Abstract
During cell division, kinetochores link chromosomes to spindle microtubules. The Ndc80 complex, a crucial microtubule binder, populates each kinetochore with dozens of copies. Whether adjacent Ndc80 complexes cooperate to promote microtubule binding remains unclear. Here we demonstrate that the Ndc80 loop, a short sequence that interrupts the Ndc80 coiled‐coil at a conserved position, folds into a more rigid structure than previously assumed and promotes direct interactions between full‐length Ndc80 complexes on microtubules. Mutations in the loop impair these Ndc80‐Ndc80 interactions, prevent the formation of force‐resistant kinetochore‐microtubule attachments, and cause cells to arrest in mitosis for hours. This arrest is not due to an inability to recruit the kinetochore‐microtubule stabilizing SKA complex and cannot be overridden by mutations in the Ndc80 tail that strengthen microtubule attachment. Thus, loop‐mediated organization of adjacent Ndc80 complexes is crucial for stable end‐on kinetochore‐microtubule attachment and spindle assembly checkpoint satisfaction.
Keywords: chromosome segregation, kinetochore, microtubule, Ndc80, spindle assembly checkpoint
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Cell Cycle; Structural Biology
Structural and biophysical analyses of a conserved loop interrupting the long Ndc80 coiled‐coil reveal its contribution to end‐on attachment to dynamic microtubule ends and explain its essentiality for mitosis.
Introduction
Establishing firm attachments between mitotic chromosomes and spindle microtubules is essential for faithful chromosome segregation and genome stability. Kinetochores, large protein platforms that assemble on centromeric DNA, mediate the attachment of chromosomes to microtubules. During mitosis, kinetochores first bind the lateral surface of microtubules to support the congression of chromosomes toward the middle of the dividing cell. They then capture the ends of dynamic microtubules to promote chromosome biorientation on the mitotic spindle (Tanaka et al, 2005; Kapoor et al, 2006; Akiyoshi et al, 2010; Magidson et al, 2011; Shrestha & Draviam, 2013; Chakraborty et al, 2019).
The widely conserved Ndc80 heterotetramer is the prime mediator of the binding between kinetochores and the ends of microtubules (Cheeseman et al, 2006; DeLuca et al, 2006; Wei et al, 2007; Ciferri et al, 2008; Guimaraes et al, 2008; Alushin et al, 2010; Sundin et al, 2011; Tooley et al, 2011). Ndc80 binds microtubules through the N‐terminal calponin homology (CH) domains of its NDC80 (also named HEC1) and NUF2 subunits, with a contribution of the unstructured NDC80 N‐terminal tail. Ndc80 docks onto the inner kinetochore through the RWD domains of its SPC24 and SPC25 subunits. The CH‐ and RWD‐domains are separated by a long coiled‐coil stalk (Musacchio & Desai, 2017).
The spindle assembly checkpoint (SAC) delays anaphase onset until all chromosomes have bioriented on the mitotic spindle. Various mitotic kinases and phosphatases control the interplay between chromosome biorientation and SAC signaling. A crucial aspect of this dynamic interplay is the ability of the kinetochore to identify and correct erroneous (e.g., syntelic) microtubule attachments. Key signaling substrates are well known (for example KNL1 and the NDC80 tail), but how they respond to kinetochore‐microtubule binding in general, and to biorientation specifically, is poorly understood (Saurin, 2018).
Tension exerted by spindle microtubules pulling on kinetochores has a crucial role in error correction (Lampson & Grishchuk, 2017), but molecular cues for tension signaling at kinetochores remain elusive. As an elongated microtubule‐binder in the outer kinetochore, the Ndc80 complex is a prime candidate to sense and signal tension. One hypothesis is that microtubule‐bound Ndc80 complexes undergo a tension‐dependent conformational change from a bent to a stretched state (the “jackknife” model) (Scarborough et al, 2019; Roscioli et al, 2020). In turn, tension‐dependent conformational changes in the Ndc80 complex may control docking sites for kinases, phosphatases, or additional microtubule binders. Ndc80 bending in the jackknife model has been proposed to require the Ndc80 loop, a sequence in the NDC80 subunit that is long known to interrupt the NDC80:NUF2 coiled coil (Wigge et al, 1998). Studies in budding yeast, fission yeast, and humans demonstrated that the Ndc80 loop is essential for the establishment of end‐on kinetochore‐microtubule attachment and proper chromosome segregation (Hsu & Toda, 2011; Maure et al, 2011; Zhang et al, 2012; Shrestha & Draviam, 2013; Tang et al, 2013; Wimbish et al, 2020).
In human cells, a bioriented kinetochore contains approximately 250 copies of the Ndc80 complex (Suzuki et al, 2015) and binds a bundle of approximately 10 microtubules, known as a k‐fiber (O'Toole et al, 2020; Kiewisz et al, 2022). Thus, a single microtubule in the k‐fiber binds multiple Ndc80 complexes (Fig 1A). Reinforcing this prediction, single Ndc80 complexes fail to bind to the ends of dynamic microtubules in vitro, but artificial multimerization overcomes this and generates robust end‐tracking (Powers et al, 2009; Volkov et al, 2018), suggesting a crucial role of Ndc80 multivalency. From a mechanistic perspective, however, it remains unclear whether the acquisition of end‐tracking reflects the uncoordinated binding of multiple individual Ndc80 complexes to the microtubule, or instead reflects additional cooperative interactions between Ndc80 complexes elicited by microtubule binding. The existing evidence for or against Ndc80 cooperativity is conflicting. In the absence of microtubules, Ndc80 complexes do not appear to form oligomeric structures (Huis in 't Veld et al, 2016), but clustering of Ndc80 complexes has been observed along a microtubule lattice (Ciferri et al, 2008; Alushin et al, 2010, 2012). Neighboring Ndc80 complexes have been proposed to align upon microtubule binding (Yoo et al, 2018; Roscioli et al, 2020), thus suggesting that they interact laterally. Conversely, a modeling study suggested that a “lawn” of non‐interacting Ndc80 complexes is ideally suited to generate robust microtubule binding (Zaytsev et al, 2015).
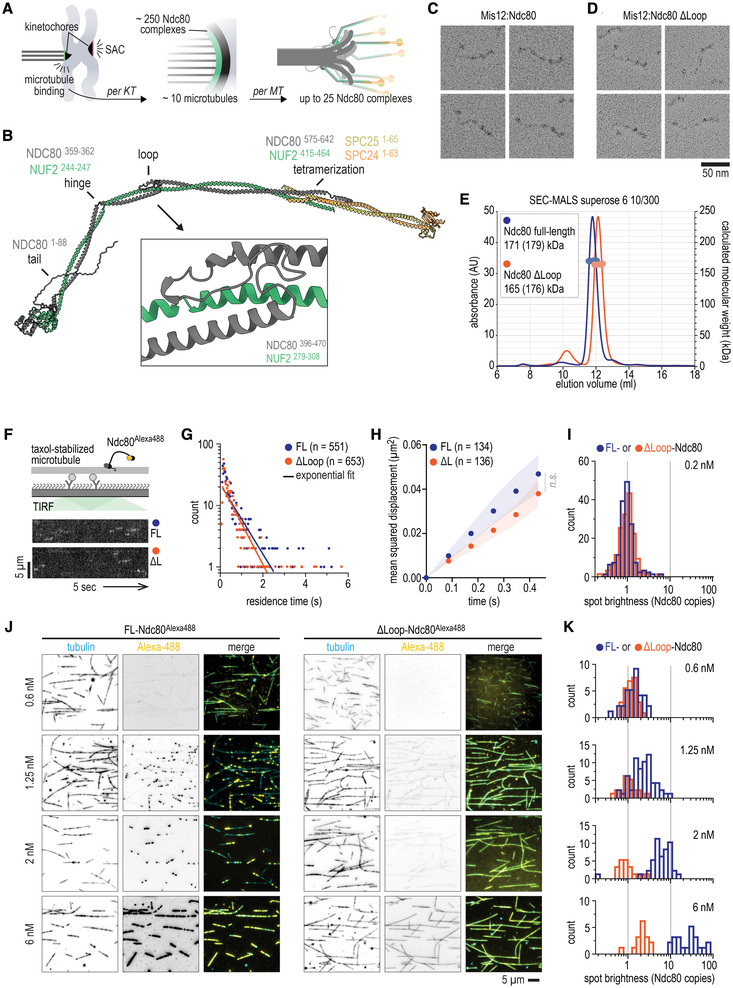
Figure 1. Structural analysis and loop‐dependent clustering of Ndc80 on microtubules.
-
ACartoon of a chromosome pair during mitosis with sister kinetochores that are attached (green) and unattached (red) to microtubules. Unattached kinetochores trigger a spindle assembly checkpoint (SAC) signal. One outer kinetochore contains a lawn of Ndc80 complexes, resulting in many Ndc80 complexes binding to a single microtubule.
-
BPrediction of the full‐length Ndc80 structure with residues that comprise the tail, the hinge, the loop, and the tetramerization domain indicated. The box shows the loop region at a 6× magnification. See Fig EV1 for more information.
-
C, DLow‐angle Pt/C shadowing of Mis12:Ndc80 (panel C) and Mis12:Ndc80Δloop (Δ431–463) (panel D) complexes. The Mis12 complex appears as a 20 nm rod‐like extension and marks the SPC24:SPC25 side of the Ndc80 complex.
-
ESize exclusion chromatography coupled with multi‐angle light scattering (SEC‐MALS) profiles of fluorescently labeled Ndc80 and Ndc80Δloop. Calculated (and theoretical) masses are indicated. See Appendix Fig S1 for more information.
-
FTotal Internal Reflection Fluorescence (TIRF) microscopy was used to investigate Ndc80Alexa488 complexes on taxol‐stabilized microtubules that were attached to a passivated glass surface. Kymographs show Ndc80 complexes at a concentration of 0.2 nM with (FL, blue) or without (ΔL, orange) the loop. Scale bars: vertical (5 μm), horizontal (5 s).
-
GQuantification of Ndc80 residence times for data as in panel (F). Solid line represents a single exponential fit.
-
HOne‐dimensional diffusion of Ndc80 complexes (with n indicated) on microtubules. Traces were split into segments of 0.5 s and averaged. Mean values (circles) and SEM (shaded area) are shown.
-
IDistribution of the initial brightness of Ndc80 complexes on stabilized microtubules.
-
JTypical fields of view showing decoration of taxol‐stabilized microtubules (cyan) incubated with full‐length or loopless Ndc80 (yellow) at the indicated concentration. Images show an average projection of 200 frames. The contrast between individual fluorescent channels (inverted grayscale) was fixed. Auto‐contrast was used for the composite images to highlight the differences in the uniformity of Ndc80 decoration.
-
KDistribution of the brightness of Ndc80 complexes at indicated concentrations on taxol‐stabilized microtubules.
Source data are available online for this figure.
Here we demonstrate that direct interactions between full‐length Ndc80 complexes on microtubules are essential for the formation of stable kinetochore‐microtubule attachments, both in a fully reconstituted system and in dividing cells. These interactions are perturbed by deletion of the Ndc80 loop, and by point mutations therein. Our study provides insight into the organization of Ndc80 complexes at kinetochores attached end‐on to microtubules, a structural model of the Ndc80 loop, and a mechanistic explanation for impaired chromosome congression in cells with Ndc80 loop mutants.
Results
The Ndc80 loop folds rigidly against NUF2 and NDC80
To gain structural understanding of Ndc80, we predicted the structure of full‐length Ndc80 using AlphaFold‐Multimer (preprint: Evans et al, 2022; Fig 1B). Except for the unstructured tail of the NDC80 subunit, the structure of full‐length Ndc80 showed high to very high confidence scores (Fig EV1), thus accurately predicting the relative positions of NDC80, NUF2, SPC25, and SPC24, including in the tetramerization region in which the coiled coils of all four subunits overlap (Fig EV1; Valverde et al, 2016). Very high confidence scores were also attributed to the Ndc80 loop, a region comprising residues 421–450 of the NDC80 subunit and interrupting the long NDC80:NUF2 coiled coil. In the predicted structure, the loop connects two slightly staggered alpha‐helices of NDC80, both of which pair with the continuous NUF2 alpha‐helix (Fig 1B). The loop makes multiple turns, but does not bulge out freely from the helical axis of the complex as previously speculated (e.g., Ciferri et al, 2008). Rather, conserved amphipathic stretches pack closely and rigidly against the NUF2 and NDC80 helices. We hypothesize that two strongly conserved cysteine residues, NUF2C289 and NDC80C449, might further stabilize this structure through a disulfide bond. The loop region itself spans 30 residues in humans and other higher eukaryotes, but exceeds 60 residues in other species, indicating remarkable variation of composition and length.
Figure EV1. Structural in silico analysis of the human Ndc80 complex.
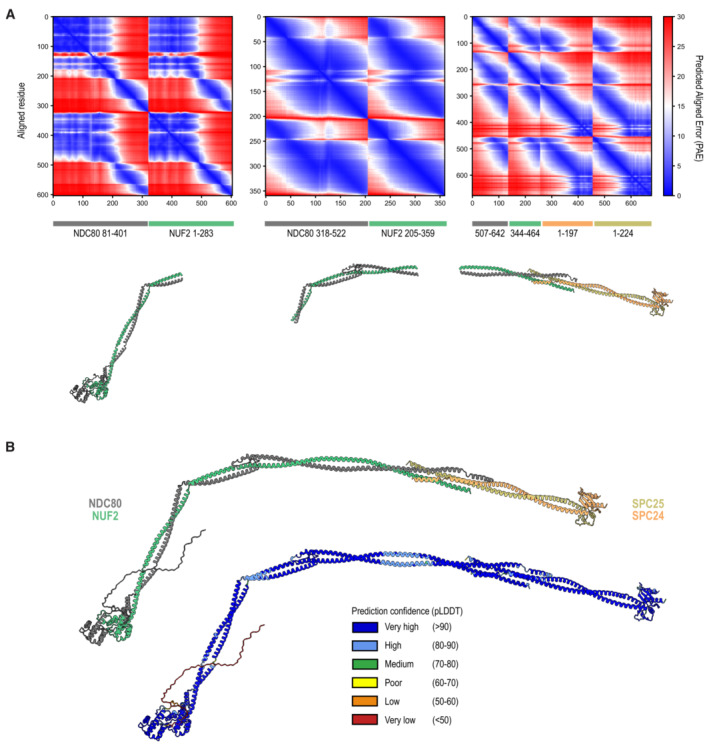
- Boundaries and Predicted Aligned Error (PAE) scores of the three Ndc80 segments that were predicted by AF2 multimer. These fragments were used to generate a composite prediction of the full‐length Ndc80 complex. More information can be found in the Materials and Methods section.
- The prediction of the full‐length Ndc80 complexes with colors representing the different subunits (as in Fig 1B) and the local prediction confidence intervals.
The loop of some species, such as Candida albicans and Dictyostelium discoideum, contain low‐complexity regions with stretches of hydrophilic residues. Despite these remarkably divergent sequences, the core folds of the predicted loop regions were comparable (Fig EV2). Thus, the loop is unlikely to be a point of bending along the Ndc80 coiled‐coils. The bending is likely to happen in a prominent hinge in the Ndc80 structure, predicted upstream from the loop around residues NDC80359–362 and NUF2244–247 (Fig 1B). The hinge is also conserved across eukaryotes and its position is in agreement with micrographs of full‐length Ndc80 complexes (Wang et al, 2008; Huis in 't Veld et al, 2016; Jenni & Harrison, 2018; Figs 1C and EV2).
Figure EV2. Alignments, phylogenetic tree, and structural conservation of the Ndc80 kink and loop.
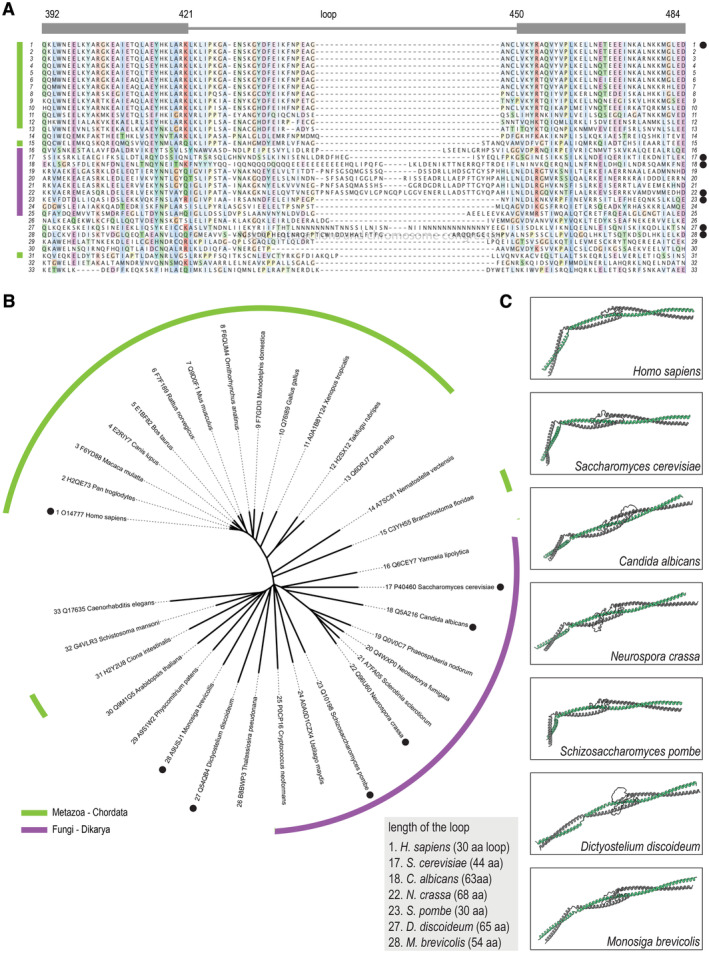
- Sequence alignment of the loop region of the NDC80 subunit in various species. Residue numbers correspond to the human NDC80.
- Unrooted phylogenetic tree that was generated with complete NDC80 sequences. Sequences in panel (A) were arranged according to this tree. Species belonging to the Chordata phylum (light green) and the Fungi kingdom (purple) are indicated. Black dots mark species for which we predicted the structure.
- Predicted structures of the NDC80:NUF2 region spanning the hinge and loop regions. Shown in similar orientations following structural alignment to the human fragment NDC80376–516:NUF2269–356.
Source data are available online for this figure.
To investigate the functional and structural relevance of the loop, we generated a recombinant human Ndc80 complex lacking NDC80 residues 431–463. Full‐length and loopless Ndc80 complexes (in complex with the Mis12 complex to mark one end) were indistinguishable in micrographs of rotary shadowed samples (Fig 1C and D). While the hinge was a prominent point of bending, both complexes displayed additional bending points along their length. SEC‐MALS and mass photometry further supported that full‐length and loopless Ndc80 complexes have comparable structural and biochemical properties (Fig 1D, Appendix Fig S1A–C). Using DSBU, a crosslinker that preferentially targets primary amines, but also reacts with proximal hydroxyls, we identified 190 unique crosslinks by mass spectrometry (Appendix Fig S2). Most crosslinks connect side‐chains that are less than 30 Å apart in our predicted structure and are consistent with previous analyses (Maiolica et al, 2007; Helgeson et al, 2018; Pan et al, 2018; Huis in 't Veld et al, 2019). High‐scoring crosslinks are predominantly found near the tetramerization domain and the Ndc80 loop and provide experimental support for the structural prediction (Appendix Fig S4). The model of the Ndc80 loop is in excellent agreement with the recently published crystal structure of the human NDC80370–509:NUF2252–347 fragment (Zahm et al, 2023).
Ndc80 complexes cluster on the microtubule lattice in a loop‐dependent manner
To test if the loop directly affects the binding of Ndc80 to microtubules, we labeled recombinant full‐length and loopless Ndc80 with AlexaFluor488 using Sortase (Hirakawa et al, 2015) and studied their binding to stabilized microtubules (Fig 1F). At sub‐nanomolar concentrations (0.2 nM), full‐length and loopless Ndc80 complexes typically resided on microtubules for 0.2–2 s with one‐dimensional diffusion (D) along the microtubule lattice of 0.10 and 0.09 μm2/s, respectively (Fig 1G–I). These values are in agreement with previously reported values for truncated Ndc80 bonsai complexes (Zaytsev et al, 2015) and budding yeast Ndc80 (Powers et al, 2009; Scarborough et al, 2019).
Full‐length and loopless Ndc80 complexes, however, bound microtubules in distinct ways at higher concentrations. Specifically, full‐length Ndc80 formed clusters on the microtubule lattice whereas loopless Ndc80 did not. The clusters increased from 3–6 Ndc80 complexes at 1.25 nM, to 5–10 complexes at 2 nM, and to 10–100 complexes at 6 nM (Fig 1J and K). Full‐length Ndc80 thus appears to bind microtubules preferably near sites already occupied by Ndc80, presumably through interactions between adjacent Ndc80 complexes. Since loopless Ndc80 binds uniformly along microtubules, we conclude that clustering of Ndc80 complexes upon microtubule binding requires an intact Ndc80 loop (Fig 1J and K). The clustering of Ndc80 is strictly microtubule‐dependent since neither full‐length Ndc80 complexes nor a NDC80:NUF2 fragment encompassing the loop form homotypic interactions in vitro in the absence of microtubules although concentrations as high as 50 μM (full‐length) and 500 μM (loop fragment) were reached (Fig 1E, Appendix Fig S3).
Loop‐dependent coordination of Ndc80‐microtubule binding
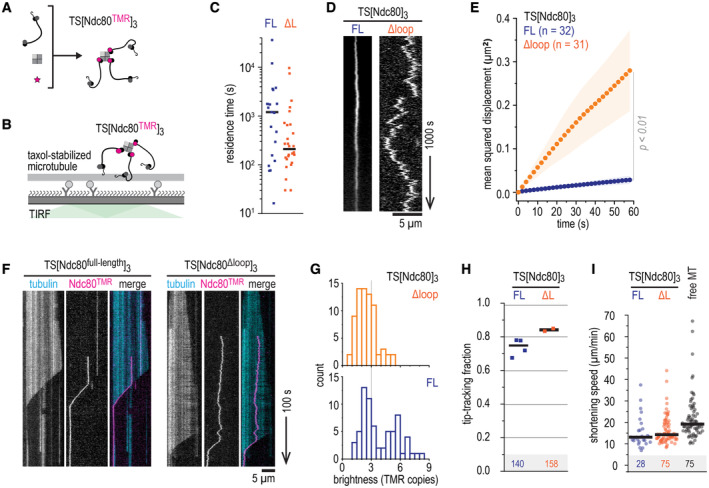
We previously used modules with three Ndc80 arms coupled to an engineered streptavidin‐derived scaffold to mimic the modular organization of Ndc80 in the outer kinetochore (Volkov et al, 2018; Huis in 't Veld et al, 2019). To understand the contribution of loop‐mediated Ndc80‐Ndc80 interactions in this context, we generated trimers of full‐length and loopless Ndc80 and studied their interactions with microtubules in vitro (Fig 2A and B, Appendix Fig S1D). Full‐length and loopless Ndc80 trimers resided stably on microtubules with residence times ranging from minutes to hours (medians of 1,000 and 200 s, respectively) (Fig 2C). Compared to the high diffusion rates of individual Ndc80 complexes on microtubules (Fig 1H), diffusion of a scaffold with three full‐length Ndc80 complexes was greatly reduced (D = 4.3 × 10−4 μm2/s), likely reflecting the presence of multiple microtubule‐binding elements in Ndc80 trimers. Interestingly, the Ndc80 loop contributed significantly to this reduction, because trimers of loopless Ndc80 diffused an order of magnitude faster (D = 36 × 10−4 μm2/s) (Fig 2D and E). Despite this difference, loopless Ndc80 trimers efficiently tracked the ends of shortening microtubules and reduced microtubule depolymerization rates, as previously observed for full‐length Ndc80 trimers (Fig 2F–I; Volkov et al, 2018; Huis in 't Veld et al, 2019). Thus, deletion of the Ndc80 loop reduced the stability of the interaction between Ndc80 trimers and the microtubule lattice, but did not grossly affect recognition of the microtubule plus end in the absence of applied forces.
Figure 2. The loop reduces diffusion of Ndc80 trimers without affecting their end‐tracking.
-
APreparation of TMR‐labeled, streptavidin‐mediated Ndc80 trimers. See Appendix Fig S1 for more information.
-
B–DTMR‐labeled Ndc80 trimers with (FL, blue) or without (ΔL, orange) the loop were added to taxol‐stabilized microtubules to measure their residence time (panel C) and one‐dimensional diffusion (panel D). Ndc80 trimers with and without the loop are shown. Scale bars: vertical (1,000 s), horizontal (5 μm).
-
EOne‐dimensional diffusion of Ndc80 trimers (with n indicated) on microtubules. Traces were split into segments of 60 s and averaged. Mean values (circles) and SEM (shaded area) are shown.
-
FKymographs of full‐length and loopless Ndc80 trimers (10 pM) that reside on dynamically growing and shortening microtubules. Trimers remain bound to the ends of shortening microtubules. Scale bars: vertical (100 s), horizontal (5 μm).
-
GDistribution of initial brightness of end‐tracking Ndc80 trimers with full‐length (blue) or loopless (orange) Ndc80.
-
HFraction of Ndc80 trimers that switches from lateral microtubule binding to tracking shortening ends. Data from four repeats (total 140 events) for full‐length Ndc80 trimers (blue), and two repeats (total 158 events) for loopless Ndc80 trimers (orange). Horizontal lines indicate average values.
-
IEnd‐tracking speed of full‐length (blue) and loopless (orange) Ndc80 trimers that follow shortening microtubule ends. Compared to Ndc80‐free shortening ends in the same field of views (gray). Horizontal lines indicate median values.
Source data are available online for this figure.
The loop is needed to stabilize end‐on Ndc80‐microtubule interactions under force
Next, we set out to compare how full‐length and loopless Ndc80 bind to the ends of shortening microtubules under force. We coated biotinylated glass beads with Ndc80 trimers and moved them with an optical tweezer to the lattice of a dynamic microtubule. When the microtubule started to depolymerize, we measured the forces exerted by the shortening end of the microtubule against the trapped bead (Fig 3A). The microtubule‐generated force typically increased until it matched the opposing force generated by the displacement of the bead from its rest position in the optical trap, a condition that stalled microtubule shortening. Such stalls were either followed by a detachment, or by a switch of the microtubule to a growing state (rescue). Detached beads rapidly moved back to the center of the trap. When a rescue allowed the bound microtubule to resume growth, beads retaining attachment to the microtubule moved back to the trap center much more gradually (Fig 3A).
Figure 3. The loop stabilizes end‐on Ndc80‐microtubule interactions under force.
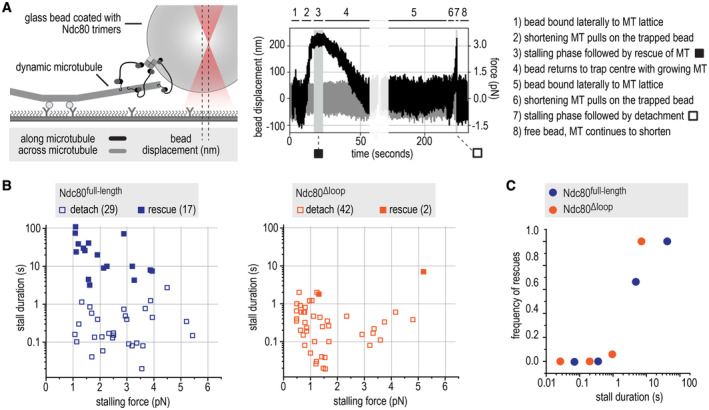
- Schematic of the optical trap experiment and a typical force trace. A glass bead coated with full‐length or loopless Ndc80 trimers is held in an optical trap near a microtubule end. The displacement of the bead (left Y axis) and the corresponding force (right Y axis) are shown along and across the microtubules axis (black and gray, respectively). Typical stages of an experiment (steps 1–8) are described.
- Correlations of stall duration and stall force. Each datapoint represents a single stall event. Filled squares: stalls resulting in microtubule rescue. Open squares: stalls resulting in bead detachment from the microtubule.
- The frequency of rescue events after a stall is plotted for full‐length (n = 46) and loopless (n = 44) Ndc80 after binning based on stall duration.
Source data are available online for this figure.
Full‐length Ndc80 trimers often stalled microtubule depolymerization for seconds and rescued microtubule shortening in 17 out of 46 cases (37%) (Fig 3B), in agreement with our previous experiments (Volkov et al, 2018; Huis in 't Veld et al, 2019). By contrast, beads coated with loopless Ndc80 trimers only rescued microtubule shortening in 2 out of 44 cases (5%) and typically detached from shortening microtubules after stalls that were shorter than 1 s (Fig 3B). We showed previously that stalls of microtubule depolymerization that last at least 1 s are much more likely to rescue microtubule shortening than shorter stalls, even with stall forces as low as 1 pN (Huis in 't Veld et al, 2019). This relation was also clearly observed with the measurements reported here, as the majority of stalls that led to rescues in presence of full‐length Ndc80 lasted more than 1 s, whereas most stalls with loopless Ndc80 were short and followed by a detachment (Fig 3B and C). Full‐length Ndc80 trimers bound to the biotinylated glass beads more efficiently than loopless trimers, possibly because the loop‐mediated Ndc80‐Ndc80 interactions described above facilitate coating. Nonetheless, beads coated with high amounts of loopless Ndc80 also failed to efficiently stall and rescue shortening microtubules (Appendix Fig S1E and F). Taken together, our single‐molecule studies suggest that loop‐mediated interactions coordinate Ndc80 complexes into a non‐diffusive microtubule‐binder that forms load‐bearing attachments with the ends of microtubules. Since a single kinetochore contains many copies of Ndc80, the loop‐mediated organization of adjacent Ndc80 complexes might be important for the attachment of kinetochores to the dynamic microtubules of the mitotic spindle.
The Ndc80 loop is essential for proper chromosome congression
To investigate how the loop influences kinetochore‐microtubule interactions in mitotic cells, we electroporated recombinant Ndc80 complexes into HeLa cells stably expressing mCherry‐H2B and depleted of endogenous Ndc80 by RNAi. This approach directly assesses if recombinant protein complexes support cell division (Alex et al, 2019). We used live‐cell imaging to follow fluorescently labeled recombinant Ndc80 complexes 18 h after they were delivered into cells (Fig 4A). Endogenous Ndc80 complexes were efficiently depleted upon exposure to a siRNA combination targeting NDC80, SPC25, and SPC24 (Fig 4B). Depletion of Ndc80 ablated the spindle assembly checkpoint (SAC) and caused cells to exit mitosis in presence of the microtubule poison nocodazole, in line with previous work (Fig 4C; McCleland et al, 2003; Kim & Yu, 2015). When recombinant full‐length Ndc80 complexes were delivered into cells depleted of endogenous Ndc80, they restored the cell cycle arrest in nocodazole‐treated cells (Fig 4C). These experiments demonstrate the efficient depletion of endogenous Ndc80 and the functionality of the electroporated recombinant Ndc80 complex.
Figure 4. Mutation of critical residues in the loop impairs chromosome congression.
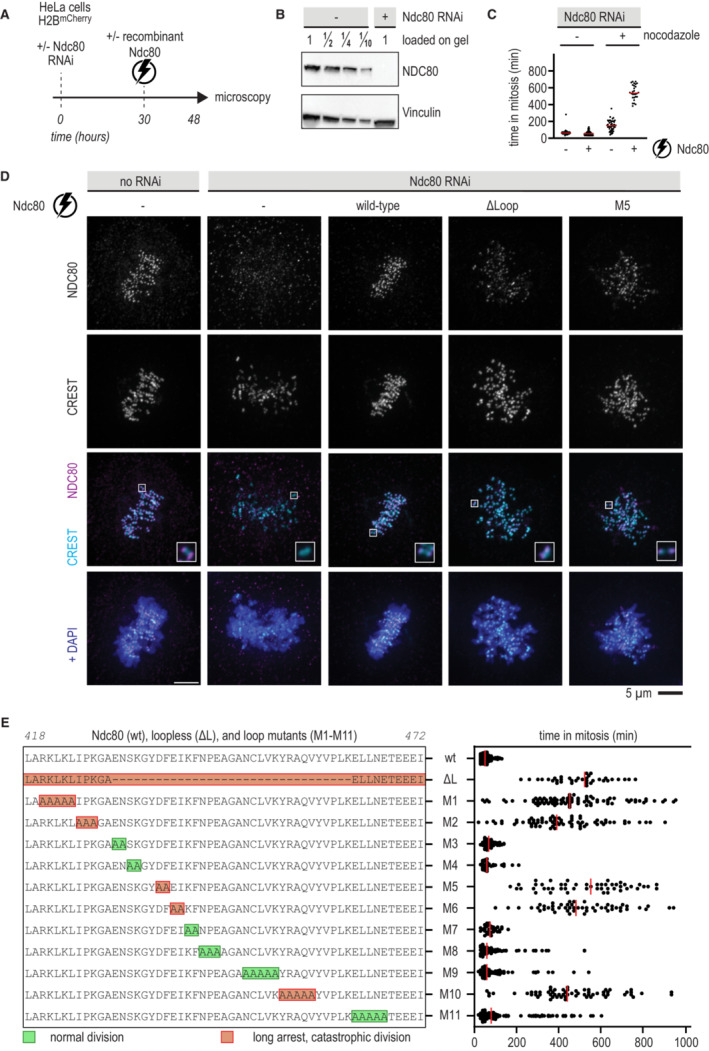
- Schematic of an electroporation experiment.
- Immunoblot of NDC80 levels following depletion of the Ndc80 complex by RNAi.
- Quantification of the time that cells spent in mitosis following various treatments. Each dot represents a cell and the red lines indicate median values. Nocodazole was added 17 h after electroporation and 1 h before microscopy. A minimum of 30 cells were analyzed for each condition.
- Immunofluorescence microscopy of mitotic cells stained for DNA (DAPI), kinetochores (CREST), and Ndc80 complexes. The Ndc80 antibody (9G3) detects endogenous Ndc80 (column 1) and electroporated recombinant Ndc80 (columns 3, 4, 5). Representative cells with a metaphase plate or uncongressed chromosomes are shown. Scale bar: 5 μm.
- Overview of mutations in the Ndc80 loop region and the effects on the time spent in mitosis following the experimental setup outlined in panel (A). Colors indicate whether cells divided normally, sometimes with delayed chromosome congression (green), or showed long arrests followed by a catastrophic division (orange). Each dot represents a cell and the red lines indicate median values. Mutation NDC80G434A‐Y435A did not support the formation of stable and soluble Ndc80 complexes. A minimum of 30 cells were analyzed for each condition.
Source data are available online for this figure.
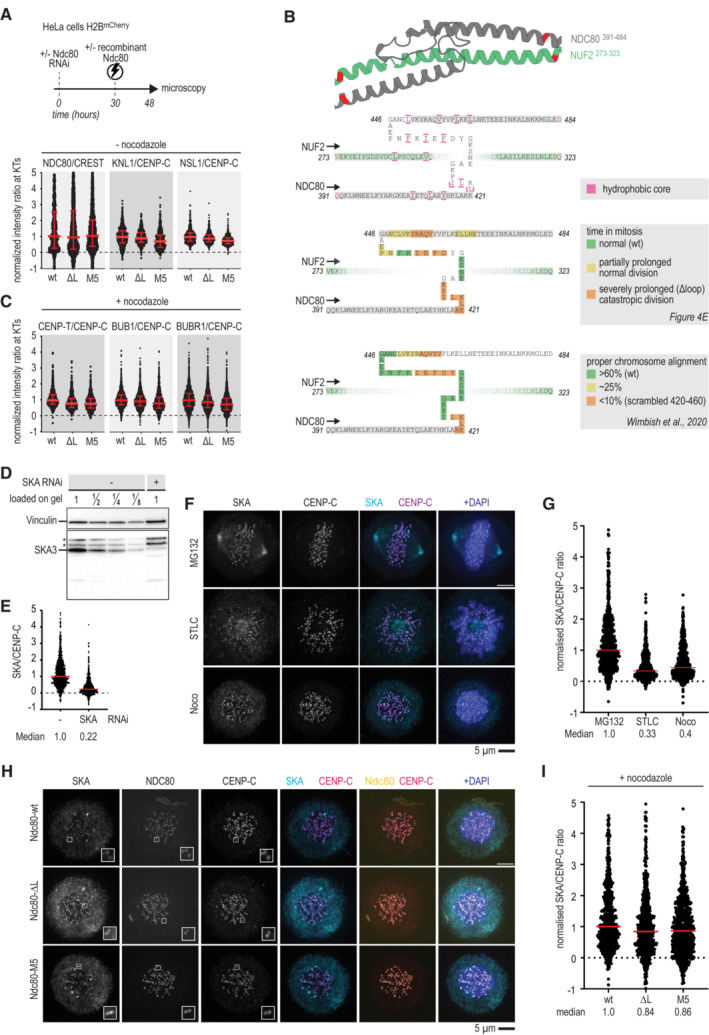
Using this assay, we established that both full‐length and loopless Ndc80 complexes were efficiently recruited to kinetochores in cells that lack endogenous Ndc80 (Fig EV3A). Whereas full‐length recombinant complexes supported the timely formation of a metaphase plate and faithful cell division (Fig 4D, 3rd column), cells electroporated with loopless complexes failed to congress their chromosomes to the midplane and remained in a prometaphase‐like state without ever fully forming a recognizable metaphase plate (Fig 4D, 4th column). Thus, the loop region is essential for chromosome alignment and bi‐orientation, as previously shown (Zhang et al, 2012; Shrestha & Draviam, 2013; Wimbish et al, 2020). In agreement with these previous studies, cells with loopless Ndc80 complexes arrested in mitosis for many hours (Fig 4E). These results indicate that cells recruiting the loopless Ndc80 mutant are impaired in chromosome congression and bi‐orientation, but sustain SAC signaling.
Figure EV3. Electroporation efficiency and a comparison of loop mutants.
-
AImmunofluorescence quantification of Ndc80, KNL1, and NSL1 at kinetochores following electroporation of different Ndc80 constructs. The number of kinetochores analyzed for NDC80: wt ‐ 758, ∆L ‐ 2,053, M5 ‐ 1,612, KNL1: wt ‐ 697, ∆L ‐ 635, M5 ‐ 602, and NSL1: wt ‐ 647, ∆L ‐ 644, M5 ‐ 611. Red lines indicate median value with interquartile range.
- B
-
CImmunofluorescence quantification of various kinetochore proteins in cells that were treated as described in panel (A), but with nocodazole added 15 h after electroporation and 3 h before fixation. The number of kinetochores analyzed for CENP‐T: wt ‐ 799, ∆L ‐ 750, M5 ‐ 855, BUB1: wt ‐ 2,226, ∆L ‐ 1,993, M5 ‐ 1,598, and BUBR1: wt ‐ 2,315, ∆L ‐ 2,112, M5 ‐ 1,549. Red lines indicate median value with interquartile range.
-
DA new anti‐SKA antibody, generated against the full‐length recombinant SKA complex, mainly recognizes SKA3 in a HeLa cell lysate using immunoblotting. Asterisks indicate non‐specific bands that are not sensitive to depletion of the SKA complex by RNAi.
-
E–GThe antibody also recognizes SKA by immunofluorescence. SKA levels are higher in MG132 arrested cells than in STLC or nocodazole arrested cells. Scale bar: 5 μm.
-
H, ISKA levels recruited to kinetochores in nocodazole‐exposed cells with various Ndc80 complexes. Scale bar: 5 μm.
Source data are available online for this figure.
To dissect the properties of the NDC80 loop in more detail, we generated recombinant full‐length Ndc80 complexes with residues of the loop mutated into Alanine. A complex carrying the NDC80G434A‐Y435A mutations was insoluble, but 11 other complexes were stable and purified to homogeneity. After adding a small fluorescent dye to these complexes, we electroporated them into cells depleted of endogenous Ndc80 complex (Fig 4E). Wild‐type Ndc80 complexes and mutants 3, 4, and 7 supported chromosome congression and segregation similarly to wild‐type Ndc80. Mutants 8, 9, and 11 also supported proper chromosome segregation, albeit often after a prolonged phase of chromosome congression. By contrast, mutants 1, 2, 5, 6, and 10 phenocopied the prometaphase‐like arrest observed with the loopless Ndc80, indicating that these mutations profoundly impair loop function (Fig 4E). The loop mutant NDC80D436A‐F437A (M5) phenocopied the deletion of the entire loop in a chromosome alignment assay (Fig 4D, 5th column). Since a role of residues NDC80436–439 in chromosome congression was also reported in a previous study (Wimbish et al, 2020; Fig EV3B), we chose to include Ndc80‐M5 for further analyses. The composition of the outer kinetochore, as assayed by quantifying the staining for components KNL1 and NSL1, was essentially identical in cells with Ndc80 full‐length, loopless, or M5. Furthermore, kinetochores with different Ndc80 mutants recruited comparable amounts of the SKA complex and SAC components in the presence of nocodazole (Fig EV3C–I).
The M5 mutant phenocopies loopless Ndc80
To further test how the Ndc80 loop promotes kinetochore‐microtubule interactions in vivo, we exposed cells electroporated with wild‐type or mutated Ndc80 to a cold shock before fixation for immunofluorescence microscopy (Fig 5A). Properly formed metaphase plates and cold‐stable kinetochore‐microtubule fibers were observed in cells with wild‐type Ndc80, but not in cells with loop mutants (Fig 5A). The frequent observation of multipolar spindles in cells with Ndc80 loop mutations further supports the idea that the loop is required for stable kinetochore‐microtubule interactions. We therefore investigated the propensity of full‐length and loopless Ndc80 monomers and trimers to cluster on microtubules. Wild‐type Ndc80 monomers, at a concentration of 0.6 nM, interacted with microtubules markedly longer when they colocalized with wild‐type Ndc80 trimers, suggesting an interaction that prolongs their residence on microtubules. Monomers of loopless Ndc80, on the other hand, were indifferent to the presence of wild‐type trimers and resided on microtubules like wild‐type monomers in the absence of trimers (Fig EV4A and B).
Figure 5. The M5 mutant phenocopies loopless Ndc80 in vivo and in vitro .
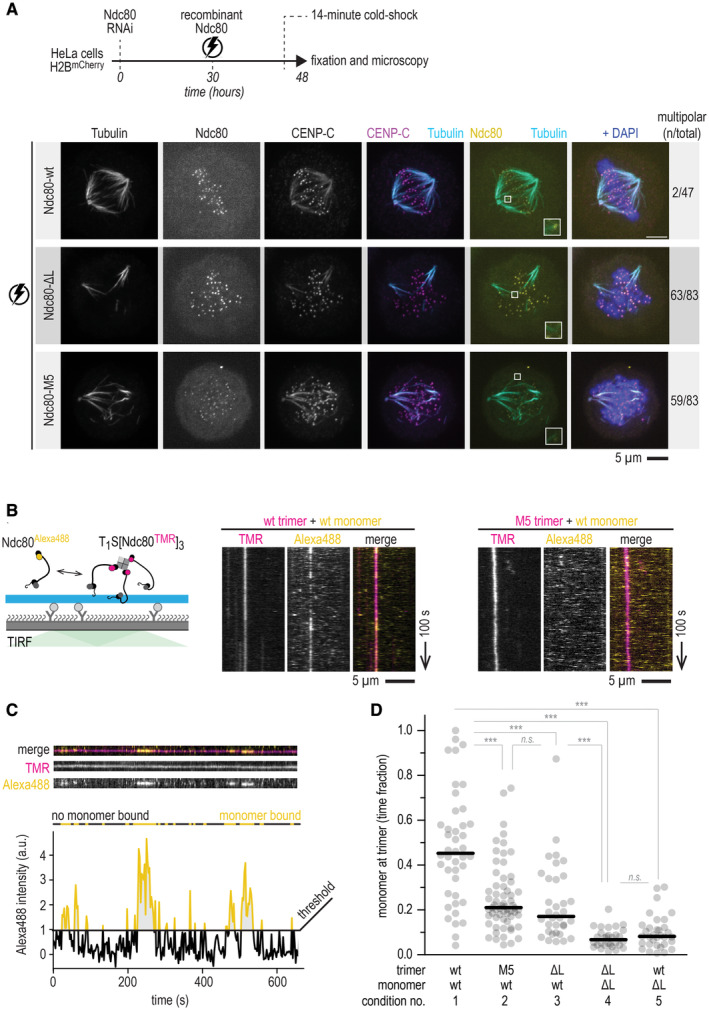
- Schematic of a cold‐shock assay following an electroporation experiment. Immunofluorescence images showing the attachment status of kinetochores to microtubules in cells electroporated with recombinant Ndc80‐wt, Ndc80‐∆L, or Ndc80‐M5 complexes. The number of cells with multipolar spindles and the total number of analyzed cells are shown. Some signal from the tubulin channel is visible in the CENP‐C channel. Scale bar: 5 μm.
- Total Internal Reflection Fluorescence (TIRF) microscopy was used to investigate Ndc80Alexa488 complexes (0.6 nM) added to trimeric Ndc80TMR (10 pM) on fluorescent taxol‐stabilized microtubules that were attached to a passivated glass surface. Typical kymographs showing virtually motionless Ndc80 trimers (magenta) and transiently binding Ndc80 monomers (yellow). Wild‐type (wt) monomers associate with wt trimers (left), but not with M5 trimers (right). Scale bars: vertical (100 s), horizontal (5 μm).
- Quantification of the intensity of the monomeric Ndc80 associating with microtubule‐bound trimeric Ndc80. A threshold for binding was set at an intensity equivalent to one Alexa488 copy. Intensities well above 1 (yellow) could thus reflects multiple monomers binding simultaneously.
- Fraction of time there was at least one monomer (added to solution at a concentration of 0.6 nM) present at the microtubule‐bound trimer (10 pM), tested in various combinations of wild‐type (wt), loopless (ΔL) and M5 monomers or trimers. All analyzed traces of Ndc80 trimers are shown (n = 42, 63, 33, 30, 34 for conditions 1–5). Horizontal lines show median values and statistical significance was determined using a two‐tailed Mann–Whitney test. P‐values: 1 (wt‐trimer + wt‐monomer) vs. 2 (M5‐trimer + wt‐monomer): 7·10−7 (***); 2 vs. 3: 0.17 (n.s.); 1 vs. 3: 1·10−6 (***); 1 vs. 4: 3·10−15 (***); 3 vs. 4: 1·10−8 (***); 1 vs. 5: 1·10−13 (***); 4 vs. 5 0.23 (n.s.).
Source data are available online for this figure.
Figure EV4. Loop‐dependent binding between Ndc80 monomers and trimers on microtubules.
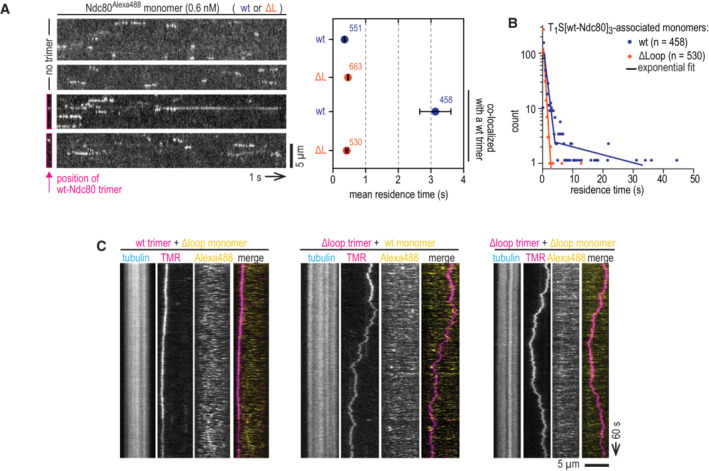
- Supplementary information for Fig 5A–D. High‐speed recordings to quantify residence time of wild‐type and loopless Ndc80 monomers. The top two kymographs show a microtubule with monomers binding and unbinding. The lower two kymographs show the binding and unbinding of Ndc80 monomers to a microtubule with a Ndc80 trimer. Since trimers are practically motionless on this timescale, only their initial location was recorded and indicated. Corresponding mean residence times and SEM. are shown. The number of analyzed events is indicated. Scale bar: 5 μm.
- Distribution of residence times of wild‐type and loopless Ndc80 monomers associating with microtubule‐bound Ndc80 trimers. A single‐exponential fit described the residence time of Ndc80Δloop, likely corresponding to Ndc80Δloop:microtubule off‐rates. Residence time of full‐length Ndc80 could be fitted with two exponents, likely corresponding to Ndc80:microtubule and Ndc80:trimer:microtubule off‐rates.
- Typical kymographs showing Ndc80 trimers (magenta) and transiently binding Ndc80 monomers (yellow). Scale bars: vertical (100 s), horizontal (5 μm).
Source data are available online for this figure.
We then analyzed the microtubule residency time of various combinations of wild‐type or mutant trimeric Ndc80TMR and monomeric Ndc80Alexa488. Wild‐type trimers bound wild‐type monomers significantly longer than M5 and loopless trimers (Fig 5B–D, compare conditions 1–3). This fraction was further reduced for loopless monomers recruited to wild‐type trimers or to loopless trimers (Fig 5B–D, conditions 4–5, and Fig EV4C). These experiments confirm that the Ndc80 loop directly contributes to Ndc80‐Ndc80 interactions on microtubules and that residues NDC80D436‐F437 contribute significantly.
Loop‐proximal Ndc80‐Ndc80 crosslinking can rescue loop deletion in vitro
Our experiments demonstrated that multivalent Ndc80 requires the loop to form load‐bearing attachments to microtubule‐ends in vitro and in vivo, presumably by clustering individual microtubule‐binding elements into a robust microtubule‐binding unit. To test this model further, we set out to crosslink Ndc80 arms artificially using polyclonal antibodies that recognize short regions adjacent to or directly in the loop (NDC80410–432/AB‐849 and NDC80429–450/AB‐850, respectively) (Figs 6A and EV5A).
Figure 6. Loop‐proximal Ndc80‐Ndc80 crosslinking rescues increased diffusivity of loopless Ndc80 trimers.
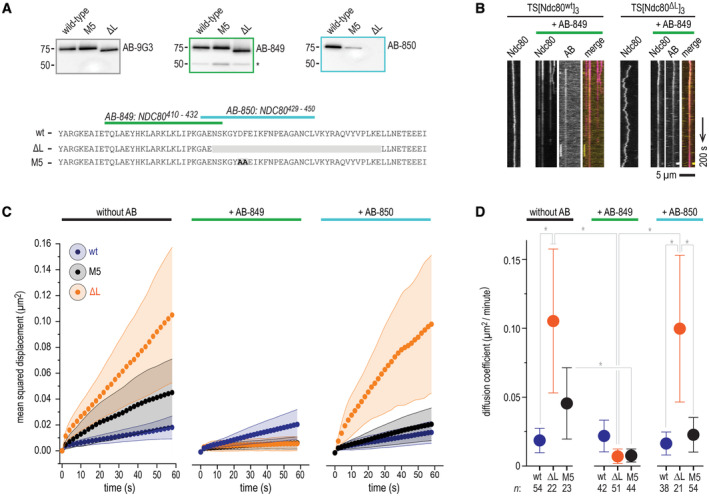
- Representation of the peptides used to raise AB‐849 and AB‐850 and immunoblots showing their recognition of wild‐type, M5 and loopless Ndc80 complexes. 9G3 is a commercially available monoclonal antibody raised against NDC8056–642, later shown to recognize NDC80200–215. Asterisk shows the non‐specific recognition of another protein, presumably NUF2.
- Diffusion of full‐length and loopless Ndc80 trimers in absence and presence of AB‐849. The primary rabbit polyclonal AB‐849 was detected using a Alexa650‐labeled anti‐rabbit secondary IgG antibody. Scale bars: vertical (100 s), horizontal (5 μm).
- One‐dimensional diffusion of full‐length (blue), loopless (orange), and M5 (black) Ndc80 trimers in presence and absence of AB‐849 and AB‐850 as described in panel (B) (see Appendix Fig S4 for more information). Traces of Ndc80 trimers (with n indicated in the legend for panel D) on microtubules were analyzed. Traces were split into segments of 60 s and averaged (see Materials and Methods). Mean (circles) and SEM (shaded areas) values are shown. We note that the omission of reducing agents, a precondition to use the antibody as a crosslinker, slightly decreased the overall diffusion of Ndc80‐modules on microtubules (compare with Fig 2E).
- Summary showing diffusion coefficients (μm2/min) that follow from the data shown in panel (C). Mean values, SEM, and number of diffusion traces (n) are indicated. Statistically significant differences were determined using a two‐tailed t‐test. P‐values: no AB, FL vs. ΔL: 0.0180 (*); ΔL, no AB vs. AB‐849: 0.0108 (*); M5, no AB vs. AB‐849: 0.0458 (*); ΔL, AB849 vs. AB‐850: 0.0156 (*); AB‐850, ΔL vs. FL: 0.0461 (*); AB‐850, ΔL vs. M5: 0.0494 (*).
Source data are available online for this figure.
Figure EV5. Characterization of AB‐849 and AB‐850 in vitro .
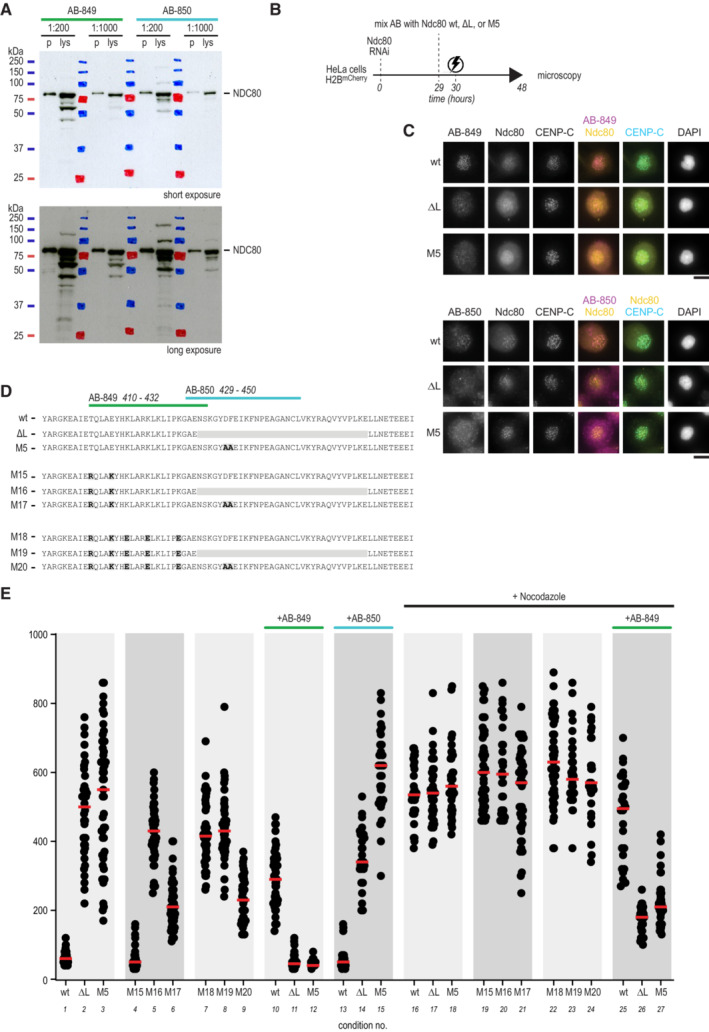
- AB‐849 and AB‐850 recognize NDC80 in a HeLa cell lysate (lys) using immunoblotting. Recombinant Ndc80 complex (p) was used as a reference (see Fig 6A) Antibody dilutions are indicated and short (top panel) and long (bottom panel) exposures are shown.
- Experimental workflow to electroporate Ndc80 complexes with AB‐849 or AB‐850 into cells.
- The localization of AB‐849, AB‐850, NDC80, CENP‐C, and DNA was analyzed using immunofluorescence microscopy. Representative cells are shown. Scale bar: 10 μm.
- Overview of mutants M15–M20, mutated in the epitope region of AB‐849 to further test putative effects of the AB on the SAC.
- Time spent in mitosis in the presence or absence of nocodazole. Cells were electroporated with various Ndc80 constructs, when indicated following mixture with AB‐849 or AB‐850. Every dot represents a cell and red lines indicate median values. Effects of AB‐849 on the time spent in mitosis were not recapitulated by any of the mutants M15–M20.
Source data are available online for this figure.
We first analyzed if antibodies altered the behavior of Ndc80 complexes on microtubules in vitro. Experiments with a fluorescently labeled secondary antibody confirmed that the loop‐directed antibodies bind Ndc80 trimers on microtubules (Fig 6B, Appendix Fig S4). The addition of AB‐849, which binds close to the loop region, to loopless trimers greatly reduced their diffusion on microtubules. Such an effect was not observed with AB‐850, which does not bind loopless Ndc80 (Fig 6C and D). Artificial Ndc80‐Ndc80 crosslinking near the loop is thus sufficient to reduce the diffusion of Ndc80 loop mutants on microtubules to wild‐type levels in a fully reconstituted system, further indicating that the loop contributes to the cooperative binding of Ndc80 ensembles to microtubules. We note however that co‐electroporating AB‐849 with Ndc80 loop mutants did not restore chromosome congression in cells (Fig EV5B–E), likely because the antibody does not recapitulate the clustering process faithfully or because it prevents further maturation of the attachment. AB‐849 also partly impaired SAC signaling, in particular when used in combination with loop mutants. However, we could not recapitulate this effect by mutating key residues in the AB‐849 epitope (M15–M20) (Fig EV5E).
Loop‐mediated mitotic arrest involves multiple phospho‐signaling pathways
To prevent chromosome mis‐segregation, a dividing cell needs to destabilize occasional syntelic and merotelic kinetochore‐microtubule misattachments. This error correction process requires Aurora B, a kinase with multiple substrates in the kinetochore (Lampson & Grishchuk, 2017). The 80‐residue unstructured N‐terminal tail of the NDC80 subunit includes several serine and threonine residues whose phosphorylation by Aurora B renders kinetochore‐microtubule attachment reversible and is critical for error correction (Krenn & Musacchio, 2015). Inhibition of multi‐site phosphorylation of the Ndc80 tail results in hyperstable kinetochore‐microtubule attachments (DeLuca et al, 2006; Guimaraes et al, 2008).
We asked if preventing phosphorylation of the NDC80 tail had the potential to override the mitotic arrest caused by mutations in the loop region. We therefore combined nine Alanine (9A) mutations in the tail (preventing its phosphorylation) with either loopless or M5 mutants. Although combining the 9A Ndc80 tail with loop mutants reduced the duration of the mitotic arrest (Fig 7, compare conditions 10 with 13 and 16 with 19), cells arrested nonetheless for several hours. Thus, artificially stabilizing microtubule attachment by preventing phosphorylation of the Ndc80 did not bypass the spindle assembly checkpoint arrest caused by tampering with the Ndc80 loop. This suggests that Ndc80 clustering has a more fundamental role in establishing stable connections with microtubules, possibly one that precedes an assessment of the quality of the attachment by the error correction pathway.
Figure 7. Synergistic contributions of the Ndc80 loop and tail to kinetochore‐microtubule binding and SAC silencing.
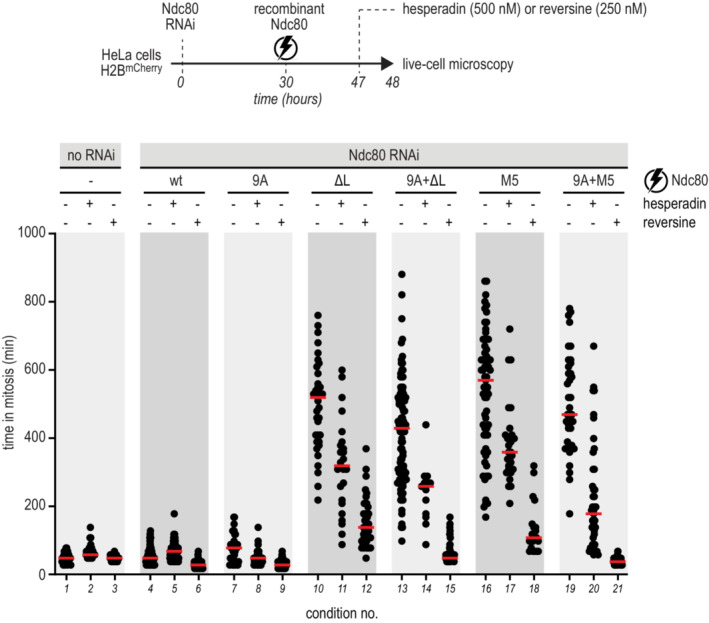
Experimental workflow to investigate the time that cells, electroporated with recombinant Ndc80 complexes, spent in mitosis in the presence and absence of mitotic kinases inhibitors. Every dot represents a cell and red lines indicate median values. At least 30 cells were analyzed for each condition.Source data are available online for this figure.
To test this hypothesis further, we exposed cells with mutations in the Ndc80 tail and/or loop to inhibitors of the mitotic kinases Aurora B and Mps1 (Krenn & Musacchio, 2015; Saurin, 2018). As expected, the addition of Hesperadin, a small molecule inhibitor of Aurora B kinase (Hauf et al, 2003), shortened mitotic arrest in cells electroporated with Ndc80 loop mutants (Fig 7, compare conditions 10 with 11 and 16 with 17). This shortening was much more evident than that caused by the Ndc80 9A error correction mutant, suggesting partial SAC abrogation, in addition to overriding the error correction pathway. This interpretation was corroborated by testing the effects of Reversine, a specific and potent inhibitor of the SAC kinase MPS1 (Santaguida et al, 2010). Reversine dramatically shortened the time that loop‐deficient cells spent in mitosis (Fig 7, conditions 12 and 18). This effect was exacerbated by the 9A mutation, likely because the enhanced microtubule binding by this mutant further facilitated checkpoint overriding by Reversine (Fig 7, compare conditions 12 with 15 and 18 with 21). This result confirms that hyperstabilization of kinetochore‐microtubule interactions with a Ndc80 tail mutant does not silence (or satisfy) the SAC, as previously suggested (Etemad et al, 2015; Tauchman et al, 2015), but rather contributes to its overriding.
NDC80 D436 and E438 promote Ndc80‐Ndc80 interactions and chromosome congression
The loop mutant M5 that by and large phenocopied the deletion of the entire loop is mutated at NDC80 D436 and F437. The side chains of these adjacent residues point in opposite directions: the acidic D436 is solvent accessible whereas F437 contributes to the hydrophobic core of the loop (this study and Zahm et al, 2023). A similar pattern is observed in loop mutant M6, mutated at residues E438 and I439. To unravel if buried hydrophobic (F437 and I439) or exposed acidic residues (E436 and D438) are required for mitotic fidelity, we generated two new mutants. Whereas cells with Ndc80F437A‐I439A (M14) congressed their chromosomes and divided successfully, cells with Ndc80D436A‐E438A (M13) did not (Fig 8A).
Figure 8. NDC80 D436 and E438 promote Ndc80 clustering on microtubules and chromosome congression.
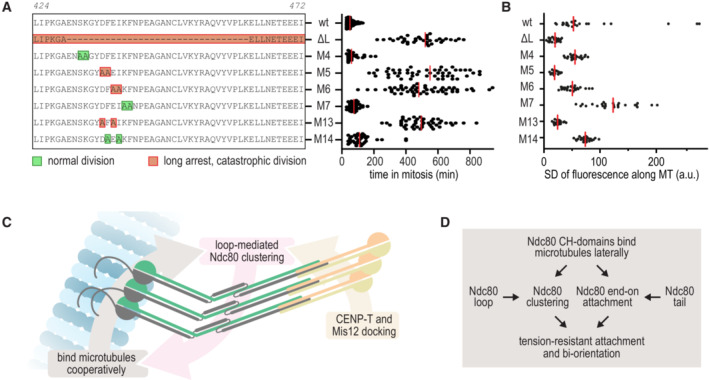
- Effects of M13 and M14 on the time spent in mitosis following the experimental setup outlined in Fig 4. All other datapoints are also shown in Fig 4E. Colors indicate whether cells divided normally, sometimes with delayed chromosome congression (green), or showed long arrests followed by a catastrophic division (orange). Each dot represents a cell and the red lines indicate median values. A minimum of 30 cells were analyzed for each condition.
- A model for the synergistic contributions of the Ndc80 loop and the Ndc80 tail to robust kinetochore‐microtubule attachments.
- A schematic representation of loop‐mediated interactions that stabilize Ndc80‐Ndc80 interactions when Ndc80 complexes align following their anchoring on multivalent receptors at both ends.
Source data are available online for this figure.
We next tested if the ability of Ndc80 mutants to support chromosome congression correlates with their loop‐dependent propensity to cluster on the microtubule lattice at low nanomolar concentrations in a reconstituted system (Fig 1). Out of eight different Ndc80 complexes, all purified to homogeneity and not showing clustering in solution at concentrations up to 50 μM, only mutants ∆Loop, M13 and, to a lesser extent, M5, did not cluster on the microtubule lattice. This experiment demonstrated the correlation between homotypic Ndc80‐Ndc80 clustering on microtubules and stable kinetochore‐microtubule interaction during mitosis and highlighted a contribution of the loop's acidic patch formed by D436 and E438 (Fig 8A, Appendix Fig S5).
Discussion
The loop region of the NDC80 subunit was previously identified for its essential role in the establishment of load‐bearing interactions between kinetochores and microtubule ends required for chromosome congression (Maure et al, 2011; Shrestha & Draviam, 2013). Our finding in a minimally reconstituted system that the loop is dispensable for microtubule end‐tracking in the absence of force, but crucial to stall and rescue microtubule shortening under force, supports this view (compare Figs 2H and I, and 3B). At a mechanistic level, however, the function of the loop has remained elusive and partly controversial, as discussed below. We reconstituted the interaction between microtubule ends and Ndc80 complexes and demonstrate that the loop is crucial to generate force‐resistant attachments. Loop‐dependent clustering of Ndc80 complexes requires binding to microtubules: full‐length Ndc80 complexes are stable monomeric complexes in solution at concentrations as high as 50 μM (~10 mg/ml) but cluster on microtubules at concentrations as low as 0.005 μM (Figs 1 and 8B). This suggests that the effects of loop‐mediated low affinity homotypic Ndc80‐Ndc80 interactions that only become manifest after microtubule binding. In this model, Ndc80 complexes (i) multimerize on their kinetochore receptors CENP‐T and Mis12, (ii) align their CH‐domains and tails on a microtubule, and (iii) stabilize this arrangement cooperatively through loop‐mediated homotypic interactions (model in Fig 8C and D). Although our structural analysis provides insight into the folding of the Ndc80 loop, a molecular explanation for loop‐dependent clustering of Ndc80 complexes on microtubules is lacking. Since point mutations phenocopy deletion of the loop both in dividing cells and in a reconstituted system, it appears likely that the loop of a microtubule‐bound Ndc80 complex directly binds an adjacent Ndc80 complex at a hitherto unidentified site.
The unstructured N‐terminal tail of the NDC80 subunit, encompassing the protein's first 80 residues, plays an important role in the coordinated binding of Ndc80 to the ends of dynamic microtubules in vivo and in vitro (Cheeseman et al, 2006; DeLuca et al, 2006, 2011, 2018; Guimaraes et al, 2008; Miller et al, 2008; Zhu et al, 2013; Long et al, 2017; Shrestha et al, 2017; Helgeson et al, 2018; Huis in 't Veld et al, 2019; Wimbish et al, 2020; Kucharski et al, 2022). We suggest that the Ndc80 tail and loop contribute to the establishment of tension‐resistant kinetochore‐microtubule attachments in a synergistic manner: the tails by coordinating the direct binding of Ndc80 complexes to the end of microtubules and the loops by stabilizing Ndc80 clusters at a distance from the microtubule‐binding site (model in Fig 8C). Cooperative microtubule binding of Ndc80 assisted by the Ndc80 tail has been demonstrated previously (Ciferri et al, 2008; Alushin et al, 2010, 2012; Janczyk et al, 2017). Notably, these studies were performed with a truncated version of the Ndc80 complex, Ndc80‐bonsai, that lacks the loop and almost the entire coiled‐coil stalk of the complex, and using Ndc80‐bonsai concentrations in the low μM range. In contrast, we observe clustering on microtubules of full‐length Ndc80 complexes at low nM concentrations. At similarly low concentrations, Ndc80‐bonsai (with or without mutations in the tail) binds microtubules in a non‐cooperative manner (Zaytsev et al, 2015).
A challenge of any reconstituted system is that it may fall short of reproducing all the layers of regulation that contribute to a biological process. With this limitation in mind, we nevertheless formulate a hypothesis on the coordination of molecular events that mark the process of bi‐orientation. Based on our findings, we surmise that force‐resistant microtubule binding by Ndc80 complexes, at its kinetochore concentration or at a realistically low concentration in vitro, requires (i) the deployment of Ndc80 loops to trigger local clustering and (ii) the deployment of Ndc80 tails at the end of the microtubule, facilitated by their dephosphorylation (Fig 8D). This model predicts that the Ndc80 complexes are oriented as parallel “pillars”, compatible with the high nematic order observed for Ndc80 in vivo (Roscioli et al, 2020). The model may also explain why hyperstabilization of kinetochore‐microtubule interactions with a non‐phosphorylatable version of the Ndc80 tail cannot rescue the dramatic effects on bi‐orientation caused by deletion of the loop: N‐terminal tails may be insufficient for clustering and coordinated microtubule binding when the loop is absent. A question for future work is how many adjacent CH‐domains and tails are required to generate a robust end‐on microtubule attachment. Furthermore, our AF2 predictions and structural analyses suggest that additional conserved elements of the Ndc80 complex may contribute to successful bi‐orientation. Most notably, and as previously observed in micrographs and in predictions, there is a prominent disruption of the coiled‐coil between the microtubule‐binding region and the loop of Ndc80 (Fig 1B and C; Jenni & Harrison, 2018; Zahm et al, 2023). We propose to name this conserved element, encompassing NDC80359–362 and NUF2244–247 in humans, the Ndc80 hinge. How the hinge contributes to Ndc80‐microtubule binding and chromosome biorientation remains to be addressed. Our analysis, however, indicates that the hinge, not the loop, is the likely primary site of bending in the first half of the Ndc80 shaft.
Importantly, the establishment of force‐bearing kinetochore‐microtubule attachments in vivo requires, in addition to the Ndc80 complex, additional microtubule binders, including the SKA and SKAP/Astrin complexes (Monda & Cheeseman, 2018). It is therefore possible that Ndc80 multivalency, in addition to having a direct effect on microtubule binding, also controls the interaction with these additional linkages. Indeed, a crucial role of the loop in kinetochore‐microtubule attachment was previously demonstrated in a range of organisms (Hsu & Toda, 2011; Maure et al, 2011; Varma et al, 2012; Zhang et al, 2012; Shrestha & Draviam, 2013; Tang et al, 2013; Scarborough et al, 2019; Wimbish et al, 2020). These studies postulated that the loop promotes stable kinetochore‐microtubule attachment through the direct binding of microtubule binding proteins such as the Ska complex, CH‐TOG/Stu2, or Cdt1 (Hsu & Toda, 2011; Varma et al, 2012; Zhang et al, 2012, 2017; Tang et al, 2013). Subsequent studies, however, questioned the notion that the loop is directly involved in these physical interactions. For instance, recruitment of Dam1 to Ndc80 complexes in budding yeast appears to require the Ndc80 loop in vivo, but not in vitro (Maure et al, 2011; Lampert et al, 2013; Jenni & Harrison, 2018). Similarly, the recruitment of the Ska complex to kinetochores requires the Ndc80 loop and the Ndc80 tail in vivo (Zhang et al, 2012; Janczyk et al, 2017), but Ska and Ndc80 form a stable stoichiometric complex in vitro in a loop‐ and tail‐independent manner, at least at micromolar concentration (Huis in 't Veld et al, 2019). Thus, lack of recruitment of certain downstream proteins to kinetochores with mutated Ndc80 loops might not reflect a direct role of the loop in these recruitments, but rather that the Ndc80 clusters assembled through the loop are stronger binding platforms for these proteins than the isolated Ndc80 complexes. This possibly reflects preferred interactions of Ndc80 clusters on microtubules with other multimers, such as the Ska and Dam1 complexes. In the future, we will ascertain the validity of this hypothesis and test why the late recruitment of certain protein to the kinetochore requires an intact loop.
Mutation of the loop increases the diffusion of Ndc80 on microtubules. Interactions between loopless Ndc80 complexes, induced with an antibody that binds near the loop, reduced the diffusion of loopless Ndc80 trimers (Fig 6). This demonstrates that the binding between adjacent Ndc80 complexes increases the grip to the microtubule lattice. Disturbing the diffusion of Ndc80 ensembles on microtubules might hinder proper chromosome congression and end‐on microtubule binding in cells. In agreement with this idea, the electroporation of antibody:Ndc80 mixtures into cells interfered with chromosome congression and checkpoint signaling (Fig EV5). These results are consistent with classic studies demonstrating that (i) antibodies against NDC80 or NUF2 injected into Xenopus cells resulted in a mitotic arrest and chromosome congression failure (McCleland et al, 2003) and (ii) a monoclonal antibody (9G3, raised against GST‐NDC8056–642, later shown to recognize NDC80200–215) interfered with mitosis by hyperstabilizing kinetochore‐microtubule connections (Chen et al, 1997; DeLuca et al, 2006). Interestingly, we found that the 9G3 antibody binds microtubules in vitro, preventing the analysis of its influence on the diffusion of Ndc80 trimers (Appendix Fig S4).
The kinetochore‐microtubule interface consists of dozens of proteins and protein complexes, all present in multiple copies. Understanding how these proteins control the attachments of chromosomes to the mitotic spindle and link this attachment state to cell cycle progression is a formidable challenge. In this study, we investigated how a short loop sequence in the outer kinetochore Ndc80 complex coordinates the formation of stable end‐on kinetochore‐microtubule attachments that support chromosome congression and bi‐orientation. To unravel the multifaceted roles of the loop, we identified and tested separation‐of‐function mutants under the controlled conditions of a fully reconstituted system and in the presence of additional proteins and regulatory pathways in cells. In the future, our approach can provide molecular and mechanistic insight into other central aspects of kinetochore biology, such as the transition from a lateral to an end‐on attachment, the role that other microtubule binders play, and the intricate feedback loops that control kinetochore‐microtubule binding.
Materials and Methods
Molecular modeling
The version AF2 Multimer 2 (preprint: Evans et al, 2022) of AlphaFold 2 (AF2) was used for all molecular modeling. Compared to the original AF2 (Jumper et al, 2021), AF2 Multimer is more sensitive to intra‐ and intermolecular interactions, resulting in a tendency to collapse extended proteins, for example, long coiled‐coil regions, into more compact structures. To avoid this, sub‐fragments with overlapping regions were modeled separately and then stitched together. Fragment length was a compromise between “long and bending” and “short with dissociated coiled coils at the ends”. Several different regions were predicted and analyzed, and we selected the longest possible fragments that did not bend. Three segments with overlapping regions were stitched together: (i) NDC80 81–401 | NUF2 1–283, (ii) NDC80 318–522 | NUF2 205–359, and (iii) NDC80 507–642 | NUF2 344–464. Models were then superimposed on their overlapping parts using PyMOL (The PyMOL Molecular Graphics System, version 2.5.2 Schrödinger, LLC.) and a residue in the middle of the best‐fitting regions was chosen as fragment boundary. The final model was comprised of (i) NDC80 81–382 | NUF2 1–267, (ii) NDC80 383–511 | NUF2 268–351, and (iii) NDC80 512–642 | NUF2 352–464 (Appendix Fig S1). The ends of the fragments tend to have higher pLDDT scores, which is expected due to the truncated multiple sequence alignment that is then used by AF2. The geometry at the stitching points was checked and optimized with COOT (Emsley & Cowtan, 2004), followed by a minimization of the complete model with PHENIX (Liebschner et al, 2019). Backbone restraints were applied to conserve the AF2‐predicted backbone positions. In the final model, 98% of the residues had pLDDT scores above 80 (high confidence) and 84% of the residues had scores above 90 (very high confidence) (Fig EV1). Corroborating the high quality of the model, the predicted alignment error (PAE) plots consistently indicated well‐defined relative positions of the monomers, including the tetramerization domain that is composed of SPC24, SPC25, and the C‐termini of NDC80 and NUF2 (Fig EV1). Omitting the structural template for the yeast tetramerization domain (PDB ID 5TCS) (Valverde et al, 2016) from the AF2 database did not change the prediction for the human tetramerization domain.
The loop region of NDC80 (residues 422–458) was predicted very reproducibly, whereas the angle of the hinge (residues 358–363 in NDC80 and 243–248 in NUF2) varied between 109° and 131° in various predictions. In addition, some predictions showed up to 15° lateral deviation of the coiled‐coil axes perpendicular to the plane of the triangle. It is unclear if this is a bona fide property of the Ndc80 complex, but it is interesting that AF2 reproduces the observation of the hinge‐like region of the Ndc80 at a distance of approximately 180–210 Å (± 30 residues) from the globular, N‐terminal NDC80/NUF2 domains (Huis in 't Veld et al, 2016; Jenni & Harrison, 2018), which corresponds well with the position of the predicted kink residues (358–363 in NDC80 and 243–248 in NUF2) in our model (approximate distance from N‐terminus 170–200 Å).
To compare the evolutionary conservation of the loop region, NDC80 sequences from 33 diverse eukaryotic organisms were selected (Boeckmann et al, 2015) and aligned and visualized using ProViz (Jehl et al, 2016) and jalview (Waterhouse et al, 2009; Fig EV2). The phylogenetic tree was generated using ITOL (Letunic & Bork, 2021) and structures were predicted using AF2 as described above (Fig EV2). All figures were made using ChimeraX (v1.2 and daily builds) (Goddard et al, 2018).
Cloning, expression, and purification of Ndc80
Expression cassettes from pLIB vectors containing NDC80, NUF2, SPC25SORT‐HIS, and SPC24 were combined on a pBIG1 vector using Gibson assembly as described (Weissmann et al, 2016; Volkov et al, 2018). Baculoviruses were generated in Sf9 insect cells and used for protein expression in Tnao38 insect cells. Between 60 and 72 h post‐infection, cells were washed in PBS and stored at −80°C. All subsequent steps were performed on ice or at 4°C. Cells were thawed and resuspended in lysis buffer (50 mM HEPES, pH 8.0, 200 or 250 mM NaCl, 10% v/v glycerol, 2 mM TCEP, 20 mM imidazole, 0.5 mM PMSF, protease‐inhibitor mix HP Plus (Serva)), lysed by sonication and cleared by centrifugation at 108,000 g for 60 min. The cleared lysate was filtered (0.8 μM) and applied to a 10 or 20 ml HisTrap FF (GE Healthcare) equilibrated in washing buffer (lysis buffer without protease inhibitors). The column was washed with approximately 20 column volumes of washing buffer and bound proteins were eluted with elution buffer (washing buffer containing 300 mM imidazole). Relevant fractions were pooled, diluted 5‐fold with buffer A (50 mM HEPES, pH 8.0, 25 mM NaCl, 5% v/v glycerol, 1 mM EDTA, 2 mM TCEP) and applied to a 25 ml Source15Q (GE Healthcare) strong anion exchange column equilibrated in buffer A. After washing with approximately 20 column volumes, bound proteins were eluted with a linear gradient from 25 to 300 mM NaCl in 180 ml. Relevant fractions were concentrated in 50 kDa molecular mass cut‐off Amicon concentrators (Millipore). Complexes were then fluorescently labeled (see next section) or directly applied to a Superdex 200 16/600 increase, a Superose 6 10/300 increase, or a Superose 6 prep grade XK 16/600 column (GE Healthcare). Columns were equilibrated in Ndc80 buffer (50 mM HEPES, pH 8.0, 250 mM NaCl, 5% v/v glycerol, 2 mM TCEP) and size‐exclusion chromatography was performed under isocratic conditions at recommended flow rates. Relevant fractions were pooled, concentrated, flash‐frozen in liquid nitrogen, and stored at −80°C.
GST‐NDC80376–517 and NUF2258–356 were cloned in a pGEX‐6P‐2rbs vector and expressed in E. coli. BL21(DE3)‐Codon‐plus‐RIPL in Terrific Broth in the presence of Chloramphenicol and Ampicillin and 1% Glucose, 0.01% Lactose, and 2 mM MgSO4 for ~24 h at 25°C. Cells were thawed and resuspended in lysis buffer (50 mM HEPES, pH 7.5, 250 mM NaCl, 10% v/v glycerol, 2 mM TCEP, 0.5 mM PMSF, protease‐inhibitor mix HP Plus (Serva)), lysed by sonication and cleared by centrifugation at 75,600 or 108,000 g for 45 min. The cleared lysate was bound to Glutathione‐Agarose resin (5 ml resin for 1 l expression culture) equilibrated in washing buffer (lysis buffer without protease inhibitors). The beads were washed extensively and protein was cleaved of the beads by overnight cleavage with 3C‐PreScission protease (generated in‐house) or eluted in washing buffer + 10 mM glutathione. The eluate was concentrated using 10 kDa molecular mass cut‐off Amicon concentrators (Millipore) and applied to a Superdex 75 10/600 column (GE Healthcare) equilibrated in 50 mM HEPES, pH 8.0, 250 mM NaCl, 2 mM TCEP, 5% v/v glycerol. Relevant fractions were pooled, concentrated, flash‐frozen in liquid nitrogen, and stored at −80°C.
Fluorescent labeling of Ndc80
The calcium‐independent Sortase 7M (Hirakawa et al, 2015) was used for the C‐terminal conjugation of a synthetic peptide to SPC25. Various synthetic peptides (Genscript) were used for this purpose: GGGGKFAM, GGGGKTMR, and GGGGCAlexa‐488. Reactions were performed at 10°C for 4–16 h in Ndc80 buffer with Sortase:Ndc80:Peptide ratios of approximately 1:5:25 with Ndc80 in the 10–20 μM range. The NDC80:NUF2 loop fragment was labeled at its N‐terminal following 3C‐cleavage using Sortase and FAMLPETGG in a similar manner. Fluorescently labeled complexes were purified using size‐exclusion chromatography, as described above.
Assembly of Ndc80 trimers
Ndc80 complexes were coupled to streptavidin‐derived T1S3 scaffolds as described (Volkov et al, 2018; Appendix Fig S1). In brief, T1S3 scaffolds were assembled from core traptavidin (T; addgene plasmid #26054) and Dead Streptavidin‐SpyCatcher (S; addgene plasmid #59547) (Chivers et al, 2010; Fairhead et al, 2014). T1S3 scaffolds were incubated with an approximate 10‐fold molar excess of Ndc80 for 12–20 h at 10°C in the presence of PMSF (1 mM) and protease inhibitor mix (Serva). Sortase labeling was achieved in the same reaction, as described above. Reaction mixtures were applied to a Superose 6 increase 10/300 column (GE Healthcare) equilibrated in 20 mM TRIS pH 8.0, 200 mM NaCl, 2% v/v glycerol, 2 mM TCEP. Size‐exclusion chromatography was performed at 4°C under isocratic conditions at recommended flow rates and the relevant fractions were pooled and concentrated using 30 kDa molecular mass cut‐off Amicon concentrators (Millipore), flash‐frozen in liquid nitrogen, and stored at −80°C.
SEC‐MALS and mass photometry
Full‐length and loopless Ndc80 complexes were analyzed by SEC‐MALS on a Dawn Heleos II System with an Optilab T‐rEX RI detector (Wyatt) and a 1260 Infinity II LC system (Agilent). The Superose 6 increase 10/300 column (GE Healthcare) was pre‐equilibrated with glycerol‐free Ndc80 buffer (50 mM HEPES pH 8.0, 250 mM NaCl, and 2 mM TCEP). Analysis was performed at room temperature with 60 μl full‐length or loopless Ndc80 complex that was diluted in running buffer to 2 mg/ml. Mass photometry was performed on a Refeyn TwoMP System (Refeyn) that was calibrated with a mixture of BSA (66.5 and 123 kDa) and Thyroglobulin (330 and 660 kDa). Ndc80 complexes were diluted to a concentration of 100 nM in glycerol‐free Ndc80 buffer and analyzed in 20 μL droplets following a 10‐fold dilution in buffer to 10 nM. Data were analyzed and plotted using the DiscoverMP software (Refeyn).
Crosslinking and mass spectrometry
Two hundred microliter of full‐length Ndc80, loopless Ndc80, and their complexes with the Mis12 complex were prepared at 7.5 μM in crosslinking buffer (50 mM HEPES pH 8, 250 mM NaCl, 5% v/v glycerol, and 2 mM TCEP), crosslinked with an approximate 500‐fold molar excess of DSBU (3.75 mM), and quenched with TRIS (100 mM) as described in Appendix Fig S2. Samples were prepared for mass spectrometry and analyzed using MeroX (version 2.0.1.4) as previously described (Arlt et al, 2016; Pan et al, 2018). Crosslinks were inspected in XiView (preprint: Graham et al, 2019) and, after setting a false‐discovery rate to 1%, ranked according to the number of matched peptides and the maximum score. Unique intra‐ and intermolecular Ndc80 crosslinks from all four datasets were identified. To distinguish frequently observed and more rarely observed crosslinked peptides, the rank was used to divide the dataset, at an arbitrary cut‐off, in two parts. All crosslinks were displayed on the predicted structure of full‐length Ndc80 and Cα‐Cα distances were plotted using ChimeraX (daily build) (Goddard et al, 2018).
Low‐angle metal shadowing and electron microscopy
Full‐length and loopless Ndc80 complexes (10 μM) were incubated with full‐length Mis12 complexes (15 μM) for approximately 1 h on ice. Mixtures were diluted 1:9 with Ndc80 buffer, diluted 1:1 with spraying buffer (200 mM ammonium acetate and 60% glycerol), and then air‐sprayed onto freshly cleaved mica (V1 quality, Plano GmbH) of approximately 2 × 3 mm. Specimens were mounted and dried in a MED020 high‐vacuum metal coater (Bal‐tec). A Platinum layer of approximately 1 nm and a 7 nm Carbon support layer were evaporated subsequently onto the rotating specimen at angles of 6–7° and 45° respectively. Pt/C replicas were released from the mica on water, captured by freshly glow‐discharged 400‐mesh Pd/Cu grids (Plano GmbH), and visualized using a LaB6 equipped JEM‐400 transmission electron microscope (JEOL) operated at 120 kV. Images were recorded at a nominal magnification of 60,000× on a 4 k × 4 k CCD camera F416 (TVIPS), resulting in 0.1890 nm per pixel. Particles were manually selected using EMAN2 (Tang et al, 2007).
Tubulin and microtubules
Pig brain tubulin, as well as TMR‐ and HiLyte488‐labeled tubulin, were purchased from a commercial source (Cytoskeleton Inc). DIG‐tubulin was made in‐house by labeling microtubules polymerized from commercial tubulin with an NHS‐ester (Sigma Aldrich) of digoxigenin according to published protocols (Hyman et al, 1991). GMPCPP‐stabilized seeds were made by two rounds of polymerization at 37°C in presence of 1 mM GMPCPP and 25 μM tubulin (40% DIG‐labeled). Seeds were sedimented in a Beckman Airfuge at 126,000 g for 5 min. After the second polymerization round, the seeds were resuspended in MRB80 buffer (80 mM K‐Pipes pH 6.9, 1 mM EGTA, 4 mM MgCl2) supplemented with 10% glycerol and aliquots were flash‐frozen in LN2.
To prepare taxol‐stabilized microtubules, 70 μM tubulin (with or without DIG‐ and fluorescent labels) was polymerized in presence of 25% glycerol and 1 mM GTP for 10 min at 37°C, and then stabilized by an addition of 25 μM taxol followed by a 20 min incubation. Microtubules were sedimented in a Beckman Airfuge at 32,000 g for 3 min, resuspended in MRB80 buffer supplemented with 40 μM taxol and stored at 25°C for up to 3 days.
Preparation of flow chambers and TIRF microscopy
Microscopy chambers were prepared using slides and coverslips treated with oxygen plasma and immediately silanized with a mixture of 2% dichlorodimethyl silane and 98% octamethylcyclooctasilane for 5 min (Gell et al, 2010; Maleki et al, 2023). Before an experiment, antibodies against DIG (Roche 11333089001) or tubulin (TU‐20, Abcam) were adsorbed to the silanized glass, followed by a passivation with 1% Pluronic F‐127 in MRB80. Taxol‐stabilized microtubules were diluted in approximately 5–7 chamber volumes of MRB80 with 10 μM taxol, washed in within 3 min, and immediately washed out with three‐chamber volumes of MRB80 with 10 μM taxol to produce microtubules mostly oriented along the chamber. Subsequently, proteins of interest were introduced into the chamber in imaging buffer containing MRB80, 0.1% methylcellulose, 40 μM taxol, 1 mg/ml κ‐casein, 4 mM DTT, 0.2 mg/ml catalase, 0.4 mg/ml glucose oxidase and 20 mM glucose. Imaging buffer was pre‐cleared in a Beckman Airfuge for 5 min at 126,000 g before the addition of Ndc80 or antibodies.
For experiments with dynamic microtubules, DIG‐labeled GMPCPP‐stabilized microtubule seeds were attached to silanized chambers containing anti‐DIG antibodies and passivated with Pluronic F‐127. Imaging buffer was prepared as described above; however, taxol was substituted with 11 μM tubulin (3–5% labeled).
Experiments reported in Figs 1 and 5 were performed using a Nikon Eclipse Ti2 microscope equipped with a Plan Apo 100 × 1.45 NA TIRF, iLas3 ring TIRF illumination system (GATACA), and an Andor iXon 897 EMCCD camera. Experiments reported in Fig 8 were prepared as described above, but were imaged using Nikon Ti‐E microscope (Visitron Systems) with a CFI 100 × 1.49 NA TIRF oil‐immersion objective, iLas2 ring TIRF module, and an Evolve 512 EMCCD camera. Images were recorded using VisiView software. Experiments reported in Figs 2 and 6 were performed with a Nikon Ti‐E microscope equipped with a Plan Apo 100 × 1.45 NA TIRF oil‐immersion objective, iLas2 ring TIRF module (Roper Scientific) and an Evolve 512 EMCCD camera (Roper Scientific). Images were recorded using MetaMorph software.
Experiments with taxol‐stabilized microtubules were performed at ambient temperature (23°C); for experiments with dynamic microtubules the objective was heated to 34°C using a custom‐made collar coupled with a thermostat, resulting in the flow chamber being heated to 30°C. Images were acquired every 300 ms, and initial 10 images were averaged for analysis of Ndc80 clustering. Clustering was expressed as standard deviation (SD) of FAM‐Ndc80 fluorescence along the microtubule length. Experiments presented in Appendix Fig S5 were performed with single blinding: MT and PJH prepared eight FAM‐labeled Ndc80 complexes, VAV then performed fluorescence microscopy measurements (repeated three times) and image analysis without knowing a complex' identity. Results were unblinded after all analysis was complete.
Image analysis
Coordinates and brightness of particles diffusing on microtubule lattice or tracking their ends were analyzed using Fiji (Schindelin et al, 2012) and Julia using custom scripts available at https://github.com/volkovdelft/kymo.jl. Kymographs were made through a reslice operation using the kymograph_(n)channel.ijm macro. Position of a particle was determined in each line of the kymograph using kymo.ipynb jupyter notebook and following in‐line comments. Oligomerization was inferred from initial brightness of a particle and the photobleaching step characteristic for a fluorophore as described previously (Volkov et al, 2018). Mean squared displacement curves to characterize one‐dimensional diffusion were generated using process_diffusion.ipynb: coordinates of each diffusing spot were iteratively split into segments of equal length with an offset of one datapoint, and subsequently aligned at their start and averaged.
To analyze co‐localization of Ndc80 monomers and trimers on microtubules, trimers were tracked in one fluorescent channel of a kymograph as above using kymo_2channel.ipynb, and then the brightness of monomers was measured in the second fluorescent channel at the coordinates determined for trimer. The resulting monomer brightness was thresholded using a value for single Alexa488 fluorophore, and the number of datapoints above the threshold was divided by the total number of datapoints to calculate the time fractions for co‐localization.
Preparation of beads and force measurements
Beads were prepared as described previously (Volkov et al, 2018). In brief, 1 μm glass COOH‐functionalized beads were coated covalently with PLL‐PEG (Poly‐L‐lysine (20 kDa) grafted with polyethyleneglycol (2 kDa), SuSoS AG, Switzerland) containing varying fraction of biotinylated PLL‐PEG of the same composition. Biotins on the beads were then saturated with streptavidin‐oligomerized Ndc80‐TMR trimers. Bead preparations were analyzed for bead brightness using at least 50 single beads.
Optical trapping was performed using a custom optical setup as described previously (Volkov et al, 2018). A bead coated with Ndc80 trimers was captured in a trap with a stiffness of 0.02–0.04 pN/nm placed near a growing end of a dynamic microtubule. Bead‐microtubule attachment was verified by switching the trap off for several seconds and monitoring bead's motion: successfully attached beads moved in an arc shape across the direction of microtubule growth and did not move significantly along it. The bead was then re‐trapped again and the experiment was initiated by simultaneous start of acquisition of DIC images of the bead‐microtubule pair, and the quadrant photo detector (QPD) readings to monitor bead's displacement from the trap center. Recording was stopped after the microtubule depolymerized and the bead detached from it, or at will after 30–40 min of data acquisition if the bead did not detach. In the latter case, the trap stiffness was increased 10 times, and the bead was ruptured from the microtubule using 100 nm steps of the piezo stage. Motions of a free bead were recorded after detachment for each bead to later use them to determine trap stiffness.
Cell electroporation
All electroporation experiments of this study were performed as described using a Neon Transfection System Kit (Thermo Fisher) (Alex et al, 2019). Cells were collected from the surface of the cell plate by trypsinization, washed two times with PBS, and counted. Between 2 and 3 million cells were used per electroporation and were resuspended in 90 μl of buffer R (Thermo Fisher). Ndc80 complexes with typical concentrations of 40 μM were spun down at 16,700 g for 10 min and then diluted with buffer R in a 1:1 ratio. For electroporation with antibody, cells were resuspended in 56 μl of buffer R and the Ndc80‐antibody mixture was diluted 1:1 in buffer R. The Ndc80‐antibody and cell suspension were mixed and then the mixture was loaded into a 100 μl Neon Pipette Tip (Thermo Fisher). Electroporation was performed at 1,005 V with two consecutive pulses of 35 msec. Cells were washed with PBS and then treated with Trypsin to digest non‐cell‐internalized proteins. After another wash with PBS, cells were plated on 6‐well plates for immunofluorescence staining or 24‐well cell‐imaging plates (Ibidi) for live‐cell imaging.
Antibody generation
Peptides C‐TQLAEYHKLARKLKLI PKGAENS‐NH 2 (NDC80410–432), and Ac‐AENSKGYDFEIKF NPEAGANCL‐NH 2 (NDC80429–450) were used to generate the antibodies AB‐849 and AB‐850, respectively. The peptides, synthesized at a minimal purity of 80% and conjugated to Keyhole Limpet Hemocyanin carrier (Mimitopes), were used for rabbit immunization (Eurogentec). Polyclonal antibodies were affinity purified from sera using peptides (elution pH 2.5) and stored at 4°C in PBS supplemented with 150 mM KCl, 0.1% BSA, and 0.05% NaN3. Recombinant full‐length SKA complex was used for rabbit immunization (Antibody Facility, MPI‐CBG). Polyclonal antibodies were affinity purified from sera using recombinant SKA (elution pH 2.5) and stored at 4°C in PBS.
Cell culture, siRNA transfection, immunoblotting
HeLa, mCherry‐H2B HeLa cell lines were cultured in DMEM (PAM Biotech) supplemented with 10% FBS (Clontech), 2 mM L‐glutamine (PAN Biotech), 1% Penicillin, Streptomycin (Gibco). Cells were grown in a humidified chamber at 37°C in the presence of 5% of CO2. Cell lines were checked regularly for mycoplasma contamination and tested negative. Depletion of endogenous Ndc80 complex was attained by RNAiMAX (Invitrogen) transfection of siRNAs targeting three of the four subunits of the Ndc80 complex as described (Kim & Yu, 2015). The siRNA oligos were GAGUAGAACUAGAAUGUGA (siNdc80‐4), GGACACGACAGUCACAAUC (siSpc24), and CUACAAGGAUUCCAUCAAA (siSpc25). To generate lysate of cells for immunoblotting, cells treated with control or Ndc80 complex siRNA were collected by trypsinization and resuspended in lysis buffer (150 mM KCl, 75 mM HEPES pH 7.5, 1.5 mM EGTA, 1.5 mM MgCl2, 10% Glycerol, and 0.075% NP‐40 supplemented with protease inhibitor cocktail (Serva)). Lysates were run in 4–12% gradient gels (NuPAGE, ThermoFisher) and proteins were transferred to a Nitrocellulose membrane for further analysis. The following antibodies were used for the immunoblot analysis of this study: anti‐Hec1 (human Ndc80; mouse clone 9G3.23 GeneTex, Inc; 1:1,000), anti‐Vinculin (mouse monoclonal; clone hVIN‐1; Sigma‐Aldrich; 1:10,000), AB‐849 and AB‐850 (rabbit polyclonal, in‐house generated, 1:500), anti‐Ska (rabbit polyclonal, in‐house generated against a recombinant Ska complex, 1:1,000).
Cell treatment, microscopy, immunofluorescence detection, and live‐cell imaging
Cells were imaged with a customized 3i Marianas system (Intelligent Imaging Innovations) equipped with an Axio Observer Z1 microscope chassis (Zeiss), a CSU‐X1 confocal scanner unit (Yokogawa Electric Corporation), Plan‐Apochromat 100×/1.4 NA objectives (Zeiss), and an Orca Flash 4.0 sCMOS Camera (Hamamatsu). Slide book software was used to acquire images as Z sections. Additional images were acquired using a DeltaVision Elite System (GE Healthcare) equipped with an IX‐71 inverted microscope (Olympus), a UPlanFLN 40 × 1.3 NA objective (Olympus), and a pco.edge sCMOS camera (PCO‐TECH Inc.). Live cell movies were taken as Z scans every 10 min interval for 16 h. The softWoRx software was used for maximal intensity projection and analysis of the movies. For immunofluorescence sample preparation, cells were grown on poly‐L‐Lysine (Millipore) coated coverslips and permeabilized with a 0.5% Triton‐X in PHEM buffer. Cells were fixed with 4% paraformaldehyde (PFA) and then blocked for 1 h with 5% boiled donkey serum. The following antibodies were used for immunostaining: anti‐Hec1 (human Ndc80; mouse clone 9G3.23 GeneTex, Inc; 1:1,000), anti‐CENP‐C (guineapig polyclonal, MBL, 1:500), anti‐CREST/anti‐centromere antibodies (human, Antibodies Inc, 1:1,000), anti‐tubulin (mouse monoclonal, Sigma, 1:5,000), Knl1 (rabbit polyclonal, in house, 1:500), Nsl1 (mouse monoclonal, in house, 1:800), CENP‐T (rabbit polyclonal, in house, 1:500), BUB1 (rabbit polyclonal, Abcam, 1:500), BubR1 (rabbit polyclonal, Abcam, 1:400), anti‐Ska (rabbit polyclonal, in‐house generated against a recombinant Ska complex, 1:5,000). DNA was visualized by staining with 0.5 μg/ml DAPI (Serva) and coverslips were mounted to slide with Mowiol mounting media. All images were processed using Fiji. Quantifications of protein signals were measured using a custom‐written script. All signals were normalized to either CREST or CENP‐C signal. To calculate times in mitosis, 16 h (960 min) long live‐cell movies were analyzed manually. Cells that entered mitosis within the first 500 min of the movie were accounted for. Cells already in mitosis at the start of the movie were not analyzed. Mitotic duration was calculated from the time that cells condensed their chromosomes until they exited mitosis. For cold shock assay, cells were kept on ice and ice‐cold media was added. Cells were kept in this condition for 14 min and then fixed with 4% PFA and proceeded to immunofluorescence staining as mentioned above. For treatment with drugs, hesperidin (500 nM), reversine (250 nM), and nocodazole (3.3 μM) were added 1 h before starting the live‐cell movies.
Author contributions
Soumitra Polley: Conceptualization; data curation; formal analysis; supervision; validation; investigation; methodology; project administration. Helen Müschenborn: Formal analysis; validation; investigation. Melina Terbeck: Resources. Anna De Antoni: Resources. Ingrid R Vetter: Data curation; software; formal analysis; validation; investigation; methodology. Marileen Dogterom: Supervision; funding acquisition; project administration. Andrea Musacchio: Conceptualization; supervision; funding acquisition; writing – original draft; project administration; writing – review and editing. Vladimir A Volkov: Conceptualization; resources; data curation; software; formal analysis; supervision; validation; investigation; visualization; methodology; writing – original draft; project administration; writing – review and editing. Pim J Huis in 't Veld: Conceptualization; resources; data curation; software; formal analysis; supervision; validation; investigation; visualization; methodology; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Acknowledgements
We thank Christina Courtis, Alicia Dammers, Farzad Khanipour, Laura Ney, Isabelle Stender, and Sabine Wohlgemuth for help with the generation of recombinant Ndc80 complexes. We are grateful to Petra Geue and Raphael Gasper‐Schoenenbruecher for assistance with SEC‐MALS and mass photometry, and to Franziska Müller, Andreas Brockmeyer, Malte Metz, and Petra Janning for mass spectrometry. VAV was supported by the European Research Council Synergy Grant 609822 to MD, and by the QMUL Startup grant (SBC8VOL2). AM acknowledges funding of this work through the European Research Council Synergy Grant SyG 951430 BIOMECANET. Open Access funding enabled and organized by Projekt DEAL.
The EMBO Journal (2023) 42: e112504
Contributor Information
Andrea Musacchio, Email: andrea.musacchio@mpi-dortmund.mpg.de.
Vladimir A Volkov, Email: v.volkov@qmul.ac.uk.
Pim J Huis in 't Veld, Email: pim.huis@mpi-dortmund.mpg.de.
Data availability
This study includes no data deposited in external repositories.
References
- Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano‐Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S (2010) Tension directly stabilizes reconstituted kinetochore‐microtubule attachments. Nature 468: 576–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alex A, Piano V, Polley S, Stuiver M, Voss S, Ciossani G, Overlack K, Voss B, Wohlgemuth S, Petrovic A et al (2019) Electroporated recombinant proteins as tools for in vivo functional complementation, imaging, and chemical biology. Elife 8: e48287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E (2010) The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature 467: 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Musinipally V, Matson D, Tooley J, Stukenberg PT, Nogales E (2012) Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat Struct Mol Biol 19: 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt C, Götze M, Ihling CH, Hage C, Schäfer M, Sinz A (2016) Integrated workflow for structural proteomics studies based on cross‐linking/mass spectrometry with an MS/MS cleavable cross‐linker. Anal Chem 88: 7930–7937 [DOI] [PubMed] [Google Scholar]
- Boeckmann B, Marcet‐Houben M, Rees JA, Forslund K, Huerta‐Cepas J, Muffato M, Yilmaz P, Xenarios I, Bork P, Lewis SE et al (2015) Quest for orthologs entails quest for tree of life: in search of the gene stream. Genome Biol Evol 7: 1988–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M, Tarasovetc EV, Zaytsev AV, Godzi M, Figueiredo AC, Ataullakhanov FI, Grishchuk EL (2019) Microtubule end conversion mediated by motors and diffusing proteins with no intrinsic microtubule end‐binding activity. Nat Commun 10: 1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson‐Kubalek EM, Desai A (2006) The conserved KMN network constitutes the core microtubule‐binding site of the kinetochore. Cell 127: 983–997 [DOI] [PubMed] [Google Scholar]
- Chen Y, Riley DJ, Chen PL, Lee WH (1997) HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol 17: 6049–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers CE, Crozat E, Chu C, Moy VT, Sherratt DJ, Howarth M (2010) A streptavidin variant with slower biotin dissociation and increased mechanostability. Nat Methods 7: 391–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P et al (2008) Implications for kinetochore‐microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED (2006) Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127: 969–982 [DOI] [PubMed] [Google Scholar]
- DeLuca KF, Lens SM, DeLuca JG (2011) Temporal changes in Hec1 phosphorylation control kinetochore‐microtubule attachment stability during mitosis. J Cell Sci 124: 622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca KF, Meppelink A, Broad AJ, Mick JE, Peersen OB, Pektas S, Lens SMA, DeLuca JG (2018) Aurora A kinase phosphorylates Hec1 to regulate metaphase kinetochore‐microtubule dynamics. J Cell Biol 217: 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Etemad B, Kuijt TEF, Kops GJPL (2015) Kinetochore‐microtubule attachment is sufficient to satisfy the human spindle assembly checkpoint. Nat Commun 6: 8987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R, O'Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J et al (2022) Protein complex prediction with AlphaFold‐Multimer. bioRxiv 10.1101/2021.10.04.463034 [PREPRINT] [DOI]
- Fairhead M, Veggiani G, Lever M, Yan J, Mesner D, Robinson CV, Dushek O, van der Merwe PA, Howarth M (2014) SpyAvidin hubs enable precise and ultrastable orthogonal nanoassembly. J Am Chem Soc 136: 12355–12363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell C, Bormuth V, Brouhard GJ, Cohen DN, Diez S, Friel CT, Helenius J, Nitzsche B, Petzold H, Ribbe J et al (2010) Microtubule dynamics reconstituted in vitro and imaged by single‐molecule fluorescence microscopy. Methods Cell Biol 95: 221–245 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE (2018) UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci 27: 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M, Combe C, Kolbowski L, Rappsilber J (2019) xiView: a common platform for the downstream analysis of crosslinking mass spectrometry data. bioRxiv 10.1101/561829 [PREPRINT] [DOI]
- Guimaraes GJ, Dong Y, McEwen BF, Deluca JG (2008) Kinetochore‐microtubule attachment relies on the disordered N‐terminal tail domain of Hec1. Curr Biol 18: 1778–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM (2003) The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore‐microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol 161: 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeson LA, Zelter A, Riffle M, MacCoss MJ, Asbury CL, Davis TN (2018) Human Ska complex and Ndc80 complex interact to form a load‐bearing assembly that strengthens kinetochore‐microtubule attachments. Proc Natl Acad Sci USA 115: 2740–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa H, Ishikawa S, Nagamune T (2015) Ca2+‐independent sortase‐A exhibits high selective protein ligation activity in the cytoplasm of Escherichia coli . Biotechnol J 10: 1487–1492 [DOI] [PubMed] [Google Scholar]
- Hsu KS, Toda T (2011) Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore‐spindle attachment in fission yeast. Curr Biol 21: 214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis in 't Veld PJ, Jeganathan S, Petrovic A, Singh P, John J, Krenn V, Weissmann F, Bange T, Musacchio A (2016) Molecular basis of outer kinetochore assembly on CENP‐T. Elife 5: e21007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis in 't Veld PJ, Volkov VA, Stender ID, Musacchio A, Dogterom M (2019) Molecular determinants of the Ska‐Ndc80 interaction and their influence on microtubule tracking and force‐coupling. Elife 8: e49539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison T (1991) Preparation of modified tubulins. Methods Enzymol 196: 478–485 [DOI] [PubMed] [Google Scholar]
- Janczyk PL, Skorupka KA, Tooley JG, Matson DR, Kestner CA, West T, Pornillos O, Stukenberg PT (2017) Mechanism of Ska recruitment by Ndc80 complexes to kinetochores. Dev Cell 41: 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehl P, Manguy J, Shields DC, Higgins DG, Davey NE (2016) ProViz—a web‐based visualization tool to investigate the functional and evolutionary features of protein sequences. Nucleic Acids Res 44: W11–W15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenni S, Harrison SC (2018) Structure of the DASH/Dam1 complex shows its role at the yeast kinetochore‐microtubule interface. Science 360: 552–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A (2006) Chromosomes can congress to the metaphase plate before biorientation. Science 311: 388–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiewisz R, Fabig G, Conway W, Baum D, Needleman D, Müller‐Reichert T (2022) Three‐dimensional structure of kinetochore‐fibers in human mitotic spindles. Elife 11: e75459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yu H (2015) Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J Cell Biol 208: 181–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn V, Musacchio A (2015) The Aurora B kinase in chromosome bi‐orientation and spindle checkpoint signaling. Front Oncol 5: 225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski TJ, Hards R, Vandal SE, Abad MA, Jeyaprakash AA, Kaye E, Al‐Rawi A, Ly T, Godek KM, Gerber SA et al (2022) Small changes in phospho‐occupancy at the kinetochore–microtubule interface drive mitotic fidelity. J Cell Biol 221: e202107107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F, Mieck C, Alushin GM, Nogales E, Westermann S (2013) Molecular requirements for the formation of a kinetochore‐microtubule interface by Dam1 and Ndc80 complexes. J Cell Biol 200: 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Grishchuk EL (2017) Mechanisms to avoid and correct erroneous kinetochore‐microtubule attachments. Biology (Basel) 6: 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49: W293–W296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung L‐W, Jain S, McCoy AJ et al (2019) Macromolecular structure determination using X‐rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75: 861–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AF, Udy DB, Dumont S (2017) Hec1 tail phosphorylation differentially regulates mammalian kinetochore coupling to polymerizing and depolymerizing microtubules. Curr Biol 27: 1692–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, O'Connell CB, Lončarek J, Paul R, Mogilner A, Khodjakov A (2011) The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 146: 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiolica A, Cittaro D, Borsotti D, Sennels L, Ciferri C, Tarricone C, Musacchio A, Rappsilber J (2007) Structural analysis of multiprotein complexes by cross‐linking, mass spectrometry, and database searching. Mol Cell Proteomics 6: 2200–2211 [DOI] [PubMed] [Google Scholar]
- Maleki AN, Huis in 't Veld PJ, Akhmanova A, Dogterom M, Volkov VA (2023) Estimation of microtubule‐generated forces using a DNA origami nanospring. J Cell Sci 136: jcs260154 [DOI] [PubMed] [Google Scholar]
- Maure JF, Komoto S, Oku Y, Mino A, Pasqualato S, Natsume K, Clayton L, Musacchio A, Tanaka TU (2011) The Ndc80 loop region facilitates formation of kinetochore attachment to the dynamic microtubule plus end. Curr Biol 21: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT (2003) The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev 17: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Johnson ML, Stukenberg PT (2008) Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1). Curr Biol 18: 1785–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monda JK, Cheeseman IM (2018) The kinetochore‐microtubule interface at a glance. J Cell Sci 131: jcs214577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Desai A (2017) A molecular view of kinetochore assembly and function. Biology (Basel) 6: 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole E, Morphew M, McIntosh JR (2020) Electron tomography reveals aspects of spindle structure important for mechanical stability at metaphase. Mol Biol Cell 31: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Brockmeyer A, Mueller F, Musacchio A, Bange T (2018) Simplified protocol for cross‐linking mass spectrometry using the MS‐cleavable cross‐linker DSBU with efficient cross‐link identification. Anal Chem 90: 10990–10999 [DOI] [PubMed] [Google Scholar]
- Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL (2009) The Ndc80 kinetochore complex forms load‐bearing attachments to dynamic microtubule tips via biased diffusion. Cell 136: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscioli E, Germanova TE, Smith CA, Embacher PA, Erent M, Thompson AI, Burroughs NJ, McAinsh AD (2020) Ensemble‐Level organization of human kinetochores and evidence for distinct tension and attachment sensors. Cell Rep 31: 107535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Tighe A, D'Alise AM, Taylor SS, Musacchio A (2010) Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol 190: 73–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AT (2018) Kinase and phosphatase cross‐talk at the kinetochore. Front Cell Dev Biol 6: 62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarborough EA, Davis TN, Asbury CL (2019) Tight bending of the Ndc80 complex provides intrinsic regulation of its binding to microtubules. Elife 8: e44489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha RL, Draviam VM (2013) Lateral to end‐on conversion of chromosome‐microtubule attachment requires kinesins CENP‐E and MCAK. Curr Biol 23: 1514–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha RL, Conti D, Tamura N, Braun D, Ramalingam RA, Cieslinski K, Ries J, Draviam VM (2017) Aurora‐B kinase pathway controls the lateral to end‐on conversion of kinetochore‐microtubule attachments in human cells. Nat Commun 8: 150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin LJ, Guimaraes GJ, Deluca JG (2011) The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore‐microtubule attachment in mitosis. Mol Biol Cell 22: 759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Badger BL, Salmon ED (2015) A quantitative description of Ndc80 complex linkage to human kinetochores. Nat Commun 6: 8161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU (2005) Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 434: 987–994 [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ (2007) EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157: 38–46 [DOI] [PubMed] [Google Scholar]
- Tang NH, Takada H, Hsu KS, Toda T (2013) The internal loop of fission yeast Ndc80 binds Alp7/TACC‐Alp14/TOG and ensures proper chromosome attachment. Mol Biol Cell 24: 1122–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchman EC, Boehm FJ, DeLuca JG (2015) Stable kinetochore–microtubule attachment is sufficient to silence the spindle assembly checkpoint in human cells. Nat Commun 6: 10036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley JG, Miller SA, Stukenberg PT (2011) The Ndc80 complex uses a tripartite attachment point to couple microtubule depolymerization to chromosome movement. Mol Biol Cell 22: 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde R, Ingram J, Harrison SC (2016) Conserved tetramer junction in the kinetochore Ndc80 complex. Cell Rep 17: 1915–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, Chandrasekaran S, Sundin LJ, Reidy KT, Wan X, Chasse DA, Nevis KR, DeLuca JG, Salmon ED, Cook JG (2012) Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore‐microtubule attachment. Nat Cell Biol 14: 593–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VA, Huis in 't Veld PJ, Dogterom M, Musacchio A (2018) Multivalency of NDC80 in the outer kinetochore is essential to track shortening microtubules and generate forces. Elife 7: e36764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E (2008) Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol 383: 894–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al‐Bassam J, Harrison SC (2007) The Ndc80/HEC1 complex is a contact point for kinetochore‐microtubule attachment. Nat Struct Mol Biol 14: 54–59 [DOI] [PubMed] [Google Scholar]
- Weissmann F, Petzold G, VanderLinden R, Huis in 't Veld PJ, Brown NG, Lampert F, Westermann S, Stark H, Schulman BA, Peters JM (2016) biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc Natl Acad Sci USA 113: E2564–E2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Jensen ON, Holmes S, Souès S, Mann M, Kilmartin JV (1998) Analysis of the Saccharomyces spindle pole by matrix‐assisted laser desorption/ionization (MALDI) mass spectrometry. J Cell Biol 141: 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimbish RT, DeLuca KF, Mick JE, Himes J, Jiménez‐Sánchez I, Jeyaprakash AA, DeLuca JG (2020) The Hec1/Ndc80 tail domain is required for force generation at kinetochores, but is dispensable for kinetochore‐microtubule attachment formation and Ska complex recruitment. Mol Biol Cell 31: 1453–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo TY, Choi J‐M, Conway W, Yu C‐H, Pappu RV, Needleman DJ (2018) Measuring NDC80 binding reveals the molecular basis of tension‐dependent kinetochore‐microtubule attachments. Elife 7: e36392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahm JA, Jenni S, Harrison SC (2023) Structure of the Ndc80 complex and its interactions at the yeast kinetochore–microtubule interface. Open Biol 13: 220378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev AV, Mick JE, Maslennikov E, Nikashin B, DeLuca JG, Grishchuk EL (2015) Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule‐binding affinity. Mol Biol Cell 26: 1829–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Kelstrup CD, Hu XW, Kaas Hansen MJ, Singleton MR, Olsen JV, Nilsson J (2012) The Ndc80 internal loop is required for recruitment of the Ska complex to establish end‐on microtubule attachment to kinetochores. J Cell Sci 125: 3243–3253 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Sivakumar S, Chen Y, Gao H, Yang L, Yuan Z, Yu H, Liu H (2017) Ska3 phosphorylated by Cdk1 binds Ndc80 and recruits Ska to kinetochores to promote mitotic progression. Curr Biol 27: 1477–1484 [DOI] [PubMed] [Google Scholar]
- Zhu T, Dou Z, Qin B, Jin C, Wang X, Xu L, Zhaoyang W, Zhu L, Liu F, Gao X et al (2013) Phosphorylation of microtubule‐binding protein Hec1 by mitotic kinase Aurora B specifies spindle checkpoint kinase Mps1 signaling at the kinetochore. J Biol Chem 288: 36149–36159 [DOI] [PMC free article] [PubMed] [Google Scholar]


