ABSTRACT
Oncogenic virus infections are estimated to cause ~15% of all cancers. Two prevalent human oncogenic viruses are members of the gammaherpesvirus family: Epstein-Barr virus (EBV) and Kaposi’s sarcoma herpesvirus (KSHV). We use murine herpesvirus 68 (MHV-68), which shares significant homology with KSHV and EBV, as a model system to study gammaherpesvirus lytic replication. Viruses implement distinct metabolic programs to support their life cycle, such as increasing the supply of lipids, amino acids, and nucleotide materials necessary to replicate. Our data define the global changes in the host cell metabolome and lipidome during gammaherpesvirus lytic replication. Our metabolomics analysis found that MHV-68 lytic infection induces glycolysis, glutaminolysis, lipid metabolism, and nucleotide metabolism. We additionally observed an increase in glutamine consumption and glutamine dehydrogenase protein expression. While both glucose and glutamine starvation of host cells decreased viral titers, glutamine starvation led to a greater loss in virion production. Our lipidomics analysis revealed a peak in triacylglycerides early during infection and an increase in free fatty acids and diacylglyceride later in the viral life cycle. Furthermore, we observed an increase in the protein expression of multiple lipogenic enzymes during infection. Interestingly, pharmacological inhibitors of glycolysis or lipogenesis resulted in decreased infectious virus production. Taken together, these results illustrate the global alterations in host cell metabolism during lytic gammaherpesvirus infection, establish essential pathways for viral production, and recommend targeted mechanisms to block viral spread and treat viral induced tumors.
IMPORTANCE Viruses are intracellular parasites which lack their own metabolism, so they must hijack host cell metabolic machinery in order to increase the production of energy, proteins, fats, and genetic material necessary to replicate. Using murine herpesvirus 68 (MHV-68) as a model system to understand how similar human gammaherpesviruses cause cancer, we profiled the metabolic changes that occur during lytic MHV-68 infection and replication. We found that MHV-68 infection of host cells increases glucose, glutamine, lipid, and nucleotide metabolic pathways. We also showed inhibition or starvation of glucose, glutamine, or lipid metabolic pathways results in an inhibition of virus production. Ultimately, targeting changes in host cell metabolism due to viral infection can be used to treat gammaherpesvirus-induced cancers and infections in humans.
KEYWORDS: Epstein-Barr virus, EBV, gammaherpesvirus, herpesvirus, Kaposi’s sarcoma herpesvirus, KSHV, lipidomics, metabolism, metabolomics, murine herpesvirus 68, MHV-68
INTRODUCTION
The oncogenic viruses Epstein-Barr virus (EBV) and Kaposi’s sarcoma herpesvirus (KSHV) are both members of the human gammaherpesvirus family. Over 90% of the human population is infected with EBV, with an estimated 137,900 to 208,700 cancer-related deaths in 2020 (1). Additionally, KSHV causes Kaposi’s sarcoma (KS) in 1 of every 200 transplant patients in the United States (2). Lytic replication studies in KSHV and EBV cell culture systems are challenging, because both viruses must be reactivated from latently infected cells to induce lytic replication and neither one can form plaques to quantify viral production (3, 4). Studying the induction of the lytic cycle during gammaherpesvirus infection is important, as it is the key to spreading virus in the host, can seed new tumors, and has implications in cancer maintenance (5).
Murine herpesvirus 68 (MHV-68), also known as murid herpesvirus 4, is a natural pathogen of wild rodents. It is a mouse gammaherpesvirus which shares ~80% of its open reading frames (ORFs) with EBV and KSHV (6). MHV-68 provides a genetically tractable model system that naturally undergoes both lytic replication and latent infection in various in vitro cell lines while also forming plaques for quantitation of virus production (7). Overall, MHV-68 infection is an ideal model system to study gammaherpesvirus lytic replication. Ultimately, MHV-68 can be used in animal models to elucidate how gammaherpesviruses influence oncogenesis within the host.
Oncogenic viral infection modifies numerous host cell mechanisms, including metabolism. Viruses are intracellular parasites which lack their own natural metabolism and therefore must commandeer the host cell metabolic machinery to generate both energy in the form of ATP and other critical biosynthetic building blocks (8). Increasing energy and central carbon metabolism to generate more amino acids, lipids, and nucleotides is essential for viral replication and the formation of new virions. Previous work by our group and others has identified major changes in central carbon metabolism during viral infection. For example, RNA viruses, such as dengue virus, influenza virus, Zika virus, and severe acute respiratory syndrome virus 2 (SARS-CoV-2), and DNA viruses, including herpes simplex virus-1 (HSV-1), human cytomegalovirus (HCMV), vaccinia virus (VACV), adenovirus, and KSHV, all induce significant changes in host cell glycolysis, glutamine metabolism, and fatty acid metabolism, revealing similarities and differences in modulating metabolism during infection (9–21). We previously showed that latent KSHV infection of endothelial cells induces glucose, glutamine, and lipid metabolism and that the induction of these metabolic pathways is necessary for the survival of latently infected cells (18–20). Studies of latent EBV infection have shown that methionine metabolism is key for maintaining latency (22). While critical alterations of metabolism during latent infection have been explored, metabolic changes during lytic infection with gammaherpesvirus are less well known (20, 23–26). A previous study of lytic KSHV infection showed that the induction of glycolysis, glutaminolysis, and fatty acid synthesis is required for viral replication; however, no global analyses of metabolic alterations were reported (21). While we know that these metabolic pathways are essential for maximal virus production, determining the viral mechanisms is important to identify potential therapeutic targets. Using MHV-68 as a strong genetically tractable system, it is possible to determine the cellular and viral mechanisms of metabolic modulation during gammaherpesvirus infections.
A few studies have begun to examine aspects of altered host metabolism during MHV-68 infection. Feeding MHV-68-infected mice a high-fat diet revealed that MHV-68 infection promotes fatty liver and elevated triglyceride levels (27). Another study showed that host cell cholesterol synthesis supports replication of MHV-68 in primary macrophages and that inhibiting cholesterol synthesis reduced virus production (28). A follow-up study in primary macrophages revealed that the regulation of cholesterol and fatty acid synthesis is in part due to the elevated expression of specific transcription factors called liver X receptors (LXRs) (29). A recent study further explored the role of hypoxia-inducible factor 1α (HIF1α) during MHV-68 infection, showing that lytic infection induces HIF1α protein expression, a global regulator of cell metabolism, and that blocking HIF1α regulation reduces virus production and reactivation in infected mouse cells (30). Overall, these previous studies highlighted individual metabolic pathways implicated in gammaherpesvirus virion production and opened the doors for further exploration of host cell metabolism changes during gammaherpesvirus lytic replication.
In this study, we define the global changes in the host cell metabolome and lipidome during gammaherpesvirus lytic replication and report that MHV-68 lytic infection induces glycolysis, glutaminolysis, lipid metabolism, and nucleotide metabolism. We also demonstrate that pharmacological inhibition of glycolysis or lipogenesis significantly reduce maximal MHV-68 viral production. Furthermore, we show that glutamine metabolism appears to be a more important pathway for virion production than glucose metabolism. Understanding how the gammaherpesvirus lytic cycle plays a role in altering global host cell metabolism can provide us with new therapeutic mechanisms to block gammaherpesvirus lytic replication and virus spread.
RESULTS
Metabolomics analysis reveals the importance of nucleotide metabolism during gammaherpesvirus lytic replication.
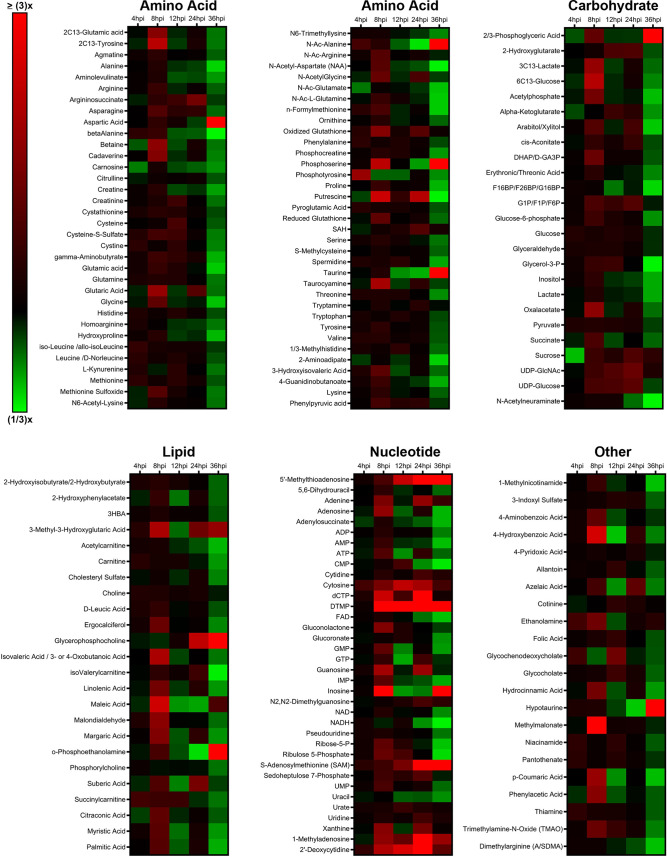
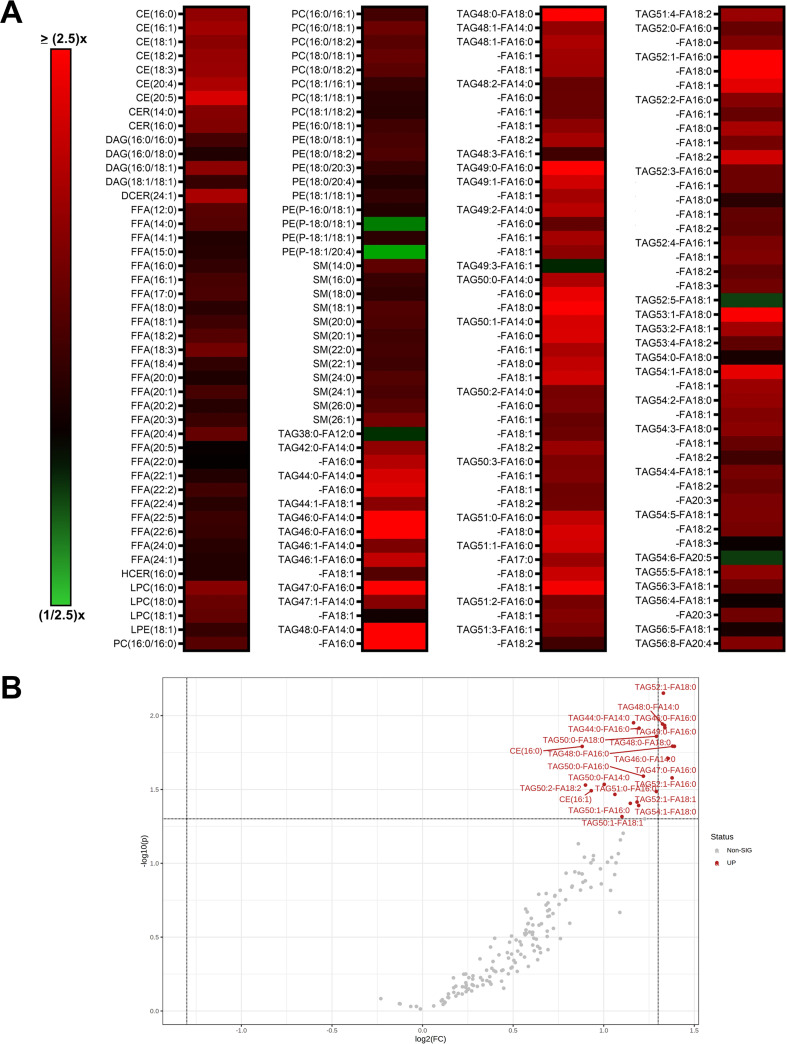
Metabolomics is a useful tool to determine global host cell changes during viral infections (9, 15, 18, 31–33). To profile the host cell metabolome during lytic MHV-68 infection, we harvested mock- and MHV-68-infected NIH 3T3 cells (multiplicity of infection [MOI] = 3) for targeted aqueous metabolomics analysis at 4, 8, 12, 24, and 36 h postinfection (hpi) in three independent experiments. A total of 176 metabolites were detected for all samples in biological triplicate at all time points; 24 lipid species, 69 amino acid species, 25 carbohydrate species, 36 nucleotide species, and 22 other species (Fig. 1; also, see Table S1 in the supplemental material). Nucleotide metabolism species contained the highest abundance of elevated metabolites throughout the virus life cycle compared to the other metabolic categories.
FIG 1.
Targeted aqueous metabolomics of mock versus MHV-68-infected cells. Mock-infected (control) and MHV-68-infected (MOI = 3) NIH 3T3 cells were harvested at 4, 8, 12, 24, and 36 hpi in three independent experiments. The relative fold change over the time course is depicted, where red shows an increase and green shows a decrease in metabolite concentration in MHV-68-infected cells compared to mock-infected cells. Metabolites are organized according to the major metabolic categories (carbohydrate, lipid, nucleotide, and amino acid) or as “other.”
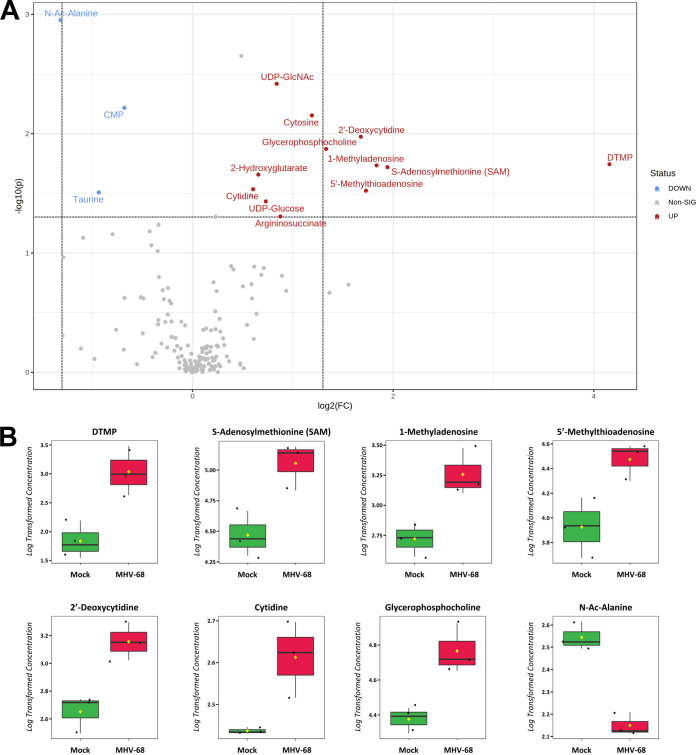
Our metabolomics analysis was performed in murine NIH 3T3 fibroblast cells. Previous studies examined the temporal changes in viral gene expression in various cell types that are lytically infected with MHV-68 (34, 35). MHV-68-infected fibroblasts were shown to contain peak amounts of viral DNA and peak mRNA expression of genes encoding viral structural proteins and viral DNA packaging proteins at 24 hpi (34). Therefore, we analyzed the top statistically significant metabolites at 24 hpi (P < 0.05 and at least a 1.5-fold change). Volcano plot analysis depicting the log2 fold change (FC) and −log10 P value in 24-hpi samples (t test, unpaired) highlighted 12 significantly elevated (Fig. 2A, red) and 3 significantly decreased (blue) metabolite species. Of the 12 elevated metabolites, 7 were nucleotide species and 2 others were nucleotide sugar species. dTMP was the most statistically significantly elevated aqueous metabolite in our metabolomics screen, reaching a 16-fold change over mock-infected cells by 24 hpi (Fig. 2A and B; Table S1). Additionally, there was elevation of other nucleotide species, such as the nucleotide cytosine, the pyrimidine nucleosides cytidine and 2′-deoxycytidine, and the purine nucleoside 1-methyladenosine (Fig. 2A and B; Table S1). S-Adenosylmethionine (SAM) and its downstream metabolite 5′-methylthioadenosine (MTA), which belong to the same class of organic compounds (5′-deoxy-5′-thionucleosides), are linked to both adenine nucleotide and polyamine metabolism (36, 37) and were both elevated in infected cells (Fig. 2A and B; Table S1). UDP-glucose and UDP-N-acetylglucosamine (UDP-GlcNAc), both pyrimidine nucleotide sugars, were elevated in infected cells (Fig. 2A; Table S1). UDP-glucose is a key intermediate in carbohydrate metabolism and a precursor to glycogen, sucrose, lipopolysaccharides, and glycosphingolipids (36). UDP-GlcNAc is a fructose-6-phosphate and glutamine derivative which is used as a substrate by glycosyltransferases to link N-acetylglucosamine to glycosaminoglycans, proteoglycans, and glycolipids (38, 39). Other elevated metabolite species in MHV-68-infected cells included 2-hydroxyglutarate (carbohydrate metabolism), glycerophosphocholine (lipid metabolism), and argininosuccinate (amino acid metabolism) (Fig. 2A and B; Table S1). Our analysis also yielded only three significantly decreased metabolite species in MHV-68-infected cells compared to mock-infected cells, including N-Ac-alanine (amino acid metabolism), CMP (nucleotide metabolism), and taurine (amino acid metabolism) (Fig. 2A and B; Table S1). Taken together, these data indicate that MHV-68-infected cells increase nucleotide metabolism during lytic replication at 24 hpi.
FIG 2.
Aqueous metabolic profiling of mock versus MHV-68-infected cells at 24 hpi. (A) Volcano plot depicting log2 fold change (FC) and −log10 P value (p) in 24-hpi samples (unpaired). The statistical threshold was set to a P value of <0.05 and at least a 1.5-fold change in MHV-68-infected cells compared to mock-infected cells using the Student’s t test. Red metabolites are significantly upregulated, and blue metabolites are significantly downregulated. (B) Box plots of the top 8 statistically significant metabolites (t test P < 0.05 and at least a 2-fold change). Black dots represent concentrations for all samples (biological triplicates). The horizontal line indicates the median, and the yellow diamond indicates the mean concentration.
MHV-68 lytic infection increases glucose metabolism.
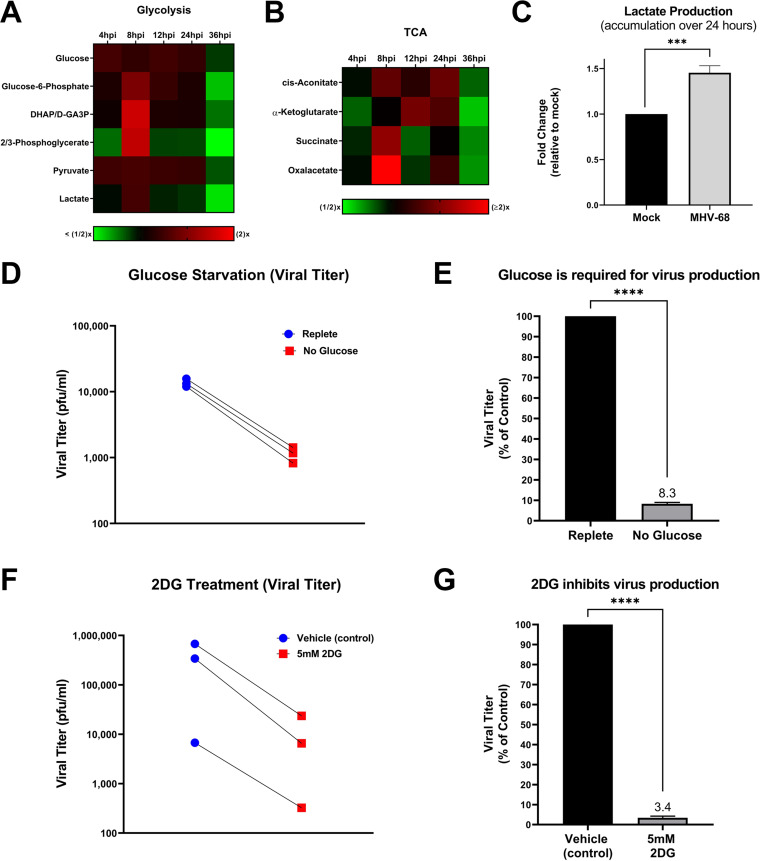
To determine if glucose metabolism is modulated during the MHV-68 lytic life cycle, we analyzed our aqueous metabolomics data for the abundance of metabolites in the glycolysis metabolic pathway and the tricarboxylic acid (TCA) cycle. Fold change was calculated as the ratio of metabolite abundance in MHV-68-infected cells to that in mock-infected cells. Our metabolomics analysis revealed that MHV-68 infection increased all detected glycolytic intermediates at 8 hpi, an early part of the viral life cycle (Fig. 3A). Most TCA cycle metabolic intermediates also increased in abundance at 8 hpi, with oxaloacetate displaying the highest fold change in abundance out of all the TCA cycle metabolites (Fig. 3B).
FIG 3.
MHV-68 increases glucose metabolism and requires glucose for infectious virion production. (A and B) Mock (control) and MHV-68-infected (MOI = 3) NIH 3T3 cells were harvested at 4, 8, 12, 24, and 36 hpi in three independent experiments. The relative fold change over the time course for glycolytic and TCA cycle intermediates is shown, where red depicts an increase and green shows a decrease in MHV-68-infected cells compared to mock-infected cells. (C) Accumulated lactate production (over 24 h) is increased in MHV-68-infected (MOI = 3) NIH 3T3 cells compared to mock-infected (control) cells. Cellular supernatant was collected at 24 hpi and accumulated lactate production was quantified using the Promega Lactate-Glo assay kit in three independent experiments. Data were normalized to cell number. (D and E) NIH 3T3 cells were MHV-68 infected (MOI = 0.1) in three independent experiments. Cells were fed replete medium (containing glucose and glutamine) for 24 h, which was replaced with fresh replete (control) or glucose-free medium for an additional 24 h. Viral titer was quantified from cell supernatants via plaque assays and graphed as PFU per milliliter or as a percentage of the control (replete medium). (F and G) NIH 3T3 cells were MHV-68 infected (MOI = 0.1) and treated with 5 mM 2DG or vehicle (control) for 48 hpi in three independent experiments. Viral titer was quantified from cell supernatants via plaque assays and graphed as PFU per milliliter or as a percentage of the control (vehicle treatment). ***, P < 0.001; ****, P < 0.0001.
To determine the overall glycolytic output in MHV-68-infected cells versus mock-infected cells, we measured the amount of accumulated lactate produced in cell supernatants over 24 h. Lactate is traditionally produced as the final product of glycolysis during anaerobic respiration, where pyruvate is converted to lactate and secreted outside the cell. However, recent data show that viral infections commonly increase lactate production under aerobic conditions, in combination with an increase in glycolysis (8, 18, 40). Mock- and MHV-68-infected cells (MOI = 3) were harvested at 24 hpi and accumulated lactate production was assessed in the cellular supernatant using an enzymatic assay. The data were normalized to the total cell number for each sample. As seen in Fig. 3C, there is an ~50% increase in accumulated lactate production in MHV-68-infected cells compared to mock-infected cells. It is important to note that the metabolomics analysis whose results are shown in Fig. 3A provides a snapshot of lactate levels inside cell samples at various time points, revealing peak lactate levels in MHV-68-infected cells compared to mock-infected cells at 8 hpi and a decrease in lactate levels in infected cells at 24 hpi. However, the lactate production assay whose results are shown in Fig. 3C measures the total accumulation of secreted lactate over 24 h (0 hpi to 24 hpi) in cellular supernatants from mock or MHV-68-infected cells. Overall, these data indicate that MHV-68-infected cells increase glycolysis during lytic infection.
MHV-68 lytic replication requires glucose for maximal virus production.
To assess if glucose, the main carbon source for cells, is important for MHV-68 virion production, we quantified viral production at 48 hpi (one to two MHV-68 replication cycles) in the presence or absence of glucose. More specifically, mock- and MHV-68-infected cells (MOI = 0.1) were first fed replete medium (containing glucose and glutamine) for 24 h to minimize glucose starvation-induced cell death. After 24 h, the replete medium was replaced with fresh replete medium (control) or glucose-free medium for an additional 24 h. At 48 hpi, viral titers in MHV-68-infected-cell supernatants were determined via plaque assays, and the number of PFU per milliliter was calculated. Cell viability and cell numbers of both mock- and MHV-68-infected cells were determined via trypan blue exclusion assays (Table 1). Glucose starvation of infected cells resulted in decreased viral titers compared to treatment of cells with replete medium (Fig. 3D and E). While glucose starvation in both mock- and MHV-68-infected cells decreased both cell viability and cell number, our data revealed an ~4-fold decrease in viral production in glucose-starved cells compared to cells in replete medium (control) when viral titer was normalized to live-cell number (Table 1). Overall, these data reveal that glucose is required for virus production.
TABLE 1.
Normalized reduction in virus production in treated compared to control MHV-68-infected NIH 3T3 cells
| Condition and treatmenta | Avg no. of cells |
% viability | Avg virus concn (PFU/mL) | Pfu to live-cell no. ratiob | Normalized reduction in virus production (fold change)c | |
|---|---|---|---|---|---|---|
| Total | Live | |||||
| Glucose starvation | ||||||
| Mock infection (replete medium) | 2,359,000 | 2,353,000 | 100 | |||
| Mock infection (no glucose) | 815,600 | 668,300 | 82 | |||
| MHV-68 (replete medium) | 1,758,000 | 1,751,000 | 100 | 13,666 | 7.81 × 10−3 | |
| MHV-68 (no glucose) | 611,500 | 519,600 | 85 | 1,141 | 2.20 × 10−3 | 4 |
| Glutamine starvation | ||||||
| Mock infection (replete medium) | 2,094,000 | 2,089,000 | 100 | |||
| Mock infection (no glutamine) | 867,500 | 858,400 | 99 | |||
| MHV-68 (replete medium) | 2,022,000 | 2,019,000 | 100 | 16,666 | 82.55 × 10−4 | |
| MHV-68 (no glutamine) | 649,700 | 647,000 | 100 | 62 | 0.95 × 10−4 | 87 |
| 2DG treatment | ||||||
| Mock infection (vehicle) | 2,652,333 | 2,642,667 | 100 | |||
| Mock infection (5 mM 2DG) | 844,333 | 837,667 | 99 | |||
| MHV-68 (vehicle) | 1,841,667 | 1,832,333 | 99 | 341,000 | 18.61 × 10−2 | |
| MHV-68 (5 mM 2DG) | 747,333 | 741,067 | 99 | 10,171 | 1.37 × 10−2 | 14 |
| TOFA treatment | ||||||
| Mock infection (vehicle) | 2,385,333 | 2,374,667 | 100 | |||
| Mock infection (10 μg/mL TOFA) | 1,222,667 | 1,219,000 | 100 | |||
| MHV-68 (vehicle) | 1,907,000 | 1,901,667 | 100 | 9,667 | 50.83 × 10−4 | |
| MHV-68 (10 μg/mL TOFA) | 1,146,000 | 1,136,333 | 99 | 57 | 0.50 × 10−4 | 102 |
NIH 3T3 cells were infected with MHV-68 (MOI = 0.1) or mock infected, and cells were treated or left untreated (replete medium or vehicle).
Average number of PFU per milliliter divided by the average live-cell number.
Calculated by dividing the ratio of the number of PFU per milliliter to live infected cell number in treated cells by the same ratio for untreated cells. Normalized fold change depicts the reduction in virus production due to each treatment type.
To assess the need for glycolysis in viral production, we treated cells with the glycolytic inhibitor 2-deoxy-d-glucose (2DG). 2DG is a glucose analog which attenuates glycolysis by competing for the active site of hexokinase, which normally converts glucose to glucose-6-phosphate. To test if interfering with glycolysis results in decreased virus production, mock- and MHV-68-infected cells (MOI = 0.1) were treated with vehicle (solvent control) or 5 mM 2DG for 48 h. Viral titers from MHV-68-infected cell supernatants were quantified via plaque assays, and cell viability and cell number were determined via trypan blue exclusion assays (Table 1). 2DG treatment significantly decreased virus production compared to control treatment (Fig. 3F and G). After normalization of viral titer to live-cell number, our data showed an ~14-fold decrease in viral production in 5 mM 2DG-treated cells compared to control-treated cells (Table 1). Taken together, these data indicate that glucose and increased glycolysis are important for viral production.
MHV-68 lytic infection increases glutamine metabolism.
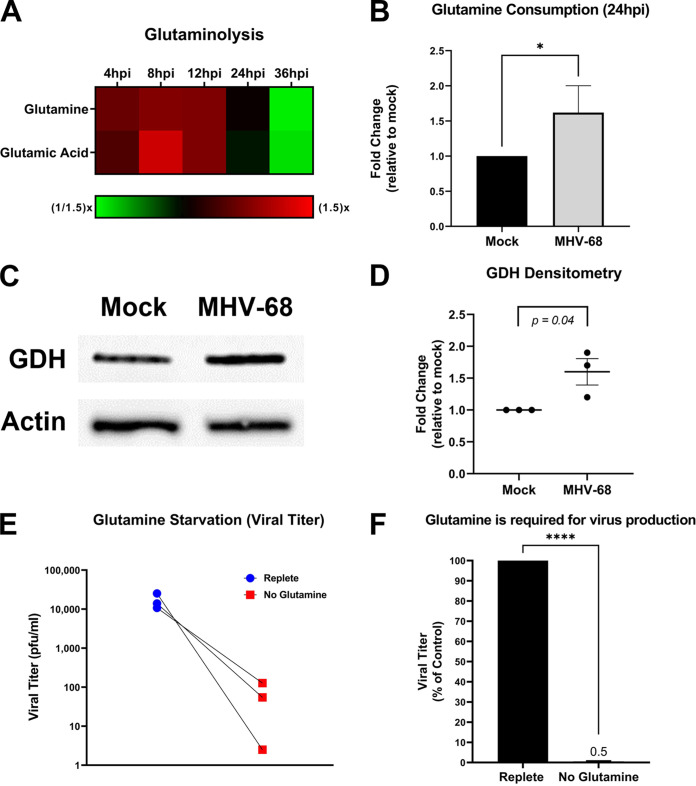
Glutamine is an alternate carbon source to glucose. During glutaminolysis, glutamine is taken in by cells and converted to glutamic acid by the enzyme glutaminase (GLS). Glutamic acid is next converted by the enzyme glutamate dehydrogenase (GDH) to α-ketoglutarate, which then enters the mitochondria. Overall, glutamine metabolism can fuel and replenish the TCA cycle, ultimately supporting increased ATP production and de novo lipid synthesis (lipogenesis). To determine if MHV-68 infection increases glutamine metabolism, we analyzed our aqueous metabolomics data for the abundance of metabolites in the glutaminolysis pathway (glutamine and glutamic acid). Fold change was calculated as a ratio of metabolite abundance in MHV-68-infected cells compared to mock-infected cells. Our metabolomics analysis revealed that MHV-68 infection increased both glutamine and glutamic acid metabolites at 4, 8, and 12 hpi (Fig. 4A). Next, we measured glutamine uptake in mock- versus MHV-68-infected cells (MOI = 3). Mock- and MHV-68-infected cells were harvested at 24 hpi, and the amount of glutamine consumption in the cellular supernatant was assessed using an enzymatic assay. The data were normalized to total cell number. As seen in Fig. 4B, there is an ~60% increase in glutamine consumption in MHV-68-infected cells compared to mock-infected cells. We then determined the expression of glutaminolysis enzymes GLS and GDH in mock- versus MHV-68-infected cells (MOI = 3) at 24 hpi. Western blot analysis showed no change in GLS expression (data not shown) but did show an average ~60% increase in GDH expression in MHV-68-infected cells compared to mock-infected cells after densitometry analysis (Fig. 4C and D). Taken together, these data indicate that MHV-68-infected cells increase glutamine metabolism during lytic infection.
FIG 4.
MHV-68 increases glutamine metabolism and requires glutamine for infectious-virion production. (A) Mock-infected (control) and MHV-68-infected (MOI = 3) NIH 3T3 cells were harvested at 4, 8, 12, 24, and 36 hpi in three independent experiments. The relative fold change over the time course for glutaminolysis intermediates is shown, where red depicts an increase and green shows a decrease in MHV-68-infected cells compared to mock-infected cells. (B) Glutamine consumption is increased in MHV-68-infected (MOI = 3) NIH 3T3 cells compared to mock-infected (control) cells. Cellular supernatant was collected at 24 hpi, and glutamine consumption was quantified using the Promega Glutamine-Glo assay kit in three independent experiments. Data were normalized to cell number. (C and D) Western blot analysis of glutamate dehydrogenase in mock versus MHV-68-infected (MOI = 3) NIH 3T3 cells at 24 hpi in three independent experiments. Densitometry analysis results are shown as fold change compared to mock-infected cells, with actin used as a loading control. A ratio paired Student’s t test was used to calculate the P value of densitometry data. (E and F) NIH 3T3 cells were MHV-68 infected (MOI = 0.1) in three independent experiments. Cells were fed replete medium (containing glucose and glutamine) or glutamine-free medium for 48 h. Viral titer was quantified from cell supernatants via plaque assays and graphed as PFU per milliliter or as a percentage of the control (replete medium). *, P < 0.05; ****, P < 0.0001.
Glutamine is important for MHV-68 virion production.
To assess the importance of glutamine for viral production, mock or MHV-68-infected cells (MOI = 0.1) were fed replete medium (containing glucose and glutamine) or glutamine-free medium for 48 h. Viral titers from MHV-68-infected cell supernatants were determined via plaque assays, and cell viability and cell number of both mock- and MHV-68-infected cells were determined via trypan blue exclusion assays (Table 1). Glutamine starvation dramatically decreased viral titer compared to treatment of cells with replete medium (Fig. 4E and F). After viral titer was normalized to live-cell number, our data showed an ~87-fold decrease in viral production in glutamine-starved cells compared to cells fed replete medium (Table 1). Overall, glutamine starvation of infected cells revealed that glutamine is essential for MHV-68 production.
Lipidomic profiling of lytic gammaherpesvirus infection reveals lipid remodeling throughout the viral life cycle.
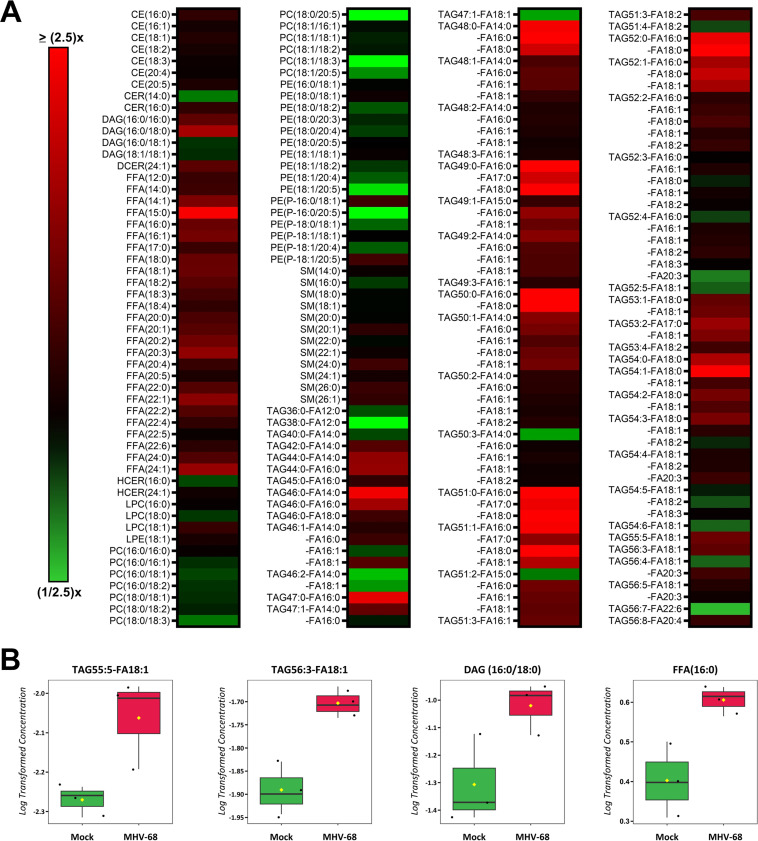
Lipids are a complex class of biological molecules which are important components of cell membranes and sources of cellular energy. Lipidomics is a bulk and quantitative analysis of lipids in cell samples which allows us to understand changes in lipid metabolism by creating a lipid profile of major lipid classes, subclasses, and individual species (41). To profile the host cell lipidome during lytic MHV-68 infection, we harvested mock- and MHV-68-infected NIH 3T3 cells (MOI = 3) for targeted lipidomics analysis at 4, 8, 12, 24, and 36 hpi in three independent experiments. Our lipidomics analysis detected ~300 unique lipid species at some point during the course of infection (Table S2). The categories of lipid species detected included cholesterol ester (CE), ceramides (CER), diacylglycerol (DAG), dihydroceramides (DCER), free fatty acids (FFA), hexosylceramides (HCER), lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), phosphatidylcholine (PC), phosphatidylethanolamine (PE), sphingomyelin (SM), and triacylglycerol (TAG). A detailed lipidomics analysis at 8 hpi, which detected 183 lipid species, revealed that MHV-68-infected cells had an increased abundance of 177 of 183 (97% of detected lipids) and decreased abundance of 54 of 183 (3% of detected lipids) compared to mock-infected cells (Fig. 5A and Table S2). We next used volcano plot analysis to uncover the top statistically significant lipid species metabolites at 8 hpi (t test P < 0.05 and at least a 1.8-fold change). This analysis highlighted 22 significantly elevated lipid species (Fig. 5B, red) and 0 significantly decreased lipid species. Of these 22 elevated lipid species, 20 were TAG species and 2 were CE lipid species. CEs are dietary lipids composed of a cholesterol molecule, fatty acids, and a hydroxyl group. TAGs, which are composed of three fatty acid tails and a glycerol backbone, can be hydrolyzed to release free fatty acids, which are a major source of energy for cells.
FIG 5.
Quantitative lipidomics of mock versus MHV-68-infected cells at 8 hpi. Mock-infected (control) and MHV-68-infected (MOI = 3) NIH 3T3 cells were harvested at 8 hpi in three independent experiments. CE, cholesterol ester; CER, ceramides; DAG, diacylglycerol; DCER, dihydroceramides; FFA, free fatty acids; HCER, hexosylceramides; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SM, sphingomyelin; TAG, triacylglycerol. (A) Relative fold change; red shows an increase and green shows a decrease in lipid concentration in MHV-68-infected cells compared to mock-infected cells. (B) Volcano plot depicting log2 fold change (FC) and −log10 P value (p) in 8-hpi samples (paired). The statistical threshold was set to a P value of <0.05 and at least a 1.8-fold change in MHV-68-infected cells compared to mock-infected cells using Student’s t test. Red metabolites are significantly upregulated.
Next, we analyzed our lipidomics data at 12 hpi, which detected 211 lipid species, and found that MHV-68-infected cells increased the abundance of 157 of 211 (74% of detected lipids) and decreased the abundance of 54 of 211 (26% detected lipids) compared to mock-infected cells (Fig. 6A and Table S2). We next used Student’s t test to identify the 4 most significant lipid species at 12 hpi (P < 0.05, paired test, and at least 1.5-fold change). Our analysis revealed a 1.9-fold increase in DAG(16:0/18:0), a 1.6-fold increase in TAG55:5-FA18:1, a 1.6-fold increase in FFA(16:0), and a 1.5-fold increase in TAG56:3-FA18:1 in MHV-68-infected cells compared to mock-infected cells (Fig. 6B and Table S2).
FIG 6.
Quantitative lipidomics of mock versus MHV-68-infected cells at 12 hpi. Mock-infected (control) and MHV-68-infected (MOI = 3) NIH 3T3 cells were harvested at 4, 8, 12, 24, or 36 hpi in three independent experiments. CE, cholesterol ester; CER, ceramides; DAG, diacylglycerol; DCER, dihydroceramides; FFA, free fatty acids; HCER, hexosylceramides; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SM, sphingomyelin; TAG, triacylglycerol. (A) Relative fold change is depicted; red shows an increase and green shows a decrease in lipid concentration in MHV-68-infected cells compared to mock-infected cells. (B) Box plots of the top four statistically significant metabolites (t test P < 0.05 and at least a 1.5-fold change). Black dots represent concentrations for all samples (biological triplicates). The horizontal line indicates the median, and yellow diamond indicates the mean concentration.
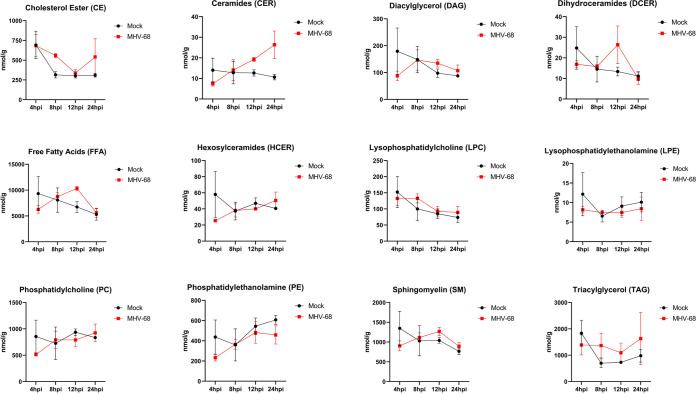
To evaluate the changes in each lipid class over the course of infection, we graphed the pooled concentrations of each lipid class at 4, 8, 12, and 24 hpi (Fig. 7). This time course analysis revealed notable increases in CE at 8 hpi and 24 hpi, CER at 12 and 24 hpi, DCER at 12 hpi, and SM at 12 and 24 hpi in MHV-68-infected cells compared to mock-infected cells (Fig. 7). Furthermore, comparison of the TAG, FFA, and DAG lipid classes in mock- versus MHV-68-infected cells over the course of infection revealed overall increases in TAGs after 8 hpi, increasing and peaking FFA at 12 hpi, and increasing and peaking DAGs at 8 to 12 hpi before decreases in concentration (Fig. 7). Interestingly, all three of these lipid species are part of the same metabolism pathway. FFA can be generated as 16-carbon fatty acids by lipogenesis and then elongated, or they can be generated by the catabolic lipolysis of TAGs. DAGs are a glycerol molecule linked to two fatty acids and can also be generated by the creation or breakdown of TAGs. Overall, these data point to an early increase in TAG lipids (8 hpi) and then an increase in FAAs and DAG at 12 hpi, by increased dietary intake of TAGs and subsequent lipolysis, lipogenesis, or both.
FIG 7.
Total lipid class concentration time course of mock-infected versus MHV-68-infected cells. Mock-infected (control) and MHV-68-infected NIH 3T3 cells were harvested at 4, 8, 12, 24, or 36 hpi in three independent experiments. CE, cholesterol ester; CER, ceramides; DAG, diacylglycerol; DCER, dihydroceramides; FFA, free fatty acids; HCER, hexosylceramides; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SM, sphingomyelin; TAG, triacylglycerol.
MHV-68 lytic infection increases fatty acid metabolism.
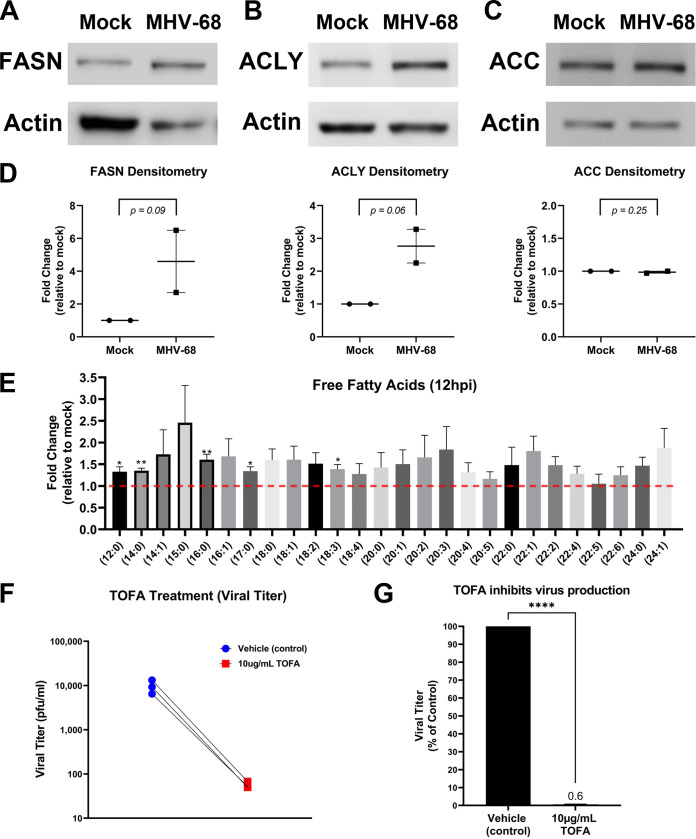
De novo fatty acid synthesis (lipogenesis) results in the generation of new fatty acids and lipid materials. During lipogenesis, citrate is exported from the TCA cycle in the mitochondria and moves into the cytoplasm, where it is converted to cytoplasmic acetyl coenzyme A (acetyl-CoA) by ATP citrate lyase (ACLY) and then malonyl-CoA by acetyl-CoA carboxylase (ACC). Acetyl-CoA and malonyl-CoA combine to form the saturated 16-carbon fatty acid palmitic acid by fatty acid synthase (FASN), and the palmitic acid is then used to make longer-chain fatty acids and lipid materials. The induction of lipogenic enzymes is a key indicator of increased fatty acid synthesis in cells. Our Western blot analysis of mock or MHV-68-infected cells (MOI = 3) at 24 hpi showed an increase in the lipogenic enzymes FASN and ACLY and no change in ACC expression (Fig. 8A to C). Densitometry analysis of these Western blot data (Fig. 8D) showed an average ~4.6-fold increase in FASN expression, ~2.8-fold increase in ACYL expression, and no change in ACC expression. Next, we analyzed our lipidomics data for the abundance of individual free fatty acids at 12 hpi (Fig. 8E). Fold change was calculated as the ratio of metabolite abundance in MHV-68-infected cells to that in mock-infected cells. Our analysis revealed that MHV-68 infection (MOI = 3) increased the abundance of all saturated and unsaturated FFA detected by this lipidomics screen (Fig. 8E). The 16-carbon saturated FFA showed the most significant change, with a 1.6-fold increase in MHV-68-infected cells compared to mock-infected cells at 12 hpi (P < 0.01). Taken together with our lipidomics data, which show overall increases in most lipid classes throughout the virus life cycle (Fig. 7), these data suggest an increase in both lipogenesis and dietary TAG uptake and lipolysis in cells with active MHV-68 lytic infection.
FIG 8.
MHV-68 increases and requires lipogenesis for infectious virion production. (A to D) Western blot analysis of fatty acid synthase (FASN), ATP-citrate lyase (ACLY), and acetyl-CoA carboxylase (ACC) in mock-infected versus MHV-68-infected (MOI = 3) NIH 3T3 cells at 24 hpi in two independent experiments. Densitometry analysis results are shown as fold change compared to mock-infected cells, with actin used as a loading control. Ratio paired Student’s t test was used to calculate the P value of densitometry data. (E) Quantitative lipidomics of free fatty acids in mock-infected (control) and MHV-68-infected (MOI = 3) NIH 3T3 cells at 12 hpi (three independent experiments). Unpaired t test (P < 0.05) data are graphed on the y axis as fold change relative to mock infection. The red dashed line at 1.0 indicates no change compared to mock-infected cells. (F and G) NIH 3T3 cells were MHV-68 infected (MOI = 0.1) and treated with 10 μg/mL TOFA or vehicle (control) for 48 hpi in three independent experiments. Viral titer was quantified from cell supernatants via plaque assays and graphed as PFU per milliliter or as a percentage of the control (vehicle treatment). ****, P < 0.0001.
Lipogenesis is required for maximal virion production during lytic MHV-68 infection.
The use of lipogenic inhibitors has been shown to block replication of several viruses, demonstrating that these metabolic alterations are an important part of the viral life cycle (21, 42–45). 5-(Tetradecyloxy)-2-furoic acid (TOFA) is a pharmacological drug that inhibits the enzyme ACC in the lipogenesis metabolism pathway. To test if induction of lipogenesis is necessary for MHV-68 virion production, we used mock- and MHV-68-infected cells (MOI = 0.1) and treated them with vehicle control or 10 μg/mL TOFA. Viral titers from MHV-68-infected-cell supernatants were determined via plaque assays, and the cell viability and cell numbers of both mock- and MHV-68-infected cells were determined via trypan blue exclusion assays (Table 1). TOFA treatment substantially decreased virus production compared to vehicle treatment (Fig. 8F and G). After normalization of viral titer to live cell number, our data show an ~102-fold decrease in viral production in cells treated with 10 μg/mL TOFA compared to controls (Table 1). These data suggest that induction of lipogenesis driven by MHV-68 is necessary for virion production and that fatty acid synthesis inhibitors are potential targets for gammaherpesvirus infections.
DISCUSSION
While metabolomics analysis of host cells during viral infection has been carried out with multiple viruses (14, 16, 19, 31–33, 46–48), this study comprehensively analyzed changes in the host cell metabolome during gammaherpesvirus lytic replication. Our targeted aqueous metabolomics analysis revealed that lytic MHV-68 infection induces many metabolic pathways, including glucose, glutamine, lipid, and nucleotide metabolism (Fig. 9). When we analyzed the top 12 statistically significant metabolites at 24 hpi, we discovered that 9 of them were nucleotide-related species (Fig. 2A). The gammaherpesviruses KSHV, EBV, and MHV-68 all encode their own thymidine kinase, which aids in the synthesis of de novo dTMP and ultimately supports robust nucleotide production for viral replication (49–51). Not surprisingly, the top elevated metabolite in our metabolomics screen was dTMP (Fig. 2). Other significant metabolites included the nucleotide cytosine and related nucleosides cytidine and 2′-deoxycytidine, key building blocks in the creation of DNA and RNA (Fig. 2). More recently, cytidine metabolism was revealed to be important during EBV-induced B cell transformation, growth, and survival (52), highlighting its importance in gammaherpesvirus induced oncogenesis. The purine nucleoside 1-methyladenosine showed a significant increase during MHV-68 infection (Fig. 2). Interestingly, during HIV-1 infection, it was revealed that HIV-1 virions contain abundant amounts of 1-methyladenosine, but this modification was shown to be linked solely to viral packaged tRNAs and not viral RNA and used to prime viral reverse transcriptase (53, 54). While gammaherpesviruses are DNA viruses and not retroviruses, it would be interesting to explore the role of 1-methyladenosine during gammaherpesvirus infection.
FIG 9.
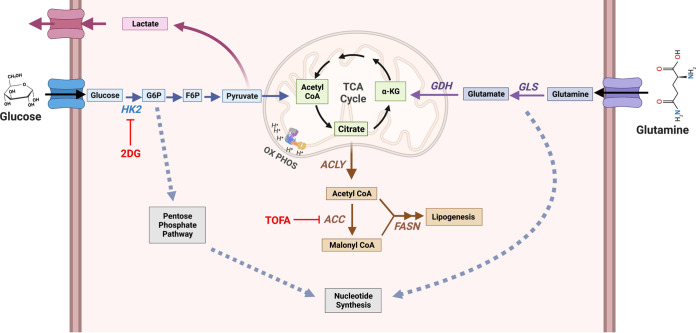
Model of host cell metabolism changes during lytic gammaherpesvirus infection. Our global metabolomics and lipidomics indicate that during MHV-68 lytic infection of host NIH 3T3 cells, metabolic intermediates found in glycolysis, glutaminolysis, lipogenesis, and nucleotide metabolism are significantly elevated over mock-infected controls. Briefly, glucose is taken up by the host cell via membrane-bound glucose transporter proteins and metabolized by hexokinase (HK) to form glucose-6-phosphate (G-6-P). Depending on cellular or viral need, G-6-P can have several fates, including (i) being further metabolized through glycolysis to form pyruvate to fuel the TCA cycle in the mitochondria, (ii) being converted from pyruvate to lactate and secreted outside the cell, or (iii) fueling the pentose phosphate pathway for nucleotide synthesis. During glutaminolysis, glutamine is taken up by the host cell via a membrane-bound glutamine transporter. Glutamine is converted to glutamate and NH3 by glutaminase (GLS) and then to α-ketoglutarate by glutamate dehydrogenase (GDH). Next, α-ketoglutarate enters and replenishes the TCA cycle. Glutamine breakdown can be used for nucleotide synthesis. Overall, downstream intermediates from both glucose and glutamine, which feed into the TCA cycle, can fuel the lipogenesis pathway. Briefly, citrate is converted to cytoplasmic acetyl-CoA by ATP citrate lyase (ACLY) and then malonyl-CoA by acetyl-CoA carboxylase (ACC). Acetyl-CoA and malonyl-CoA combine to form palmitic acid by fatty acid synthase (FASN), which is then used as a backbone to make longer-chain fatty acids, triglycerides, phospholipids, and other lipid materials. During MHV-68 lytic infection, the expression of key enzymes in these pathways, including GDH, ACLY, and FASN, are elevated. Figure created with BioRender.com.
Metabolomics analysis of two different herpesviruses, HCMV, which has an ~96-h life cycle, and HSV-1, which has an ~24-h life cycle, revealed divergent effects on cellular metabolism (14). While HSV-1 increased metabolic influx through the TCA cycle and pyrimidine nucleotide biosynthesis, HCMV increased glycolysis and the TCA cycle to support fatty acid synthesis. Because there is evidence that viruses trigger distinct metabolome changes in their host cells but overall often target central carbon metabolism, it is important to metabolically profile viral families and their target host cell types to understand which points in cellular metabolism would provide the best therapeutic targets. This approach will reveal both similarities and differences between host-pathogen interactions across herpesviruses.
Glucose, the main central carbon source of cells, feeds into glycolysis and subsequently fuels cellular ATP production. Glucose is also an important carbon source to fuel other metabolic pathways, such as the pentose phosphate pathways for nucleotide production and lipogenesis through the TCA cycle for de novo fat production (Fig. 9) (55). The catabolism of glutamine, an alternative carbon source to glucose, replenishes the TCA cycle. This is particularly important when the TCA cycle is depleted due to glucose being metabolized to lactate instead of pyruvate and citrate being shuttled out of the TCA cycle to support fatty acid synthesis (Fig. 9). Therefore, glutamine breakdown and fueling of the TCA cycle ultimately supports increased ATP production and lipid synthesis (Fig. 9). Glutamine breakdown also provides an important nitrogen source for nucleotide synthesis (56, 57). Interestingly, increased glucose and glutamine metabolism have been shown to be hallmarks of cancer and are often required for induction of lipogenesis (58, 59). We and others have previously investigated whether glucose or glutamine metabolism is induced by infection or required for viral production. For example, we showed that latent KSHV infection requires both glucose and glutamine for the maintenance of latently infected endothelial cells (18, 20). Furthermore, we showed that induction of lytic replication during KSHV infection was reduced when host cells were starved of glucose or glutamine, with glucose being more important than glutamine for virion production (21). However, studies with HCMV and VACV show that glutamine but not glucose is required for infectious-virus production (16, 60). These studies reveal that the need for glucose or glutamine during infection is virus dependent, and understanding which viruses prefer glucose versus glutamine can help provide clues for more selective viral therapeutics.
Our metabolomics data show an increase in glycolytic and TCA intermediates at 8 hpi (Fig. 3 and Table S1). We also show an increase in accumulated lactate production, an output of glycolysis, at 24 hpi (Fig. 3C). We and others have shown that 2DG treatment, which attenuates glycolysis, inhibits MHV-68 production (Fig. 3F and G; Table 1) (30, 61), demonstrating that glycolysis is required for viral production. Additionally, our metabolomics data show an increase in glutaminolysis intermediates at 4 hpi, 8 hpi, and 12 hpi (Fig. 4A). Furthermore, we observed an increase in glutamine consumption and glutamate dehydrogenase expression at 24 hpi, highlighting an overall increase in glutamine metabolism during MHV-68 lytic replication (Fig. 4B to D). Interestingly, we demonstrated that glucose starvation of host cells led to an ~4-fold decrease in infectious virus production, while glutamine starvation of host cells led to an ~87-fold decrease in virus production (Table 1). While our data demonstrate that MHV-68-infected cells increase both glucose and glutamine metabolism, they also indicate that virus production is more sensitive to glutamine loss than glucose loss. It is possible that glutamine may be necessary to feed multiple host cell metabolic pathways, including the TCA cycle for energy production and lipogenesis and nucleotide metabolism for the synthesis of de novo DNA and RNA (Fig. 9). Further labeled-glutamine flux experiments are needed to track glutamine utilization to ultimately determine its role during MHV-68 lytic infection and its necessity during viral production.
Essential alterations in the cellular lipid profile have been observed for many tumor models (62–65). Induced lipid metabolism fills energy stores for the cell as well as gives rise to biosynthetic material for cellular membranes and other lipid intermediates. Lipid metabolism has been proven to be critical for tumorigenesis and tumor cell survival (64). As with human gammaherpesvirus, one of the most important implications of studying MHV-68 infection is understanding virus-induced oncogenesis and tumor formation. Our lipidomics data reveal that a clear gammaherpesvirus lipid profile is established during lytic gammaherpesvirus infection (Fig. 5 to 7; Table S2). We show that pharmacological inhibition of lipogenesis blocks virus production, indicating that gammaherpesvirus virion production is dependent on this modulation in lipid metabolism (Fig. 8F and G; Table 1).
Previous work has shown that other herpesviruses also depend on alterations of lipid metabolism (14, 19, 21, 29, 66–69). Inhibition of lipogenesis, specifically, synthesis of phosphatidylethanolamine (PE), has been shown to inhibit viral production and budding in HSV-1 (70). While we did not detect a change in PE in our data, we did measure an increase in CE detection at 8 h after MHV-68 infection (Fig. 7). This suggests that distinct lipid requirements may be critical for dissemination of different herpesviruses. Additionally, HCMV induces adipocyte-like conditions by significantly inducing lipogenesis (66). Specifically, sterol regulatory element-binding transcription factor 1a (SREBP1a) and SREB1c are involved in mediating this adipocyte-like phenotype and the inhibition of SREBP1 leads to impaired HCMV growth. It is possible that SREBP1 contributes to the mechanism by which gammaherpesviruses induce lipogenesis, as we saw a significant increase in numerous lipid species in our data. Finally, several RNA viruses also hijack host lipid metabolism, including dengue virus, hepatitis C virus, influenza virus, and SARS-CoV-2 (45, 71, 72). Positive-sense, single-stranded RNA viruses enhance lipid droplet formation during early infection and hijack membranes during replication (73). Inhibiting lipid biosynthesis and formation of lipid droplets negatively impacts viral replication of these viruses. Exploring the role of lipid droplets and lipid droplet composition during lytic gammaherpesvirus infection could further indicate whether these structures and their components are essential for viral replication and virion production.
Overall, previous work has shown that central carbon metabolism is essential for the replication of KSHV, a related gammaherpesvirus (21). We showed that inhibition of glycolysis, glutamine metabolism, and fatty acid synthesis blocked KSHV replication at distinct steps of viral gene transcription, translation, and virion assembly. While it is possible that additional metabolic alterations are required for maximal KSHV replication and virion production, a global analysis of metabolic changes was not included. Our global metabolomics and lipidomics data obtained with MHV-68, a model gammaherpesvirus, demonstrate that gammaherpesvirus replication is highly dependent on induced central carbon metabolism. We show that glucose, glutamine, and lipid metabolism are essential for maximal infectious virus production, and inhibition of these pathways via nutrient depletion or drug inhibition results in a significant reduction of virus production (Fig. 3, 4, and 8). Determining that these key metabolic pathways are critical for the production of infectious virions is the first step in selecting molecular targets for pharmacological inhibition. Our findings provide a framework for future in vivo experiments in which high-priority metabolic inhibitors can be tested to block virus infection and cancer progression in mice infected with MHV-68. Ultimately, the translation of this mouse model system can be used to treat gammaherpesvirus-induced cancers and infections in humans.
MATERIALS AND METHODS
Cell lines and reagents.
NIH 3T3 cells (ATCC CRL-1658) or Vero cells (ATCC CCL-81) were maintained at 37°C in 5% CO2 in as monolayer cultures in Dulbecco’s modified Eagle medium (DMEM) with l-glutamine and sodium pyruvate (Genesee no. 25-500). NIH 3T3 cell medium was supplemented with 10% newborn calf serum (NBCS) and 1% penicillin streptomycin (P/S). Vero cell medium was supplemented with 10% fetal bovine serum (FBS) and 1% P/S. Stocks of 2DG (2 M) (Sigma no. D6134) were diluted in distilled water (dH2O), and 5-mg/mL stocks of TOFA (SCBT no. 200653) were diluted in dimethyl sulfoxide (DMSO).
Viruses, infection, and plaque assays.
To propagate virus stocks, NIH 3T3 or Vero cells were infected with MHV-68 (ATCC no. VR-1465) at an MOI of 0.01 to 0.05. When ~80% of the cells detached from the plate, the cells and media were freeze-thawed and pelleted at 3,000 rpm for 10 min at 4°C. The cleared viral supernatant was concentrated by centrifugation at 20,500 × g at 4°C for 90 min. Viral pellets were resuspended in serum-free (SF) DMEM. Viral infections were performed at an MOI of 0.1 or 3 in DMEM for 2 h, after which the medium was replaced with fresh DMEM. Viral titers were determined using traditional plaque assays. Briefly, 4 h prior to infection, 12-well plates were seeded with Vero cells (250,000 cells/well). Virus samples were serial diluted (100 to 106) in SF DMEM and Vero cells were infected for 1.5 h. Viral dilutions were then aspirated, and cells were overlaid with DMEM–glucose–glutamine–pyruvate (Thermo Fisher no. 12800017)–1% methylcellulose (MC) (Sigma no. M0387)–sodium bicarbonate (Sigma no. S5761)–2.5 μg/mL Amphotericin B (Thermo Fisher no. 15290026)–5% FBS–1% P/S for 6 days. Cells were fixed in 10% formalin, the medium overlay was removed, and cells were stained with 1% crystal violet (Sigma no. C0775) in 20% methanol and rinsed with water before drying.
Western blot analysis.
Mock- and MHV-68-infected NIH 3T3 cells (MOI = 3) and media were harvested at 24 h postinfection (hpi) using a cell scraper and pelleted at 200 × g for 5 min at 4°C. Cell pellets were washed in cold phosphate-buffered saline (PBS) and centrifuged at 13,000 rpm at 4°C. Cell pellets were resuspended in radioimmunoprecipitation assay (RIPA) lysis buffer (Sigma no. R0278)–protease inhibitor cocktail (Sigma no. S8820)–phosphatase inhibitor cocktail (Sigma no. 524629) on ice for 30 min. Protein concentrations were determined by a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher no. 23225). Ten to twenty micrograms of protein was separated by a sodium dodecyl sulfate polyacrylamide (SDS-PAGE) gel, transferred to a polyvinylidene difluoride or nitrocellulose membrane, and blotted with the appropriate primary antibody (1:1,000 mouse antiactin [SCBT no. 47778], 1:2,500 rabbit anti-actin [~43 kDa] [Cell Signaling no. 4970], 1:1,000 anti-glutamate dehydrogenase 1 and 2 [~50 to 55 kDa] [SCBT no. 515542], 1:500 anti-fatty acid synthase [~270 kDa] [SCBT no. 20140], 1:1,000 anti-ATP-citrate lyase [~125 kDa] [Cell Signaling no. 4332]) and subsequently with horseradish peroxidase (HRP)-conjugated 1:5,000 goat-anti-mouse (LI-COR no. 926-80010) or 1:5,000 goat-anti-rabbit (LI-COR no. 926-80011) or infrared conjugated 1:10,000 goat-anti-mouse-680 (LI-COR no. 926-68070) or 1:5,000 goat-anti-rabbit-800 (LI-COR no. 926-32211). HRP-conjugated proteins were incubated in WesternSure chemiluminescent substrate (LI-COR no. 926-95000). Immunoreactive proteins were detected using the LI-COR Odyssey Fc or C-DiGit imager. Densitometry analysis of protein expression was determined using the LI-COR Image Studio or LI-COR Empiria Studio software, with actin set as the loading control.
Glucose starvation, glutamine starvation, and drug assays.
NIH 3T3 cells were seeded at a density of 760,000 cells in 6-cm dishes approximately 4 h prior to infection. MHV-68-infected cells were infected at an MOI of 0.1 for 2 h. For both glucose and glutamine starvation assays, a base medium of DMEM (no glucose, no glutamine, and no phenol red) (Thermo Fisher no. 11960044) was used. Medium was supplemented with 10% newborn calf serum, 1% penicillin and streptomycin, 2 mM l-glutamine (Genesee no. 25-509), and/or 4.5 g/L glucose (Thermo Fisher no. A2494001). After mock- or MHV-68 infection, cells were washed three times with PBS and then fed replete (glucose and glutamine), glucose-free (with glutamine), or glutamine-free (with glucose) medium. During glucose starvation experiments, cells were fed replete medium for 24 h and washed three times with PBS, and then the medium was replaced with replete or glucose-free medium for the final 24 h. For 2DG and TOFA experiments, the media of mock- and MHV-68-infected cells were replaced with complete DMEM (containing 2 mM glutamine, 1% penicillin-streptomycin, and 10% newborn calf serum) and their respective drug treatment. 2DG was added to the cell medium at a final concentration of 5 mM in DMSO. TOFA was added to cell culture medium at a final concentration of 10 μg/mL in water. Cell supernatants were cleared by centrifugation and used to determine viral titers via plaque assays, and cell pellets were harvested to determine cell number and viability via trypan blue exclusion assays.
Cell number and viability.
Cell supernatants and trypsinized cells from 6-cm dishes were centrifuged for 5 min. Cell pellets were resuspended in 500 μL of complete DMEM. Cell number and viability were determined via trypan blue exclusion assays using the Bio-Rad TC20 automated cell counter (Bio-Rad no. 1450102). Briefly, 1:1 ratios of cells to trypan blue were mixed and loaded on dual-chambered slides (Bio-Rad no. 1450003). Total-cell numbers, live-cell numbers, and percent viability were calculated with a Bio-Rad TC20 cell counter.
Glutamine consumption and lactate production.
NIH 3T3 cells were seeded at a density of 700,000 cells in 6-cm dishes approximately 4 h prior to infection. MHV-68-infected cells were infected at an MOI of 3 for 2 h. Mock- and MHV-68-infected cells were washed with PBS and replaced with DMEM (no glucose, no glutamine, and no phenol red) (Thermo Fisher no. 11960044) supplemented with 10% dialyzed fetal bovine serum (Thermo Fisher no. A3382001), 1% penicillin and streptomycin, 2 mM l-glutamine (Thermo Fisher #25030149), and 5 mM glucose (Thermo Fisher no. A2494001). At 24 hpi, cell supernatants were cleared by centrifugation and cell pellets were harvested to determine cell number and viability via trypan blue exclusion assays using the Bio-Rad TC20 cell counter. Cell supernatants were assayed for glutamine consumption (Promega no. J8021) and accumulated lactate production (Promega no. J5921) according to the kit specifications. Glutamine consumption and lactate production were normalized to cell number.
Aqueous metabolomics and lipidomics experimental setup.
NIH 3T3 cells were seeded in 10-cm dishes (~1.5 million cells) or 6-cm dishes (~720,000 cells) approximately 4 h prior to infection. MHV-68-infected cells were infected at an MOI of 3 for 2 h. After infection, mock- and MHV-68-infected cells were fed complete DMEM (containing 2 mM glutamine, 1% penicillin-streptomycin, and 10% newborn calf serum). In biological triplicates at 4, 8, 12, 24, and 36 hpi, cell metabolism was quenched by harvesting and rinsing with −80°C chilled methanol on ice with cell scrapers. Cells were centrifuged for 5 min at 125 × g (4°C), and the supernatant was aspirated. Cell pellets were flash frozen in liquid nitrogen and transported to the University of Washington Northwest Metabolomics Research Center (NW-MRC) for analysis. The NW-MRC provided metabolic profiling of samples using targeted aqueous metabolomics and quantitative lipidomics platforms (see supplementary methods document).
Metabolomics data analysis.
For each metabolite (lipid or aqueous), the concentration was divided by total protein to scale for the variable numbers of cells in each sample, as follows: S = M/P, where S is the scaled value, M is the absolute lipid or aqueous metabolite concentration, and P is the protein concentration in the sample. For each time point in each run, the virally infected sample’s scaled value was divided by that of the corresponding control (i.e., mock infection), as follows: R = Strv/Strm, where R is the ratio of virus scaled value to mock infection scale value for a given lipid or aqueous metabolite, Strv is the scaled value for a given time point and run in the virally infected replicate, and Strm is the equivalent value for the mock infection replicate. Mean R was found for each time point by averaging across all 3 runs (biological triplicate). Metabolomics heat maps were generated using GraphPad Prism 8 software, and results are shown as fold change in MHV-68-infected cells compared to mock-infected cells. Volcano plots and box plots were generated using the freeware software MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/home.xhtml). Normalized metabolite concentrations (mock- versus MHV-68-infected cells) were uploaded on the Metaboanalyst website under the Statistical Analysis (one-factor) module. Data were analyzed in Metaboanalyst using Student’s (two-tailed) t test. Graphs were generated to show data as MHV-68/mock, with P values set to 0.05 or lower and fold change cutoffs to 1.5-fold or higher. Data were log transformed, and volcano plot or box plots were downloaded for this publication. Volcano plots were graphed as log2 fold change on the x axis and −log2 P on the y axis. Medians were determined with 95% confidence, defined as ±1.58 times the interquartile range (IQR) divided by square root (n).
Statistical analysis and graphs.
All bar and line graphs were generated using GraphPad Prism 8 software. Statistical analysis of bar graphs was performed using the Prism software. Standard errors of the mean (SEM) are shown, and P values were analyzed using Student’s (two-tailed) t test. A P value of <0.05 was considered statistically significant. Western blot densitometry data were plotted with SEM, and P values were calculated using a ratio-paired Student’s t test (log transformed, one-tailed, and paired) using GraphPad Prism 8 software.
ACKNOWLEDGMENTS
The Delgado lab metabolomics and lipidomics project was sponsored by a grant from the Supporting Structures: Innovative Partnerships to Enhance Bench Science at CCCU Member Institutions program, run by Scholarship and Christianity in Oxford, the UK subsidiary of the Council for Christian Colleges and Universities, with funding by the John Templeton Foundation and the MJ Murdock Charitable Trust. Other Delgado lab funding sources include the Murdock College Research Program for Natural Sciences grant, Seattle Pacific University Faculty Research and Scholarship Grants, and the Northwest University Ray and Shirley Clark Faculty Research Grant. Osvaldo Kevin Moreno was supported through a Genentech Foundation Scholarship. We thank Michael Lagunoff for support. We also acknowledge the collaborative services of The Northwest Metabolomics Research Center at the University of Washington, Seattle and NIH S10 grant 1S10OD021562-01, which funded a purchase of the Sciex mass spectrometry platform that was used to acquire metabolomics (quantitative lipidomics) data. Finally, we thank Delgado lab undergraduate researchers Glenwood Ray Clark and Aubrey Massman for helping set up general lab protocols and Tyler Ptacek and Anna Miller for setting up preliminary experiments not included in this article.
Conceived and designed the experiments: S.A.C., A.V., K.F., M.K.S., H.S., A.K.B., and T.D. Performed the experiments: S.A.C., A.V., K.F., M.K.S., H.S., A.K.B., S.-J.B., M.M., E.R.O., and T.D. Analyzed the data: S.A.C., A.V., K.F., M.K.S., H.S., A.K.B., M.R., S.-B., O.K.M., T.A.N., E.L.S., and T.D. Performed statistical analysis: T.A.N., T.D., M.R., and S.-J.B. Wrote the manuscript: T.D., E.L.S., O.K.M., and S.-J.B. Conceptualized the study and was the PI: T.D.
Footnotes
Supplemental material is available online only.
Contributor Information
Erica L. Sanchez, Email: erica.sanchez@utdallas.edu.
Tracie Delgado, Email: delgadot@spu.edu.
Jae U. Jung, Lerner Research Institute, Cleveland Clinic
REFERENCES
- 1.Wong Y, Meehan MT, Burrows SR, Doolan DL, Miles JJ. 2022. Estimating the global burden of Epstein–Barr virus-related cancers. J Cancer Res Clin Oncol 148:31–46. doi: 10.1007/s00432-021-03824-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Cancer Society. Key statistics about Kaposi sarcoma. https://www.cancer.org/cancer/types/kaposi-sarcoma/about/what-is-key-statistics.html. Accessed 16 January 2023.
- 3.Kenney SC. 2007. Reactivation and lytic replication of EBV. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, England. [PubMed] [Google Scholar]
- 4.Gao S-J, Deng J-H, Zhou F-C. 2003. Productive lytic replication of a recombinant Kaposi’s sarcoma-associated herpesvirus in efficient primary infection of primary human endothelial cells. J Virol 77:9738–9749. doi: 10.1128/jvi.77.18.9738-9749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quinlivan EB, Zhang C, Stewart PW, Komoltri C, Davis MG, Wehbie RS. 2002. Elevated virus loads of Kaposi’s sarcoma-associated human herpesvirus 8 predict Kaposi’s sarcoma disease progression, but elevated levels of human immunodeficiency virus type 1 do not. J Infect Dis 185:1736–1744. doi: 10.1086/340652. [DOI] [PubMed] [Google Scholar]
- 6.Virgin HW, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol 71:5894–5904. doi: 10.1128/JVI.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leang RS, Wu T-T, Hwang S, Liang LT, Tong L, Truong JT, Sun R. 2011. The anti-interferon activity of conserved viral dUTPase ORF54 is essential for an effective MHV-68 infection. PLoS Pathog 7:e1002292. doi: 10.1371/journal.ppat.1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez EL, Lagunoff M. 2015. Viral activation of cellular metabolism. Virology 479–480:609–618. doi: 10.1016/j.virol.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontaine KA, Sanchez EL, Camarda R, Lagunoff M. 2015. Dengue virus induces and requires glycolysis for optimal replication. J Virol 89:2358–2366. doi: 10.1128/JVI.02309-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allonso D, Andrade IS, Conde JN, Coelho DR, Rocha DCP, da Silva ML, Ventura GT, Silva EM, Mohana-Borges R. 2015. Dengue virus NS1 protein modulates cellular energy metabolism by increasing glyceraldehyde-3-phosphate dehydrogenase activity. J Virol 89:11871–11883. doi: 10.1128/JVI.01342-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thaker SK, Chapa T, Garcia G, Gong D, Schmid EW, Arumugaswami V, Sun R, Christofk HR. 2019. Differential metabolic reprogramming by Zika virus promotes cell death in human versus mosquito cells. Cell Metab 29:1206–1216.E4. doi: 10.1016/j.cmet.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullen PJ, Garcia G, Purkayastha A, Matulionis N, Schmid EW, Momcilovic M, Sen C, Langerman J, Ramaiah A, Shackelford DB, Damoiseaux R, French SW, Plath K, Gomperts BN, Arumugaswami V, Christofk HR. 2021. SARS-CoV-2 infection rewires host cell metabolism and is potentially susceptible to mTORC1 inhibition. 1. Nat Commun 12:1876. doi: 10.1038/s41467-021-22166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abrantes JL, Alves CM, Costa J, Almeida FCL, Sola-Penna M, Fontes CFL, Souza TML. 2012. Herpes simplex type 1 activates glycolysis through engagement of the enzyme 6-phosphofructo-1-kinase (PFK-1). Biochim Biophys Acta 1822:1198–1206. doi: 10.1016/j.bbadis.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 14.Vastag L, Koyuncu E, Grady SL, Shenk TE, Rabinowitz JD. 2011. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog 7:e1002124. doi: 10.1371/journal.ppat.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. 2006. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog 2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontaine KA, Camarda R, Lagunoff M. 2014. Vaccinia virus requires glutamine but not glucose for efficient replication. J Virol 88:4366–4374. doi: 10.1128/JVI.03134-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prusinkiewicz MA, Mymryk JS. 2019. Metabolic reprogramming of the host cell by human adenovirus infection. Viruses 11:141. doi: 10.3390/v11020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delgado T, Carroll PA, Punjabi AS, Margineantu D, Hockenbery DM, Lagunoff M. 2010. Induction of the Warburg effect by Kaposi’s sarcoma herpesvirus is required for the maintenance of latently infected endothelial cells. Proc Natl Acad Sci USA 107:10696–10701. doi: 10.1073/pnas.1004882107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delgado T, Sanchez EL, Camarda R, Lagunoff M. 2012. Global metabolic profiling of infection by an oncogenic virus: KSHV induces and requires lipogenesis for survival of latent infection. PLoS Pathog 8:e1002866. doi: 10.1371/journal.ppat.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez EL, Carroll PA, Thalhofer AB, Lagunoff M. 2015. Latent KSHV infected endothelial cells are glutamine addicted and require glutaminolysis for survival. PLoS Pathog 11:e1005052. doi: 10.1371/journal.ppat.1005052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez EL, Pulliam TH, Dimaio TA, Thalhofer AB, Delgado T, Lagunoff M. 2017. Glycolysis, glutaminolysis, and fatty acid synthesis are required for distinct stages of Kaposi’s sarcoma-associated herpesvirus lytic replication. J Virol 91:e02237-16. doi: 10.1128/JVI.02237-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo R, Liang JH, Zhang Y, Lutchenkov M, Li Z, Wang Y, Trujillo-Alonso V, Puri R, Giulino-Roth L, Gewurz BE. 2022. Methionine metabolism controls the B cell EBV epigenome and viral latency. Cell Metab 34:1280–1297.E9. doi: 10.1016/j.cmet.2022.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yogev O, Lagos D, Enver T, Boshoff C. 2014. Kaposi’s sarcoma herpesvirus microRNAs induce metabolic transformation of infected cells. PLoS Pathog 10:e1004400. doi: 10.1371/journal.ppat.1004400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmes DL, Vogt DT, Lagunoff M. 2020. A CRISPR-Cas9 screen identifies mitochondrial translation as an essential process in latent KSHV infection of human endothelial cells. Proc Natl Acad Sci USA 117:28384–28392. doi: 10.1073/pnas.2011645117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veettil MV, Dutta D, Bottero V, Bandyopadhyay C, Gjyshi O, Sharma-Walia N, Dutta S, Chandran B. 2014. Glutamate secretion and metabotropic glutamate receptor 1 expression during Kaposi’s sarcoma-associated herpesvirus infection promotes cell proliferation. PLoS Pathog 10:e1004389. doi: 10.1371/journal.ppat.1004389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Y, Li T, Ramos da Silva S, Lee J-J, Lu C, Eoh H, Jung JU, Gao S-J. 2017. A critical role of glutamine and asparagine γ-nitrogen in nucleotide biosynthesis in cancer cells hijacked by an oncogenic virus. mBio 8:e01179-17. doi: 10.1128/mBio.01179-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao L, Chen Y-X, Varghese Z, Huang A-L, Tang R-K, Jia B, Moorhead JF, Gong J-P, Ruan XZ. 2012. Murine gamma herpes virus 68 infection promotes fatty liver formation and hepatic insulin resistance in C57BL/6J mice. Hepatol Int 6:520–530. doi: 10.1007/s12072-011-9283-x. [DOI] [PubMed] [Google Scholar]
- 28.Lange PT, Darrah EJ, Vonderhaar EP, Mboko WP, Rekow MM, Patel SB, Sidjanin DJ, Tarakanova VL. 2016. Type I interferon counteracts antiviral effects of statins in the context of gammaherpesvirus infection. J Virol 90:3342–3354. doi: 10.1128/JVI.02277-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange PT, Schorl C, Sahoo D, Tarakanova VL. 2018. Liver X receptors suppress activity of cholesterol and fatty acid synthesis pathways to oppose gammaherpesvirus replication. mBio 9:e01115-18. doi: 10.1128/mBio.01115-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.López-Rodríguez DM, Kirillov V, Krug LT, Mesri EA, Andreansky S. 2019. A role of hypoxia-inducible factor 1 alpha in murine gammaherpesvirus 68 (MHV68) lytic replication and reactivation from latency. PLoS Pathog 15:e1008192. doi: 10.1371/journal.ppat.1008192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birungi G, Chen SM, Loy BP, Ng ML, Li SFY. 2010. Metabolomics approach for investigation of effects of dengue virus infection using the EA.hy926 cell line. J Proteome Res 9:6523–6534. doi: 10.1021/pr100727m. [DOI] [PubMed] [Google Scholar]
- 32.Roe B, Kensicki E, Mohney R, Hall WW. 2011. Metabolomic profile of hepatitis C virus-infected hepatocytes. PLoS One 6:e23641. doi: 10.1371/journal.pone.0023641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian X, Zhang K, Min J, Chen C, Cao Y, Ding C, Liu W, Li J. 2019. Metabolomic analysis of influenza A virus A/WSN/1933 (H1N1) infected A549 cells during first cycle of viral replication. Viruses 11:1007. doi: 10.3390/v11111007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Guzman D, Rickabaugh T, Wu T-T, Brown H, Cole S, Song MJ, Tong L, Sun R. 2003. Transcription program of murine gammaherpesvirus 68. J Virol 77:10488–10503. doi: 10.1128/jvi.77.19.10488-10503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu T-T, Usherwood EJ, Stewart JP, Nash AA, Sun R. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J Virol 74:3659–3667. doi: 10.1128/jvi.74.8.3659-3667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart DS, Guo A, Oler E, Wang F, Anjum A, Peters H, Dizon R, Sayeeda Z, Tian S, Lee BL, Berjanskii M, Mah R, Yamamoto M, Jovel J, Torres-Calzada C, Hiebert-Giesbrecht M, Lui VW, Varshavi D, Varshavi D, Allen D, Arndt D, Khetarpal N, Sivakumaran A, Harford K, Sanford S, Yee K, Cao X, Budinski Z, Liigand J, Zhang L, Zheng J, Mandal R, Karu N, Dambrova M, Schiöth HB, Greiner R, Gautam V. 2022. HMDB 5.0: the Human Metabolome Database for 2022. Nucleic Acids Res 50:D622–D631. doi: 10.1093/nar/gkab1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avila MA, García-Trevijano ER, Lu SC, Corrales FJ, Mato JM. 2004. Methylthioadenosine. Int J Biochem Cell Biol 36:2125–2130. doi: 10.1016/j.biocel.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 38.Milewski S, Gabriel I, Olchowy J. 2006. Enzymes of UDP-GlcNAc biosynthesis in yeast. Yeast 23:1–14. doi: 10.1002/yea.1337. [DOI] [PubMed] [Google Scholar]
- 39.Hanover JA. 2001. Glycan-dependent signaling: O-linked N-acetylglucosamine. FASEB J 15:1865–1876. doi: 10.1096/fj.01-0094rev. [DOI] [PubMed] [Google Scholar]
- 40.Thaker SK, Ch’ng J, Christofk HR. 2019. Viral hijacking of cellular metabolism. BMC Biol 17:59. doi: 10.1186/s12915-019-0678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han X. 2016. Lipidomics for studying metabolism. Nat Rev Endocrinol 12:668–679. doi: 10.1038/nrendo.2016.98. [DOI] [PubMed] [Google Scholar]
- 42.Merino-Ramos T, Vázquez-Calvo Á, Casas J, Sobrino F, Saiz J-C, Martín-Acebes MA. 2016. Modification of the host cell lipid metabolism induced by hypolipidemic drugs targeting the acetyl coenzyme A carboxylase impairs West Nile virus replication. Antimicrob Agents Chemother 60:307–315. doi: 10.1128/AAC.01578-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greseth MD, Traktman P. 2014. De novo fatty acid biosynthesis contributes significantly to establishment of a bioenergetically favorable environment for vaccinia virus infection. PLoS Pathog 10:e1004021. doi: 10.1371/journal.ppat.1004021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaunt ER, Cheung W, Richards JE, Lever A, Desselberger U. 2013. Inhibition of rotavirus replication by downregulation of fatty acid synthesis. J Gen Virol 94:1310–1317. doi: 10.1099/vir.0.050146-0. [DOI] [PubMed] [Google Scholar]
- 45.Heaton NS, Perera R, Berger KL, Khadka S, LaCount DJ, Kuhn RJ, Randall G. 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci USA 107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang F, Yan S, He Y, Wang F, Song S, Guo Y, Zhou Q, Wang Y, Lin Z, Yang Y, Zhang W, Sun S. 2008. Expression of hepatitis B virus proteins in transgenic mice alters lipid metabolism and induces oxidative stress in the liver. J Hepatol 48:12–19. doi: 10.1016/j.jhep.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 47.Baniasadi H, Gowda GAN, Gu H, Zeng A, Zhuang S, Skill N, Maluccio M, Raftery D. 2013. Targeted metabolic profiling of hepatocellular carcinoma and hepatitis C using LC-MS/MS. Electrophoresis 34:2910–2917. doi: 10.1002/elps.201300029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin S, Liu N, Yang Z, Song W, Wang P, Chen H, Lucio M, Schmitt-Kopplin P, Chen G, Cai Z. 2010. GC/MS-based metabolomics reveals fatty acid biosynthesis and cholesterol metabolism in cell lines infected with influenza A virus. Talanta 83:262–268. doi: 10.1016/j.talanta.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 49.Coleman HM, de Lima B, Morton V, Stevenson PG. 2003. Murine gammaherpesvirus 68 lacking thymidine kinase shows severe attenuation of lytic cycle replication in vivo but still establishes latency. J Virol 77:2410–2417. doi: 10.1128/jvi.77.4.2410-2417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gáspár G, De Clercq E, Neyts J. 2002. Human herpesvirus 8 gene encodes a functional thymidylate synthase. J Virol 76:10530–10532. doi: 10.1128/jvi.76.20.10530-10532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng Q, Hagemeier SR, Fingeroth JD, Gershburg E, Pagano JS, Kenney SC. 2010. The Epstein-Barr virus (EBV)-encoded protein kinase, EBV-PK, but not the thymidine kinase (EBV-TK), is required for ganciclovir and acyclovir inhibition of lytic viral production. J Virol 84:4534–4542. doi: 10.1128/JVI.02487-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang JH, Wang C, Yiu SPT, Zhao B, Guo R, Gewurz BE. 2021. Epstein-Barr virus induced cytidine metabolism roles in transformed B-cell growth and survival. mBio 12:e01530-21. doi: 10.1128/mBio.01530-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Šimonová A, Svojanovská B, Trylčová J, Hubálek M, Moravčík O, Zavřel M, Pávová M, Hodek J, Weber J, Cvačka J, Pačes J, Cahová H. 2019. LC/MS analysis and deep sequencing reveal the accurate RNA composition in the HIV-1 virion. 1. Sci Rep 9:8697. doi: 10.1038/s41598-019-45079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fukuda H, Chujo T, Wei F-Y, Shi S-L, Hirayama M, Kaitsuka T, Yamamoto T, Oshiumi H, Tomizawa K. 2021. Cooperative methylation of human tRNA3Lys at positions A58 and U54 drives the early and late steps of HIV-1 replication. Nucleic Acids Res 49:11855–11867. doi: 10.1093/nar/gkab879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stine ZE, Altman BJ, Hsieh AL, Gouw AM, Dang CV. 2014. Deregulation of the cellular energetics of cancer cells, p 444–455. In McManus LM, Mitchell RN (ed), Pathobiology of human disease. Academic Press, San Diego, CA. [Google Scholar]
- 56.Cory JG, Cory AH. 2006. Critical roles of glutamine as nitrogen donors in purine and pyrimidine nucleotide synthesis: asparaginase treatment in childhood acute lymphoblastic leukemia. In Vivo 20:587–589. [PubMed] [Google Scholar]
- 57.Wang Y, Bai C, Ruan Y, Liu M, Chu Q, Qiu L, Yang C, Li B. 2019. Coordinative metabolism of glutamine carbon and nitrogen in proliferating cancer cells under hypoxia. Nat Commun 10:201. doi: 10.1038/s41467-018-08033-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu J, Thompson CB. 2019. Metabolic regulation of cell growth and proliferation. Nat Rev Mol Cell Biol 20:436–450. doi: 10.1038/s41580-019-0123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chambers JW, Maguire TG, Alwine JC. 2010. Glutamine metabolism is essential for human cytomegalovirus infection. J Virol 84:1867–1873. doi: 10.1128/JVI.02123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leung HJ, Duran EM, Kurtoglu M, Andreansky S, Lampidis TJ, Mesri EA. 2012. Activation of the unfolded protein response by 2-deoxy-d-glucose inhibits Kaposi’s sarcoma-associated herpesvirus replication and gene expression. Antimicrob Agents Chemother 56:5794–5803. doi: 10.1128/AAC.01126-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Svensson RU, Parker SJ, Eichner LJ, Kolar MJ, Wallace M, Brun SN, Lombardo PS, Van Nostrand JL, Hutchins A, Vera L, Gerken L, Greenwood J, Bhat S, Harriman G, Westlin WF, Harwood HJ, Saghatelian A, Kapeller R, Metallo CM, Shaw RJ. 2016. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat Med 22:1108–1119. doi: 10.1038/nm.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calvisi DF, Wang C, Ho C, Ladu S, Lee SA, Mattu S, Destefanis G, Delogu S, Zimmermann A, Ericsson J, Brozzetti S, Staniscia T, Chen X, Dombrowski F, Evert M. 2011. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology 140:1071–1083.E5. doi: 10.1053/j.gastro.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Snaebjornsson MT, Janaki-Raman S, Schulze A. 2020. Greasing the wheels of the cancer machine: the role of lipid metabolism in cancer. Cell Metab 31:62–76. doi: 10.1016/j.cmet.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 65.Fernández LP, Gómez de Cedrón M, Ramírez de Molina A. 2020. Alterations of lipid metabolism in cancer: implications in prognosis and treatment. Front Oncol 10. doi: 10.3389/fonc.2020.577420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu Y, Maguire TG, Alwine JC. 2012. Human cytomegalovirus infection induces adipocyte-like lipogenesis through activation of sterol regulatory element binding protein 1. J Virol 86:2942–2949. doi: 10.1128/JVI.06467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhatt AP, Jacobs SR, Freemerman AJ, Makowski L, Rathmell JC, Dittmer DP, Damania B. 2012. Dysregulation of fatty acid synthesis and glycolysis in non-Hodgkin lymphoma. Proc Natl Acad Sci USA 109:11818–11823. doi: 10.1073/pnas.1205995109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lo AK-F, Lung RW-M, Dawson CW, Young LS, Ko C-W, Yeung WW, Kang W, To K-F, Lo K-W. 2018. Activation of sterol regulatory element-binding protein 1 (SREBP1)-mediated lipogenesis by the Epstein–Barr virus-encoded latent membrane protein 1 (LMP1) promotes cell proliferation and progression of nasopharyngeal carcinoma. J Pathol 246:180–190. doi: 10.1002/path.5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lange PT, Lagunoff M, Tarakanova VL. 2019. Chewing the fat: the conserved ability of DNA viruses to hijack cellular lipid metabolism. Viruses 11:119. doi: 10.3390/v11020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arii J, Fukui A, Shimanaka Y, Kono N, Arai H, Maruzuru Y, Koyanagi N, Kato A, Mori Y, Kawaguchi Y. 2020. Role of phosphatidylethanolamine biosynthesis in herpes simplex virus 1-infected cells in progeny virus morphogenesis in the cytoplasm and in viral pathogenicity in vivo. J Virol 94:e01572-20. doi: 10.1128/JVI.01572-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Q, Pène V, Krishnamurthy S, Cha H, Liang TJ. 2013. Hepatitis C virus infection activates an innate pathway involving IKK-α in lipogenesis and viral assembly. Nat Med 19:722–729. doi: 10.1038/nm.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baek Y-B, Kwon H-J, Sharif M, Lim J, Lee I-C, Ryu YB, Lee J-I, Kim J-S, Lee Y-S, Kim D-H, Park S-I, Kim D-K, Kim J-S, Choy HE, Lee S, Choi H-S, Osborne TF, Jeon T-I, Cho K-O. 2022. Therapeutic strategy targeting host lipolysis limits infection by SARS-CoV-2 and influenza A virus. Signal Transduct Target Ther 7:367. doi: 10.1038/s41392-022-01223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Camus G, Vogt DA, Kondratowicz AS, Ott M. 2013. Lipid droplets and viral infections. Methods Cell Biol 116:167–190. doi: 10.1016/B978-0-12-408051-5.00009-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download jvi.00506-23-s0001.xlsx, XLSX file, 0.1 MB (92.3KB, xlsx)
Table S2. Download jvi.00506-23-s0002.xlsx, XLSX file, 0.1 MB (53.8KB, xlsx)
Supplemental methods and legends of Tables S1 and S2. Download jvi.00506-23-s0003.docx, DOCX file, 0.2 MB (16KB, docx)