Abstract
The RNA lariat debranching enzyme is the sole enzyme responsible for hydrolyzing the 2′–5′ phosphodiester bond in RNA lariats produced by the spliceosome. Here we test the ability of Dbr1 to hydrolyze branched RNAs (bRNAs) which contain a 2′–5′-phosphorothioate linkage, a modification commonly used to resist degradation. We attempted to co-crystallize a phosphorothioate branched RNA (PS-bRNA) with wild-type Entamoeba histolytica Dbr1 (EhDbr1) but observed in-crystal hydrolysis of the phosphorothioate bond. The crystal structure revealed EhDbr1 in a product-bound state, with the hydrolyzed 2′–5′ fragment of the PS-bRNA mimicking the binding mode of the native bRNA substrate. These findings suggest that product inhibition may contribute to the kinetic mechanism of Dbr1. We show that Dbr1 enzymes cleave phosphorothioate linkages at rates ~10,000-fold more slowly than native phosphate linkages. This new product-bound crystal structure offers atomic details which can aid inhibitor design. Dbr1 inhibitors could be therapeutic or investigative compounds for human diseases such as HIV, ALS, cancer, and viral encephalitis.
Keywords: RNA, lariats, introns, debranching enzyme, metalloenzymes, metallophosphodiesterase, phosphorothioate, branched RNA, crystal structure
Graphical Abstract
Introduction.
In eucaryotic organisms, genes are composed of protein-coding exons and intervening intronic sequences. The introns are removed by the spliceosome in the production of mature messenger RNA. Due to the chemical reactions catalyzed by the spliceosome, the excised introns have a lariat configuration, where the 5′ end of the intron is covalently bonded via a 2′–5′-phosphodiester linkage to a branchpoint-A residue ~50 nts away from the 3′ end of the intron 1, 2. A single enzyme is responsible for hydrolyzing the 2′–5′-linkage, the RNA lariat debranching enzyme Dbr1 3. Dbr1 is implicated in several biological processes, and loss of Dbr1 in humans can cause serious disease. Class-switch recombination of immunoglobulin genes requires Dbr1, HIV replication is impaired when Dbr1 in knocked-down 4, Dbr1 loss is oncogenic 5, and Dbr1 mutations in humans impair cell-intrinsic innate immunity in the brainstem 6. Most cases of amyotrophic lateral sclerosis (ALS) are characterized by aggregates of the tar-DNA binding protein, TDP-43 7, 8. Ablation of the DBR1 gene can reduce the toxicity of TDP-43 in disease models 9. Considering these important roles in human health and disease, understanding of the molecular determinants of Dbr1 activity will aid efforts to design potent Dbr1 inhibitors. Such compounds could be used to study the biological role of Dbr1 and may have therapeutic benefits for diseases such as ALS or conditions where the innate immune response is problematic.
The identification of a crystallizable Dbr1 homolog from Entamoeba histolytica (EhDbr1) enabled structural studies of Dbr110. Subsequent work demonstrated that yeast and amoeba Dbr1 require Fe2+, or Fe2+/Zn2+ for efficient debranching, and described methods for producing highly active enzymes that co-purify with Fe2+ and Zn2+ co-factors11{Clark, 2016 #3, 12. A number of high-resolution Dbr1 structures are now available13, including structures of inactive Dbr1 variants in complex with branched RNA (bRNA) substrates 12.
Phosphorothiate modified nucleic acids are more resistant to hydrolysis than native phosphate-linked nucleic acids, and most therapeutic oligonucleotides contain phosphorothioate modifications 14–16. Previous studies found that phosphorothioate-modified bRNAs were not hydrolyzed by Dbr1. Partially purified Dbr1 from HeLa cell extracts was unable to cleave pure Rp-2′–5′ phosphorothioate containing RNA lariats 17. A subsequent study demonstrated that recombinant yDbr1 had no activity against Rp branched tri-nucleotides, and weak activity towards Sp branched tri-nucleotides, with only a small fraction (~<10%) of the substrate hydrolyzed in 24 hours 18. Based on these studies, we hypothesized that we could obtain a structure of catalytically active EhDbr1 in complex with a 2′–5′ phosphorothioate linked bRNA (PS-bRNA). Surprisingly, the PS-bRNA was hydrolyzed in-crystal, and the hydrolyzed 5′ product was clearly visible in electron density maps (Figures 1-3), providing the first structure of Dbr1 in complex with a product of hydrolysis. We measured the rate of phosphorothioate hydrolysis by EhDBr1 and human Dbr1 (hDbr1, Figure 4) and found that Dbr1 enzymes hydrolyze phosphorothioates 4-orders of magnitudes more slowly than phosphodiesters. The new crystal structure provides new atomic details which will inform the design of structure-based Dbr1 inhibitors.
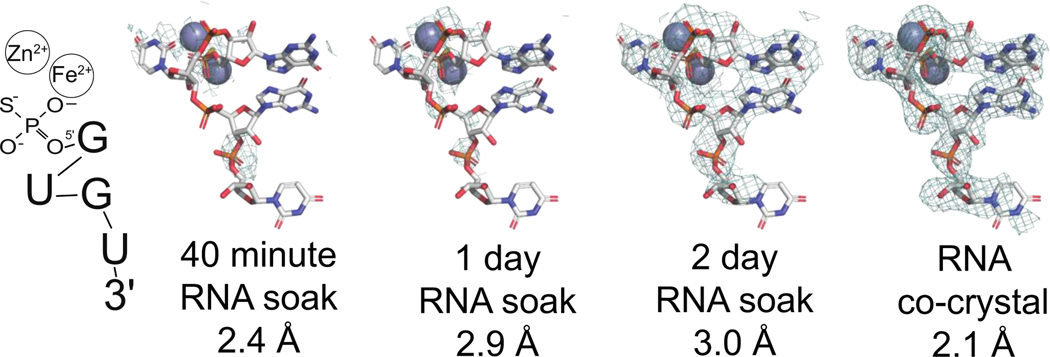
Figure 1.
in-crystal cleavage of PS-bRNA. PS-bRNA was soaked into EhDbr1 crystals for 40 minutes, 1 day, or 2 days. Clear density for the cleaved 5′ arm of PS-bRNA appeared after 48 hours. A co-crystallization experiment produced a higher-resolution structure of the PS-bRNA product (PDB 8DZK). The resolved cleavage product 5′PS-GUGU-3′ is illustrated on the left. Electron density maps contoured at 1 σ. The active site metal ions are gray spheres.
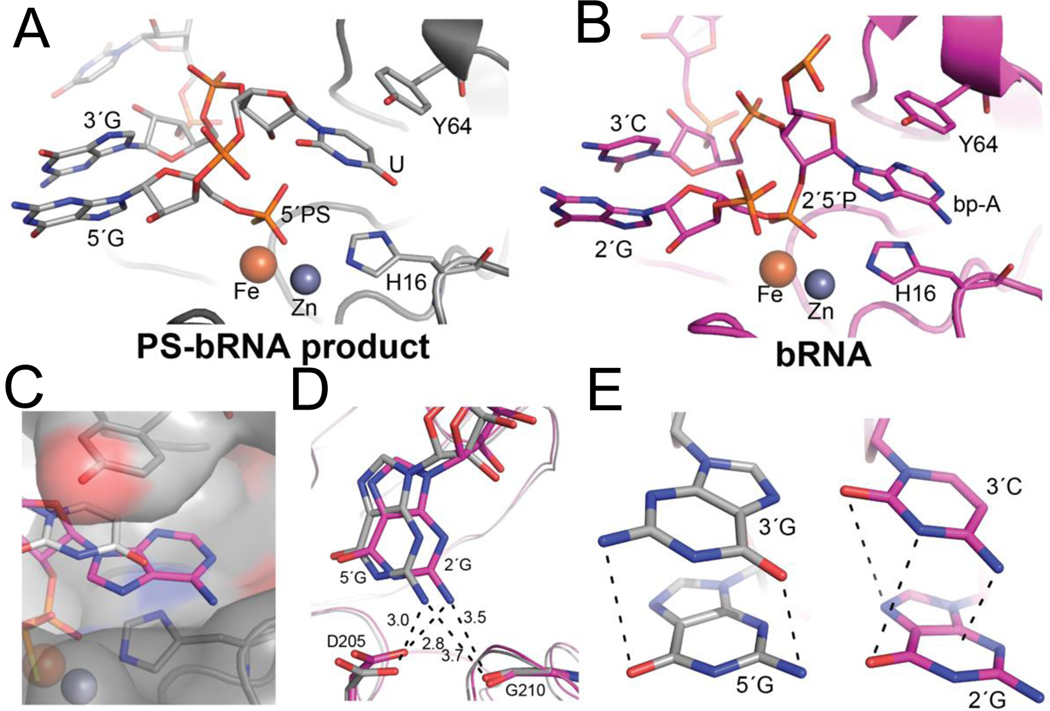
Figure 3.
Active site interactions between PS-bRNA product (A, gray, PDB 8DZK) and bRNA (B, pink, PDB 5K78). A,B) EhDbr1 is shown as a ribbon, and the branchpoint binding pocket formed by Tyr 64 and His 16 shown as sticks. The Zn ion is a gray sphere, the Fe is orange. The PS-bRNA product is in A, and the co-crystal structure with native bRNA is in B. C) For the PS-bRNA product, the U(gray) only partially fills the BBP when compared to the A in bRNA (magenta). D) Hydrogen bonds between D205 and G210 and amino group of G observed in PS-bRNA product and bRNA structures. E) the G-G stack observed in the 5′ -PS-GUGUG crystal structure(left) compared to the G-C stack in the bRNA crystal structure (right). In both structures, the stacking interactions are such that partial dipoles of opposing charges are positioned opposite each other
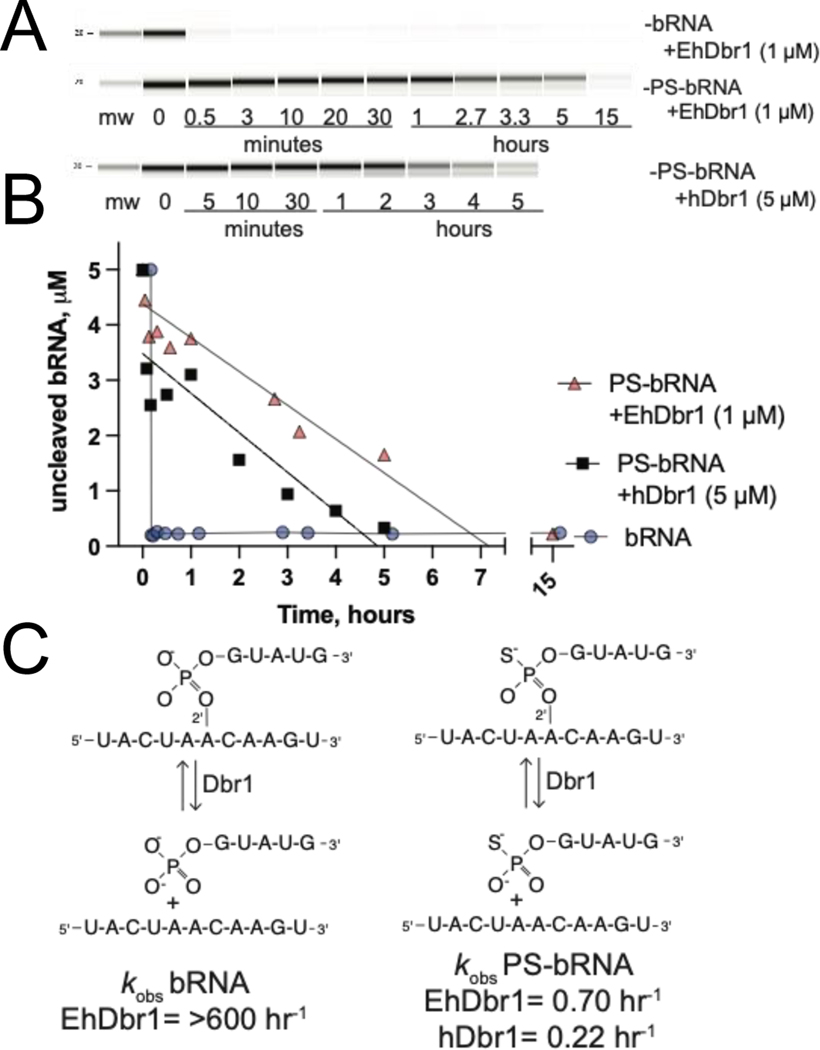
Figure 4.
Kinetics of PS-bRNA and bRNA hydrolysis by EhDbr1 and hDbr1 A) Bioanalyzer gel image showing time course of PS-bRNA or bRNA cleavage by EhDbr1 and hDbr1 B) quantification of uncleaved, intact bRNA as a function of time. C) Schematic of the hydrolysis reactions of bRNA (left) and PS-bRNA (right), with kobs calculated from the plot in A.
Materials and Methods:
Synthesis of PS-bRNA and bRNA molecules.
The sequences used for crystallization (Figure 1) were based on the metazoan consensus 5′ splice site and branchpoint sequences 19. Solid-phase synthesis of PS-bRNAs and bRNA were described previously20. The sequences of the bRNAs used in crystallization are shown in Figures 2C,D, and those used enzymatic assays are shown in Figure 4C. The PS-bRNA used for co-crystallization is based on the metazoan consensus sequence (with a G instead of an A at the 3rd position from the 5’ termini, Figure 2B). Both the PS-bRNA and bRNA molecules used in the cleavage assays are based on the yeast consensus sequence (with an A at the 3rd position from the 5’ termini, Figure 4C, 21).
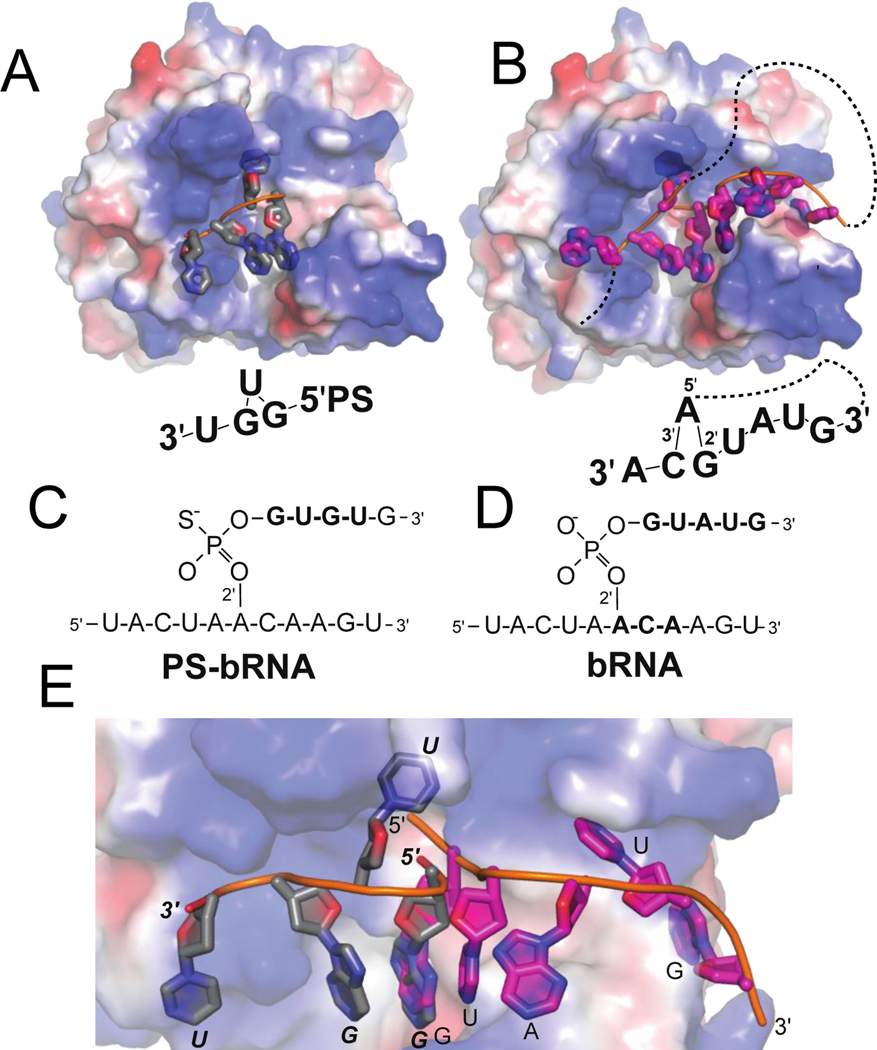
Figure 2.
Comparison of the cleaved PS-bRNA co-crystal structure (A, gray, PDB 8DZK, wt-Dbr1), with intact bRNA co-crystal structure (B, pink, PDB 5K78, catalytically inactive H91A-Dbr1). A,B) Surface view of the entire RNA and binding surface. The surface is colored by electrostatic potential to illustrate positively charged regions (blue) that interact with the phosphate backbone of RNA. RNAs are shown as cartoons. C,D) Schematic view of the synthetic RNAs used in crystallization experiments. The nucleotides that are resolved in the crystal structures(bold) are drawn beneath the surface representation in the approximate orientations. The lariat connectivity is illustrated with a dashed line E). Overlay of the 5′ fragments illustrates the different binding modes, colored as in A-B
Protein expression and purification:
The EhDbr1 expression plasmid (Uniprot C4M1P9) was synthesized as a codon-optimized gene (Genscript), and subcloned into a modified pET 32 vector. The open reading frame consisted of an 8x HIS tag, followed by a tobacco etch virus protease site, and the EhDbr1 open reading frame. The plasmid was transformed into Bl21 E. coli cells. Cells were cultured at 30 deg C. in 12×1L flasks using auto-induction media22. Cells were lysed in Ni-A buffer (50 mM Tris-HCL pH8, 500 mM NaCl, 20 mM imidazole, 1 mM TCEP) with sonication, clarified by centrifugation, applied to a 25 ml Ni-Sepharose FF column (GE Healthcare), and eluted with a shallow gradient of Ni-B buffer (=Ni-A buffer + 400 mM imidazole). Pure fractions were pooled in a dialysis bag, mixed with 1/50 volume of 1 mg/ml TEV protease, and dialyzed against 50 mM NaCl, 20 mM HEPES pH7, 1 mM EDTA, and 1 mM TCEP. After dialysis, the protein was diluted 1:5 in 10 mM sodium phosphate pH7, applied to a 5 ml Heparin Hi-Trap column (GE-Healthcare), and eluted with a gradient of 2 M NaCl. Pure fractions were diluted 10-fold into 20 mM HEPES pH7, 1 mM TCEP, and concentrated to 18 mg/ml for crystallization.
For hDbr1 (Uniprot Q9UK59), the expression plasmid was synthesized as a codon-optimized gene (DNA2.0) and contained a non-cleavable C-terminal 8x HIS tag. Lysis and purification were as described above, except a Q-column was used instead of a Heparin column (buffer A= 50 mM Tris pH 8, 50 mM NaCl, 1 mM TCEP; buffer B=A+1M NaCl), and there was no TEV-protease step.
Protein crystallization:
EhDbr1 (2 uL of 18 mg/ml) and crystallization solution (2 uL of 15% PEG3350, 0.4 M LiSO4, 0.1 M bis-tris pH 5.5) were combined in a hanging drop plate (Nextal) over 0.25 mL of reservoir solution. After 24 hours, the drops were streak-seeded from crystals that appeared in drops containing higher concentrations of PEG3350 using a cat-whisker. 3–4 days after seeding single crystals appeared that were suitable for X-ray diffraction studies. Crystals were moved to a soak solution containing 20% PEG3350, 0.4 M LiSO4 , 0.1 M bis-tris pH 5.5, 7.5% glycerol, 1 mM ZnSO4, and 3 mM PS-bRNA (See Figure 2C for sequence). Crystals were removed and frozen with LN2 after 40 minutes, 24, and 48 hours. For the co-crystallization experiment, 1 uL of protein, 1 uL of reservoir solution, and 1 uL of soak solution were mixed and equilibrated in a hanging drop plate, and streak-seeded 24-hours later.
X-ray data collection, processing, and model building:
Initial diffraction data were collected in-house with a Rigaku 007 source. The best crystals were sent to the Advanced Light Source (40-min, 1-day, 2-day soaks), or Advanced Photon Source (co-crystal). Diffraction data were processed with XDS23. As the crystals were isomorphous with previous EhDbr1 crystals, initial maps were generated by rigid-body fitting with PDB 5K73 12 using PHENIX24. Based on initial maps, models were re-built manually in Coot25, and RNA was built using the R-crane plugin26. The data from the co-crystal structure and final model are deposited with the RCSB, PDB ID 8DZK. The RNA molecule is modeled in all 5 chains in the asymmetric unit, but the density is strongest in chains A, B, and D. As the 40-minute, 1-day, and 2-day soaks were lower resolution, and provided no additional structural insights relative to the high-resolution co-crystal structure, we submitted only the co-crystal structure and data to PDB. All data are available upon request. Figures were generated with PyMol (Schrodinger, LLC). Statistics for data collection and refinement are in Table S1.
Branched RNA cleavage assay:
To measure the kinetics of Dbr1 cleavage, we used 16-mer PS-bRNA and native bRNA molecules. The sequences for these RNA molecules are presented in Figure 4C. The PS-bRNA used here differed from the one used in crystallization by a single G->A substitution in the 2′ arm. EhDbr1 (1 μM) or hDbr1 (5 μM) were combined with either bRNA (5 μM) or PS-bRNA (5 μM) were combined in assay buffer (50 mM HEPES pH7, 100 mM NaCl, 1 mM TCEP) at room temperature. At the indicated timepoints, a 3 μL aliquot was removed, added to 12 μL of water, and frozen at −80 deg C. Timepoints were heated at 90 deg C for 2 minutes to inactivate Dbr1. 1 μL of the diluted reactions was analyzed with a Bioanalyzer using the small RNA kit (Agilent 5067–1548). The Bioanalyzer uses microfluidic chips to perform electrophoresis in the presence of fluorescent dyes to detect separated RNAs from 6–150 nts. The concentration of uncleaved bRNA was quantified with the instrument software (Agilent 2100 Expert), and the observed rates were estimated from plotting the resulting data in Prism and fitting with a linear regression (Graphpad). The disappearance of full-length bRNA was plotted as a function of time to estimate the rates of hydrolysis. Uncropped pseudo-electrophoretograms are presented in Figure S1. Although we were unable to test other microcapillary electrophoresis instruments (such as the Agilent Tapestation, or PerkinElmer GX Touch), we suspect that these instruments would be equally suitable.
Results and Discussion
Crystal structure of EhDbr1 and PS-bRNA-1
Our initial hypothesis was that the presence of a phosphorothioate linkage would prevent hydrolysis of the PS-bRNA by EhDbr1 during structure determination. This PS-bRNA inhibits Dbr1 with an IC50 of 220 nM 20 and we predicted that it would form a Dbr1:PS-bRNA complex suitable for X-ray diffraction studies. All previous structures of Dbr1:bRNA to date used catalytically inactive EhDbr1 12, and we hypothesized that the PS-bRNA would remain intact when soaked into crystals of active Fe2+/Zn2+, offering a view of active-Dbr1 in complex with a bRNA. Pre-grown EhDbr1 crystals were transferred to a solution of PS-bRNA and harvested after 40 min, 24 hour, or 48 hours of soaking. Complete diffraction data were collected for each time point. Electron density maps displayed strong density active site density at the 48-hour timepoint (Figure 1), however this density did not support a branched RNA model. We modeled the 2′–5′ hydrolysis product of the PS-bRNA and found good agreement with the unmodeled density. A higher-resolution co-crystal structure provided a clearer view of this fragment in complex with EhDbr1, confirming that the PS-bRNA was hydrolyzed in crystal. Four of the five nucleotides from the 5′-PS-GUGUG-3′ fragment were clearly resolved in 3 of the 5 EhDbr1 monomers present in the asymmetric unit (Figure 1). This structure is the first example of Dbr1 bound to a cleaved RNA fragment.
The binding mode of the product mimics key features of bRNA:EhDbr1 crystal structures. A comparison of Dbr1 with the PS-bRNA product (PDB 8DZK, Figure 2A,C) with native bRNA substrate (PDB 5K7812, Figure 2 B,D) illustrates how positively charged residues interact with the phosphate backbone (Figure 2 A,B). The PS-bRNA product and bRNA substrate interact with the same RNA binding surfaces, but with different, incompatible binding modes (Figure 2E). In the bRNA substrate, 5K78, the 3’ terminus moves away from the catalytic center (towards the right side of the image), and in 8DZK the 3’ terminus is on the opposite side of the enzyme (towards the left side of the image), blocking the catalytic center. For the PS-bRNA product, the U nucleotide (second from the 5′ end of cleaved product), is flipped into the branchpoint-binding pocket (BBP, discussed below), and the adjacent G residues form a G-G stack (Figure 2A). In bRNA, the branchpoint-forming A is in the BBP, and adjacent C-G residues stack (Figure 2B). The PS-bRNA 5’ product occupies the region on the active site where catalysis would occur, and we hypothesize that this may represent an inhibited-state. This binding mode would not be possible for the uncleaved PS-bRNA, as the 5’-UACUAACAAGU-3’ fragment would be attached to the 5’ termini (Figure 2E) resulting in a steric clash with the enzyme.
The active site of EhDbr1 features two metal ions, a Zn2+ and Fe2+, which bind the scissile 2′–5′-phosphodiester linkage. The 5′ phosphorothioate group binds to both metals in the same position occupied by the 2′–5′-phosphate in bRNA (Figure 3A, B). The branchpoint-binding pocket (BBP) is formed by His 16 (a Zn ligand) on the bottom, and a conserved Tyr (64) on the top (Figure 3A, B). In a native bRNA:EhDBr1(H91A) structure (PDB 5K78, 12, the branchpoint A flips ~180 degrees (relative to the other nucleobases in the bRNA) into the BBP (Figure 3B). The adjacent 2′G and 3′C residues in the bRNA form an aromatic base-stacking interaction. The PS-bRNA product, 5′ -PS-GUGUG-3′, places a U into the BBP, and the adjacent 5′ and 3′ G residues form the stacking interaction (Figure 2A, 3A). However, the U does not fully occupy the branchpoint binding pocket (Figure 3C), as the PS-bRNA has only 3′–5′-phosphodiester linkages, which cannot occupy the same positions as the native bRNA substrate (compare phosphate positions in Figures 3A, 3C).
Intron sequences begin with a 5′ GUA in yeast and a GU(G/A) in metazoa 21. The PS-bRNA used in crystallization is based on the metazoa sequence (Figure 2C), while the bRNA used in 5K78 is based on the yeast sequence. The only sequence-specific hydrogen bonds observed in Dbr1 crystal structures are between the 5′ G, and the branchpoint A to which it is covalently liked though a 5’2’ phosphodiester bond. Presumably, this lack of extensive hydrogen bonding allows Dbr1 enzymes to recognize lariat substrates with diverse sequences that share a common topology conferred by the branchpoint. Interestingly, the 5′-Gs in both structures (PS-bRNA product PDB 8DZK, and bRNA substrate PDB 5K78), occupy the same space and form the same guanine-NH2 Asp205, Gly210 hydrogen bonds (Figure 2D).
The stacking interactions in both the PS-bRNA product and bRNA appear to be favorable as they position partial positive and negative dipoles opposite each other. In PS-bRNA product G-G stack (Figure 3E, left), the carbonyl and amino group of the nucleobases are opposite to each other, which may allow the partial dipoles on these groups to cancel out. For the bRNA G-C stack (Figure 3E, right) the amino group of the C is positioned at the center of the 6-member aromatic ring, and carbonyl groups are opposite the cyclic amine groups.
In summary, the product mimics four key features of native bRNA binding: i) placing a flipped nucleotide in the branchpoint binding pocket; ii) favorable intramolecular stacking of nucleobases adjacent to the flipped nucleotide; iii) ionic interactions between positively charged protein surface and negatively charged phosphate backbone, and iv) hydrogen bonds between the 5′ G (of PS-bRNA-1) or 2′–5′ G (of bRNA) and residues D205 and G210 (Figures 2, 3).
Enzymatic hydrolysis of PS-bRNA and bRNA in solution.
The crystal-soaking time course demonstrated that Dbr1 can hydrolyze PS-bRNA, but we could not estimate a rate of cleavage from these experiments. Although we knew the PS-bRNA concentration (3 mM), it is difficult to estimate the enzyme concentration when crystals are transferred into a drop of substrate, and to estimate the specific activity of crystalline enzyme. Additionally, the crystallization conditions are non-physiological, with a pH of 5.5 and high concentrations of Li2SO4 and polyethylene glycol. Therefore, we used a bRNA cleavage assay to measure how quickly EhDbr1 and hDbr1 could hydrolyze PS-bRNA and bRNA under physiological conditions.
Traditionally, bRNA cleavage assays are performed with 32P-labeled samples and urea-PAGE 18, 27, 28. However, these assays are laborious, and require a considerable investment of time and laboratory resources, so we developed a non-radioactive bRNA cleavage assay. We synthesized and purified the bRNA and PS-bRNA using solid-phase synthesis, as these molecules are not commercially available. Initially we tested if we could detect these bRNAs in urea-PAGE gels using SYBR-Gold stain, silver staining, and UV-shadowing. However, none of those approaches could detect the bRNAs at reasonable concentrations (~20 pmols/lane, or 10 μL of at 5 μM bRNA solution). Next, we tested microcapillary electrophoresis using a Bioanalyzer instrument. The Bioanalyzer reliably detected intact 16-mer bRNAs at 5 μM, and we used the disappearance of intact bRNA to measure Dbr1 cleavage rates. The 11- and 5-mer products were not well resolved (Figure S1). We found that the bRNA was cleaved very quickly by EhDbr1, with 100% cleaved at our first timepoint (30 sec, Figure 4A). In contrast, the PS-bRNA was cleaved very slowly, with 100% cleavage occurring between the 5- and 15- hour timepoints. We quantified intact substrate vs time and based on the slope of that curve (Figure 4B), we estimate that 100% PS-bRNA cleavage occurred around 7 hours for EhDbr1, and 5 hours with hDbr1. From the concentration of enzyme (1 uM EhDbr1, 5 uM hDbr1 on account of lower hDbr1 specific activity) and substrate (5 uM), we calculated turnover rates of >600 hr−1 for bRNA by EhDbr1, 0.70 hr−1 for EhDbr1 and PS-bRNA, and 0.22 hr−1 for hDbr1 and PS-bRNA. The bRNA rate was likely faster than 600 hr−1 (equivalent to 0.2 sec−1), as rates of 2 sec−1 and 5.6 sec−1 for EhDbr1 and yDbr1 were measured in a continuous assay with a fluorescent substrate 11, 12. The high enzyme concentrations necessary to achieve cleavage of the PS-bRNA in a reasonable timeframe limited our ability to measure rates > 0.2 sec-1.
The PS-bRNA used in this assay is based on the yeast consensus sequence, while the PS-bRNA used in crystallization is based on the metazoan consensus sequence and differs by a single G->A substitution at the 3rd position of the 5′ arm (compare Figure 2A to 4C, 21). We used the metazoan substrate for crystallization experiments because we had a larger quantity after synthesis and purification. The crystallization experiments consumed our entire supply of the metazoan substrate, and we switched to the yeast substrate for the hydrolysis assays. Unfortunately, we were unable to systematically test both metazoa and yeast sequences in this assay.
Conclusions.
Here we report that EhDbr1 and hDbr1 enzymes cleave phosphorothioate linked bRNA substrates 3–4-orders of magnitude more slowly than similar phosphate linked bRNA substrates. Dbr1 is a member of the metallophosphoesterase family of enzymes, a family of nucleases and phosphatases that use a common catalytic mechanism and share highly-conserved active site residues 29. Therefore, we hypothesize that like Dbr1, other MPEs may cleave phosphorothioate substrates.
We report a non-radioactive bRNA cleavage assay assisted by capillary electrophoresis analysis. Our results demonstrate that Dbr1 enzymes cleave both Rp and Sp phosphorothioates (Figure 4B), as our PS-bRNA was a mixture diastereomers and was 100% cleaved. This contrasts with previous reports in which Dbr1 samples were unable to hydrolyze PS-bRNA 17 18. The crystal structure of EhDbr1 with the 5′-PS-GUGUG-3′ product (PDB 8DZK) is the first atomic structure of a Dbr1 enzyme in complex with a product of its hydrolysis reaction. Because the binding of product occupies the catalytic center and is incompatible with substrate binding (Figure 2,3), we hypothesize that the product of Dbr1 cleavage may be inhibitory, and that product inhibition may be relevant to the kinetic mechanism of Dbr1. However further experiments would be required to test this hypothesis. The structure we report here compliments previous structures of Dbr1 at different stages of catalysis 10, 12, and provide atomic details for structure-based Dbr1 inhibitor design. For example, tri-nucleotide motifs bind the catalytic center and branchpoint binding pockets in both the product and bRNA crystal structures (PDB 8DZK, 5K78), and small molecules that mimic these structures could be potent inhibitors. Also, as we demonstrate that phosphorothioate linkages are not as resistant to Dbr1 hydrolysis as previously believed, phosphoramidate 30 or phosphonate linkages may be preferable for Dbr1 inhibitors. We hope these results aid the development of Dbr1 inhibitors that may illuminate the biological roles of Dbr1 in normal physiology and diseases such as viral encephalitis, cancer, and ALS 5, 6, 9, 31, 32.
Supplementary Material
Acknowledgements.
This work is based upon research conducted in the Structural Biology Core Facilities, a part of the Institutional Research Cores at the University of Texas Health Science Center at San Antonio supported by the Office of the Vice President for Research and the Mays Cancer Center Drug Discovery and Structural Biology Shared Resource (NIH P30 CA054174). The Rigaku HyPix-6000HE Detector, Universal Goniometer and VariMax-VHF Optic instrumentation in the Structural Biology Core Facilities are funded by NIH-ORIP SIG Grant S10OD030374.
This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Eiger 16M detector on 24-ID-E is funded by a NIH-ORIP HEI grant (S10OD021527). This research used resources of the Advanced Photon Source; a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Grant support.
M.J.D. was supported by a Discovery Grant from the National Science and Engineering Council of Canada (NSERC).
W.G.F was supported by NIH grants R01GM105681 and R01GM127472.
Footnotes
Uniprot accession numbers
human Dbr1: Q9UK5
Entamoeba histolytica Dbr1: C4M1P
Supporting Information: Supporting Information available free of charge:
Table of X-ray data and refinement statistics, and uncropped gel images of branched RNA cleavage assay.
Literature cited
- [1].Ruskin B, Krainer AR, Maniatis T, and Green MR (1984) Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro, Cell 38, 317–331. [DOI] [PubMed] [Google Scholar]
- [2].Padgett RA, Konarska MM, Grabowski PJ, Hardy SF, and Sharp PA (1984) Lariat RNA’s as intermediates and products in the splicing of messenger RNA precursors, Science 225, 898–903. [DOI] [PubMed] [Google Scholar]
- [3].Garrey SM, Katolik A, Prekeris M, Li X, York K, Bernards S, Fields S, Zhao R, Damha MJ, and Hesselberth JR (2014) A homolog of lariat-debranching enzyme modulates turnover of branched RNA, Rna 20, 1337–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zheng S, Vuongs BQ, Vaidyanathan B, Lin JY, Huang FT, and Chaudhuri J. (2015) Non-coding RNA Generated following Lariat Debranching Mediates Targeting of AID to DNA, Cell 161, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Han B, Park HK, Ching T, Panneerselvam J, Wang H, Shen Y, Zhang J, Li L, Che R, Garmire L, and Fei P. (2017) Human DBR1 modulates the recycling of snRNPs to affect alternative RNA splicing and contributes to the suppression of cancer development, Oncogene, 5382–5391. [DOI] [PMC free article] [PubMed]
- [6].Zhang SY, Clark NE, Freije CA, Pauwels E, Taggart AJ, Okada S, Mandel H, Garcia P, Ciancanelli MJ, Biran A, Lafaille FG, Tsumura M, Cobat A, Luo J, Volpi S, Zimmer B, Sakata S, Dinis A, Ohara O, Garcia Reino EJ, Dobbs K, Hasek M, Holloway SP, McCammon K, Hussong SA, DeRosa N, Van Skike CE, Katolik A, Lorenzo L, Hyodo M, Faria E, Halwani R, Fukuhara R, Smith GA, Galvan V, Damha MJ, Al-Muhsen S, Itan Y, Boeke JD, Notarangelo LD, Studer L, Kobayashi M, Diogo L, Fairbrother WG, Abel L, Rosenberg BR, Hart PJ, Etzioni A, and Casanova JL (2018) Inborn Errors of RNA Lariat Metabolism in Humans with Brainstem Viral Infection, Cell 172, 952–965 e918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mackenzie IR, Bigio EH, Ince PG, Geser F, Neumann M, Cairns NJ, Kwong LK, Forman MS, Ravits J, Stewart H, Eisen A, McClusky L, Kretzschmar HA, Monoranu CM, Highley JR, Kirby J, Siddique T, Shaw PJ, Lee VM, and Trojanowski JQ (2007) Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations, Annals of neurology 61, 427–434. [DOI] [PubMed] [Google Scholar]
- [8].Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, and Lee VM (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis, Science 314, 130–133. [DOI] [PubMed] [Google Scholar]
- [9].Armakola M, Higgins MJ, Figley MD, Barmada SJ, Scarborough EA, Diaz Z, Fang X, Shorter J, Krogan NJ, Finkbeiner S, Farese RV Jr., and Gitler AD (2012) Inhibition of RNA lariat debranching enzyme suppresses TDP-43 toxicity in ALS disease models, Nature genetics 44, 1302–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Montemayor EJ, Katolik A, Clark NE, Taylor AB, Schuermann JP, Combs DJ, Johnsson R, Holloway SP, Stevens SW, Damha MJ, and Hart PJ (2014) Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1, Nucleic Acids Res 42, 10845–10855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Clark N, Katolik A, Taggart AJ, Buerer L, Holloway S, Miller N, Phillips JD, Farrell CP, Damha MJ, and Fairbrother W. (2022) Metal content and kinetic properties of yeast RNA lariat debranching enzyme Dbr1, Rna 28, 927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Clark NE, Katolik A, Roberts KM, Taylor AB, Holloway SP, Schuermann JP, Montemayor EJ, Stevens SW, Fitzpatrick PF, Damha MJ, and Hart PJ (2016) Metal dependence and branched RNA cocrystal structures of the RNA lariat debranching enzyme Dbr1, Proc Natl Acad Sci U S A 113, 14727–14732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ransey E, Paredes E, Dey SK, Das SR, Heroux A, and Macbeth MR (2017) Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation, FEBS Lett 591, 2003–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Eckstein F. (2014) Phosphorothioates, essential components of therapeutic oligonucleotides, Nucleic Acid Ther 24, 374–387. [DOI] [PubMed] [Google Scholar]
- [15].Volk DE, and Lokesh GLR (2017) Development of Phosphorothioate DNA and DNA Thioaptamers, Biomedicines 5, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Corey DR, Damha MJ, and Manoharan M. (2022) Challenges and Opportunities for Nucleic Acid Therapeutics, Nucleic Acid Ther 32, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Maschhoff KL, and Padgett RA (1993) The stereochemical course of the first step of pre-mRNA splicing, Nucleic Acids Res 21, 5456–5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mourani R, and Damha MJ (2006) Synthesis, characterization, and biological properties of small branched RNA fragments containing chiral (Rp and Sp) 2’,5’-phosphorothioate linkages, Nucleosides Nucleotides Nucleic Acids 25, 203–229. [DOI] [PubMed] [Google Scholar]
- [19].Nam K, Hudson RH, Chapman KB, Ganeshan K, Damha MJ, and Boeke JD (1994) Yeast lariat debranching enzyme. Substrate and sequence specificity, Journal of Biological Chemistry 269, 20613–20621. [PubMed] [Google Scholar]
- [20].Katolik A, Clark NE, Tago N, Montemayor EJ, Hart PJ, and Damha MJ (2017) Fluorescent Branched RNAs for High-Throughput Analysis of Dbr1 Enzyme Kinetics and Inhibition, ACS Chem Biol 12, 622–627. [DOI] [PubMed] [Google Scholar]
- [21].Li C, and Tarn W-Y (2006) Splicing, In Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine, pp 1788–1792, Springer Berlin Heidelberg, Berlin, Heidelberg. [Google Scholar]
- [22].Studier FW (2005) Protein production by auto-induction in high density shaking cultures, Protein Expr Purif 41, 207–234. [DOI] [PubMed] [Google Scholar]
- [23].Kabsch W. (2010) Xds, Acta Crystallogr D Biol Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, and Adams PD (2019) Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix, Acta Crystallogr D Struct Biol 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Emsley P, Lohkamp B, Scott WG, and Cowtan K. (2010) Features and development of Coot, Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Keating KS, and Pyle AM (2010) Semiautomated model building for RNA crystallography using a directed rotameric approach, Proc Natl Acad Sci U S A 107, 8177–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Khalid MF, Damha MJ, Shuman S, and Schwer B. (2005) Structure-function analysis of yeast RNA debranching enzyme (Dbr1), a manganese-dependent phosphodiesterase, Nucleic Acids Res 33, 6349–6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nam K, Hudson RH, Chapman KB, Ganeshan K, Damha MJ, and Boeke JD (1994) Yeast lariat debranching enzyme. Substrate and sequence specificity, J Biol Chem 269, 20613–20621. [PubMed] [Google Scholar]
- [29].Matange N, Podobnik M, and Visweswariah SS (2015) Metallophosphoesterases: structural fidelity with functional promiscuity, Biochem J 467, 201–216. [DOI] [PubMed] [Google Scholar]
- [30].Tago N, Katolik A, Clark NE, Montemayor EJ, Seio K, Sekine M, Hart PJ, and Damha MJ (2015) Design, Synthesis, and Properties of Phosphoramidate 2’,5’-Linked Branched RNA: Toward the Rational Design of Inhibitors of the RNA Lariat Debranching Enzyme, J Org Chem 80, 10108–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Galvis AE, Fisher HE, Fan H, and Camerini D. (2017) Conformational Changes in the 5’ End of the HIV-1 Genome Dependent on the Debranching Enzyme DBR1 during Early Stages of Infection, J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ye Y, De Leon J, Yokoyama N, Naidu Y, and Camerini D. (2005) DBR1 siRNA inhibition of HIV-1 replication, Retrovirology 2, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.