ABSTRACT
We identified neutralizing monoclonal antibodies against severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) variants (including Omicron variants BA.5 and BA.2.75) from individuals who received two doses of mRNA vaccination after they had been infected with the D614G virus. We named them MO1, MO2, and MO3. Among them, MO1 showed particularly high neutralizing activity against authentic variants: D614G, Delta, BA.1, BA.1.1, BA.2, BA.2.75, and BA.5. Furthermore, MO1 suppressed BA.5 infection in hamsters. A structural analysis revealed that MO1 binds to the conserved epitope of seven variants, including Omicron variants BA.5 and BA.2.75, in the receptor-binding domain of the spike protein. MO1 targets an epitope conserved among Omicron variants BA.1, BA.2, and BA.5 in a unique binding mode. Our findings confirm that D614G-derived vaccination can induce neutralizing antibodies that recognize the epitopes conserved among the SARS-CoV-2 variants.
IMPORTANCE Omicron variants of SARS-CoV-2 acquired escape ability from host immunity and authorized antibody therapeutics and thereby have been spreading worldwide. We reported that patients infected with an early SARS-CoV-2 variant, D614G, and who received subsequent two-dose mRNA vaccination have high neutralizing antibody titer against Omicron lineages. It was speculated that the patients have neutralizing antibodies broadly effective against SARS-CoV-2 variants by targeting common epitopes. Here, we explored human monoclonal antibodies from B cells of the patients. One of the monoclonal antibodies, named MO1, showed high potency against broad SARS-CoV-2 variants including BA.2.75 and BA.5 variants. The results prove that monoclonal antibodies that have common neutralizing epitopes among several Omicrons were produced in patients infected with D614G and who received mRNA vaccination.
KEYWORDS: severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2), Omicron variants, human monoclonal antibody, broad neutralizing activity, spike, receptor-binding domain, cryoelectron microscopy, common epitope, vaccine
INTRODUCTION
Severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) caused the devastating coronavirus disease 2019 (COVID-19) pandemic that began in late 2019; as of 15 February 2023, SARS-CoV-2 had infected over 750 million people worldwide and is responsible for more than 6.8 million deaths (https://covid19.who.int/). The Omicron variant (BA.1; B.1.1.529) of SARS-CoV-2 was first reported in South Africa in November 2021 and quickly spread thereafter (https://www.cdc.gov/coronavirus/2019-ncov/variants/Omicron-variant.html) (1). The spread of the Omicron variant caused a further increase in COVID-19 infections, and the Omicron BA.5 variant in particular has spread worldwide (2). Furthermore, several new variants have appeared all over the world. BQ.1 and BQ.1.1 variants are sublineages of BA.5, first reported in Nigeria in July 2022. XBB variant is recombinant of BA.2.10.1 and BA.2.75, and XBB.1.5 has an additional F486P mutation (3). WHO is tracking four subvariants of BF.7, BQ.1, BA.2.75, and XBB under monitoring as of 16 January 2023 (https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---19-january-2023; 19 January 2023). BQ and XBB subvariants have been reported to have an immune evasion capacity higher than BA.5 in mRNA-vaccinated patients (4). We and other research groups have shown that three doses of the mRNA vaccine can protect individuals against the BA.1 and BA.2 variants of SARS-CoV-2 (5–7). We have also observed that neutralizing antibodies against BA.1 increased in the sera of individuals who received a two-dose mRNA vaccine after having been infected with D614G SARS-CoV-2 (8). We therefore screened those individuals’ B cells, as we suspected that it is possible that multiply challenged B cells could produce antibodies that have broad neutralizing activity against SARS-CoV-2 variants including Omicrons. In the present study, we identified a monoclonal antibody (MAb), “MO1,” that is broadly effective against SARS-CoV-2 variants including Omicron BA.5; this MAb has structural features and binding sites that differ from those of all other MAbs against SARS-CoV-2 reported to date.
RESULTS
B cells that produce antibodies with broad neutralizing activity against SARS-CoV-2 were obtained from the peripheral blood mononuclear cells of SARS-CoV-2-infected and subsequently two-dose-vaccinated patients.
We performed the following experiments to identify human-neutralizing antibodies that could recognize epitopes conserved among all the variants ever identified. To this end, we performed a series of experiments as summarized in Fig. 1. To isolate MAbs targeting common epitopes of SARS-CoV-2 variants, we searched for antibody genes from peripheral blood mononuclear cells (PBMCs) of infected and subsequently vaccinated patients (Table 1). We used PBMCs isolated from three patients who showed 256 or higher neutralizing titers against the D614G, Delta, and Omicron BA.1 variants as previously reported (8). In the present study, single B cells were isolated from a mixture of the PBMCs of three patients, and fragments antigen binding (Fabs) derived from the antibody variable region genes of each single cell were screened by enzyme-linked immunosorbent assay (ELISA) (see Fig. 1, step 3) using the Ecobody technology (9). The top 10 candidates were selected based on their reactivity to the spike antigen (Fig. S1 in the supplemental material). The 10 selected variable region genes were used to make recombinant immunoglobulin (IgG) expression constructs, and expressed MAbs were purified for the following analysis. The 10 MAbs were examined for neutralizing activity, and we identified the three (which we named MO1, MO2, and MO3) that showed neutralizing activity against the Omicron BA.1 variant. The VH/VL gene pairs of MO1, MO2, and MO3 were as follows: MO1: IGHV3-9*01/IGKV1-9*01, MO2: IGHV3-11*05/IGKV3-15*01, and MO3: IGHV3-30*18/IGKV1-33*01.
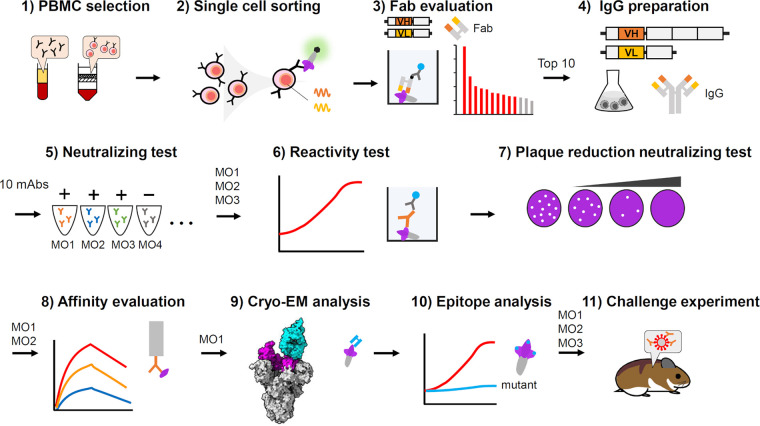
FIG 1.
Flow chart of experimental procedures for the neutralizing antibody analysis in this study. (1) PBMCs isolated from three patients who showed high neutralizing activity for Omicron BA.1 (Table 1), were selected, mixed, and used for the study. (2) Memory B cells were isolated from the PBMCs, sorted by fluorescently labeled spike antigen, and then dispensed as single cells. (3) Antibody variable region genes amplified from each single cell were used to express fragment antigen binding (Fab) according to the protocol of the Ecobody technology (9). Each Fab was tested for reactivity against spike antigens by ELISA, and the top 10 candidates were selected based on the results. (4) The antibody variable region genes were used to construct recombinant IgG antibody expression plasmids. Each MAb secreted from culture cells transfected by the plasmids was purified for subsequent analysis. (5) 10 MAbs were subjected to the neutralization assay against the SARS-CoV-2 Omicron BA.1, and were labeled MO1 to MO10 according to the potency. Only three antibodies (MO1, MO2, and MO3) showed neutralizing activity against BA.1; therefore, these 3 antibodies were further analyzed. (6) Reactivities of MO1, MO2, and MO3 to spike antigen from several variants were evaluated by ELISA. (7) The neutralizing activities of MO1, MO2, and MO3 were quantitatively evaluated by a plaque reduction neutralizing test. (8) The affinity of MO1 and MO2 to the spike receptor binding domain (RBD) was analyzed by biolayer interferometry assay. (9) The best neutralizing antibody, MO1, was subjected to cryo-EM to analyze the binding mode to the spike antigen. (10) Spike residues at the MO1-binding site were examined for their importance by ELISA using mutant spike proteins. (11) Animal challenge experiment was performed to evaluate the neutralizing activities of MO1, MO2, and MO3 against Omicron BA.5 in a hamster infection model.
TABLE 1.
Selected PBMCs from the patients who showed high neutralizing activity against SARS-CoV-2-infected
| Age | Sex | Severity | Time of infection | Months from onset to 2 doses of vaccine | Months from onset to blood collection | Neutralizing antibody titera |
|||
|---|---|---|---|---|---|---|---|---|---|
| D614G | Delta | BA.1 | |||||||
| Donor 1 | 45 | Female | Mild | July 2020 | 12 | 13 | 2048 | 1024 | 512 |
| Donor 2 | 57 | Male | Severe | October 2020 | 9 | 12 | 1024 | 256 | 256 |
| Donor 3 | 51 | Male | Severe | November 2020 | 10 | 12 | 512 | 512 | 256 |
Broadly neutralizing MAbs against SARS-CoV-2 variants.
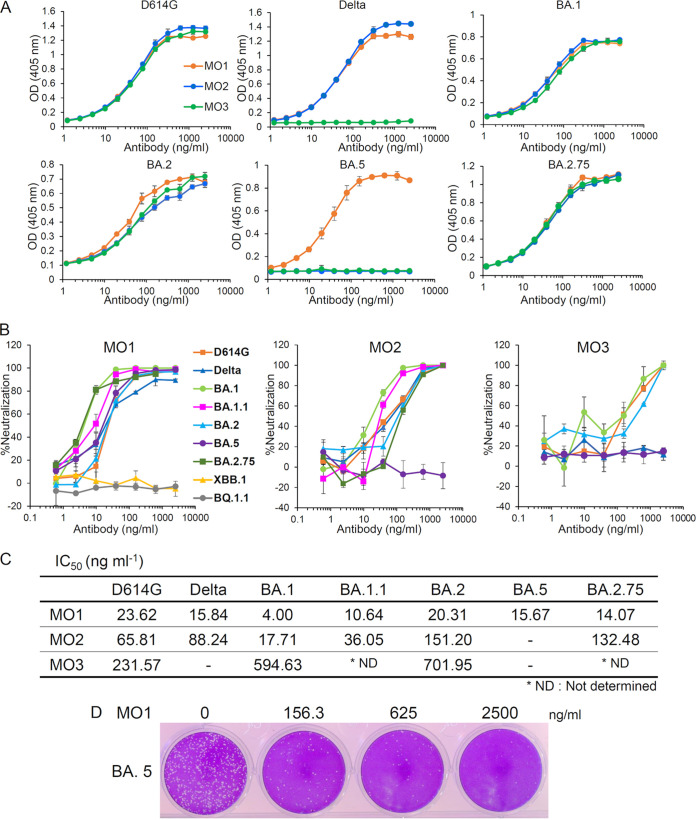
As depicted in Fig. 2 and Fig. S2, we observed that the three selected neutralizing MAbs for D614G, i.e., MO1, MO2, and MO3, recognized the SARS-CoV-2 spike protein of D614G. MO1 recognized the spike protein of nine variants: D614G, Delta, BA.1, BA.1.1, BA.2, BA.2.75, BA.5, BA.4.6, or BF.7. MO2 recognized the spike protein of six of these variants (not BA.5, BA.4.6, and BF.7). MO3 recognized five of the variants (not Delta, BA.5, BA.4.6, or BF.7). We next assessed whether these three MAbs had neutralizing activity against SARS-CoV-2 variants. As shown in Fig. 2B to D, the MAb MO1 inhibited all variants examined in this study (D614G, Delta, BA.1, BA.1.1, BA.2, BA.5, and BA.2.75) with the following IC50 values: D614G (23.62 ng/mL), Delta (15.84 ng/mL), BA.1 (4.0 ng/mL), BA.1.1 (10.64 ng/mL), BA.2 (20.31 ng/mL), BA.5 (15.67 ng/mL), and BA.2.75 (14.1 ng/mL). MO2 inhibited six variants (BA.5 was the exception), with the following IC50 values: D614G (65.81 ng/mL), Delta (88.24 ng/mL), BA.1 (17.71 ng/mL), BA.1.1 (36.05 ng/mL), BA.2 (151.2 ng/mL), and BA.2.75 (132.5 ng/mL). MO3 suppressed three variants (Delta and BA.5 were the exceptions; neutralizing activity against BA.1.1 and BA.2.75 were not determined), with the following IC50 values: D614G (231.57 ng/mL), BA.1 (594.63 ng/mL), and BA.2 (701.95 ng/mL). However, MO1, MO2, and MO3 could not recognize the spike of BQ.1.1 variant in the ELISA, indicating that the recognition sites of the MAbs were mutated (Fig. S2 and S6). In addition, MO1 lost neutralizing activity against BQ.1.1 and XBB.1 (Fig. 2B).
FIG 2.
Identification of broadly neutralizing MAbs against SARS-CoV-2 variants. (A) The binding of three MAbs to the SARS-CoV-2 spike ectodomains of the D614G, Delta, BA.1, BA.2, BA.5, and BA.2.75 variants as revealed by respective ELISAs. (B) The neutralizing activity of MAbs MO1, MO2, and MO3 against D614G, Delta, BA.1, BA.1.1, BA.2, BA.5, BA.2.75, BQ.1.1, or XBB.1 as evaluated by the plaque reduction neutralization test (PRNT). (C) The 50% inhibitory concentrations (IC50) of MAbs MO1, MO2, and MO3 against the SARS-CoV-2 variants calculated from the above neutralization data (B) are shown. (D) A presentation of plaque reduction in MO1’s PRNT test against BA.5.
The neutralizing MAb MO1 has a high affinity against the SARS-CoV-2 BA.5 spike protein.
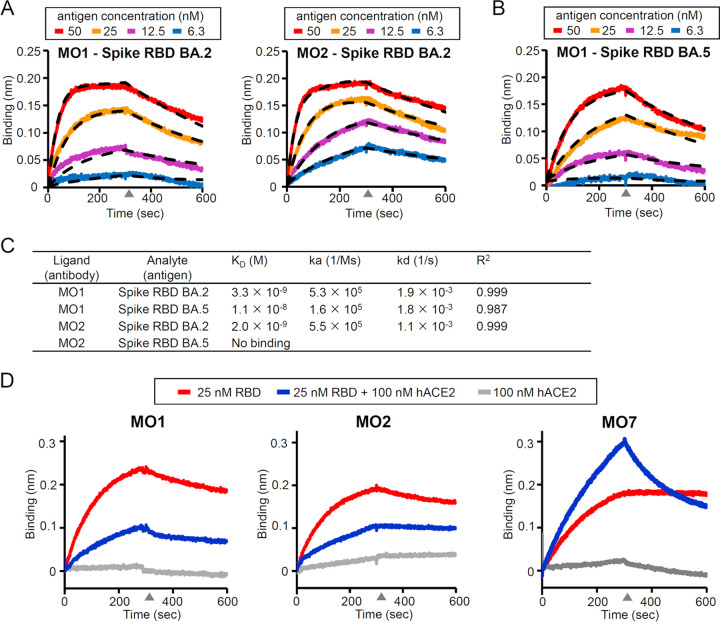
Next, the affinities of both MO1 and MO2 MAbs with the SARS-CoV-2 BA.2 spike protein were evaluated by the biolayer interferometry (BLI) method. We assumed that these MAbs target the receptor-binding domain (RBD) of the spike protein and analyzed the interaction kinetics between the MAbs on the sensor tip and RBD in the solvent. Both MO1 and MO2 showed a high affinity with the BA.2 spike RBD, with the dissociation constants (KD) of 3.3 nM and 2.0 nM, respectively (Fig. 3A and C). MO1 also had a high affinity with the BA.5 spike RBD, with a KD of 11 nM (Fig. 3B and C), whereas MO2 showed no binding to the BA.5 spike RBD, which is consistent with its lack of neutralizing activity for BA.5.
FIG 3.
Analysis of the affinity between the three MAbs and spike antigens by biolayer interferometry (BLI). (A) The sensorgram for the BA.2 spike RBD’s binding to the MAb MO1 or MO2. Dashed lines: the fitting curves. (B) The same BLI analysis as in panel A between the BA.5 spike RBD and MO1. (C) Summary of the BLI kinetics evaluated from the curve fitting. (D) Competition between MAbs and human ACE. A nonneutralizing antibody MO7 was used as a competition-negative control.
To investigate the neutralizing mechanism of MO1 and MO2, a competition assay on spike binding between the antibodies and the receptor of SARS-CoV-2, that is, human angiotensin-converting enzyme 2 (ACE2), was performed. The binding of both MO1 and MO2 to the BA.2 RBD was suppressed when ACE2 was mixed with BA.2 spike RBD, indicating a competition between these antibodies and hACE2 (Fig. 3D). A nonneutralizing antibody MO7 binds to BA.2 spike RBD and showed an increased response when RBD is mixed with ACE2, indicating MO7’s ability to bind to the RBD-hACE2 complex without competition (Fig. 3D).
Recognition of the conserved epitope by neutralizing MAb MO1 was revealed by cryoelectron microscopy.
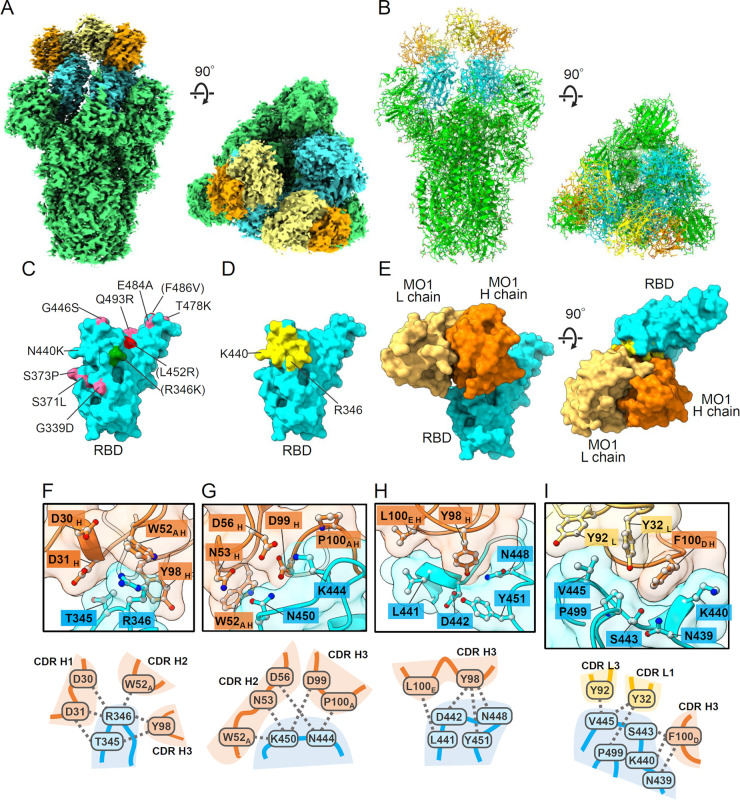
We performed a cryoelectron microscopy (cryo-EM) analysis to investigate the target sites and binding mode of MAb MO1. The prefusion-stabilized BA.1 spike ectodomain and Fab domain of MO1 were copurified by chromatography (Fig. S3) and used for the analysis. The cryo-EM density map for the MO1-Fab complex was obtained at 2.7-Å global resolution (Fig. 4A and B; Fig. S4C to E). Three MO1 Fabs were interpretable in the map of the conformation 2 of the three observed conformations with the best sphericity in the three-dimensional (3D) FSC analysis (Fig. S4C and S5), binding to one “up” and two “down” RBDs. The identity of the Fab that bound to the up RBD was not entirely clear compared to the other two down-binding Fabs, probably due to the movable nature of the RBD conformation; thus, the following observations were based on the Fabs bound to the down RBDs.
FIG 4.
The binding mode of MAb MO1 with the BA.1 spike trimer. (A and B) The cryo-EM density map (A) and of the atomic model (B) of the MO1 Fab and prefusion BA.1 spike trimer. The coloring is as follows. MO1 heavy chain, orange; light chain, yellow; spike RBD, cyan; spike NTD, and S2, green. (C) Map of the Omicron BA.1 or BA.2 mutation sites on the RBD viewed from the MO1 binding site. The Omicron BA.5 mutations F486V and L452R, and Omicron BA.1.1 mutation R346K are also indicated. (D) The residues involved in the MO1 footprint are yellow. The view is the same as that in panel C. (E) The MO1 binding mode on the RBD. The left image is the same view as in panel C and D. (F to I) Notable interactions between MO1 and the RBD at the interface. Three-dimensional arrangements of the protein residues (top) and their schematic illustrations (bottom) are shown.
MO1 binds near the “right shoulder” (10) of the spike RBD (Fig. 4A) as do other antibodies such as S309 (11). Although the number of mutations on this face of the RBD is relatively small, several such mutations are present in the Omicron variants, especially in the BA.5 variant (Fig. 4C). Nonetheless, the MO1 footprint on the RBD neatly avoids the mutation sites, except for the R346 and K440 sites at the outer rim (Fig. 4C to E; Fig. S6). The complementarity determining region (CDR) H3 of the MO1 heavy chain is positioned close to the loops 344 to 349 and 442 to 452 of RBD, and the CDRs H1, H2, L1, and L3 surround the contact site. The footprint of MO1 is compact with a moderate buried surface area (638 Å2) and high shape complementarity (0.746) (12). The footprint does not overlap the area of the ACE2-binding site (Fig. S7A) (13). A simulation model of the Spike RBD/ACE2/MO1 tertiary complex indicates that MO1 binds to RBD without large steric hindrance with ACE2, although the glycan at residue N53 of ACE2 makes slight contact with the L chain of MO1 (Fig. S7B).
The interactions that we observed between MO1 and the RBD are illustrated in Fig. 4F to I and summarized in Table 2 with information about the conservation among the major SARS-CoV-2 strains. All of the residues in the footprint were conserved among the major SARS-CoV-2 variants with the exceptions of N440K found in Omicron variants and R346K and R346T in the BA.1.1 and BA.4.6/BF.7 variants, respectively. The N440K mutation site of the BA.1 spike is located near the F100D of the MO1 heavy chain, but the N440K sidechain extends in a direction away from the MO1-spike interface (Fig. 4I and Table 2). Thus, it seems reasonable that the amino acid change, N440K of Omicron variants at this position, has little effect on the MO1 binding. Notably, the R346 of the RBD is located near the sidechain of W52A of MO1, implying an interaction via cation–pi stacking between R346 and W52A (Fig. 4F). In addition, the MO1 heavy-chain residues Y98, D30, and D31 had the sidechains near the R346 of the RBD.
TABLE 2.
SARS-CoV-2 spike RBD residues involved in the footprinta of the MO1
| WT spike residues | Conservation among SARS-CoV-2 variants | MO1 residues located nearbyc | Possible interaction | Note | Figure |
|---|---|---|---|---|---|
| T345 | Conserved | D31 H, Y98 H | Van der Waals | Fig. 4F | |
| R346 | K in BA.1.1 | W52A H | Cation-pi | Fig. 4F | |
| T in BA.4.6, BF.7, | Y98 H | Van der Waals | |||
| BQ.1.1, and XBB.1 | D30 H, D31 H | Electrostatic | |||
| N439 | Conserved | F100D H | Van der Waals | N439 sidechain faces away from the antibody | Fig. 4I |
| N440b | K in Omicron variants | F100D H | Van der Waals | Fig. 4I | |
| L441 | Conserved | L100E H | Van der Waals | L441 sidechain faces away from the antibody | Fig. 4H |
| D442 | Conserved | Y98 H | Van der Waals | Fig. 4H | |
| S443 | Conserved | F100D H | Van der Waals | S443 sidechain faces away from the antibody | Fig. 4I |
| K444 | T in BQ.1 and BQ.1.1 | D56 H | Electrostatic | Fig. 4G | |
| P100A H | Van der Waals | ||||
| V445 | P in XBB.1 | Y32 L, Y92 L | Van der Waals | Fig. 4I | |
| N448 | Conserved | Y98 H | Hydrogen bond | Fig. 4H | |
| N450 | Conserved | N53 H, D99 H, W52A H | Hydrogen bond | Fig. 4G | |
| Y451 | Conserved | Y98 H | Van der Waals | Fig. 4H | |
| P499 | Conserved | Y32 L, F100DH | Van der Waals | Fig. 4I |
The footprint is defined as the residues that contain atoms within 4 Å from atoms of the MAb MO1.
The BA.1 cryoEM structure contains the N440K mutation.
The subscripts are used for Kabat numbering of antibody residues and the subscripts H or L represent heavy chain or light chain, respectively.
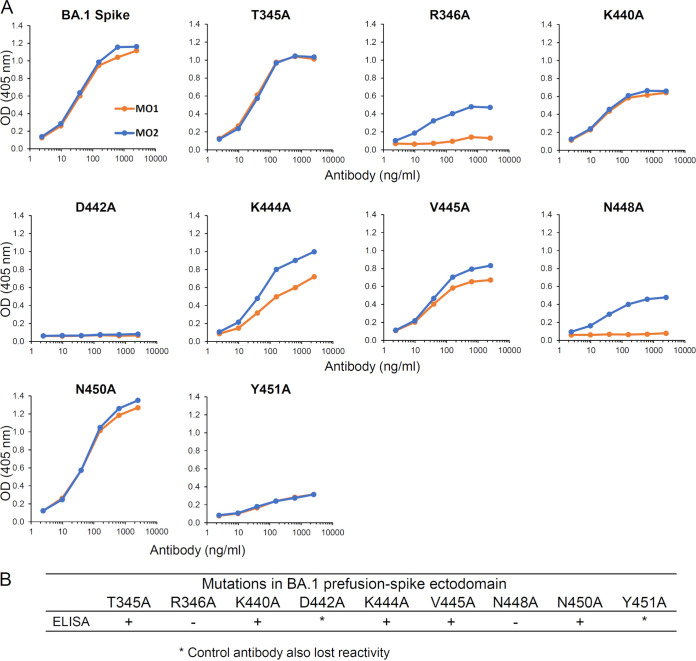
We analyzed the contributions of the possible epitope residues, i.e., T345, R346, K440, D442, K444, V445, N448, N450, and Y451, in the MO1 footprint by using site-directed alanine mutations of the BA.1 spike ectodomain. As shown in Fig. 5A, the antigen reactivity of MO1 was tested by ELISA, and a reduction of reactivity was observed for the R346A and N448A mutant antigens; MO1 clearly reacted to the spike with other substitutions. In summary, the antibody MO1 recognizes R346 and N448 as key contacts (Fig. 5B).
FIG 5.
MO1 recognized R346 and N448 as key epitopes. (A) Binding of MO1 to T345, R346, K440, D442, K444, V445, N448, N450, and Y451 was analyzed by ELISA using site-specific alanine mutations in the BA.1 spike ectodomain. (B) Summarized results of site-specific alanine mutations in the BA.1 spike ectodomain.
The neutralizing MAb MO1 effectively neutralizes Omicron BA.5 in vivo.
Syrian hamsters have been used to evaluate the efficacies of monoclonal antibodies against SARS-CoV-2 (14). To evaluate the efficacy of the MO1 antibody in vivo, we evaluated the effect of passive immunization with MO1 and its isotype controls, MO2 and MO3, at preventing infection with SARS-CoV-2 omicron subvariant BA.5 in hamsters. The experimental scheme is shown in Fig. 6A. The weights of MO1-administered hamsters did not decrease without infection, so there was no apparent toxicity of this antibody (Fig. 6B). After the viral challenge, MO2- or MO3-administered hamsters lost significant body weight, unlike mock-treated hamsters. On the other hand, no significant weight loss was observed in MO1-inoculated hamsters (Fig. 6C). Furthermore, infectious viruses in lung homogenates and nasal wash specimens were significantly reduced in hamsters treated with MO1 antibodies compared to those in MO2- or MO3-treated hamsters (Fig. 6D and E). These results suggest that the MO1 antibody is effective against the SARS-CoV-2 omicron subvariant BA.5 in vivo without toxicity.
FIG 6.
(A) Schematic representation of the evaluation of the toxicity and efficacy of monoclonal antibodies in vivo. (B) The weight changes of hamsters after MO1 administration (n = 5) or mock treatment (n = 5). The averages of weight changes were plotted with symbols, and error bars represent SDs. Two-way ANOVA analysis was performed for statistical analysis (n.s.: not significant). (C) The weight changes after challenge with SARS-CoV-2 omicron subvariant BA.5 to hamsters administered monoclonal antibodies (n = 10). The average weight changes are plotted with symbols, and error bars represent SDs. Two-way ANOVA analysis was performed for statistical analysis to compare to mock-treated hamsters (n = 5, identical to the data shown in Fig. 6B; n.s., not significant; ***, P < 0.001; ****, P < 0.0001). (D and E) Infectious virus titers in lung homogenates (D) and nasal wash specimens (E) at 4 days postinfection. The individual viral titers are represented with symbols. Medians are shown with bars and 95% CIs are indicated with error bars. The limit of detections was 101.5 PFU/g (D) and 101.3 PFU/mL (E), respectively. One-way ANOVA analysis was used for statistical analysis after logarithmic conversion, and Tukey’s test was used as a post hoc analysis (n.s., not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001).
DISCUSSION
The emergence of SARS-CoV-2 variants has prolonged the COVID-19 pandemic. The neutralizing MAb that we identified in the present study and named MO1 has strong neutralizing potency against SARS-CoV-2 variants including the Omicron BA.5 variant. The Omicron variants BA.1 and BA.2 are able to escape from the majority of the known neutralizing antibodies (15, 16); meanwhile, the escape ability of BA.5 is strengthened by additional mutations such as F486V around the binding site of class 1 antibodies and the mutation site L452R around the binding site of class 3 antibodies (17). Indeed, the neutralizing antibody MO2 identified herein, which can strongly neutralize both BA.1 and BA.2, lacked neutralizing activity for BA.5. In contrast, MO1 (which recognizes the conserved epitopes among SARS-CoV-2 variants) has maintained its neutralizing activity against BA.5 and the conserved epitopes by avoiding the position of the L452R mutation site, accounting for this antibody’s broad neutralizing activity.
MO1 does not share the footprint on RBD with ACE2 (Fig. 4; Fig. S7A). A simulation model indicated that there is no large overlap between MO1 and ACE2 on the RBD; however, a slight contact at the glycan of ACE2 N53 is possible (Fig. S7B). The BLI competition assay indicated that MO1 and ACE2 do not bind to RBD simultaneously (Fig. 3D). Thus, it is suggested that the binding of Spike to the ACE2 is interfered with due to the steric clash at the glycan if MO1 occupies its binding site. A simulation model extending the N53 glycan (Fig. S7C and D), which is more natural for ACE2 molecules, showed an obvious steric clash with MO1 light chain, supporting the assumption about competition. Because the MO1 biding site is similar to the binding sites of class 3 antibodies, which neutralize SARS-CoV-2 without inhibiting the Spike-ACE2 interaction, MO1 may also have other mechanisms of neutralization, i.e., steric hindrance or aggregation of virions or inhibition of an undefined step, as suggested for the MAb sotrovimab developed from the antibody S309 (https://www.fda.gov/media/149534/download) (11).
It has been reported that the antibodies bebtelovimab (LY-CoV1404; reference 16) and cilgavimab (AZD1061; reference 18) have neutralizing activity against Omicron BA.1, BA.2, and BA.5 and that cilgavimab showed reduced activity against BA.5 (19). The binding affinity and neutralizing function of MO1 against BA.5 are high (Fig. 2 and 3). Interestingly, both have a different but overlapping footprint with MO1 (Fig. S8). Because the right shoulder of the RBD is distant from both the key mutation site L452R and the F486V site of the Omicron BA.5 variant, it is reasonable that broadly neutralizing antibodies that are effective against variants up to BA.5 target this area of the RBD. Both bebtelovimab and cilgavimab are class 2 antibodies that block the ACE2 binding by covering that footprint (16, 18) (Fig. S8), and MO1 also inhibits spike-ACE2 interaction as discussed above. One of the recently reported BA.5 neutralizing antibodies, 002-S21F2, also binds to overlapping site on RBD (20), suggesting a common target site for antibodies for effective neutralization against the BA.5 variant.
Other antibodies that have been reported to recognize a location of the RBD that is similar to that recognized by MO1 have different binding modes from that of MO1, as none of them has an identical set of residues to MO1 in the footprint on RBD (Fig. S9). The VH/VL gene pair of MO1, IGHV3-9*01/IGKV1-9*01, is not shared with these antibodies (Table S2) and with reported antibodies including the 002-S21F2, bebtelvimab, cilgavimab, and so on (20). Thus, it is suggested that MO1 is a broadly effective neutralizing antibody with a unique sequence and structure to recognize the shared target site on the RBD.
The spike residue R346 is a key residue for MO1 to recognize (Table 2 and Fig. 5), and most of the other antibodies targeting similar sites on the RBD also include R346 in their footprint (Fig. S9A). One of the major variants, Omicron BA.1.1, which has also spread worldwide (21), contains an R346K mutation, but MO1 can potently neutralize this variant (Fig. 2). Isolation of the broadly neutralizing MAbs in this study may explain the insubstantial effect of the R346K mutation for virus neutralization sensitivity against sera from vaccinated people (22). Of note, the next variants BA.4.6 and BF.7, which emerged as the threat (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports), both possess an R346T mutation in the spike in addition to the BA.5 mutations (see Fig. S6). Despite the mutation at the epitope residue, MO1 recognized the SARS-CoV-2 spike protein of BA.4.6 and BF.7 (Fig. S2). However, unfortunately, MO1 did not neutralize the new Omicrons, BQ.1.1 and XBB.1, which have mutations in the recognition sites that emerged during the manuscript submission. It has been reported that bebtelovimab was also unable to neutralize these Omicrons (23). BQ.1.1 and XBB.1 have two mutations in the MO1 recognition site, R346T and K444T for BQ.1.1 and R346T and V445P for XBB.1. These mutations are assumed to cause MO1 to lose its binding ability and neutralizing activity. The momentum of the virus evolution is amazing, and it is splendidly avoiding immunity. However, not a few neutralizing antibodies against these variants were present in the individuals’ sera in this study (data not shown), and it cannot be said that the viruses completely evade immunity. Further screening may find monoclonal antibodies capable of neutralizing these variants. In addition, so far, the BA.5 lineage is the mainstream in our country, Japan as of 11 January 2023 (Current Situation of Infection, 11 January 2023, https://www.niid.go.jp/niid/en/2019-ncov-e/11761-covid19-ab113th-en.html), and different BA5-derived derivatives may become prevalent in the future. MO1 may also have an effect if the other subtypes occur. The MAbs that can neutralize the new variants may be obtained in those individuals by further screening. We are currently working on it.
MO1 is derived from PBMCs of individuals who were infected with a SARS-CoV-2 variant, presumably the D614G with only the D614G mutation in the spike, and then received two doses of an mRNA vaccine encoding the spike gene of the ancestral SARS-CoV-2 (8). Our finding that MO1 can broadly neutralize the early SARS-CoV-2 variants and the Omicron BA.1, BA.1.1, BA.2, and BA.5 variants demonstrates that immunity against the spike with the ancestral SARS-CoV-2 RBD sequence can protect humans by inducing neutralizing antibodies, like MO1, that recognize conserved epitopes. Indeed, our serological study regarding the sustainability of the neutralizing antibodies after early SARS-CoV-2 (D614G) infection indicated that cross-reactive neutralizing antibodies are sustained for >6 months, although neutralizing antibodies specific for SARS-CoV-2 (D614G) decline (8, 24). It is also noteworthy that the booster dose (third-dose) vaccination using the mRNA vaccine based on D614G SARS-CoV-2 can induce neutralizing antibodies against Omicron BA.1 and BA.2 (6, 7). Because MO1 recognizes a common epitope in the spike proteins of SARS-CoV-2 virus and vaccine, it is possible that infection resulted in MO1 antibody-producing B cells that were boosted by vaccination.
The conserved epitope recognized by MO1 demonstrates that room for target sites remains available for neutralizing antibodies and the key task to developing effective immunity against a broad range of SARS-CoV-2 variants is to determine how to effectively induce antibodies against this conserved epitope. One of our previous studies revealed that patients who were infected with D614G SARS-CoV-2 and then received a two-dose mRNA vaccination acquired high neutralizing antibody titers against Omicron BA.1 (8), and the neutralizing antibodies that were isolated in the present study were derived from donors with this background. Repeated exposure to the SARS-CoV-2 spike protein, irrespective of the variation in the sequence, may induce broadly neutralizing antibodies.
Cao et al. (17) reported that BA.1-derived vaccine boosters may not achieve broad-spectrum protection against new Omicron variants because Omicron may evolve mutations to evade the humoral immunity elicited by BA.1. In addition, all neutralizing antibodies, MO1, MO2, and MO3, obtained in this study were found to be able to neutralize the BA.2.75 variant (Fig. 2) which has some unique mutations (24). The MO1 epitope is conserved in the BA.2.75 variant explaining for the susceptibility of this variant (Fig. S6).
Our present findings demonstrate that the booster vaccination for the D614G can induce neutralizing antibodies that recognize common epitopes that are conserved among SARS-CoV-2 variants up to BA.5. It can also be speculated that a three- or four-dose vaccination based on the D614G may stimulate memory B cells that can produce antibodies that have common epitopes conserved, showing broad neutralizing activity, and that booster vaccinations should thus be required even for previously infected individuals.
MATERIALS AND METHODS
Collection of human samples.
Blood samples were collected at Hyogo Prefectural Kakogawa Medical Center (Kakogawa, Japan) from patients who were infected with COVID-19 during the 5-month period of July to November 2020 and then received a two-dose messenger (m)RNA vaccine. Patients whose COVID-19 onset occurred during this period were infected mainly with the D614G variant of SARS-CoV-2, according to the epidemiological data in the Japan Variant Report (https://outbreak.info/location-reports?loc=JPN). For each of the patient’s samples, sera and PBMCs were separated. In our earlier investigation (8), we identified the sera that had high neutralizing activity against SARS-CoV-2, and in the present study we used those patients’ (donors’) sera and data.
Isolation of human antibodies’ genes from the donors’ PBMCs.
Isolation of human antibody genes from the above-described donor PBMCs was performed according to the protocols of Ecobody technology (9) (iBody inc, Nagoya, Japan). Briefly, the recombinant SARS-CoV-2 spike protein was fluorescently labeled with Alexa Fluor 633 NHS ester (succinimidyl ester) according to the manufacturer’s protocol (ThermoFisher Scientific) and employing the labeled protein, memory B cells expressing anti-SARS-CoV-2 spike MAbs were screened and sorted as single cells by FACS SH800S (Sony, Japan). Antibody variable region genes of the light chain and heavy chain were PCR amplified from the cDNA of each single cell and used to make Fab expression construct for Escherichia coli cell-free protein synthesis (PUREfrex 2.0, GeneFrontier, Japan). The translation mixture was diluted with Block ACE solution (KAC, Japan), and the reactivity against the spike antigen was tested for each expressed Fab by ELISA. The 10 candidates that showed the highest reactivities were selected for the subsequent analyses.
The sequences of the selected antibody variable region genes were determined by the Sanger method and then subcloned into human IgG MAb-expressing vectors as follows. The heavy-chain and light-chain V-region sequences were amplified. With the use of the EcoRI-XhoI sites and EcoRI-BsiWI sites, the sequences were then inserted into the pFUSEss-CHIg-hG1 and pFUSE2ss-CLIg-hK expression vectors (InvivoGen, San Diego, CA), respectively. We used a capillary electrophoresis (CE) sequencer (model DS3000, Hitachi High-Tech, Tokyo) to confirm the sequences. This study was approved by the ethical committees of Kobe University Graduate School of Medicine (approval code: B200200).
Antibody expression and purification.
With polyethyleneimine and the two plasmids containing the heavy-chain and light-chain sequences of the antibodies, recombinant MAbs were expressed in HEK293T (human kidney) cells by transfection. The cells were cultured in 10% FBS (fetal bovine serum) and added to Dulbecco’s modified Eagle’s medium (DMEM) for 3 days in a CO2 incubator (5% CO2) maintained at 37°C. The Expi293F Expression System (Thermo Fisher Scientific) was also used to express MAbs, in accordance with the manufacturer's protocol.
For the purification of the antibodies, we added rProtein A Sepharose (Cytiva, Marlborough, MA) to the culture supernatant, and the mixture was gently rocked for 10 to 12 h at 4°C. The resulting resin was then centrifuged at 500 × g at 4°C for 5 min and subsequently washed with cold phosphate-buffered saline (PBS) five times. The trapped MAbs were eluted with sodium citrate buffer (i.e., 40 mM trisodium citrate, pH 3.4). The solution was then immediately neutralized to ~pH 7.0 by the addition of 1 M Tris-HCl buffer.
Ultrafiltration with an Amicon Ultra centrifugal filter (Sigma Chemicals, St. Louis, MO) with the molecular-weight cutoff of 50,000 Da was conducted to replace the solvent with PBS, and each MAb was concentrated. We performed sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to determine the purity of each MAb, and we used a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA) to determine the concentration of each antibody.
Plasmid construction and the expression of SARS-CoV-2 spike proteins.
One of our earlier studies describes the preparation of the prefusion spike ectodomain with the D614G mutation (24). Briefly, to express the prefusion-stabilized spike ectodomain trimer, we used the plasmid pCAGGS (25) that codes the amino acid (aa) residues of spike 1 to 1213 including the mutations D614G, R682del, R683del, R685del, F817P, A892P, A899P, A942P, K986P, and V987P (26). This construct also contains a T4 foldon sequence and a His6 tag at the C-terminal side. We used an overlap PCR to prepare the spike ectodomain with the Delta mutations. The GeneArt Gene Synthesis service (Thermo Fisher Scientific) synthesized the spike sequences of Omicron BA.1 and BA.2; the BA.1.1 sequence was prepared by an overlap PCR based on the BA.1 sequence. The BA.2.75, BA.5, BA.4.6, BF.7, and BQ.1.1 sequences were prepared based on the BA.2 sequence. In the present study, we defined the RBD of the spike as residues 334 to 528.
Next, each variant’s RBD coding sequence was subcloned into plasmid pCAGGS with the spike signal sequence at the N-terminal side and the His6 tag at the C-terminal side. Overlap PCRs were performed to achieve site-directed substitutions. All sequences were confirmed by the DS3000 CE sequencer mentioned above. The spike ectodomain or the RBD was expressed in HEK293T cells or the Expi293F expression system mentioned above. The culture supernatants were collected at 4 to 5 days posttransfection. The affinity chromatography matrix nickel-charged nitriloacetic acid (Ni-NTA) agarose (Qiagen, Hilden, Germany) was used to purify the target proteins. After a protein was concentrated by an Amicon Ultra centrifugal filter, its purity was evaluated by SDS-PAGE, and its concentration was determined with the NanoDrop spectrophotometer.
Plasmid construction and the expression of human ACE2 ectodomain.
The human ACE2 gene was synthesized by the GeneArt Gene Synthesis service (Thermo Fisher Scientific), and the ectodomain region (amino acid residue 1 to 614) was subcloned into the PCAGGS plasmid with His6 tag sequence. Expression and purification of the human ACE2 ectodomain were done in the same way for spike ectodomain described above.
Enzyme-linked immunosorbent assay.
Our earlier study (24) also describes the ELISA performed herein; briefly, prefusion-stabilized spike protein as the antigen was added to the wells (at 100 or 200 ng/well) of 96-well ELISA plates (Corning, New York, NY), followed by incubation for >12 h at 4°C. Each well was then washed twice with PBS-0.1% Tween 20 (PBST) before reagents were added for the subsequent steps.
The wells were incubated for 2 h at 4°C with blocking buffer (1% bovine serum albumin with PBST), and then the selected antibody was reacted for 1 h at 37°C. Horseradish peroxidase (HRP)-conjugated goat anti-human IgG (1:10,000 dilution, Abcam, Cambridge, MA) was used to detect bound antibodies, and the reaction was continued for 1 h at 37°C. To start the enzyme reaction, we added ABTS solution (Roche Diagnostics, Indianapolis, IN). After 40-min incubation in the dark at room temperature, we stopped the reaction by adding 1.5% (wt/vol) oxalic acid dehydrate solution. As the final step, the optical density was measured at a wavelength of 405 nm (OD405) by a microplate photometer (Multiskan FC, Thermo Fisher Scientific).
Viruses.
BIKEN Innovative Vaccine Research Alliance Laboratories (Osaka, Japan) provided the SARS-CoV-2 strain that contains the spike D614G mutation (DNA Data Bank of Japan [DDBJ]: accession no. LC644163), and in the present study we refer to this strain as strain D614G. Japan’s National Institute of Infectious Disease (Tokyo) provided the SARS-CoV-2 strains of the Pango lineage AY.122 (EPI_ISL_2158617), which we used as the Delta variant; BA.1.18 (EPI_ISL_7418017), which we used as the Omicron BA.1 variant; BA.1.1 (EPI_ISL_7571618), which we used as the BA.1.1 variant; BA.2 (EPI_ISL_9595859), which we used as the BA.2 variant; BA.2.75 (EPI_ISL_13969765), which we used as the BA.2.75 variant; and BA.5 (EPI_13241867); BQ.1.1 (EPI_ISL_15579783), which we used as the BQ.1.1 variant; and XBB.1 (EPI_ISL_15669344), which we used as the Omicron XBB.1 variant. We propagated the viruses by the infection of Vero E6 (TMPRSS2) cells in 2% FBS containing DMEM (27) to create a stock of each virus (28).
Plaque reduction neutralization test.
To conduct a plaque reduction neutralization test (PRNT) and determine the neutralization percentage for each virus strain, we seeded Vero E6/TMPRSS2 cells (2 × 105 cells/well) on 12-well plates (Corning) and cultivated them for 24 h with 5% CO2 at 37°C. The cell monolayers were then washed one time with DMEM (without FBS). Each antibody diluted in DMEM (without FBS) was mixed with 100 PFU of SARS-CoV-2 and incubated for 1 h at 37°C. We then added the virus-antibody mixture to the Vero E6/TMPRSS2 cells and cultured for 1 h with 5% CO2 at 37°C. After inoculum was removed, the infected cells were washed twice with PBS and incubated for 3 to 6 days at 37°C with 5% CO2 together with DMEM containing 2% FBS and 1.6% methylcellulose.
The culture medium was removed, and cells were washed twice with PBS and fixed with 80% methanol for 1 h at room temperature. The remaining cells were stained with 1% crystal violet in 50% methanol for the visualization of plaques, which were counted manually. The ratio of neutralization was obtained by dividing the number of plaques obtained without the antibody by the number of plaques obtained with the antibody.
Biolayer interferometry.
The affinity between each antibody and spike protein was measured by biolayer interferometry (BLI) with a BLItz Biolayer Interferometer System (Sartorius, Göettingen, Germany). The streptavidin sensor was coated by Capture Select Biotin Anti-IgG-Fc (Multispecies) Conjugate (Thermo Fisher Scientific) to capture the antibodies tested in this study. After each antibody was loaded on the sensor as the ligand, the sensor was soaked in solution containing the RBD of each SARS-CoV-2 variant as the analyte. The obtained kinetics data were fitted using standard equations of a BLI association and dissociation model assuming a 1:1 interaction. Competition between ACE2 and the antibodies was assessed by comparing the response for the BA.2 RBD and the BA.2 RBD/ACE2 mixture. At least two repeated measurements were performed for all ligand-analyte combinations; representative data are presented.
Cryoelectron microscopy.
The Fab domain of the antibody was prepared by papain digestion. The antibody solution was mixed with an immobilized papain agarose (Thermo Fisher Scientific) supplemented with 10 mM cysteine, and the pH was adjusted to approximately pH 7 by adding 1 M Tris-HCl, pH 8.8. After incubation at 37°C for 48 h, the papain agarose was filtrated, and the Fc and uncleaved antibodies were removed by passing through rProtein A Sepharose resin (Cytiva, Marlborough, MA). The complex of MO1 Fab and BA.1 prefusion-stabilized spike ectodomain was purified by a Sephacryl S-300 HR column (Cytiva, Marlborough, MA) equilibrated with the running buffer (20 mM Tris-HCl pH 8.0 and 150 mM sodium chloride) using the ÄKTA pure system (Cytiva, Marlborough, MA). The purified complex was concentrated by ultrafiltration using an Amicon Ultra centrifugal filter (Sigma) with the molecular weight cutoff of 100,000 Da to 9.4 mg/mL. The sample was applied to a freshly glow-discharged Quantifoil holey carbon grid (R0.6/1.0, Cu, 300 mesh), using a Vitrobot Mark IV (FEI) at 8°C with a blot force of 15 and a blotting time of 4 s under 100% humidity conditions, and then the grids were plunge-frozen in liquid ethane. The prepared grids were transferred to a CRYO ARM 300 (JEOL, Tokyo), running at 300 kV and equipped with a Cold Field Emission Gun, an Omega-type in-column energy filter, and a Gatan K3 camera (Gatan AMETEK, CA USA) in a correlated double-sampling, counting mode. Imaging was performed at a nominal magnification of ×60,000, corresponding to a calibrated pixel size of 0.752 Å/pix at the specimen (EM01CT at SPring-8). The incident electron beam was set to parallel illumination condition with the dose rate of 10 e-/pix/s at the detector. Each exposure was recorded as a movie of 50 frames for 2.26 sec with an accumulated dose of 50 e-/Å2 at the specimen. The data were automatically collected by SerialEM ver. 4.0beta6 (Fig. S4A) (29) with custom scripts provided by JEOL using a defocus range set at a range of −1.2 to −1.6 μm from the center of the image-shift matrix of 7 × 7 × 1 at each stage shift. The stage shift was refined by using the software yoneoLocr (30) at the center of the hole. Movies of 6,174 were acquired and subjected to beam-induced motion correction, using RELION-4.0-beta2 (31), and the contrast transfer function (CTF) parameters were estimated using CTFFIND4.1 (32).
Particles were picked using crYOLO ver. 1.8.2 (33) and extracted with a rescaled pixel size of 3.008 Å/pix and a box size of 120. Image processing as described in detail below was performed with RELION-4.0-beta2. Particles of 176,322 were divided into 6 groups and 1 of the subsets was subjected to two-dimensional classification with the VDAM algorithm and only good classes (rlnClassScore >0.295) (Fig. S4B) were selected for initial model building. Initial single models were built by RELION-4.0-beta2 for a single model with a circular mask diameter of 270 Å and then aligned with C3 symmetry for further 3D image processing. All the particles from subset 1 were subjected to 3D classification with 8 classes, and only good-looking classes showing 3 Fabs were selected. The selected good classes were subjected to the 3D autorefinement with the reference model from one of the selected classes from the 3D classification with the circular mask diameter of 280 Å. The rest of the subsets were then processed like the first subset, and the refined particles were joined and extracted with an original pixel size of 0.752 Å/pix and Box size of 512. This particle set was subjected to 3D autorefinement with a circular mask diameter of 320 Å to obtain the consensus map. A solvent mask was generated for the consensus map and applied in postprocessing to obtain a sharpened map and to calculate the gold-standard FSC. Then, the refined particles were subjected to 3D classification into 4 classes without image alignment with a tau value of 30 to classify particles with small differences in conformation. One of the classes showing distinct a difference in the angle of one of the Fabs in the horizontal position was selected (conformation 2). Then, the rest of three that looked similar in terms of the horizontal Fab position were subjected to further 3D classification into two classes (conformation 1 and 3). These three conformations of the horizontal Fab bound to the spike proteins are shown in Fig. S4C. Those three were subjected to further refinement for CTF and beam aberration and a Bayesian polishing process. The final maps were obtained with another round of beam aberration refinement only for BeamTilt and 3D auto refinement. The resolution of each conformation was calculated with map sharpening (Fig. S4C), and the final maps of conformation 1, conformation 2, and conformation 3 are shown in Fig. S4C.
A cryo-EM map of conformation 2 was used for the atomic-model building of the MO1 Fab and BA.1 prefusion-stabilized spike ectodomain complex because of the clear visibility of residues near the interface between MO1 and the BA.1 prefusion-stabilized spike ectodomain. The structure of the Omicron spike trimer (PDBID: 7WPE; reference 34) for the BA.1 prefusion-stabilized spike ectodomain and the homology model for the MO1 Fab constructed using the AlphaFold 2.2.0 (35) were first manually fitted into the map using UCSF Chimera 1.15 (36) and then inspected and manually adjusted with Coot 0.9.8.1 (37). The complete structure was refined with phenix.real_space_refine of PHENIX 1.20.1 (38) with geometric restraints for the protein-NAG coordination. The statistics for all data collection and structural refinements are summarized in Table S1. The software UCSF ChimeraX 1.3 (39) was used to make the molecular illustrations.
Data and materials availability.
The cryo-EM map and coordinates have been deposited at the Electron Microscopy Data Bank and Protein Data Bank with the following accession numbers: conformation 1 MO1-BA.1 prefusion-stabilized spike ectodomain, EMD-34469 and PDB 8H3M; conformation 2 MO1-BA.1 prefusion-stabilized spike ectodomain, EMD-34470 and PDB 8H3N; and conformation 3 BA.1 prefusion-stabilized spike ectodomain, EMD-34488.
Evaluation of toxicity and efficacy of monoclonal antibodies in vivo.
Four-week-old male Syrian hamsters were purchased from Japan SLC, Inc. Hamsters were allowed free access to food and water. After 1 week of housing, monoclonal antibodies were administered intraperitoneally at a dose of 10 mg/kg. SARS-CoV-2 Omicron BA.5 3 × 105 PFU was administered intranasally at a dose of 100 μL under anesthetic conditions at 12 h after inoculation of the antibodies. The weights of the hamsters were measured every day for up to 4 days postinfection. The animals were euthanized by loss of blood under anesthesia conditions, and we collected lungs and nasal wash specimens. The lungs were cut and mashed with a Biomasher II (Kanto Chemical Co., Inc.) and suspended in DMEM. After centrifugation at 100 × g, the supernatants were collected as lung homogenates. Infectious virus titers of lung homogenates and nasal wash specimens were evaluated by plaque formation assay with veroE6/TMPRSSII cells (Japanese Cancer Research Resources Bank [JCRB]). Briefly, diluted samples were inoculated to confluent veroE6/TMPRSSII cells on 6-well plates, and incubated at 37°C for 1 h for absorption. Then, the cells were washed with d-PBS and layered with DMEM + 1% SeaPlaque agarose (Lonza). After incubation for 3 days, the cells were fixed with formalin and stained with crystal violet solution. Visible plaques were counted for the calculation of infectious virus titers.
Quantification and statistical analysis.
All data for animal experiments are represented as means ± SD or medians ± 95% confidence intervals (CI). Two-way ANOVA was used for the weight change over time. One-way ANOVA was performed for the viral titers in the lung homogenates and nasal wash specimens, and Tukey’s test was used for post hoc analysis. The value of limit of detection was applied for statistical analyses for the samples below the limit. All analyses were performed using the GraphPad Prism software (n.s., not significant: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001).
Ethics statement.
The Kobe University Graduate School of Medicine's Ethics Committee approved the collection and use of the COVID-19 patients’ blood samples (approval code: B200200). The patients’ written informed consent for this use was obtained. All animal experimental protocols, including anesthesia conditions, endpoints for infection, and euthanasia methods, were reviewed and approved by the Osaka University Animal Experiment Committee (approval no. R02-10-0).
ACKNOWLEDGMENTS
We thank Kazuro Sugimura for his full support of this study. We express our gratitude for the cooperation of researchers of the Division of Respiratory Medicine, Department of Internal Medicine, Kobe University Graduate School of Medicine, and medical staff of Hyogo Prefectural Kakogawa Medical Center. We thank BIKEN Innovative Vaccine Research Alliance Laboratories for providing the SARS-CoV-2 B2 strain used as the D614G variant herein. We also thank the National Institute of Infectious Disease Japan for providing the SARS-CoV-2 of Pango lineages AY.122, BA.1.18, BA.1.1, BA.2, BA.2.75, BA.5, BQ.1.1, and XBB.1 used here as the Delta, Omicron (B.1.159) BA.1, BA.1.1, BA.2, BA.2.75, BA.5, BQ.1.1, and XBB.1 variants, respectively.
This work is partially supported by the Research Support Project for Life Science and Drug Discovery (Basis for Supporting Innovative Drug Discovery and Life Science Research: BINDS) from the Japan Agency for Medical Research and Development (AMED) under grant number JP22ama121001. Y.K. was supported by the Taniguchi Memorial Scholarship program provided by the BIKEN Foundation. S. Sutandhio, L.H.T., M.I.M., and G.B.E. were supported by Japanese Government (Monbukagakusho: MEXT) Scholarships.
This work was also supported by the Hyogo Prefectural Government and a grant from the Kobayashi Foundation. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication. Molecular graphics and analyses performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases.
Conceptualization, H.I., M.N., L.H.T., and Y.M.; Methodology, H.I., M.N., H.S., K.K. S.O., and Y.M.; Sample Collection, T.N., S.N., S. Sano, and S.I.; Formal analysis, H.I., M.N., L.H.T., S. Sutandhio, M.I.M., G.B.E., H.S., K.K., N.H., K.A., Y.K., K.F., M.S., T.N., J.A., S.O., and Y.M.; Funding Acquisition, Y.M.; Project Administration, H.I., M.N., and Y.M.; Supervision, Y.M.; Writing – Original draft, H.I., M.N., and Y.M.; Writing – Review & Editing, H.I., M.N., and Y.M.
S.O. is employed by BIKEN foundation. The other authors declare no conflicts of interest related to this research.
Footnotes
Supplemental material is available online only.
Contributor Information
Yasuko Mori, Email: ymori@med.kobe-u.ac.jp.
Mark T. Heise, University of North Carolina at Chapel Hill
REFERENCES
- 1.Dejnirattisai W, Huo J, Zhou D, Zahradník J, Supasa P, Liu C, Duyvesteyn HME, Ginn HM, Mentzer AJ, Tuekprakhon A, Nutalai R, Wang B, Dijokaite A, Khan S, Avinoam O, Bahar M, Skelly D, Adele S, Johnson SA, Amini A, Ritter TG, Mason C, Dold C, Pan D, Assadi S, Bellass A, Omo-Dare N, Koeckerling D, Flaxman A, Jenkin D, Aley PK, Voysey M, Costa Clemens SA, Naveca FG, Nascimento V, Nascimento F, Fernandes da Costa C, Resende PC, Pauvolid-Correa A, Siqueira MM, Baillie V, Serafin N, Kwatra G, Da Silva K, Madhi SA, Nunes MC, Malik T, Openshaw PJM, Baillie JK, Semple MG, ISARIC4C Consortium ., et al. 2022. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tegally H, Moir M, Everatt J, Giovanetti M, Scheepers C, Wilkinson E, Subramoney K, Moyo S, Amoako DG, Baxter C, Althaus CL, Anyaneji UJ, Kekana D, Viana R, Giandhari J, Lessells RJ, Maponga T, Maruapula D, Choga W, Matshaba M, Mayaphi S, Mbhele N, Mbulawa MB, Msomi N, Naidoo Y, Pillay S, Sanko TJ, San JE, Scott L, Singh L, Magini NA, Smith-Lawrence P, Stevens W, Dor G, Tshiabuila D, Wolter N, Preiser W, Treurnicht FK, Venter M, Davids M, Chiloane G, Mendes A, McIntyre C, O’Toole A, Ruis C, Peacock TP, Roemer C, Williamson C, Pybus OG, Bhiman J, et al. 2022. Continued emergence and evolution of Omicron in South Africa: new BA.4 and BA.5 lineages. medRxiv. doi: 10.1101/2022.05.01.22274406. [DOI] [Google Scholar]
- 3.Anonymous. In press. COVID variants to watch, and more–this week's best science graphics. Nature. doi: 10.1038/d41586-022-03533-7. [DOI] [PubMed] [Google Scholar]
- 4.Kurhade C, Zou J, Xia H, Liu M, Chang HC, Ren P, Xie X, Shi PY. 2023. Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nat Med 29:344–347. doi: 10.1038/s41591-022-02162-x. [DOI] [PubMed] [Google Scholar]
- 5.Muik A, Lui BG, Wallisch A-K, Bacher M, Mühl J, Reinholz J, Ozhelvaci O, Beckmann N, Güimil Garcia RdlC, Poran A, Shpyro S, Finlayson A, Cai H, Yang Q, Swanson KA, Türeci Ö, Şahin U. 2022. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science 375:678–680. doi: 10.1126/science.abn7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa K, Tjan LH, Kurahashi Y, Sutandhio S, Nishimura M, Arii J, Mori Y. 2022. Assessment of neutralizing antibody response against SARS-CoV-2 variants after 2 to 3 doses of the BNT162b2 mRNA COVID-19 vaccine. JAMA Netw Open 5:e2210780. doi: 10.1001/jamanetworkopen.2022.10780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tjan LH, Furukawa K, Kurahashi Y, Sutandhio S, Nishimura M, Arii J, Mori Y. 2022. Induction of high neutralizing activity against both Omicron BA.2 and Omicron BA.1 by coronavirus disease 2019 messenger RNA booster vaccination. J Infect Dis 226:1481–1483. doi: 10.1093/infdis/jiac159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurahashi Y, Furukawa K, Sutandhio S, Tjan LH, Iwata S, Sano S, Tohma Y, Ohkita H, Nakamura S, Nishimura M, Arii J, Kiriu T, Yamamoto M, Nagano T, Nishimura Y, Mori Y. 2022. Cross-neutralizing activity against Omicron could be obtained in SARS-CoV-2 convalescent patients who received two doses of mRNA vaccination. J Infect Dis 226:1391–1395. doi: 10.1093/infdis/jiac178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ojima-Kato T, Nagai S, Nakano H. 2017. Ecobody technology: rapid monoclonal antibody screening method from single B cells using cell-free protein synthesis for antigen-binding fragment formation. Sci Rep 7:13979. doi: 10.1038/s41598-017-14277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nutalai R, Zhou D, Tuekprakhon A, Ginn HM, Supasa P, Liu C, Huo J, Mentzer AJ, Duyvesteyn HME, Dijokaite-Guraliuc A, Skelly D, Ritter TG, Amini A, Bibi S, Adele S, Johnson SA, Constantinides B, Webster H, Temperton N, Klenerman P, Barnes E, Dunachie SJ, Crook D, Pollard AJ, Lambe T, Goulder P, Paterson NG, Williams MA, Hall DR, Mongkolsapaya J, Fry EE, Dejnirattisai W, Ren J, Stuart DI, Screaton GR, OPTIC consortium, ISARIC4C consortium . 2022. Potent cross-reactive antibodies following Omicron breakthrough in vaccinees. Cell 185:2116–2131.e18. doi: 10.1016/j.cell.2022.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, Peter A, Guarino B, Spreafico R, Cameroni E, Case JB, Chen RE, Havenar-Daughton C, Snell G, Telenti A, Virgin HW, Lanzavecchia A, Diamond MS, Fink K, Veesler D, Corti D. 2020. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence MC, Colman PM. 1993. Shape complementarity at protein/protein interfaces. J Mol Biol 234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 13.Geng Q, Shi K, Ye G, Zhang W, Aihara H, Li F. 2022. Structural basis for human receptor recognition by SARS-CoV-2 omicron variant BA.1. J Virol 96:e0024922. doi: 10.1128/jvi.00249-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers TF, Zhao F, Huang D, Beutler N, Burns A, He WT, Limbo O, Smith C, Song G, Woehl J, Yang L, Abbott RK, Callaghan S, Garcia E, Hurtado J, Parren M, Peng L, Ramirez S, Ricketts J, Ricciardi MJ, Rawlings SA, Wu NC, Yuan M, Smith DM, Nemazee D, Teijaro JR, Voss JE, Wilson IA, Andrabi R, Briney B, Landais E, Sok D, Jardine JG, Burton DR. 2020. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamasoba D, Kimura I, Nasser H, Morioka Y, Nao N, Ito J, Uriu K, Tsuda M, Zahradnik J, Shirakawa K, Suzuki R, Kishimoto M, Kosugi Y, Kobiyama K, Hara T, Toyoda M, Tanaka YL, Butlertanaka EP, Shimizu R, Ito H, Wang L, Oda Y, Orba Y, Sasaki M, Nagata K, Yoshimatsu K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Kuramochi J, Seki M, Fujiki R, Kaneda A, Shimada T, Nakada TA, Sakao S, Suzuki T, Ueno T, Takaori-Kondo A, Ishii KJ, Schreiber G, Sawa H, Saito A, Irie T, Tanaka S, Matsuno K, Fukuhara T, Ikeda T, Sato K, Genotype to Phenotype Japan (G2P-Japan) Consortium . 2022. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell 185:2103–2115.e19. doi: 10.1016/j.cell.2022.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westendorf K, Žentelis S, Wang L, Foster D, Vaillancourt P, Wiggin M, Lovett E, van der Lee R, Hendle J, Pustilnik A, Sauder JM, Kraft L, Hwang Y, Siegel RW, Chen J, Heinz BA, Higgs RE, Kallewaard NL, Jepson K, Goya R, Smith MA, Collins DW, Pellacani D, Xiang P, de Puyraimond V, Ricicova M, Devorkin L, Pritchard C, O'Neill A, Dalal K, Panwar P, Dhupar H, Garces FA, Cohen CA, Dye JM, Huie KE, Badger CV, Kobasa D, Audet J, Freitas JJ, Hassanali S, Hughes I, Munoz L, Palma HC, Ramamurthy B, Cross RW, Geisbert TW, Menachery V, Lokugamage K, Borisevich V, et al. 2022. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep 39:110812. doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, Du S, Wang J, Li Q, Chen X, Yu Y, Wang P, Zhang Z, Liu P, An R, Hao X, Wang Y, Wang J, Feng R, Sun H, Zhao L, Zhang W, Zhao D, Zheng J, Yu L, Li C, Zhang N, Wang R, Niu X, Yang S, Song X, Chai Y, Hu Y, Shi Y, Zheng L, Li Z, Gu Q, Shao F, Huang W, Jin R, Shen Z, Wang Y, Wang X, Xiao J, Xie XS. 2022. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong J, Zost SJ, Greaney AJ, Starr TN, Dingens AS, Chen EC, Chen RE, Case JB, Sutton RE, Gilchuk P, Rodriguez J, Armstrong E, Gainza C, Nargi RS, Binshtein E, Xie X, Zhang X, Shi PY, Logue J, Weston S, McGrath ME, Frieman MB, Brady T, Tuffy KM, Bright H, Loo YM, McTamney PM, Esser MT, Carnahan RH, Diamond MS, Bloom JD, Crowe JE. Jr, 2021. Genetic and structural basis for SARS-CoV-2 variant neutralization by a two-antibody cocktail. Nat Microbiol 6:1233–1244. doi: 10.1038/s41564-021-00972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamasoba D, Kosugi Y, Kimura I, Fujita S, Uriu K, Ito J, Sato K, Genotype to Phenotype Japan (G2P-Japan) Consortium . 2022. Neutralisation sensitivity of SARS-CoV-2 omicron subvariants to therapeutic monoclonal antibodies. Lancet Infectious Diseases 22:942–943. doi: 10.1016/S1473-3099(22)00365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, Patel A, Lai L, Chakravarthy C, Valanparambil R, Reddy ES, Gottimukkala K, Davis-Gardner ME, Edara VV, Linderman S, Nayak K, Dixit K, Sharma P, Bajpai P, Singh V, Frank F, Cheedarla N, Verkerke HP, Neish AS, Roback JD, Mantus G, Goel PK, Rahi M, Davis CW, Wrammert J, Godbole S, Henry AR, Douek DC, Suthar MS, Ahmed R, Ortlund E, Sharma A, Murali-Krishna K, Chandele A. 2022. Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody. Sci Adv 8:eadd2032. doi: 10.1126/sciadv.add2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iketani S, Liu L, Guo Y, Liu L, Chan JF-W, Huang Y, Wang M, Luo Y, Yu J, Chu H, Chik KK-H, Yuen TT-T, Yin MT, Sobieszczyk ME, Huang Y, Yuen K-Y, Wang HH, Sheng Z, Ho DD. 2022. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans JP, Zeng C, Qu P, Faraone J, Zheng YM, Carlin C, Bednash JS, Zhou T, Lozanski G, Mallampalli R, Saif LJ, Oltz EM, Mohler PJ, Xu K, Gumina RJ, Liu SL. 2022. Neutralization of SARS-CoV-2 Omicron sub-lineages BA.1, BA.1.1, and BA.2. Cell Host Microbe 30:1093–1102.e3. doi: 10.1016/j.chom.2022.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Iketani S, Li Z, Liu L, Guo Y, Huang Y, Bowen AD, Liu M, Wang M, Yu J, Valdez R, Lauring AS, Sheng Z, Wang HH, Gordon A, Liu L, Ho DD. 2023. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186:279–286.e8. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren Z, Nishimura M, Tjan LH, Furukawa K, Kurahashi Y, Sutandhio S, Aoki K, Hasegawa N, Arii J, Uto K, Matsui K, Sato I, Saegusa J, Godai N, Takeshita K, Yamamoto M, Nagashima T, Mori Y. 2022. Large-scale serosurveillance of COVID-19 in Japan: acquisition of neutralizing antibodies for Delta but not for Omicron and requirement of booster vaccination to overcome the Omicron's outbreak. PLoS One 17:e0266270. doi: 10.1371/journal.pone.0266270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199. doi: 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh CL, Goldsmith JA, Schaub JM, DiVenere AM, Kuo HC, Javanmardi K, Le KC, Wrapp D, Lee AG, Liu Y, Chou CW, Byrne PO, Hjorth CK, Johnson NV, Ludes-Meyers J, Nguyen AW, Park J, Wang N, Amengor D, Lavinder JJ, Ippolito GC, Maynard JA, Finkelstein IJ, McLellan JS. 2020. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, Nagata N, Sekizuka T, Katoh H, Kato F, Sakata M, Tahara M, Kutsuna S, Ohmagari N, Kuroda M, Suzuki T, Kageyama T, Takeda M. 2020. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci USA 117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furukawa K, Tjan LH, Sutandhio S, Kurahashi Y, Iwata S, Tohma Y, Sano S, Nakamura S, Nishimura M, Arii J, Kiriu T, Yamamoto M, Nagano T, Nishimura Y, Mori Y. 2021. Cross-neutralizing activity against SARS-CoV-2 variants in COVID-19 patients: comparison of 4 waves of the pandemic in Japan. Open Forum Infect Dis 8:ofab430. doi: 10.1093/ofid/ofab430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Yonekura K, Maki-Yonekura S, Naitow H, Hamaguchi T, Takaba K. 2021. Machine learning-based real-time object locator/evaluator for cryo-EM data collection. Commun Biol 4:1044. doi: 10.1038/s42003-021-02577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH. 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohou A, Grigorieff N. 2015. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol 192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner T, Merino F, Stabrin M, Moriya T, Antoni C, Apelbaum A, Hagel P, Sitsel O, Raisch T, Prumbaum D, Quentin D, Roderer D, Tacke S, Siebolds B, Schubert E, Shaikh TR, Lill P, Gatsogiannis C, Raunser S. 2019. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol 2:218. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin W, Xu Y, Xu P, Cao X, Wu C, Gu C, He X, Wang X, Huang S, Yuan Q, Wu K, Hu W, Huang Z, Liu J, Wang Z, Jia F, Xia K, Liu P, Wang X, Song B, Zheng J, Jiang H, Cheng X, Jiang Y, Deng S-J, Xu HE. 2022. Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody. Science 375:1048–1053. doi: 10.1126/science.abn8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD. 2019. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. 2021. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci 30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S9 and Tables S1 and S2. Download jvi.00286-23-s0001.pdf, PDF file, 2.1 MB (2.1MB, pdf)