Abstract
Background and aim:
Since December 2019, the Coronavirus disease 2019, caused by severe acute respiratory syndrome coronavirus 2 (Sars-CoV-2), has spread from China, becoming a pandemic. Bacterial and fungal co-infections may lead to increase in COVID-19 severity with a decrease in patients survive. The aim of this work was to evaluate bacterial and fungal co-infections in COVID-19 patients admitted to ICU in comparison with patients recovered in ICU in pre-COVID-19 era in order to understand whether the pandemic had changed the incidence of overinfections in patients admitted to ICU. In fact, the epidemiological data should guide the choice of empirical therapy.
Methods:
During pandemic, AOUC Policlinico of Bari organized dedicated ICUs for patient with SARS-CoV-2. Blood cultures, urine, and tracheobronchial aspirate were included in the analysis.
Results:
Specimens of 1905 patients were analysed in this work. Comparing clinical isolates prevalence by material and COVID-19 vs. non-COVID-19 patients statistically significant differences were detected for A. baumannii complex, Aspergillus fumigatus, Escherichia coli, Haemophilus influenzae and Serratia marcescens isolated from tracheobronchial aspirates; C. albicans from urine samples, A. baumannii complex, Enterococcus faecalis and Enterococcus faecium isolated from blood culture.
Conclusions:
Although the organisms isolated in COVID-19 patients are consistent with those frequently associated with healthcare associated infection, our data suggest a particular prevalence in COVID-19 patients of A. baumannii, Stenotrophomonas maltophilia and Aspergillus spp. in the respiratory tract, C. albicans in urine and A. baumannii, E. faecalis and E. faecium in blood cultures. (www.actabiomedica.it)
Keywords: COVID-19, coinfection, epidemiology
Introduction
Since December 2019, the Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (Sars-CoV-2), has spread from its epicentre in Wuhan, across mainland China and became a global threat. In Italy, Sars-CoV-2 has infected more than 14 million people and caused more than 150.000 deaths (1).
As the COVID-19 pandemic proceeds, many studies have been published to collect epidemiological and clinical data on the disease. Current reports revealed that bacterial or fungal co-infections are observed among COVID-19 patients. Systematic reviews found that a small percentage of hospitalized patients had a secondary infection, although co-infection rates increase in patients admitted to the ICU (2-4).
Bacterial and fungal co-infections may lead to increase in disease severity with a decrease in survive of COVID-19 patients, in particular a secondary infection was reported in 50% of non-survivors and only 1% of survivors (5). For patients admitted to ICU, deaths related to co-infections by bacteria, fungi and other viruses occurred in half of them (6).
Healthcare associated infections (HAIs) issue is already well known all over the world and ICUs are the hospital wards with the highest prevalence. Indeed, according to European Centre for Disease Prevention and Control (ECDC), 8.3% of the patients who stayed in ICUs for more than two days were affected by at least once acquired pneumonia, bloodstream infection, or urinary tract infection. In Italian ICUs, the most frequently isolated microorganisms in pre-COVID era were multidrug-resistance (MDR) gram negative bacteria, such as Pseudomonas aeruginosa, Klebsiella spp., and Escherichia coli, coagulase-negative Staphylococci and Staphylococcus aureus (7).
Considering that most of the HAIs are considered preventable and associated with the use of invasive devices, understanding their burden and incidence, also in COVID-19 patients admitted in ICUs, should help the implementation of appropriate infection control and antimicrobial stewardship activities.
This study explored bacterial and fungal co-infections in COVID-19 patients admitted to ICU in comparison with patients recovered in ICU in pre-COVID-19 era. Bacterial and fungal over-infection can occur in ICU patients and is associated with morbidity and mortality. Data on severe COVID-19 infection and concomitant over-infection are lacking. The aim was to describe the incidence and nature of co-infection in COVID-19 patients admitted to ICU and to understand whether the pandemic changed the incidence of over-infections in these critically ill patients. The goal was to assess whether Sars-cov-2 was a risk factor for specific superinfections to use epidemiological data to guide choice of empirical therapy.
Patients and methods
Patients eligibility and microbiological data
All patients admitted to both the non-COVID-19 UTIs (January 2018 to the end of December 2019) and the COVID-19 UTIs (beginning of March 2020 to the end of December 2021) subjected to microbiological cultural analysis were retrospectively included in the study. The microbiological cultural data were limited to following samples: tracheobronchial aspirate, urine, and blood cultures. Only the first clinical isolate from each sample per patient was retained for final analysis.
Due to the retrospective nature and the eligibility criteria of the study a formal sample size calculation was not performed.
This study follows the 305 ethical standards of the institutional and/or national research committee and the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Sample and patients’ information (date of sampling, ward, type of specimen, testing results, sex) have been recorded in an anonymous database by transforming sensitive data into alphanumeric codes. No clinical data associated with these specimens were available. This was designed as a retrospective study, so specific approval by the Ethics Committee was not required by the Italian laws.
Statistical analysis
The Independence of categorical variables was assessed by two-tailed Fisher’s exact test or Chi Squared test as appropriate. In particular, the comparison of all the prevalence rates of the clinical isolates per material was performed by the two-tailed Fisher’s exact test with Benjamini and Hochberg’s correction with FDR<0.1. Moreover, evaluation of the effect size was performed by Cramer’s V. Cramer’s V is a measure of association between two nominal random variables. The coefficient ranges between 0 (no relationship) and 1 (perfect relationship). The equation to calculate the Cramer’s V on a kxr table is:
χ^2=Chi-squared value
k,r=number of columns and rows, respectively
n=number of observations
The comparison of the age of the patients was evaluated per sex and ward by Kruskal-Wallis test and the epsilon squared (implemented in the R package r-companion, version 2.4.1) was calculated as effect size.
Shannon’s Index and Shannon’s Equability Index were used to evaluate the evenness of the isolates both total and per material. The Shannon’s Index values of the COVID-19 and non-COVID-19 wards were compared by the two-tailed Hutcheson’s test.
Shannon’s Index was calculated as:
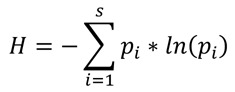
Shannon’s Equability Index was calculated as:
![]()
S=Number of species
pi=Proportion of the specie i
Shannon’s Equability Index normalizes the Shannon’s Index to a value comprised between 0 (maximum diversity) and 1 (minimum diversity).
The calculations of all statistical tests were performed by the open-source environment R 4.0.3 (8,9).
Results
A total of 1905 patients were reviewed. In particular, 619 were COVID-19 patients and 1286 were non-COVID-19 patients. Demographically, 724 patients were females and 1181 were males (F/M 0.613). In COVID-19 ICUs the female and male patients were 32.96% (204/619) and 67.04% (415/619), respectively, while in non-COVID-19 ICUs they were 40.43% (520/1286) and 59.56% (766/1286) (Chi Squared test p-value: 0.002, Cramer’s V: 0.072). The median age of female and male patients in COVID-19 ICUs was 66 (Interquantile Range [IQR]: 58-74) and 66 (IQR: 56-73) while in non-COVID-19 ICUs it was 65 (IQR: 48-74) and 64 (49-75), respectively. Comparison of the age of the four groups (Females COVID-19+, Males COVID-19+, Females COVID-19- and Males COVID-19-) by Kruskal test was not statistically significant (p-value: 0.059) with an epsilon squared value=0.0038.
From COVID-19 patients were analysed 1619 samples (558 blood cultures, 476 tracheobronchial aspirates and 585 urines) while 3400 samples from non-COVID-19 patients (1068 blood cultures, 1126 tracheobronchial aspirates and 1206 urines). The overall prevalence of clinical isolates from tracheobronchial aspirates in COVID-19 and non-COVID-19 patients was 13.45% (64/476) and 17.41% (196/1126) (Fisher’s exact test p-value: 0.0538, Cramer’s V: 0.049), and from urines 11.45% (67/585) and 9.37% (113/1206), respectively (Fisher’s exact test p-value: 0.180, Cramer’s V: 0.032). Nevertheless, the prevalence of clinical isolates from blood culture was statistically lower in COVID-19 (10.04%, 56/558) than in non-COVID-19 patients (17.32%, 185/1068) (Fisher’s exact test p-value: <0.001, Cramer’s V: 0.097).
The analysis of the Shannon’s Equability index by material and ward highlighted a value of 0.773 in the COVID-19 UTIs and 0.879 in the non-COVID-19 UTIs (Table 1).
Table 1.
Shannon’s Index and Shannon’s Equability Index by ward and material.
| Ward | Material | Shannon’s Index / Shannon’s Equability Index |
P value (Hutcheson’s test) |
|---|---|---|---|
| Covid-19 ICU | 2.425 / 0.7733 | <0.001 | |
| Non Covid-19 ICU | 2.588 / 0.879 | ||
| Covid-19 ICU | Tracheobronchial Aspirate | 2.101 / 0.680 | <0.001 |
| Non Covid-19 ICU | Tracheobronchial Aspirate | 2.429 / 0.841 | |
| Covid-19 ICU | Urine | 2.180 / 0.805 | <0.001 |
| Non Covid-19 ICU | Urine | 2.376 / 0.877 | |
| Covid-19 ICU | Blood | 1.765 / 0.636 | 0.091 |
| Non Covid-19 ICU | Blood | 1.889 / 0.667 |
In particular, the Shannon’s Equitabily index values obtained from isolates of the tracheobronchial aspirates in the COVID-19 UTIs and non-COVID-19 UTIs were 0.680 and 0.841 (p-value <0.001), respectively.
Globally, the most prevalent clinical isolates were coagulase negative Staphylococci (588, 15.63%), Acinetobacter baumannii complex (581, 15.44%), Candida albicans (438, 11.64%), Klebsiella pneumoniae (382, 10.15%) and Pseudomonas aeruginosa (289, 7.68%).
By comparing clinical isolates prevalence by material and patients (COVID-19 Vs. non- COVID-19) several statistically significant differences were detected. However, it’s important to highlight the small values of the Cramer’s V except for A. baumannii complex (0.332), Aspergillus fumigatus (0.139), Escherichia coli (0.111), Haemophilus influenzae (0.118) and Serratia marcescens (0.106) isolated from tracheobronchial aspirates (Table 2), C. albicans (0.169) from urine samples (Table 3), A. baumannii complex (0.217), Enterococcus faecalis (0.124) and Enterococcus faecium (0.168) isolated from blood culture (Table 4).
Table 2.
Comparison of prevalence of the clinical isolates collected from tracheobronchial.
| Microrganism | TA Covid-19 (%) | TA Non Covid-19 (%) | Cramer’s V | Corrected P-values (Significant) |
|---|---|---|---|---|
| Acinetobacter baumannii group | 45.59 | 14.65 | 0.332 | TRUE |
| Aspergillus flavus/oryzae | 0.42 | 0.0 | 0.054 | FALSE |
| Aspergillus fumigatus | 4.2 | 0.44 | 0.139 | TRUE |
| Aspergillus nidulans | 0.21 | 0.0 | 0.038 | FALSE |
| Aspergillus niger | 0.42 | 0.09 | 0.035 | FALSE |
| Aspergillus spp. | 0.42 | 0.0 | 0.054 | FALSE |
| Aspergillus terreus | 0.84 | 0.0 | 0.077 | TRUE |
| Candida albicans | 13.66 | 12.79 | 0.012 | FALSE |
| Candida glabrata | 4.62 | 3.46 | 0.028 | FALSE |
| Candida parapsilosis | 0.21 | 0.44 | 0.018 | FALSE |
| Candida tropicalis | 1.68 | 1.42 | 0.01 | FALSE |
| Coagulase negative Staphylococcus | 0.0 | 0.0 | NA | NA |
| Enterobacter cloacae complex | 1.89 | 2.75 | 0.025 | FALSE |
| Enterococcus faecalis | 0.21 | 0.27 | 0.005 | FALSE |
| Enterococcus faecium | 0.21 | 0.44 | 0.018 | FALSE |
| Escherichia coli | 0.84 | 5.77 | 0.111 | TRUE |
| Haemophilus influenzae | 0.63 | 5.86 | 0.118 | TRUE |
| Klebsiella pneumoniae | 8.82 | 13.5 | 0.065 | TRUE |
| Proteus mirabilis | 1.26 | 1.51 | 0.01 | FALSE |
| Pseudomonas aeruginosa | 6.93 | 13.23 | 0.091 | TRUE |
| Serratia marcescens | 2.52 | 8.26 | 0.106 | TRUE |
| Staphylococcus aureus | 6.3 | 10.92 | 0.072 | TRUE |
| Stenotrophomonas maltophilia | 7.98 | 4.53 | 0.069 | TRUE |
aspirates (TA). TRUE: Statistically significant after Benjamini and Hochberg’s correction. FALSE: Not statistically significant after Benjamini and Hochberg’s correction. NA: Not available.
Table 3.
Comparison of prevalence of the clinical isolates collected from urines.
| Microrganism | Urine Covid-19 (%) |
Urine Non Covid-19 (%) |
Cramer’s V | Corrected P-values (Significant) |
|---|---|---|---|---|
| Acinetobacter baumannii group | 4.27 | 1.99 | 0.066 | TRUE |
| Aspergillus flavus/oryzae | 0.0 | 0.0 | NA | NA |
| Aspergillus fumigatus | 0.0 | 0.0 | NA | NA |
| Aspergillus nidulans | 0.0 | 0.0 | NA | NA |
| Aspergillus niger | 0.0 | 0.0 | NA | NA |
| Aspergillus spp. | 0.0 | 0.0 | NA | NA |
| Aspergillus terreus | 0.0 | 0.0 | NA | NA |
| Candida albicans | 18.97 | 7.55 | 0.169 | TRUE |
| Candida glabrata | 8.03 | 4.15 | 0.081 | TRUE |
| Candida parapsilosis | 2.39 | 1.33 | 0.039 | FALSE |
| Candida tropicalis | 1.88 | 0.91 | 0.041 | FALSE |
| Coagulase negative Staphylococcus | 0.85 | 0.91 | 0.003 | FALSE |
| Enterobacter cloacae complex | 0.51 | 0.58 | 0.004 | FALSE |
| Enterococcus faecalis | 8.72 | 7.21 | 0.026 | FALSE |
| Enterococcus faecium | 2.39 | 1.08 | 0.051 | TRUE |
| Escherichia coli | 6.67 | 7.88 | 0.022 | FALSE |
| Haemophilus influenzae | 0.0 | 0.0 | NA | NA |
| Klebsiella pneumoniae | 5.3 | 5.89 | 0.012 | FALSE |
| Proteus mirabilis | 0.68 | 2.32 | 0.058 | TRUE |
| Pseudomonas aeruginosa | 2.74 | 3.9 | 0.03 | FALSE |
| Serratia marcescens | 0.17 | 3.23 | 0.097 | TRUE |
| Staphylococcus aureus | 0.0 | 0.0 | NA | NA |
| Stenotrophomonas maltophilia | 0.17 | 0.08 | 0.012 | FALSE |
TRUE: Statistically significant after Benjamini and Hochberg’s correction. FALSE: Not statistically significant after Benjamini and Hochberg’s correction. NA: Not available
Table 4.
Comparison of prevalence of the clinical isolates collected from blood coltures.
| Microrganism | Blood Covid-19 (%) |
Blood Non Covid-19 (%) |
Cramer’s V | Corrected P-values (Significant) |
|---|---|---|---|---|
| Acinetobacter baumannii group | 17.92 | 4.68 | 0.217 | TRUE |
| Aspergillus flavus/oryzae | 0.0 | 0.0 | NA | NA |
| Aspergillus fumigatus | 0.18 | 0.0 | 0.034 | FALSE |
| Aspergillus nidulans | 0.0 | 0.0 | NA | NA |
| Aspergillus niger | 0.0 | 0.0 | NA | NA |
| Aspergillus spp. | 0.0 | 0.0 | NA | NA |
| Aspergillus terreus | 0.0 | 0.0 | NA | NA |
| Candida albicans | 1.79 | 1.59 | 0.007 | FALSE |
| Candida glabrata | 0.18 | 0.19 | 0.001 | FALSE |
| Candida parapsilosis | 1.43 | 1.22 | 0.009 | FALSE |
| Candida tropicalis | 0.0 | 0.09 | 0.018 | FALSE |
| Coagulase negative Staphylococcus | 40.14 | 32.58 | 0.075 | TRUE |
| Enterobacter cloacae complex | 1.25 | 0.75 | 0.025 | FALSE |
| Enterococcus faecalis | 7.35 | 2.25 | 0.124 | TRUE |
| Enterococcus faecium | 6.99 | 0.94 | 0.168 | TRUE |
| Escherichia coli | 0.54 | 3.65 | 0.093 | TRUE |
| Haemophilus influenzae | 0.0 | 0.09 | 0.018 | FALSE |
| Klebsiella pneumoniae | 3.41 | 6.27 | 0.061 | TRUE |
| Proteus mirabilis | 0.36 | 1.03 | 0.036 | FALSE |
| Pseudomonas aeruginosa | 1.97 | 3.09 | 0.033 | FALSE |
| Serratia marcescens | 0.54 | 3.09 | 0.082 | TRUE |
| Staphylococcus aureus | 1.97 | 3.46 | 0.042 | FALSE |
| Stenotrophomonas maltophilia | 1.08 | 0.37 | 0.043 | FALSE |
TRUE: Statistically significant after Benjamini and Hochberg’s correction. FALSE: Not statistically significant after Benjamini and Hochberg’s correction. NA: Not available
Discussion
Patients admitted to ICUs are more prone to infections for multiple reasons: the primary diagnosis and the resulting immunosuppression state, the invasive medical devices used for monitoring patients’ status, the medical therapies that compromise natural defence mechanisms, and the close contact of the patients with the hospital staff, favouring, therefore, the cross contamination between patients in the same ward. In addition, the selective pressure exerted by exposure to antibiotic therapies encourage the selection of resistant pathogens.
A recent meta-analysis indicated that 7% of hospitalized COVID-19 patients had a bacterial co-infection, increasing to 14% in studies that included only ICU patients. However, it is noteworthy that the authors of the meta-analysis reported a high heterogeneity of the studies in terms of populations (ICU and non-ICU patients), microbiological samplings and bacterial isolates (2).
Data on COVID-19 infection and concomitant overinfection were discordant so this study described the incidence and nature of bacterial and fungal co-infections in COVID-19 patients admitted to ICU compared to patients recovered in ICU in the pre-COVID-19 era. The aim was to assess whether Sars-cov-2 was a risk factor for specific superinfections to use epidemiological data to guide choice of empirical therapy.
Regarding the pathogens responsible for coinfections in COVID-19 patients, the most common bacteria isolated were Klebsiella species, Staphylococcus aureus, and E. coli (10). As regards respiratory coinfections, Sharifipour et al. reported A. baumannii and S. aureus as the main responsible (11), while Temperoni et al. detected a high prevalence of Carbapenem-resistant A. baumannii, and, among gram-positive bacteria, S. aureus, E. faecalis and MDR E. faecium (12).
Mazzariol et al. reported that the most frequently isolated bacterial species in bronchial aspirate samples collected from mechanically ventilated patients with severe COVID-19 was carbapenem resistant P. aeruginosa (13).
Nori et al. found that most frequently isolated organisms from blood cultures were gram-positive bacteria, in particular S. aureus as the main one. For gram-negative bacteria, E. coli and P. aeruginosa were the most frequent. Also, Gil et al. described a similar situation with a prevalence of S. aureus and gram-positive bacteria as the main responsible of bacteraemia in COVID-19 patients (14,15).
Sepulveda et al. evaluated the organisms responsible of bacteraemia in COVID-19 and non-COVID-19 patients revealing that the rate of bacteraemia was significantly lower among the former than in the latter. Among COVID-19 patients, the most common causes of bacteraemia were E. coli, S. aureus, K. pneumoniae, and E. cloacae complex but none of these organisms were overrepresented comparing to non-COVID-19 patients (16). Also our data confirmed the decrease in the prevalence of clinical blood culture isolates.
Our purpose was to evaluate bacterial and/or fungal co-infections and assess whether the pandemic changed the incidence of pathogens affecting patients admitted to ICUs by comparing the results with pre-pandemic data. The objective was to understand if epidemiological features had changed in order to provide a thorough representation of the situation and evaluate prevention activities. Outbreaks or changes in the infectious disease distribution pattern should be investigated to detect trends. In fact, in our case, the incidence of co-infections remained the same, but the distribution of cases changed.
In particular, the analysis of blood culture data revealed an increase in the isolation of Enterococcus spp., and mainly A. baumannii as the gram-negative bacteria responsible for bacteraemia in COVID-19 patients with a tripled prevalence compared to pre-COVID-19 era. Even tracheobronchial aspirates analysis showed an increase of A. baumannii isolates, and a decrease of E. coli, H. influenzae and S. marcescens isolates. The situation remained rather stable in urine samples data except for an increase in the isolation of Candida spp. In addition, our analysis highlighted an increased risk for COVID-19 critical patients to develop respiratory coinfection with Aspergillus spp. in accordance with current literature (17,18). In fact, Sars-CoV-2 directly damages airway epithelium, enabling Aspergillus invasion and clinicians should consider Aspergillus spp. co-infection, which is likely to further increase mortality rates.
Limitations
These data underline the importance of microbiological surveillance to detect variations in pathogens distribution. Providing clinicians with useful information to set up empirical therapies could improve patients’ outcomes and reduce unnecessary administration of antibiotics.
The purely epidemiological nature of our analysis is the main limitation of this study. In fact, the absence of clinical data of patient did not permit any kind of stratification, which could affect the results. The question that naturally arises is why there is such a difference in microbiological isolates of some bacteria compared to others during COVID-19 and pre-COVID-19 era. Are there perhaps predisposing factors during Sars-CoV-2 infection or is it depends on a different patient management by healthcare professionals? Moreover, an important limitation of this study is the retrospective nature of the analysis that may have problems in recalling previous environmental exposures. The differences in prevalence could be due to temporal trends without any association with Covid-19.
Conclusion
To our knowledge, this is the first study that compares microbiological data of COVID-19 and pre-COVID-19 era patients admitted to ICUs. Summing up, although the organisms isolated in COVID-19 patients are consistent with those frequently associated with HAIs, our data suggest a particular prevalence in COVID-19 patients of A. baumannii, Stenotrophomonas maltophilia and Aspergillus spp. in the respiratory tract, C. albicans in urine and A. baumannii, E. faecalis and E. faecium in blood cultures.
Despite these limitations, epidemiological studies are remarkably productive in clarifying etiological factors and prevention through monitoring of disease prevalence.
For these reasons, it would be interesting to investigate the environment and evaluating the patients management by healthcare professionals may help to understand if there are predisposing factors that contribute to bacterial and fungal outbreak.
Conflict of Interest:
Each author declares that he or she has no commercial associations (e.g. consultancies, stock ownership, equity interest, patent/licensing arrangement etc.) that might pose a conflict of interest in connection with the submitted article.
References
- Task force COVID-19 del Dipartimento Malattie Infettive e Servizio di Informatica, Istituto Superiore di Sanità. Epidemia COVID-19, Aggiornamento nazionale: 18 novembre 2020 [Google Scholar]
- Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81:266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattorini L, Creti R, Palma C, Pantosti A. Bacterial coinfections in COVID-19: an underestimated adversary. Ann Ist Super Sanità. 2020;56:359–364. doi: 10.4415/ANN_20_03_14. [DOI] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–62. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Hu C, Luo L, et al. Clinical features and short-term outcomes of 221 patients with COVID-19 in Wuhan, China. J Clin Virol. 2020;127:104364. doi: 10.1016/j.jcv.2020.104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control. Stockholm: ECDC; 2019. Healthcare-associated infections acquired in intensive care units. In: ECDC. Annual epidemiological report for 2017. [Google Scholar]
- Mangiafico S. The Comprehensive R Archive Network: Vienna, Austria; 2021. (2021). Rcompanion: Functions to Support Extension Education Program Evaluation. [Google Scholar]
- R Core Team. R Foundation for Statistical Computing, Vienna, Austria; 2020. (2020). R: A language and environment for statistical computing. [Google Scholar]
- Mahmoudi H. Bacterial co-infections and antibiotic resistance in patients with COVID-19. GMS Hyg Infect Control. 2020;15:Doc35. doi: 10.3205/dgkh000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifipour E, Shams S, Esmkhani M, et al. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect Dis. 2020;20:1–7. doi: 10.1186/s12879-020-05374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperoni C, Caiazzo L, Barchiesi F. High prevalence of antibiotic resistance among opportunistic pathogens isolated from patients with COVID-19 under mechanical ventilation: results of a single-center study. Antibiotics. 2021;10:1080. doi: 10.3390/antibiotics10091080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzariol A, Benini A, Unali I, et al. Dynamics of SARS-CoV2 infection and multi-drug resistant bacteria superinfection in patients with assisted mechanical ventilation. Front Cell Infect Microbiol. 2021;12:683409. doi: 10.3389/fcimb.2021.683409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nori P, Cowman K, Chen V, et al. Bacterial and fungal coinfections in COVID-19 patients hospitalized during the New York City pandemic surge. Infect Control Hosp Epidemiol. 2021;42:84–88. doi: 10.1017/ice.2020.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil E, Martyn E, Rokadiya S, Jain S, Chin TL. Bacterial Coinfection in COVID-19. Clin Infect Dis. 2021;73:e843–e845. doi: 10.1093/cid/ciaa1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda J, Westblade LF, Whittier S, Satlin MJ, Greendyke WG, Aaron JG, Zucker J, Dietz D, Sobieszczyk M, Choi JJ, Liu D, Russell S, Connelly C, Green DA. Bacteremia and Blood Culture Utilization during COVID-19 Surge in New York City. J Clin Microbiol. 2020;58:e00875–20. doi: 10.1128/JCM.00875-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Arkel ALE, Rijpstra TA, Belderbos HNA, van Wijngaarden P, Verweij PE, Bentvelsen RG. COVID-19-associated Pulmonary Aspergillosis. Am J Respir Crit Care Med. 2020;202:132–135. doi: 10.1164/rccm.202004-1038LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler P, Cornely OA, Böttiger BW, et al. COVID-19 associated pulmonary aspergillosis. Mycoses. 2020;63:528–34. doi: 10.1111/myc.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]


