Abstract
Introduction
The glomerular filtration rate (GFR) is crucial for chronic kidney disease (CKD) diagnosis and therapy. Various studies have sought to recognize ideal endogenous markers to improve the estimated GFR for clinical practice. To screen out potential novel metabolites related to GFR (mGFR) measurement in CKD patients from the Chinese population, we identified more biomarkers for improving GFR estimation.
Methods
Fifty-three CKD participants were recruited from the Third Affiliated Hospital of Sun Yat-sen University in 2020. For each participant, mGFR was evaluated by utilizing the plasma clearance of iohexol and collecting serum samples for untargeted metabolomics analyses by ultrahigh-performance liquid chromatography-tandem mass spectroscopy. All participants were divided into four groups according to mGFR. The metabolite peak area data were uploaded to MetaboAnalyst 5.0 for one-way analysis of variance, principal component analysis, and partial least squares-discriminant analysis and confirmed the metabolites whose levels increased or decreased with mGFR and variable importance in projection (VIP) values >1. Metabolites were ranked by correlation with the original values of mGFR, and metabolites with a correlation coefficient >0.8 and VIP >2 were identified.
Results
We screened out 198 metabolites that increased or decreased with mGFR decline. After ranking by correlation with mGFR, the top 50 metabolites were confirmed. Further studies confirmed the 10 most highly correlated metabolites.
Conclusion
We screened out the metabolites that increased or decreased with mGFR decline in CKD patients from the Chinese population, and 10 of them were highly correlated. They are potential novel metabolites to improve GFR estimation.
Keywords: Metabolomics, Glomerular filtration rate, Chronic kidney diseases, China
Introduction
Chronic kidney disease (CKD) is a very common disease. There are more than 100 million CKD patients in China [1]. The glomerular filtration rate (GFR) is a very important indicator in the diagnosis of CKD and assesses global renal function [2]. GFR can not only help evaluate the severity of CKD in the clinic but also be an important basis for adjusting drug doses. GFR is the rate at which the kidney removes a substance from the plasma and can be measured by clearance or serum levels of filtration markers. Measuring GFR (mGFR) with exogenous glomerular filtration agents is most accurate, for example, inulin, iohexol, or iothalamate. However, these methods are complex and difficult to routinely use in clinical practice. Therefore, the estimated GFR (eGFR), which is estimated by endogenous filtration marker-based (most commonly creatinine) equations, is preferred for use in clinical applications [3]. The ideal endogenous marker for eGFR should appear in the plasma at a constant, filter at the glomerulus freely, be neither secreted nor reabsorbed by the renal tubule, and not undergo extra-renal elimination. Despite intensive attempts, no such ideal endogenous marker has been identified. eGFRcr equations require specification of age, sex, and race. The accuracy of eGFRcr equations may be impacted by other factors, such as muscle mass and diet [4]. Urea, as an endogenous marker, is also affected by diet [5].
Cystatin C is less influenced by muscle and diet than creatinine, so it is considered an alternative endogenous filtration marker [6]. β-Trace protein and β-2-microglobulin are low molecular weight serum proteins, the serum levels of which are less dependent upon muscle mass and diet [7]. However, the accuracy of GFR estimated by these three substances is not more than eGFRcr [6, 8]. Except for the evaluation of low molecular weight proteins, with the development of metabolite profiling, there are other studies identifying metabolites to improve eGFR.
Toyohara et al. [9] discovered 22 cations and 30 anions that accumulated substantially as the eGFR declined. Kobayashi et al. [10] further predicted CKD stage by multivariate analyses with more accuracy. Sekula et al. [11] evaluated the associations of metabolomics with eGFR, which was based on creatinine (eGFRcr) and cystatin C levels in the Cooperative Health Research in the Region of Augsburg (KORA) F4 study and replicated in the Twins UK Registry. Then, six kidney function-associated metabolites were certified to help improve the estimation of GFR. Titan et al. [12] used the Progredir Cohort (a CKD cohort in Sao Paulo, Brazil), the Diabetic Nephropathy (DN) study, and the Baependi Heart Study (BHS) as the derivation study and identified four metabolites as potential biomarkers of renal function. Coresh et al. [4] selected African American Study of Kidney (AASK) and Multi-Ethnic Study of Atherosclerosis (MESA) participants with mGFR and identified 15 candidate metabolites by metabolomic profiling to improve the estimated GFR. Ehrich et al. [13] evaluated serum samples from 320 individuals from Lyon (France), Gothenburg (Sweden), and Berlin (Germany) using nuclear magnetic resonance spectroscopy and demonstrated that myo-inositol, valine, and dimethyl sulfone in combination with creatinine reflected GFR as well as CKD-associated renal dysfunction.
The above studies are from Japan, Europe, America, Brazil, and so on. By comparison, the Chinese population has a significant difference in muscle mass and diet. In this article, we aimed to screen out potential novel metabolites related to mGFR in CKD patients from the Chinese population and identify more biomarkers for improving GFR estimation.
Materials and Methods
Participants and Measurement for mGFR
The participants were recruited from the Third Affiliated Hospital of Sun Yat-sen University in 2020. All 53 participants were consistent with the diagnosis of CKD based on the Kidney Disease Outcome Quality Initiative (K/DOQI) guidelines established by the National Kidney Foundation (NKF). Ethical approval was obtained from the Local Ethics Committee. Age, sex, weight, height, blood pressure, medication history, and so on were recorded for each participant, and their serum creatinine was measured. To evaluate mGFR by utilizing the plasma clearance of iohexol [14], after collecting additional serum samples from participants for metabolomic analysis, iohexol was injected over 2 min (300 mg/mL, GE Healthcare, Shanghai, China). At 120 and 240 min after iohexol administration, 6 mL plasma was collected from the contralateral upper extremity to detect the iohexol concentration (by HPLC). The plasma collection times would change to 120 and 300 min if the participants had an eGFR<30 min/mL/1.73 m2, computed from the Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation [7]. All serum and plasma samples were stored at −80°C until analysis.
Metabolomics Analysis
The Dian Calibra-92 Metabolon Joint Metabolomics Laboratory (Hangzhou, China) completed all untargeted metabolomics analyses. For each sample, four different ultrahigh-performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) assays of small molecule metabolites were conducted [15]. On a Hamilton automated MicroLab STAR® system (Hamilton, Switzerland), each sample was analyzed for automated liquid transfer. Each sample was added to a methanol-based sample extraction solution and then placed in a GenoGrinder 2010 (Spex SamplePrep, USA), and proteins in the sample were precipitated with vigorous shaking for 2 min. The proteins and other debris were then centrifuged into pellets. The extracted metabolites existed in the resulting supernatant and were then divided into four fractions: one fraction for reversed-phase/UPLC-MS/MS analysis in negative ion electrospray ionization (ESI); one fraction for hydrophilic interaction chromatography/UPLC-MS/MS under negative ion ESI; and the last two fractions for reversed-phase UPLC-MS/MS analyses in positive ion ESI mode in two different chromatography programs. To remove the organic solvent, each fraction was dried under nitrogen gas. To be appropriate for injection into each corresponding UPLC-MS/MS system, the dried extract was dissolved in a reconstitution solution. Software developed in-house was used to process, extract, and peak-identify the raw mass spectrometry data and identify metabolites by comparison of experimental ion features to library entries of purified reference standards. The library entries include the mass to charge ratio, retention time/index (RI), and MS/MS spectral data of reference standards.
Metabolomics Statistical Analysis
According to mGFR, all participants were divided into four groups: group A (mGFR<30 mL/min/1.73 m2), group B (30 mL/min/1.73 m2≤ mGFR<60 mL/min/1.73 m2), group C (60 mL/min/1.73 m2≤ mGFR <90 mL/min/1.73 m2), and group D (mGFR ≥90 mL/min/1.73 m2). After grouping, the metabolite peak area data were uploaded to MetaboAnalyst 5.0 (https://www.metaboanalyst.ca) in comma-separated values format. All missing values, zeros, and negative values were replaced by half of the minimum positive value found within the data. After data normalization, MetaboAnalyst 5.0 provided one-way analysis of variance (ANOVA), which was determined using a threshold of p < 0.05 as statistical significance. In MetaboAnalyst 5.0, principal component analysis and partial least squares-discriminant analysis (PLS-DA) were processed for the metabolites. Variable importance in projection (VIP) is a weighted sum of squares of the PLS loadings taking into account the amount of explained Y-variation (mGFR) in each dimension. VIP scores were calculated for each metabolite using a threshold of VIP >1.0 as statistical significance.
The processing of basic characteristics of participants and the correlation analysis of metabolites and mGFR were analyzed by SPSS26.0. If both metabolite levels and mGFR were normally distributed after log transformation, Pearson correlation analysis was used; otherwise, Spearman’s rank correlation was calculated to assess the correlation between mGFR and the levels of the metabolites. A value of p < 0.05 was accepted as indicating statistical significance.
Results
Characteristics of Participants
Participants in this study were aged 27–79 years. According to the mGFR, 22 participants were assigned to group A, 11 to group B, 15 to group C, and 5 to group D. Table 1 presents the basic characteristics of the participants in the four groups. Age, weight, body mass index, systolic blood pressure, and diastolic blood pressure were symmetrically distributed. The arithmetic mean, standard deviation, and p value test results of one-way ANOVA are provided. Creatinine, eGFR, and mGFR followed an asymmetric distribution, and consequently, median and interquartile ranges were shown. The p values of the Kruskal-Wallis test are presented. Sex, current drinking, current smoking, hypertension, diabetes, stroke, coronary heart disease, and a medical history of antihypertensive agent, hypoglycemic agent, antiplatelet drugs, antilipemic agent, hyperuricemia, uric acid reduction medicine, immunosuppressor were dichotomous variables. Positive quantities and frequencies in each group are illustrated as appropriate. The p values of χ2 tests are indicated.
Table 1.
Basic characteristics of participants
| Variables | Group | p value | |||
|---|---|---|---|---|---|
| A | B | C | D | ||
| Age, years | 56.45±11.80 | 49.00±13.74 | 42.80±14.63 | 37.20±13.76 | 0.01 |
| Sex (M/F) | 15/7 | 2/9 | 9/6 | 1/4 | 0.11 |
| Height, cm | 160.68±7.13 | 165.09±7.61 | 165.53±9.93 | 159.80±5.40 | 0.20 |
| Weight, kg | 60.43±9.22 | 65.83±9.62 | 62.43±8.71 | 56.26±4.02 | 0.20 |
| BMI, cm/kg2 | 23.35±2.86 | 24.02±2.16 | 22.74±2.29 | 22.03±1.06 | 0.41 |
| Systolic blood pressure, mm Hg |
153.95±15.72 |
133.36±25.47 |
133.33±17.03 |
126.20±17.54 |
<0.01 |
| Diastolic blood pressure, mm Hg |
87.32±11.31 |
84.45±11.21 |
86.80±11.86 |
79.20±10.43 |
0.55 |
| Creatinine, mg per 100 mL | 464.35 (326.06, 692.93) | 155.5 (117.00, 205.90) | 82.00 (63.00, 98.20) | 55.00 (45.50, 70.90) | <0.01 |
| eGFR, mL min–1 per 1.73 m2 | 10.96 (6.54,15.79) | 41.06 (30.29, 72.17) | 98.18 (76.21, 107.26) | 119.95 (100.32, 128.13) | <0.01 |
| mGFR, mL min–1 per 1.73 m2 | 15.28 (8.42, 18.31) | 39.53 (37.15, 52.45) | 74.53 (66.33, 81.98) | 97.11 (92.83, 104.42) | <0.01 |
| Current drinking, n (%) | 1 (4.55) | 3 (27.27) | 1 (6.67) | 0 | 0.15 |
| Current smoking, n (%) | 5 (22.73) | 1 (9.09) | 1 (6.67) | 0 | 0.36 |
| Hypertension, n (%) | 20 (90.91) | 8 (72.73) | 4 (26.67) | 1 (20.00) | <0.01 |
| Diabetes, n (%) | 15 (68.18) | 6 (54.55) | 1 (6.67) | 1 (20.00) | <0.01 |
| Stroke, n (%) | 2 (9.09) | 0 | 0 | 0 | 0.40 |
| Coronary heart disease, n (%) | 2 (9.09) | 0 | 0 | 0 | 0.40 |
| Antihypertensive agent, n (%) | 22 (100.00) | 10 (90.91) | 12 (80.00) | 4 (80.00) | 0.18 |
| Hypoglycemic agent, n (%) | 16 (72.73) | 6 (54.55) | 1 (6.67) | 1 (20.00) | <0.01 |
| Antiplatelet drugs, n (%) | 6 (27.27) | 1 (9.09) | 0 | 0 | 0.07 |
| Antilipemic agent, n (%) | 12 (54.55) | 3 (27.27) | 4 (26.67) | 4 (80.00) | 0.08 |
| Hyperuricemia, n (%) | 8 (36.36) | 3 (27.27) | 1 (6.67) | 0 | 0.11 |
| Uric acid reduction medicine, n (%) | 10 (45.45) | 3 (27.27) | 1 (6.67) | 0 | 0.03 |
| Immunosuppressor, n (%) | 0 | 2 (18.18) | 2 (13.33) | 2 (40.00) | 0.06 |
One-Way ANOVA for Metabolites
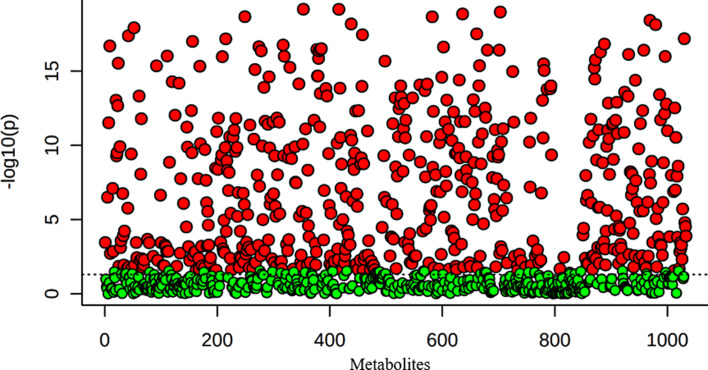
A total of 1,094 metabolites were detected in this study. The uploaded data file contains 53 samples in a 1,094 metabolite data matrix. ANOVA estimates whether the metabolites in each group are significant or not using a threshold of p < 0.05 as statistical significance in MetaboAnalyst 5.0. A total of 517 metabolites were identified. Figure 1 presents the important features identified by ANOVA. Table 2 shows the top 50 metabolites with details of the F value, p value, −log10(p), and FDR.
Fig. 1.
Important features identified by ANOVA plot (p < 0.05).
Table 2.
Top 50 features identified by one-way ANOVA
| Metabolites | f value | p value | −log10(p) | FDR | |
|---|---|---|---|---|---|
| 1 | Erythronate | 89.45 | 6.93E−20 | 19.16 | 3.63E−17 |
| 2 | Gulonate | 89.30 | 7.04E−20 | 19.15 | 3.63E−17 |
| 3 | Arabitol/xylitol | 87.57 | 1.08E−19 | 18.97 | 3.70E−17 |
| 4 | N-acetyltaurine | 86.32 | 1.44E−19 | 18.84 | 3.73E−17 |
| 5 | 1-Methylguanidine | 84.57 | 2.20E−19 | 18.66 | 3.85E−17 |
| 6 | Mannonate | 84.49 | 2.24E−19 | 18.65 | 3.85E−17 |
| 7 | 3-Hydroxy-2-methylpyridine sulfate | 82.24 | 3.88E−19 | 18.41 | 5.73E−17 |
| 8 | 1-Ribosyl-imidazoleacetate | 80.02 | 6.77E−19 | 18.17 | 8.63E−17 |
| 9 | Hydroxy-N6,N6,N6-trimethyllysine | 79.61 | 7.52E−19 | 18.12 | 8.63E−17 |
| 10 | Gluconate | 77.70 | 1.23E−18 | 17.91 | 1.27E−16 |
| 11 | Arabonate/xylonate | 74.12 | 3.16E−18 | 17.50 | 2.97E−16 |
| 12 | N-acetylserine | 73.62 | 3.62E−18 | 17.44 | 3.12E−16 |
| 13 | Creatinine | 73.04 | 4.24E−18 | 17.37 | 3.37E−16 |
| 14 | N,N-dimethyl-pro-pro | 71.39 | 6.68E−18 | 17.18 | 4.71E−16 |
| 15 | Glucuronate | 71.31 | 6.83E−18 | 17.17 | 4.71E−16 |
| 16 | 4-Acetamidobutanoate | 69.90 | 1.02E−17 | 16.99 | 6.55E−16 |
| 17 | N,N,N-trimethyl-alanylproline betaine (TMAP) | 68.45 | 1.53E−17 | 16.81 | 9.32E−16 |
| 18 | 1-Methyl-4-imidazoleacetate | 67.92 | 1.79E−17 | 16.75 | 1.03E−15 |
| 19 | 3-Hydroxy-3-methylglutarate | 67.44 | 2.06E−17 | 16.69 | 1.12E−15 |
| 20 | Homocitrulline | 67.03 | 2.32E−17 | 16.64 | 1.19E−15 |
| 21 | Gamma-CEHC glucuronide | 66.87 | 2.43E−17 | 16.62 | 1.19E−15 |
| 22 | 3-(3-Amino-3-carboxypropyl)uridine | 66.00 | 3.14E−17 | 16.50 | 1.47E−15 |
| 23 | 5,6-Dihydrouridine | 65.64 | 3.49E−17 | 16.46 | 1.52E−15 |
| 24 | N1-methylinosine | 65.59 | 3.54E−17 | 16.45 | 1.52E−15 |
| 25 | Umbelliferone sulfate | 65.27 | 3.90E−17 | 16.41 | 1.52E−15 |
| 26 | C-glycosyltryptophan | 65.25 | 3.92E−17 | 16.41 | 1.52E−15 |
| 27 | 2,3-Dihydroxy-5-methylthio-4-pentenoate (DMTPA) | 65.22 | 3.97E−17 | 16.40 | 1.52E−15 |
| 28 | Orotidine | 64.86 | 4.41E−17 | 16.36 | 1.57E−15 |
| 29 | Ribonate | 64.86 | 4.41E−17 | 16.36 | 1.57E−15 |
| 30 | Hydroxyasparagine | 64.21 | 5.36E−17 | 16.27 | 1.84E−15 |
| 31 | N-acetylhomocitrulline | 63.06 | 7.60E−17 | 16.12 | 2.53E−15 |
| 32 | Pseudouridine | 62.26 | 9.75E−17 | 16.01 | 3.15E−15 |
| 33 | Guanidinosuccinate | 62.13 | 1.01E−16 | 15.99 | 3.18E−15 |
| 34 | Pentose acid | 62.02 | 1.05E−16 | 15.98 | 3.18E−15 |
| 35 | Suberate (C8-DC) | 61.94 | 1.08E−16 | 15.97 | 3.18E−15 |
| 36 | N6-carbamoylthreonyladenosine | 61.06 | 1.42E−16 | 15.85 | 4.07E−15 |
| 37 | Glucuronide of C14H26O4 (1) | 60.36 | 1.77E−16 | 15.75 | 4.94E−15 |
| 38 | 5-Methylthioribose | 59.76 | 2.14E−16 | 15.67 | 5.82E−15 |
| 39 | S-adenosylhomocysteine (SAH) | 58.72 | 2.99E−16 | 15.52 | 7.93E−15 |
| 40 | Furaneol sulfate | 58.42 | 3.30E−16 | 15.48 | 8.53E−15 |
| 41 | Vanillactate | 57.57 | 4.37E−16 | 15.36 | 1.10E−14 |
| 42 | Urea | 57.48 | 4.49E−16 | 15.35 | 1.10E−14 |
| 43 | N-acetylneuraminate | 57.31 | 4.76E−16 | 15.32 | 1.14E−14 |
| 44 | N-acetylasparagine | 56.79 | 5.64E−16 | 15.25 | 1.33E−14 |
| 45 | Glucuronide of C12H22O4 (1) | 56.56 | 6.10E−16 | 15.22 | 1.40E−14 |
| 46 | 4-Hydroxybenzoate | 55.76 | 7.99E−16 | 15.10 | 1.79E−14 |
| 47 | Ascorbic acid 2-sulfate | 55.34 | 9.20E−16 | 15.04 | 2.02E−14 |
| 48 | 2-Methylcitrate/homocitrate | 54.87 | 1.08E−15 | 14.97 | 2.32E−14 |
| 49 | N4-acetylcytidine | 52.85 | 2.17E−15 | 14.66 | 4.50E−14 |
| 50 | N2,N2-dimethylguanosine | 52.85 | 2.18E−15 | 14.66 | 4.50E−14 |
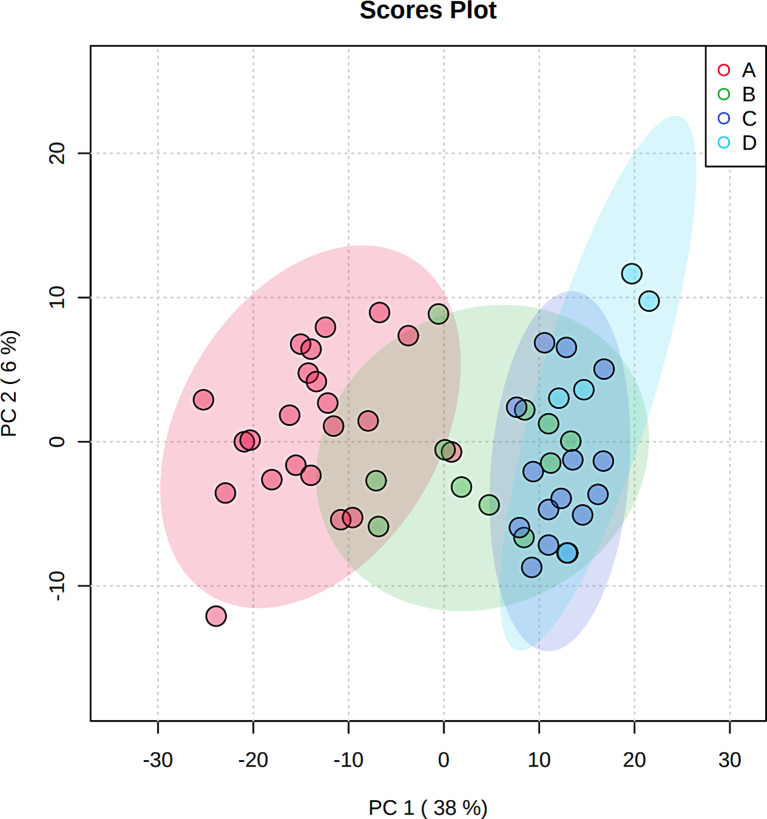
Principal Component Analysis
Principal component analysis is an unsupervised method aiming to identify the directions that best explain the metabolite variance in a dataset (X) without referring to class labels (Y). The data are summarized into much fewer variables called scores, which are weighted averages of the original variables. Figure 2 presents the 2-D score plot between selected PCs. Figure 2 illustrates a separation trend among each group, indicating that each group had a unique metabolic spectrum.
Fig. 2.
Scores plot between the selected PCs. The explained variances are shown in brackets.
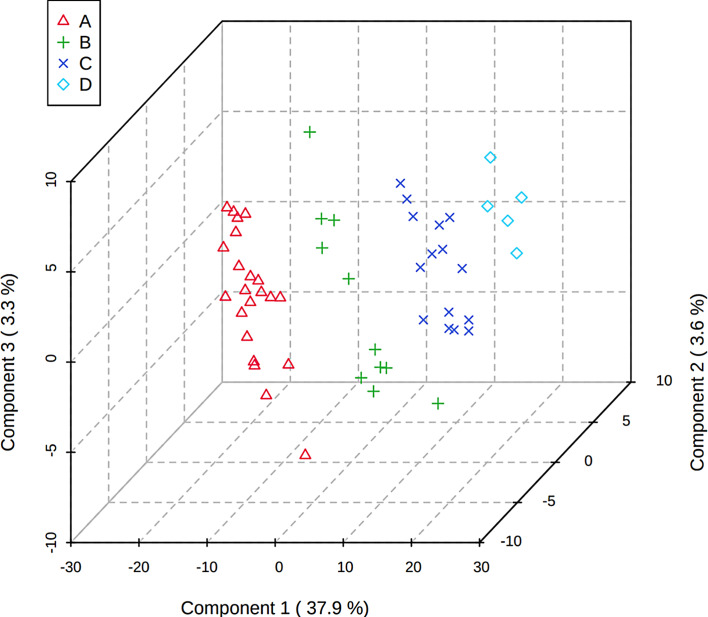
Partial Least Squares-Discriminant Analysis
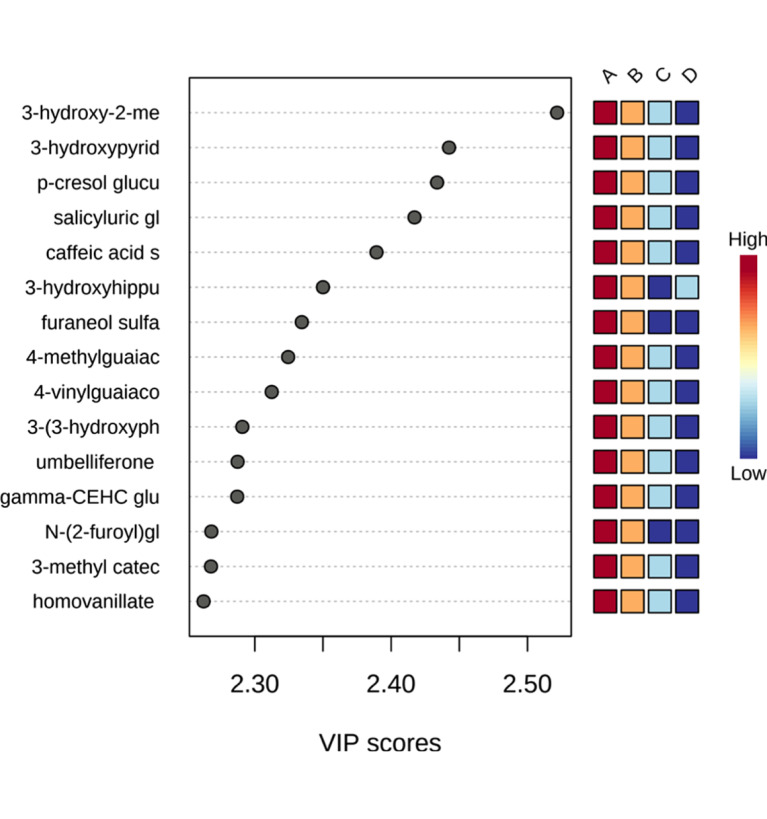
PLS is a supervised method that uses multivariate regression techniques to extract via linear combination of original metabolite variables (X) the information that can predict the class membership (Y). Figure 3 shows the 3-D score plot between the selected components, and the four groups can be well distinguished by PLS-DA. We also calculated the VIP value of each metabolite by MetaboAnalyst 5.0, and 347 metabolites had VIP values >1. Figure 4 shows important features of partial metabolites identified by PLS-DA, including VIP value and its level variation trend in each group. As shown in Figure 4, the levels of some metabolites did not increase or decrease with mGFR decline. Excluding these metabolites, the levels of 198 metabolites with VIP values >1 increased or decreased with mGFR decline.
Fig. 3.
3D scores plot between the selected PCs. The explained variances are shown in brackets.
Fig. 4.
Important features identified by PLS-DA. The colored boxes on the right indicate the relative levels of the corresponding metabolite in each group under study.
Correlation Analysis
The 198 metabolites, the levels of which with VIP values >1 increased or decreased with mGFR decline, screened by PLS-DA were selected. Metabolite levels were log transformed, and metabolites were ranked by correlation with logmGFR. Table 3 showed the top 50 metabolites that were in correlation with mGFR and the VIP value of each metabolite. LogmGFR followed an asymmetric distribution (Shapiro-Wilk test: p = 0.03); Table 3 shows the correlation coefficient and p value of Spearman’s rank correlation. Table 4 shows the metabolites with a correlation coefficient >0.8 and VIP >2.
Table 3.
Top 50 metabolites correlation with mGFR
| Metabolites | R | p value | VIP | |
|---|---|---|---|---|
| 1 | Hydroxyasparagine | −0.96 | <0.01 | 1.34 |
| 2 | N6-carbamoylthreonyladenosine | −0.96 | <0.01 | 1.46 |
| 3 | 3-(3-Amino-3-carboxypropyl)uridine | −0.96 | <0.01 | 1.51 |
| 4 | Gulonate | −0.96 | <0.01 | 1.76 |
| 5 | 2,3-Dihydroxy-5-methylthio-4-pentenoate (DMTPA) | −0.95 | <0.01 | 1.48 |
| 6 | C-glycosyltryptophan | −0.95 | <0.01 | 1.44 |
| 7 | Pseudouridine | −0.95 | <0.01 | 1.32 |
| 8 | N-acetylserine | −0.95 | <0.01 | 1.32 |
| 9 | Arabonate/xylonate | −0.94 | <0.01 | 1.59 |
| 10 | S-adenosylhomocysteine (SAH) | −0.94 | <0.01 | 1.43 |
| 11 | Glucuronate | −0.93 | <0.01 | 1.56 |
| 12 | 4-Acetamidobutanoate | −0.93 | <0.01 | 1.58 |
| 13 | N-acetylhistidine | −0.93 | <0.01 | 1.35 |
| 14 | 1-Methyl-4-imidazoleacetate | −0.93 | <0.01 | 1.46 |
| 15 | Gamma-CEHC glucuronide | −0.93 | <0.01 | 2.14 |
| 16 | N,N-dimethyl-pro-pro | −0.93 | <0.01 | 1.34 |
| 17 | N2,N2-dimethylguanosine | −0.93 | <0.01 | 1.38 |
| 18 | Quinolinate | −0.93 | <0.01 | 1.34 |
| 19 | Arabitol/xylitol | −0.93 | <0.01 | 1.44 |
| 20 | Creatinine | −0.93 | <0.01 | 1.10 |
| 21 | Homocitrulline | −0.93 | <0.01 | 1.66 |
| 22 | N1-methylinosine | −0.93 | <0.01 | 1.56 |
| 23 | 5-Methylthioribose | −0.92 | <0.01 | 1.28 |
| 24 | N,N,N-trimethyl-alanylproline betaine (TMAP) | −0.92 | <0.01 | 1.36 |
| 25 | Vanillactate | −0.92 | <0.01 | 1.96 |
| 26 | Erythronate | −0.92 | <0.01 | 1.36 |
| 27 | Gluconate | −0.92 | <0.01 | 1.53252 |
| 28 | Hydroxy-N6,N6,N6-trimethyllysine | −0.92 | <0.01 | 1.48 |
| 29 | Orotidine | −0.92 | <0.01 | 1.53 |
| 30 | O-sulfo-l-tyrosine | −0.92 | <0.01 | 1.34 |
| 31 | 5-(Galactosylhydroxy)-l-lysine | −0.91 | <0.01 | 1.39 |
| 32 | N4-acetylcytidine | −0.91 | <0.01 | 1.40 |
| 33 | Heptenedioate (C7:1-DC) | −0.91 | <0.01 | 1.50 |
| 34 | N-formylmethionine | −0.91 | <0.01 | 1.13 |
| 35 | Pimeloylcarnitine/3-methyladipoylcarnitine (C7-DC) | −0.91 | <0.01 | 1.62 |
| 36 | 1-Ribosyl-imidazoleacetate | −0.91 | <0.01 | 1.66 |
| 37 | 2-Methylcitrate/homocitrate | −0.91 | <0.01 | 1.28 |
| 38 | N-acetyl-2-aminoadipate | −0.91 | <0.01 | 1.56 |
| 39 | Kynurenate | −0.90 | <0.01 | 1.36 |
| 40 | Glutamine conjugate of C6H10O2 (1) | −0.90 | <0.01 | 1.65 |
| 41 | N-acetylmethionine | −0.90 | <0.01 | 1.19 |
| 42 | N-acetylalanine | −0.90 | <0.01 | 1.00 |
| 43 | Mannonate | −0.90 | <0.01 | 1.42 |
| 44 | N-acetyl-isoputreanine | −0.90 | <0.01 | 1.30 |
| 45 | Guaiacol sulfate | −0.90 | <0.01 | 2.01 |
| 46 | Alpha-CEHC glucuronide | −0.89 | <0.01 | 2.02 |
| 47 | Suberate (C8-DC) | −0.89 | <0.01 | 1.72 |
| 48 | 8-Methoxykynurenate | −0.89 | <0.01 | 1.89 |
| 49 | 2-Hydroxysebacate | −0.89 | <0.01 | 1.86 |
| 50 | 3-Hydroxy-3-methylglutarate | −0.89 | <0.01 | 1.32 |
Table 4.
Metabolites with correlation coefficient >0.8 and VIP >2
| Metabolites | r | p value | VIP | |
|---|---|---|---|---|
| 1 | Gamma-CEHC glucuronide | −0.93 | <0.01 | 2.14 |
| 2 | Guaiacol sulfate | −0.90 | <0.01 | 2.01 |
| 3 | Alpha-CEHC glucuronide | −0.90 | <0.01 | 2.01 |
| 4 | 4-Methylguaiacol sulfate | −0.88 | <0.01 | 2.22 |
| 5 | 3-Hydroxypyridine sulfate | −0.88 | <0.01 | 2.28 |
| 6 | Homovanillate sulfate | −0.87 | <0.01 | 2.10 |
| 7 | Caffeic acid sulfate | −0.86 | <0.01 | 2.23 |
| 8 | p-Cresol glucuronide | −0.86 | <0.01 | 2.30 |
| 9 | 3-Methyl catechol sulfate (1) | −0.84 | <0.01 | 2.16 |
| 10 | N-acetylalliin | −0.82 | <0.01 | 2.02 |
Discussion
In this study, serum metabolic profiles of the Chinese population were used to screen out potential novel metabolites related to mGFR in CKD patients by metabonomic analysis. We used PLS-DA to screen out 347 metabolites that were statistically significant (VIP >1), and 198 of them increased or decreased with mGFR decline. Furthermore, 198 metabolites were ranked by correlation with mGFR, containing the 10 most highly correlated metabolites (|r | > 0.8 and VIP > 2).
Kobayashi et al. [10] showed a high correlation between N6-carbamoylthreonyladenosine and 1/eGFR by a simple linear regression model, which was in the top 50 metabolites of our study. Sekula et al. [11] highlighted six metabolites significantly associated with eGFR, containing pseudouridine, C-mannosyltryptophan, N-acetylalanine, erythronate, myo-inositol, and N-acetylcarnosine. Pseudouridine, N-acetylalanine, and erythronate were also among the top 50 metabolites correlated with mGFR in our study. Titan et al. [12] found correlations of D-threitol, myo-inositol, 4-deoxyerythronic acid, and galacturonic acid with eGFR in the Progredir Cohort, DN Study, and BHS. Moreover, pseudouridine was strongly associated with eGFR in the Progredir and DN Study but not in the BHS. Among these five metabolites, only pseudouridine was found to be associated with mGFR in our study. Coresh et al. [4] listed the 50 most highly positively and negatively correlated metabolites, which were ranked by correlation in AASK. In this study, the top 50 metabolites were all negatively correlated with mGFR, and 16 of the top 50 metabolites were the same as those in Coresh et al. [4]: N6-carbamoylthreonyladenosine, C-glycosyltryptophan, pseudouridine, N-acetylserine, arabonate/xylonate, glucuronate, 4-acetamidobutanoate, gamma-CEHC glucuronide, N2,N2-dimethylguanosine, arabitol, creatinine, homocitrulline, erythronate, N-formylmethionine, and N-acetylalanine. Nierenberg et al. [16] further identified 12 metabolites that have not been reported by previous kidney function metabolomics studies in both Bogalusa Heart Study (BHS) and MESA-Kidney participants. Gulonate, gamma-CEHC glucuronide, and quinolinate were also the top 50 metabolites correlated with mGFR in our study. Compared with the above studies, the obvious correlation between the metabolite pseudouridine and eGFR or mGFR was mentioned in three studies. The obvious correlation between N-acetylalanine was also found in the study of Sekula et al. [11] and Coresh et al. [4], The obvious correlation between N6-carbamoylthreonyladenosine and N2,N2-dimethylguanosine was also found in the studies by Kobayashi et al. [10] and Coresh et al. [4]. In addition, the metabolite gamma-CEHC glucuronide was the most relevant metabolite in this study, which is also one of the top 50 negatively correlated metabolites in the study of Coresh et al. [4] and a novel identified metabolite in the study of Nierenberg et al. [16]. Myo-inositol showed an obvious correlation in three studies but not in this study. In contrast to the result of the study by Toyohara et al. [9], only creatinine existed in the top 50 metabolites correlated with mGFR in our study.
In our study, CKD patients were selected for metabolic analysis, and correlation analysis was conducted with mGFR. In addition to the studies of Coresh et al. [4] and Nierenberg et al. [16], all previous studies were related to eGFR correlation analysis, and only Toyohara et al. [9] and Titan et al. [12] enrolled patients in cohorts of CKD patients. In addition, regional and ethnic differences lead to different results.
Without using demographic characteristics, Coresh et al. [4] found that pseudouridine and N-acetylserine performed as well as or better in root mean square error than creatinine alone in both studies, but arabitol and N-acetylalanine performed better in the MESA only. Bernert et al. [17] found that the clearance of [3H]pseudouridine in rats was lower, 0.78, than inulin, indicating that pseudouridine would not be suitable for estimation of the GFR. When controlled for age, sex, and race, erythronate and N6-carbamoylthreonyladenosine showed stronger associations with mGFR than Cr in cirrhosis patients [18]. The levels of N2,N2-dimethylguanosine (involved in nucleotide metabolism) and homovanillate sulfate (involved in amino acid metabolism) were consistently higher in polycystic kidney disease than in glomerular disease and CKD of other causes [19]. Pseudouridine, N-acetylalanine, and N6-carbamoylthreonyladenosine were also associated with renal function decline and time to ESRD in patients with type 1 diabetes.
There are 10 highly correlated metabolites in this study. Both gamma-CEHC glucuronide [20] and alpha-CEHC glucuronide [21] are metabolites related to vitamin E metabolism. Lower levels of gamma‐CEHC glucuronide were associated with an increased hazard of liver‐related events [20]. Urinary alpha-CEHC glucuronide was identified as a good biomarker of intestinal inflammation [21]. Guaiacol sulfate is a phenolic uremic toxin and decreases in CKD cats after consumption of scFOS (4-hydroxyhippurate, catechol sulfate, 3-methylcatechol sulfate [1], 3-[4-hydroxyphenyl] propionate) [22]. Metabolites are secreted into the urine after oral aldehydes, mainly as glucuronide and/or sulfate conjugates, and 4-methylguaiacol is reduced from vanillic in rats [23]. 3-Hydroxypyridine sulfate is a trace metabolite of vitamin B6 and is positively correlated with coffee consumption [24]. Caffeic acid is one of the most representative phenolic acids in the diet and is present in humans predominantly in conjugated forms such as glucuronide or sulfate [25]. P-cresol glucuronide is an end product of tyrosine biotransformation that is generated by anaerobic intestinal bacteria [26, 27]. P-cresol glucuronide levels rise with the decline in GFR and are associated with total and cardiovascular mortality [28]. 3-Methyl catechol sulfate, which is a serum metabolite associated with coffee consumption, is a xenobiotic involved in benzoate metabolism and may be harmful to kidney health [29]. The level of serum N-acetylalliin correlates with garlic intake [30]. Previous studies on the 10 metabolites were limited in their metabolic pathways and some pathological functions. In this research, we discovered that they were markedly correlated with mGFR. Nonetheless, whether they can improve the accuracy of eGFR equation requires further verification by targeted metabolomics analyses and repeated in other populations.
Conclusion
We screened out 198 metabolites that increased or decreased with mGFR decline by metabonomic analysis in the Chinese population. After ranking by correlation with mGFR, N6-carbamoylthreonyladenosine, pseudouridine, gamma-CEHC glucuronide, N2,N2-dimethylguanosine, erythronate, and N-acetylalanine not only ranked in the top 50 metabolites in this study but were also identified to be associated with GFR in two or more previous studies. Further studies found that 10 metabolites were highly correlated with mGFR: gamma-CEHC glucuronide, guaiacol sulfate, alpha-CEHC glucuronide, 4-methylguaiacol sulfate, 3-hydroxypyridine sulfate, homovanillate sulfate, caffeic acid sulfate, p-cresol glucuronide, 3-methyl catechol sulfate (1), and N-acetylalliin. They are potential novel metabolites to improve GFR estimation.
Statement of Ethics
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University, number: [2018]02-227-01, and written informed consent was obtained for participation.
Conflict of Interest Statement
The authors declare no conflicts of interest.
Funding Sources
This study was supported by the Science and Technology Development Fund, Macau SAR (File no.0032/2018/A1), the National Natural Science Foundation of China (Grant No. 81873631, 81370866, 81070612), and the Guangzhou Science and technology planning project (Grant No. 202002020047).
Author Contributions
All authors have read and approved the manuscript. Xinghua Guo and Hongquan Peng: conceptualization, methodology, and writing – original draft preparation. Hongquan Peng: funding acquisition. Peijia Liu: visualization. Leile Tang: investigation. Jia Fang: software and validation. Chiwa Aoieong: supervision. Tou Tou and Tsungyang Tsai: data curation. Hongquan Peng and Xun Liu: writing – review, funding, and editing.
Funding Statement
This study was supported by the Science and Technology Development Fund, Macau SAR (File no.0032/2018/A1), the National Natural Science Foundation of China (Grant No. 81873631, 81370866, 81070612), and the Guangzhou Science and technology planning project (Grant No. 202002020047).
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding author.
References
- 1. Zhang L, Wang F, Wang L, Wang W, Liu B, Liu J, et al. Prevalence of chronic kidney disease in China: a cross-sectional survey. Lancet. 2012;379(9818):815–22. 10.1016/S0140-6736(12)60033-6. [DOI] [PubMed] [Google Scholar]
- 2. Charles C, Ferris AH. Chronic kidney disease. Prim Care. 2020;47(4):585–95. 10.1016/j.pop.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 3. Inker LA, Titan S. Measurement and estimation of GFR for use in clinical practice: core curriculum 2021. Am J Kidney Dis. 2021;78(5):736–49. 10.1053/j.ajkd.2021.04.016. [DOI] [PubMed] [Google Scholar]
- 4. Coresh J, Inker LA, Sang Y, Chen J, Shafi T, Post WS, et al. Metabolomic profiling to improve glomerular filtration rate estimation: a proof-of-concept study. Nephrol Dial Transpl. 2019;34(5):825–33. 10.1093/ndt/gfy094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weiner ID, Mitch WE, Sands JM. Urea and ammonia metabolism and the control of renal nitrogen excretion. Clin J Am Soc Nephrol. 2015;10(8):1444–58. 10.2215/CJN.10311013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Levey AS, Inker LA, Coresh J. GFR estimation: from physiology to public health. Am J Kidney Dis. 2014;63(5):820–34. 10.1053/j.ajkd.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Juraschek SP, Coresh J, Inker LA, Levey AS, Köttgen A, Foster MC, et al. Comparison of serum concentrations of β-trace protein, β2-microglobulin, cystatin C, and creatinine in the US population. Clin J Am Soc Nephrol. 2013;8(4):584–92. 10.2215/CJN.08700812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Inker LA, Tighiouart H, Coresh J, Foster MC, Anderson AH, Beck GJ, et al. GFR estimation using β-trace protein and β2-microglobulin in CKD. Am J Kidney Dis. 2016;67(1):40–8. 10.1053/j.ajkd.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Toyohara T, Akiyama Y, Suzuki T, Takeuchi Y, Mishima E, Tanemoto M, et al. Metabolomic profiling of uremic solutes in CKD patients. Hypertens Res. 2010;33(9):944–52. 10.1038/hr.2010.113. [DOI] [PubMed] [Google Scholar]
- 10. Kobayashi T, Yoshida T, Fujisawa T, Matsumura Y, Ozawa T, Yanai H, et al. A metabolomics-based approach for predicting stages of chronic kidney disease. Biochem Biophys Res Commun. 2014;445(2):412–6. 10.1016/j.bbrc.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 11. Sekula P, Goek ON, Quaye L, Barrios C, Levey AS, Römisch-Margl W, et al. A metabolome-wide association study of kidney function and disease in the general population. J Am Soc Nephrol. 2016;27(4):1175–88. 10.1681/ASN.2014111099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Titan SM, Venturini G, Padilha K, Tavares G, Zatz R, Bensenor I, et al. Metabolites related to eGFR: evaluation of candidate molecules for GFR estimation using untargeted metabolomics. Clin Chim Acta. 2019;489:242–8. 10.1016/j.cca.2018.08.037. [DOI] [PubMed] [Google Scholar]
- 13. Ehrich J, Dubourg L, Hansson S, Pape L, Steinle T, Fruth J, et al. Serum myo-inositol, dimethyl sulfone, and valine in combination with creatinine allow accurate assessment of renal insufficiency-A proof of concept. Diagnostics. 2021;11(2):234. 10.3390/diagnostics11020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shah VO, Townsend RR, Feldman HI, Pappan KL, Kensicki E, Vander Jagt DL. Plasma metabolomic profiles in different stages of CKD. Clin J Am Soc Nephrol. 2013;8(3):363–70. 10.2215/CJN.05540512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182(1):59–72.e15. 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nierenberg JL, He J, Li C, Gu X, Shi M, Razavi AC, et al. Novel associations between blood metabolites and kidney function among Bogalusa Heart Study and Multi-Ethnic Study of Atherosclerosis participants. Metabolomics. 2019;15(12):149. 10.1007/s11306-019-1613-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bernert JT, Jr, Bell CJ, Guntupalli J, Hannon WH. Pseudouridine is unsuitable as an endogenous renal clearance marker. Clin Chem. 1988;34(6):1011–7. 10.1093/clinchem/34.6.1011. [DOI] [PubMed] [Google Scholar]
- 18. Mindikoglu AL, Opekun AR, Putluri N, Devaraj S, Sheikh-Hamad D, Vierling JM, et al. Unique metabolomic signature associated with hepatorenal dysfunction and mortality in cirrhosis. Transl Res. 2018;195:25–47. 10.1016/j.trsl.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grams ME, Tin A, Rebholz CM, Shafi T, Köttgen A, Perrone RD, et al. Metabolomic alterations associated with cause of CKD. Clin J Am Soc Nephrol. 2017;12(11):1787–94. 10.2215/CJN.02560317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wegermann K, Howe C, Henao R, Wang Y, Guy CD, Abdelmalek MF, et al. Serum bile acid, vitamin E, and serotonin metabolites are associated with future liver-related events in nonalcoholic fatty liver disease. Hepatol Commun. 2021;5(4):608–17. 10.1002/hep4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pope SAS, Burtin GE, Clayton PT, Madge DJ, Muller DPR. Synthesis and analysis of conjugates of the major vitamin E metabolite, alpha-CEHC. Free Radic Biol Med. 2002;33(6):807–17. 10.1016/s0891-5849(02)00974-7. [DOI] [PubMed] [Google Scholar]
- 22. Hall JA, Jackson MI, Dennis EJ, Eden E. Chronic kidney disease in cats alters response of the plasma metabolome and fecal microbiome to dietary fiber. PLoS One. 2020;15(7):e235480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strand LP, Scheline RR. The metabolism of vanillin and isovanillin in the rat. Xenobiotica. 1975;5(1):49–63. 10.3109/00498257509056093. [DOI] [PubMed] [Google Scholar]
- 24. Chau Y-P, Au PCM, Li GHY, Sing CW, Cheng VKF, Tan KCB, et al. Serum metabolome of coffee consumption and its association with bone mineral density: the Hong Kong osteoporosis study. J Clin Endocrinol Metab. 2020;105(3):dgz210. 10.1210/clinem/dgz210. [DOI] [PubMed] [Google Scholar]
- 25. Piazzon A, Vrhovsek U, Masuero D, Mattivi F, Mandoj F, Nardini M. Antioxidant activity of phenolic acids and their metabolites: synthesis and antioxidant properties of the sulfate derivatives of ferulic and caffeic acids and of the acyl glucuronide of ferulic acid. J Agric Food Chem. 2012;60(50):12312–23. 10.1021/jf304076z. [DOI] [PubMed] [Google Scholar]
- 26. Curtius HC, Mettler M, Ettlinger L. Study of the intestinal tyrosine metabolism using stable isotopes and gas chromatography-mass spectrometry. J Chromatogr. 1976;126:569–80. 10.1016/s0021-9673(01)84102-9. [DOI] [PubMed] [Google Scholar]
- 27. De Smet R, Van Kaer J, Van Vlem B, De Cubber A, Brunet P, Lameire N, et al. Toxicity of free p-cresol: a prospective and cross-sectional analysis. Clin Chem. 2003;49(3):470–8. 10.1373/49.3.470. [DOI] [PubMed] [Google Scholar]
- 28. Liabeuf S, Glorieux G, Lenglet A, Diouf M, Schepers E, Desjardins L, et al. Does p-cresylglucuronide have the same impact on mortality as other protein-bound uremic toxins? PLoS One. 2013;8(6):e67168. 10.1371/journal.pone.0067168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He WJ, Chen J, Razavi AC, Hu EA, Grams ME, Yu B, et al. Metabolites associated with coffee consumption and incident chronic kidney disease. Clin J Am Soc Nephrol. 2021;16(11):1620–9. 10.2215/CJN.05520421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y, Gapstur SM, Carter BD, Hartman TJ, Stevens VL, Gaudet MM, et al. Untargeted metabolomics identifies novel potential biomarkers of habitual food intake in a cross-sectional study of postmenopausal women. J Nutr. 2018;148(6):932–43. 10.1093/jn/nxy027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding author.