ABSTRACT
Bathyarchaeota, known as key participants of global elements cycling, is highly abundant and diverse in the sedimentary environments. Bathyarchaeota has been the research spotlight on sedimentary microbiology; however, its distribution in arable soils is far from understanding. Paddy soil is a habitat similar to freshwater sediments, while the distribution and composition of Bathyarchaeota in paddy soils have largely been overlooked. In this study, we collected 342 in situ paddy soil sequencing data worldwide to illuminate the distribution patterns of Bathyarchaeota and explore their potential ecological functions in paddy soils. The results showed that Bathyarchaeota is the dominant archaeal lineage, and Bathy-6 is the most predominant subgroup in paddy soils. Based on random forest analysis and construction of a multivariate regression tree, the mean annual precipitation and mean annual temperature are identified as the factors significantly influencing the abundance and composition of Bathyarchaeota in paddy soils. Bathy-6 was abundant in temperate environments, while other subgroups were more abundant in sites with higher rainfall. There are highly frequent associations between Bathyarchaeota and methanogens and ammonia-oxidizing archaea. The interactions between Bathyarchaeota and microorganisms involved in carbon and nitrogen metabolism imply a potential syntrophy between these microorganisms, suggesting that members of Bathyarchaeota could be important participants of geochemical cycle in paddy soils. These results shed light on the ecological lifestyle of Bathyarchaeota in paddy soils, and provide some baseline for further understanding Bathyarchaeota in arable soils.
IMPORTANCE
Bathyarchaeota, the dominant archaeal lineage in sedimentary environments, has been the spotlight of microbial research due to its vital role in carbon cycling. Although Bathyarchaeota has been also detected in paddy soils worldwide, its distribution in this environment has not yet been investigated. In this study, we conducted a global scale meta-analysis and found that Bathyarchaeota is also the dominant archaeal lineage in paddy soils with significant regional abundance differences. Bathy-6 is the most predominant subgroup in paddy soils, which differs from sediments. Furthermore, Bathyarchaeota are highly associated with methanogens and ammonia-oxidizing archaea, suggesting that they may be involved in the carbon and nitrogen cycle in paddy soil. These interactions provide insight into the ecological functions of Bathyarchaeota in paddy soils, which will be the foundation of future studies regarding the geochemical cycle in arable soils and global climate change.
KEYWORDS: Bathyarchaeota, archaea, paddy soil, meta-analysis, co-occurrence network
INTRODUCTION
Bathyarchaeota, formerly known as Miscellaneous Crenarchaeotal Group (MCG), were firstly discovered in hot spring (1) and have been characterized by high intragroup diversity containing 25 subgroups (2). Based on phylogenetic analyses, MCG was considered a novel archaeal phylum and proposed to name “Bathyarchaeota” (3, 4). Alternatively, the Genome Taxonomy Database (GTDB) has reclassified the phylum Bathyarchaeota to a class-level clade and renamed as Bathyarchaeia within the phylum of Thermoproteota. In addition to hot springs, Bathyarchaeota is widespread and abundant in various anoxic environments, such as sediments (5, 6), acid-sulfate springs (7), termite guts (8), and bioreactors (9).
Due to its highly abundant in various environments, Bathyarchaeota has been widely recognized as an important player in global geochemical cycling, especially in carbon cycling, which is closely linked to global climate change (2). Bathyarchaeotal multiple metabolic pathways have been inferred depending on genome information and isotope culture experiments. Evans et al. (10) recovered two bathyarchaeotal genomes belonging to Bathy-3 and Bathy-8, which contained genes encoding the methyl-coenzyme M reductase (MCR) complex, implying that Bathyarchaeota might be involved in methane metabolism. Bathyarchaeota are supposed to be capable to utilize various complex organic substrates, such as detrital proteins, aromatic compounds, carbohydrates, etc (11, 12). Through single-cell sequencing, genes encoding extracellular protein degrading enzymes were found in the incomplete bathyarchaeotal genomes obtained from marine subsea floor, indicating potential protein degrading capability (13). He et al. (3) recovered six Bathyarchaeota bins from a marine sediment sample collected from the Guaymas Basin in the Gulf of California, and the key genes involved in the reductive acetyl-CoA (Wood–Ljungdahl, WL) pathway were found in five bins, indicating that Bathyarchaeota had the potential of inorganic carbon fixation and acetate generation. In addition, genomic analysis demonstrated that Bathyarchaeota could degrade proteins, cellulose, chitin, and aromatic compounds for their own growth (3). Isotope culture experiments further demonstrated the important roles of Bathyarchaeota in sediment carbon cycle (14 - 16). Yin et al. (15) confirmed that Bathy-15 were active catabolic archaeal protein degraders in marine sediments. Moreover, Bathy-8 could utilize lignin as an energy source and bicarbonate as a carbon source, suggesting an organoautotrophic lifestyle of Bathyarchaeota (16). Our previous study found that adding fulvic acid to paddy soil significantly enriched Bathyarchaeota, which indicated that Bathyarchaeota could grow on fulvic acid or its metabolites as carbon sources (17). All these studies suggested that Bathyarchaeota might have diverse carbon metabolic pathways, including heterotrophic and autotrophic lifestyles, and different subgroups might be active degraders of different organic matters. In addition to carbon metabolism, Bathyarchaeota might also be involved in dissimilatory nitrite reduction to ammonium (18) and dissimilatory sulfate reduction (19). These studies greatly expanded our knowledge of ecological roles of Bathyarchaeota in sediments.
Besides diverse metabolic pathways, different bathyarchaeotal subgroups have specific environment preferences. Fillol et al. (20) collected bathyarchaeotal sequences and constructed a multivariate regression tree, indicating that salinity was the best explanatory variable for dissimilarity of bathyarchaeotal community. Bathy-1 and Bathy-8 dominated in saline sediments, while Bathy-11 and Bathy-5b dominated in freshwater sediments. Pan et al. (21) found that pH is the most important factor affecting the bathyarchaeotal community structure in mangrove wetlands and inferred that Bathy-6 preferred slightly acidic, high TOC, and subsurface environments. While Fillol et al. (22) demonstrated that Bathy-6 present in both planktonic and benthic habitats with a wide range of reducing conditions. Bathy-18 was found predominantly within the reduced environments, while Bathy-10 is the opposite (23). All these studies demonstrate that the habitat conditions shape bathyarchaeotal communities.
However, all these research progress on Bathyarchaeota mainly focuses on sedimentary environments. Paddy soil is usually flooded and similar to freshwater sediments that inhabit abundant Bathyarchaeota, while little attention has been paid to this distinctive habitat of Bathyarchaeota. Therefore, we hypothesized that Bathyarchaeota might be widespread and be important players for the biogeochemical cycle in paddy soils. Previously, some studies have found a high relative abundance of Bathyarchaeota in Italian paddy soils (24, 25). In contrast, a continental-scale survey of paddy soil in eastern China reported a low relative abundance of Bathyarchaeota (26). So far, the distribution and community composition of Bathyarchaeota in paddy soils has yet to be revealed. In addition, compared with sediments, the microbial community could be strongly influenced by plant growth and anthropogenic activities in paddy soil; thus, the distribution patterns of Bathyarchaeota and its subgroup might differ from that in sediments. Therefore, in this study, we collected in situ paddy soil sequencing data around the world, and aimed to illustrate the distribution patterns and subgroups compositions of Bathyarchaeota in paddy soils; to determine the environmental factors shaping the bathyarchaeotal community structure in paddy soils; to infer the potential functions of Bathyarchaeota in paddy soils according to the associations between Bathyarchaeota and other microorganisms.
MATERIALS AND METHODS
Publications collection and data set construction
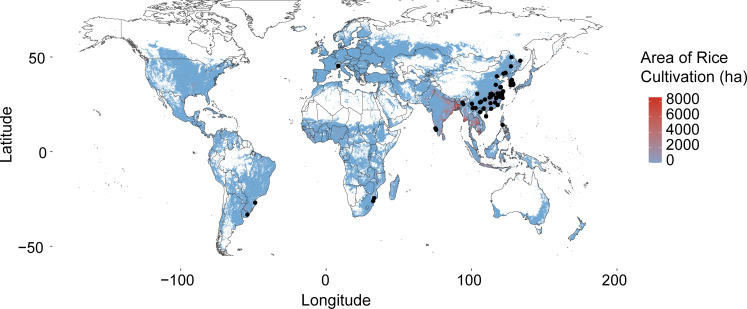
To investigate the distribution of Bathyarchaeota in paddy soil, we collected the publications about paddy soil microbial community by searching Google Scholar, PubMed, and Web of Science using “paddy soil” AND “microbial community” OR “microbial structure” as keywords (published before 1 September 2020). Then, we filtered the publications according to the following criteria: the soil samples should be collected from in situ soil or field trial, excluding microcosmic experiments and pot experiments; the amplicon primers should target archaea or both bacteria and archaea; raw data is available and can be downloaded from public databases according to the access number provided in the publications. After filtering, 23 publications (containing 342 paddy fields from nine countries) were included in this meta-analysis (Table 1). The sampling sites were mainly concentrated in Asia (China, South Korea, Philippines, Vietnam, and India), which cover the main rice production regions around the world (including Asia, Americas, Africa) (https://www.fao.org/faostat/en/#data/QCL/visualize) (Fig. 1, Table 1). Additionally, the samples used for the analysis included different water management phases. Details of these publication were shown in supporting information (Table S1). Climatic data for sampling sites was collected from WorldClim database (version 2.1, https://worldclim.org/) and sampling condition and soil physicochemical properties of each sample were collected from corresponding publication (Table S1).
TABLE 1.
Sample sites and corresponding publications involved in this study
Fig 1.
The location of sampling sites in this study. The black dots represented the sampling sites. The color of different areas on the map represented the area of rice cultivation. The closer the color to red meant the larger the area of rice cultivation. Data on area of rice cultivation came from https://www.mapspam.info/data/.
Bioinformatics analysis
The raw data of each study was downloaded and independently processed with the same pipeline as the previous study (48) (Fig. S1). Briefly, nonbiological sequences were removed by Cutadapt (version 3.7). The trimmed sequences were denoised using DADA2 plugin in the Qiime2 (version 2022.2) (49). The feature table and amplicon sequence variant (ASV) sequences were obtained. Then the taxonomy classification of ASVs was performed using a pretrained Naive Bayes classifier and the q2-feature-classifier plugin based on the Silva 138 database. The unassigned ASVs and the ASVs belonging to mitochondria, chloroplasts, or eukaryotes were discarded. After data processing, samples with a sequence number of archaea of more than 500 were included in the meta-analysis. The relative abundance of archaeal lineages in these samples was tabulated as data set 1 (Table S2). The rarefaction of samples in data set 1 was performed in R (version 4.2.2) using “vegan” package. After rarefaction, the relative abundance of bathyarchaeotal subgroups in samples with Bathyarchaeota reads > 50 constituted the data set 2 (Table S3) and was included in the downstream analysis.
Phylogenetic analysis
To investigate bathyarchaeotal community structure in paddy soil, the phylogenetic analysis was conducted to classify the Bathyarchaeota ASVs into different subgroups. The 2,030 ASVs belonging to Bathyarchaeota of data set 2 were dereplicated at 90% identity threshold and clustered into 80 operational taxonomic units (OTUs) with VSEARCH (version 2.7.0) (50). OTUs and reference sequences from Zhou et al. (2) were aligned by MAFFT (version 7.490) and trimmed by trimAl (version 1.4) (51). The remaining OTUs were classified into a reference tree (2) by RAxML (version 8.2.12) using “-f v -m GTRGAMMA” and the best tree was visualized by iTOL (https://itol.embl.de/).
Statistical analysis
All statistical analyses and visualization were conducted in R (version 4.2.2) and Origin 2023. Species Abundance Distribution (SAD) was performed to investigate the abundance and pattern of archaeal lineages. The index of dispersion for each archaeal lineage was calculated as the ratio of the variance to the mean relative abundance multiplied by the occurrence to reflect the dispersion pattern of archaeal lineages (20). To investigate the effect of environmental factors on the microbial community, the samples were classified into different groups based on the mean annual precipitation (MAP) and the mean annual temperature (MAT). For MAP, the samples were divided into three groups (>2000 mm, >1000 mm and <2000 mm, <1000 mm). For MAT, the samples were divided into three groups (>20℃, >10℃ and <20℃, <10℃). Based on this classification, principal Coordinates Analysis (PCoA) based on Bray–Curtis distances and permutational multivariate analysis of variance (PERMANOVA) were carried out using “vegan” package. Random forest analysis was performed to rank the importance of factors influencing the abundance of Bathyarchaeota using “rfPermute” package with the option “ntree = 500, nrep = 1000”. And Wilcoxon rank sum test was used to further explore the preferred environment for Bathyarchaeota. A multivariate regression tree was constructed using “mvpart” and “MVPARTwrap” packages to identify the factors affecting the abundance of bathyarchaeotal subgroups with the option “xv = pick, xvmult = 100” (52).
Co-occurrence network construction
Interactions between Bathyarchaeota and other microorganisms were inferred from the cooccurrence network. OTUs (at 90% identity threshold) with more than five sequences were used to calculate checkerboard score (C-score) and standardized effect size (SES) to analyze the randomness of the species distribution through “EcoSimR” package. Then, the Spearman correlations were calculated to construct cooccurrence network using “psych” package (53). The edges with spearman correlation > 0.6 and Benjamini–Hochberg adjusted P < 0.01 remained and used to construct the network by “igraph” package (54). The robust network was imported into Gephi (version 0.9) for visualization and property characterization.
RESULTS
The relative abundance of Bathyarchaeota in paddy soils
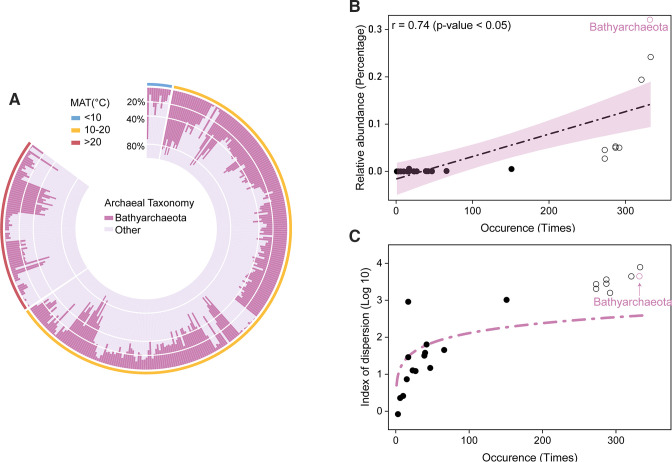
In this meta-analysis, a total of 10,549 ASVs belonging to 24 archaeal lineages were obtained. Among these 24 archaeal lineages, Bathyarchaeota was the most frequently occurring lineage with the highest average relative abundance in paddy soils. Bathyarchaeota was detected in 332 samples, and its relative abundance ranged from 0.38% to 92.13% to the total relative abundance of archaea, with the average relative abundance of 32.07% (Fig. 2A and B). In addition, the relationship between the average relative abundance and occurrence frequency of archaeal lineages was analyzed to determine the ecological importance of Bathyarchaeota (Fig. 2). Results showed that there was a significant positive relationship between the average relative abundance against occurrence frequency (r = 0.74, P < 0.05). The index of dispersion against occurrence frequency plot were drawn to test whether these lineages followed a stochastic distribution (Fig. 2C). The point representing Bathyarchaeota fell above of the line depicted the 2.5% confidence limit of the χ2 distribution, indicating the distribution of Bathyarchaeota in paddy soils follows a nonstochastic process. In addition to Bathyarchaeota, Nitrososphaeria and Methanosarcinia were dominant archaea lineages in paddy soils with the relative abundances of 24.17% and 19.39% within archaea, respectively.
Fig 2.
Distribution patterns of Bathyarchaeota in paddy soils. (A) Circular barplot showed the relative abundance of Bathyarchaeota within archaea in all samples. The samples were divided into three groups according to the annual mean temperature of the sample sites (> 20℃, > 10℃ and < 20℃, < 10℃. (B) The relationship between the relative abundance of all the archaeal lineages with their occurrence frequency. Hollow circles represent core lineages. (C) The index of dispersion against occurrence frequency. The line depicts the 2.5% confidence limit of the χ2 distribution. The point fell above of the line indicating a nonstochastic distribution.
The distribution patterns of Bathyarchaeota in paddy soils
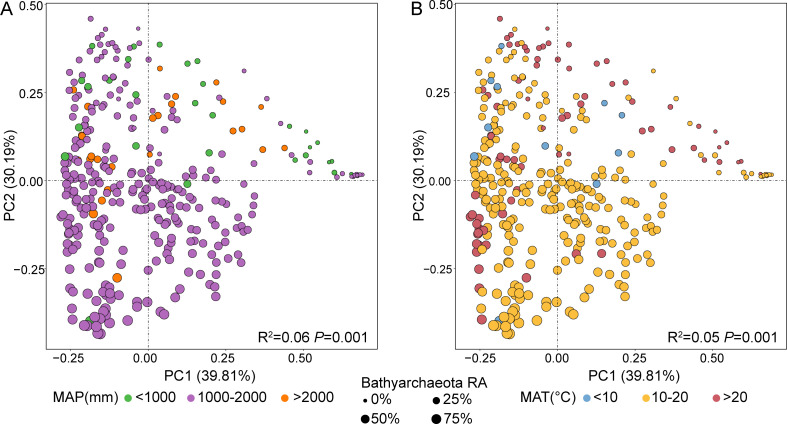
To illustrate the influence of environmental factors on archaeal community structure, we classified all the samples into different categories according to climatic data and sampling site conditions (Table S1 and S2). Principal Coordinates Analysis based on Bray–Curtis distances was performed to investigate the difference in archaeal community structure among different samples (Fig. 3). PC1 and PC2 explained 39.81% and 30.19% of the total variance, respectively. As shown in Fig. 3, the archaeal community of all samples significantly separated in paddy soils with different annual precipitation, and also separated in paddy soils with different temperatures. In addition, the relative abundance of Bathyarchaeota also significantly separated according to annual precipitation and temperature. The samples with high relative abundance of Bathyarchaeota were concentrated in the regions with moderate temperature or medium rainfall (Fig. 3). The result of PERMANOVA indicated that the MAP explained the most variation (R2 = 0.06, P = 0.001) and followed by the MAT (R2 = 0.05, P = 0.001). Additionally, results illustrated that climatic factors more significantly influenced the archaeal community structure than agricultural activities, such as planting (R2 = 0.03) or flooding (R2 = 0.01).
Fig 3.
PCoA based on Bray–Curtis distances and PERMANOVA of archaeal community. Figure (A) and (B) colored the points according to the MAP and MAT, respectively. Samples were classified into different groups according to theMAP and the MAT. For MAP, samples were divided into three groups (>2000 mm, >1000 mm and <2000 mm, <1000 mm). For MAT, samples were divided into three groups (>20℃, >10℃ and < 20℃, <10℃).
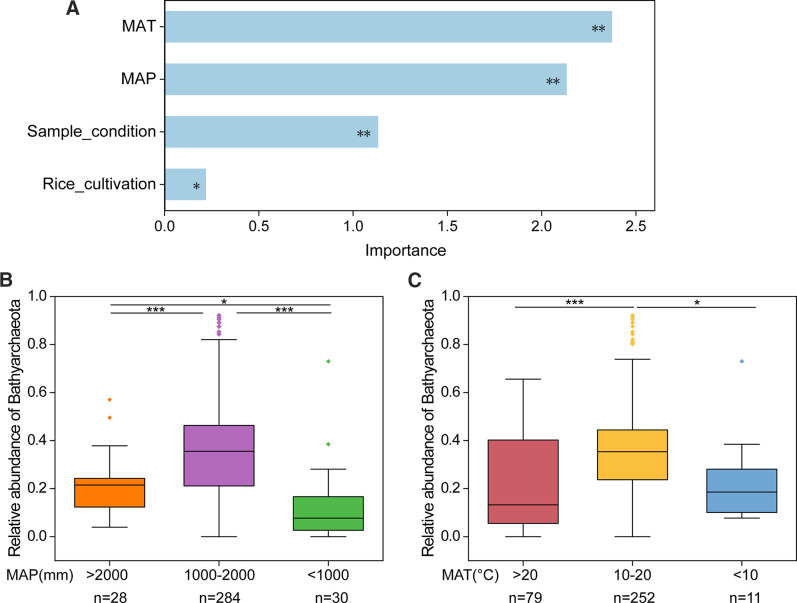
To identify the key drivers for the abundance of Bathyarchaeota, random forest analysis was carried out to evaluate the importance of predictors on the relative abundance of Bathyarchaeota (Fig. 4A). MAT was found to be the most important predictor (IncNodePurity = 2.37) followed by MAP (IncNodePurity = 2.13). Compared with climatic factors, the agricultural activities, such as rice cultivation and sampling condition had less effects on the abundance of Bathyarchaeota (Fig. 4A). This result was consistent with the conclusion obtained from the previous PCoA analysis (Fig. 3). Wilcoxon rank sum test was performed to further characterize the environmental preference of Bathyarchaeota. Based on MAP classification, the relative abundance of Bathyarchaeota was significantly higher in samples with MAP between 1000 mm and 2000 mm (32.72%) than in those with MAP higher than 2000 mm (20.96%) and less than 1000 mm (12.30%) (P < 0.001) (Fig. 4B). For MAT, the relative abundance of Bathyarchaeota was highest under moderate temperature conditions (>10℃ and <20℃) (35.82%) than under high (>20℃) or low (< 10℃) temperature conditions (P < 0.01) (Fig. 4C, Fig. 2A). These results indicate that Bathyarchaeota prefer temperate environments.
Fig 4.
Environmental factors driving the abundance of Bathyarchaeota. (A) Random Forest Analysis revealed the importance of predictors on the relative abundance of Bathyarchaeota. (B, C) Wilcoxon rank sum test determined the significance of the difference in the relative abundance of Bathyarchaeota between different groups. The groupings of samples in Figure (B) and (C) were based on MAP and MAT, respectively.
The biogeography of Bathyarchaeota subgroups in paddy soils
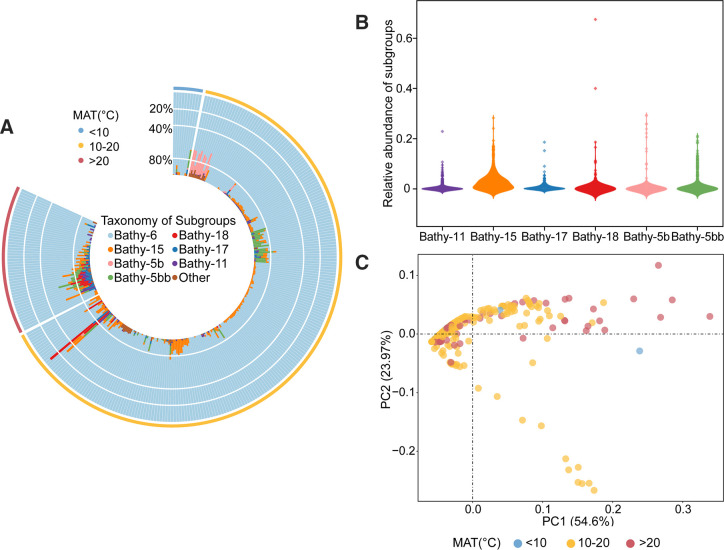
There were 80 OTUs (clustered by 2,030 ASVs at the threshold of 97% similarity) belonging to Bathyarchaeota, belonging to 11 subgroups according to the reference tree constructed by Zhou et al. (2) (Fig. S2). Among these 80 OTUs, 40 OTUs were identified as Bathy-6, which was the overwhelmingly dominant subgroup in paddy soil, accounting for 91.67% of the whole bathyarchaeotal community with a relative abundance of 7.23% ~ 100% (Fig. 5A). In addition to Bathy-6, there were six other subgroups with relative abundance higher than 0.5%, accounting for 5.3% of bathyarchaeotal community (Fig. 5B). Bathy-15 was the second abundant subgroup which was detected in 192 samples with an average relative abundance of 3.01%.
Fig 5.
The composition pattern of bathyarchaeotal subgroups in paddy soils. (A) The relative abundance of bathyarchaeotal subgroups within Bathyarchaeota. (B) The abundance of the remaining subgroups except Bathy-6. (C) PCoA based on Bray–Curtis distances and PERMANOVA of bathyarchaeotal community. The points colored according to MAT.
PCoA analysis illustrated that all samples were mainly distributed along PC1 axis which explained 54.6% of the total variance (Fig. 5C). Both MAT (R2 = 0.02, P = 0.012) and MAP (R2 = 0.02, P = 0.004) significantly influenced the bathyarchaeotal community structure. We further conducted multivariate regression tree analysis to explore the effect of climatic factors and soil properties on distribution patterns of bathyarchaeotal subgroups. The result was presented as a four-leaf tree, explaining 43.7% of the total variance (Fig. S3A), and MAT was further identified as the key driver shaping bathyarchaeotal subgroups. Since MAT might covaries with the other environmental variables and a high proportion of Bathy-6 might weaken the abundance changes of the other subgroups, the specific drivers for different subgroups were further explored (Fig. S3B, Fig. S4). As shown in Fig. S3B, the relative abundance of Bathy-6 was significantly influenced by MAT (R2 = 0.282), and the samples were clustered into two groups according to temperature 27.18℃. Bathy-6 was more abundant (with an average abundance of 93.35%) when MAT lower than 27.18℃ (n = 274), this result is confirmed by Wilcoxon sum test (P < 0.001). MAP was another important factor for the composition of bathyarchaeotal subgroups. Bathy-11, Bathy-17, Bathy-18, and Bathy-5bb were more abundant in paddy soils with higher MAP (Fig. S4). In addition to climatic factors, soil pH significantly influenced the abundance of Bathy-5b, Bathy-5bb, and Bathy-15.
Interactions between Bathyarchaeota and other microorganisms
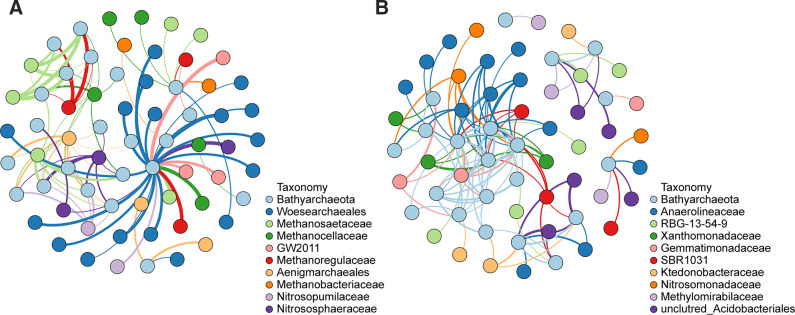
We constructed robust co-occurrence networks based on Spearman correlations (a coefficient > 0.6 and PFDR < 0.01) to preliminarily understand the potential interactions between Bathyarchaeota and other microorganisms. Firstly, the C-score was calculated to evaluate the co-occurrence patterns and stochasticity of archaeal community (54). We observed a C-score value of 278.68, which was significantly higher than simulated C-score (C-scoresim = 257.61, P < 0.001). The result rejected the null model hypothesis and confirmed that the archaeal community was a nonrandom and segregated co-occurrence pattern. The archaeal network was composed of 181 nodes and 1,052 undirected edges, which was classified into 18 modules with a module index of 0.56 (values > 0.4 suggest that the network has a modular structure) (Fig. S5A). Among all the archaeal lineages, Bathyarchaeota was the second most abundant node (16.02%), occurring in six major modules (Fig. S5B). To more clearly demonstrate the potential ecological niches of Bathyarchaeota, we filtered and retained the edges between Bathyarchaeota and the top 10 archaeal lineages (Fig. 6A). Within this network, Bathyarchaeota exhibited the closest connection with Woesearchaeales. In addition, methanogens, such as Methanosaetaceae, Methanocellaceae, Methanoregulaceae, and Methanobacteriaceae, had nonrandom associations with Bathyarchaeota. Moreover, Bathyarchaeota also co-occurred with ammonia-oxidizing archaea (Nitrosopumilaceae and Nitrososphaeraceae). After calculating the C-score, a nonrandom co-occurrence network between bathyarchaeotal and bacterial OTUs was also constructed (Fig. 6B). There were 377 edges connecting Bathyarchaeota and bacteria. Among them, Anaerolineaceae showed the most associations with Bathyarchaeota, followed by RBG-13-54-9.
Fig 6.
Co-occurrence network analysis based on Spearman correlations. (A) Co-occurrence network between Bathyarchaeota and other archaea. (B) Co-occurrence network between Bathyarchaeota and bacteria. Top nine microorganisms connected with Bathyarchaeota were shown in the network.
DISCUSSION
Bathyarchaeota is widespread and abundant in paddy soils
Paddy soil is one of the major types of global land use (Fig. 1), and long-term flooding during rice cultivation makes it an important source of methane emissions (55). At the same time, paddy soil is an important carbon sink with long-term stable carbon sequestration (56). Hence, the carbon cycling in paddy soil has been a hot research topic. Bathyarchaeota, known as important players in the carbon cycle of environments with high carbon content, such as marine ecosystems and terrestrial permafrost (57), has been found to be abundant in paddy soil (24, 58). We thus inferred that members of Bathyarchaeota might be involved in paddy soil carbon metabolism. Therefore, we collected sequencing data from 342 in situ paddy soil samples worldwide to provide a primary understanding of the distribution and composition patterns of Bathyarchaeota at the global scale.
We analyzed the SAD (species abundance distribution) of the observed 24 archaeal lineages in paddy soil (Fig. 2B and C), which could be divided into two groups based on their frequency of occurrence and average abundance (20). One group was rare/less abundant satellite lineages, which were detected in less than 160 samples; another group was persistent/abundant core lineages consisting of 8 lineages which were detected in more than 270 samples. Bathyarchaeota were included in the persistent/abundant group, and which was the most frequently occurring lineages with the occurrence of 97% (observed in 332 samples of total 342 samples analyzed) (Fig. 2B). In addition, Bathyarchaeota shows high abundance in paddy soils, with an average relative abundance of 32.07%, which is similar to the abundance in the sediments (20). These results suggest that Bathyarchaeota is the core archaeal lineage in paddy soils. In this study, high abundance of Bathyarchaeota is observed in both flooded and unflooded paddy soils, with average relative abundance of 27.78% and 37.07% (Fig. 2), respectively. These results suggest that Bathyarchaeota was also dominant in aerobic paddy soil, and broadened previous prescription that Bathyarchaeota are generalists in anaerobic environments such as sediments (5), peatland (59), and anaerobic digestion system (9), which will expand our understanding of Bathyarchaeota.
The high diversity of Bathyarchaeota makes it interesting to understand the distribution patterns of different subgroups, which can contribute to revealing ecological functions of Bathyarchaeota. We identified 2,030 bathyarchaeotal ASVs in this study, and which were classified into 11 subgroups. Among these subgroups, Bathy-6 was the most predominant subgroup in all samples, this result is consistent with previously observed paddy soil archaeal community of the Yunhe terrace (58). The high abundance of Bathy-6 in paddy soils might result from the presence of genes encoding superoxide dismutase in genomes of Bathy-6 recovered from terrestrial environments (60). These findings suggest that the lifestyle of Bathyarchaeota might have changed in terrestrial and marine ecosystems and imply the metabolic pathway of Bathy-6 in paddy soils might be different from other members in marine sediments. In addition, Bathy-15 and Bathy-5bb were two other abundant subgroups in paddy soils with average relative abundance more than 1% (Fig. 5B), while their abundance was much lower compared to those detected in freshwater sediments (5, 59). However, Bathy-1 and Bathy-8, which are Bathyarchaeota indicator lineages of marine sediments (20), were almost undetected in paddy soils analyzed in this study. Based on these results, our study demonstrated that bathyarchaeotal community structure in paddy soils are distinctive to those in sediments.
Climatic factors significantly influence bathyarchaeotal community
The relative abundance of Bathyarchaeota ranged from 0.38% to 92.13% within archaea, and this excessive variation among different samples made it is interesting to investigate the variation mechanisms and to identify the key factors regulating bathyarchaeotal abundance. Compared with sediments, the microbial community in paddy soil is more susceptible to agricultural activities, plant roots and climatic factors. Therefore, we collected the climatic data of sampling sites, soil properties and soil condition at the time of sampling from corresponding studies (Table S1) to perform random forest analysis and multivariate regression tree analysis. Results demonstrated that MAT and MAP strongly affected the archaeal community structure. These climatic factors have a long-term regulation of microbial activity and elements cycle, and have been generally considered to be important factors affecting microbial assembly (26, 61). Bathyarchaeota, as the core archaeal lineage in paddy soils, was also strongly influenced by MAT and MAP. We conjectured that the effect of MAT on the abundance of Bathyarchaeota was partly caused by variation in the abundance of Bathy-6, which is the most predominated subgroup in all the samples. Among all these factors, the proportion of Bathy-6 was only influenced by MAT (Fig. 3B). This result is in agreement with previous study (62), which found that high abundance was observed in samples with moderate temperature. Bathy-11, Bathy-17, Bathy-18, and Bathy-5bb were mainly influenced by MAP (Fig. S4). Surprisingly, the abundance of these subgroups, which are abundant in anoxic environments, was not affected by water management in paddy soils. In flooded paddy soil, rice aerenchyma can transport atmospheric oxygen to the rhizosphere, resulting in a microoxic environment. Thus either flooded or unflooded paddy soils are not anoxic environments; however, no genes involved in oxygen-dependent pathways have been detected in these subgroups so far. Thus, the abundance of these subgroups might depend on the gradient in reduction caused by abundant rainfall (34).
Bathyarchaeota highly connects with microorganisms involved in carbon and nitrogen metabolism
Bathyarchaeota have been identified as complex organic matter degraders and participants in methane metabolism in sediments (10). However, the ecological roles and niches of Bathyarchaeota in paddy soil are still unclear. We performed co-occurrence network analysis to reveal the syntrophic interactions between Bathyarchaeota and other microorganisms, through which we can infer the ecological roles of Bathyarchaeota in paddy soils (Fig. 6).
Interestingly, highly frequent associations between Bathyarchaeota and Woesearchaeales were found in the co-occurrence network of archaeal lineages. Recent studies illustrated that Woesearchaeales are widespread in both terrestrial and marine ecosystems which are classified in the GTDB database as Woesearchaeota (63, 64). According to the analysis of metagenome-assembled genomes, Woesearchaeales exhibit conspicuous metabolic deficiencies, suggesting an anaerobic syntrophic lifestyle of Woesearchaeales (64, 65). This syntrophic lifestyle is consistent with our result, that Woesearchaeales have the frequent connections with other archaea (Fig. 5B). The co-occurrence of Bathyarchaeota and Woesearchaeales implied that Woesearchaeales might obtain intermediate products from the degradation of organic matter by Bathyarchaeota. For example, detrital proteins may be degraded to amino acids by extracellular peptidases encoded by Bathyarchaeota (13), which could be then used by Woesearchaeales for their own growth (64). In addition, Bathyarchaeota have close relationship with methanogens, such as Methanosaetaceae (known as Methanotrichaceae), Methanocellaceae, Methanoregulaceae, and Methanobacteriaceae. This result is consistent with the findings in the marine sediment (66) and terrestrial environments (62). Among these methanogens, Methanosaetaceae can directly use acetate as substrate, while hydrogen-consuming methanogenic archaea (Methanocellaceae, Methanoregulaceae and Methanobacteriaceae) prefer to utilize oxidation products of acetate (53, 67) to produce methane. Recent studies demonstrated that Bathyarchaeotal genomes encode a series of phosphate acetyltransferase (Pta) and acetate kinase (Ack) (3) or acetyl-CoA synthetase (Acd) for acetate production (18). Potential acetogeneic pathway of Bathyarchaeota might explain its close association with methanogens. Furthermore, the co-occurrence patterns can also result from overlapping ecological niches between microorganisms instead of syntrophy (68). Previous study revealed the presence of genes encoding the methyl-coenzyme M reductase (MCR) complex essential for methanogenesis (10), suggesting that Bathyarchaeota might produce methane as well as methanogens. Since the relationships between Bathyarchaeota and methanogens recurred in different modules of the network (Fig. S5), we preferred to consider a syntrophy between these two microorganisms. Besides archaeal network, we also constructed the network between Bathyarchaeota and bacteria (Fig. 6B). We found that Anaerolineaceae was the bacterial lineage that had the most frequent nonrandom associations with Bathyarchaeota (Fig. 6B). It has been reported that members of Anaerolineaceae are typical complex organic matter degraders under anoxic condition (69), implying that Bathyarchaeota might be involved in the carbon cycle in paddy soils.
In addition, the co-occurrence associations between Bathyarchaeota and ammonia-oxidizing archaea (Nitrosopumilaceae and Nitrososphaeraceae) occurred in several modules, implying a syntrophy relationship between Bathyarchaeota and nitrogen-cycling microorganisms. Ammonia-oxidizing archaea are keystone members that convert ammonia to nitrite and significantly influence nitrogen cycle in paddy soil (70). Bathyarchaeota harbor a series of genes related to the conversion of different nitrogen compounds to ammonium (60), and then ammonium can be further used as substrates for ammonia-oxidizing archaea. In addition, Genes involved in urea production also were observed in Bathyarchaeota genomes, and genes involved in urea degradation were found in the genomes of Nitrososphaeraceae (71). These findings indicate the interactions between Bathyarchaeota and nitrogen cycling microorganisms through urea and ammonium transformation. These co-occurrence patterns imply that Bathyarchaeota might be involved in nitrogen cycle in paddy soils. However, due to lack of pure culture, so far, the research on the metabolism of Bathyarchaeota are mainly based on genomic information. The specific metabolic pathways of Bathyarchaeota in paddy soil should be further confirmed by clear experimental evidences.
Conclusion
Through a meta-analysis on the global in situ paddy soils sequencing data, we reveal a wide distribution and high abundance of Bathyarchaeota in paddy soils, and Bathy-6 is the predominant group. In addition, MAT and MAP are the main factors affecting the relative abundance of Bathyarchaeota and shaping bathyarchaeotal community structure. Recurrence of the associations between Bathyarchaeota and microorganisms involved in carbon and nitrogen metabolism suggested that Bathyarchaeota might be involved in diverse biogeochemical cycling in paddy soils. This study provides novel understanding of Bathyarchaeota in paddy soils at a global scale, and extends the research on Bathyarchaeota in arable soils other than sediments.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (grant no. 31970105, 41977038, and 41991332).
Shu-Dan Xue: Investigation, Data Curation, Writing - Original Draft; Xing-Yun Yi: Data Curation; Hui-Ling Cui: Data Curation; Meng Li: Writing - Review & Editing; Jing-Jing Peng: Data Curation, Writing - Original Draft; Yong-Guan Zhu: Conceptualization, Writing - Review & Editing; Gui-Lan Duan: Conceptualization, Resources, Funding acquisition; Writing - Review & Editing, Supervision.
The authors declare that they have no competing financial interests.
Contributor Information
Gui-Lan Duan, Email: duangl@rcees.ac.cn.
Nick Bouskill, E O Lawrence Berkeley National Laboratory, Berkeley, California, USA .
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00143-23.
Tables S1 to S3.
Fig. S1 to S5.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Barns SM, Delwiche CF, Palmer JD, Pace NR. 1996. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci U S A 93:9188–9193. doi: 10.1073/pnas.93.17.9188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou ZC, Pan J, Wang FP, Gu JD, Li M. 2018. Bathyarchaeota: globally distributed metabolic generalists in anoxic environments. FEMS Microbiol Rev 42:639–655. doi: 10.1093/femsre/fuy023 [DOI] [PubMed] [Google Scholar]
- 3. He Y, Li M, Perumal V, Feng X, Fang J, Xie J, Sievert SM, Wang F. 2016. Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat Microbiol 1:16035. doi: 10.1038/nmicrobiol.2016.35 [DOI] [PubMed] [Google Scholar]
- 4. Meng J, Xu J, Qin D, He Y, Xiao X, Wang F. 2014. Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. ISME J 8:650–659. doi: 10.1038/ismej.2013.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fan X, Xing P. 2016. Differences in the composition of archaeal communities in sediments from contrasting zones of lake Taihu. Front Microbiol 7: 1510. doi: 10.3389/fmicb.2016.01510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kubo K, Lloyd KG, F Biddle J, Amann R, Teske A, Knittel K. 2012. Archaea of the miscellaneous crenarchaeotal group are abundant, diverse and widespread in marine sediments. ISME J 6:1949–1965. doi: 10.1038/ismej.2012.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McKay LJ, Hatzenpichler R, Inskeep WP, Fields MW. 2017. Occurrence and expression of novel methyl-coenzyme M reductase gene (mcrA) variants in hot spring sediments. Sci Rep 7:7252. doi: 10.1038/s41598-017-07354-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loh HQ, Hervé V, Brune A. 2020. Metabolic potential for reductive acetogenesis and a novel energy-converting [NiFe] hydrogenase in bathyarchaeia from termite guts-a genome-centric analysis. Front Microbiol 11:635786. doi: 10.3389/fmicb.2020.635786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu X, Lee C, Kim JY. 2023. Comparison of mesophilic and thermophilic anaerobic digestions of thermal hydrolysis pretreated swine manure: process performance, microbial communities and energy balance. J Environ Sci (China) 126:222–233. doi: 10.1016/j.jes.2022.03.032 [DOI] [PubMed] [Google Scholar]
- 10. Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, Tyson GW. 2015. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350:434–438. doi: 10.1126/science.aac7745 [DOI] [PubMed] [Google Scholar]
- 11. Feng X, Wang Y, Zubin R, Wang F. 2019. Core metabolic features and hot origin of Bathyarchaeota. Engineering 5:498–504. doi: 10.1016/j.eng.2019.01.011 [DOI] [Google Scholar]
- 12. Qi Y-L, Evans PN, Li Y-X, Rao Y-Z, Qu Y-N, Tan S, Jiao J-Y, Chen Y-T, Hedlund BP, Shu W-S, Hua Z-S, Li W-J. 2021. Comparative genomics reveals thermal adaptation and a high metabolic diversity in "Candidatus Bathyarchaeia." mSystems 6: e0025221. doi: 10.1128/mSystems.00252-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD, Stepanauskas R, Richter M, Kleindienst S, Lenk S, Schramm A, Jørgensen BB. 2013. Predominant archaea in marine sediments degrade detrital proteins. Nature 496:215–218. doi: 10.1038/nature12033 [DOI] [PubMed] [Google Scholar]
- 14. Seyler LM, McGuinness LM, Kerkhof LJ. 2014. Crenarchaeal heterotrophy in salt marsh sediments. ISME J 8:1534–1543. doi: 10.1038/ismej.2014.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yin XR, Zhou GW, Cai MW, Zhu QZ, Richter-Heitmann T, Aromokeye DA, Liu Y, Nimzyk R, Zheng QF, Tang XY, Elvert M, Li M, Friedrich MW. 2022. Catabolic protein degradation in marine sediments confined to distinct archaea. ISME J 16:1617–1626. doi: 10.1038/s41396-022-01210-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu T, Wu W, Liang W, Lever MA, Hinrichs K-U, Wang F. 2018. Growth of sedimentary Bathyarchaeota on lignin as an energy source. Proc Natl Acad Sci U S A 115:6022–6027. doi: 10.1073/pnas.1718854115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yi X-Y, Yang Y-P, Yuan H-Y, Chen Z, Duan G-L, Zhu Y-G. 2019. Coupling metabolisms of arsenic and iron with humic substances through microorganisms in paddy soil. J Hazard Mater 373:591–599. doi: 10.1016/j.jhazmat.2019.03.113 [DOI] [PubMed] [Google Scholar]
- 18. Lazar CS, Baker BJ, Seitz K, Hyde AS, Dick GJ, Hinrichs K-U, Teske AP. 2016. Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ Microbiol 18:1200–1211. doi: 10.1111/1462-2920.13142 [DOI] [PubMed] [Google Scholar]
- 19. Zhang W, Ding W, Yang B, Tian R, Gu S, Luo H, Qian P-Y. 2016. Genomic and transcriptomic evidence for carbohydrate consumption among microorganisms in a cold seep brine pool. Front Microbiol 7:1825. doi: 10.3389/fmicb.2016.01825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fillol M, Auguet J-C, Casamayor EO, Borrego CM. 2016. Insights in the ecology and evolutionary history of the miscellaneous crenarchaeotic group lineage. ISME J 10:665–677. doi: 10.1038/ismej.2015.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pan J, Chen Y, Wang Y, Zhou Z, Li M. 2019. Vertical distribution of bathyarchaeotal communities in mangrove wetlands suggests distinct niche preference of Bathyarchaeota subgroup 6. Microb Ecol 77:417–428. doi: 10.1007/s00248-018-1309-7 [DOI] [PubMed] [Google Scholar]
- 22. Fillol M, Sànchez-Melsió A, Gich F, Borrego CM. 2015. Diversity of miscellaneous crenarchaeotic group archaea in freshwater karstic lakes and their segregation between planktonic and sediment habitats. FEMS Microbiol Ecol 91: fiv020. doi: 10.1093/femsec/fiv020 [DOI] [PubMed] [Google Scholar]
- 23. Yu T, Liang Q, Niu M, Wang F. 2017. High occurrence of Bathyarchaeota (MCG) in the deep-sea sediments of South China Sea quantified using newly designed PCR primers. Environ Microbiol Rep 9:374–382. doi: 10.1111/1758-2229.12539 [DOI] [PubMed] [Google Scholar]
- 24. Hester ER, Vaksmaa A, Valè G, Monaco S, Jetten MSM, Lüke C. 2022. Effect of water management on microbial diversity and composition in an Italian rice field system. FEMS Microbiol Ecol 98: fiac018. doi: 10.1093/femsec/fiac018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vaksmaa A, van Alen TA, Ettwig KF, Lupotto E, Valè G, Jetten MSM, Lüke C. 2017. Stratification of diversity and activity of methanogenic and methanotrophic microorganisms in a nitrogen-fertilized Italian paddy soil. Front Microbiol 8:2127. doi: 10.3389/fmicb.2017.02127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiao S, Xu Y, Zhang J, Lu Y. 2019. Environmental filtering drives distinct continental atlases of soil archaea between dryland and wetland agricultural ecosystems. Microbiome 7:15. doi: 10.1186/s40168-019-0630-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ahn J-H, Song J, Kim B-Y, Kim M-S, Joa J-H, Weon H-Y. 2012. Characterization of the bacterial and archaeal communities in rice field soils subjected to long-term fertilization practices. J Microbiol 50:754–765. doi: 10.1007/s12275-012-2409-6 [DOI] [PubMed] [Google Scholar]
- 28. Kim H, Jeon J, Lee KK, Lee Y-H. 2021. Compositional shift of bacterial, archaeal, and fungal communities is dependent on trophic lifestyles in rice paddy soil. Front Microbiol 12:719486. doi: 10.3389/fmicb.2021.719486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ahn J-H, Choi M-Y, Kim B-Y, Lee J-S, Song J, Kim G-Y, Weon H-Y. 2014. Effects of water-saving irrigation on emissions of greenhouse gases and prokaryotic communities in rice paddy soil. Microb Ecol 68:271–283. doi: 10.1007/s00248-014-0371-z [DOI] [PubMed] [Google Scholar]
- 30. Ezeokoli OT, Nuaila VNA, Obieze CC, Muetanene BA, Fraga I, Tesinde MN, Ndayiragije A, Coutinho J, Melo AMP, Adeleke RA, Ribeiro-Barros AI, Fangueiro D. 2021. Assessing the impact of rice cultivation and off-season period on dynamics of soil enzyme activities and bacterial communities in two agro-ecological regions of Mozambique. Agronomy 11:694. doi: 10.3390/agronomy11040694 [DOI] [Google Scholar]
- 31. Serbent MP, Dos Anjos Borges LG, Quadros A, Marconatto L, Tavares LBB, Giongo A. 2021. Prokaryotic and microeukaryotic communities in an experimental rice plantation under long-term use of pesticides. Environ Sci Pollut Res Int 28:2328–2341. doi: 10.1007/s11356-020-10614-5 [DOI] [PubMed] [Google Scholar]
- 32. Imchen M, Kumavath R, Vaz ABM, Góes-Neto A, Barh D, Ghosh P, Kozyrovska N, Podolich O, Azevedo V. 2019. 16S rRNA gene amplicon based metagenomic signatures of rhizobiome community in rice field during various growth stages. Front Microbiol 10: 2103. doi: 10.3389/fmicb.2019.02103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nguyen SG, Guevarra RB, Kim J, Ho CT, Trinh MV, Unno T. 2015. Impacts of initial fertilizers and irrigation systems on paddy methanogens and methane emission. Water Air Soil Pollut 226:309. doi: 10.1007/s11270-015-2501-8 [DOI] [Google Scholar]
- 34. Li H-Y, Wang H, Wang H-T, Xin P-Y, Xu X-H, Ma Y, Liu W-P, Teng C-Y, Jiang C-L, Lou L-P, Arnold W, Cralle L, Zhu Y-G, Chu J-F, Gilbert JA, Zhang Z-J. 2018. The chemodiversity of paddy soil dissolved organic matter correlates with microbial community at continental scales. Microbiome 6:187. doi: 10.1186/s40168-018-0561-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shao J, He Y, Zhang H, Chen A, Lei M, Chen J, Peng L, Gu J-D. 2016. Silica fertilization and nano-mno₂ Amendment on bacterial community composition in high arsenic paddy soils. Appl Microbiol Biotechnol 100:2429–2437. doi: 10.1007/s00253-015-7131-y [DOI] [PubMed] [Google Scholar]
- 36. Ali I, Yuan P, Ullah S, Iqbal A, Zhao Q, Liang H, Khan A, Zhang H, Wu X, Wei S, Gu M, Jiang L. 2022. Biochar Amendment and nitrogen fertilizer contribute to the changes in soil properties and microbial communities in a paddy field. Front Microbiol 13:834751. doi: 10.3389/fmicb.2022.834751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li P, Liu M, Ma X, Wu M, Jiang C, Liu K, Liu J, Li Z. 2020. Responses of microbial communities to a gradient of pig manure Amendment in red paddy soils. Sci Total Environ 705:135884. doi: 10.1016/j.scitotenv.2019.135884 [DOI] [PubMed] [Google Scholar]
- 38. Wang Y, Shi X, Huang X, Huang C, Wang H, Yin H, Shao Y, Li P. 2022. Linking microbial community composition to farming pattern in selenium-enriched region: potential role of microorganisms on se geochemistry. J Environ Sci (China) 112:269–279. doi: 10.1016/j.jes.2021.05.015 [DOI] [PubMed] [Google Scholar]
- 39. Song J, Shen Q, Shi J, Xu J, Brookes PC, Liu X. 2021. Changes in microbial community structure due to chronic trace element concentrations in different sizes of soil aggregates. Environ Pollut 268:115933. doi: 10.1016/j.envpol.2020.115933 [DOI] [PubMed] [Google Scholar]
- 40. Zhang J, Jiao S, Lu Y. 2018. Biogeographic distribution of bacterial, archaeal and methanogenic communities and their associations with methanogenic capacity in Chinese wetlands. Sci Total Environ 622–623:664–675. doi: 10.1016/j.scitotenv.2017.11.279 [DOI] [PubMed] [Google Scholar]
- 41. Zhao X, Huang J, Zhu X, Chai J, Ji X. 2020. Ecological effects of heavy metal pollution on soil microbial community structure and diversity on both sides of a river around a mining area. Int J Environ Res Public Health 17: 5680. doi: 10.3390/ijerph17165680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pu Q, Zhang K, Poulain AJ, Liu J, Zhang R, Abdelhafiz MA, Meng B, Feng X. 2022. Mercury drives microbial community assembly and ecosystem multifunctionality across a Hg contamination gradient in rice paddies. J Hazard Mater 435:129055. doi: 10.1016/j.jhazmat.2022.129055 [DOI] [PubMed] [Google Scholar]
- 43. Li W, Kuzyakov Y, Zheng Y, Li P, Li G, Liu M, Alharbi HA, Li Z. 2022. Depth effects on bacterial community assembly processes in paddy soils. Soil Biol Biochem 165:108517. doi: 10.1016/j.soilbio.2021.108517 [DOI] [Google Scholar]
- 44. Yuan C-L, Zhang L-M, Wang J-T, Teng W-K, Hu H-W, Shen J-P, He J-Z. n.d. Limited effects of depth (0-80 cm) on communities of archaea, bacteria and fungi in paddy soil profiles. Eur J Soil Sci. doi: 10.1111/ejss.12921 [DOI] [Google Scholar]
- 45. Yuan J, Yuan Y, Zhu Y, Cao L. 2018. Effects of different fertilizers on methane emissions and methanogenic community structures in paddy rhizosphere soil. Sci Total Environ 627:770–781. doi: 10.1016/j.scitotenv.2018.01.233 [DOI] [PubMed] [Google Scholar]
- 46. Breidenbach B, Blaser MB, Klose M, Conrad R. 2016. Crop rotation of flooded rice with upland maize impacts the resident and active methanogenic microbial community. Environ Microbiol 18:2868–2885. doi: 10.1111/1462-2920.13041 [DOI] [PubMed] [Google Scholar]
- 47. Pereira-Mora L, Terra JA, Fernández-Scavino A. 2022. Methanogenic community linked to organic acids fermentation from root exudates are affected by rice intensification in rotational soil systems. Applied Soil Ecology 176:104498. doi: 10.1016/j.apsoil.2022.104498 [DOI] [Google Scholar]
- 48. Simonin M, Briand M, Chesneau G, Rochefort A, Marais C, Sarniguet A, Barret M. 2022. Seed microbiota revealed by a large-scale meta-analysis including 50 plant species. New Phytol 234:1448–1463. doi: 10.1111/nph.18037 [DOI] [PubMed] [Google Scholar]
- 49. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4: e2584. doi: 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cai M, Richter-Heitmann T, Yin X, Huang W-C, Yang Y, Zhang C, Duan C, Pan J, Liu Y, Liu Y, Friedrich MW, Li M. 2021. Ecological features and global distribution of asgard archaea. Sci Total Environ 758:143581. doi: 10.1016/j.scitotenv.2020.143581 [DOI] [PubMed] [Google Scholar]
- 53. Peng J, Wegner C-E, Bei Q, Liu P, Liesack W. 2018. Metatranscriptomics reveals a differential temperature effect on the structural and functional organization of the anaerobic food web in rice field soil. Microbiome 6:169. doi: 10.1186/s40168-018-0546-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ju F, Xia Y, Guo F, Wang Z, Zhang T. 2014. Taxonomic relatedness shapes bacterial assembly in activated sludge of globally distributed wastewater treatment plants. Environ Microbiol 16:2421–2432. doi: 10.1111/1462-2920.12355 [DOI] [PubMed] [Google Scholar]
- 55. Carlson KM, Gerber JS, Mueller ND, Herrero M, MacDonald GK, Brauman KA, Havlik P, O’Connell CS, Johnson JA, Saatchi S, West PC. 2017. Greenhouse gas emissions intensity of global croplands. Nature Clim Change 7:63–68. doi: 10.1038/nclimate3158 [DOI] [Google Scholar]
- 56. Pan G, Li L, Wu L, Zhang X. 2004. Storage and sequestration potential of topsoil organic carbon in China’s paddy soils. Glob Chang Biol 10:79–92. doi: 10.1111/j.1365-2486.2003.00717.x [DOI] [Google Scholar]
- 57. Winkel M, Mitzscherling J, Overduin PP, Horn F, Winterfeld M, Rijkers R, Grigoriev MN, Knoblauch C, Mangelsdorf K, Wagner D, Liebner S. 2018. Anaerobic methanotrophic communities thrive in deep submarine permafrost. Sci Rep 8:1291. doi: 10.1038/s41598-018-19505-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang Q, Li Y, Xing J, Brookes PC, Xu J. 2019. Soil available phosphorus content drives the spatial distribution of archaeal communities along elevation in acidic terrace paddy soils. Science of The Total Environment 658:723–731. doi: 10.1016/j.scitotenv.2018.12.144 [DOI] [PubMed] [Google Scholar]
- 59. Xiang X, Wang H, Man B, Xu Y, Gong L, Tian W, Yang H. 2023. Diverse bathyarchaeotal lineages dominate archaeal communities in the acidic dajiuhu peatland, central China. Microb Ecol 85:557–571. doi: 10.1007/s00248-022-01990-1 [DOI] [PubMed] [Google Scholar]
- 60. Pan J, Zhou Z, Béjà O, Cai M, Yang Y, Liu Y, Gu J-D, Li M. 2020. Genomic and transcriptomic evidence of light-sensing, porphyrin biosynthesis, Calvin-Benson-Bassham cycle, and urea production in bathyarchaeota. Microbiome 8:43. doi: 10.1186/s40168-020-00820-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li H-Y, Wang H, Tao X-H, Wang X-Z, Jin W-Z, Gilbert JA, Zhu Y-G, Zhang Z-J. 2021. Continental-scale paddy soil bacterial community structure, function, and biotic interaction. mSystems 6: e0136820. doi: 10.1128/mSystems.01368-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xiang X, Wang R, Wang H, Gong L, Man B, Xu Y. 2017. Distribution of bathyarchaeota communities across different terrestrial settings and their potential ecological functions. Sci Rep 7: 45028. doi: 10.1038/srep45028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kuroda K, Narihiro T, Shinshima F, Yoshida M, Yamaguchi H, Kurashita H, Nakahara N, Nobu MK, Noguchi TQP, Yamauchi M, Yamada M. 2022. High-rate cotreatment of purified terephthalate and dimethyl terephthalate manufacturing wastewater by a mesophilic upflow anaerobic sludge blanket reactor and the microbial ecology relevant to aromatic compound degradation. Water Res 219:118581. doi: 10.1016/j.watres.2022.118581 [DOI] [PubMed] [Google Scholar]
- 64. Liu X, Li M, Castelle CJ, Probst AJ, Zhou Z, Pan J, Liu Y, Banfield JF, Gu J-D. 2018. Insights into the ecology, evolution, and metabolism of the widespread woesearchaeotal lineages. Microbiome 6:102. doi: 10.1186/s40168-018-0488-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang W-C, Liu Y, Zhang X, Zhang C-J, Zou D, Zheng S, Xu W, Luo Z, Liu F, Li M. 2021. Comparative genomic analysis reveals metabolic flexibility of woesearchaeota. Nat Commun 12: 5281. doi: 10.1038/s41467-021-25565-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zou D, Pan J, Liu Z, Zhang C, Liu H, Li M. 2020. The distribution of bathyarchaeota in surface sediments of the pearl river estuary along salinity gradient. Front Microbiol 11:285. doi: 10.3389/fmicb.2020.00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Karakashev D, Batstone DJ, Trably E, Angelidaki I. 2006. Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of methanosaetaceae. Appl Environ Microbiol 72:5138–5141. doi: 10.1128/AEM.00489-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Barberán A, Bates ST, Casamayor EO, Fierer N. 2012. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6:343–351. doi: 10.1038/ismej.2011.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang Y, Chen M, Zhao Y-Y, Zhang A-Y, Peng D-H, Lu F, Dai C-C. 2021. Destruction of the soil microbial ecological environment caused by the over-utilization of the rice-crayfish co-cropping pattern. Sci Total Environ 788:147794. doi: 10.1016/j.scitotenv.2021.147794 [DOI] [PubMed] [Google Scholar]
- 70. Bose H, Sahu RP, Sar P. 2022. Impact of arsenic on microbial community structure and their metabolic potential from rice soils of West Bengal, India. Sci Total Environ 841:156486. doi: 10.1016/j.scitotenv.2022.156486 [DOI] [PubMed] [Google Scholar]
- 71. Wu X, Peng J, Liu P, Bei Q, Rensing C, Li Y, Yuan H, Liesack W, Zhang F, Cui Z. 2021. Metagenomic insights into nitrogen and phosphorus cycling at the soil aggregate scale driven by organic material amendments. Sci Total Environ 785:147329. doi: 10.1016/j.scitotenv.2021.147329 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3.
Fig. S1 to S5.