Abstract

There is an increasing prevalence of diabetes mellitus throughout the world, and new compounds are necessary to combat this. The currently available antidiabetic therapies are long-term complicated and side effect-prone, and this has led to a demand for more affordable and more effective methods of tackling diabetes. Research is focused on finding alternative medicinal remedies with significant antidiabetic efficacy as well as low adverse effects. In this research work, we have focused our efforts to synthesize a series of 1,2,4-triazole-based bis-hydrazones and evaluated their antidiabetic properties. In addition, the precise structures of the synthesized derivatives were confirmed with the help of various spectroscopic techniques including 1H-NMR, 13C-NMR, and HREI-MS. To find the antidiabetic potentials of the synthesized compounds, in vitro α-glucosidase and α-amylase inhibitory activities were characterized using acarbose as the reference standard. From structure–activity (SAR) analysis, it was confirmed that any variation found in inhibitory activities of both α-amylase and α-glucosidase enzymes was due to the different substitution patterns of the substituent(s) at variable positions of both aryl rings A and B. The results of the antidiabetic assay were very encouraging and showed moderate to good inhibitory potentials with IC50 values ranging from 0.70 ± 0.05 to 35.70 ± 0.80 μM (α-amylase) and 1.10 ± 0.05 to 30.40 ± 0.70 μM (α-glucosidase). The obtained results were compared to those of the standard acarbose drug (IC50 = 10.30 ± 0.20 μM for α-amylase and IC50 = 9.80 ± 0.20 μM for α-glucosidase). Specifically, compounds 17, 15, and 16 were found to be significantly active with IC50 values of 0.70 ± 0.05, 1.80 ± 0.10, and 2.10 ± 0.10 μM against α-amylase and 1.10 ± 0.05, 1.50 ± 0.05, and 1.70 ± 0.10 μM against α-glucosidase, respectively. These findings reveal that triazole-containing bis-hydrazones act as α-amylase and α-glucosidase inhibitors, which help develop novel therapeutics for treating type-II diabetes mellitus and can act as lead molecules in drug discovery as potential antidiabetic agents.
1. Introduction
Diabetes mellitus is associated with a group of metabolic disorders, mainly hyperglycemia, which is characterized by an elevated level of blood glucose, caused by failure of insulin secretion, its action, or both.1,2 Type-2 diabetes is more important than type-1 diabetes because it is considered a preventable disease. Type-2 diabetes is brought about by an imbalance between glucose ingestion and insulin discharge. Controlling blood sugar levels is the basic way for preventing type-2 diabetes.3 Dynamic insulin release can be used to accomplish this goal with careful food management and medication.4−6 The UK Project Diabetes Research (UKPDS) Group, in 1998, recommended that diet control can be used for the protection and corresponding treatment of diabetes. The list of food sources that depend on blood sugar is currently one of the most advocated health remedies. It has been determined that nutritional therapy can be used along with other clinical medications to maximize the impact.7−9 In any case, this has the disadvantage of restricting the sorts and amounts of food consumed. Another possible arrangement is diminishing the pace of glucose ingestion from the small digestive tract by delaying the processing of dietary starch, the significant dietary goodwill of glucose.6 In order to avoid paying for testing, making accommodations, and disclosing results, this practice is believed to be more effective than regulating insulin discharge.5 The inhibition of enzymes that convert dietary starch into glucose, such as α-amylase and α-glucosidase, has received attention as a technique to regulate blood sugar levels.10−12 α-Amylase catalyzes the hydrolysis of (1,4)-glycosidic linkages and produces maltose and glucose from starch,13,14 while α-glucosidase discharges glucose from maltose or potentially sucrose.15,16 The two of them are discharged in the little digestive tract, while just α-amylase is found in salivation. By limiting these two proteins, the retention of glucose in the circulatory system can be deferred to reduce type-2 diabetes. These efforts have been made to distinguish α-amylase and also α-glucosidase inhibitors that can be utilized as food or food additive substances. However, the evidence17,18 demonstrated that several sugar-like phenolic substances have the potential to block α-glucosidase. The majority of research on glucosidase and amylase inhibitors has been concentrated on phenolic substances.19,20 According to research, bis-hydrazone derivatives show an antidiabetic effect, while heterocyclic compounds have long been known to have this potential. The in vivo analysis supports the hydrazone compounds’ safety.21
1,2,4-Triazole motif-bearing scaffolds are known to exhibit a diverse range of biological and pharmaceutical profiles such as anticonvulsant, anti-inflammatory, antiproliferative, and antifungal activities.22−24 In addition, 1,2,4-triazole constitutes pharmacologically active scaffolds that play an essential role as drug candidates such as anastrozole, vorozole, and letrozole and is reported to be effective aromatase inhibitors and finds application in the treatment of breast cancer.25 Furthermore, 1,2,4-triazole-bearing bioactive drugs were found to possess anticancer (letrozole), antibacterial (tazobactam), antifungal (isavuconazole), antidiabetic (sitagliptin), seizure disorder (rufinamide), and antiviral (ribavirin) activities26−35 (Figure 1).
Figure 1.
Bioactive drugs containing the triazole nucleus.
Molecular hybridization is a new concept in drug design and development based on the combination of two or more than two bioactive moieties to produce a new hybrid compound with improved biological activities, when compared to the parent drugs. In the past, researchers had synthesized bis-hydrazone37,38 and triazole analogues36,39 as α-glucosidase and α-amylase inhibitors individually, but no one had incorporated both these bioactive entities in the same molecules to enhance the biological potentials. Therefore, this research work was carried out to incorporate both triazole and bis-hydrazone entities in the same compounds to further enhance the α-amylase and α-glucosidase activities as the more the heteroatoms in the bulk structure of the compound, the more will be its chance of interactions with the active sites of the targeted enzymes, which in turn enhances the biological profile. The result obtained showed that the synthesized compounds were found to be significantly active against α-glucosidase and α-amylase enzymes in comparison to the previously reported individual triazole and bis-hydrazone analogues, and hence, these synthesized analogues could be considered lead molecules for the development of more potential and improved antidiabetic agents (Figure 2).
Figure 2.
Newly synthesized triazole-based bis-hydrazone analogues.
2. Results and Discussion
2.1. Chemistry
This work is based on a multistep reaction procedure. The targeted compounds (1–19) were obtained using methods outlined in Scheme 1. In the first step, a p-NO2-substituted benzoyl chloride was reacted with thiosemicarbazide (I) to obtain a substrate (II). The reaction was carried out in N,N-dimethyl formamide (DMF) in the presence of triethylamine (Et3N) as a catalyst. The substrate (II) underwent cyclization with 2% sodium hydroxide solution followed by neutralization with dilute hydrochloric acid, affording a 3-mercapto-1,2,4-triazole intermediate (III), which was further reacted with different-substituted phenacyl bromide in ethanol and triethylamine, and the reaction mixture was stirred for 3 h to access the S-substituted triazole substrate (IV). In the next step, excess of hydrazine hydrate was added dropwise to the solution of substrate (IV) being stirred in methanol and glacial acetic acid, and the resulting residue was stirred until the conversion was complete (progress of the reaction was monitored by TLC, reflux, 4 h) to yield a triazole-based hydrazone intermediate (V). Finally, the intermediate (V) was treated with various substituted benzaldehyde in methanol and acetic acid, affording the targeted triazole-based bis-hydrazone derivatives (1–19) in appropriate yields. Reaction completion was periodically observed by thin-layer chromatography (TLC) (hexane/ethyl acetate 8:2). The solvent was evaporated under reduced pressure after completion of the reaction. Work-up and crystallization afforded pure products. Structural elucidation of all synthetic compounds was done using distinct spectroscopic techniques such as 1H-NMR, 13C-NMR, and HREI-MS (Scheme 1).
Scheme 1. Synthetic Route for the Synthesis of Derivatives (1–19): (a) P-NO2 Benzoyl Chloride, DMF, Et3N, and Reflux 3 h; (b) 2% NaOH, Reflux 12 h, and Dil HCl; (c) Phenacyl Bromide, Et3N, EtOH, and Reflux 3 h; (d) N2H4·H2O, MeOH/CH3COOH, and Reflux 4 h; and (e) Diversely Substituted Benzaldehyde, MeOH/CH3COOH, and Reflux 4 h.
2.2. In Vitro α-Amylase and α-Glucosidase Inhibitory Activities
The synthesized scaffolds based on triazole-bearing bis-hydrazones (1–19) were subjected to in vitro α-amylase and α-glucosidase inhibitory activities. All the newly afforded analogues showed α-amylase and α-glucosidase inhibitory activities in the range of IC50 = 0.70 ± 0.05–35.70 ± 0.80 μM (against α-amylase) and 1.10 ± 0.05–30.40 ± 0.70 μM (α-glucosidase). The results obtained were compared to those of the standard acarbose drug (IC50 = 10.30 ± 0.20 μM for α-amylase and IC50 = 9.80 ± 0.20 μM for α-glucosidase). Among the synthesized analogues, the analogues 17 (IC50 = 0.70 ± 0.05 and 1.10 ± 0.05 μM), 15 (IC50 = 1.80 ± 0.10 and 1.50 ± 0.05 μM), and 16 (IC50 = 2.10 ± 0.10 and 1.70 ± 0.10 μM) emerged to be significantly potent, even manifold more active than the standard acarbose drug, whereas the remaining analogues demonstrated moderate α-amylase and α-glucosidase inhibitory activities.
2.2.1. Structure–Activity Relationship (SAR) Studies for α-Amylase and α-Glucosidase Inhibitory Activities
By inspecting the influence of varying moieties (R1 and R2) on α-amylase and α-glucosidase activities, the structure–activity relationship (SAR) studies were established. Among the series, compound 17 bearing 3,4-diCl substitutions on both rings B and C emerged to be the most potent inhibitors of α-amylase and α-glucosidase enzymes, even manifold more active than the standard acarbose drug. This enhanced inhibitory potential of compound 17 was due to the EW nature of the attached −Cl groups. Furthermore, the comparison of compound 15 (bearing 3,4-diCl moieties at ring B and the 4-NO2 group at ring C) with compound 16 (holds 3,4-diCl groups at ring B and the 3-NO2 moiety at ring C) demonstrated that compound 15 showed better α-amylase and α-glucosidase inhibitory activities than its counterpart 16, even though both these compounds hold the same substituent(s) at variable positions of both rings B and C. The discrepancy found in the activities of these analogues was due to the different orientations of the −NO2 group toward the active sites of the targeted enzymes. However, the α-amylase and α-glucosidase inhibitory activities were affected by bringing alteration in the number/s of attached substituents around aryl ring C; therefore, analogue 17 (bearing di-Cl moieties at the 3,4-position of both aryl rings B and C) was more active than analogue 19 (holds di-Cl moieties at the 3,4-position of ring B and the 4-Cl group on ring C), indicating that incorporation of −Cl substitutions was more effective for α-amylase and α-glucosidase activities (Figure 3 and Table 1).
Figure 3.
SAR studies of analogues 15, 16, 17, and 19.
Table 1. Different Substituent(s) of Triazole-Based bis-Hydrazone Derivatives (1-19) and Their In Vitro α-Amylase and α-Glucosidase Inhibitory Activitiesa.
Standard error mean (SEMa); acarboseb (standard inhibitor for α-amylase and α-glucosidase activities).
Compounds that hold the −NO2 group at the 3-position of ring B and other EW groups, either −NO2 or −Cl, on varied positions of ring C were found to be better competitors of both α-amylase and α-glucosidase enzymes. Among these analogues, analogue 3 showed remarkable potency owing to the attachment of di-Cl moieties at the 2,4-position of ring C along with meta-nitro substitution at ring B. However, the inhibitory potential of analogue 3 was reduced sharply by deattachment of the ortho–Cl moiety at ring C as in the case of compound 5, indicating that incorporation of the −Cl group uplifts the α-amylase and α-glucosidase inhibitory potential. Furthermore, the comparison of compound 1 (that holds the −NO2 substitution at the 3-position of both rings B and C) with compound 2 (bearing 3-NO2 at ring B and 4-NO2 at ring C) demonstrated that compound 2 was found to be significantly active, even threefold more potent than the standard acarbose drug. This discrepancy in activity was due to the different orientations of −NO2 groups at ring C toward the active sites of α-amylase and α-glucosidase enzymes (Figure 4 and Table 1).
Figure 4.
SAR studies of analogues 1, 2, 3, and 5.
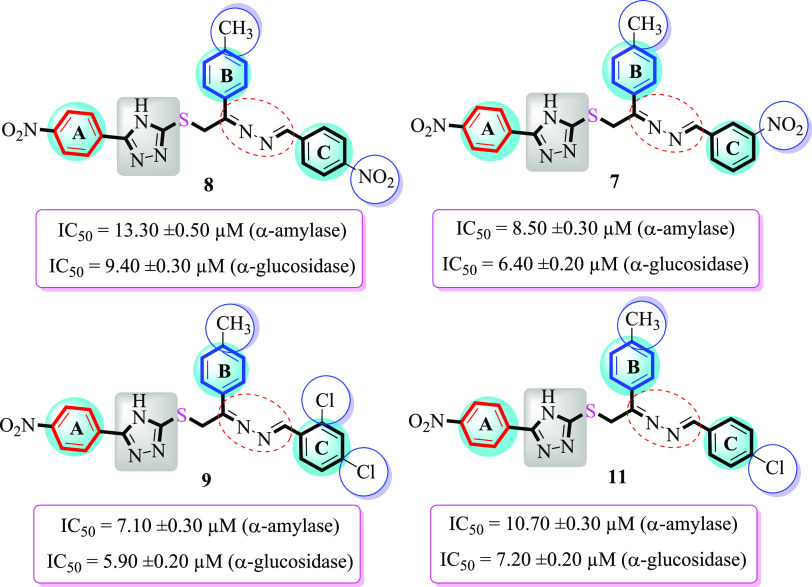
It was noteworthy that incorporation of EW groups such as −NO2 and −Cl moieties at varied positions of ring C in addition to the ED −CH3 moiety at the 4-position of ring B has significant impact on the inhibition profile of targeted α-amylase and α-glucosidase enzymes. Analogue 9 (bearing a −CH3 group at the 4-position of ring B and di-Cl moieties at the 2,4-position of ring C) was identified as a better competitor of α-amylase and α-glucosidase enzymes and displayed more potency than that of the standard acarbose drug. In addition, di-Cl substitutions can inhibit α-amylase and α-glucosidase enzymes more effectively than −NO2 substitution. Therefore, the comparison of derivatives 7 (holds a −NO2 moiety at the 3-position of ring C) and 8 (bearing a −NO2 group at the 4-position of ring C) with derivative 9 (bearing di-Cl substitutions at the 2,4-position of ring C) illustrated that di-Cl moiety-bearing derivative 9 exhibited enhanced α-amylase and α-glucosidase inhibitory potential, even found to be more active than the standard acarbose drug. Likewise, derivative 11 (bearing a −Cl moiety at the 4-position of ring C) exhibited superior inhibitory potential to that of derivative 8 (bearing a −NO2 moiety at the 4-position of ring C) but was less potent when compared to its counterpart 7 (holds a −NO2 moiety at the 3-position of ring C). This activity comparison shows that not only the nature of the attached substituent affects the inhibitory potential but also their positions play an important role in the inhibition of α-amylase and α-glucosidase enzymes (Figure 5 and Table 1).
Figure 5.
SAR studies of analogues 7, 8, 9, and 11.
Compounds 4, 10, and 18 had a di-methylamino group at the 4-position of ring C along with a variety of other groups such as −Cl, −NO2, and −CH3 at variable positions of ring B and showed moderate inhibitory activities against α-amylase and α-glucosidase enzymes. Analogue 18 (bearing a −CH3 moiety at the 4-position of ring B along with a 4-dimethylamino group on ring C) showed slightly decreased activities than those of −NO2 and −Cl group-bearing analogues 4 and 18. The moderate α-amylase and α-glucosidase inhibitory activities revealed by compounds 4, 10, and 18 might be due to the bulky di-methylamino group, which may create steric hindrance during binding with the active pocket of the targeted enzymes. However, the inhibitory potential of analogue 10 was further decreased by introduction of either naphthalene or para-benzyloxy substitutions of more bulky nature at ring C as in the case of compounds 14 and 6. It is interesting to note that compound 13 with a para-formyl group on ring C showed decreased inhibitory potential compared to compound 10 with a para-dimethylamino group on ring C. Likewise, derivatives 18 and 4 showed close inhibitory activities, which revealed that the di-methylamino group at the 4-position of ring C does not really affect the inhibitory activities. In addition, the inferior inhibitory activities of derivatives 18 and 4 compared to compounds 17 and 3 might be due to steric hindrance created by the bulky di-methylamino group while binding with the active sites of targeted enzymes as well as smaller susceptibility of the dimethylamino group itself to inhibit α-amylase and α-glucosidase enzymes compared to electronegative −Cl and −NO2 groups (Figure 6 and Table 1).
Figure 6.
SAR studies of analogues 4, 10, 13, and 18.
The SAR showed that electron-withdrawing groups such −Cl and −NO2 groups play an important role in significant α-amylase and α-glucosidase inhibitory activities of the synthesized compounds. Additionally, other groups also demonstrated significant activities when placed in a certain number/s and positions.
2.3. In Silico Molecular Docking Studies
Molecular docking was performed in order to explore the binding mode of the synthesized compounds against the targeted enzymes, i.e., α-amylase and α-glucosidase. Both the enzyme crystallographic coordinates were retrieved from www.rcsb.org. The docking procedure revealed that the selected compounds had numerous potentials when tested against the targeted enzymes. Among the tested series, most of the compounds possess varied functional groups such as chloro and nitro substitutions with a superposed surface complex. These different substituted ring structures were docked and observed for their binding modalities against the selected enzymes. In this regard, chloro- and nitro-substituted compounds exhibited better potential against these enzymes. Specifically, In the case of efficacious compounds 17 and 15, the protein–ligand interaction (PLI) profile not only listed the best potential (in silico) but also demonstrated better potency in vitro study. The most potent compound 17 displayed remarkable behavior due to various PLI profiles. This compound bearing dichloro-moieties at the 3,4-position of both aromatic rings B and C have significant interactive residues for α-amylase such as His201 (hb), Glu233 (hb), Leu162 (π-Alkyl), His305 (hb), His299 (π-Alkyl), and Trp62 (Alkyl) as shown in (Figure 7), while against α-glucosidase, they include Ser505 (hb), Ser497 (hb), Asp232 (π-anion), ASP568 (π-Anion), Trp329 (π-π Stacked), Phe601 (π-π Stacked), His626 (Alkyl), Met470 (Alkyl), Ala628 (Alkyl), etc., as shown in (Figure 8).
Figure 7.
Protein–ligand interaction (PLI) profile of the most potent analogue 17 against the α-amylase enzyme and its 3D (left) and 2D (right) diagrams.
Figure 8.
Protein–ligand interaction (PLI) profile of the most potent analogue 17 against the α-glucosidase enzyme and its 3D (left) and 2D (right) diagrams.
The protein–ligand interaction profile for compound 15 (having dichloro groups at the 3,4-position of ring B and a para-nitro group at ring C) showed different interactive residues for α-amylase as shown in (Figure 9) such as His-305 (donor-interactions), Leu-165 (π-alkyl), Trp-59(π-cation), Tyr-62(π-π), and Asp (hb). Similarly, for α-glucosidase, the interactive residues are Ala-602 (π-Alkyl), Ala-234 (π-Alkyl), ASP-568 (Ala-602) (π-cation), MET470 (π-alkyl), Trp470 (π-π), Trp329 (π-π), His626 (hb), PHE601 (Alkyl), and Asp232 (hb) as shown in (Figure 10).
Figure 9.
Protein–ligand interaction (PLI) profile of the most potent analogue 15 against the α-amylase enzyme and its 3D (left) and 2D (right) diagrams.
Figure 10.
Protein–ligand interaction (PLI) profile of the most potent analogue 15 against the α-glucosidase enzyme and its 3D (left) and 2D (right) diagrams.
The only differences found in both compounds 17 and 15 are the attached substituents and PLI profile; in both the cases, chloro and nitro are attached to the para position of the aromatic ring, in which the chloro group increases the nucleophilic character of the ring, while nitro being an electron-withdrawing moiety decreases the nucleophilic character; therefore, with moieties a very weak interaction, however, the presence of triazole and extended benzene ring moieties facilitates a strong interaction with the enzyme active site, while in the case of the chloro-substituted analogue, with the presence of a more nucleophilic π-system, the interaction of the compounds was observed to be maximum. It seemed from SAR studies that scaffolds bearing di-Cl and −NO2 groups showed better inhibitory activities because the oxygen of the −NO2 group plays an effective role in the inhibition of both of these enzymes through formation of H-bonding with the enzyme active site; therefore, compound 17 was recognized as more potent owing to the involvement of oxygen of the −NO2 group in hydrogen bonding with the active site of both targeted enzymes. In addition, in the docking results of the selected compounds when compared to the standard drug acarbose, the binding energy was few-fold better than that of the standard drug and the interaction of the heteroatom in the synthesized moiety was found to be more significant. Overall docking studies show that scaffolds 17 and 15 having electron-donating and -withdrawing moieties showed better results in both the targeted enzymes. The interaction of scaffolds was observed by hydrogen bonding, pi–pi interaction, pi–sulfur, and pi–cation. Scaffolds showed all the mentioned interactions due to various ring moieties and heteroatoms.
3. Conclusions
Triazole-containing bis-hydrazone derivatives (1–19) were synthesized. The novel synthesized compounds were characterized by spectral (1H-NMR, 13C-NMR, and HREI-MS) data and were screened for α-amylase and α-glucosidase activities. The results are correlated with the docking studies. The molecular docking data provided positive correlation with in vitro α-amylase and α-glucosidase activities, and hence, results revealed that these compounds can act as potential inhibitors. The core nucleus triazole-bearing bis-hydrazones (1–19) were efficiently synthesized by treating triazole-based hydrazide with different-substituted benzaldehyde. This hydrazone formation reaction proceeded very smoothly, in a short reaction time with an excellent yield. Triazole-bearing bis-hydrazone compounds (1–19) showed good binding interactions with the target enzyme with the least binding energies. The degree of activity and docking studies displayed by the novel innovative structural hybrids of triazole and bis-hydrazone moieties make these compounds new active leads and promising candidates for the development of antidiabetic agents.
4. Experimental Section
4.1. Materials and Methods
Analytical-grade reagents and solvents were purchased from Sigma-Aldrich and used as received. Thin-layer chromatography (TLC) was performed on precoated silica gel aluminum plates (Kieselgel 60, 254, E. Merck, Germany). TLC chromatograms were visualized under ultraviolet light at 254 and 366 nm. Melting points of the compounds were determined on Stuart SMP10 melting point apparatus. Mass spectra were recorded by electron impact (EI) on MAT 312 and MAT 113D mass spectrometers. The 1H- and 13C-NMR spectra were recorded on Advance Bruker AM spectrometers, operating at 600 and 150 MHz. The chemical shift values are presented in ppm (δ), relative to tetramethylsilane (TMS) as an internal standard, and the coupling constants (J) are in Hz. Multiplicities are reported as singlet (s), doublet (d), triplet (t), doublet of doublets (dd), doublet of triplets (dt), quartet (q), or multiplet (m).
4.2. General Procedure for the Synthesis of Triazole-Bearing bis-Hydrazone Derivatives (1–19)
4.2.1. Synthesis of 2-(4-Nitrobenzoyl)hydrazine-1-carbothioamide II
Thiosemicarbazide (I, 1 equivalent) and 4-nitrobenzoyl chloride (1 equivalent) were taken in DMF (10 mL) and stirred at room temperature. Then, triethylamine was added dropwise and stirred for 3 h. After completion, the reaction mixture was filtered and dried and preserved for the next reaction.40
4.2.2. Synthesis of 5-(4-Nitrophenyl)-4H-1,2,4-triazole-3-thiol III
2% sodium hydroxide solution (10 mL) was taken in a round-bottom flask, and then, 2-(4-nitrobenzoyl)hydrazine-1-carbothioamide (II) was added and refluxed for 12 h. After completion, the excess solvent was removed. The product (III) was precipitated in dil hydrochloric acid, filtered, dried, and recrystallized.41
4.2.3. Synthesis of Triazole-Based Hydrazone Substrate V
An equimolar amount of product (III) and different-substituted phenacyl bromide were taken in ethanol (10 mL), and Et3N was added dropwise while keeping under reflux for 3 h. The substituted 2-[{5-(4-nitrophenyl)-4H-1,2,4-triazol-3-yl}thio]-1-phenylethan-1-one product (IV) so obtained was filtered and dried and then further mixed with hydrazine hydrate (5 mL) in methanol (10 mL) along with dropwise addition of glacial acetic acid. The reaction mixture was refluxed for 4 h and then quenched with water. The product (V) precipitated was filtered, washed, and dried.
4.2.4. Synthesis of Triazole-Based bis-Hydrazones (1–19)
The targeted compounds (1–19) were finally synthesized by refluxing an equimolar amount of substrate (V) with different-substituted benzaldehyde for 4 h catalyzed using 4–6 drops of glacial acetic acid. After completion, the product was precipitated in cold water, filtered, washed, and dried (Scheme 1).
3-((2-(((E)-3-Nitrobenzylidene)hydrazono)-2-(3-nitrophenyl)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (1)
Yield: 61%; m.p.: 191–192 °C; 1H-NMR (600 MHz, DMSO-d6): δ 11.16 (s, 1H, NH), 8.50 (s, 1H, Ar-H), 8.44 (s, 1H, CH), 8.40 (s, 1H, Ar-H), 8.30 (dd, J = 7.8, 1.9Hz, 1H, Ar-H), 8.25 (d, J = 7.6Hz, 2H, Ar-H), 8.18 (dd, J = 7.3, 1.5Hz, 1H, Ar-H), 8.13 (dd, J = 7.3, 1.4Hz, 1H, Ar-H), 8.04 (dd, J = 7.9, 2.1Hz, 1H, Ar-H), 8.01 (d, J = 7.4Hz, 2H, Ar-H), 7.65 (t, J = 7.5Hz, 1H, Ar-H), 7.61 (t, J = 6.9Hz, 1H, Ar-H), 3.73 (s, 2H, CH2), 13C-NMR (150 MHz, DMSO-d6): δ177.6, 164.2, 158.4, 157.3, 149.1, 147.9, 147.6, 138.2, 134.5, 134.1, 134.0, 131.3, 131.3, 129.5, 126.8, 126.8, 126.0, 126.0, 124.0, 124.0, 121.3, 31.5.; HREI-MS: m/z calcd for C23H16N8O6S [M]+532.0914; found; 532.0884.
3-((2-(((E)-4-Nitrobenzylidene)hydrazono)-2-(3-nitrophenyl)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (2)
Yield: 63%; m.p.: 189–190 °C; 1H-NMR (600 MHz, DMSO-d6): δ 11.14 (s, 1H, NH), 8.49 (s, 1H, Ar-H), 8.38 (s, 1H, CH), 8.30 (dd, J = 7.9, 1.7Hz, 1H, Ar-H), 8.29 (d, J = 7.9Hz, 2H, Ar-H),8.24 (d, J = 7.8Hz, 2H, Ar-H), 8.11 (dd, J = 7.5, 1.6Hz, 1H, Ar-H), 8.10(d, J = 7.6Hz, 2H, Ar-H), 8.01 (d, J = 7.4Hz, 2H, Ar-H), 7.60 (t, J = 7.3Hz, 1H, Ar-H), 3.75 (s, 2H, CH2), 13C-NMR (150 MHz, DMSO-d6): δ164.4, 158.5, 157.4, 149.0, 147.6, 141.5, 138.4, 134.5, 134.1, 131.4, 131.4, 127.5, 127.5, 126.0, 126.9, 126.9, 125.3, 124.2, 124.2, 123.6, 123.6, 121.4, 31.4.; HREI-MS: m/z calcd for C23H16N8O6S [M]+532.0914; found;532.0884.
3-((2-(((E)-2,4-Dichlorobenzylidene)hydrazono)-2-(3-nitrophenyl)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (3)
Yield: 66%; m.p.: 194–195 °C; 1H-NMR (600 MHz, DMSO-d6): δ 11.11 (s, 1H, NH), 8.60 (s, 1H, CH),8.50 (s, 1H, Ar-H),8.31 (dd, J = 7.8, 1.6Hz, 1H, Ar-H), 8.25 (d, J = 7.8Hz, 2H, Ar-H), 8.19 (d, J = 7.7Hz, 1H, Ar-H),8.10 (dd, J = 7.6, 1.9Hz, 1H, Ar-H), 8.00 (d, J = 7.8Hz, 2H, Ar-H), 7.68(s, 1H, Ar-H), 7.65 (t, J = 7.4Hz, 1H, Ar-H), 7.49 (d, J = 7.9Hz, 1H, Ar-H), 3.74 (s, 2H, CH2), 13C-NMR (150 MHz, DMSO-d6): δ164.3, 158.4, 157.3, 150.4, 147.3, 138.5, 134.4, 134.2, 134.0, 131.6, 131.6, 129.0, 128.9, 128.1, 126.8, 126.8, 126.4, 126.0, 125.4, 124.2, 124.2, 31.6.; HREI-MS: m/z calcd for C23H15Cl2N7O4S [M]+555.0283; found; 555.0178.
N,N-Dimethyl-4-((1E)-((1-(3-nitrophenyl)-2-((5-(4-nitrophenyl)-4H-1,2,4-triazol-3-yl)thio)ethylidene)hydrazono)methyl)aniline (4)
Yield: 69%; m.p.: 193–194 °C; 1H-NMR (600 MHz, DMSO-d6): δ 11.15 (s, 1H, NH), 8.41 (s, 1H, Ar-H), 8.36 (s, 1H, CH), 8.28 (dd, J = 7.8, 1.6Hz, 1H, Ar-H), 8.22 (d, J = 7.6Hz, 2H, Ar-H), 8.07 (dd, J = 7.7, 2.1Hz, 1H, Ar-H),7.99 (d, J = 7.5Hz, 2H, Ar-H), 7.91 (d, J = 7.5Hz, 2H, Ar-H),7.63 (t, J = 8.0Hz, 1H, Ar-H), 7.28 (d, J = 7.5Hz, 2H, Ar-H),3.71 (s, 2H, CH2), 3.00 (t, 6H, CH3),13C-NMR (150 MHz, DMSO-d6): δ164.5, 158.6, 157.2, 153.2, 149.2, 147.5, 138.5, 134.3, 134.1, 131.2, 131.2, 131.0, 131.0, 126.7, 126.7, 126.0, 125.4, 124.9, 124.9, 121.6, 111.3, 111.3, 41.0, 41.0, 31.8.; HREI-MS: m/z calcd for C25H22N8O4S [M]+530.1485; found; 530.1393.
3-((2-(((E)-4-Chlorobenzylidene)hydrazono)-2-(3-nitrophenyl)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (5)
Yield: 62%; m.p.: 196–197 °C; 1H-NMR (600 MHz, DMSO-d6): δ 11.11 (s, 1H, NH), 8.50 (s, 1H, Ar-H), 8.41 (s, 1H, CH), 8.31 (dd, J = 8.0, 1.9Hz, 1H, Ar-H), 8.25 (d, J = 7.9Hz, 2H, Ar-H), 8.13 (dd, J = 7.6, 1.3Hz, 1H, Ar-H), 8.05 (d, J = 7.7Hz, 2H, Ar-H), 7.85 (d, J = 7.8Hz, 2H, Ar-H), 7.65 (t, J = 7.5Hz, 1H, Ar-H), 7.45 (d, J = 7.8Hz, 2H, Ar-H),3.73 (s, 2H, CH2), 13C-NMR (150 MHz, DMSO-d6): δ164.1, 158.3, 157.2, 149.2, 147.65 138.3, 136.3, 134.6, 134.0, 133.3, 131.3, 131.3, 128.5, 128.5, 128.0, 128.0, 126.3, 125.8, 125.8, 124.6, 123.6, 123.6, 31.3.; HREI-MS: m/z calcd for C23H16ClN7O4S [M]+521.0673; found; 521.0543.
3-((2-(((E)-4-(Benzyloxy)benzylidene)hydrazono)-2-(p-tolyl)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (6)
Yield: 64%; m.p.: 198–199 °C; 1H-NMR (600 MHz, DMSO-d6): δ 11.14 (s, 1H, NH), 8.45 (s, 1H, CH), 8.26 (d, J = 8.1Hz, 2H, Ar-H), 8.01 (d, J = 8.0Hz, 2H, Ar-H),7.75 (d, J = 7.9Hz, 2H, Ar-H), 7.67 (d, J = 7.7Hz, 2H, Ar-H), 7.42 (dd, J = 7.6, 1.8Hz, 2H, Ar-H), 7.35 (m, 2H, Ar-H), 7.27 (t, J = 7.8Hz, 1H, Ar-H), 7.21 (d, J = 7.9Hz, 2H, Ar-H),7.09 (d, J = 7.5Hz, 2H, Ar-H), 5.06 (s, 2H, CH2), 3.71 (s, 2H, CH2),2.30 (s, 3H, CH3),13C-NMR (150 MHz, DMSO-d6): δ164.5, 161.1, 158.6, 157.2, 149.1, 147.6, 140.0, 138.3, 136.5, 136.4, 131.2, 129.0, 129.0, 128.6, 128.6, 127.8, 127.4, 127.1, 127.1, 126.6, 126.6, 126.3, 124.1, 124.1, 114.2, 114.2, 70.2, 31.5, 21.0.; HREI-MS: m/z calcd for C31H26N6O3S [M]+562.1787; found; 562.1703.
3-((2-(((E)-3-Nitrobenzylidene)hydrazono)-2-(p-tolyl)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (7)
Yield: 70%; m.p.: 190–191 °C; 1H-NMR (600 MHz, DMSO-d6): δ 11.10 (s, 1H, NH), 8.43 (s, 1H, CH),8.41 (s, 1H, Ar-H),8.21 (d, J = 7.9Hz, 2H, Ar-H),8.12 (dd, J = 7.8, 1.6Hz, 1H, Ar-H), 8.05 (dd, J = 7.6, 1.8Hz, 1H, Ar-H), 8.00 (d, J = 7.6Hz, 2H, Ar-H),7.68 (t, J = 7.7Hz, 1H, Ar-H), 7.65 (d, J = 7.7Hz, 2H, Ar-H), 7.21 (d, J = 8.0Hz, 2H, Ar-H), 3.72 (s, 2H, CH2),2.39 (s, 3H, CH3),13C-NMR (150 MHz, DMSO-d6): δ 164.1, 158.0, 157.1, 149.2, 147.9, 147.6, 140.0, 138.4, 135.1, 134.3, 131.1, 129.5, 128.6, 128.6, 126.8, 126.8, 126.4, 126.4, 125.9, 124.2, 121.1, 31.3, 21.6.; HREI-MS: m/z calcd for C24H19N7O4S [M]+501.1219; found; 501.1115.
3-((2-(((E)-4-Nitrobenzylidene)hydrazono)-2-(p-tolyl)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (8)
Yield: 71%; m.p.: 195–196 °C; 1H-NMR (600 MHz, DMSO-d6): δ 12.53 (s, 1H, NH), 9.68 (s, 1H, CH),8.10 (d, J = 8.2Hz, 2H, Ar-H), 7.70 (d, J = 8.2Hz, 2H, Ar-H), 7.61 (d, J = 7.2Hz, 2H, Ar-H), 7.59 (d, J = 8.2Hz, 2H, Ar-H), 7.36 (d, J = 8.2Hz, 2H, Ar-H), 7.07 (d, J = 7.3Hz, 2H, Ar-H), 5.27 (s, 2H, CH2), 4.30 (s, 3H, CH3), 13C-NMR (150 MHz, DMSO-d6): δ192.0, 174.1, 172.2, 171.6, 170.8, 169.3, 169.2, 168.9, 168.5, 149.1, 148.7, 148.6, 136.4, 135.6, 131.8, 131.1, 130.3, 130.1, 129.9, 128.4, 121.4, 121.1, 49.5, 33.4.; HREI-MS: m/z calcd for C24H19N7O4S [M]+501.1219; found; 501.1115.
3-((2-(((E)-2,4-Dichlorobenzylidene)hydrazono)-2-(p-tolyl)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (9)
Yield: 68%; m.p.: 188–189 °C; 1H-NMR (600 MHz, DMSO-d6): δ 11.17 (s, 1H, NH), 8.45 (s, 1H, CH),8.25 (d, J = 7.8Hz, 2H, Ar-H), 8.20 (d, J = 7.7Hz, 1H, Ar-H), 8.06 (d, J = 8.0Hz, 2H, Ar-H),7.69 (s,1H, Ar-H), 7.66 (d, J = 7.5Hz, 2H, Ar-H), 7.49 (d, J = 7.9Hz, 1H, Ar-H), 7.21 (d, J = 7.4Hz, 2H, Ar-H), 3.70 (s, 2H, CH2),2.38 (s, 3H, CH3), 13C-NMR (150 MHz, DMSO-d6): δ161.1, 158.2, 157.4, 150.3, 147.5, 140.1, 138.4, 134.2, 131.2, 131.1, 129.3, 129.0, 129.0, 128.9, 128.1, 126.7, 126.5, 126.5, 126.2, 126.2, 124.2, 124.2, 31.5, 21.0.; HREI-MS: m/z calcd for C24H18Cl2N6O2S [M]+524.0589; Found; 524.0516.
N,N-Dimethyl-4-((1E)-((2-((5-(4-nitrophenyl)-4H-1,2,4-triazol-3-yl)thio)-1-(p-tolyl)ethylidene)hydrazono)methyl)aniline (10)
Yield: 73%; m.p.: 187–188 °C; 1H-NMR (600 MHz, DMSO-d6): δ 12.20 (s, 1H, NH), 9.49 (s, 1H, CH), 8.31 (d, J = 7.0Hz, 2H, Ar-H), 8.04 (d, J = 6.0Hz, 2H, Ar-H), 7.91 (d, J = 8.4Hz, 2H, Ar-H), 7.58 (d, J = 6.0Hz, 2H, Ar-H), 7.33 (d, J = 6.1Hz, 2H, Ar-H), 7.19 (d, J = 7.5Hz, 2H, Ar-H), 5.25 (s, 2H, CH2), 1.99 (s, 6H, CH3), 1.23 (s, 3H, CH3), 13C-NMR (150 MHz, DMSO-d6): δ190.8, 178.2, 177.4, 168.0, 151.3, 149.9, 137.2, 137.1, 136.8, 136.3, 136.2, 135.6, 135.1, 132.2, 132.0, 131.9, 131.4, 130.4, 130.2, 128.6, 128.4, 127.6, 40.7, 40.0, 30.6, 25.1.; HREI-MS: m/z calcd for C26H25N7O2S [M]+499.1790; found; 499.1773.
3-((2-(((E)-4-Chlorobenzylidene)hydrazono)-2-(p-tolyl)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (11)
Yield: 60%; m.p.: 191–192 °C; 1H-NMR (600 MHz, DMSO-d6): δ 12.20 (s, 1H, NH), 9.49 (s, 1H, CH), 8.31 (d, J = 7.1Hz, 2H, Ar-H), 8.04 (d, J = 7.8Hz, 2H, Ar-H), 7.90 (d, J = 8.3Hz, 2H, Ar-H), 7.56 (d, J = 6.0Hz, 2H, Ar-H), 7.31 (d, J = 7.2Hz, 2H, Ar-H), 7.17 (d, J = 6.2Hz, 2H, Ar-H), 5.21 (s, 2H, CH2), 4.76 (s, 3H, CH3), 13C-NMR (150 MHz, DMSO-d6): δ190.8, 178.2, 177.4, 168.0, 151.3, 149.9, 149.4, 137.2, 137.1, 136.8, 136.3, 136.2, 135.6, 135.1, 132.2, 132.0, 131.9, 131.4, 131.2, 130.4, 130.2, 128.6, 40.7, 25.1.; HREI-MS: m/z calcd for C24H19ClN6O2S [M]+490.0979; found; 490.0929.
3-((2-(((E)-4-(Benzyloxy)benzylidene)hydrazono)-2-(o-tolyl)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (12)
Yield: 65%; m.p.: 194–195 °C; 1H-NMR (600 MHz, DMSO-d6): δ 11.14 (s, 1H, NH), 8.45 (s, 1H, CH), 8.26 (d, J = 8.1Hz, 2H, Ar-H), 8.01 (d, J = 8.0Hz, 2H, Ar-H), 7.75 (d, J = 7.9Hz, 2H, Ar-H), 7.42 (dd, J = 7.6, 1.8Hz, 2H, Ar-H), 7.39 (d, J = 7.7Hz, 2H, Ar-H), 7.35 (m, 2H, Ar-H), 7.27 (t, J = 7.8Hz, 1H, Ar-H), 7.21 (d, J = 7.9Hz, 2H, Ar-H), 7.09 (d, J = 7.5Hz, 2H, Ar-H), 5.06 (s, 2H, CH2), 3.71 (s, 2H, CH2), 2.30 (s, 3H, CH3), 13C-NMR (150 MHz, DMSO-d6): δ 164.4, 161.0, 158.5, 157.1, 149.1, 147.5, 140.1, 138.2, 136.4, 136.4, 131.2, 129.0, 129.0, 128.6, 128.6, 127.8, 127.4, 127.1, 127.1, 126.6, 126.6, 126.3, 124.1, 124.1, 114.2, 114.2, 70.2, 31.4, 21.1.; HREI-MS: m/z calcd for C31H26N6O3S [M]+562.1767; found; 562.1703.
4-((1E)-((2-((5-(4-Nitrophenyl)-4H-1,2,4-triazol-3-yl)thio)-1-(p-tolyl)ethylidene)hydrazono)methyl)benzaldehyde (13)
Yield: 69%; m.p.: 201–202 °C; 1H-NMR (600 MHz, DMSO-d6): δ 11.03 (s, 1H, NH), 8.41 (s, 1H, CH),8.21 (d, J = 7.6Hz, 2H, Ar-H), 8.08 (d, J = 8.1Hz, 2H, Ar-H),8.00 (d, J = 7.7Hz, 2H, Ar-H), 7.90 (d, J = 7.5Hz, 2H, Ar-H), 7.67 (d, J = 7.7Hz, 2H, Ar-H), 7.19 (d, J = 8.0Hz, 2H, Ar-H), 9.80 (s, 1H, CO-H), 3.70 (s, 2H, CH2),2.36 (s, 3H, CH3), 13C-NMR (150 MHz, DMSO-d6): δ189.7,164.3, 158.2, 157.5, 149.2, 147.5, 141.2, 140.4, 139.0, 138.3, 130.9, 130.2, 130.2, 129.5, 129.5, 129.0, 129.0, 126.9, 126.9, 126.6, 126.6, 124.2, 124.2, 31.5, 21.0.; HREI-MS: m/z calcd for C25H20N6O3S [M]+484.1318; found; 484.1280.
3-((2-(((E)-Naphthalen-2-ylmethylene)hydrazono)-2-(p-tolyl)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (14)
Yield: 68%; m.p.: 203–204 °C;1H-NMR (600 MHz, DMSO-d6): δ 11.09 (s, 1H, NH), 8.59 (s, 1H, Ar-H), 8.44 (s, 1H, CH), 8.39 (d, J = 7.8Hz, 1H, Ar-H),8.22 (d, J = 8.0Hz, 2H, Ar-H), 8.07 (d, J = 7.9Hz, 2H, Ar-H),8.03 (d, J = 7.5Hz, 2H, Ar-H), 7.97 (d, J = 7.8Hz, 1H, Ar-H), 7.91 (dd, J = 7.7, 1.9Hz, 1H, Ar-H), 7.83 (dd, J = 8.1, 2.0Hz, 1H, Ar-H), 7.55-7.52 (m, 1H, Ar-H), 7.51-7.48 (m, 1H, Ar-H), 7.20 (d, J = 7.4Hz, 2H, Ar-H), 3.72 (s, 2H, CH2),2.35 (s, 3H, CH3), 13C-NMR (150 MHz, DMSO-d6): δ164.4, 158.7, 157.3, 149.2, 147.6, 140.5, 138.6, 136.0, 133.6, 131.0, 129.0, 129.0, 128.4, 128.0, 128.0, 127.1, 126.9, 126.9, 126.5, 126.5, 126.1, 125.8, 125.8, 124.1, 124.1, 31.3, 21.6.; HREI-MS: m/z calcd for C28H22N6O2S [M]+506.1525; found; 506.1456.
3-((2-(3,4-Dichlorophenyl)-2-(((E)-4-nitrobenzylidene)hydrazono)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (15)
Yield: 74%; m.p.: 206–207 °C;1H-NMR (600 MHz, DMSO-d6): δ 11.15 (s, 1H, NH), 8.48(s, 1H, CH), 8.30 (d, J = 7.7Hz, 2H, Ar-H),8.23 (d, J = 7.8Hz, 2H, Ar-H),8.11 (d, J = 7.9Hz, 2H, Ar-H), 8.02 (d, J = 7.6Hz, 2H, Ar-H),7.83 (d, J = 7.8Hz, 1H, Ar-H),7.81 (s, 1H, Ar-H), 7.60(d, J = 7.5Hz, 1H, Ar-H), 3.75 (s, 2H, CH2), 13C-NMR (150 MHz, DMSO-d6): δ164.4, 158.5, 157.4, 149.0, 147.6, 141.5, 138.4,135.3, 133.0, 130.2, 130.1, 127.5, 127.1, 127.1, 126.2, 124.2, 124.2, 123.7, 123.7, 31.1.; HREI-MS: m/z calcd for C23H15Cl2N7O4S [M]+555.0283; found; 555.0178.
3-((2-(3,4-Dichlorophenyl)-2-(((E)-3-nitrobenzylidene)hydrazono)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (16)
Yield: 76%; m.p.: 201-202°C;1H-NMR (600 MHz, DMSO-d6): δ11.15 (s, 1H, NH), 8.48(s, 1H, CH), 8.41 (s, 1H, Ar-H), 8.33 (d, J = 7.8Hz, 2H, Ar-H), 8.25 (dd, J = 7.8, 1.9Hz, 1H, Ar-H),8.01 (dd, J = 7.8, 1.6Hz, 1H, Ar-H), 7.96 (d, J = 7.6Hz, 2H, Ar-H), 7.83 (d, J = 7.8Hz, 1H, Ar-H), 7.81 (s, 1H, Ar-H), 7.69(t, J = 7.8Hz, 1H, Ar-H), 7.50 (d, J = 6.9Hz, 1H, Ar-H), 3.75 (s, 2H, CH2), 13C-NMR (150 MHz, DMSO-d6): δ164.4, 158.5, 157.4, 149.0, 147.6, 141.5, 138.4, 137.4, 137.1, 135.3, 133.0, 130.2, 130.1, 127.5, 127.1, 127.1, 126.2, 125.7, 124.2, 124.2, 123.7, 123.7, 31.1.; HREI-MS: m/z calcd for C23H15Cl2N7O4S [M]+555.0283; found; 555.0178.
3-((2-(((E)-3,4-Dichlorobenzylidene)hydrazono)-2-(3,4-dichlorophenyl)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (17)
Yield: 72%; m.p.: 207–208 °C;1H-NMR (600 MHz, DMSO-d6): δ 11.05 (s, 1H, NH), 8.48(s, 1H, CH), 8.41 (s, 1H, Ar-H), 8.26 (s, 1H, Ar-H), 8.21 (dd, J = 7.7, 1.7Hz, 1H, Ar-H),8.01 (d, J = 7.5Hz, 2H, Ar-H),7.85 (d, J = 7.7 Hz, 1H, Ar-H), 7.82 (d, J = 7.9Hz, 1H, Ar-H),7.80 (s, 1H, Ar-H), 7.67(d, J = 7.5Hz, 1H, Ar-H), 7.65 (d, J = 7.8Hz, 1H, Ar-H), 3.75 (s, 2H, CH2), 13C-NMR (150 MHz, DMSO-d6): δ164.5, 158.4, 157.3, 149.1, 147.4, 141.3, 138.2,135.1, 133.3, 130.4, 130.4, 127.5, 127.1, 127.1, 126.2, 124.2, 124.2, 123.7, 123.7, 31.1.; HREI-MS: m/z calcd for C23H14Cl4N6O2S [M]+579.9624; found; 579.9523.
4-((1E)-((1-(3,4-Dichlorophenyl)-2-((5-(4-nitrophenyl)-4H-1,2,4-triazol-3-yl)thio)ethylidene)hydrazono)methyl)-N,N-dimethylaniline (18)
Yield: 77%; m.p.: 210-211°C;1H-NMR (600 MHz, DMSO-d6): δ11.15 (s, 1H, NH), 8.47(s, 1H, CH), 8.22 (d, J = 7.7Hz, 2H, Ar-H), 8.01 (d, J = 7.8Hz, 2H, Ar-H), 7.82 (d, J = 7.8Hz, 1H, Ar-H), 7.80 (s, 1H, Ar-H),7.62 (d, J = 7.7Hz, 2H, Ar-H),7.59 (d, J = 7.5Hz, 1H, Ar-H), 6.70 (d, J = 7.9Hz, 2H, Ar-H), 3.75 (s, 2H, CH2), 3.01 (s, 6H, CH3),13C-NMR (150 MHz, DMSO-d6): δ164.3, 158.6, 157.4, 153.2, 149.1, 147.6, 138.2, 135.7, 133.3, 131.1, 131.0, 130.4, 130.1, 127.0, 127.0, 126.1, 124.2, 124.2, 121.1, 111.8, 111.8, 41.1, 41.1, 31.5.; HREI-MS: m/z calcd for C25H21Cl2N7O2S [M]+553.0854; found; 553.0783.
3-((2-(((E)-4-Chlorobenzylidene)hydrazono)-2-(3,4-dichlorophenyl)ethyl)thio)-5-(4-nitrophenyl)-4H-1,2,4-triazole (19)
Yield: 65%; m.p.: 200-201°C; 1H-NMR (600 MHz, DMSO-d6): δ11.14 (s, 1H, NH), 8.47(s, 1H, CH), 8.24 (d, J = 7.8Hz, 2H, Ar-H), 8.01 (d, J = 7.6Hz, 2H, Ar-H), 7.82 (d, J = 7.9Hz, 2H, Ar-H), 7.81 (d, J = 7.8Hz, 1H, Ar-H), 7.79 (s, 1H, Ar-H), 7.59 (d, J = 7.5Hz, 1H, Ar-H), 7.49 (d, J = 7.7Hz, 2H, Ar-H), 3.75 (s, 2H, CH2), 13C-NMR (150 MHz, DMSO-d6): δ164.4, 158.5, 157.4, 149.0, 147.6, 141.5, 138.4,135.3, 133.0, 130.2, 130.1, 127.5, 127.1, 127.1, 126.2, 124.2, 124.2, 123.7, 123.7, 31.1.; HREI-MS: m/z calcd for C23H15Cl3N6O2S [M]+544.0043; found; 544.0015.
4.3. α-Glucosidase Inhibition Assay
To find out the α-glucosidase inhibition, kinetic studies were carried out using different concentrations of the inhibitors (0.0624, 0.3, 0.125, 0.4 mM), and different concentrations of the substrate inhibitor (p-nitrophenol R-D maltoside NMP) were prepared. 0.1, 0.2, 0.4, 0.8, and 1.0 mM (0.2 mg/mL) concentrations were prepared in deionized water with the enzyme. PIPES buffer was used for adjusting the pH of the solution. This solution was incubated for 30 min at 25 °C, and the absorbance was recorded on an ELISA reader in 96-well plates. Kinetic parameters such as Vmax, AICs, Km, and R2 were calculated using Sigmaplot Enzyme Kinetics software.42,43
4.4. α-Amylase Inhibition Assay
The Kwon and Apostolidis methods were used for the determination of α-amylase inhibition.44 500 μL (0.5 mg/mL) of α-amylase was prepared in phosphate buffer, and 500 μL of the sample (100, 200, 400, 800, 1000 μg/mL) was also prepared. Both solutions were incubated at 25°C for 10 min. 1% starch solution (500 μL) and 0.02 M sodium phosphate buffer were added and incubated for 10 min. Dinitrosalisylic acid was added as a color agent, incubated in boiling water for 5 minutes, cooled, and diluted using distilled water. The percentage inhibition was recorded from the absorbance using the formula
4.5. Molecular Docking Protocol
Molecular docking study was performed by using Discovery Studio Visualizer (DSV) and Autodoc Tools 1.5.7. The synthesized compounds were docked against α-amylase and α-glucosidase, and their structures were obtained from the Protein Data bank (PDB) by searching codes such as 1b2y and 3w37. Initially, protein was prepared using DSV to maintain the structure by removing water, and both the structure protein as well as the selected analogue was saved in the PDB format. Both the structures were open in Autodoc Tools by adding polar hydrogen to protein as well as Kollman and Gasteiger charges. Ligand preparation was done by using Torsion Tree to detect the root and saved in the PDBQT format. The configuration file was generated along with X, Y, and Z axis, and the protein structures were saved in PDBQT. In order to generate different poses of molecules with varied energy, a command prompt was employed. The top-ranking molecules were docked with the protein PDBQT format in DSV. The protein–ligand interaction (PLI) of molecules with the active site of enzymes is summarized in Figures 7A, 8B, 9C, and 10D.
Acknowledgments
The authors extend their appreciation to the Researchers Supporting Project number (RSPD2023R628), King Saud University, Riyadh, Saudi Arabia, for funding this project. The authors also extend their appreciation to Princess Nourah bint Abdulrahman University Researcher supporting project number (PNURSP2023R342), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia for funding this project.
Data Availability Statement
Data will be made available on request.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c00702.
1H-NMR and 13C-NMR data of compounds (PDF)
Author Contributions
I.K., W.R., F.R., R.H., S.K., L.R., M.M.A., A.S.A., and M.H.A.
The authors declare no competing financial interest.
Supplementary Material
References
- Aguiar M. M. G. B. D.; Albuquerque R. P. D.; Marinho D. S.; Braga B. R. S.; Dornelas C. B.; Oliveira A.; Sousa V. P. D.; Torres S. R.; Alviano D. S.; Alviano C. S.; Cabral L. M.; Holandino C. Oral sustained release nystatin tablets for the treatment of oral candidiasis: formulation development and validation of UV spectrophotometric analytical methodology for content determination. Drug Dev. Ind. Pharm. 2010, 36, 594–600. 10.3109/03639040903384729. [DOI] [PubMed] [Google Scholar]
- Fall J. A.; Holen D. L.; Davis B.; Krieg T.; Koster D.. Subsistence harvests and uses of wild resources in Iliamna, Newhalen, Nondalton, Pedro Bay, and Port Alsworth, Alaska, Alaska Department of Fish and Game Division of Subsistence Technical Paper 2006, p 302.
- Baron A. D. Postprandial hyperglycaemia and α-glucosidase inhibitors. Diabetes Res. Clin. Pract. 1998, 40, 51–55. 10.1016/S0168-8227(98)00043-6. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L.; Goldberg A. L. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998, 8, 397–403. 10.1016/S0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- Porte D.; Kahn S. E. beta-cell dysfunction and failure in type 2 diabetes: potential mechanisms. Diabetes 2001, 50, S160 10.2337/diabetes.50.2007.S160. [DOI] [PubMed] [Google Scholar]
- Rabasa-Lhoret R.; Chiasson J. L.. α-Glucosidase inhibitors International textbook of diabetes mellitus 2003.
- Albright A.; Franz M.; Hornsby G.; Kriska A.; Marrero D.; Ullrich I.; Verity L. S. American College of Sports Medicine position stand. Exercise and type 2 diabetes. Med. Sci. Sports Exerc. 2000, 32, 1345–1360. 10.1097/00005768-200007000-00024. [DOI] [PubMed] [Google Scholar]
- Jenkins D. J.; Wolever T. M.; Taylor R. H.; Barker H.; Fielden H.; Baldwin J. M.; Goff D. V.; et al. Glycemic index of foods: a physiological basis for carbohydrate exchange. Am. J. Clin. Nutr. 1981, 34, 362–366. 10.1093/ajcn/34.3.362. [DOI] [PubMed] [Google Scholar]
- Wolever T. M.; Katzman-Relle L.; Jenkins A. L.; Vuksan V.; Josse R. G.; Jenkins D. J. Glycaemic index of 102 complex carbohydrate foods in patients with diabetes. Nutr. Res. 1994, 14, 651–669. 10.1016/S0271-5317(05)80201-5. [DOI] [Google Scholar]
- Ali S.; Stone M. A.; Peters J. L.; Davies M. J.; Khunti K. The prevalence of co-morbid depression in adults with Type 2 diabetes: a systematic review and meta-analysis. Diabet. Med. 2006, 23, 1165–1173. 10.1111/j.1464-5491.2006.01943.x. [DOI] [PubMed] [Google Scholar]
- Geng P.; Qiu F.; Zhu Y.; Bai G. Four acarviosin-containing oligosaccharides identified from Streptomyces coelicoflavus ZG0656 are potent inhibitors of α-amylase. Carbohydr. Res. 2008, 343, 882–892. 10.1016/j.carres.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Ahrén B. O.; Landin-Olsson M.; Jansson P. A.; Svensson M.; Holmes D.; Schweizer A. Inhibition of dipeptidyl peptidase-4 reduces glycemia, sustains insulin levels, and reduces glucagon levels in type 2 diabetes. J. Clin. Endocr. Metab. 2004, 89, 2078–2084. 10.1210/jc.2003-031907. [DOI] [PubMed] [Google Scholar]
- Svensson B.; Søgaard M. Mutational analysis of glycosylase function. J. Biotechnol. 1993, 29, 1–37. 10.1016/0168-1656(93)90038-O. [DOI] [Google Scholar]
- Takkinen K.; Laukkanen M. L.; Sizmann D.; Alfthan K.; Immonen T.; Vanne L.; Teeri T. T. An active single-chain antibody containing a cellulase linker domain is secreted by Escherichia coli Protein Eng. Des. Sel. 1991, 4, 837–841. 10.1093/protein/4.7.837. [DOI] [PubMed] [Google Scholar]
- Mohan S.; Sim L.; Rose D. R.; Pinto B. M. Synthesis of S-alkylated sulfonium-ions and their glucosidase inhibitory activities against recombinant human maltase glucoamylase. Carbohydr. Res. 2007, 342, 901–912. 10.1016/j.carres.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Anderson G. G.; Palermo J. J.; Schilling J. D.; Roth R.; Heuser J.; Hultgren S. J. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 2003, 301, 105–107. 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- Seo M. J.; Suh S. Y.; Bae Y. C.; Jung J. S. Differentiation of human adipose stromal cells into hepatic lineage in vitro and in vivo. Biochem. Biophys. Res. Commun. 2005, 328, 258–264. 10.1016/j.bbrc.2004.12.158. [DOI] [PubMed] [Google Scholar]
- Wakita T.; Pietschmann T.; Kato T.; Date T.; Miyamoto M.; Zhao Z.; Liang T. J.; et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 2005, 11, 791–796. 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari M. R.; Jong-Anurakkun N.; Hong G.; Kawabata J. α-Glucosidase and α-amylase inhibitory activities of Nepalese medicinal herb Pakhanbhed (Bergenia ciliata, Haw.). Food Chem. 2008, 106, 247–252. 10.1016/j.foodchem.2007.05.077. [DOI] [Google Scholar]
- Hermeking H. P53 enters the microRNA world. Cancer Cell 2007, 12, 414–418. 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- Hussaina F.; Khan Z.; Saeed J. M.; Ahmad S.; Ahmad A.; Rashid U.; Ullah F.; Ayaz M.; Sadiq A. Synthesis, in-vitro α-glucosidase inhibition, antioxidant, in-vivo antidiabetic and molecular docking studies of pyrrolidine-2,5-dione and thiazolidine-2,4-dione derivatives. Bioorg. Chem. 2019, 91, 103128 10.1016/j.bioorg.2019.103128. [DOI] [PubMed] [Google Scholar]
- Jahani R.; Abtahi S. R.; Nematpour M.; Dastjerdi H. F.; Chamanara M.; Hami Z.; Paknejad B. Design, synthesis, and pharmacological evaluation of novel 1,2,4-triazol-3- amine derivatives as potential agonists of GABAA subtype receptors with anticonvulsant and hypnotic effects. Bioorg. Chem. 2020, 104, 104212 10.1016/j.bioorg.2020.104212. [DOI] [PubMed] [Google Scholar]
- Fadaly W. A. A.; Elshaier Y. A. M. M.; Hassanein E. H. M.; Abdellatif K. R. A. New 1,2,4-triazole/pyrazole hybrids linked to oxime moiety as nitric oxide donor celecoxib analogs: Synthesis, cyclooxygenase inhibition antiinflammatory, ulcerogenicity, anti-proliferative activities, apoptosis, molecular modeling and nitric oxide release studies. Bioorg. Chem. 2020, 98, 103752 10.1016/j.bioorg.2020.103752. [DOI] [PubMed] [Google Scholar]
- Joshi R.; Kumari A.; Singh K.; Mishra H.; Pokharia S. Triorganotin(IV) complexes of Schiff base derived from 1,2,4-triazole moiety: Synthesis, spectroscopic investigation, DFT studies, antifungal activity and molecular docking studies. J. Mol. Struct. 2020, 1206, 127639 10.1016/j.molstruc.2019.127639. [DOI] [Google Scholar]
- Ammazzalorso A.; Gallorini M.; Fantacuzzi M.; Gambacorta N.; Filippis B. D.; Giampietro L.; Maccallini C.; Nicolotti O.; Cataldi A.; Amoroso R. Design, synthesis and biological evaluation of imidazole and triazole-based carbamate as novel aromatase inhibitors. Eur. J. Med. Chem. 2020, 211, 113115 10.1016/j.ejmech.2020.113115. [DOI] [PubMed] [Google Scholar]
- Mallikarjuna B. P.; Sastry B. S.; Kumar G. V. S.; Rajendraprasad Y.; Chandrashekar S. M.; Sathisha K. Synthesis of new 4-isopropylthiazole hydrazide analogs and some derived clubbed triazole, oxadiazole ring systems-A novel class of potential antibacterial, antifungal and antitubercular agents. Eur. J. Med. Chem. 2009, 44, 4739–4746. 10.1016/j.ejmech.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Pitucha M.; Janeczko M.; Klimek K.; Fornal E.; Wos M.; Stec A. P.; Ginalska G.; Kaczor A. A. 1,2,4-Triazolin-5-thione derivatives with anticancer activity as CK1γ kinase inhibitors. Bioorg. Chem. 2020, 99, 103806 10.1016/j.bioorg.2020.103806. [DOI] [PubMed] [Google Scholar]
- a Taha M.; Ullah H.; Al-Muqarrabun L. M. R.; Khan M. N.; Rahim F.; Ahmat N.; Ali M.; Perveen S. Synthesis of bis-indolylmethanes as new potential inhibitors of β-glucuronidase and their molecular docking studies. Eur. J. Med. Chem. 2018, 143, 1757–1767. 10.1016/j.ejmech.2017.10.071. [DOI] [PubMed] [Google Scholar]; b Noreen T.; Taha M.; Imran S.; Chigurupati S.; Rahim F.; Selvaraj M.; Ismail N. H.; Mohammad J. I.; Ullah J. I.; Javid H.; Nawaz F.; Irshad M.; Ali M. Synthesis of Alpha Amylase Inhibitors Based on Privileged Indole Scaffold. Bioorg. Chem. 2017, 72, 248–255. 10.1016/j.bioorg.2017.04.010. [DOI] [PubMed] [Google Scholar]
- a Taha M.; Sultan S.; Nuzar H. A.; Rahim F.; Imran S.; Ismail N. H.; Naz H.; Ullah H. Synthesis and biological evaluation of novel N-arylidenequinoline-3-carbohydrazides as potent β-glucuronidase inhibitors. Bioorg. Med. Chem. 2016, 24, 3696–3704. 10.1016/j.bmc.2016.06.008. [DOI] [PubMed] [Google Scholar]; b Taha M.; Imran S.; Ismail N. H.; Selvaraj M.; Rahim F.; Chigurupati S.; Ullah H.; Khan F.; Salar U.; Javid M. T.; Vijayabalan S.; Zaman K.; Khan K. M. Biology-oriented drug synthesis (BIODS) of 2-(2-methyl-5-nitro-1Himidazol-1-yl)ethylaryl ether derivatives, in vitro α-amylase inhibitory activity and in silico studies. Bioorg. Chem. 2017, 74, 1–9. 10.1016/j.bioorg.2017.07.001. [DOI] [PubMed] [Google Scholar]
- a Rahim F.; Javid M. T.; Ullah H.; Wadood A.; Taha M.; Ashraf M.; Aine Q. U.; Khan M. A.; Khan F.; Mirza S.; Khan K. M. Synthesis, Molecular Docking, Acetylcholinesterase and Butyrylcholinesterase Inhibitory Potential of Thiazole Analogs as New Inhibitors for Alzheimer Disease. Bioorg. Chem. 2015, 62, 106–116. 10.1016/j.bioorg.2015.08.002. [DOI] [PubMed] [Google Scholar]; b Taha M.; Ismail N. H.; Imran S.; Rahim F.; Wadood A.; Khan H.; Ullah H.; Salar U.; Khan K. M. Synthesis, β-Glucuronidase Inhibition and Molecular Docking Studies of Hybrid Bisindole-Thiosemicarbazides Analogs. Bioorg. Chem. 2016, 68, 56–63. 10.1016/j.bioorg.2016.07.008. [DOI] [PubMed] [Google Scholar]
- a Rahim F.; Ullah H.; Taha M.; Wadood A.; Javid M. T.; Rehman W.; Nawaz M.; Ashraf M.; Ali M.; Sajid M.; Ali F.; Khan M. N.; Khan K. M. Synthesis and in vitro Acetylcholinesterase and Butyrylcholinesterase Inhibitory Potential of Hydrazide based Schiff Bases. Bioorg. Chem. 2016, 68, 30–40. 10.1016/j.bioorg.2016.07.005. [DOI] [PubMed] [Google Scholar]; b Taha M.; Javid M. T.; Imran S.; Selvaraj M.; Chigurupati S.; Ullah H.; Rahim F.; Khan F.; Mohammad J. I.; Khan K. M. Synthesis and study of the a-amylase inhibitory potential of thiadiazole quinoline derivatives. Bioorg. Chem. 2017, 74, 179–186. 10.1016/j.bioorg.2017.08.003. [DOI] [PubMed] [Google Scholar]
- a Rahim F.; Ullah K.; Ullah H.; Wadood A.; Taha M.; Rehman A.; Ashraf I. U.; Shaukat M.; Rehman A.; Hussain W. S.; Khan K. M. Triazinoindole analogs as potent inhibitors of α-glucosidase: Synthesis, biological evaluation and molecular docking studies. Bioorg. Chem. 2015, 58, 81–87. 10.1016/j.bioorg.2014.12.001. [DOI] [PubMed] [Google Scholar]; b Taha M.; Rahim F.; Imran S.; Ismail N. H.; Ullah H.; Selvaraj M.; Javid M. T.; Salar U.; Ali M.; Khan K. M. Synthesis, α-glucosidase inhibitory activity and in silico study of tris-indole hybrid scaffold with oxadiazole ring: As potential leads for the management of type-II diabetes mellitus. Bioorg. Chem. 2017, 74, 30–40. 10.1016/j.bioorg.2017.07.009. [DOI] [PubMed] [Google Scholar]
- a Rahim F.; Ullah H.; Javid M. T.; Wadood A.; Taha M.; Ashraf M.; Shaukat A.; Junaid M.; Hussain S.; Rehman W.; Mehmood R.; Sajid M.; Khan M. N.; Khan K. M. Synthesis, in vitro evaluation and molecular docking studies of thiazole derivatives as new inhibitors of α-glucosidase. Bioorg. Chem. 2015, 62, 15–21. 10.1016/j.bioorg.2015.06.006. [DOI] [PubMed] [Google Scholar]; b Rahim F.; Malik F.; Ullah H.; Wadood A.; Khan F.; Javid M. T.; Taha M.; Rehman W.; Rehman A. U.; Khan K. M. Isatin based Schiff bases as inhibitors of α-glucosidase: Synthesis, characterization, in vitro evaluation and molecular docking studies. Bioorg. Chem. 2015, 60, 42–48. 10.1016/j.bioorg.2015.03.005. [DOI] [PubMed] [Google Scholar]
- a Rahim F.; Ali M.; Ullah S.; Rashid U.; Ullah H.; Taha M.; Javed M. T.; Rehman W.; Abid O. U. R.; Khan A. A.; Bilal M. Development of bis-Thiobarbiturates as Successful Urease Inhibitors and their Molecular Modelling Studies. Chin. Chem. Lett. 2016, 27, 693–697. 10.1016/j.cclet.2015.12.035. [DOI] [Google Scholar]; b Rashid U.; Rahim F.; Taha M.; Arshad M.; Ullah H.; Mahmood T.; Ali M. Synthesis of 2-Acylated and Sulfonated 4-hydroxycoumarins: In vitro Urease Inhibition and Molecular Docking Studies. Bioorg. Chem. 2016, 66, 111–116. 10.1016/j.bioorg.2016.04.005. [DOI] [PubMed] [Google Scholar]
- a Rahim F.; Zaman K.; Ullah H.; Taha M.; Wadood A.; Javed M. T.; Rehman W.; Ashraf M.; Uddin R.; Uddin I.; Asghar H.; Khan A. A.; Khan K. M. Synthesis of 4-thiazolidinone Analogs as potent in vitro Anti-Urease Agents. Bioorg. Chem. 2015, 63, 123–131. 10.1016/j.bioorg.2015.10.005. [DOI] [PubMed] [Google Scholar]; b Taha M.; Ullah H.; Al-Muqarrabun L. M. R.; Khan M. N.; Rahim F.; Ahmat N.; Javid M. T.; Ali M.; Khan K. M. Bisindolylmethanethiosemicarbazides as potential inhibitors of urease: Synthesis and molecular modelling studies. Bioorg. Med. Chem. 2018, 26, 152–160. 10.1016/j.bmc.2017.11.028. [DOI] [PubMed] [Google Scholar]
- Yeye E. O.; Khan K. M.; Chigurupati S.; Wadood A.; Rehman A. U.; Perveen S.; Maharajan M. K.; Shamim S.; Hameed S.; Aboaba S. A.; Taha M. Syntheses, in vitro α-amylase and α-glucosidase dual inhibitory activities of 4-amino-1, 2, 4-triazole derivatives their molecular docking and kinetic studies. Bioorg. Med. Chem. 2020, 28, 115467 10.1016/j.bmc.2020.115467. [DOI] [PubMed] [Google Scholar]
- Rahim F.; Zaman K.; Taha M.; Ullah H.; Ghufran M.; Wadood A.; Rehman W.; Uddin N.; Shah S. A. A.; Sajid M.; Nawaz F.; Khan K. M. Synthesis, in vitro alpha-glucosidase inhibitory potential of benzimidazole bearing bis-Schiff bases and their molecular docking study. Bioorg. Chem. 2020, 94, 103394 10.1016/j.bioorg.2019.103394. [DOI] [PubMed] [Google Scholar]
- Khan I.; Rehman W.; Rahim F.; Hussain R.; Khan S.; Fazil S.; Rasheed L.; Taha M.; Shah S.A.A.; Abdellattif M. H.; Farghaly T. A. Synthesis, In Vitro α-Glucosidase Inhibitory Activity and Molecular Docking Study of New Benzotriazole-Based Bis-Schiff Base Derivatives. Pharmaceuticals 2023, 16, 17 10.3390/ph16010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousaf K.; Shahid I.; Mazloom S.; Aneela M.; Rafaqat H.; Shoaib K.; Imran K.; Rami P.; Adel A.; Eman F.; Abd-ElAziem A. M.; Issa A.; Hisham S. M. New quinoline-based triazole hybrid analogs as effective inhibitors of α-amylase and α-glucosidase: Preparation, in vitro evaluation, and molecular docking along with in silico studies. Front. Chem. 2022, 10, 995820 10.3389/fchem.2022.995820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mioc M.; Soica C.; Bercean V.; Avram S.; Balan-Porcarasu M.; Coricovac D.; Ghiulai R.; Muntean D.; Andrica F.; Dehelean C.; Spandidos D. A.; et al. Design, synthesis and pharmaco-toxicological assessment of 5-mercapto-1, 2, 4-triazole derivatives with antibacterial and antiproliferative activity. Int. J. Oncol. 2017, 50, 1175–1183. 10.3892/ijo.2017.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui N.; Ahsan W. Triazole incorporated thiazoles as a new class of anticonvulsants: Design, synthesis and in vivo screening. Eur. J. Med. Chem. 2010, 45, 1536–1543. 10.1016/j.ejmech.2009.12.062. [DOI] [PubMed] [Google Scholar]
- Hussain R.; Iqbal S.; Shah M.; Rehman W.; Khan S.; Rasheed L.; Rahim F.; Dera A. A.; Kehili S.; Elkaeed E. B.; Awwad N.S. Synthesis of Novel Benzimidazole-Based Thiazole Derivatives as Multipotent Inhibitors of α-Amylase and α-Glucosidase: In Vitro Evaluation along with Molecular Docking Study. Molecules 2022, 27, 6457 10.3390/molecules27196457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Escudero M.; Gimeno-Perez M.; Gonzalez B.; Linde D.; Merdzo Z.; Lobato M. F.; Sanz-Aparicio J. Structural analysis of β-fructofuranosidase from Xanthophyllomycesdendrorhous reveals unique features and the crucial role of N-glycosylation in oligomerization and activity. J. Biol. Chem. 2016, 291, 6843–6857. 10.1074/jbc.M115.708495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salar U.; Khan K. M.; Chigurupati S.; Taha M.; Wadood A.; Vijayabalan S.; Perveen S. New hybrid hydrazinyl thiazole substituted chromones: as potential α-amylase inhibitors and radical (DPPH & ABTS) scavengers. Sci. Rep. 2017, 7, 16980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.