ABSTRACT
Pseudorabies virus (PRV) is a double-stranded DNA virus that causes Aujeszky’s disease and is responsible for economic loss worldwide. Transmembrane protein 41B (TMEM41B) is a novel endoplasmic reticulum (ER)-localized regulator of autophagosome biogenesis and lipid mobilization; however, the role of TMEM41B in regulating PRV replication remains undocumented. In this study, PRV infection was found to upregulate TMEM41B mRNA and protein levels both in vitro and in vivo. For the first time, we found that TMEM41B could be induced by interferon (IFN), suggesting that TMEM41B is an IFN-stimulated gene (ISG). While TMEM41B knockdown suppressed PRV proliferation, TMEM41B overexpression promoted PRV proliferation. We next studied the specific stages of the virus life cycle and found that TMEM41B knockdown affected PRV entry. Mechanistically, we demonstrated that the knockdown of TMEM41B blocked PRV-stimulated expression of the key enzymes involved in lipid synthesis. Additionally, TMEM41B knockdown played a role in the dynamics of lipid-regulated PRV entry-dependent clathrin-coated pits (CCPs). Lipid replenishment restored the CCP dynamic and PRV entry in TMEM41B knockdown cells. Together, our results indicate that TMEM41B plays a role in PRV infection via regulating lipid homeostasis.
IMPORTANCE PRV belongs to the alphaherpesvirus subfamily and can establish and maintain a lifelong latent infection in pigs. As such, an intermittent active cycle presents great challenges to the prevention and control of PRV disease and is responsible for serious economic losses to the pig breeding industry. Studies have shown that lipids play a crucial role in PRV proliferation. Thus, the manipulation of lipid metabolism may represent a new perspective for the prevention and treatment of PRV. In this study, we report that the ER transmembrane protein TMEM41B is a novel ISG involved in PRV infection by regulating lipid synthesis. Therefore, our findings indicate that targeting TMEM41B may be a promising approach for the development of PRV vaccines and therapeutics.
KEYWORDS: pseudorabies virus, TMEM41B, interferon-stimulated gene, clathrin-coated pits, lipid homeostasis
INTRODUCTION
Aujeszky’s disease is a notable infectious disease of pigs that causes economic losses to the pig industry worldwide. Pseudorabies virus (PRV) is the causative pathogen responsible for Aujeszky’s disease and is a member of the subfamily alphaherpesvirinae of the family herpesviridae (1). The PRV genome is approximately 143 kb and encodes at least 70 open reading frames (2). PRV can infect pigs, sheep, and cattle, causing severe clinical symptoms and death in a variety of mammals (3). Since PRV is an important infectious disease in many countries, scientists have strived to develop diagnostics and vaccines. Several recent reports suggest that PRV can cause human endophthalmitis and encephalitis (4–6). These findings indicate that PRV infection is not limited to the swine industry and represents a potential public health risk. During entry, the glycoproteins gC, gB, gD, gH, and gL are responsible for PRV attachment to the host cell surface and the subsequent fusion of the viral envelope with the plasma membrane (PM) (2). In some cell types, PRV enters via clathrin-mediated endocytosis, followed by fusion of the viral envelope with the endosomal membrane (7, 8).
TMEM41B has been previously identified as a novel autophagy factor that modulates autophagosome formation (9–11). In addition, TMEM41B-deficient cells exhibit larger lipid droplets (LDs), a phenomenon that may be caused by a lipid buildup due to a disruption in the release of free fatty acids (FFAs) from LDs to other organelles (e.g., mitochondria) (9, 10). TMEM41B has been characterized as an ER scramblase for lipoprotein biogenesis and lipid homeostasis that can shuttle phospholipids between bilayer membrane leaflets (12–14). More recently, TMEM41B has been characterized as a host factor for viral replication in a variety of viruses, including pan-flavivirus (15), coronaviruses (16), and severe acute respiratory syndrome coronavirus (SARS-CoV-2) (17).
Lipids are a source of energy molecules and are one of the key components that determine membrane fluidity and structure (18). Moreover, some viruses manipulate lipid metabolism to facilitate viral reproduction (18–20). In particular, the entry of a variety of viruses, including SARS-CoV-2 (21), herpes simplex virus (22), murine leukemia virus (23), and PRV (7, 20), has been associated with sphingolipid and cholesterol-enriched lipid raft microdomains in the PM. Notably, TMEM41B plays a role in the formation of viral double-membrane vesicles during β-coronavirus infection (24). Moreover, a TMEM41B knockdown can inhibit the internalization and early stage replication of coronaviruses (16). Here, we examined the effects of TMEM41B on PRV replication. Knockdown of TMEM41B resulted in decreased lipid levels and inhibited PRV proliferation. Additionally, knockdown of TMEM41B dramatically decreased PRV entry by interfering with lipid-dependent clathrin-coated pit (CCP) dynamics. We further studied the effect of TMEM41B on PRV replication. We demonstrated that knockdown of TMEM41B could inhibit a PRV-induced increase in lipid synthesis, thereby inhibiting PRV proliferation. In addition, our study emphasized the importance of TMEM41B for PRV infection both in vivo and in vitro, suggesting that TMEM41B may be a suitable target for the development of anti-PRV therapeutics.
RESULTS
PRV infection upregulates TMEM41B expression.
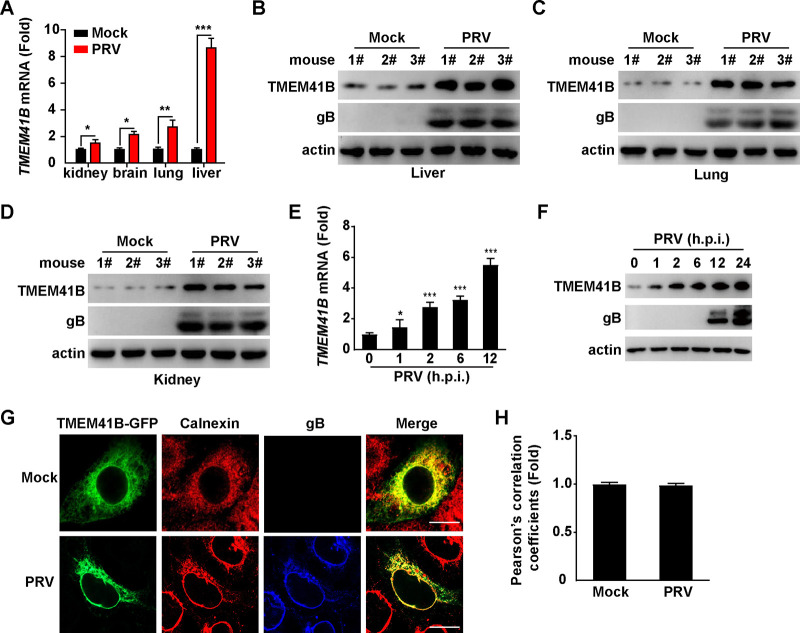
To determine the role of TMEM41B in PRV infection, we evaluated the expression of TMEM41B following PRV challenge in vivo. Mice were mock infected or intranasally infected with the PRV isolate QXX (PRV-QXX) for 3 days. The kidney, brain, lung, and liver were assessed for the level of TMEM41B mRNA and protein expression by quantitative real-time PCR (qRT-PCR) and immunoblotting analysis. PRV infection resulted in increased TMEM41B mRNA expression compared to that of the mock-infected kidney, brain, lung, and liver (Fig. 1A). The levels of TMEM41B protein expression were similarly increased (Fig. 1B to D). We next verified whether PRV enhanced TMEM41B expression in vitro. PK-15 cells were infected with PRV-QXX and processed to measure the mRNA and protein levels of TMEM41B. PRV infection caused an increase in TMEM41B mRNA expression in PK-15 cells (Fig. 1E). Consistent with the mRNA levels, TMEM41B protein levels in PK-15 cells were also upregulated in response to PRV infection (Fig. 1F). TMEM41B is an ER transmembrane protein. Immunofluorescence analysis indicated that PRV infection did not alter the subcellular localization of TMEM41B in the ER (Fig. 1G and H). These results suggest that PRV promotes TMEM41B expression both in vivo and in vitro.
FIG 1.
TMEM41B expression is unregulated by PRV infection. (A) Mice were mock infected or intranasally infected with PRV-QXX (5 × 103 TCID50/50 μL per mouse) for 3 days. The levels of TMEM41B mRNA expression in the kidney, brain, lung, and liver were assessed by qRT-PCR analysis (n = 3 per group). *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B to D) Mice were treated as described in A. TMEM41B and PRV gB in the liver (B), lung (C), and kidney (D) were assessed by immunoblotting analysis (n = 3 per group). (E) PK-15 cells were infected with PRV-QXX (MOI = 1) for 0 to 12 h; h.p.i., hours postinfection. The level of TMEM41B mRNA expression was assessed by qRT-PCR analysis. *, P < 0.05; ***, P < 0.001. (F) PK-15 cells were infected with PRV-QXX (MOI = 1) for 0 to 24 h. TMEM41B and PRV gB were assessed by immunoblotting analysis. (G) PK-15 cells were transfected with the TMEM41B-GFP plasmid for 12 h and then mock infected or infected with PRV-QXX (MOI = 1) for 24 h. The colocalization of TMEM41B-GFP with the ER (calnexin, red) was analyzed by immunofluorescence analysis. Scale bar = 10 μm. (H) Quantification of the colocalization of TMEM41B with calnexin from panel G by ImageJ (n = 15).
TMEM41B is an interferon-stimulated gene.
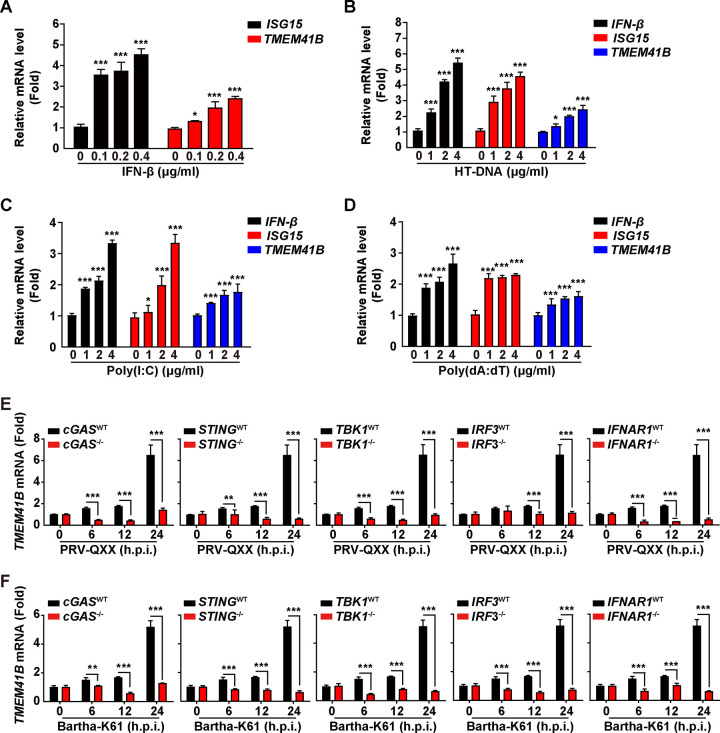
Since we found that PRV infection upregulated TMEM41B expression, we next aimed to determine whether TMEM41B expression was dependent on the type I interferon (IFN) signaling pathway. We treated PK-15 cells with IFN-β, HT-DNA, poly(I·C), and poly(dA:dT) and found that all of the treatments increased the mRNA levels of TMEM41B and IFN-stimulated gene 15 (ISG15) (Fig. 2A to D). The results of our previous study suggested that porcine cyclic GMP-AMP (cGAMP) synthase (cGAS) and its downstream effectors, such as stimulator of interferon genes protein (STING), TANK-binding kinase 1 (TBK1), and IFN regulatory factor 3 (IRF3), were responsible for PRV-induced type I IFN activation (25). Infection of PK-15 cells with PRV-QXX or Bartha-K61 induced TMEM41B expression in a time-dependent manner (Fig. 2E and F). However, the induction of TMEM41B expression was abolished in cGAS−/−, STING−/−, TBK1−/−, IRF3−/−, and IFNAR1−/− PK-15 cells (Fig. 2E and F). These results indicate that PRV-induced TMEM41B expression relies on the innate cGAS/STING/TBK1/IRF3 innate immune and IFN signaling pathways.
FIG 2.
TMEM41B expression is dependent on IFN and cGAS innate immune pathways. (A) PK-15 cells were treated with IFN-β (0 to 0.4 μg/mL) for 24 h. The levels of ISG15 and TMEM41B mRNA expression were assessed by qRT-PCR analysis. *, P < 0.05; ***, P < 0.001. (B to D) PK-15 cells were treated with HT-DNA (0 to 4 μg/mL) (B), poly(I·C) (0 to 4 μg/mL) (C), and poly(dA:dT) (0 to 4 μg/mL) (D) for 24 h. The levels of IFN-β, ISG15, and TMEM41B mRNA expression were assessed by qRT-PCR analysis. *, P < 0.05; ***, P < 0.001. (E and F) cGAS−/−, STING−/−, TBK1−/−, IRF3−/− and IFNAR1−/− PK-15 cells were infected with PRV-QXX (E; MOI = 5) or Bartha-K61 (F; MOI = 5) for 0 to 24 h. The level of TMEM41B mRNA expression was assessed by qRT-PCR analysis. **, P < 0.01; ***, P < 0.001.
TMEM41B knockdown inhibits PRV infection.
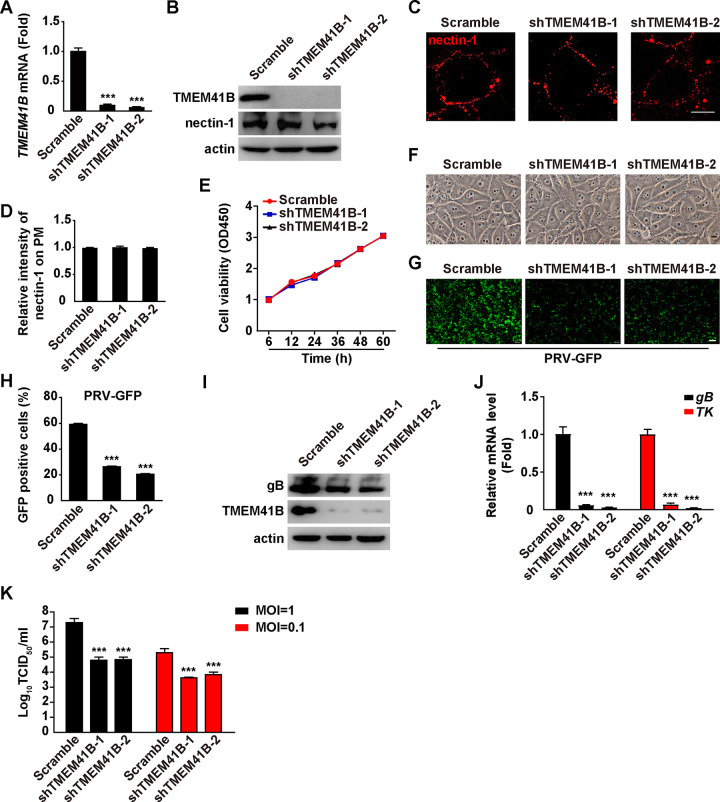
To determine whether TMEM41B was involved in PRV infection, we depleted TMEM41B using short-hairpin RNA (shRNA)-mediated RNA interference and infected cells with PRV green fluorescent protein (PRV-GFP) and PRV-QXX. Both the qRT-PCR and immunoblotting analyses showed that the two shRNAs against TMEM41B exhibited significant knockdown efficiency in PK-15 cells (Fig. 3A and B). Interference of TMEM41B had no effect on nectin-1 expression and membrane distribution, which participated in PRV entry (Fig. 3B to D). Moreover, a TMEM41B knockdown did not affect cellular viability or morphology (Fig. 3E and F).
FIG 3.
TMEM41B knockdown inhibits PRV infection. (A) The level of TMEM41B mRNA expression was assessed in scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells by qRT-PCR analysis. ***, P < 0.001. (B) TMEM41B and nectin-1 were assessed in scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells by immunoblotting analysis. (C) Membrane distribution of nectin-1 in scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells was analyzed by a nonpermeabilizing immunofluorescence analysis. Scale bar = 10 μm. (D) Quantification of the relative intensity of nectin-1 on the PM from panel C by ImageJ (n = 15). The intensity of nectin-1 on the PM from scramble PK-15 cells was normalized to 1. (E) Cell viability of scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells was assessed by a CCK-8 assay for 0 to 60 h. (F) Morphology of scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells. Scale bar = 10 μm. (G) Scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells were infected with PRV-GFP (MOI = 1) for 20 h. Viral replication was analyzed by fluorescence microscopy of GFP expression. Scale bar = 100 μm. (H) GFP-positive cells from E were analyzed by flow cytometry. ***, P < 0.001. (I) Scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells were infected with PRV-QXX (MOI = 1) for 24 h. TMEM41B and PRV gB were assessed by immunoblotting analysis. (J) Scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells were infected with PRV-QXX (MOI = 1) for 24 h. The level of PRV gB and TK mRNA expression was assessed by qRT-PCR analysis. ***, P < 0.001. (K) Scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells were infected with PRV-QXX (MOI = 0.1 and 1) for 24 h. Viral titers were assessed by a TCID50 assay. ***, P < 0.001.
We subsequently infected scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells with PRV-GFP and examined viral replication by fluorescence microscopy and a flow cytometry assay. As shown in Fig. 3G and H, a significant decrease in the GFP signal was detected in shTMEM41B-1 and shTMEM41B-2 PK-15 cells compared to that in the control cells, indicating that knockdown of TMEM41B inhibited PRV-GFP replication. Scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells were infected with PRV-QXX, and viral replication was examined by immunoblotting of PRV gB and qRT-PCR analysis of the level of PRV gB and thymidine kinase (TK) mRNA expression. The level of PRV gB protein expression was lower in shTMEM41B-1 and shTMEM41B-2 cells than in the scramble (Fig. 3I). Moreover, a knockdown of TMEM41B decreased the levels of PRV gB and TK mRNA expression (Fig. 3J). Additionally, we infected cells with PRV-QXX and found that viral titer was also lower in the shTMEM41B-1 and shTMEM41B-2 cells compared to that in the scramble (Fig. 3K). Collectively, these data indicate that knockdown of TMEM41B inhibits PRV infection.
TMEM41B overexpression promotes PRV infection.
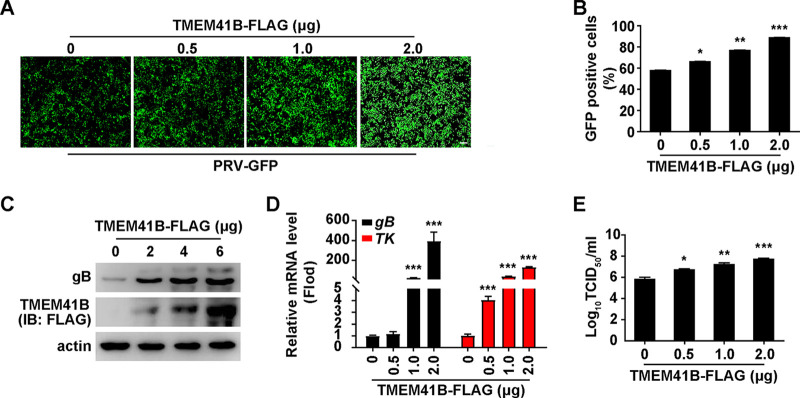
To confirm the role of TMEM41B in PRV infection, PK-15 cells were transfected with an empty vector or various concentrations of a plasmid encoding TMEM41B-FLAG for 24 h, followed by an infection with PRV-GFP. Fluorescent microscopy and flow cytometry assays showed that the GFP intensity was higher in TMEM41B-overexpressing cells than in control cells, suggesting that TMEM41B overexpression promoted PRV-GFP infection (Fig. 4A and B). We also examined the effect of TMEM41B overexpression on PRV gB expression with an immunoblotting analysis. As shown in Fig. 4C, PRV gB expression was increased due to TMEM41B-FLAG expression. PK-15 cells expressing TMEM41B-FLAG generated higher levels of PRV gB and TK mRNA than control cells did (Fig. 4D). The viral titer assay indicated that TMEM41B overexpression enhanced viral production (Fig. 4E). These results demonstrate that overexpression of TMEM41B promotes PRV infection.
FIG 4.
Overexpression of TMEM41B enhances PRV infection. (A) PK-15 cells were transfected with the TMEM41B-FLAG plasmid (0 to 2 μg) for 12 h and then infected with PRV-GFP (MOI = 1) for 20 h. Viral replication was analyzed by fluorescence microscopy of GFP expression. Scale bar = 100 μm. (B) GFP-positive cells from panel A were analyzed by flow cytometry. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) PK-15 cells were transfected with the TMEM41B-FLAG plasmid (0 to 6 μg) for 12 h and infected with PRV-QXX (MOI = 1) for 24 h. PRV gB and TMEM41B-FLAG were assessed by immunoblotting (IB) analysis. (D) PK-15 cells were transfected with the TMEM41B-FLAG plasmid (0 to 2 μg) for 12 h and then infected with PRV-QXX (MOI = 1) for 24 h. The level of PRV gB and TK mRNA expression was assessed by qRT-PCR analysis. ***, P < 0.001. (E) PK-15 cells were transfected with plasmid TMEM41B-FLAG (0 to 2 μg) for 12 h and infected with PRV-QXX (MOI = 1) for 24 h. Viral titers were assessed by a TCID50 assay. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
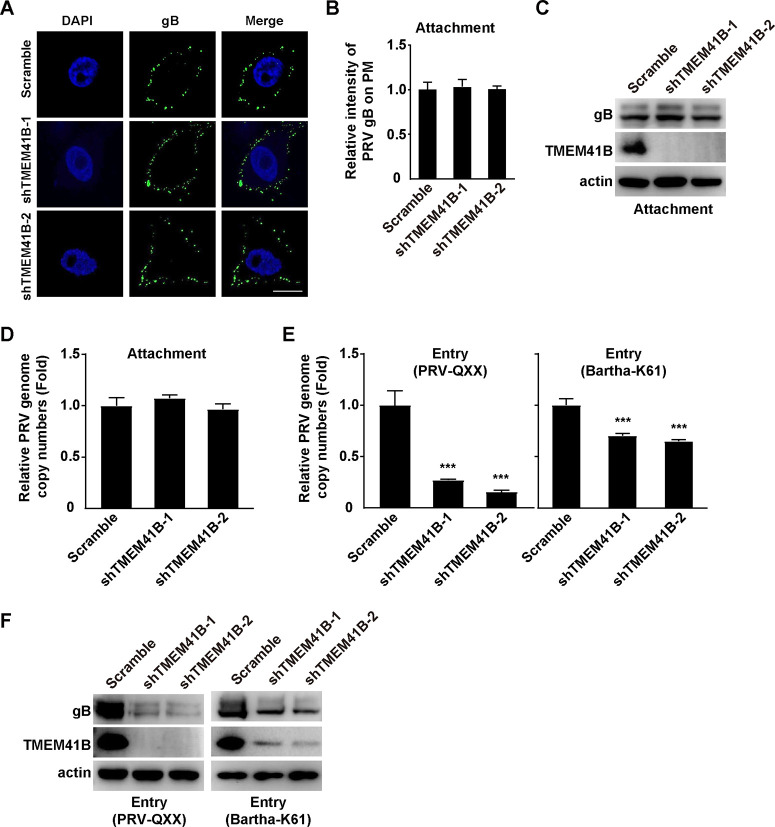
TMEM41B knockdown inhibits PRV entry.
Next, we sought to determine which stage of the viral life cycle was influenced by TMEM41B. We incubated scramble and shTMEM41B-1 cells with PRV-QXX for 2 h at 4°C and washed them three times with ice-cold PBS. Immunofluorescence analysis of PRV gB indicated that knockdown of TMEM41B did not affect PRV attachment to the PM (Fig. 5A and B). The results of the immunoblotting analysis showed that much the same PRV gB were detected on the PM of scramble and TMEM41B knockdown cells (Fig. 5C). We also analyzed viral attachment by quantification of the PRV genome copy number and found that the knockdown of TMEM41B did not affect PRV attachment to cells (Fig. 5D). We next performed a viral entry assay by quantification of the PRV genome copy number. qRT-PCR analysis indicated that the TMEM41B knockdown inhibited PRV entry (Fig. 5E). We also assessed PRV entry by performing an immunoblotting analysis of PRV gB. The level of PRV gB was decreased in the TMEM41B knockdown cells, which further indicated that a TMEM41B knockdown inhibited PRV entry (Fig. 5F). These results suggest that a knockdown of TMEM41B can inhibit PRV entry.
FIG 5.
TMEM41B is involved in PRV entry. (A) A viral attachment assay was used to assess scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells by nonpermeabilizing immunofluorescence analysis of PRV gB on the PM. Scale bar = 10 μm. (B) Quantification of the relative intensity of PRV gB on the PM from panel A by ImageJ (n = 15). The intensity of PRV gB on the PM from scramble PK-15 cells was normalized to 1. (C) Viral attachment assay was assessed in scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells by immunoblotting analysis of PRV gB on the PM. (D) Viral attachment assay was assessed in scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells by a qRT-PCR analysis of PRV genome copy numbers on the PM. PRV genome copy numbers on the PM from scramble PK-15 cells were normalized to 1. (E) A viral entry assay was used to assess scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells in a qRT-PCR analysis of PRV genome copy numbers in the cells. ***, P < 0.001. PRV genome copy numbers in scramble PK-15 cells were normalized to 1. (F) Viral entry assay was assessed in scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells by immunoblotting analysis of PRV gB in the cells.
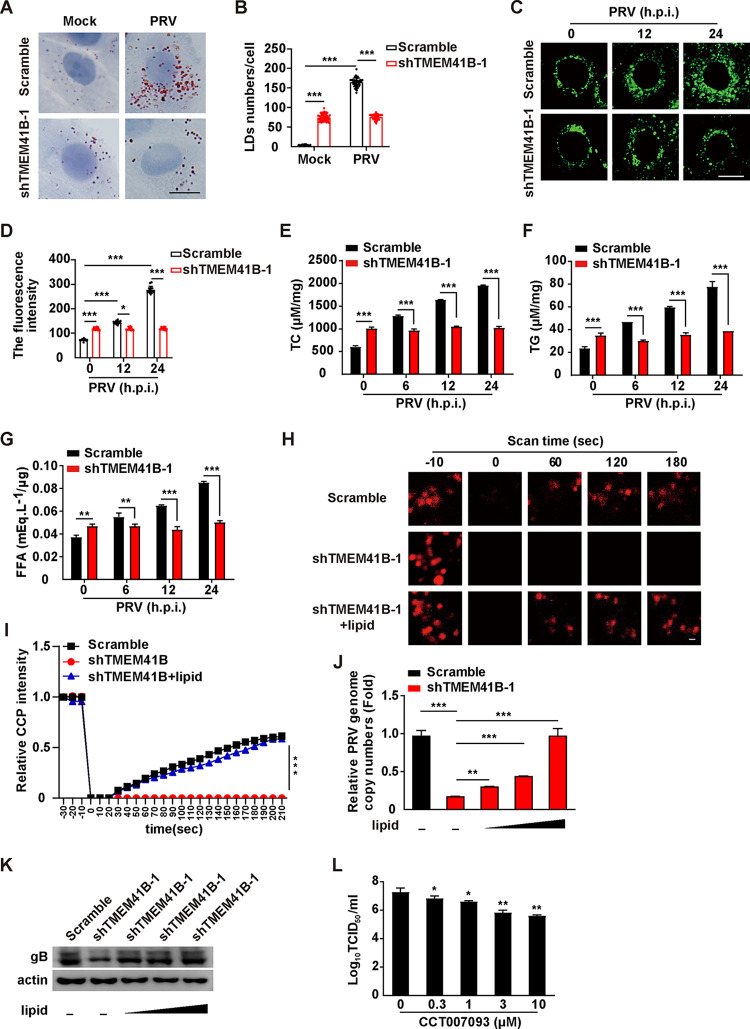
TMEM41B knockdown reduces the cellular lipid content induced by PRV.
Previous reports indicate that TMEM41B-deficient cells display enlarged LDs (10). Oil Red O and BODIPY staining indicated that there were increased numbers of LDs in TMEM41B knockdown PK-15 cells compared to that in the scramble cells (Fig. 6A to D). PRV infection further enhanced the number of LDs in the scramble cells, but not in TMEM41B knockdown cells (Fig. 6A to D). We measured the lipid content in scramble and TMEM41B knockdown cells infected with PRV-QXX for 0 to 24 h. PRV infection enhanced the intracellular levels of total cholesterol (TC), triglyceride (TG), and FFAs (Fig. 6E to G). However, a TMEM41B knockdown prevented the generation of these lipids, suggesting that knockdown of TMEM41B might inhibit lipid synthesis (Fig. 6E to G). These results demonstrate that a knockdown of TMEM41B can inhibit the number of PRV-enhanced LDs and lipid content.
FIG 6.
TMEM41B participates in PRV-induced lipid metabolism. (A) Scramble and shTMEM41B-1 PK-15 cells were infected with PRV-QXX (MOI = 1) for 24 h. LDs were detected by Oil Red O staining. Scale bar = 10 μm. (B) Quantification of the number of LDs per cell from panel A by ImageJ (n = 50). ***, P < 0.001. (C) Scramble and shTMEM41B-1 PK-15 cells were infected with PRV-QXX (MOI = 1) for 0 to 24 h. LDs were detected by BODIPY staining. Scale bar = 10 μm. (D) Quantification of the fluorescence intensity of LDs from panel C by ImageJ (n = 15). *, P < 0.05; ***, P < 0.001. (E to G) Scramble and shTMEM41B-1 PK-15 cells were infected with PRV-QXX (MOI = 1) for 0 to 24 h. Cellular lipids were extracted, and the amount of TC (E), TG (F), and FFAs (G) was quantified using biochemical kits. **, P < 0.01; ***, P < 0.001. (H) Scramble and shTMEM41B-1 PK-15 cells were transfected with AP2B1-mCherry plasmid for 24 h and were treated with lipid (0.003 μg/mL cholesterol and 150 μM oleic acid) for 8 h. The CCP dynamic was assessed by FRAP analysis. Scale bar = 1 μm. (I) Quantification of the relative fluorescent intensity of AP2B1 puncta in the FRAP region over time from panel H by ImageJ (n = 15). ***, P < 0.001. (J) A viral entry assay was used to assess the scramble and shTMEM41B-1 PK-15 cells treated with lipid (0 to 0.003 μg/mL cholesterol and 0 to 150 μM oleic acid) as indicated by qRT-PCR analysis of the PRV genome copy numbers in the cells. **, P < 0.01; ***, P < 0.001. (K) Cells were treated as in panel I. A viral entry assay was assessed by immunoblotting analysis. (L) PK-15 cells were infected with PRV-QXX (MOI = 1) for 1 h and then treated with CCT007093 (0 to 150 μM) for 24 h. Viral titers were assessed by a TCID50 assay. *, P < 0.05; **, P < 0.01.
Our previous study suggested that Niemann-Pick C1 (NPC1) deficiency attenuates PRV entry by decreasing the abundance of cholesterol in the PM and inhibiting CCP dynamics (7). Therefore, we next examined whether TMEM41B could act via a similar mechanism. To corroborate the role of TMEM41B in CCP dynamics, fluorescence recovery after photobleaching (FRAP) analysis was performed for CCP adaptor-related protein complex 2 subunit beta 1 (AP2B1) using live-cell confocal microscopy imaging. Following a 20-s bleaching laser pulse, AP2B1-mCherry fluorescence rapidly recovered to about 50% of its initial value at approximately 180 s in the scramble cells (Fig. 6H and I), whereas the TMEM41B knockdown cells did not show a recovery in AP2B1-mCherry fluorescence (Fig. 6H and I). Exogenous cholesterol and oleic acid were able to complement the inhibitory effect of the TMEM41B deficiency on the recovery of AP2B1-mCherry fluorescence (Fig. 6H and I). Moreover, we examined whether cholesterol and oleic acid replenishment could rescue PRV entry in TMEM41B knockdown cells. qRT-PCR analysis indicated that the PRV genome copy number in TMEM41B knockdown cells gradually increased with an increased concentration of cholesterol and oleic acid (Fig. 6J). Exogenous supplementation of cholesterol and oleic acid in TMEM41B knockdown cells restored the internalization of PRV gB, as indicated by immunoblotting analysis (Fig. 6K). We next assessed whether inhibition of LD formation affected PRV replication. CCT007093 is a chemical inhibitor of Ser/Thr phosphatase PPM1D, which regulates LD formation via dephosphorylation of perilipin 1 (26). The 50% tissue culture infective dose (TCID50) assay indicated that treatment of PK-15 cells with CCT007093 affected PRV replication (Fig. 6L). These data suggest that TMEM41B knockdown affects cellular lipid synthesis to inhibit PRV entry.
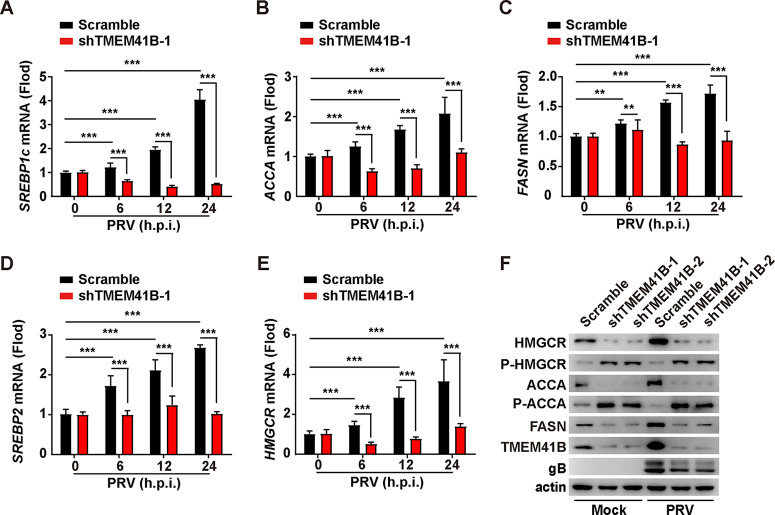
TMEM41B knockdown inhibits the expression of the key lipid synthesis-related factors.
We sought to determine whether knockdown of TMEM41B could inhibit lipid synthesis. We infected scramble and TMEM41B knockdown cells with PRV-QXX and subsequently examined the transcription of the key lipid synthesis-related factors, including sterol regulatory element-binding protein 1c (SREBP1c), fatty acid synthase (FASN), acetyl-CoA carboxylase alpha (ACCA), sterol regulatory element-binding protein 2 (SREBP2), and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). qRT-PCR analysis indicated that PRV failed to upregulate the levels of SREBP1c, ACCA, FASN, SREBP2, and HMGCR mRNA expression in the TMEM41B knockdown cells compared to that in the control cells (Fig. 7A to E). An immunoblotting analysis indicated that PRV infection upregulated the expression of ACCA, HMGCR, and FASN (Fig. 7F). However, a knockdown of TMEM41B eliminated PRV-enhanced expression of ACCA, HMGCR, and FASN (Fig. 7F). HMGCR and ACCA can be inactivated by phosphorylation (27). Moreover, enhanced phosphorylation of HMGCR and ACCA was observed in TMEM41B knockdown PK-15 cells either mock infected or infected with PRV-QXX, suggesting that HMGCR and ACCA were inactivated by the TMEM41B knockdown (Fig. 7F). These results suggest that knockdown of TMEM41B inhibited PRV-stimulated lipid synthesis.
FIG 7.
TMEM41B is contributed to lipid synthesis. (A to E) Scramble and shTMEM41B-1 PK-15 cells were infected with PRV-QXX (MOI = 1) for 0 to 24 h. The mRNA levels of SREBP1c (A), ACCA (B), FASN (C), SREBP2 (D), and HMGCR (E) were assessed by qRT-PCR analysis. **, P < 0.01; ***, P < 0.001. (F) Scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells were mock infected or infected with PRV-QXX (MOI = 1) for 24 h. HMGCR, P-HMGCR, ACCA, P-ACCA, FASN, TMEM41B, and PRV gB were assessed by immunoblotting analysis.
DISCUSSION
Viruses rely on host cells to complete their life cycle. Moreover, lipid metabolism has been shown to be involved in various stages of the viral life cycle (18). We have previously shown that porcine reproductive and respiratory syndrome virus (PRRSV) infection enhances lipid synthesis and promotes lipolysis through lipophagy to promote optimal viral replication (28, 29). However, how lipids are involved in the entire life cycle of PRV remains unclear. In this study, we demonstrated that PRV infection could enhance lipid synthesis through upregulation of TMEM41B through the cGAS-STING innate immune and IFN signaling pathways.
A key event in the innate immune response to viral infection is the recognition of cytoplasmic DNA by DNA sensors, which can induce the production of type I IFN, cytokines, and chemokines. The production of these factors is mediated through cascaded amplification reactions, thereby exerting antiviral innate immune functionality (30). DNA-dependent activators of IFN-regulatory factors are the first DNA sensors thought to be required to respond to viral DNA (31, 32). DNA sensors were subsequently discovered to recognize cytoplasmic DNA, including DEAD box helicase 41 (33), RNA polymerase III (34), IFN-γ-inducible protein 16 (35), and high mobility group box proteins (36). However, whether these DNA sensors are involved in TMEM41B expression requires further investigation.
It has previously been reported that there is a close relationship between lipid metabolism and antiviral innate immunity (37). Cholesterol-25-hydroxylase, an ISG, can inhibit viral entry to mediate a wide range of antiviral effects through 25-hydroxycholesterol (38, 39). Moreover, 7-dehydrocholesterol can exert various antiviral effects by regulating the production of type I IFN through regulating the activation of AKT serine/threonine kinase 3 (37). TMEM41B functions as a host factor in SARS-CoV-2 (17, 40, 41) and pan-flavivirus infection (15). We found that TMEM41B was an ISG that could facilitate PRV infection, indicating a novel mechanism through which ISG can participate in viral replication.
Our previous studies have demonstrated that NPC1- and liver X receptor (LXR)-mediated lipid metabolism can influence PRV entry (7, 42). Moreover, EGCC can restrict PRRSV replication and assembly by disturbing lipid metabolism (28). In addition, N-myc downstream-regulated gene 1 plays a negative role in PRRSV replication and viral assembly by suppressing autophagy and LD degradation (29). For some viruses, cholesterol is critical to facilitate viral entry into host cells, such as PRV (20, 38), foot-and-mouth disease virus (43), and human rhinovirus type 2 (44). Furthermore, cholesterol consumption can significantly inhibit the entry and infection of viruses. Lipids provide a source of energy molecules and are one of the key components that determine membrane fluidity and structure. Membrane fusion is widespread in eukaryotic cell survival and the entry of enveloped viruses into host cells (45). In addition, lipid composition can regulate membrane organization and dynamics, as well as the membrane protein structure and conformation (46). Previous reports have demonstrated that high membrane tension can inhibit endocytosis (47) and some viruses can induce changes in the PM structure or use lipid synthetase to promote viral proliferation during infection (18).
TMEM41B is involved in LD kinetics and plays an important role in lipid metabolism (48). Important findings suggest that TMEM41B is an ER-disrupting enzyme required for lipoprotein biogenesis and lipid homeostasis. Moreover, TMEM41B can mediate the synthesis and shuttling of phospholipids in cytoplasmic lobules and promotes the balance of the phospholipid bilayer (12). Multiple studies have demonstrated that lipid mobilization is impaired in TMEM41B knockout cells (48–50). We hypothesize that the knockdown of TMEM41B inhibits PRV infection by disrupting lipid homeostasis. PRV has been observed to enter cells through clathrin-mediated endocytosis (7, 51), and NPC1 defects have been found to inhibit CCP dynamics by reducing cholesterol transportation to the PM (7). In the present study, impaired CCP dynamics and reduced internalized virions were observed in TMEM41B knockdown cells. Our results further suggest that TMEM41 plays a key role in CCP dynamics and viral entry.
In summary, our results suggest that TMEM41B represents a host factor for PRV replication, suggesting its important potential as a therapeutic target for drug and vaccine development to inhibit PRV replication.
MATERIALS AND METHODS
Reagents.
SYBR Premix Ex Taq (RR420A) and TRIzol Reagent (D9108B) were purchased from TaKaRa (Otsu, Shiga, Japan); BODIPY 493/503 (D3922) was purchased from Thermo Fisher Scientific (MA, USA); Oil Red O (O0625) was purchased from Sigma-Aldrich (MO, USA); a lab assay nonessential fatty acid (294-63601) assay kit for FFAs was purchased from Wako Bioproducts (VA, USA); and a TG assay kit (E1013) and TC assay kit (E1015) were purchased from Applygen Technologies Inc. (Beijing, China). CCT007093 (HY-15880) was purchased from MedChemExpress (NJ, USA).
Antibodies.
Anti-ACCA (no. 3676) and anti-phosphorylated ACCA (no. 11818) antibodies were purchased from Cell Signaling Technology (MA, USA); anti-calnexin (10427-2-AP), anti-nectin-1 (24713-1-AP), anti-TMEM41B (292701-1-AP), and anti-β-actin (20536-1-AP) were purchased from Proteintech (Wuhan, China); anti-FASN (ab22759) was ordered from Abcam (MA, USA); anti-HMGCR (MABS1233) was ordered from Sigma-Aldrich (MO, USA); anti-phosphorylated HMGCR (bs-4063R) was ordered from Bioss (MA, USA); horseradish peroxidase (HRP)-conjugated donkey anti-mouse IgG (715-035-150) and anti-rabbit IgG (711-035-152) were purchased from Jackson ImmunoResearch Laboratories (PA, USA); and anti-mouse IgG labeled with Alexa Fluor 555 (A21424), Alexa Fluor 488 (A21429), and anti-rabbit IgG labeled with Alexa Fluor 488 (A11034) were purchased from Thermo Fisher Scientific. Antiserum against PRV gB was generated by immunizing mice with purified recombinant gB. The antibodies were used at dilutions of 1:500 for immunofluorescence staining and 1:1,000 for immunoblotting analysis.
Cells, viruses, and plasmids.
Porcine kidney epithelial PK-15 cells (CCL-33; ATCC, MD, USA) and HE293T cells (CRL-11268; ATCC) were grown in monolayers at 37°C under 5% CO2 in DMEM (10566-016; Gibco, NY, USA) supplemented with 10% FBS (10099141C; Gibco, MA, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin sulfate (B540732; Sangon, Shanghai, China). cGAS−/−, STING−/−, TBK1−/−, IRF3−/−, and IFNAR1−/− PK-15 cells were used as previously described (52). Briefly, lentiviruses-mediated CRISPR-Cas9 technology was used to generate gene knockout cell lines. PK15 cells were infected with the lentiviruses and subsequently selected with puromycin (4 μg/mL) for 7 days. Single clonal knockout cells were obtained by serial dilution and verified by Sanger sequencing and immunoblotting analysis.
The PRV strain Bartha-K61 was used as previously described (25). The virulent PRV isolate QXX (PRV-QXX) was kindly donated by Yong-Tao Li from the College of Veterinary Medicine, Henan Agricultural University (53). The recombinant PRV strain of PRV-GFP, derived from the PRV Hubei strain with the TK gene replaced by a GFP expression cassette from the pEGFP-N1 plasmid, was kindly donated by Han-Zhong Wang from Wuhan Institute of Virology, Chinese Academy of Sciences (54). Viral titers were determined using the TCID50 assay, which was calculated via the Reed-Muench method.
Full-length porcine TMEM41B and AP2B1 cDNA was amplified by PCR. TMEM41B cDNA was cloned into pEGFP-N1 and p3×Flag-CMV-14 expression plasmids to generate TMEM41B-GFP and TMEM41B-FLAG. AP2B1 cDNA was cloned into an mCherry-N1 expression plasmid to generate AP2B1-mCherry. All plasmids were transfected with Lipofectamine 3000 (L3000015; Invitrogen, NY, USA) according to the manufacturer’s instructions. The primer sequences used for gene amplification were used as follows: TMEM41B-forward (Fw): 5′-ATGACGAAAGGCGGAGTCGTCGAAA-3′ and TMEM41B-reverse (Rv): 5′-CTCAAATTTCTGCTTCAGCTTTTTTTG-3′; and AP2B1-Fw: 5′-ATGACTGACTCCAAGTACTTCACAA-3′ and AP2B1-Rv: 5′-GTTTTTCAAAATGCTGTCGTAGACC-3′.
Mice.
Female 6- to 8-week-old BALB/c mice were purchased from the Center of Experimental Animals of Zhengzhou University (Zhengzhou, China) and maintained in a specific pathogen-free animal facility according to the Guide for the Care and Use of Laboratory Animals and the related ethical regulations instilled at Henan Agricultural University.
Cell viability assay.
Cell viability was evaluated using a cell counting kit-8 (CCK-8) according to the manufacturer’s instructions (GK3607; Dingguo, Beijing). Briefly, the cells were seeded into 96-well plates at a density of 0.8 × 104 per well for 24 h. CCK-8 (10 μL) was then added to each well, and the cells were incubated at 37°C for 3 h. The absorbance was detected at 450 nm with a microplate reader (Varioskan Flash; Thermo Fisher Scientific).
Flow cytometry assay.
A flow cytometry assay was performed as previously described (55). Briefly, PK-15 cells were infected with PRV-GFP (multiplicity of infection [MOI] = 1) for 20 h. Cells were digested with trypsin-EDTA (25200072; Gibco, MA, USA), collected by centrifugation, and suspended in PBS. The percentage of GFP-positive cells was measured by flow cytometry using a Beckman CytoFLEX instrument. All data were analyzed with CytExpert software 2.0.
qRT-PCR.
Total RNA was extracted using TRIzol Reagent (9108; TaKaRa) and reverse transcribed with a PrimeScript RT reagent kit (RR047A; TaKaRa). qRT-PCR was performed in triplicate using SYBR Premix Ex Taq (RR820A; TaKaRa). Data were normalized to the expression of the control gene encoding β-actin. The relative changes in expression were calculated using the 2−ΔΔCT method.
Quantification of the genome copy number of PRV was performed as previously described (55). Briefly, the PRV gH PCR product (187 bp) was cloned into the pGEM-T vector. A standard curve was prepared from serial 10-fold dilutions of the plasmid for the quantification of PRV genomic DNA. The following primers were used for qRT-PCR analysis: porcine ACTB-Fw: 5′-GCACAGAGCCTCGCCTT-3′ and porcine ACTB-Rv: 5′-CCTTGCACATGCCGGAG-3′; porcine TMEM41B-Fw: 5′-GTCGAAAGATCGCAAACGGG-3′ and porcine TMEM41B-Rv: 5′-CTCTCCTTTAGGCGGTCTCC-3′; porcine FASN-Fw: 5′-GCACACTTACGTACTGGCCT-3′ and porcine FASN-Rv: 5′-TGATGATTAGGTCCACGGCG-3′; porcine ACACA-Fw: 5′-CCGCTTGCCTGTCTTTTGAT-3′ and porcine ACACA-Rv: 5′-ACGTTATCCCCAAACCCAGG-3′; porcine HMGCR-Fw: 5′-ATTGTGTGCGGGACCGTAAT-3′ and porcine HMGCR-Rv: 5′-AATGCCCGTGTTCCAGTTCA-3′; porcine SREBP1c-Fw: 5′-GACGAGCCACCCTTCAGCAA-3′ and porcine SREBP1c-Rv: 5′-GCATGTCTTCGAACGTGCAAT-3′; porcine SREBP2-Fw: 5′-TTGTCGGGTGTCATGGGCG-3′ and porcine SREBP2-Rv: 5′-ATTGCAGCATCTCGTCGATGT-3′; PRV gB-Fw: 5′-CTCGCCATCGTCAGCAAPRV-3′ and PRV gB-Rv: 5′-GCTGCTCCTCCATGTCCTT-3′; and PRV gH-Fw: 5′-CTCGCCATCGTCAGCAA-3′ and PRV gH-Rv: 5′-GCTGCTCCTCCATGTCCTT-3′.
Immunoblotting analysis.
The cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and 2 mM MgCl2) supplemented with a protease and phosphatase inhibitor cocktail (HY-K0010 and HY-K0022; MedChemExpress). Protein samples were separated by SDS-PAGE and transferred to polypropylene fluoride membranes (C3117; Millipore, MA, USA). After a 30-min incubation in 5% nonfat milk (A600669; Sangon Biotech), the membrane was incubated with the primary antibody overnight at 4°C followed by incubation with the appropriate HRP-conjugated secondary antibody for 1 h at room temperature. The target proteins were detected with Luminata Crescendo immunoblotting HRP substrate (WBLUR0500; Millipore) on a GE AI600 imaging system.
Immunofluorescence analysis.
Cells grown on coverslips were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. Cells were nonpermeabilized or permeabilized with 0.2% Triton X-100 for 20 min. Cells were then blocked with PBS/10% FBS for 1 h at room temperature. Cells were washed three times with PBS and incubated with the PRV gB antiserum for 1 h at room temperature. Cells were then incubated with the appropriate Alexa-Fluor-conjugated secondary antibodies for 1 h at room temperature. The images were captured using a Zeiss LSM 800 confocal microscope (Oberkochen, Germany).
RNA interference.
Lentivirus-mediated gene silencing was conducted as previously described (29). Briefly, shRNAs (scramble: GCCACAACGTCTATATCATGG; shTMEM41B-1: GTCGTCGAAAGATCGCAAACG; and shTMEM41B-2: GAAGCTGGGTCAGCAAGAATG) were synthesized as double-stranded oligonucleotides, cloned into the pLKO.1 vector, and cotransfected with packaging plasmids pMD2.G and psPAX2 into HE293T cells. Lentiviruses were harvested at 48 h posttransfection and used to infect cells that were then selected with puromycin (4 μg/mL) for 7 days. Knockdown efficiency was determined by qRT-PCR or immunoblotting analysis.
Viral attachment assays.
Scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells were incubated with PRV-QXX (MOI = 100) at 4°C for 2 h. After three extensive washes in ice-cold PBS, viral attachment assays were performed by immunofluorescence and immunoblotting analyses of PRV gB or by qRT-PCR analysis of the PRV genome copy numbers on the PM.
Viral entry assays.
Scramble, shTMEM41B-1, and shTMEM41B-2 PK-15 cells were incubated with PRV-QXX (MOI = 100) at 4°C for 2 h. Cells were extensively washed three times with ice-cold PBS and then shifted to 37°C for 15 min to allow entry. After being washed with trypsin (1 mg/mL) to remove any residual virions on the PM, viral entry was detected by qRT-PCR analysis of viral genome copy numbers, or by intracellular PRV gB immunoblotting.
Determination of intracellular FFAs, TG, and TC.
The cell lysates were extracted with a syringe needle in 250 μL RIPA buffer and centrifuged at 12,000 × g for 5 min at 4°C. The total lipids in 200 μL lysate were extracted by the addition of 100 μL chloroform-methanol (2:1, vol/vol) mixture. The extract was evaporated until dry and dissolved in 50 μL of TRB (100 mM KH2PO4, 100 mM K2HPO4, 5 mM sodium cholate, 50 mM NaCl, and 0.1% Triton X-100, pH 7.4) for the FFA, TG, and TC assays. FFA (Wako), TG (Applygen), and TC (Applygen) were measured with biochemical assay kits in accordance with the manufacturer’s protocols. The values were normalized to the total cellular protein content.
Oil Red O staining.
The cells were fixed in 4% paraformaldehyde for 30 min and incubated in Oil Red O (3 μg/mL) for 15 min. The cells were washed with 70% alcohol for 5 s to remove any background stain, rinsed in double-distilled Millipore water, counterstained with Harris hematoxylin, washed, mounted, and observed under a light microscope. The LD number was determined using the ImageJ “analyze particles” function (areas of particles <0.01 mm2 were excluded).
BODIPY staining.
Cells were fixed in 4% PFA for 30 min and incubated with 2 μg/mL BODIPY 493/503 (493-nm excitation/503-nm emission) for 30 min. Digital images were obtained with a Zeiss LSM800 confocal microscope. The fluorescence intensity was determined using ImageJ software.
FRAP.
Cells were transfected with the AP2B1-mCherry plasmid at 37°C for 24 h. The cells were then bleached at maximum laser intensity for 30 s in a region of 7 × 7 μm2 at 37°C and imaged every 60 s for 5 min. CCP dynamics were defined by the fluorescence recovery of AP2B1-mCherry, which was performed using a Zeiss LSM 800 confocal microscope.
Statistical analysis.
Data are representative of at least three independent experiments for quantitative analyses and expressed as the means ± standard errors of the means. All statistical analyses were performed with a two-tailed Student's t test. Significant differences relative to the corresponding controls were accepted at P < 0.05.
ACKNOWLEDGMENTS
This research was supported by grants from the National Natural Science Foundation of China (32272961), the National Key R&D Program of China (2021YFD1301200), the Doctoral Foundation of Henan Agricultural University (30501221), and Henan Province Higher Education Teaching Reform Research and Practice Project (2021SJGLX351).
We declare that we have no competing interests.
Contributor Information
Kai Zhong, Email: zhongkai.henau@gmail.com.
Bei-Bei Chu, Email: chubeibei@henau.edu.cn.
Jiang Wang, Email: wangjiang@henau.edu.cn.
Anna Ruth Cliffe, University of Virginia.
REFERENCES
- 1.Mettenleiter TC. 2000. Aujeszky’s disease (pseudorabies) virus: the virus and molecular pathogenesis. Vet Res 31:99–115. doi: 10.1051/vetres:2000110. [DOI] [PubMed] [Google Scholar]
- 2.Pomeranz LE, Reynolds AE, Hengartner CJ. 2005. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev 69:462–500. doi: 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woźniakowski G, Samorek-Salamonowicz E. 2015. Animal herpesviruses and their zoonotic potential for cross-species infection. Ann Agric Environ Med 22:191–194. doi: 10.5604/12321966.1152063. [DOI] [PubMed] [Google Scholar]
- 4.Ai JW, Weng SS, Cheng Q, Cui P, Li YJ, Wu HL, Zhu YM, Xu B, Zhang WH. 2018. Human endophthalmitis caused by pseudorabies virus infection, China, 2017. Emerg Infect Dis 24:1087–1090. doi: 10.3201/eid2406.171612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang X, Guan H, Li C, Li Y, Wang S, Zhao X, Zhao Y, Liu Y. 2019. Characteristics of human encephalitis caused by pseudorabies virus: a case series study. Int J Infect Dis 87:92–99. doi: 10.1016/j.ijid.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Wong G, Lu J, Zhang W, Gao GF. 2019. Pseudorabies virus: a neglected zoonotic pathogen in humans? Emerg Microbes Infect 8:150–154. doi: 10.1080/22221751.2018.1563459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G, Su B, Fu P, Bai Y, Ding G, Li D, Wang J, Yang G, Chu B. 2022. NPC1-regulated dynamic of clathrin-coated pits is essential for viral entry. Sci China Life Sci 65:341–361. doi: 10.1007/s11427-021-1929-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Minnebruggen G, Favoreel HW, Nauwynck HJ. 2004. Internalization of pseudorabies virus glycoprotein B is mediated by an interaction between the YQRL motif in its cytoplasmic domain and the clathrin-associated AP-2 adaptor complex. J Virol 78:8852–8859. doi: 10.1128/JVI.78.16.8852-8859.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita K, Hama Y, Izume T, Tamura N, Ueno T, Yamashita Y, Sakamaki Y, Mimura K, Morishita H, Shihoya W, Nureki O, Mano H, Mizushima N. 2018. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J Cell Biol 217:3817–3828. doi: 10.1083/jcb.201804132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moretti F, Bergman P, Dodgson S, Marcellin D, Claerr I, Goodwin JM, DeJesus R, Kang Z, Antczak C, Begue D, Bonenfant D, Graff A, Genoud C, Reece-Hoyes JS, Russ C, Yang Z, Hoffman GR, Mueller M, Murphy LO, Xavier RJ, Nyfeler B. 2018. TMEM 41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep 19:e45889. doi: 10.15252/embr.201845889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shoemaker CJ, Huang TQ, Weir NR, Polyakov NJ, Schultz SW, Denic V. 2019. CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol 17:e2007044. doi: 10.1371/journal.pbio.2007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang D, Xu B, Liu L, Wu L, Zhu Y, Ghanbarpour A, Wang Y, Chen FJ, Lyu J, Hu Y, Kang Y, Zhou W, Wang X, Ding W, Li X, Jiang Z, Chen J, Zhang X, Zhou H, Li JZ, Guo C, Zheng W, Zhang X, Li P, Melia T, Reinisch K, Chen XW. 2021. TMEM41B acts as an ER scramblase required for lipoprotein biogenesis and lipid homeostasis. Cell Metab 33:1655–1670.e8. doi: 10.1016/j.cmet.2021.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Li YE, Wang Y, Du X, Zhang T, Mak HY, Hancock SE, McEwen H, Pandzic E, Whan RM, Aw YC, Lukmantara IE, Yuan Y, Dong X, Don A, Turner N, Qi S, Yang H. 2021. TMEM41B and VMP1 are scramblases and regulate the distribution of cholesterol and phosphatidylserine. J Cell Biol 220:e202103105. doi: 10.1083/jcb.202103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang T, Li YE, Yuan Y, Du X, Wang Y, Dong X, Yang H, Qi S. 2021. TMEM41B and VMP1 are phospholipid scramblases. Autophagy 17:2048–2050. doi: 10.1080/15548627.2021.1937898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann HH, Schneider WM, Rozen-Gagnon K, Miles LA, Schuster F, Razooky B, Jacobson E, Wu X, Yi S, Rudin CM, MacDonald MR, McMullan LK, Poirier JT, Rice CM. 2021. TMEM41B is a pan-flavivirus host factor. Cell 184:133–148.e20. doi: 10.1016/j.cell.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L, Zhao C, Fu Z, Fu Y, Su Z, Li Y, Zhou Y, Tan Y, Li J, Xiang Y, Nie X, Zhang J, Liu F, Zhao S, Xie S, Peng G. 2021. Genome-scale CRISPR screen identifies TMEM41B as a multi-function host factor required for coronavirus replication. PLoS Pathog 17:e1010113. doi: 10.1371/journal.ppat.1010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider WM, Luna JM, Hoffmann HH, Sanchez-Rivera FJ, Leal AA, Ashbrook AW, Le Pen J, Ricardo-Lax I, Michailidis E, Peace A, Stenzel AF, Lowe SW, MacDonald MR, Rice CM, Poirier JT. 2021. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell 184:120–132.e14. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorizate M, Krausslich HG. 2011. Role of lipids in virus replication. Cold Spring Harb Perspect Biol 3:a004820. doi: 10.1101/cshperspect.a004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Targett-Adams P, Boulant S, Douglas MW, McLauchlan J. 2010. Lipid metabolism and HCV infection. Viruses 2:1195–1217. doi: 10.3390/v2051195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desplanques AS, Nauwynck HJ, Vercauteren D, Geens T, Favoreel HW. 2008. Plasma membrane cholesterol is required for efficient pseudorabies virus entry. Virology 376:339–345. doi: 10.1016/j.virol.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, Jiang C. 2008. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res 18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bender FC, Whitbeck JC, Ponce de Leon M, Lou H, Eisenberg RJ, Cohen GH. 2003. Specific association of glycoprotein B with lipid rafts during herpes simplex virus entry. J Virol 77:9542–9552. doi: 10.1128/jvi.77.17.9542-9552.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Yang C, Tong S, Weidmann A, Compans RW. 2002. Palmitoylation of the murine leukemia virus envelope protein is critical for lipid raft association and surface expression. J Virol 76:11845–11852. doi: 10.1128/jvi.76.23.11845-11852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji M, Li M, Sun L, Zhao H, Li Y, Zhou L, Yang Z, Zhao X, Qu W, Xue H, Zheng Z, Li Y, Deng H, Zhao YG. 2022. VMP1 and TMEM41B are essential for DMV formation during β-coronavirus infection. J Cell Biol 221:e202112081. doi: 10.1083/jcb.202112081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Chu B, Du L, Han Y, Zhang X, Fan S, Wang Y, Yang G. 2015. Molecular cloning and functional characterization of porcine cyclic GMP-AMP synthase. Mol Immunol 65:436–445. doi: 10.1016/j.molimm.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Kamada R, Uno S, Kimura N, Yoshimura F, Tanino K, Sakaguchi K. 2022. Lipid droplet formation is regulated by Ser/Thr phosphatase PPM1D via dephosphorylation of perilipin 1. Int J Mol Sci 23:12046. doi: 10.3390/ijms231912046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Day EA, Ford RJ, Smith BK, Houde VP, Stypa S, Rehal S, Lhotak S, Kemp BE, Trigatti BL, Werstuck GH, Austin RC, Fullerton MD, Steinberg GR. 2021. Salsalate reduces atherosclerosis through AMPKbeta1 in mice. Mol Metab 53:101321. doi: 10.1016/j.molmet.2021.101321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu P-W, Fu P-F, Zeng L, Qi Y-L, Li X-Q, Wang Q, Yang G-Y, Li H-W, Wang J, Chu B-B, Wang M-D. 2022. EGCG restricts PRRSV proliferation by disturbing lipid metabolism. Microbiol Spectr 10:e227621. doi: 10.1128/spectrum.02276-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Liu JY, Shao KY, Han YQ, Li GL, Ming SL, Su BQ, Du YK, Liu ZH, Zhang GP, Yang GY, Chu BB. 2019. Porcine reproductive and respiratory syndrome virus activates lipophagy to facilitate viral replication through downregulation of NDRG1 expression. J Virol 93:e00526-19. doi: 10.1128/JVI.00526-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keating SE, Baran M, Bowie AG. 2011. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol 32:574–581. doi: 10.1016/j.it.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, Choi M, Ban T, Yanai H, Negishi H, Lu Y, Tamura T, Takaoka A, Nishikura K, Taniguchi T. 2008. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci USA 105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. 2011. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiu YH, Macmillan JB, Chen ZJ. 2009. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, Nakasato M, Lu Y, Hangai S, Koshiba R, Savitsky D, Ronfani L, Akira S, Bianchi ME, Honda K, Tamura T, Kodama T, Taniguchi T. 2009. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 462:99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- 37.Xiao J, Li W, Zheng X, Qi L, Wang H, Zhang C, Wan X, Zheng Y, Zhong R, Zhou X, Lu Y, Li Z, Qiu Y, Liu C, Zhang F, Zhang Y, Xu X, Yang Z, Chen H, Zhai Q, Wei B, Wang H. 2020. Targeting 7-dehydrocholesterol reductase integrates cholesterol metabolism and IRF3 activation to eliminate infection. Immunity 52:109–122.e6. doi: 10.1016/j.immuni.2019.11.015. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Zeng L, Zhang L, Guo ZZ, Lu SF, Ming SL, Li GL, Wan B, Tian KG, Yang GY, Chu BB. 2017. Cholesterol 25-hydroxylase acts as a host restriction factor on pseudorabies virus replication. J Gen Virol 98:1467–1476. doi: 10.1099/jgv.0.000797. [DOI] [PubMed] [Google Scholar]
- 39.Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, Guo H, Nusbaum R, Zack JA, Freiberg AN, Su L, Lee B, Cheng G. 2013. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei J, Alfajaro MM, DeWeirdt PC, Hanna RE, Lu-Culligan WJ, Cai WL, Strine MS, Zhang SM, Graziano VR, Schmitz CO, Chen JS, Mankowski MC, Filler RB, Ravindra NG, Gasque V, de Miguel FJ, Patil A, Chen H, Oguntuyo KY, Abriola L, Surovtseva YV, Orchard RC, Lee B, Lindenbach BD, Politi K, van Dijk D, Kadoch C, Simon MD, Yan Q, Doench JG, Wilen CB. 2021. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell 184:76–91.e13. doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang R, Simoneau CR, Kulsuptrakul J, Bouhaddou M, Travisano KA, Hayashi JM, Carlson-Stevermer J, Zengel JR, Richards CM, Fozouni P, Oki J, Rodriguez L, Joehnk B, Walcott K, Holden K, Sil A, Carette JE, Krogan NJ, Ott M, Puschnik AS. 2021. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell 184:106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Li GL, Qi YL, Li LY, Wang LF, Wang CR, Niu XR, Liu TX, Wang J, Yang GY, Zeng L, Chu BB. 2022. Pseudorabies virus inhibits expression of liver X receptors to assist viral infection. Viruses 14:514. doi: 10.3390/v14030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin-Acebes MA, Gonzalez-Magaldi M, Sandvig K, Sobrino F, Armas-Portela R. 2007. Productive entry of type C foot-and-mouth disease virus into susceptible cultured cells requires clathrin and is dependent on the presence of plasma membrane cholesterol. Virology 369:105–118. doi: 10.1016/j.virol.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 44.Snyers L, Zwickl H, Blaas D. 2003. Human rhinovirus type 2 is internalized by clathrin-mediated endocytosis. J Virol 77:5360–5369. doi: 10.1128/jvi.77.9.5360-5369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ketter E, Randall G. 2019. Virus impact on lipids and membranes. Annu Rev Virol 6:319–340. doi: 10.1146/annurev-virology-092818-015748. [DOI] [PubMed] [Google Scholar]
- 46.Joardar A, Pandia S, Chakraborty H. 2023. Effect of polyunsaturated free fatty acids on the membrane fusion mechanism. Soft Matter 19:733–742. doi: 10.1039/d2sm01474b. [DOI] [PubMed] [Google Scholar]
- 47.Dai J, Ting-Beall HP, Sheetz MP. 1997. The secretion-coupled endocytosis correlates with membrane tension changes in RBL 2H3 cells. J Gen Physiol 110:1–10. doi: 10.1085/jgp.110.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morel E, Codogno P. 2018. A novel regulator of autophagosome biogenesis and lipid droplet dynamics. EMBO Rep 19:e46858. doi: 10.15252/embr.201846858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trimarco JD, Heaton BE, Chaparian RR, Burke KN, Binder RA, Gray GC, Smith CM, Menachery VD, Heaton NS. 2021. TMEM41B is a host factor required for the replication of diverse coronaviruses including SARS-CoV-2. PLoS Pathog 17:e1009599. doi: 10.1371/journal.ppat.1009599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang ZB, Moutsatsos I, Moretti F, Bergman P, Zhang X, Nyfeler B, Antczak C. 2020. Fluopack screening platform for unbiased cellular phenotype profiling. Sci Rep 10:2097. doi: 10.1038/s41598-020-58861-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu YT, Shivakoti S, Jia F, Tao CL, Zhang B, Xu F, Lau P, Bi GQ, Zhou ZH. 2020. Biphasic exocytosis of herpesvirus from hippocampal neurons and mechanistic implication to membrane fusion. Cell Discov 6:2. doi: 10.1038/s41421-019-0134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, Wang CF, Ming SL, Li GL, Zeng L, Wang MD, Su BQ, Wang Q, Yang GY, Chu BB. 2020. Porcine IFITM1 is a host restriction factor that inhibits pseudorabies virus infection. Int J Biol Macromol 151:1181–1193. doi: 10.1016/j.ijbiomac.2019.10.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Chang H, Yang X, Zhao Y, Chen L, Wang X, Liu H, Wang C, Zhao J. 2015. Antiviral activity of porcine interferon regulatory factor 1 against swine viruses in cell culture. Viruses 7:5908–5918. doi: 10.3390/v7112913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu N, Zhang Z-F, Wang L, Gao B, Pang D-W, Wang H-Z, Zhang Z-L. 2012. A microfluidic platform for real-time and in situ monitoring of virus infection process. Biomicrofluidics 6:034122. doi: 10.1063/1.4756793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Li GL, Ming SL, Wang CF, Shi LJ, Su BQ, Wu HT, Zeng L, Han YQ, Liu ZH, Jiang DW, Du YK, Li XD, Zhang GP, Yang GY, Chu BB. 2020. BRD4 inhibition exerts anti-viral activity through DNA damage-dependent innate immune responses. PLoS Pathog 16:e1008429. doi: 10.1371/journal.ppat.1008429. [DOI] [PMC free article] [PubMed] [Google Scholar]