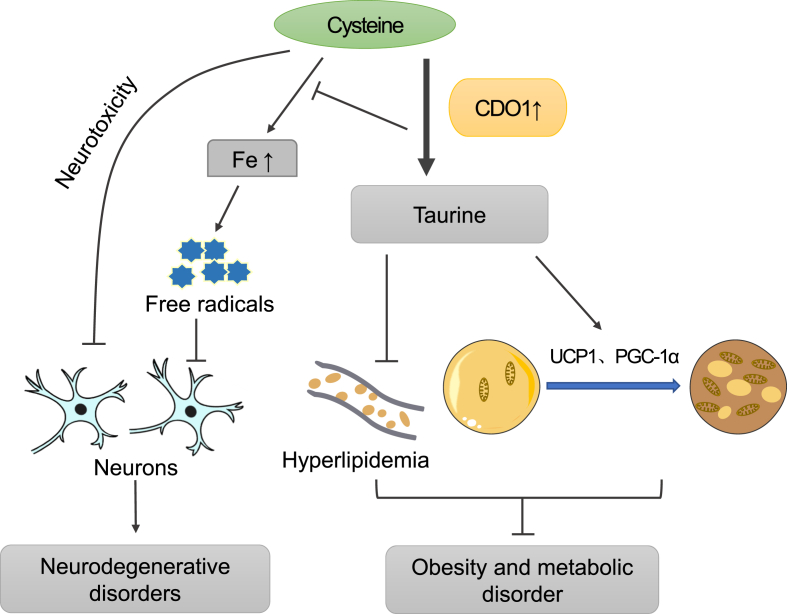
Obesity and metabolic disorders are serious health problems throughout the world.101, 102, 103, 104, 105 Taurine plays a beneficial role in the prevention of diabetes and metabolic disorders.106,107 In the HFD-induced obese mice model, long-term taurine supplementation causes weight loss, most likely by inhibiting adipogenesis in WAT.108 Related studies have shown that adipogenic genes PPAR-α, PPAR-γ, C/EBP-α and C/EBP-β are down-regulated in WAT after taurine supplementation,2 but the underlying mechanism is poorly defined. Similarly, intraperitoneal treatment of mice with taurine alleviated HFD-induced obesity and increased insulin sensitivity, which was partially attributable to taurine-mediated enhancement of energy expenditure and adaptive thermogenesis in mice.109 Meanwhile, the study also showed that taurine can promote the expression of the peroxisome proliferator activated receptor γ coactivator 1-α (PGC-1α), uncoupling protein 1 (UCP1) and other thermogenic genes in inguinal white adipose tissue (iWAT), and then significantly induce browning of iWAT.109 Furthermore, dietary supplementation of taurine ameliorated high blood pressure, liver damage, and hypercholesterolemia in mammals.110 Taurine significantly decreased serum levels of lipids such as triglyceride, cholesterol, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, and leptin level, which protects against obesity and improves insulin sensitivity.111 In mammals, taurine is obtained via two pathways. The first is dietary ingestion and the second is de novo synthesis. The key enzyme in the taurine biosynthetic pathway is CDO1, which can regulate the synthesis of taurine. In mice, CDO1 can promote taurine production in parametrial WAT.2 However, HFD-induced obesity is associated with decreased CDO1 expression in WAT and decreased blood taurine concentration. Similarly, CDO1 expression in WAT is reduced in genetically obese and diabetic mice.2 Thus, CDO1 can promote taurine production and taurine could promote the browning of white fat, inhibit fat production and alleviate hyperlipidemia, thus exerting beneficial effects in the combat against obesity and related metabolic disorders (Fig. 6). Consequently, future research can be carried out to investigate the functional role and mechanism of CDO1 in regulating the homeostasis of lipid and carbohydrate metabolism.
Figure 6.
The effect of CDO1 on neurodegenerative disorders, obesity and metabolic disorders. Aberrant accumulation of cysteine can lead to neurotoxicity. Furthermore, accumulated cysteine may chelate iron, leading to the local increase in iron content. The combined excess of cysteine and ferrous iron would generate free radicals that damage neurons. Therefore, CDO1 can improve neurodegenerative disorders by promoting cysteine catabolism. On the other hand, CDO1 can improve obesity and metabolic disorders by promoting taurine production, which in turn inhibits plasma lipid content and facilitates the browning of white fat. Fe, local iron level.
Neurodegenerative disorders
The elevated level of cysteine can lead to neurotoxicity and cytotoxicity.3 Elevated cysteine to sulfate ratio was observed in patients with MND, PD and AD. The abnormally low expression of CDO1 in the human brain was proposed to have an etiological role in the pathogenesis of these diseases. CDO1 protein and mRNA was expressed in the neurons of the brain, and their expression levels vary in different parts of the brain, with high expression levels in the hippocampus, the dentate gyrus, the outer cortices of the brain, and the substantia nigra.112 Aberrant accumulation of cysteine can lead to neurotoxicity, so the expression of CDO1 can control the level of intracellular cysteine to protect the neurons.112 In Hallervorden–Spatz disease, a rare progressive extrapyramidal dysfunction and dementia, the activity of CDO1 was reduced and cysteine accumulates locally in the globus pallidus.4 Accumulated cysteine may chelate iron, accounting for the local increase in iron content.4 The combined excess of cysteine and ferrous iron may generate free radicals that damage neuronal membranes to cause the typical morphological changes observed in this disorder.4 Therefore, in the brain, CDO1 may protect neurons by promoting the catabolism of cysteine and reducing cysteine-mediated cytotoxicity and production of free radicals, which needs to be further verified (Fig. 6).
Concluding remarks
CDO1 is a mammalian non-heme Fe(II) dioxygenases enzyme, which is expressed mainly in adipose tissue, liver, brain, small intestine, lung, and kidney.1,2 Studies in cellular and animal models indicate that CDO1 is involved in a variety of physiological processes. A deeper understanding of the mechanisms underlying the action of CDO1 is required. CDO1 mainly catabolizes cysteine to produce taurine and sulphate, and affects the reproductive ability by regulating the levels of taurine and sulfate. In addition, CDO1 could be involved in lipid metabolism, and it can facilitate adipocyte differentiation and inhibit osteoblastic differentiation. CDO1 indirectly regulates the level of glutathione and leads to oxidative stress, which contributes to its anti-tumor effect. Further research is also needed to assess whether CDO1 activation could result in any adverse effects in normal cells. Besides, CDO1-mediated cysteine desulfurization plays a critical role in maintaining appropriate levels of H2S in the body and guaranteeing a functional role of H2S as a signal molecule. Furthermore, CDO1 can affect bile acids metabolism by regulating taurine biosynthesis, and the effect of CDO1 on enterohepatic circulation of bile acid merits further investigation. In physiological conditions, the activity and expression of CDO1 is regulated by multiple mechanisms, including the positive feedback of its substrate (cysteine) and ubiquitination. Because of the important physiological roles of CDO1, identification of the pathways and critical factors that regulate the activity, expression and stability of CDO1 gene and protein may help to better regulate CDO1-mediated physiological processes.
In pathophysiological conditions, CDO1 silencing or abnormal expression may be associated with the occurrence and development of variety of diseases. Promoter methylation-mediated CDO1 silencing can promote the initiation and progression of cancers. Additionally, DNA methylation on CDO1 promoter is considered to be a hallmark in the early stage of cancer, and elevating the expression level of CDO1 is proved to promote the ferroptosis of cancer cells. Therefore, these results above would provide more information for patients regarding outcome predictions and targeting CDO1 might provide a new route of treating cancer. Besides, taurine could promote WAT browning and inhibit fat production, thus improving metabolic health in obesity and metabolic disorders. Consequently, future research can be carried out to verify the function of CDO1 in regulating lipid and carbohydrate metabolism. Furthermore, deficiency of CDO1 would lead to neurodegenerative disorders possibly owing to dysregulated cysteine metabolism. However, how CDO1-mediated cysteine metabolism protects against neurodegenerative disorders remains to be defined. Therefore, further elucidation of the mechanisms underlying the functional role of CDO1 would provide a clearer view of the therapeutic potential and possible risks of targeting this important regulatory protein.
Author contributions
The search and collection of literatures was performed by M.C., J.Z., W.M. and L.G. The first draft of the manuscript was written by M.C. and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Conflict of interests
The authors declare no competing interests.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32070751 and 31871435) to L.G.
Acknowledgements
We apologize for incomplete citations due to space limitations.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Hirschberger L.L., Daval S., Stover P.J., et al. Murine cysteine dioxygenase gene: structural organization, tissue-specific expression and promoter identification. Gene. 2001;277(1–2):153–161. doi: 10.1016/s0378-1119(01)00691-6. [DOI] [PubMed] [Google Scholar]
- 2.Tsuboyama-Kasaoka N., Shozawa C., Sano K., et al. Taurine (2-aminoethanesulfonic acid) deficiency creates a vicious circle promoting obesity. Endocrinology. 2006;147(7):3276–3284. doi: 10.1210/en.2005-1007. [DOI] [PubMed] [Google Scholar]
- 3.Parsons R.B., Waring R.H., Ramsden D.B., et al. Toxicity of cysteine and cysteine sulphinic acid to human neuronal cell-lines. J Neurol Sci. 1997;152(Suppl 1):S62–S66. doi: 10.1016/s0022-510x(97)00246-3. [DOI] [PubMed] [Google Scholar]
- 4.Perry T.L., Norman M.G., Yong V.W., et al. Hallervorden-Spatz disease: cysteine accumulation and cysteine dioxygenase deficiency in the globus pallidus. Ann Neurol. 1985;18(4):482–489. doi: 10.1002/ana.410180411. [DOI] [PubMed] [Google Scholar]
- 5.Brait M., Ling S., Nagpal J.K., et al. Cysteine dioxygenase 1 is a tumor suppressor gene silenced by promoter methylation in multiple human cancers. PLoS One. 2012;7(9):e44951. doi: 10.1371/journal.pone.0044951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pau M.Y., Lipscomb J.D., Solomon E.I. Substrate activation for O2 reactions by oxidized metal centers in biology. Proc Natl Acad Sci U S A. 2007;104(47):18355–18362. doi: 10.1073/pnas.0704191104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaesi E.J., Fox B.G., Brunold T.C. Spectroscopic and computational investigation of the H155A variant of cysteine dioxygenase: geometric and electronic consequences of a third-sphere amino acid substitution. Biochemistry. 2015;54(18):2874–2884. doi: 10.1021/acs.biochem.5b00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmons C.R., Liu Q., Huang Q., et al. Crystal structure of mammalian cysteine dioxygenase. A novel mononuclear iron center for cysteine thiol oxidation. J Biol Chem. 2006;281(27):18723–18733. doi: 10.1074/jbc.M601555200. [DOI] [PubMed] [Google Scholar]
- 9.McCoy J.G., Bailey L.J., Bitto E., et al. Structure and mechanism of mouse cysteine dioxygenase. Proc Natl Acad Sci U S A. 2006;103(9):3084–3089. doi: 10.1073/pnas.0509262103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunwell J.M., Khuri S., Gane P.J. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol Mol Biol Rev. 2000;64(1):153–179. doi: 10.1128/mmbr.64.1.153-179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arjune S., Schwarz G., Belaidi A.A. Involvement of the Cys-Tyr cofactor on iron binding in the active site of human cysteine dioxygenase. Amino Acids. 2015;47(1):55–63. doi: 10.1007/s00726-014-1843-7. [DOI] [PubMed] [Google Scholar]
- 12.Tsuboyama-Kasaoka N., Hosokawa Y., Kodama H., et al. Human cysteine dioxygenase gene: structural organization, tissue-specific expression and downregulation by phorbol 12-myristate 13-acetate. Biosci Biotechnol Biochem. 1999;63(6):1017–1024. doi: 10.1271/bbb.63.1017. [DOI] [PubMed] [Google Scholar]
- 13.McCann K.P., Akbari M.T., Williams A.C., et al. Human cysteine dioxygenase type I: primary structure derived from base sequencing of cDNA. Biochim Biophys Acta. 1994;1209(1):107–110. doi: 10.1016/0167-4838(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 14.Tsuboyama N., Hosokawa Y., Totani M., et al. Structural organization and tissue-specific expression of the gene encoding rat cysteine dioxygenase. Gene. 1996;181(1–2):161–165. doi: 10.1016/s0378-1119(96)00496-9. [DOI] [PubMed] [Google Scholar]
- 15.Dominy J.E., Jr., Hwang J., Guo S., et al. Synthesis of amino acid cofactor in cysteine dioxygenase is regulated by substrate and represents a novel post-translational regulation of activity. J Biol Chem. 2008;283(18):12188–12201. doi: 10.1074/jbc.M800044200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stipanuk M.H., Londono M., Hirschberger L.L., et al. Evidence for expression of a single distinct form of mammalian cysteine dioxygenase. Amino Acids. 2004;26(1):99–106. doi: 10.1007/s00726-003-0001-4. [DOI] [PubMed] [Google Scholar]
- 17.Stipanuk M.H., Ueki I., Dominy J.E., Jr., et al. Cysteine dioxygenase: a robust system for regulation of cellular cysteine levels. Amino Acids. 2009;37(1):55–63. doi: 10.1007/s00726-008-0202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Driggers C.M., Kean K.M., Hirschberger L.L., et al. Structure-based insights into the role of the cys-Tyr crosslink and inhibitor recognition by mammalian cysteine dioxygenase. J Mol Biol. 2016;428(20):3999–4012. doi: 10.1016/j.jmb.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Yan L., Li X., et al. Formation mechanism of cofactor cys-Tyr in the cysteine dioxygenases (CDO and F2-CDO) and its influence on catalysis: a QM/MM study. Inorg Chem. 2021;60(11):7844–7856. doi: 10.1021/acs.inorgchem.1c00340. [DOI] [PubMed] [Google Scholar]
- 20.Dominy J.E., Jr., Hirschberger L.L., Coloso R.M., et al. Regulation of cysteine dioxygenase degradation is mediated by intracellular cysteine levels and the ubiquitin-26 S proteasome system in the living rat. Biochem J. 2006;394(Pt 1):267–273. doi: 10.1042/BJ20051510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominy J.E., Jr., Hirschberger L.L., Coloso R.M., et al. In vivo regulation of cysteine dioxygenase via the ubiquitin-26S proteasome system. Adv Exp Med Biol. 2006;583:37–47. doi: 10.1007/978-0-387-33504-9_4. [DOI] [PubMed] [Google Scholar]
- 22.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 23.Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 24.Herman J.G., Baylin S.B. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 25.Yamashita K., Upadhyay S., Osada M., et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2(6):485–495. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y., Li J., Liu A. Oxygen activation by mononuclear nonheme iron dioxygenases involved in the degradation of aromatics. J Biol Inorg Chem. 2017;22(2–3):395–405. doi: 10.1007/s00775-017-1436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueki I., Roman H.B., Valli A., et al. Knockout of the murine cysteine dioxygenase gene results in severe impairment in ability to synthesize taurine and an increased catabolism of cysteine to hydrogen sulfide. Am J Physiol Endocrinol Metab. 2011;301(4):E668–E684. doi: 10.1152/ajpendo.00151.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stipanuk M.H., Ueki I. Dealing with methionine/homocysteine sulfur: cysteine metabolism to taurine and inorganic sulfur. J Inherit Metab Dis. 2011;34(1):17–32. doi: 10.1007/s10545-009-9006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho J., Woo J., Nam W. A chromium(III)-superoxo complex in oxygen atom transfer reactions as a chemical model of cysteine dioxygenase. J Am Chem Soc. 2012;134(27):11112–11115. doi: 10.1021/ja304357z. [DOI] [PubMed] [Google Scholar]
- 30.Tiranti V., Zeviani M. Altered sulfide (H(2)S) metabolism in ethylmalonic encephalopathy. Cold Spring Harbor Perspect Biol. 2013;5(1):a011437. doi: 10.1101/cshperspect.a011437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ripps H., Shen W. Review: taurine: a "very essential" amino acid. Mol Vis. 2012;18:2673–2686. [PMC free article] [PubMed] [Google Scholar]
- 32.Ueki I., Roman H.B., Hirschberger L.L., et al. Extrahepatic tissues compensate for loss of hepatic taurine synthesis in mice with liver-specific knockout of cysteine dioxygenase. Am J Physiol Endocrinol Metab. 2012;302(10):E1292–E1299. doi: 10.1152/ajpendo.00589.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niewiadomski J., Zhou J.Q., Roman H.B., et al. Effects of a block in cysteine catabolism on energy balance and fat metabolism in mice. Ann N Y Acad Sci. 2016;1363(1):99–115. doi: 10.1111/nyas.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaker M., Pascarelli K.M., Plantinga M.J., et al. Differential expression of cysteine dioxygenase 1 in complex karyotype liposarcomas. Biomarkers Cancer. 2014;6:1–10. doi: 10.4137/BIC.S14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun L., Goff L.A., Trapnell C., et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110(9):3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng P., Chen Y., Ji N., et al. Cysteine dioxygenase type 1 promotes adipogenesis via interaction with peroxisome proliferator-activated receptor gamma. Biochem Biophys Res Commun. 2015;458(1):123–127. doi: 10.1016/j.bbrc.2015.01.080. [DOI] [PubMed] [Google Scholar]
- 37.Montaigne D., Butruille L., Staels B. PPAR control of metabolism and cardiovascular functions. Nat Rev Cardiol. 2021;18(12):809–823. doi: 10.1038/s41569-021-00569-6. [DOI] [PubMed] [Google Scholar]
- 38.Fajas L., Fruchart J.C., Auwerx J. Transcriptional control of adipogenesis. Curr Opin Cell Biol. 1998;10(2):165–173. doi: 10.1016/s0955-0674(98)80138-5. [DOI] [PubMed] [Google Scholar]
- 39.Madsen M.S., Siersbæk R., Boergesen M., et al. Peroxisome proliferator-activated receptor γ and C/EBPα synergistically activate key metabolic adipocyte genes by assisted loading. Mol Cell Biol. 2014;34(6):939–954. doi: 10.1128/MCB.01344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen S.S., Chen J.F., Johnson P.F., et al. C/EBPβ, when expressed from the C/ebp α gene locus, can functionally replace C/EBPα in liver but not in adipose tissue. Mol Cell Biol. 2000;20(19):7292–7299. doi: 10.1128/mcb.20.19.7292-7299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deponte M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim Biophys Acta. 2013;1830(5):3217–3266. doi: 10.1016/j.bbagen.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 42.Xie Y., Hou W., Song X., et al. Ferroptosis: process and function. Cell Death Differ. 2016;23(3):369–379. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stipanuk M.H., Dominy J.E., Jr., Lee J.I., et al. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J Nutr. 2006;136(6 Suppl):1652s–1659s. doi: 10.1093/jn/136.6.1652S. [DOI] [PubMed] [Google Scholar]
- 44.Dominy J.E., Jr., Hwang J., Stipanuk M.H. Overexpression of cysteine dioxygenase reduces intracellular cysteine and glutathione pools in HepG2/C3A cells. Am J Physiol Endocrinol Metab. 2007;293(1):E62–E69. doi: 10.1152/ajpendo.00053.2007. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y., Li J., Matye D., et al. Bile acids regulate cysteine catabolism and glutathione regeneration to modulate hepatic sensitivity to oxidative injury. JCI Insight. 2018;3(8):e99676. doi: 10.1172/jci.insight.99676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asano A., Roman H.B., Hirschberger L.L., et al. Cysteine dioxygenase is essential for mouse sperm osmoadaptation and male fertility. FEBS J. 2018;285(10):1827–1839. doi: 10.1111/febs.14449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rezaee-Tazangi F., Zeidooni L., Rafiee Z., et al. Taurine effects on Bisphenol A-induced oxidative stress in the mouse testicular mitochondria and sperm motility. JBRA Assist Reprod. 2020;24(4):428–435. doi: 10.5935/1518-0557.20200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dawson P.A. Sulfate in fetal development. Semin Cell Dev Biol. 2011;22(6):653–659. doi: 10.1016/j.semcdb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 49.Rakoczy J., Zhang Z., Bowling F.G., et al. Loss of the sulfate transporter Slc13a4 in placenta causes severe fetal abnormalities and death in mice. Cell Res. 2015;25(11):1273–1276. doi: 10.1038/cr.2015.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rakoczy J., Lee S., Weerasekera S.J., et al. Placental and fetal cysteine dioxygenase gene expression in mouse gestation. Placenta. 2015;36(8):956–959. doi: 10.1016/j.placenta.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Neve A., Corrado A., Cantatore F.P. Osteoblast physiology in normal and pathological conditions. Cell Tissue Res. 2011;343(2):289–302. doi: 10.1007/s00441-010-1086-1. [DOI] [PubMed] [Google Scholar]
- 52.Zhao X., Deng P., Feng J., et al. Cysteine dioxygenase type 1 inhibits osteogenesis by regulating Wnt signaling in primary mouse bone marrow stromal cells. Sci Rep. 2016;6:19296. doi: 10.1038/srep19296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilkinson L.J., Waring R.H. Cysteine dioxygenase: modulation of expression in human cell lines by cytokines and control of sulphate production. Toxicol Vitro. 2002;16(4):481–483. doi: 10.1016/s0887-2333(02)00031-0. [DOI] [PubMed] [Google Scholar]
- 54.Jurkowska H., Roman H.B., Hirschberger L.L., et al. Primary hepatocytes from mice lacking cysteine dioxygenase show increased cysteine concentrations and higher rates of metabolism of cysteine to hydrogen sulfide and thiosulfate. Amino Acids. 2014;46(5):1353–1365. doi: 10.1007/s00726-014-1700-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Predmore B.L., Lefer D.J., Gojon G. Hydrogen sulfide in biochemistry and medicine. Antioxidants Redox Signal. 2012;17(1):119–140. doi: 10.1089/ars.2012.4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas C., Gioiello A., Noriega L., et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metabol. 2009;10(3):167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jansen P.L. A new life for bile acids. J Hepatol. 2010;52(6):937–938. doi: 10.1016/j.jhep.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 58.Shonsey E.M., Sfakianos M., Johnson M., et al. Bile acid coenzyme A: amino acid N-acyltransferase in the amino acid conjugation of bile acids. Methods Enzymol. 2005;400:374–394. doi: 10.1016/S0076-6879(05)00022-4. [DOI] [PubMed] [Google Scholar]
- 59.Setchell K.D., Heubi J.E., Shah S., et al. Genetic defects in bile acid conjugation cause fat-soluble vitamin deficiency. Gastroenterology. 2013;144(5):945–955. doi: 10.1053/j.gastro.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y., Matye D., Nguyen N., et al. HNF4α regulates CSAD to couple hepatic taurine production to bile acid synthesis in mice. Gene Expr. 2018;18(3):187–196. doi: 10.3727/105221618X15277685544442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stipanuk M.H., Jurkowska H., Niewiadomski J., et al. Identification of taurine-responsive genes in murine liver using the Cdo1-null mouse model. Adv Exp Med Biol. 2017;975(Pt 1):475–495. doi: 10.1007/978-94-024-1079-2_38. [DOI] [PubMed] [Google Scholar]
- 62.Fujiyama Y., Kumamoto Y., Nishizawa N., et al. Promoter DNA hypermethylation of the cysteine dioxygenase 1 (CDO1) gene in intraductal papillary mucinous neoplasm (IPMN) Ann Surg Oncol. 2020;27(10):4007–4016. doi: 10.1245/s10434-020-08291-2. [DOI] [PubMed] [Google Scholar]
- 63.Igarashi K., Yamashita K., Katoh H., et al. Prognostic significance of promoter DNA hypermethylation of the cysteine dioxygenase 1 (CDO1) gene in primary gallbladder cancer and gallbladder disease. PLoS One. 2017;12(11):e0188178. doi: 10.1371/journal.pone.0188178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kojima K., Nakamura T., Ohbu M., et al. Cysteine dioxygenase type 1 (CDO1) gene promoter methylation during the adenoma-carcinoma sequence in colorectal cancer. PLoS One. 2018;13(5):e0194785. doi: 10.1371/journal.pone.0194785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vedeld H.M., Andresen K., Eilertsen I.A., et al. The novel colorectal cancer biomarkers CDO1, ZSCAN18 and ZNF331 are frequently methylated across gastrointestinal cancers. Int J Cancer. 2015;136(4):844–853. doi: 10.1002/ijc.29039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yokoi K., Harada H., Yokota K., et al. Epigenetic status of CDO1 gene may reflect chemosensitivity in colon cancer with postoperative adjuvant chemotherapy. Ann Surg Oncol. 2019;26(2):406–414. doi: 10.1245/s10434-018-6865-z. [DOI] [PubMed] [Google Scholar]
- 67.Yamashita K., Waraya M., Kim M.S., et al. Detection of methylated CDO1 in plasma of colorectal cancer; a PCR study. PLoS One. 2014;9(12):e113546. doi: 10.1371/journal.pone.0113546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakamoto S., Kumamoto Y., Igarashi K., et al. Methylated promoter DNA of CDO1 gene and preoperative serum CA19-9 are prognostic biomarkers in primary extrahepatic cholangiocarcinoma. PLoS One. 2018;13(10):e0205864. doi: 10.1371/journal.pone.0205864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu B., Filho J.R., Mallisetty A., et al. Detection of promoter DNA methylation in urine and plasma aids the detection of non-small cell lung cancer. Clin Cancer Res. 2020;26(16):4339–4348. doi: 10.1158/1078-0432.CCR-19-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wrangle J., Machida E.O., Danilova L., et al. Functional identification of cancer-specific methylation of CDO1, HOXA9, and TAC1 for the diagnosis of lung cancer. Clin Cancer Res. 2014;20(7):1856–1864. doi: 10.1158/1078-0432.CCR-13-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kang Y.P., Torrente L., Falzone A., et al. Cysteine dioxygenase 1 is a metabolic liability for non-small cell lung cancer. Elife. 2019;8:e45572. doi: 10.7554/eLife.45572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dietrich D., Krispin M., Dietrich J., et al. CDO1 promoter methylation is a biomarker for outcome prediction of anthracycline treated, estrogen receptor-positive, lymph node-positive breast cancer patients. BMC Cancer. 2010;10:247. doi: 10.1186/1471-2407-10-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Minatani N., Waraya M., Yamashita K., et al. Prognostic significance of promoter DNA hypermethylation of cysteine dioxygenase 1 (CDO1) gene in primary breast cancer. PLoS One. 2016;11(1):e0144862. doi: 10.1371/journal.pone.0144862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanaka Y., Kosaka Y., Waraya M., et al. Differential prognostic relevance of promoter DNA methylation of CDO1 and HOPX in primary breast cancer. Anticancer Res. 2019;39(5):2289–2298. doi: 10.21873/anticanres.13345. [DOI] [PubMed] [Google Scholar]
- 75.Jeschke J., O'Hagan H.M., Zhang W., et al. Frequent inactivation of cysteine dioxygenase type 1 contributes to survival of breast cancer cells and resistance to anthracyclines. Clin Cancer Res. 2013;19(12):3201–3211. doi: 10.1158/1078-0432.CCR-12-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deckers I.A., Schouten L.J., Van Neste L., et al. Promoter methylation of CDO1 identifies clear-cell renal cell cancer patients with poor survival outcome. Clin Cancer Res. 2015;21(15):3492–3500. doi: 10.1158/1078-0432.CCR-14-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kojima K., Yamashita K., Ushiku H., et al. The clinical significance of cysteine dioxygenase type 1 methylation in Barrett esophagus adenocarcinoma. Dis Esophagus. 2017;30(3):1–9. doi: 10.1093/dote/dow001. [DOI] [PubMed] [Google Scholar]
- 78.Ushiku H., Yamashita K., Katoh H., et al. Promoter DNA methylation of CDO1 gene and its clinical significance in esophageal squamous cell carcinoma. Dis Esophagus. 2017;30(2):1–9. doi: 10.1111/dote.12496. [DOI] [PubMed] [Google Scholar]
- 79.Harada H., Hosoda K., Moriya H., et al. Cancer-specific promoter DNA methylation of Cysteine dioxygenase type 1 (CDO1) gene as an important prognostic biomarker of gastric cancer. PLoS One. 2019;14(4):e0214872. doi: 10.1371/journal.pone.0214872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harada H., Soeno T., Nishizawa N., et al. Prospective study to validate the clinical utility of DNA diagnosis of peritoneal fluid cytology test in gastric cancer. Cancer Sci. 2021;112(4):1644–1654. doi: 10.1111/cas.14850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ushiku H., Yamashita K., Ema A., et al. DNA diagnosis of peritoneal fluid cytology test by CDO1 promoter DNA hypermethylation in gastric cancer. Gastric Cancer. 2017;20(5):784–792. doi: 10.1007/s10120-017-0697-6. [DOI] [PubMed] [Google Scholar]
- 82.Kojima K., Nakamura T., Ooizumi Y., et al. Clinical significance of cancer specific methylation of the CDO1 gene in small bowel cancer. PLoS One. 2019;14(1):e0211108. doi: 10.1371/journal.pone.0211108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choi J.I., Cho E.H., Kim S.B., et al. Promoter methylation of cysteine dioxygenase type 1:gene silencing and tumorigenesis in hepatocellular carcinoma. Ann Hepatobiliary Pancreat Surg. 2017;21(4):181–187. doi: 10.14701/ahbps.2017.21.4.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liew P.L., Huang R.L., Wu T.I., et al. Combined genetic mutations and DNA-methylated genes as biomarkers for endometrial cancer detection from cervical scrapings. Clin Epigenet. 2019;11(1):170. doi: 10.1186/s13148-019-0765-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang P., Zhao H., Shi R., et al. The role of plasma CDO1 methylation in the early diagnosis of lung cancer. Zhongguo Fei Ai Za Zhi. 2020;23(5):314–320. doi: 10.3779/j.issn.1009-3419.2020.102.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ooki A., Maleki Z., Tsay J.J., et al. A panel of novel detection and prognostic methylated DNA markers in primary non-small cell lung cancer and serum DNA. Clin Cancer Res. 2017;23(22):7141–7152. doi: 10.1158/1078-0432.CCR-17-1222. [DOI] [PubMed] [Google Scholar]
- 87.Meller S., Zipfel L., Gevensleben H., et al. CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients. Epigenetics. 2016;11(12):871–880. doi: 10.1080/15592294.2016.1241931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakamura H., Takada K. Reactive oxygen species in cancer: current findings and future directions. Cancer Sci. 2021;112(10):3945–3952. doi: 10.1111/cas.15068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Okon I.S., Zou M.H. Mitochondrial ROS and cancer drug resistance: implications for therapy. Pharmacol Res. 2015;100:170–174. doi: 10.1016/j.phrs.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ramundo V., Giribaldi G., Aldieri E. Transforming growth factor-β and oxidative stress in cancer: a crosstalk in driving tumor transformation. Cancers (Basel) 2021;13(12):3093. doi: 10.3390/cancers13123093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8(7):579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 92.Dixon S.J., Lemberg K.M., Lamprecht M.R., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Conrad M., Friedmann Angeli J.P. Glutathione peroxidase 4 (Gpx4) and ferroptosis: what’s so special about it? Mol Cell Oncol. 2015;2(3):e995047. doi: 10.4161/23723556.2014.995047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang W.S., SriRamaratnam R., Welsch M.E., et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hao S., Yu J., He W., et al. Cysteine dioxygenase 1 mediates erastin-induced ferroptosis in human gastric cancer cells. Neoplasia. 2017;19(12):1022–1032. doi: 10.1016/j.neo.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang R.H., Chu Y.H., Lin K.T. The hidden role of hydrogen sulfide metabolism in cancer. Int J Mol Sci. 2021;22(12):6562. doi: 10.3390/ijms22126562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Blachier F., Davila A.M., Mimoun S., et al. Luminal sulfide and large intestine mucosa: friend or foe? Amino Acids. 2010;39(2):335–347. doi: 10.1007/s00726-009-0445-2. [DOI] [PubMed] [Google Scholar]
- 98.Srinivasan S., Guha M., Dong D.W., et al. Disruption of cytochrome c oxidase function induces the Warburg effect and metabolic reprogramming. Oncogene. 2016;35(12):1585–1595. doi: 10.1038/onc.2015.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dong D.W., Srinivasan S., Guha M., Avadhani N.G. Defects in cytochrome c oxidase expression induce a metabolic shift to glycolysis and carcinogenesis. Genom Data. 2015;6:99–107. doi: 10.1016/j.gdata.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Prabhu A., Sarcar B., Kahali S., et al. Cysteine catabolism: a novel metabolic pathway contributing to glioblastoma growth. Cancer Res. 2014;74(3):787–796. doi: 10.1158/0008-5472.CAN-13-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Luan X., Tian X., Zhang H., et al. Exercise as a prescription for patients with various diseases. J Sport Health Sci. 2019;8(5):422–441. doi: 10.1016/j.jshs.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mu W.J., Zhu J.Y., Chen M., et al. Exercise-mediated browning of white adipose tissue: its significance, mechanism and effectiveness. Int J Mol Sci. 2021;22(21):11512. doi: 10.3390/ijms222111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Peng W.Q., Xiao G., Li B.Y., et al. L-theanine activates the browning of white adipose tissue through the AMPK/α-ketoglutarate/Prdm16 axis and ameliorates diet-induced obesity in mice. Diabetes. 2021;70(7):1458–1472. doi: 10.2337/db20-1210. [DOI] [PubMed] [Google Scholar]
- 104.Guo S., Huang Y., Zhang Y., et al. Impacts of exercise interventions on different diseases and organ functions in mice. J Sport Health Sci. 2020;9(1):53–73. doi: 10.1016/j.jshs.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang R., Tian H., Guo D., et al. Impacts of exercise intervention on various diseases in rats. J Sport Health Sci. 2020;9(3):211–227. doi: 10.1016/j.jshs.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.De Carvalho F.G., Brandao C.F.C., Munoz V.R., et al. Taurine supplementation in conjunction with exercise modulated cytokines and improved subcutaneous white adipose tissue plasticity in obese women. Amino Acids. 2021;53(9):1391–1403. doi: 10.1007/s00726-021-03041-4. [DOI] [PubMed] [Google Scholar]
- 107.Liu P.J., Liu Y., Ma L., et al. The relationship between plasma taurine levels in early pregnancy and later gestational diabetes mellitus risk in Chinese pregnant women. Sci Rep. 2021;11(1):7993. doi: 10.1038/s41598-021-87178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim K.S., Jang M.J., Fang S., et al. Anti-obesity effect of taurine through inhibition of adipogenesis in white fat tissue but not in brown fat tissue in a high-fat diet-induced obese mouse model. Amino Acids. 2019;51(2):245–254. doi: 10.1007/s00726-018-2659-7. [DOI] [PubMed] [Google Scholar]
- 109.Guo Y.Y., Li B.Y., Peng W.Q., et al. Taurine-mediated browning of white adipose tissue is involved in its anti-obesity effect in mice. J Biol Chem. 2019;294(41):15014–15024. doi: 10.1074/jbc.RA119.009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen W., Guo J.X., Chang P. The effect of taurine on cholesterol metabolism. Mol Nutr Food Res. 2012;56(5):681–690. doi: 10.1002/mnfr.201100799. [DOI] [PubMed] [Google Scholar]
- 111.Kim K.S., Oh D.H., Kim J.Y., et al. Taurine ameliorates hyperglycemia and dyslipidemia by reducing insulin resistance and leptin level in Otsuka Long-Evans Tokushima fatty (OLETF) rats with long-term diabetes. Exp Mol Med. 2012;44(11):665–673. doi: 10.3858/emm.2012.44.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Parsons R.B., Waring R.H., Williams A.C., et al. Cysteine dioxygenase: regional localisation of protein and mRNA in rat brain. J Neurosci Res. 2001;65(1):78–84. doi: 10.1002/jnr.1130. [DOI] [PubMed] [Google Scholar]