ABSTRACT
Rift Valley fever virus (RVFV) (family Phenuiviridae) can cause severe disease, and outbreaks of this mosquito-borne pathogen pose a significant threat to public and animal health. Yet many molecular aspects of RVFV pathogenesis remain incompletely understood. Natural RVFV infections are acute, characterized by a rapid onset of peak viremia during the first days post-infection, followed by a rapid decline. Although in vitro studies identified a major role of interferon (IFN) responses in counteracting the infection, a comprehensive overview of the specific host factors that play a role in RVFV pathogenesis in vivo is still lacking. Here, the host in vivo transcriptional profiles in the liver and spleen tissues of lambs exposed to RVFV are studied using RNA sequencing (RNA-seq) technology. We validate that IFN-mediated pathways are robustly activated in response to infection. We also link the observed hepatocellular necrosis with severely compromised organ function, which is reflected as a marked downregulation of multiple metabolic enzymes essential for homeostasis. Furthermore, we associate the elevated basal expression of LRP1 in the liver with RVFV tissue tropism. Collectively, the results of this study deepen the knowledge of the in vivo host response during RVFV infection and reveal new insights into the gene regulation networks underlying pathogenesis in a natural host.
IMPORTANCE Rift Valley fever virus (RVFV) is a mosquito-transmitted pathogen capable of causing severe disease in animals and humans. Outbreaks of RVFV pose a significant threat to public health and can result in substantial economic losses. Little is known about the molecular basis of RVFV pathogenesis in vivo, particularly in its natural hosts. We employed RNA-seq technology to investigate genome-wide host responses in the liver and spleen of lambs during acute RVFV infection. We show that RVFV infection drastically decreases the expression of metabolic enzymes, which impairs normal liver function. Moreover, we highlight that basal expression levels of the host factor LRP1 may be a determinant of RVFV tissue tropism. This study links the typical pathological phenotype induced by RVFV infection with tissue-specific gene expression profiles, thereby improving our understanding of RVFV pathogenesis.
KEYWORDS: host-pathogen interactions, pathogenesis, RNA-seq, Rift Valley fever virus
INTRODUCTION
Rift Valley fever virus (RVFV) (family Phenuiviridae) is a single-stranded, three-segmented, negative-sense RNA virus transmitted by mosquitoes (1–3). RVFV mainly affects ruminants such as sheep, goats, and cattle but can also affect camelids and humans (4–6). Infected animals generally present with fever, anorexia, diarrhea, and overall weakness. Epizootic outbreaks are commonly characterized by abortion storms in sheep flocks and high mortality rates among newborns (1, 7). In humans, the clinical presentation of the disease is characterized by symptoms such as fever, headache, nausea, vomiting, abdominal pain, and diarrhea. In a minority of cases, the infection may progress to severe disease leading to hepato-renal failure, encephalitis, retinitis, and/or hemorrhagic manifestations (8–10).
In both animals and humans, the liver is the primary site of RVFV replication. Histopathological examinations of infected liver tissues revealed that diseased animals exhibit multifocal lesions and necrotic hepatitis (11, 12). The spleen is also commonly targeted during RVFV infection, with microscopic examination revealing various degrees of necrosis. Apart from the liver and the spleen, RVFV can occasionally be found in the kidneys, lungs, skin, brain, and placenta (11, 13–15).
RVFV is endemic to most countries on the African Continent and the Arabian Peninsula. Nevertheless, as competent mosquito vectors already inhabit other geographical regions and are still expanding their territory, facilitated by climate change and international trade and transport, it is likely that the virus will be capable of invading previously unaffected areas (16–19). RVFV outbreaks represent a significant threat to public health and can result in substantial economic losses (4, 20, 21), yet many key aspects of RVFV infection cycle and the molecular mechanisms underlying its pathogenesis are poorly comprehended.
High viremia peaking at 2 to 3 days post-infection followed by an abrupt decline from 4 to 5 days post-infection onward is a signature feature of acute RVFV infections in ruminants (22). Generally, infected animals either succumb or fully recover during these days. Previous studies investigating RVFV virulence factors and pathogenesis revealed the predominant role of the non-structural protein NSs as an innate immune response antagonist. NSs acts primarily through the blockage of type I interferon (IFN) (IFN-α/β) production by inhibiting host cell transcription (23–25) and by inducing the degradation of the double-stranded RNA-dependent protein kinase (PKR) (26–28).
Recently, RNA sequencing (RNA-seq) technologies have enabled the transcriptome-wide analysis of host responses during virus infections. RNA-seq studies benefit from an exhaustive approach to investigate the expression of the whole transcriptome using high-throughput sequencing instead of focusing on a short list of predicted genes of interest (29, 30). This broad analysis allows the detection of novel host factors playing an important role in the infection cycle that have escaped the radar of single-pathway-oriented investigations.
Two previous RVFV-mammalian host RNA-seq studies have been carried out with samples derived from infected cell cultures. The first study investigated the cellular response upon infection with the attenuated MP-12 strain and the virulent ZH548 strain in human small airway epithelial cells. In that study, the top pathways altered in response to infection with both RVFV strains included the regulation of the antiviral response, mitochondrial dysfunction, the DNA damage response, and integrin-linked kinase (ILK) signaling (31). The second study investigated the response induced by MP-12 in HEK293 human embryonic kidney cells. The activation of innate immune signaling pathways and the upregulation of pro-inflammatory cytokines were observed, as were alterations in pathways associated with fatty acid metabolism and extracellular matrix receptor signaling (32).
Although these in vitro studies confirmed that IFN-mediated responses are crucial for fighting RVFV infection, our understanding of the tissue-specific host factors that play a role in in vivo pathogenesis is still very limited. To date, only one in vivo transcriptomic study on a RVFV-infected mammalian (surrogate) host has been performed. Investigation of the immune response in the brain of mice intranasally infected with RVFV revealed a protective response mediated by the mitochondrial antiviral-signaling protein (MAVS) (33). A better understanding of the virus-host interface, especially in a natural RVFV host, would benefit the development of more effective outbreak control strategies.
In this work, we used RNA-seq technology to study in vivo the host transcriptional profiles in the liver and spleen tissues of lambs during RVFV peak viremia. We compared the gene expression profiles of infected lambs at 2 and 4 days post-infection with those of uninfected lambs and performed a genome-wide pathway analysis to identify biological processes that are affected during an acute infection. We revealed gene expression signatures underlying the observed histopathology phenotype, in addition to confirming the transcriptional responses to RVFV previously identified by in vitro experiments. Thus, this study improves our understanding of the in vivo host response to RVFV infection and uncovers new molecular features of RVFV pathogenesis.
RESULTS
Selection of RVFV-infected ovine tissue samples for transcriptome analysis.
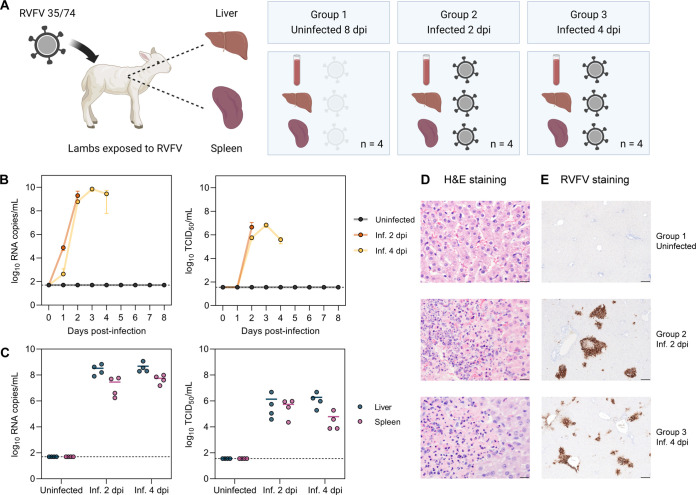
Since sheep are the primary natural hosts of RVFV, and the liver and spleen are the two main target organs of the virus, we selected liver and spleen samples from lambs with moderate to high levels of viral RNA for host transcriptome analysis. For comparison, we selected samples from uninfected lambs. All the tissue samples were obtained from an experiment with Texel-Swifter lambs that were exposed to RVFV either via intravenous injection or via bites from infected mosquitoes (34) (Fig. 1A; see also Materials and Methods for additional details). Samples from group 1 belonged to lambs that were exposed to a low number of infected mosquitoes. These lambs did not develop any signs of disease, nor did they show detectable levels of infectious virus or viral RNA in the blood or the target organs (Fig. 1B and C). Group 1 was thus considered the control group. Samples from group 2 and group 3 belonged to lambs necropsied on days 2 and 4 post-infection, respectively, that presented with high levels of viral RNA (ranging between 107 and 1010 copies/mL on average) and infectious virus (ranging between 104 and 107 median tissue culture infectious doses [TCID50]/mL on average) in the blood and the target organs (Fig. 1B and C). Noteworthy, the RNA copy numbers and infectious titers were slightly higher in the liver samples than in the spleen samples, without appreciable differences between lambs of groups 2 and 3.
FIG 1.
Selection of biologically relevant RVFV-infected samples. (A) Schematic representation of the selected animal samples. Liver and spleen samples of lambs exposed to RVFV strain 35/74 were selected from another study (34). Group 1 consists of uninfected (non-responsive) lambs necropsied at 8 days post-exposure. Group 2 and group 3 consist of infected lambs necropsied at 2 and 4 days post-infection (dpi), respectively (n = 4 samples per group). (B and C) Viral RNA copy numbers and infectious titers in the blood (B) and target organs (C). Viral RNA was quantified by M-segment-specific RT-qPCR, and infectious titers were determined by an endpoint dilution virus isolation assay (34). In panel B, graphs show the means with standard deviations (SD) at each time point. In panel C, dots represent individual replicates, and the horizontal lines represent the means. Dashed lines indicate the limits of detection (50 RNA copies/mL for RT-qPCR and 35.5 median tissue culture infectious doses [TCID50]/mL for the virus isolation assay). (D) Hematoxylin and eosin (H&E) staining of liver tissue sections. Group 1 shows no histological alterations in the liver, while groups 2 and 3 show acute hepatitis with necrosis of hepatocytes and an influx of polymorphonuclear cells. Bars, 20 μm. (E) Immunohistochemical detection of RVFV antigen in liver tissue. RVFV Gn glycoprotein (brown) was detected with antibody 4-D4 (61) in combination with HRP-conjugated secondary antibodies. Bars, 200 μm. Inf., infected.
Histological examination confirmed the infection status.
Upon histological examination of tissue sections of the liver, acute necrosis of hepatocytes and an influx of neutrophils were observed in lambs from groups 2 and 3, clear indicators of acute hepatitis. In contrast, no histological alterations were observed in the liver tissues of the control lambs (group 1) (Fig. 1D). In line with the histological observations, immunohistochemistry detection of RVFV antigen revealed foci of infected cells in the livers of lambs from groups 2 and 3. The majority of the infected cells corresponded to necrotic hepatocytes, and a minority corresponded to endothelial cells. No virus-specific staining was observed in samples from group 1 (Fig. 1E).
RVFV infection status determines characteristic gene expression profiles.
To determine the genome-wide gene expression profile in response to RVFV infection in the liver and spleen, total RNA from frozen tissue samples preserved in RNAlater was isolated and subjected to poly(A)-enriched RNA-seq-based host transcriptome analysis. The integrity of the isolated RNA was confirmed by assessment of the RNA quality number (RQN) or the RNA integrity number (RIN) (35), with RQN or RIN values >6 being found for the majority of the samples (see Table S1 in the supplemental material). At least 20 million high-quality reads (average Phred score >30) per sample were obtained and mapped (ranging from 61.1% to 82.4% alignment) to the sheep (Ovis aries) NCBI reference genome (Fig. S1A to C). A summary of the next-generation sequencing run and general statistics are provided in Table S2. Initially, a total of 26,200 genes were detected across the different samples, of which 18,005 genes were retained for analysis after applying a filter to keep only genes with counts of 3 or higher in at least 2 of the samples. Of the input genes retained for the analysis, 83.8% were annotated in the reference genome (Fig. S1D).
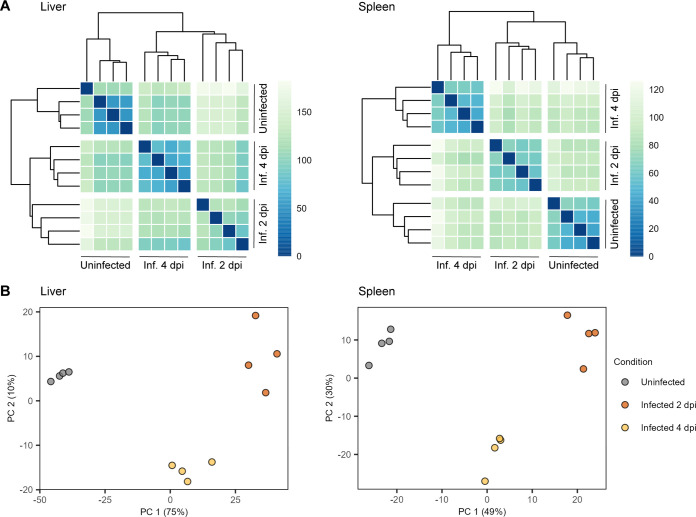
Separation mostly between liver and spleen samples was observed by both heat maps of the Euclidean distances and principal component analysis when all the samples were analyzed together (Fig. S2A and B). As expected, this separation indicates that the gene expression profiles depend first on the host tissue rather than the infection status. We next performed similar analyses of each tissue separately. Heat maps of the Euclidean distances between samples revealed groupings into three distinct clusters (Fig. 2A). This clustering was also evident in the principal component analysis, where PC1 and PC2 represented 75% and 10% of the variance in the liver samples and 49% and 30% of the variance in the spleen samples, respectively (Fig. 2B). Notably, each cluster corresponded to either the uninfected lambs (group 1), the lambs at 2 days post-infection (group 2), or the lambs at 4 days post-infection (group 3), indicating that the infection status of the lamb determines a characteristic gene expression profile in each tissue.
FIG 2.
RVFV infection status determines characteristic gene expression profiles. (A) Heat maps of the Euclidean distances between liver (left) and spleen (right) tissue samples. Based on the calculated distance, each cell is color-coded in shades of a sequential gradient ranging from dark blue (close distance, implying similarity) to light green (far distance, implying dissimilarity). (B) Principal component analyses of liver (left) and spleen (right) tissue samples. Within each tissue type, samples cluster into three distinct well-defined clusters corresponding to a particular infection status. Abbreviations: Inf., infected; dpi, days post-infection.
RVFV infection induces extensive changes in liver and spleen gene expression.
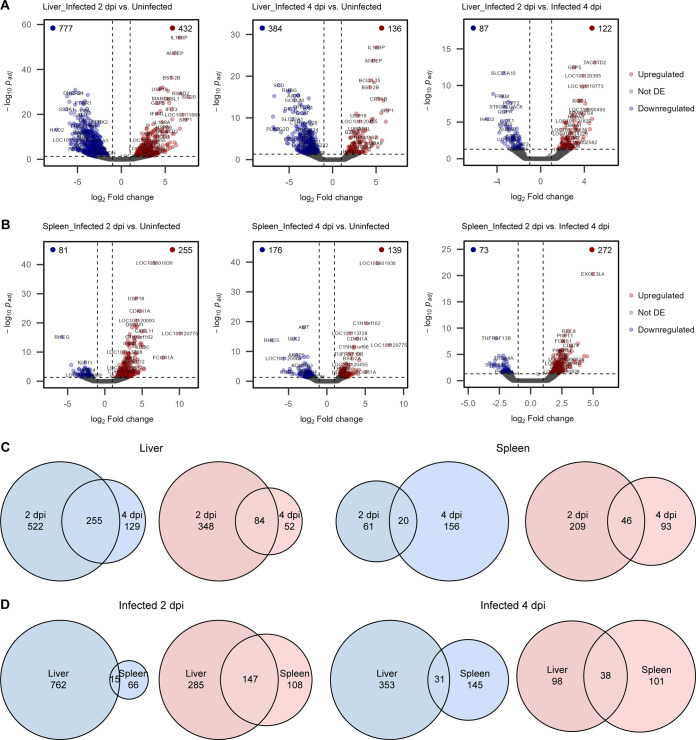
To examine how the infection status influenced the host liver and spleen responses upon RVFV infection, we performed pairwise differential gene expression analysis between the samples of each tissue type. Genes with an absolute log2 fold change of 1 and an adjusted P value <0.05 were considered significantly differentially expressed. In all of the paired comparisons, tens to hundreds of genes were differentially expressed (down- or upregulated), confirming that RVFV infection induced significant changes in the host liver and spleen transcriptomes (Fig. 3A and B). Lists of all of the differentially expressed genes with their corresponding log2 fold changes and adjusted P values are provided in Table S3.
FIG 3.
RVFV-induced changes in liver and spleen gene expression. (A and B) Volcano plots of differential gene expression analysis in liver (A) and spleen (B) tissues. Uninfected (group 1) and infected (groups 2 and 3) samples were compared pairwise. Dots represent individual genes. Genes with an absolute log2 fold change of 1 and a Wald test P value (adjusted by the Benjamini-Hochberg method) <0.05 were considered significantly differentially expressed (DE). The vertical and horizontal dashed lines indicate the log2 fold changes and adjusted P value thresholds, respectively. The numbers of significantly downregulated (blue) and upregulated (red) genes are indicated at the top corners of each plot. (C and D) Euler diagrams representing the numbers of shared (intersection) and unique downregulated (blue) and upregulated (red) genes between infected groups 2 and 3 (compared to control group 1). Comparisons of time points for the same tissue type (C) and comparisons of tissues at the same time point (D) are shown. Abbreviation: dpi, days post-infection.
RVFV infection leads to immune response activation and decreased tissue-specific function.
Within each tissue type, a fraction of differentially expressed genes were common to infected samples (groups 2 and 3) compared to uninfected samples (group 1), as depicted by the genes belonging to the intersection (overlap) of the corresponding gene lists (Fig. 3C and Table S4). This commonality between the infected groups indicates that some genes remain either down- or upregulated during the peak phase (day 2) and the start of virus clearance (day 4) of an acute infection. Interestingly, when comparing samples between tissue types but at the same time points, there was a substantial intersection of genes upregulated in both the liver and the spleen. This intersection of upregulated genes in both tissues is mainly due to genes involved in the host’s immune response against viral infection. On the contrary, the intersection of downregulated genes between both tissue types was minor, suggesting that downregulated gene expression changes are tissue specific (Fig. 3D and Table S4).
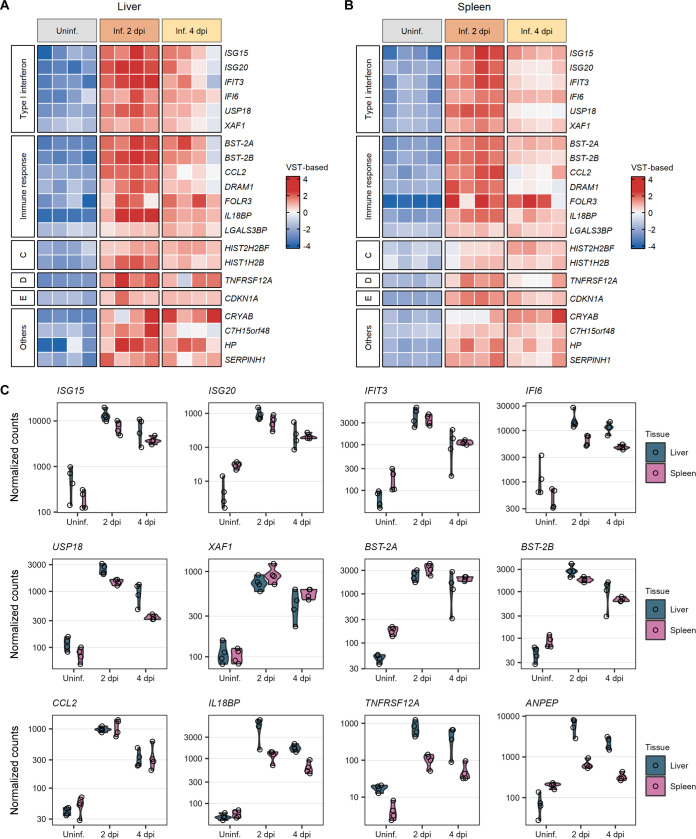
A selection of the top 50 most variably expressed genes in the liver and spleen not only separated the different groups of samples based on infection status but also showed divergent patterns of expression in specific subsets of genes (Fig. S3). In both liver and spleen tissues, the largest differences were observed between the uninfected (group 1) and the 2 days post-infection (group 2) samples. Remarkably, some of the most variably expressed genes showing the largest divergences in expression patterns belonged to a set of 21 genes that were upregulated in the liver and spleen across all the infected samples (2 and 4 days post-infection) (Fig. 4A to C). Most of these genes, including ISG15, ISG20, IFIT3, IFI6, USP18, XAF1, BST-2A, BST-2B, CCL2, DRAM1, FOLR3, IL18BP, and LGALS3BP, are involved in the immune response of the host against viral infection, mainly via IFN-mediated signaling pathways. The remaining genes are involved in, among others, nucleosome assembly, apoptosis, and collagen biosynthesis.
FIG 4.
Commonly upregulated genes in the liver and spleen during peak RVFV infection. (A and B) Heat maps of a set of 21 genes commonly upregulated after RVFV infection in liver (A) and spleen (B) tissues. Samples were clustered based on their infection status, as indicated on top of the heat maps. Genes were clustered into categories based on their molecular function or biological process, as indicated at the left of the heat maps. To represent the magnitude of the log2 fold change of each gene compared to the mean gene expression level, cells are color-coded in shades of a gradient ranging from dark blue (low) to dark red (high). (C) Expression profiles of 12 selected genes commonly upregulated in RVFV-infected (Inf.) samples at 2 days post-infection (dpi) (group 2) and 4 days post-infection (group 3) compared to uninfected (Uninf.) samples (group 1). Dots represent individual normalized gene counts (on a log10 scale), and the shaded area shows the distribution of the samples within each group. Abbreviations: VST, variance-stabilizing transformation; C, nucleosome assembly; D, cytokine receptor; E, apoptosis; Others, a chaperone and genes involved in metabolic processes such as collagen biosynthesis.
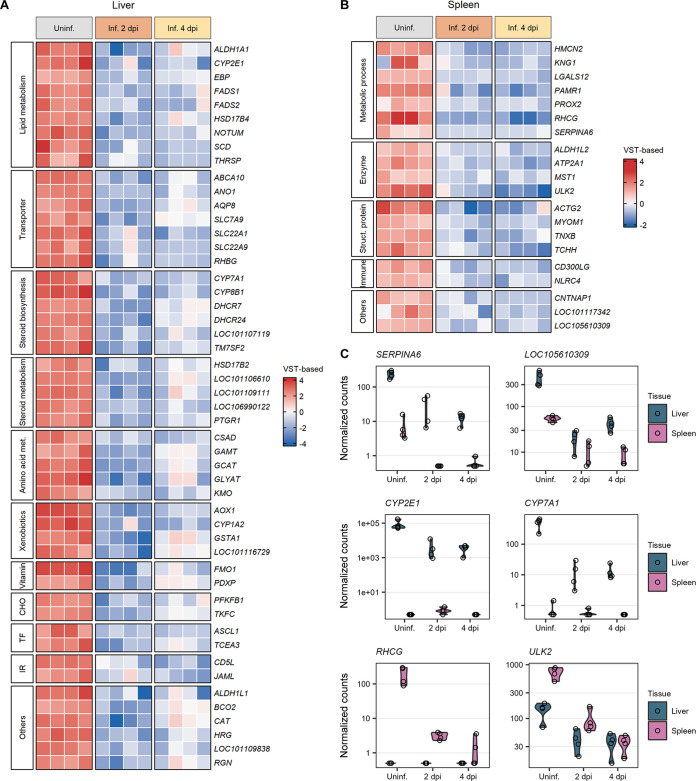
As initially noticed in the Euler diagram (Fig. 3D), the majority of the downregulated genes in RVFV-infected samples compared to uninfected samples are not common between the liver and spleen. The biological processes in which these genes are involved depend on the specific functions of these organs (Fig. 5A to C). The majority of the genes downregulated in the liver upon RVFV infection code for transporters (ABCA10, AQP8, RHBG, and genes encoding proteins of the SLC solute carrier family) and hepatic enzymes. These hepatic enzymes are involved in diverse metabolic processes, including the metabolism of lipids (ALDH1A1, CYP2E1, FADS1, NOTUM, and SCD), steroid hormones (CYP7A1, CYP8B1, DHCR7, HSD17B2, and TM7SF2), amino acids (CSAD, GAMT, GCAT, and GLYAT), vitamins (FMO1 and PDXP), carbohydrates (PFKB1 and TKFC), and xenobiotics (AOX1, CYP1A2, and GSTA1) (Fig. 5A). Genes downregulated in the spleen after RVFV infection were mostly those encoding structural proteins (ACTG2, MYOM1, TNXB, and TCHH), enzymes (ATP2A1, ALDH1L2, and ULK2), and proteins with ligand binding activity (HMCN2, LGALS12, and PAMR1) (Fig. 5B). In both the liver and spleen tissues, the downregulation trend of the underexpressed genes was maintained during both phases of the infection (2 and 4 days post-infection).
FIG 5.
Downregulated genes in the liver and spleen during peak RVFV infection. (A and B) Heat maps of sets of genes significantly downregulated after RVFV infection in liver (A) and spleen (B) tissues. Samples were clustered based on their infection status, as indicated on top of the heat maps. Genes were clustered into categories based on their molecular function or biological process, as indicated at the left of the heat maps. To represent the magnitude of the log2 fold change of each gene compared to the mean gene expression level, cells are color-coded in shades of a gradient ranging from dark blue (low) to dark red (high). (C) Expression profiles of 6 selected genes significantly downregulated in RVFV-infected (Inf.) liver or spleen samples at 2 days post-infection (dpi) (group 2) and 4 days post-infection (group 3) compared to uninfected (Uninf.) samples (group 1). Dots represent individual normalized gene counts (on a log10 scale), and the shaded area shows the distribution of the samples within each group. Abbreviations: VST, variance-stabilizing transformation; met., metabolism; CHO, carbohydrate metabolism; TF, transcription factor; IR, immune response; Struc., structural; Others, genes involved in folate metabolism, carotenoid metabolism, purine metabolism, signaling receptor activity, cell growth, hemostasis, and senescence.
Host immune responses and metabolic pathways are markedly influenced by RVFV infection.
Besides inspecting the profiles of individual differentially expressed genes, we also performed a functional analysis of gene sets of interest based on the Gene Ontology (GO) database. Over-representation analysis (ORA) determines if known biological processes or molecular functions are significantly over-represented in a particular list of differentially expressed genes (36), while gene set enrichment analysis (GSEA) determines if an a priori-defined set of genes shows significantly coordinated differences (i.e., detection of small but consistent changes in the same direction) (37).
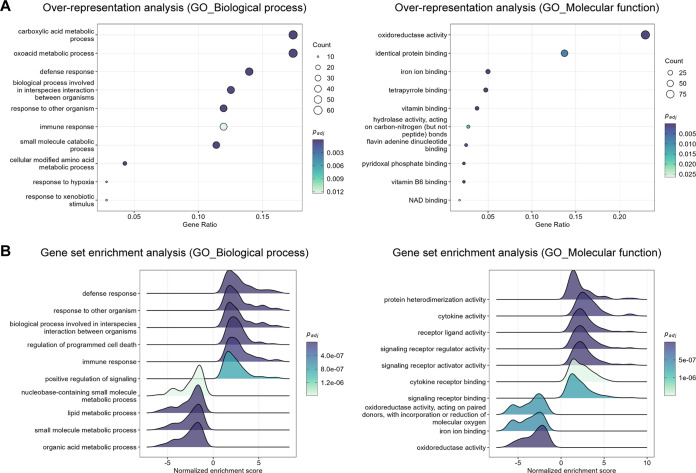
Specifically for the liver, in terms of biological processes, ORA and GSEA showed significant enrichment for GO terms related to the immune response, positive regulation of signaling, regulation of programmed cell death, and the metabolism of diverse molecules (e.g., lipids, organic acids, and nucleobase-containing molecules) at the peak of the infection (2 days post-infection). Molecular functions associated with these processes included cytokine activity, cytokine receptor binding, binding to other ligands (e.g., iron and vitamins), and enzymatic activity (e.g., oxidoreductase and hydrolase) (Fig. 6A and B and Table S5). ORA and GSEA revealed very similar results for the 4 days post-infection time point (Fig. S4A and B and Table S5). Ridge plots in Fig. 6B depict that gene sets involved in the immune response were upregulated (positive normalized enrichment score), whereas gene sets involved in metabolic processes were downregulated (negative normalized enrichment score).
FIG 6.
Top significantly enriched pathways altered in the liver in response to RVFV infection (2 days post-infection). (A) Gene Ontology (GO) biological process (left) and molecular function (right) over-representation analysis of genes differentially expressed in RVFV-infected liver tissue. The dot size represents the number of enriched genes associated with each GO term. Dots are color-coded according to their P values (adjusted by the Benjamini-Hochberg method). (B) GO biological process (left) and molecular function (right) gene set enrichment analysis in RVFV-infected liver tissue. Gene sets with positive normalized enrichment scores are upregulated, whereas gene sets with negative normalized enrichment scores are downregulated. Ridges are color-coded according to their P values (adjusted by the Benjamini-Hochberg method). The cutoff for significance for all the analyses was set to an adjusted P value <0.05. The top significantly enriched pathways altered in the liver in response to RVFV infection at 4 days post-infection are presented in Fig. S4 in the supplemental material.
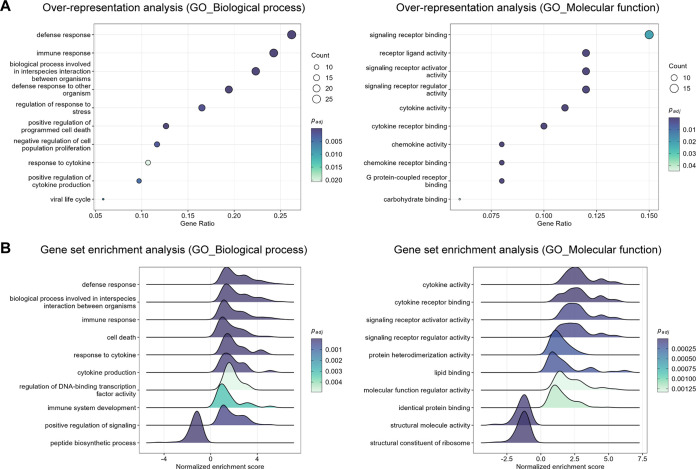
Similar to the liver, ORA and GSEA of the spleen data at the level of biological processes identified significant positive enrichment for GO terms involved in the immune response, regulation of the response to stress, regulation of programmed cell death, cytokine production, and the response to cytokines at 2 days post-infection. Regarding molecular functions, GO terms significantly enriched included cytokine activity, chemokine activity, signaling receptor binding, binding to other ligands (e.g., lipids and carbohydrates), and structural molecule activity (Fig. 7A and B and Table S5). Furthermore, GSEA identified GO terms related to the upregulation of apoptotic signaling, protein heterodimerization activity, translation regulator activity, and the downregulation of ion transmembrane transporter activity specifically at the 4 days post-infection time point (Fig. S5 and Table S5).
FIG 7.
Top significantly enriched pathways altered in the spleen in response to RVFV infection (2 days post-infection). (A) Gene Ontology (GO) biological process (left) and molecular function (right) over-representation analysis of genes differentially expressed in RVFV-infected spleen tissue. The dot size represents the number of enriched genes associated with each GO term. Dots are color-coded according to their P values (adjusted by the Benjamini-Hochberg method). (B) GO biological process (left) and molecular function (right) gene set enrichment analysis in RVFV-infected spleen tissues. Gene sets with positive normalized enrichment scores are upregulated, whereas gene sets with negative normalized enrichment scores are downregulated. Ridges are color-coded according to their P values (adjusted by the Benjamini-Hochberg method). The cutoff for significance in all the analyses was set to an adjusted P value <0.05. The top significantly enriched gene sets altered in the spleen in response to RVFV infection at 4 days post-infection are presented in Fig. S5 in the supplemental material.
Additionally, ORA based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database corroborated the host’s most influenced pathways upon RVFV infection. Pathways related to the metabolism of diverse molecules (e.g., xenobiotics, fatty acids, retinol, and arachidonic acid), peroxisome proliferator-activated receptor (PPAR) signaling, and the biosynthesis of steroid hormones and cofactors were identified in the liver samples (Fig. S6A and Table S5). Pathways involved in viral infection, cytokine receptor interaction, NF-κB signaling, NOD-like receptor signaling, protein digestion, and apoptosis were identified in the spleen samples (Fig. S6B and Table S5).
Levels of the host factor LRP1 correlate with RVFV tissue tropism.
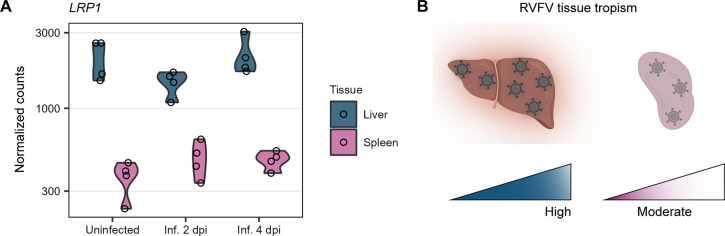
Recently, a genome-wide CRISPR screen identified low-density lipoprotein receptor-related protein 1 (LRP1) as a receptor for RVFV entry into host cells (38). We took advantage of our in-depth RNA-seq analysis to investigate LRP1 expression. We observed that the levels of LRP1 expression in lambs clearly differed between organs but were not affected by the infection status. LRP1 was more abundantly expressed in the liver than in the spleen (Fig. 8A). Such high basal expression levels of the entry factor LRP1 in the liver correlated with the preference of RVFV for targeting this organ (Fig. 8B).
FIG 8.
LRP1 expression correlates with RVFV tissue tropism. (A) Expression profile of LRP1 in the liver and spleen of lambs with different infection statuses. Dots represent individual normalized gene counts (on a log10 scale), and the shaded area shows the distribution of the samples within each group. (B) Schematic representation of RVFV tissue tropism. The elevated basal levels of LRP1 in the liver correlated with the preference of RVFV for targeting this organ. Abbreviations: Inf., infected; dpi, days post-infection.
DISCUSSION
The acute onset of clinical signs and rapid progression to severe disease are features of RVFV infection (39). Experimental infections in rodents, ferrets, ruminants, and non-human primates have advanced our understanding of RVFV pathogenesis (22, 40–45). However, our knowledge of the underlying mechanisms is still rudimentary. Previous RNA-seq studies of in vitro infections and infections of non-target animals have unveiled important genes and pathways that are affected during RVFV infection (31–33). Nevertheless, RNA-seq studies with primary target animals and tissues have not yet been performed. Here, we examined the in vivo genome-wide transcriptional responses in target organs (liver and spleen) of RVFV-infected lambs. This work provides a broad view of the host factors affected by RVFV to better understand the molecular basis of RVFV pathogenesis.
Our study harnessed preserved tissue samples from a previous animal trial in which lambs were exposed to RVFV (34). Since this trial was not initially designed with the aim of performing a subsequent time course transcriptomic analysis, our study carries two inherent limitations. First, we did not have a conventional negative control group of uninfected lambs. To overcome this, we selected a specific set of samples from a group of lambs that were in theory exposed to the virus (through low-level exposure to mosquitoes) but that did not develop any signs of clinical disease and from which we could not detect infectious virus or viral RNA copies in the blood or the organs. In essence, these lambs had the same characteristics as those of lambs that have not been exposed to the virus. Arguably, these samples also represent a good control for the potential effects of mosquito bites.
As a second limitation, lambs from the two RVFV-infected groups were exposed to the virus via different routes. Lambs from group 2 were infected via intravenous injection, while lambs from group 3 were infected via the bites of infected mosquitoes. While it can be argued that different routes of infection may lead to distinct infection outcomes, we previously observed that RVFV infection of lambs via intravenous injection or mosquito bites leads to highly similar levels of infectious virus in the blood, comparable histopathological changes, and the same clinical outcomes (22). We are thus confident that these two above-mentioned limitations do not call into question the validity of our reported observations.
Previous studies by others on the host transcriptomic responses following RVFV infection reported the induction of defense responses (31–33). Here, we confirmed that upon infection, RVFV induced a strong activation of innate immune and inflammatory responses in both the liver and spleen tissues of lambs, principally through the sharp upregulation of IFN-induced antiviral host restriction factors and cytokine-mediated signaling pathways. The immune and inflammatory activation explains the typical high fever steadily observed through the critical 2- to 4-day-post-infection phase, reflecting the quick and sustained counteraction of the host for clearing the virus.
Among the top upregulated genes, ISG20 and ISG15 are of special interest. Indeed, an in vitro IFN-stimulated gene (ISG) expression screening identified potent antibunyaviral effects of ISG20 against a diverse panel of members of the Peribunyaviridae, Hantaviridae, and Nairoviridae families through its RNase activity (46). However, several members of the Phenuiviridae family (including RVFV) were described to resist the effects of ISG20 (46). We observed a significant upregulation of ISG20 in infected tissues. This could mean either that RVFV may be susceptible to ISG20-mediated inhibition in vivo or that despite the high levels of ISG20, RVFV is indeed resistant to its antiviral effects (e.g., due to delayed kinetics of ISG20 induction). ISG15 has also been shown to exert antiviral activity against a wide range of viruses (47, 48), including the bunyavirus Crimean-Congo hemorrhagic fever virus (CCHFV) (family Nairoviridae) (49, 50). The over-expression of ISG15 during RVFV infection likely reflects its antiviral role.
Several other top upregulated genes in both RVFV-infected liver and spleen tissues have also previously been implicated as key factors in the host antiviral response. For instance, IFIT3 has been associated with the inhibition of adenovirus (51), rabies virus (52), Japanese encephalitis virus, herpesviruses (53), and even RVFV (31). IFI6 was reported to block flavivirus replication (54), and USP18 was reported to counteract infection by Sendai virus and encephalomyocarditis virus (55). Similar broad antiviral activities have been described for XAF1 (56) and LGALS3BP (57). It is conceivable to think that the upregulation of these genes upon RVFV infection is the result of host cellular responses trying to limit viral replication. Thus, our results validate the role of commonly characterized antiviral host factors and expand the range of viruses against which they are known to exert restrictive activity.
The liver is the main target organ of RVFV, and infection generally leads to severe cellular damage that progresses to hepatic necrosis, often followed by inflammation with an infiltration of neutrophils (13, 39). Despite this tropism for the liver, we observed that only a small fraction of the tissue (approximately 10 to 20%) was infected in our samples. Most likely, RVFV-infected hepatocytes signal the viral threat to uninfected neighboring cells, inducing a strong antiviral state that counteracts the infection but compromises liver homeostasis for several days. Dozens of genes coding for enzymes involved in the metabolism of lipids, carbohydrates, proteins, nucleic acids, vitamins, and xenobiotics were markedly downregulated during peak viremia. In animals that recover from the infection, the generalized antiviral state, including transcriptional changes from uninfected neighboring cells, could explain the fast clearance of the virus. Once the levels of the virus decline, unaffected hepatocytes rapidly restore normal liver function.
The downregulation of oxidoreductases, hydrolases, transferases, and transporters, and overall liver failure are clear consequences of the observed tissue necrosis. Such a downregulation of hepatic enzymes responsible for essential metabolic processes possibly explains, for example, the icterus sometimes observed in RVFV-infected animals. Similarly, hemorrhage and coagulation disorders are likely to appear secondary to liver dysfunction due to the damaged liver’s incapacity to synthesize proteins crucial for blood homeostasis.
Interestingly, previous in vitro RNA-seq studies investigating the mammalian host response upon RVFV infection did not decidedly identify the downregulation of many important metabolic enzymes (31, 32). This apparent discrepancy is most probably explained by the fact that those studies employed small airway epithelial cells and embryonic kidney cells instead of more biologically relevant cells. One additional difference relates to the expression levels of genes involved in ion transport. Two solute carriers (SLC24A2 and SLC1A3) were reported to be among the top upregulated genes upon RVFV infection (31), whereas we consistently found that several transporters from the same family (e.g., SLC7A9, SLC22A1, and SLC22A9) were significantly downregulated.
The spleen is a secondary target organ of RVFV. Necrosis of the spleen has been described as a common characteristic in naturally infected young lambs, often noticeable in both the red and white pulp (13). Although less evident than in the liver, the downregulation of genes coding for cellular structural proteins and genes involved in metabolic processes also denotes that the function of the spleen was seriously compromised as a consequence of RVFV infection. Importantly, it might be that the transcriptomic changes observed in the spleen are a combination of direct effects on infected splenocytes and affected immune cells that trafficked to the spleen.
LRP1, a newly identified host entry factor for RVFV (38), is ubiquitously expressed in numerous human tissues, but its expression levels are higher in the liver, placenta, brain, adipose tissue, and fibroblasts (data available from https://www.proteinatlas.org/ENSG00000123384-LRP1) (58). High expression levels of the LRP1 protein in these tissues can be associated with its natural function as a key molecule in intracellular lipoprotein metabolism (e.g., central cholesterol metabolism in the liver and progesterone biosynthesis in the placenta). In line with the human tissue-specific gene expression data, we found that the levels of LRP1 expression in lambs were much higher in the liver than in the spleen. The high basal expression level of LRP1 in the liver possibly explains, at least partially, the marked tropism of RVFV for human and ovine hepatocytes. RVFV also displays preferential tropism for placental cells in pregnant ewes (59), which normally leads to abortion storms during outbreaks (14). Whether there also is elevated basal LRP1 expression in the sheep placenta and whether this favors placental infection remain to be investigated. Confirmation of a good correlation between LRP1 gene expression and its protein levels in lambs awaits further investigation as well.
In summary, the transcriptomic analysis of RVFV-infected liver and spleen tissues of lambs presented here revealed the most important genes and pathways that mediate the strong innate immune response upon infection and the drastic decrease in tissue-specific function. Our results validated that IFN-mediated signaling pathways are key regulators of RVFV infection. Moreover, we uncovered the severely damaged liver metabolic function underlying the acute hepatitis and necrosis induced by RVFV. In addition, we highlighted the elevated basal expression level of LRP1 in liver cells as a potential factor influencing RVFV tissue tropism. Altogether, the results of this study shed light on the extensive perturbance of the in vivo host transcriptome during critical phases of RVFV infection and provide new insights to better comprehend RVFV pathogenesis at the molecular level. Future studies could expand the current knowledge by concomitantly quantifying host and viral gene expression in individual cells. Transcriptomic profiling at single-cell resolution would make it possible to discriminate the innate immune responses of RVFV-infected cells from those of uninfected neighboring cells, ultimately improving our understanding of the molecular mechanisms employed by RVFV to overcome the host response.
MATERIALS AND METHODS
Tissue samples.
Liver and spleen samples were selected from another study aimed at comparing RVFV infection in Texel-Swifter lambs after low-exposure (3 mosquitoes) and high-exposure (28 to 31 mosquitoes) challenges (34). Mosquitoes used for the challenge were initially fed on lambs 2 days after the intravenous injection of 105 median tissue culture infectious doses (TCID50) of RVFV strain 35/74 (60). For the purpose of the present study, 4 samples per group were selected from (i) non-responsive lambs (that did not develop viremia or disease) necropsied 8 days after low-level mosquito exposure (group 1), (ii) lambs presenting with high viremia necropsied 2 days after intravenous injection (group 2), and (iii) lambs presenting with high viremia necropsied 4 days after high-level mosquito exposure (group 3). A summary of the different groups is presented in Table 1. The group numbers and descriptions presented in Table 1 belong exclusively to the present study, but to enable traceability, the original identification numbers of the selected samples are included. Within each group, samples were selected based on similar profiles of infectious titers and viral RNA copy numbers in the blood, liver, and spleen. The infectious titers and viral RNA copy numbers were determined by an endpoint dilution virus isolation assay and M-segment-specific quantitative reverse transcription PCR (RT-qPCR), respectively, as described previously (34). Liver and spleen samples were long-term preserved in RNAlater (Invitrogen) at ≤ −65°C.
TABLE 1.
Description, route of exposure, identification number, sex, and age of the lambs from a previous animal studya selected for transcriptomic analysis
| Group | Descriptionb | Route of exposure | Animal ID | Sex | Age (at challenge) (wks) |
|---|---|---|---|---|---|
| 1 | Uninfected | Mosquito bite (low exposure) | 276 | Female | 10 |
| 277 | Male | ||||
| 278 | Female | ||||
| 279 | Female | ||||
| 2 | Infected 2 dpi | Intravenous injection | 271 | Female | 10 |
| 272 | Female | ||||
| 273 | Male | ||||
| 274 | Female | ||||
| 3 | Infected 4 dpi | Mosquito bite (high exposure) | 4618 | Male | 10 |
| 4619 | Female | ||||
| 4624 | Male | ||||
| 4642 | Female | ||||
See reference 34.
dpi, days post-infection.
Histology and immunohistochemistry.
Tissue samples were fixed in 10% neutral buffered formalin for 48 h and processed routinely into paraffin blocks. Sections were cut on silane-coated glass slides and dried in a 37°C incubator for at least 48 h. After deparaffinization in xylene and rehydration in a series of graded alcohols, sections were stained routinely with hematoxylin and eosin (H&E) or immunostained for RVFV antigen. For immunostaining, heat-induced epitope retrieval was applied by autoclaving the sections for 15 min at 121°C in citrate buffer (pH 6) (Vector Laboratories). Monoclonal antibody 4-D4 (61), directed against the Gn glycoprotein, was used as the primary antibody. Goat polyclonal anti-mouse horseradish peroxidase (HRP)-conjugated IgG (Invitrogen) was used as the secondary antibody, with diaminobenzidine (DAB+) (Dako, Agilent) as the substrate. Sections were counterstained with Mayer’s hematoxylin and mounted permanently. Images were taken with an Olympus BX51 microscope equipped with a high-resolution digital camera.
RNA isolation.
Following thawing, liver and spleen tissue fragments of approximately 80 to 260 mg were washed once in 500 μL of Dulbecco’s phosphate-buffered saline (DPBS) (Gibco) to remove excess RNAlater and then placed into Lysing Matrix D tubes (MP Biomedicals) with 1 mL of TRIzol (Invitrogen). Tissue fragments were homogenized with at least 2 cycles in a FastPrep-24 homogenizer (MP Biomedicals) at 6 m/s for 50 s each cycle until complete homogenization was achieved. When required, to improve the homogenization of complex samples, 2 glass beads per tube were added. Homogenates were centrifuged at 20,800 relative centrifugal force (rcf) (Eppendorf 5417R) for 40 min at 4°C, and the cleared supernatants were kept. Total RNA extractions of 350 μL of lysed samples were performed with the Direct-zol RNA MiniPrep kit (Zymo Research), according to the manufacturer’s instructions, with an additional centrifugation step after the addition of RNA wash buffer. Lysed preparations were treated with 30 U of DNase I for 15 min. Total RNA was eventually eluted in 30 μL of DNase/RNase-free water. The RNA concentration and purity were measured on a NanoDrop One instrument (Thermo Fisher Scientific), and samples were stored at ≤ −65°C until further use.
RNA-seq.
Isolated RNA was sent for next-generation sequencing to GenomeScan B.V. (Leiden, The Netherlands). The RNA integrity was determined using a Fragment Analyzer system (Agilent) or a 2100 Bioanalyzer (Agilent). Samples were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (catalog number E7760S/L; New England BioLabs [NEB]) according to the instructions of the manufacturer. Briefly, host poly(A) mRNA was isolated from total RNA using oligo(dT) magnetic beads. After the fragmentation of the mRNA, cDNA was synthesized, sequencing adapters were ligated, and the fragments were PCR amplified. The quality and yield of the sequencing library were determined using a Fragment Analyzer (Agilent). The resulting products had a size distribution with a broad peak between 300 and 500 bp. Clustering and sequencing of 1.1 nM DNA samples were performed using an Illumina NovaSeq 6000 instrument with NovaSeq control software NCS version 1.7, according to the instructions of the manufacturer. The sequence length of the short reads ranged between 151 and 159 bp.
RNA-seq data preprocessing.
Raw sequencing reads were processed according to the nf-core/RNA-seq pipeline (62) version 3.4 (https://doi.org/10.5281/zenodo.1400710). Quality control was performed with FastQC (63) version 0.11.9 and MultiQC (64) version 1.10.1. Based on the high mean sequence quality (average Phred score >30) (see Fig. S1 in the supplemental material), only soft trimming was required. Adapter sequences were removed with Cutadapt (65) version 3.4. Trimmed reads had an average length ranging between 131 and 150 bp. Reads were deduplicated based on unique molecular identifiers (UMIs) using UMI-tools (66) version 1.1.2. Unique reads were aligned to the sheep (Ovis aries) NCBI reference genome version 4.0 (Oar_v4.0) (https://www.ncbi.nlm.nih.gov/assembly/GCF_000298735.2/) with HISAT2 (67) version 2.2.0 and quantified using featureCounts (68) (within the Rsubread package version 2.4.3). Unannotated genes were automatically assigned a LOC prefix gene identification nomenclature designation based on genomic position. However, the corresponding gene name, when available, was manually assigned for the plots presented in Fig. 4A to C and Fig. 5A to C.
Transcriptome analysis.
The data set was filtered to remove low-count genes (at least 2 samples should have counts of 3 or higher for a gene to be kept). Pairwise differential gene expression analysis was performed with DESeq2 (69) version 1.38.3 in R (70) version 4.2.2, using an absolute log2 fold change threshold of 1 and an alpha value of 0.05. DESeq2 models the count data with a negative binomial distribution using a generalized linear model. Wald test P values <0.05 (adjusted with the Benjamini-Hochberg method) were considered significant. For the initial exploration of the data, Euclidean distances between samples were calculated, and principal component analysis was conducted after applying a variance-stabilizing transformation to the data. For visualization and ranking purposes, the log2 fold change was shrunken using the adaptive shrinkage estimator from the ashr package (71) version 2.2-54. Heat maps were created with the R package pheatmap version 1.0.12 (https://CRAN.R-project.org/package=pheatmap) or ComplexHeatmap (72) version 2.14.0. Euler diagrams were plotted with eulerr (73) version 6.1.1. Volcano plots were generated using EnhancedVolcano (74) version 1.16.0.
Functional ORA and GSEA were performed with clusterProfiler (75, 76) version 4.6.0, based on the GO and KEGG databases. When required, the clusterProfiler function simplify was used to reduce the redundancy of enriched GO terms. P values <0.05 (adjusted by the Benjamini-Hochberg method) of a one-sided version of Fisher’s exact test were considered significant for ORA. P values <0.05 (adjusted by the Benjamini-Hochberg method) of a permutation test were considered significant for GSEA.
Data analysis and visualization.
Prism 9 (GraphPad Software) was used to generate graphs of infectious titer and viral RNA copy number data. Transcriptomic data were analyzed and plotted in R (70) version 4.2.2, using the above-mentioned R packages. Statistical tests differed per analysis and are indicated in the description of each analysis and the corresponding figure legends. P values ≥0.05 were considered not significant.
Ethics statement.
The animal experiment carried out within the scope of another study (34), from which organ samples were obtained for analysis, was conducted in accordance with European regulations (European Union directive 2010/63/EU) and the Dutch Law on Animal Experiments (Wod, identification number BWBR0003081). Permissions were granted by the Dutch Central Authority for Scientific Procedures on Animals (permit numbers AVD4010020185564 and AVD4010020187168). Specific procedures were approved by the Animal Ethics Committees of Wageningen Research.
Data availability.
Raw sequencing data were deposited at the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA935986.
ACKNOWLEDGMENTS
E.B.-M. is a grateful recipient of scholarships from the Graduate School of Production Ecology & Resource Conservation (PE&RC) and Universidad de Costa Rica (OAICE-031-2019). This work was supported in part by funds from the Rob Goldbach Fund to E.B.-M. for a laboratory training visit.
We thank Quirine ten Bosch (Quantitative Veterinary Epidemiology, Wageningen University & Research) for authorizing the utilization of samples from the RVFV animal trial (34). We thank Ingrid Cárdenas Rey for proofreading. The illustrations in Fig. 1A and Fig. 8B were created with BioRender.com.
P.J.W.S., E.B.-M., and J.K. conceived the project. E.B.-M. and S.v.d.W. performed the RNA isolation for RNA-seq. E.B.-M. and P.A. performed the RNA-seq data processing and bioinformatic analysis. E.B.-M., P.A., and P.J.W.S. interpreted the RNA-seq data with contributions from B.R., G.P.P., A.C., and J.K. L.v.K. performed the histology and immunohistochemistry experiments. P.J.W.S., J.K., A.C., and G.P.P. supervised the project. E.B.-M. wrote the manuscript with contributions from P.J.W.S. and L.v.K. E.B.-M. made the figures with contributions from P.A., L.v.K., and P.J.W.S. All authors reviewed the manuscript and provided feedback.
Footnotes
Supplemental material is available online only.
Contributor Information
Paul J. Wichgers Schreur, Email: paul.wichgersschreur@wur.nl.
Rebecca Ellis Dutch, University of Kentucky College of Medicine.
REFERENCES
- 1.Flick R, Bouloy M. 2005. Rift Valley fever virus. Curr Mol Med 5:827–834. doi: 10.2174/156652405774962263. [DOI] [PubMed] [Google Scholar]
- 2.Pepin M, Bouloy M, Bird BH, Kemp A, Paweska J. 2010. Rift Valley fever virus (Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet Res 41:61. doi: 10.1051/vetres/2010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abudurexiti A, Adkins S, Alioto D, Alkhovsky SV, Avšič-Županc T, Ballinger MJ, Bente DA, Beer M, Bergeron É, Blair CD, Briese T, Buchmeier MJ, Burt FJ, Calisher CH, Cháng C, Charrel RN, Choi IR, Clegg JCS, de la Torre JC, de Lamballerie X, Dèng F, Di Serio F, Digiaro M, Drebot MA, Duàn X, Ebihara H, Elbeaino T, Ergünay K, Fulhorst CF, Garrison AR, Gāo GF, Gonzalez J-PJ, Groschup MH, Günther S, Haenni A-L, Hall RA, Hepojoki J, Hewson R, Hú Z, Hughes HR, Jonson MG, Junglen S, Klempa B, Klingström J, Kòu C, Laenen L, Lambert AJ, Langevin SA, Liu D, Lukashevich IS, et al. 2019. Taxonomy of the order Bunyavirales: update 2019. Arch Virol 164:1949–1965. doi: 10.1007/s00705-019-04253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nanyingi MO, Munyua P, Kiama SG, Muchemi GM, Thumbi SM, Bitek AO, Bett B, Muriithi RM, Njenga MK. 2015. A systematic review of Rift Valley fever epidemiology 1931–2014. Infect Ecol Epidemiol 5:28024. doi: 10.3402/iee.v5.28024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark MHA, Warimwe GM, Di Nardo A, Lyons NA, Gubbins S. 2018. Systematic literature review of Rift Valley fever virus seroprevalence in livestock, wildlife and humans in Africa from 1968 to 2016. PLoS Negl Trop Dis 12:e0006627. doi: 10.1371/journal.pntd.0006627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anywaine Z, Lule SA, Hansen C, Warimwe G, Elliott A. 2022. Clinical manifestations of Rift Valley fever in humans: systematic review and meta-analysis. PLoS Negl Trop Dis 16:e0010233. doi: 10.1371/journal.pntd.0010233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daubney R, Hudson JR, Garnham PC. 1931. Enzootic hepatitis or Rift Valley fever. An undescribed virus disease of sheep cattle and man from East Africa. J Pathol 34:545–579. doi: 10.1002/path.1700340418. [DOI] [Google Scholar]
- 8.Madani TA, Al-Mazrou YY, Al-Jeffri MH, Mishkhas AA, Al-Rabeah AM, Turkistani AM, Al-Sayed MO, Abodahish AA, Khan AS, Ksiazek TG, Shobokshi O. 2003. Rift Valley fever epidemic in Saudi Arabia: epidemiological, clinical, and laboratory characteristics. Clin Infect Dis 37:1084–1092. doi: 10.1086/378747. [DOI] [PubMed] [Google Scholar]
- 9.Al-Hazmi M, Ayoola EA, Abdurahman M, Banzal S, Ashraf J, El-Bushra A, Hazmi A, Abdullah M, Abbo H, Elamin A, Al-Sammani E-T, Gadour M, Menon C, Hamza M, Rahim I, Hafez M, Jambavalikar M, Arishi H, Aqeel A. 2003. Epidemic Rift Valley fever in Saudi Arabia: a clinical study of severe illness in humans. Clin Infect Dis 36:245–252. doi: 10.1086/345671. [DOI] [PubMed] [Google Scholar]
- 10.Hassan OA, Ahlm C, Sang R, Evander M. 2011. The 2007 Rift Valley fever outbreak in Sudan. PLoS Negl Trop Dis 5:e1229. doi: 10.1371/journal.pntd.0001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odendaal L, Clift SJ, Fosgate GT, Davis AS. 2019. Lesions and cellular tropism of natural Rift Valley fever virus infection in adult sheep. Vet Pathol 56:61–77. doi: 10.1177/0300985818806049. [DOI] [PubMed] [Google Scholar]
- 12.Wright D, Kortekaas J, Bowden TA, Warimwe GM. 2019. Rift Valley fever: biology and epidemiology. J Gen Virol 100:1187–1199. doi: 10.1099/jgv.0.001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odendaal L, Davis AS, Fosgate GT, Clift SJ. 2020. Lesions and cellular tropism of natural Rift Valley fever virus infection in young lambs. Vet Pathol 57:66–81. doi: 10.1177/0300985819882633. [DOI] [PubMed] [Google Scholar]
- 14.Odendaal L, Clift SJ, Fosgate GT, Davis AS. 2020. Ovine fetal and placental lesions and cellular tropism in natural Rift Valley fever virus infections. Vet Pathol 57:791–806. doi: 10.1177/0300985820954549. [DOI] [PubMed] [Google Scholar]
- 15.El-Din Abdel-Wahab KS, El Baz LM, El Tayeb EM, Omar H, Moneim Ossman MA, Yasin W. 1978. Rift Valley fever virus infections in Egypt: pathological and virological findings in man. Trans R Soc Trop Med Hyg 72:392–396. doi: 10.1016/0035-9203(78)90134-7. [DOI] [PubMed] [Google Scholar]
- 16.Turell MJ, Dohm DJ, Mores CN, Terracina L, Wallette DL, Hribar LJ, Pecor JE, Blow JA. 2008. Potential for North American mosquitoes to transmit Rift Valley fever virus. J Am Mosq Control Assoc 24:502–507. doi: 10.2987/08-5791.1. [DOI] [PubMed] [Google Scholar]
- 17.Turell MJ, Britch SC, Aldridge RL, Kline DL, Boohene C, Linthicum KJ. 2013. Potential for mosquitoes (Diptera: Culicidae) from Florida to transmit Rift Valley fever virus. J Med Entomol 50:1111–1117. doi: 10.1603/me13049. [DOI] [PubMed] [Google Scholar]
- 18.Brustolin M, Talavera S, Nuñez A, Santamaría C, Rivas R, Pujol N, Valle M, Verdún M, Brun A, Pagès N, Busquets N. 2017. Rift Valley fever virus and European mosquitoes: vector competence of Culex pipiens and Stegomyia albopicta (= Aedes albopictus). Med Vet Entomol 31:365–372. doi: 10.1111/mve.12254. [DOI] [PubMed] [Google Scholar]
- 19.Kraemer MUG, Reiner RC, Brady OJ, Messina JP, Gilbert M, Pigott DM, Yi D, Johnson K, Earl L, Marczak LB, Shirude S, Davis Weaver N, Bisanzio D, Perkins TA, Lai S, Lu X, Jones P, Coelho GE, Carvalho RG, Van Bortel W, Marsboom C, Hendrickx G, Schaffner F, Moore CG, Nax HH, Bengtsson L, Wetter E, Tatem AJ, Brownstein JS, Smith DL, Lambrechts L, Cauchemez S, Linard C, Faria NR, Pybus OG, Scott TW, Liu Q, Yu H, Wint GRW, Hay SI, Golding N. 2019. Past and future spread of the arbovirus vectors Aedes aegypti and Aedes albopictus. Nat Microbiol 4:854–863. doi: 10.1038/s41564-019-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rich KM, Wanyoike F. 2010. An assessment of the regional and national socio-economic impacts of the 2007 Rift Valley fever outbreak in Kenya. Am J Trop Med Hyg 83:52–57. doi: 10.4269/ajtmh.2010.09-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peyre M, Chevalier V, Abdo-Salem S, Velthuis A, Antoine-Moussiaux N, Thiry E, Roger F. 2015. A systematic scoping study of the socio-economic impact of Rift Valley fever: research gaps and needs. Zoonoses Public Health 62:309–325. doi: 10.1111/zph.12153. [DOI] [PubMed] [Google Scholar]
- 22.Wichgers Schreur PJ, Vloet RPM, Kant J, van Keulen L, Gonzales JL, Visser TM, Koenraadt CJM, Vogels CBF, Kortekaas J. 2021. Reproducing the Rift Valley fever virus mosquito-lamb-mosquito transmission cycle. Sci Rep 11:1477. doi: 10.1038/s41598-020-79267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouloy M, Janzen C, Vialat P, Khun H, Pavlovic J, Huerre M, Haller O. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J Virol 75:1371–1377. doi: 10.1128/JVI.75.3.1371-1377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Billecocq A, Spiegel M, Vialat P, Kohl A, Weber F, Bouloy M, Haller O. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J Virol 78:9798–9806. doi: 10.1128/JVI.78.18.9798-9806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le May N, Dubaele S, De Santis LP, Billecocq A, Bouloy M, Egly J-M. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541–550. doi: 10.1016/s0092-8674(04)00132-1. [DOI] [PubMed] [Google Scholar]
- 26.Habjan M, Pichlmair A, Elliott RM, Överby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. 2009. NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J Virol 83:4365–4375. doi: 10.1128/JVI.02148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. 2009. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog 5:e1000287. doi: 10.1371/journal.ppat.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kainulainen M, Lau S, Samuel CE, Hornung V, Weber F. 2016. NSs virulence factor of Rift Valley fever virus engages the F-box proteins FBXW11 and β-TRCP1 to degrade the antiviral protein kinase PKR. J Virol 90:6140–6147. doi: 10.1128/JVI.00016-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stark R, Grzelak M, Hadfield J. 2019. RNA sequencing: the teenage years. Nat Rev Genet 20:631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- 31.Pinkham C, Dahal B, de la Fuente CL, Bracci N, Beitzel B, Lindquist M, Garrison A, Schmaljohn C, Palacios G, Narayanan A, Campbell CE, Kehn-Hall K. 2017. Alterations in the host transcriptome in vitro following Rift Valley fever virus infection. Sci Rep 7:14385. doi: 10.1038/s41598-017-14800-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Havranek KE, White LA, Lanchy J-M, Lodmell JS. 2019. Transcriptome profiling in Rift Valley fever virus infected cells reveals modified transcriptional and alternative splicing programs. PLoS One 14:e0217497. doi: 10.1371/journal.pone.0217497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hum NR, Bourguet FA, Sebastian A, Lam D, Phillips AM, Sanchez KR, Rasley A, Loots GG, Weilhammer DR. 2022. MAVS mediates a protective immune response in the brain to Rift Valley fever virus. PLoS Pathog 18:e1010231. doi: 10.1371/journal.ppat.1010231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bron GM, Wichgers Schreur PJ, de Jong MCM, van Keulen L, Vloet RPM, Koenraadt CJM, Kortekaas J, ten Bosch QA. 2023. Quantifying Rift Valley fever virus transmission efficiency in a lamb-mosquito-lamb model. bioRxiv. doi: 10.1101/2023.04.25.538218. [DOI] [PMC free article] [PubMed]
- 35.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. 2004. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganaie SS, Schwarz MM, McMillen CM, Price DA, Feng AX, Albe JR, Wang W, Miersch S, Orvedahl A, Cole AR, Sentmanat MF, Mishra N, Boyles DA, Koenig ZT, Kujawa MR, Demers MA, Hoehl RM, Moyle AB, Wagner ND, Stubbs SH, Cardarelli L, Teyra J, McElroy A, Gross ML, Whelan SPJ, Doench J, Cui X, Brett TJ, Sidhu SS, Virgin HW, Egawa T, Leung DW, Amarasinghe GK, Hartman AL. 2021. Lrp1 is a host entry factor for Rift Valley fever virus. Cell 184:5163–5178.e24. doi: 10.1016/j.cell.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terasaki K, Makino S. 2015. Interplay between the virus and host in Rift Valley fever pathogenesis. J Innate Immun 7:450–458. doi: 10.1159/000373924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scharton D, Wettere AJV, Bailey KW, Vest Z, Westover JB, Siddharthan V, Gowen BB. 2015. Rift Valley fever virus infection in Golden Syrian hamsters. PLoS One 10:e0116722. doi: 10.1371/journal.pone.0116722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang Y, Henningson J, Jasperson D, Li Y, Lee J, Ma J, Li Y, Cao N, Liu H, Wilson W, Richt J, Ruder M, McVey S, Ma W. 2016. Mouse model for the Rift Valley fever virus MP12 strain infection. Vet Microbiol 195:70–77. doi: 10.1016/j.vetmic.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 42.Albe JR, Boyles DA, Walters AW, Kujawa MR, McMillen CM, Reed DS, Hartman AL. 2019. Neutrophil and macrophage influx into the central nervous system are inflammatory components of lethal Rift Valley fever encephalitis in rats. PLoS Pathog 15:e1007833. doi: 10.1371/journal.ppat.1007833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbeau DJ, Albe JR, Nambulli S, Tilston-Lunel NL, Hartman AL, Lakdawala SS, Klein E, Duprex WP, McElroy AK. 2020. Rift Valley fever virus infection causes acute encephalitis in the ferret. mSphere 5:e00798-20. doi: 10.1128/mSphere.00798-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson WC, Davis AS, Gaudreault NN, Faburay B, Trujillo JD, Shivanna V, Sunwoo SY, Balogh A, Endalew A, Ma W, Drolet BS, Ruder MG, Morozov I, McVey DS, Richt JA. 2016. Experimental infection of calves by two genetically-distinct strains of Rift Valley fever virus. Viruses 8:145. doi: 10.3390/v8050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith DR, Bird BH, Lewis B, Johnston SC, McCarthy S, Keeney A, Botto M, Donnelly G, Shamblin J, Albariño CG, Nichol ST, Hensley LE. 2012. Development of a novel nonhuman primate model for Rift Valley fever. J Virol 86:2109–2120. doi: 10.1128/JVI.06190-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng J, Wickenhagen A, Turnbull ML, Rezelj VV, Kreher F, Tilston-Lunel NL, Slack GS, Brennan B, Koudriakova E, Shaw AE, Rihn SJ, Rice CM, Bieniasz PD, Elliott RM, Shi X, Wilson SJ. 2018. Interferon-stimulated gene (ISG)-expression screening reveals the specific antibunyaviral activity of ISG20. J Virol 92:e02140-17. doi: 10.1128/JVI.02140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao C, Collins MN, Hsiang T-Y, Krug RM. 2013. Interferon-induced ISG15 pathway: an ongoing virus-host battle. Trends Microbiol 21:181–186. doi: 10.1016/j.tim.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perng Y-C, Lenschow DJ. 2018. ISG15 in antiviral immunity and beyond. Nat Rev Microbiol 16:423–439. doi: 10.1038/s41579-018-0020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scholte FEM, Zivcec M, Dzimianski JV, Deaton MK, Spengler JR, Welch SR, Nichol ST, Pegan SD, Spiropoulou CF, Bergeron É. 2017. Crimean-Congo hemorrhagic fever virus suppresses innate immune responses via a ubiquitin and ISG15 specific protease. Cell Rep 20:2396–2407. doi: 10.1016/j.celrep.2017.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neogi U, Elaldi N, Appelberg S, Ambikan A, Kennedy E, Dowall S, Bagci BK, Gupta S, Rodriguez JE, Svensson-Akusjärvi S, Monteil V, Vegvari A, Benfeitas R, Banerjea A, Weber F, Hewson R, Mirazimi A. 2022. Multi-omics insights into host-viral response and pathogenesis in Crimean-Congo hemorrhagic fever viruses for novel therapeutic target. Elife 11:e76071. doi: 10.7554/eLife.76071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chikhalya A, Dittmann M, Zheng Y, Sohn S-Y, Rice CM, Hearing P. 2021. Human IFIT3 protein induces interferon signaling and inhibits adenovirus immediate early gene expression. mBio 12:e02829-21. doi: 10.1128/mBio.02829-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chai B, Tian D, Zhou M, Tian B, Yuan Y, Sui B, Wang K, Pei J, Huang F, Wu Q, Lv L, Yang Y, Wang C, Fu Z, Zhao L. 2021. Murine Ifit3 restricts the replication of rabies virus both in vitro and in vivo. J Gen Virol 102:001619. doi: 10.1099/jgv.0.001619. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W, Li Y, Xin S, Yang L, Jiang M, Xin Y, Wang Y, Cao P, Zhang S, Yang Y, Lu J. 2023. The emerging roles of IFIT3 in antiviral innate immunity and cellular biology. J Med Virol 95:e28259. doi: 10.1002/jmv.28259. [DOI] [PubMed] [Google Scholar]
- 54.Richardson RB, Ohlson MB, Eitson JL, Kumar A, McDougal MB, Boys IN, Mar KB, De La Cruz-Rivera PC, Douglas C, Konopka G, Xing C, Schoggins JW. 2018. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat Microbiol 3:1214–1223. doi: 10.1038/s41564-018-0244-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou J, Han L, Zhao Z, Liu H, Zhang L, Ma C, Yi F, Liu B, Zheng Y, Gao C. 2021. USP18 positively regulates innate antiviral immunity by promoting K63-linked polyubiquitination of MAVS. Nat Commun 12:2970. doi: 10.1038/s41467-021-23219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han Y, Bai X, Liu S, Zhu J, Zhang F, Xie L, Liu G, Jiang X, Zhang M, Huang Y, Wang J, Li D, Zhang H, He Y, He S, Xia Y, Xu X, Xu F, Ma F. 2022. XAF1 protects host against emerging RNA viruses by stabilizing IRF1-dependent antiviral immunity. J Virol 96:e00774-22. doi: 10.1128/jvi.00774-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu G, Xia Z, Deng F, Liu L, Wang Q, Yu Y, Wang F, Zhu C, Liu W, Cheng Z, Zhu Y, Zhou L, Zhang Y, Lu M, Liu S. 2019. Inducible LGALS3BP/90K activates antiviral innate immune responses by targeting TRAF6 and TRAF3 complex. PLoS Pathog 15:e1008002. doi: 10.1371/journal.ppat.1008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. 2015. Tissue-based map of the human proteome. Science 347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 59.Oymans J, Wichgers Schreur PJ, van Keulen L, Kant J, Kortekaas J. 2020. Rift Valley fever virus targets the maternal-foetal interface in ovine and human placentas. PLoS Negl Trop Dis 14:e0007898. doi: 10.1371/journal.pntd.0007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barnard BJH. 1979. Rift Valley fever vaccine—antibody and immune response in cattle to a live and an inactivated vaccine. J S Afr Vet Assoc 50:155–157. [PubMed] [Google Scholar]
- 61.Keegan K, Collett MS. 1986. Use of bacterial expression cloning to define the amino acid sequences of antigenic determinants on the G2 glycoprotein of Rift Valley fever virus. J Virol 58:263–270. doi: 10.1128/JVI.58.2.263-270.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, Garcia MU, Di Tommaso P, Nahnsen S. 2020. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 38:276–278. doi: 10.1038/s41587-020-0439-x. [DOI] [PubMed] [Google Scholar]
- 63.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 64.Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 66.Smith TS, Heger A, Sudbery I. 2017. UMI-tools: modelling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res 27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 69.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R Core Team. 2022. R: A language and environment for statistical computing (4.2.1). R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 71.Stephens M. 2017. False discovery rates: a new deal. Biostatistics 18:275–294. doi: 10.1093/biostatistics/kxw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu Z. 2022. Complex heatmap visualization. Imeta 1:e43. doi: 10.1002/imt2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Larsson J. 2021. eulerr: Area-proportional Euler and Venn Diagrams with Ellipses (version 6.1.1). R package. https://CRAN.R-project.org/package=eulerr.
- 74.Blighe K, Rana S, Myles L. 2022. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling (version 1.8.0). R package. https://github.com/kevinblighe/EnhancedVolcano.
- 75.Yu G, Wang L-G, Han Y, He Q-Y. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. 2021. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation 2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S6 Tables S1 and S2. Download jvi.00415-23-s0001.docx, DOCX file, 2.1 MB (2.1MB, docx)
Table S3. Download jvi.00415-23-s0002.xlsx, XLSX file, 0.7 MB (721KB, xlsx)
Table S4. Download jvi.00415-23-s0003.xlsx, XLSX file, 0.03 MB (27.4KB, xlsx)
Table S5. Download jvi.00415-23-s0004.xlsx, XLSX file, 0.1 MB (111.5KB, xlsx)
Data Availability Statement
Raw sequencing data were deposited at the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA935986.