ABSTRACT
Plant viruses depend on a number of host factors for successful infection. Deficiency of critical host factors confers recessively inherited viral resistance in plants. For example, loss of Essential for poteXvirus Accumulation 1 (EXA1) in Arabidopsis thaliana confers resistance to potexviruses. However, the molecular mechanism of how EXA1 assists potexvirus infection remains largely unknown. Previous studies reported that the salicylic acid (SA) pathway is upregulated in exa1 mutants, and EXA1 modulates hypersensitive response-related cell death during EDS1-dependent effector-triggered immunity. Here, we show that exa1-mediated viral resistance is mostly independent of SA and EDS1 pathways. We demonstrate that Arabidopsis EXA1 interacts with three members of the eukaryotic translation initiation factor 4E (eIF4E) family, eIF4E1, eIFiso4E, and novel cap-binding protein (nCBP), through the eIF4E-binding motif (4EBM). Expression of EXA1 in exa1 mutants restored infection by the potexvirus Plantago asiatica mosaic virus (PlAMV), but EXA1 with mutations in 4EBM only partially restored infection. In virus inoculation experiments using Arabidopsis knockout mutants, EXA1 promoted PlAMV infection in concert with nCBP, but the functions of eIFiso4E and nCBP in promoting PlAMV infection were redundant. By contrast, the promotion of PlAMV infection by eIF4E1 was, at least partially, EXA1 independent. Taken together, our results imply that the interaction of EXA1-eIF4E family members is essential for efficient PlAMV multiplication, although specific roles of three eIF4E family members in PlAMV infection differ.
IMPORTANCE The genus Potexvirus comprises a group of plant RNA viruses, including viruses that cause serious damage to agricultural crops. We previously showed that loss of Essential for poteXvirus Accumulation 1 (EXA1) in Arabidopsis thaliana confers resistance to potexviruses. EXA1 may thus play a critical role in the success of potexvirus infection; hence, elucidation of its mechanism of action is crucial for understanding the infection process of potexviruses and for effective viral control. Previous studies reported that loss of EXA1 enhances plant immune responses, but our results indicate that this is not the primary mechanism of exa1-mediated viral resistance. Here, we show that Arabidopsis EXA1 assists infection by the potexvirus Plantago asiatica mosaic virus (PlAMV) by interacting with the eukaryotic translation initiation factor 4E family. Our results imply that EXA1 contributes to PlAMV multiplication by regulating translation.
KEYWORDS: host factor, plant virus, potexvirus, resistance
INTRODUCTION
The genomes of plant viruses contain a limited number of genes; as such, infection by these viruses is heavily dependent on host factors. Mutations or deletions in genes encoding these host factors may result in viral resistance due to loss of susceptibility. This type of resistance is inherited recessively and is thus called recessive resistance. Recessive resistance genes make up about half of the natural resistance alleles found in crops (1) and are widely used in breeding because they confer durable resistance. Thus, studies on host factors can contribute to understanding the mechanism of viral infection and provide potential strategies for controlling viral diseases.
The most widely exploited recessive resistance genes are eukaryotic translation initiation factor 4E (eIF4E) family members, which account for roughly half of the identified recessive resistance genes (1). eIF4E family members are cap-binding proteins that are conserved among eukaryotes, with three types of eIF4E family members present in plants: eIF4E, eIFiso4E, and novel cap-binding protein (nCBP) (2, 3). eIF4E and eIFiso4E interact with eIF4G and eIFiso4G, respectively, to form the translation initiation complexes required for cap-dependent mRNA translation (4). Resistance mediated by mutations or deletions of eIF4E or eIFiso4E has been identified in various plants for a wide range of viruses of the genera Potyvirus, Bymovirus, Polerovirus, Gammacarmovirus, Umbravirus, and Cucumovirus (5–11). eIF4E and eIFiso4E are implicated in different stages of viral infection, including viral genome translation, viral replication, cell-to-cell movement, and systemic infection (12–16). Arabidopsis nCBP, another eIF4E family member, interacts with wheat eIFiso4G in yeast and modestly promotes the translation of reporter mRNAs in vitro (17). Although the molecular function of nCBP in planta has been poorly characterized, we previously showed that nCBP deficiency in Arabidopsis thaliana delays infection by plantago asiatica mosaic virus (PlAMV; genus Potexvirus, family Alphaflexiviridae), most likely due to reduced accumulation of the viral proteins required for cell-to-cell movement (18). In addition, disruption of nCBP-1 and nCBP-2 in cassava by genome editing suppresses infection by cassava brown streak virus (genus Ipomovirus, family Potyviridae) (19). Thus, while eIF4E family members are involved in plant virus infection, their roles are not fully understood.
The genus Potexvirus comprises a group of positive-sense, single-stranded RNA viruses belonging to the family Alphaflexiviridae and include viruses that cause serious damage to crop production, such as PlAMV, pepino mosaic virus (PepMV), cymbidium mosaic virus (CymMV), and potato virus X (PVX) (20, 21). We previously identified a susceptibility gene for potexviruses, Essential for poteXvirus Accumulation 1 (EXA1) in A. thaliana (22). EXA1 is conserved among a wide range of plant species and is required for efficient accumulation of many potexviruses, including PlAMV, PepMV, CymMV, and PVX and the closely related lolium latent virus (LoLV; genus Lolavirus, family Alphaflexiviridae), in A. thaliana and in Nicotiana benthamiana (22, 23). This implies that the essential role of EXA1 in viral infection is conserved among these viruses. Accordingly, elucidating the role of EXA1 in potexvirus infection will enable the identification of determinants of successful infection by this group of viruses and in turn the development of strategies to control them effectively. While cellular accumulation of PlAMV is significantly reduced in EXA1-deficient A. thaliana plants compared to in wild-type plants (22), the mechanism responsible for this trait is unknown.
Previous studies have shown that EXA1/MUSE11/PSIG1 is involved in the regulation of plant immune responses. The salicylic acid (SA) pathway is upregulated in exa1 mutants compared to in wild-type plants (24, 25). Following bacterial inoculation, cell death via effector-triggered immunity (ETI) is enhanced in exa1 mutants compared to in wild-type plants in an SA-independent manner (24). Both the SA and the ETI pathways are involved in plant viral defenses (26), including against potexviruses (27, 28). These findings suggest that enhanced immune responses in exa1 mutants contributes to exa1-mediated resistance against potexviruses.
Plant EXA1 has two conserved motifs (22): a GYF domain that binds to proline-rich sequences (29) and an eIF4E-binding motif (4EBM) that binds to eIF4E family members (30, 31). This implies that EXA1 interacts with eIF4E family members, including nCBP, a susceptibility factor for PlAMV described in our previous study (18). Indeed, Arabidopsis EXA1 has been reported to interact with eIF4E1 and eIF4E1b in planta (25).
In this study, we analyzed the association between plant immune pathways and the eIF4E family and exa1-mediated viral resistance. We show that the enhancement of immune responses in exa1 mutants is not the primary mechanism of exa1-mediated resistance. Rather, the 4EBM of EXA1 is required for both efficient PlAMV accumulation and the interaction between EXA1 and eIF4E family members. In addition, eIF4E family members assist PlAMV infection in a dependent or independent manner with EXA1. Our results thus reveal the roles of EXA1 and eIF4E family members in PlAMV infection.
RESULTS
exa1-mediated viral resistance is independent of plant immune pathways.
The role of SA pathway activation in exa1-mediated viral resistance was explored using the Arabidopsis mutants sid2-2 (32), eds5-1 (33, 34), and NahG (35, 36), in which SA accumulation is suppressed, and the double mutants exa1-1 sid2-2 (24), exa1-1 eds5-1, and exa1-1 NahG. These mutants were mechanically inoculated with PlAMV-GFP (Fig. 1a), a green fluorescent protein (GFP)-expressing derivative of PlAMV (18). Quantitative reverse transcription-PCR (qRT-PCR) showed that expression of PR1, a marker gene of the SA pathway, was significantly higher in exa1-1 than in wild-type ecotype Columbia-0 (Col-0). PR1 expression levels in eds5-1, sid2-2, and NahG and in their double mutants with exa1-1 were significantly lower than in Col-0 (Fig. 1b), implying that the SA pathway was not activated in these double mutants. GFP fluorescence representing foci of viral infection was observed on the inoculated leaves of Col-0, sid2-2, eds5-1, and NahG at 6 days post-inoculation (dpi) but not in the inoculated leaves of exa1-1, exa1-1 sid2-2, exa1-1 eds5-1, and exa1-1 NahG (Fig. 1a). qRT-PCR confirmed that viral RNA levels in the double mutants were comparable to those in exa1-1 and significantly lower than those in either Col-0 or the SA-deficient mutants (Fig. 1c). These results demonstrate that exa1-mediated viral resistance is not compromised by deficiency in the SA pathway.
FIG 1.
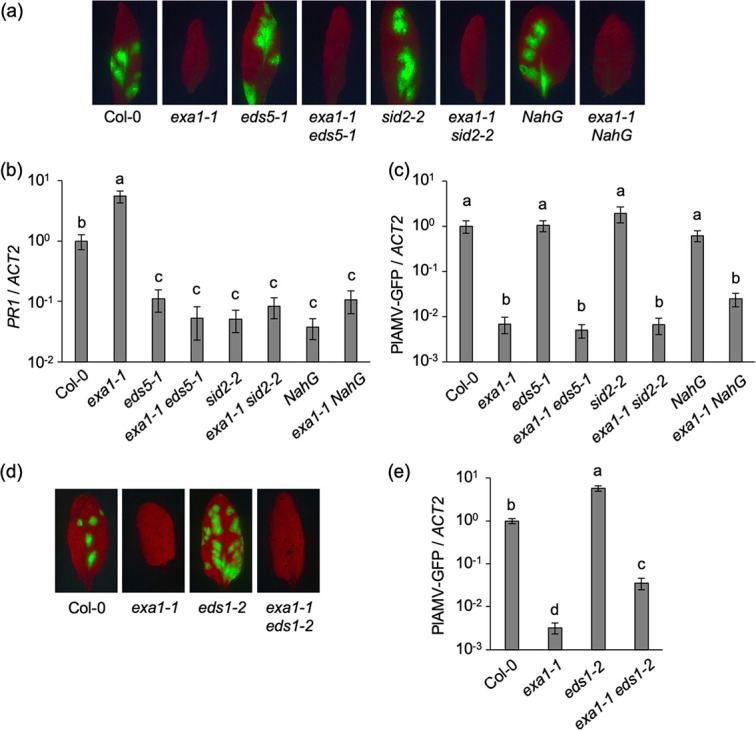
exa1-mediated viral resistance is independent of the salicylic acid (SA) and EDS1 pathways. (a) GFP fluorescence of PlAMV-GFP in the inoculated leaves of SA- and EXA1-deficient A. thaliana mutant lines. The plants were mechanically inoculated with PlAMV-GFP. Representative fluorescence images of the inoculated leaves at 6 days post-inoculation (dpi) are shown. (b and c) PR1 gene expression levels (b) and viral RNA levels (c) in PlAMV-GFP-inoculated leaves. The samples shown in panel a were subjected to qRT-PCR to quantify RNA levels at 6 dpi. The data are presented as the mean ± standard error (SE) obtained from at least three independent repeat experiments for each experimental plot. The mean in Col-0 was set as the standard (1.0). Statistically significant differences are indicated by different letters (Steel-Dwass test, P < 0.05). (d) GFP fluorescence of PlAMV-GFP in the inoculated leaves of EDS1- and EXA1-deficient A. thaliana mutant lines. The plants were mechanically inoculated with PlAMV-GFP. Representative fluorescence images of the inoculated leaves at 6 dpi are shown. (e) Viral RNA levels in PlAMV-GFP-inoculated leaves. The samples shown in panel d were subjected to qRT-PCR to quantify the RNA levels at 6 dpi. The data are presented as the mean ± SE obtained from at least two independent repeat experiments for each experimental plot. The mean in Col-0 was set as the standard (1.0). Statistically significant differences are indicated by different letters (Steel-Dwass test, P < 0.05).
We then investigated the relationship between exa1-mediated viral resistance and EDS1, which regulates the ETI pathway and basal defense (37). eds1-2 (38), a null mutant of EDS1, and exa1-1 eds1-2 (24) were mechanically inoculated with PlAMV-GFP. The fluorescence observed in the inoculated leaves at 6 dpi indicated that Col-0 and eds1-2 were susceptible to PlAMV, whereas exa1-1 and exa1-1 eds1-2 were not (Fig. 1d). qRT-PCR showed significantly higher viral RNA levels in eds1-2 than in Col-0 (Fig. 1e). This result implies that the EDS1 pathway reduces PlAMV accumulation, similar to the previously reported negative effect of EDS1 on the accumulation of bamboo mosaic virus (genus Potexvirus) in A. thaliana (39). Viral RNA levels in exa1-1 eds1-2 were also significantly higher than those in exa1-1 but still significantly lower than those in eds1-2 (Fig. 1e). Thus, although the EDS1 pathway negatively regulated PlAMV accumulation in exa1-1, exa1-mediated viral resistance was not compromised by deficiency in the EDS1 pathway. Based on these results, the primary mechanism of exa1-mediated viral resistance is independent of the enhancement of plant immune responses reported in previous studies (24, 25).
EXA1 interacts with the three eIF4E family members through the eIF4E-binding motif.
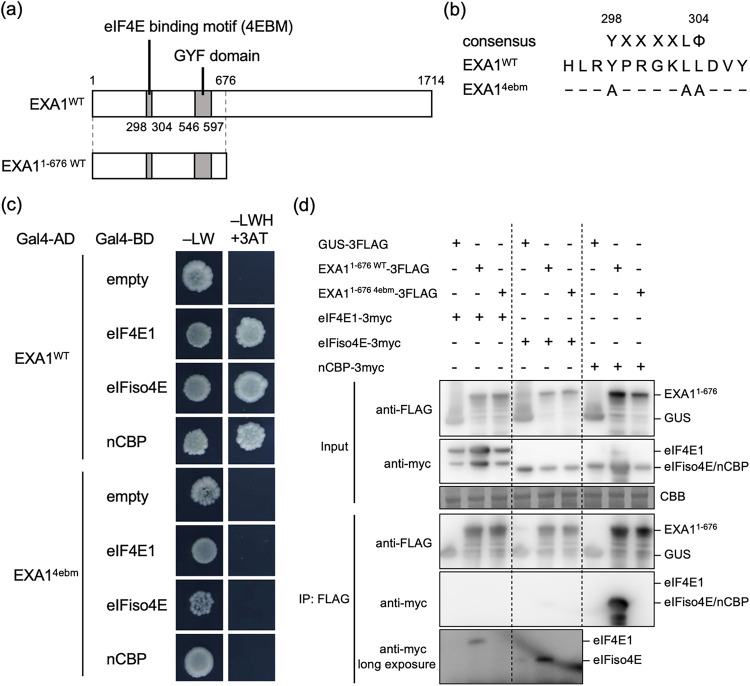
We next investigated the association of the eIF4E family with exa1-mediated viral resistance. A. thaliana has five eIF4E proteins: eIF4E1, eIFiso4E, and nCBP and the eIF4E1-diverged Brassicaceae-specific eIF4E1b and eIF4E1c (17, 40). Expression of eIF4E1b and eIF4E1c is at low levels in most tissues, but eIF4E1b is upregulated in pollen and developing embryos, implying its involvement in reproduction (40). Here, we focused on the interaction of Arabidopsis EXA1 with three eIF4E family members conserved throughout plants, eIF4E1, eIFiso4E, and nCBP. In a yeast two-hybrid (Y2H) assay, the obvious growth of yeast cells transformed with AD-EXA1WT and BD-eIF4E1, BD-eIFiso4E, or BD-nCBP indicated that EXA1 interacts with all three eIF4E family members (Fig. 2c). However, when an EXA1 mutant containing mutations in the 4EBM (EXA14ebm) (Fig. 2a and b) was used in the same assay, the yeast transformants did not grow (Fig. 2c), indicating that EXA1 interacts with the three eIF4E family members through the 4EBM in yeast.
FIG 2.
EXA1 interacts with the three eIF4E family members through the eIF4E-binding motif (4EBM). (a) Schematic structures of EXA1WT and EXA11-676 WT. White and gray boxes indicate the EXA1 coding region and motifs, respectively. The numbers represent the amino acid positions. (b) Sequence alignment of the predicted 4EBM of EXA1. The consensus sequence of 4EBM, the sequence of wild-type EXA1 (EXA1WT), and the sequence of the EXA1 mutant with alanine substitutions in the 4EBM (EXA14ebm) are shown. X indicates any amino acid, and Φ indicates a hydrophobic amino acid. Hyphens indicate that the amino acid at that position was unchanged. The numbers represent the amino acid positions. (c) Yeast two-hybrid (Y2H) assay of EXA1 and the three eIF4E family members. The yeast AH109 strain was cotransformed with Gal4-AD and Gal4-BD fusions of the proteins shown on the left. Equal amounts of transformed yeast cells were spotted on –LW or –LWH + 3AT. This experiment was replicated twice, with similar results. (d) Coimmunoprecipitation (co-IP) assay of EXA1 and the three eIF4E family members in planta. The constructs shown at the top were transiently coexpressed with P19 in N. benthamiana and subjected to co-IP. The input and anti-FLAG antibody immunoprecipitates were analyzed by Western blotting with anti-Myc and anti-FLAG antibodies. Coomassie brilliant blue (CBB) staining is shown as the loading control. The experiment was replicated four times and consistently yielded similar results, although in some replicates, eIFiso4E was not detected specifically in the immunoprecipitates of EXA11-676 WT-3FLAG.
The interaction between EXA1 and the eIF4E family members in planta was further examined in a coimmunoprecipitation (co-IP) assay. Due to difficulties in detecting full-length EXA1 protein under the denaturing conditions suitable for immunoprecipitation, the N-terminal region of EXA1 (EXA11-676 WT), which still contains the 4EBM and the GYF domain, was used instead (Fig. 2a). EXA11-676 WT fused to a 3FLAG tag (EXA11-676 WT-3FLAG), its 4EBM mutant (Fig. 2b) fused to a 3FLAG tag (EXA11-676 4ebm-3FLAG), or β-glucuronidase (GUS) with a 3FLAG tag as a negative control was transiently coexpressed with each eIF4E family member fused to a 3myc tag in N. benthamiana leaves and immunoprecipitated using an anti-FLAG antibody. While nCBP was clearly detected in the immunoprecipitates of EXA11-676 WT-3FLAG, this was not the case with EXA11-676 4ebm-3FLAG (Fig. 2d), indicating that nCBP interacts with EXA1 through the 4EBM in planta. eIF4E1 and eIFiso4E were also specifically detected in the immunoprecipitates of EXA11-676 WT-3FLAG, but the signal was weaker than that of nCBP (Fig. 2d), although eIFiso4E was not detected in some replicates. These results demonstrate that eIF4E1 and eIFiso4E can interact with EXA1 in planta.
The 4EBM of EXA1 is required for efficient PlAMV accumulation.
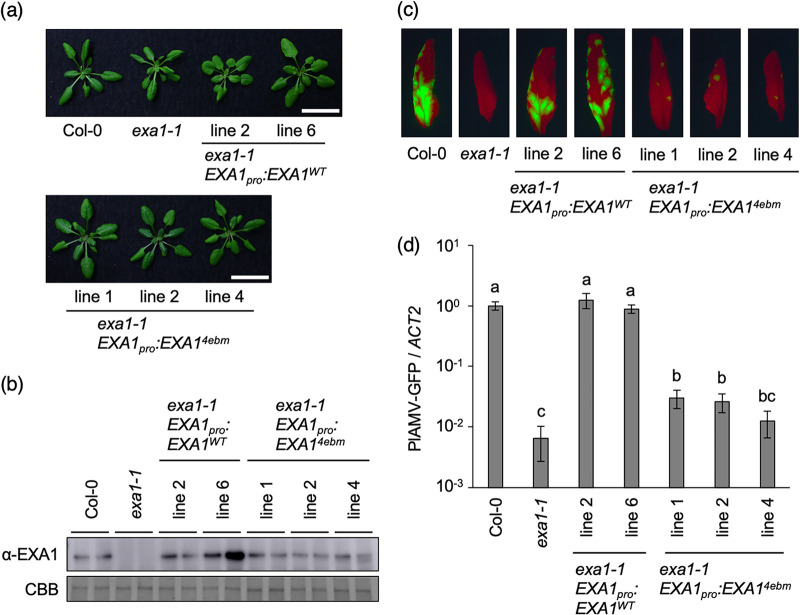
Both the Y2H and the co-IP assays showed that the 4EBM of EXA1 was responsible for the interaction with the three eIF4E family members. To evaluate the importance of the 4EBM of EXA1 in PlAMV infection, Col-0 mutant exa1-1 was transformed with a genomic DNA fragment of EXA1 containing the promoter region (EXA1pro:EXA1WT) or a 4EBM mutant (EXA1pro:EXA14ebm) (Fig. 2b). As previously reported (24, 25), exa1-1 showed a mildly dwarfed phenotype compared to Col-0 (Fig. 3a). Complementation with either EXA1pro:EXA1WT or EXA1pro:EXA14ebm restored the growth of exa1-1, although restoration of the growth of exa1-1 EXA1pro:EXA1WT in line 2 was weak (Fig. 3a). Western blotting showed sufficient accumulation of EXA1 protein in all of the complemented lines (Fig. 3b). The complemented lines were mechanically inoculated with PlAMV-GFP. Fluorescent foci representing viral infection were clearly observed on the inoculated leaves of exa1-1 EXA1pro:EXA1WT lines and resembled those present on Col-0, whereas the fluorescent foci of exa1-1 EXA1pro:EXA14ebm lines were significantly smaller (Fig. 3c). qRT-PCR also showed comparable levels of viral RNA in the exa1-1 EXA1pro:EXA1WT lines and Col-0, whereas viral RNA levels in the exa1-1 EXA1pro:EXA14ebm lines were significantly lower than those in Col-0 and slightly higher than those in exa1-1 (Fig. 3d). Together, these results imply that the 4EBM of EXA1 is crucial for successful PlAMV infection.
FIG 3.
The 4EBM of EXA1 is required for efficient PlAMV accumulation. (a) Representative photos of 4-week-old EXA1-complemented lines. Scale bars, 2 cm. (b) Protein accumulation of EXA1 in 4-week-old Col-0, exa1-1, and EXA1-complemented lines. Total protein was extracted from rosette leaves of each mutant line and analyzed by Western blotting using specific antibodies. CBB staining is shown as the loading control. The experiment was replicated three times with similar results. (c) GFP fluorescence of PlAMV-GFP in the inoculated leaves of EXA1-complemented lines. The plants were mechanically inoculated with PlAMV-GFP. Representative fluorescence images of the inoculated leaves at 6 dpi are shown. (d) Viral RNA levels in PlAMV-GFP-inoculated leaves. The samples shown in panel c were subjected to qRT-PCR to quantify the RNA levels at 6 dpi. The data are presented as the mean ± SE obtained from three independent repeat experiments. The mean in Col-0 was set as the standard (1.0). Statistically significant differences are indicated by different letters (Steel-Dwass test, P < 0.05).
Loss of eIF4E family members inhibits PlAMV infection.
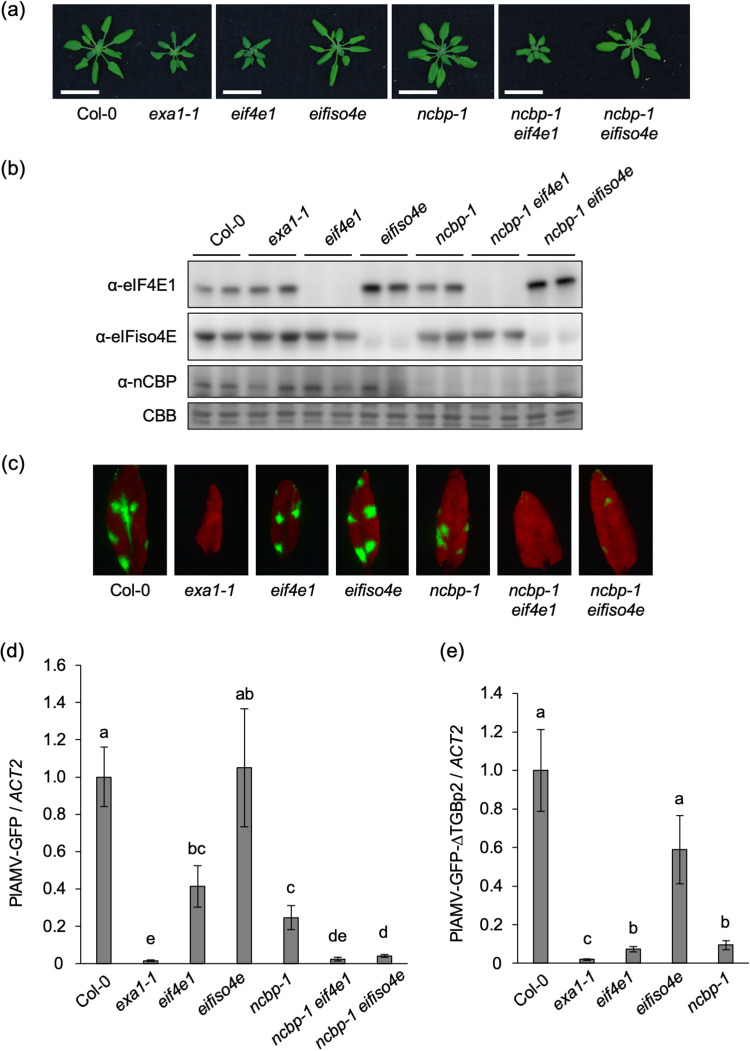
Given the significance of the 4EBM of EXA1 in PlAMV infection, the role of each eIF4E family member in PlAMV infection was examined using the respective single or double mutants, with the exception of a double mutant of eIF4E1 and eIFiso4E, which could not be tested because of its lethality (40, 41). Both eif4e1 and ncbp-1 eif4e1 showed dwarfed phenotypes compared to Col-0, while the growth of eifiso4e, ncbp-1, and ncbp-1 eifiso4e was similar to that of Col-0 (Fig. 4a). Western blotting confirmed that the corresponding eIF4E family protein did not accumulate in each A. thaliana mutant line (Fig. 4b). Increased accumulation of eIF4E1 protein in the eifiso4e mutant compared to Col-0 was observed, as previously reported (42, 43), which was also the case in the ncbp-1 eifiso4e mutant. No obvious compensation for the loss of eIF4E1 or nCBP by other eIF4E family members was observed.
FIG 4.
Loss of eIF4E family members inhibits PlAMV infection. (a) Representative photos of 4-week-old eIF4E family-deficient mutant lines of A. thaliana. Scale bars, 2 cm. (b) Protein accumulation of eIF4E family members in 4-week-old Col-0, exa1-1, and eIF4E family-deficient mutant lines. Total protein was extracted from rosette leaves of each mutant line and analyzed by Western blotting using specific antibodies. CBB staining is shown as the loading control. The experiment was replicated twice with similar results. (c) GFP fluorescence of PlAMV-GFP in the inoculated leaves of eIF4E family-deficient A. thaliana mutant lines. The plants were mechanically inoculated with PlAMV-GFP. Representative fluorescence images of the inoculated leaves at 6 dpi are shown. (d) Viral RNA levels in PlAMV-GFP-inoculated leaves. The samples shown in panel c were subjected to qRT-PCR to quantify the RNA levels at 6 dpi. The data are presented as the mean ± SE obtained from at least four independent repeat experiments for each experimental plot. The mean in Col-0 was set as the standard (1.0). Statistically significant differences are indicated by different letters (Steel-Dwass test, P < 0.05). (e) Viral RNA levels in PlAMV-GFP-ΔTGBp2-inoculated leaves of eIF4E family-deficient A. thaliana mutant lines. Indicated plants were mechanically inoculated with PlAMV-GFP, and RNA levels at 4 dpi were quantified by qRT-PCR. The data are presented as the mean ± SE obtained from three independent repeat experiments. The mean in Col-0 was set as the standard (1.0). Statistically significant differences are indicated by different letters (Steel-Dwass test, P < 0.05).
The wild-type and mutant plants were mechanically inoculated with PlAMV-GFP. Fluorescent foci of infection of similar sizes were observed at 6 dpi on the inoculated leaves of Col-0, eif4e1, and eifiso4e, whereas the fluorescent foci on the inoculated leaves of ncbp-1 and ncbp-1 eifiso4e were noticeably smaller than those of Col-0 (Fig. 4c). Furthermore, few fluorescent foci were visible on ncbp-1 eif4e1. qRT-PCR analysis showed significantly lower viral RNA levels in eif4e1 and ncbp-1 than in Col-0 (Fig. 4d). Because the results obtained with the eif4e1 mutant differed from those reported in a previous study (18), another eif4e1 mutant with a transfer DNA (T-DNA) insertion (SALK_145583C) (Fig. S1a in the supplemental material) was inoculated with PlAMV-GFP. Viral RNA levels in that eif4e1 mutant were also significantly lower than those in Col-0, supporting the involvement of eIF4E1 in PlAMV infection (Fig. S1b). The significantly lower viral RNA levels in ncbp-1 eif4e1 than in the ncbp-1 and eif4e1 mutants (Fig. 4d) can be explained by the additive reduction of PlAMV accumulation incurred by the loss of both nCBP and eIF4E1. Viral RNA levels in eifiso4e and Col-0 were comparable (Fig. 4d), implying that eIFiso4E is less important for PlAMV infection, although it also remains possible that increased eIF4E1 protein accumulation in eifiso4e (Fig. 4b) complements loss of eIFiso4E to support viral accumulation. Notably, viral RNA levels were significantly lower in ncbp-1 eifiso4e than in ncbp-1 (Fig. 4d). Because the viral RNA levels in eifiso4e were similar to those in Col-0 (Fig. 4d), the contribution of eIFiso4E to PlAMV infection may increase in response to loss of nCBP. This eIFiso4E contribution is not associated with changes in eIF4E1 protein accumulation because the increase in eIF4E1 protein accumulation was observed not only in eifiso4e but also in ncbp-1 eifiso4e (Fig. 4b). These results indicate that all three eIF4E family members contribute to PlAMV infection.
Because EXA1 is essential for PlAMV accumulation in plant cells (22), we examined whether eIF4E family members, as interaction partners of EXA1, are also important in this process. In this experiment, PlAMV-GFP-ΔTGBp2, which lacks triple gene block protein 2 (TGBp2) (44), a movement protein required for viral cell-to-cell movement, was used. Deficiency in cell-to-cell movement by PlAMV-GFP-ΔTGBp2 was confirmed by following the development of fluorescent foci of infection on the inoculated leaves (Fig. S2). eIF4E family-deficient mutant lines inoculated with PlAMV-GFP-ΔTGBp2 were subjected to qRT-PCR, which showed that viral RNA levels in the inoculated leaves of eifiso4e at 4 dpi did not differ significantly from those in Col-0, whereas the viral RNA levels in eif4e1 and ncbp-1 were significantly lower than those in Col-0 but not as low as those in exa1-1 (Fig. 4e), indicating the contribution of eIF4E1 and nCBP to the cellular accumulation of PlAMV. By contrast, in a previous study, there was no significant difference between PlAMV accumulation in protoplasts prepared from ncbp-1 and Col-0 (18). To reevaluate the importance of nCBP in the cellular accumulation of PlAMV, PlAMV-GFP-ΔTGBp2 was inoculated into nCBP-complemented lines. The viral RNA levels in those lines were significantly higher than those in ncbp-1 and did not differ significantly from those in Col-0 (Fig. S3), confirming that nCBP is required for the efficient cellular accumulation of PlAMV.
Relationship between EXA1-mediated and eIF4E family-mediated viral resistance.
To investigate the relationship between EXA1 and three eIF4E family members in PlAMV infection, exa1-1 was crossed with the eif4e1, eifiso4e, and ncbp-1 mutants. The growth of exa1-1 eifiso4e and exa1-1 ncbp-1 was similar to that of exa1-1, while the growth of exa1-1 eif4e1 was dwarfed compared to that of exa1-1 (Fig. 5a). When the mutants were inoculated with PlAMV-GFP, the qRT-PCR analysis showed that the viral RNA levels in the inoculated leaves of exa1-1 eifiso4e at 6 dpi were comparable to those in exa1-1 (Fig. 5b). In contrast to the lower viral RNA levels in ncbp-1 than in Col-0 (Fig. 4d), the viral RNA levels in exa1-1 ncbp-1 and exa1-1 were not significantly different (Fig. 5b), implying that the contribution of nCBP to PlAMV infection is EXA1 dependent. By contrast, the viral RNA levels in exa1-1 eif4e1 were significantly lower than those in exa1-1 (Fig. 5b). This result implies that loss of EXA1 and eIF4E1 additively reduce PlAMV accumulation, and that the contribution of eIF4E1 to PlAMV infection is, at least partially, independent of EXA1.
FIG 5.
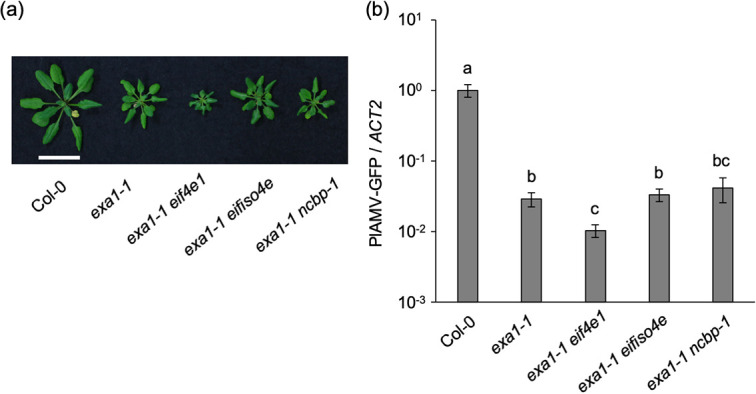
Relationship between EXA1-mediated and eIF4E family-mediated viral resistance. (a) Representative photos of 5-week-old A. thaliana mutant lines deficient in eIF4E family members and EXA1. Scale bar, 2 cm. (b) Viral RNA levels in PlAMV-GFP-inoculated leaves of A. thaliana mutant lines deficient in eIF4E family members and EXA1. The plants were mechanically inoculated with PlAMV-GFP, and RNA levels were quantified by qRT-PCR at 6 dpi. The data are presented as the mean ± SE obtained from three independent repeat experiments. The mean in Col-0 was set as the standard (1.0). Statistically significant differences are indicated by different letters (Steel-Dwass test, P < 0.05).
DISCUSSION
This study showed that exa1-mediated viral resistance is independent of the enhancement of plant immune responses and that EXA1 interacts with the three eIF4E family members. Those interactions appear to be crucial for PlAMV infection. eIF4E family members promote the cellular multiplication of PlAMV in a dependent or independent manner with EXA1. Our results thus reveal the roles of EXA1 and eIF4E family members in promoting PlAMV infection.
Functional relationship between EXA1 and eIF4E family members in PlAMV infection.
In previous studies, we identified EXA1 and nCBP as susceptibility factors for PlAMV infection (18, 22). Here, we showed that EXA1 interacts with the three eIF4E family members through the 4EBM (Fig. 2c and d) and that the 4EBM of EXA1 is required for efficient PlAMV accumulation (Fig. 3c and d). These findings strongly suggest that the interaction between EXA1 and eIF4E family members is required for PlAMV infection. In support of this hypothesis, PlAMV accumulation was reduced by loss of nCBP in the Col-0 background (Fig. 4d) but not by loss of nCBP in the exa1-1 background (Fig. 5b), implying that nCBP contributes to PlAMV infection in an EXA1-dependent manner. Despite comparable PlAMV accumulation in eifiso4e and Col-0, PlAMV accumulation was significantly lower in ncbp-1 eifiso4e than in ncbp-1 (Fig. 4d), which indicates a larger contribution of eIFiso4E to PlAMV infection following the loss of nCBP. Thus, the functions of eIFiso4E and nCBP are partially redundant in their interactions with EXA1. We also found that PlAMV accumulation was reduced by loss of eIF4E1 in both the Col-0 background (Fig. 4d) and the exa1-1 background (Fig. 5b), indicating that the contribution of eIF4E1 to PlAMV infection is, at least partially, independent of EXA1. Because eIF4E1 is able to interact with EXA1 (Fig. 2c and d), it may be that the functions of not only eIFiso4E but also eIF4E1 overlap with those of nCBP. Our results thus provide evidence for the redundancy and the dependent or independent roles with EXA1 of eIF4E family members during PlAMV infection (Fig. 6).
FIG 6.
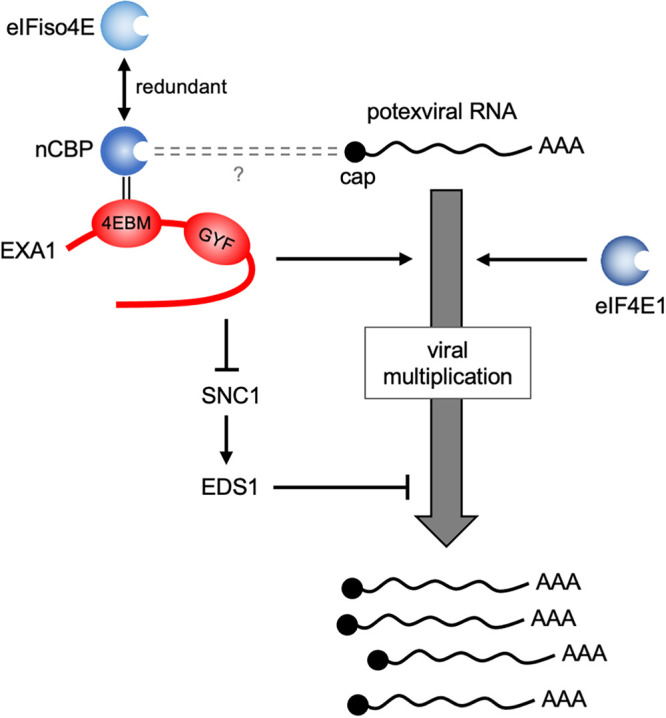
A proposed model for the roles of Arabidopsis EXA1 and the three eIF4E family members in PlAMV infection. nCBP assists PlAMV multiplication in concert with EXA1, possibly via binding to the 5′ cap structure of viral RNA. The functions of eIFiso4E and nCBP in their interaction with EXA1 are partially redundant. eIF4E1 assists PlAMV multiplication in an EXA1-independent manner. In addition, EXA1 suppresses the SNC1-EDS1 pathway, which negatively regulates PlAMV accumulation, although this is not the primary role of EXA1 in PlAMV infection.
The necessity of nCBP and eIF4E1 in the efficient cellular accumulation of PlAMV.
The reduction in PlAMV-GFP-ΔTGBp2 RNA levels following the loss of nCBP (Fig. 4e) indicates that nCBP assists the cellular accumulation of PlAMV. However, a previous study showed that loss of nCBP delays cell-to-cell movement by PlAMV-GFP but does not significantly affect PlAMV accumulation at the single-cell level (18). This discrepancy can be explained by differences in experimental systems. In the previous study, plasmids containing PlAMV expressed under the control of the constitutive 35S promoter were introduced into protoplasts of A. thaliana (18), which may have resulted in higher inoculation pressure than was the case following the mechanical inoculation of PlAMV performed in this study. The necessity of nCBP in the efficient cellular accumulation of PlAMV was confirmed by the recovery of PlAMV-GFP-ΔTGBp2 accumulation in the nCBP-complemented lines (Fig. S3 in the supplemental material). Based on the results of the present study, the delayed cell-to-cell movement of PlAMV-GFP observed in the previous study (18) may have reflected reduced cellular accumulation of PlAMV. However, the accumulation of TGBp2 and TGBp3, both of which are required for the cell-to-cell movement of PlAMV, was less in ncbp-1 than in Col-0 (18), implying that suppression of cell-to-cell movement also occurred in ncbp-1.
The reduction in RNA levels of PlAMV-GFP and PlAMV-GFP-ΔTGBp2 following the loss of eIF4E1 (Fig. 4d and e) indicates that eIF4E1 also contributes to the cellular accumulation of PlAMV. However, in a previous study, there was no significant difference between PlAMV-GFP accumulation in eif4e1 and Col-0 (18). This discrepancy may have been caused by differences in experimental conditions. Mechanical inoculation of PlAMV-GFP was performed on 3-week-old eif4e1 and Col-0 plants in the previous study (18) and on 4-week-old eif4e1 and Col-0 plants in the present study. In general, susceptibility to pathogens, including viruses, varies depending on plant age (45). Differences in the age of the plants used for virus inoculation may have affected the results.
The roles of EXA1 and eIF4E family members in mRNA translation.
eIF4E1 and eIFiso4E play essential roles in cap-dependent mRNA translation via their interaction with eIF4G and eIFiso4G, respectively (4). Additionally, eIF4E1 and eIFiso4E interact with CERES and form a noncanonical translation initiation complex to boost mRNA translation when intracellular metabolic and translational conditions are favorable (46). Since Arabidopsis nCBP interacts with wheat eIFiso4G in yeast and modestly promotes translation in vitro (17), nCBP might play some role in translation initiation. However, the double loss of Arabidopsis eIF4E1 and eIFiso4E is lethal, whereas loss of either of them is not lethal (40, 41), which indicates the essential and redundant roles of these two isoforms in plant growth and the distinct role of nCBP.
As discussed above, our results suggest that the function of nCBP is dependent on EXA1, at least during PlAMV infection, and that eIFiso4E and possibly eIF4E1 are redundant with nCBP in their interaction with EXA1. Because eIF4E family members are cap-binding proteins, eIF4E family members interacting with EXA1 may bind to the cap structure of mRNA. Indeed, both EXA1 and eIF4E family members are enriched by affinity purification using m7GTP-Sepharose, an analog of the cap structure (47). In addition, EXA1 has been shown to interact with a ribosomal protein, suggesting that EXA1 is involved in translational regulation (25). As shown in Fig. 2c and d, the 4EBM of EXA1 is required for interaction between EXA1 and eIF4E family members. Because eIF4G and eIFiso4G also have the 4EBM (48, 49), EXA1 may compete with eIF4G and eIFiso4G in its interaction with the eIF4E family, just as human eIF4E-binding proteins (4E-BPs) have the 4EBM and compete with eIF4G in their binding to eIF4E (30, 50). If so, there are two possible functions of EXA1 in translational regulation. One is that EXA1 may promote mRNA translation initiation by forming a noncanonical translation initiation complex, such as that formed by CERES (46). Another possibility is that EXA1, together with the eIF4E family, inhibits mRNA translation initiation by competing with the eIF4E-eIF4G and eIFiso4E-eIFiso4G complexes in binding to the mRNA cap structure. A previous study has shown that loss of EXA1 in A. thaliana increases the protein accumulation of immune receptors but does not affect their mRNA levels, suggesting that EXA1 may suppress immune receptor accumulation through translational repression (25). Thus, EXA1 appears to repress translation rather than promote it.
Further support for the function of Arabidopsis EXA1 with the eIF4E family in translational regulation comes from studies in human cells. eIF4E homologous protein (4EHP), an nCBP ortholog in human cells (3), interacts with Grb10-interacting GYF protein 2 (GIGYF2) (51), a GYF domain protein homologous to EXA1 (22, 25). GIGYF2 interacting with 4EHP binds to the cap structure of specific mRNAs to induce translational repression and mRNA decay (51–54).
Possible functions of EXA1 and eIF4E family members in PlAMV infection.
EXA1 represses the translation of SNC1 (25), a positive regulator of the EDS1 pathway (55, 56). In this study, the EDS1 pathway was shown to negatively regulate PlAMV accumulation (Fig. 1e). These findings suggest that loss of EXA1 promotes the SNC1-EDS1 pathway, thereby preventing PlAMV infection. However, the absence of GFP fluorescence associated with viral infection in exa1-1 eds1-2 (Fig. 1d) and the significant reduction in viral RNA levels in exa1-1 eds1-2 compared to eds1-2 (Fig. 1e) indicate that the primary mechanism of exa1-mediated viral resistance is independent of EDS1. Besides SNC1, some genes encoding immune receptors involved in the NDR1-dependent ETI pathway may be regulated by EXA1 (25). The possibility that these or other unknown plant genes regulated by EXA1 indirectly suppress PlAMV infection remains to be investigated.
Similar to plant mRNAs, genomic/subgenomic RNAs of potexviruses, including PlAMV, have a cap structure at their 5′ end and a poly(A) tail at their 3′ end (57, 58). Thus, the eIF4E family interacting with EXA1 may bind to genomic/subgenomic RNAs of PlAMV. We previously showed that the accumulation of TGBp2 and TGBp3, membrane proteins translated from subgenomic RNA1 of PlAMV (59), is reduced in ncbp-1 compared to in Col-0 (18). Therefore, EXA1 and nCBP may regulate the accumulation of TGBp2 and TGBp3. In addition, EXA1 assists another stage of PlAMV infection that does not require TGBp2 and TGBp3 because exa1-mediated viral resistance is effective against 53U-RdRp, a PlAMV replicon that encodes only RNA-dependent RNA polymerase (RdRp) (22, 23). Because EXA1 is likely to be involved in translational regulation, as discussed above, EXA1 may regulate genomic RNA translation during the translation-replication cycle of 53U-RdRp. The genomic RNA of positive-stranded RNA viruses, including potexvirus, serves as a template for both translation and replication. Although the replication protein must first be translated from the genomic RNA, ribosomes translating in the 5′-to-3′ direction inhibit minus-strand RNA synthesis in the 3′-to-5′ direction by the replication protein (60). Thus, translation must be properly terminated before viral replication. As discussed above, EXA1, which interacts with the eIF4E family, may compete with the eIF4E-eIF4G and eIFiso4E-eIFiso4G complexes for binding to the cap structure, thereby inhibiting translation initiation. EXA1 may allow PlAMV genomic RNA to switch from translation to replication by inhibiting translation initiation of genomic RNA.
Our results indicate a partial and EXA1-independent contribution of eIF4E1 to PlAMV infection (Fig. 5b). In viral infections of other genera, eIF4E and eIFiso4E are implicated not only in viral genome translation but also in replication, cell-to-cell movement, and systemic infection (12–16). Since loss of eIF4E1 reduced accumulation of PlAMV-GFP-ΔTGBp2 (Fig. 4e), eIF4E1 assists PlAMV infection at least in the process before cell-to-cell movement, the mechanism of which remains to be analyzed in the future.
MATERIALS AND METHODS
Plant materials and growth conditions.
A. thaliana ecotype Columbia-0 (Col-0) was used as the wild-type control. The Arabidopsis mutants exa1-1 (SALK_005994C) (22), eds5-1 (CS3735) (33, 34), eif4e1 (cum1-1) (11), eif4e1 (SALK_145583C) (61), and ncbp-1 (SALK_131503C) (18) were purchased from the Arabidopsis Biological Resource Center (ABRC; The Ohio State University, Columbus, OH, USA). sid2-2 (32) seeds were kindly provided by Kohki Yoshimoto (Meiji University, Japan), and eds1-2 (38) seeds were provided by Shigeyuki Betsuyaku (Ryukoku University, Japan) and Jane Parker (Max Planck Institute for Plant Breeding Research, Germany). eifiso4e (42), ncbp-1 eif4e1 (cum1-1), and ncbp-1 eifiso4e seeds were kindly provided by Karen Browning and Laura Mayberry (University of Texas at Austin, USA). NahG-expressing plants were generated by transforming Col-0 with NahG cloned into the pFAST02 vector (62), and exa1-1 eds5-1, exa1-1 NahG, exa1-1 eif4e1 (cum1-1), exa1-1 eifiso4e, and exa1-1 ncbp-1 plants were generated by crossing each mutant. The nCBP-complemented lines 1A and 3F were described previously (18), as were exa1-1 sid2-2 and exa1-1 eds1-2 (24). A. thaliana and N. benthamiana were grown on soil under 16-h light/8-h dark conditions at 22°C and 25°C, respectively.
Plasmid construction.
A GFP-expressing derivative of PlAMV (PlAMV-GFP) (18) and PlAMV-GFP lacking TGBp2 (PlAMV-GFPΔTGBp2) (44) were used for virus inoculation.
For the Y2H assay, cDNA fragments encoding eIF4E1, eIFiso4E, and nCBP were PCR amplified using primer pairs pBK-eIF4E-F/pGBKT7-eIF4E-R2, pBK-eIFiso4E-F/pGBKT7-eIFiso4E-R2, and pGBKT7-AtnCBP-F/pGBKT7-AtnCBP-R, respectively. The amplified fragments were assembled with NdeI- and EcoRI-digested pGBKT7 vector (Clontech) using Gibson Assembly master mix (New England Biolabs [NEB], Ipswich, MA, USA) to obtain constructs expressing each protein fused with the Gal4-binding domain (pGBKT7-eIF4E1, pGBKT7-eIFiso4E, and pGBKT7-nCBP). EXA1 fused with the Gal4-activating domain (pGADT7-EXA1WT) was obtained by amplifying the cDNA fragment encoding EXA1 using primers Sm-AT5G42950-1F and Cl-AT5G42950-5145R. The amplified fragment was digested with XmaI and ClaI and inserted into the pGADT7 AD vector (Clontech) digested with XmaI and ClaI. A pGADT7-EXA1WT mutant with alanine substitutions in the 4EBM (pGADT7-EXA14ebm) was generated by introducing a mutation into the plasmid containing the cDNA fragment of EXA1 by PCR using primers At5G42950-Y298A-303LLAA-F and At5G42950-Y298A-303LLAA-R, followed by a second PCR using primers pAD-EXA1-1F and pAD-EXA1-5145R. The amplified fragment was assembled using NEBuilder HiFi DNA assembly master mix (NEB) with the DNA fragment of the pGADT7 AD vector (Clontech) and PCR amplified using the primer pair pAD-EXA1-invF and pAD-EXA1-invR.
For the co-IP assay, cDNA fragments encoding eIF4E1, eIFiso4E, and nCBP were PCR amplified using primer pairs Kp-At4E-1F/Nt-At4E-705R, Kp-Atiso4E-1F/Nt-Atiso4E-594R, and Kp-nCBP-1F/Nt-nCBP-663R, respectively. The amplified fragments were digested with KpnI and NotI and inserted into the Gateway Entry vector pENTA (63) digested with KpnI and NotI. The DNA sequence of GUS in the pBI121 vector (Clontech) was PCR amplified using primers pENTA-GUS1F and EcV-GUS1809R and subcloned into pENTA. The partial cDNA fragment of EXA1 (EXA11-676 WT) and its mutant in the 4EBM (EXA11-676 4ebm) were PCR amplified using the primers SalI-EXA1-1F and Nt-At5G42950-2028R. The amplified fragments were digested with SalI and NotI and inserted into pENTA digested with SalI and NotI. Next, pENTA-cloned GUS, eIF4E1, eIFiso4E, nCBP, EXA11-676 WT, and EXA11-676 4ebm were subcloned into pEarlyGateC3myc (a pEarlyGate-based vector modified to express C-terminal triple c-Myc-tagged proteins) (64) or pEarlyGateC3FLAG (a pEarlyGate-based vector modified to express C-terminal triple FLAG-tagged proteins) using the Gateway LR Clonase II enzyme mix (Thermo Fisher Scientific, Waltham, MA, USA). Efficient protein expression was achieved using p19, an RNA-silencing suppressor from tomato bushy stunt virus cloned into pE7133-GW (44).
Plants were transformed using a genomic fragment of EXA1 containing the promoter region cloned into pENTA (pENTA-EXA1pro:EXA1WT) (22). The 4EBM alanine substitution mutant (pENTA-EXA1pro:EXA14ebm) was obtained by PCR amplifying two DNA fragments from pENTA-EXA1pro:EXA1WT using primer pairs pENTR-insertF/At5G42950-Y298A-303LLAA-R and At5G42950-Y298A-303LLAA-F/pENTR-insertR2 followed by assembly using NEBuilder HiFi DNA assembly master mix (NEB). The NahG-expressing construct was generated by PCR amplifying NahG from Pseudomonas putida using primers NahG_Fopt and NahG_Ropt; the resulting fragment was inserted into pENTA using a GeneArt seamless cloning and assembly kit (Thermo Fisher Scientific). The inserts in pENTA were subcloned into pFAST01 or pFAST02 vectors (62) using Gateway LR Clonase II enzyme mix (Thermo Fisher Scientific) to generate pFAST01-EXA1pro:EXA1WT, pFAST01-EXA1pro:EXA14ebm, and pFAST02-NahG. The primers used for molecular cloning are listed in Table 1.
TABLE 1.
Primers used in this study
| Primer name | Primer sequence (5′ to 3′) |
|---|---|
| pBK-eIF4E-F | TCAGAGGAGGACCTGCATATGATGGCGGTAGAAGACACTCCCAAATCTG |
| pGBKT7-eIF4E-R2 | TCGACGGATCCCCGGGAACTATCAAGCGGTGTAAGCGTTCTTTGC |
| pBK-eIFiso4E-F | TCAGAGGAGGACCTGCATATGATGGCGACCGATGATGTGAACGAGCCTC |
| pGBKT7-eIFiso4E-R2 | TCGACGGATCCCCGGGAACTATCAGACAGTGAACCGGCTTCTTCTG |
| pGBKT7-AtnCBP-F | GAGGAGGACCTGCATATGGAGGTTTTGGATAGG |
| pGBKT7-AtnCBP-R | ACGGATCCCCGGGAACTATCCTCTCAGCCATGTG |
| Sm-AT5G42950-1F | TCCCCCGGGGATGGCTAACTCTTCCGCTGG |
| Cl-AT5G42950-5145R | CCATCGATTCAGTCCTCAATTGTCTGAAT |
| At5G42950-Y298A-303LLAA-F | ACCTCCCCATCTGAGAGCTAGCAGAATGAAAGCGGCGGATGTGTACAGG |
| At5G42950-Y298A-303LLAA-R | CCTGTACACATCCGCCGCTTTCATTCTGCTAGCTCTCAGATGGGGAGGT |
| pAD-EXA1-1F | GTGAATTCCACCCGGGGATGGCTAACTCTTCCGC |
| pAD-EXA1-5145R | CGATGGATCCCGTATCGATTCAGTCCTCAATTGTCTG |
| pAD-EXA1-invR | GCGGAAGAGTTAGCCATCCCCGGGTGGAATTCAC |
| pAD-EXA1-invF | CAGACAATTGAGGACTGAATCGATACGGGATCCATCG |
| Kp-At4E-1F | CGGGGTACCGAATGGCGGTAGAAGACACTCC |
| Nt-At4E-705R | ATAGTTTAGCGGCCGCGAAGCGGTGTAAGCGTTCTTTG |
| Kp-Atiso4E-1F | CGGGGTACCGAATGGCGACCGATGATGTGAA |
| Nt-Atiso4E-594R | ATAGTTTAGCGGCCGCGAGACAGTGAACCGGCTTCTTC |
| Kp-nCBP-1F | CGGGGTACCGAATGGAGGTTTTGGATAGGAGA |
| Nt-nCBP-663R | ATAGTTTAGCGGCCGCGATCCTCTCAGCCATGTGTTTC |
| pENTA-GUS1F | AATTCAGTCGACTGGATCATGTTACGTCCTGTAGAAACC |
| EcV-GUS1809R | AAGCTGGGTCTAGATATCTTTGTTTGCCTCCCTGCTGCG |
| SalI-EXA1-1F | CGGAATTCGTCGACAATGGCTAACTCTTCCGCTGG |
| Nt-At5G42950-2028R | ATAGTTTAGCGGCCGCGAAGATGAATTGGTCAGACCC |
| pENTR-insertF | AGTTACTTAAGCTCGGGCCC |
| pENTR-insertR2 | GGGCCCGAGCTTAAGTAACT |
| NahG_Fopt | AAGGAACCAATTCAGTCATGAAGAATAACAAGCTTGGACTCAG |
| NahG_Ropt | AAGCTGGGTCTAGATTCAGCCTTGTCTTAACGCTCCTCCT |
| PlRep-F3 | AATCCCCAGACTTCCATGAGCACC |
| PlRep-R3 | TTTTCTTTGCGCCGAGCTTCTC |
| Actin2_F | GCACCCTGTTCTTCTTACCG |
| Actin2_R | AACCCTCGTAGATTGGCACA |
| PR-1_qF | TCACAACCAGGCACGAGGAG |
| PR-1_qR | CACCGCTACCCCAGGCTAAG |
Virus inoculation.
PlAMV-GFP and PlAMV-GFP-ΔTGBp2 were mechanically inoculated as described previously (65). Agrobacterium tumefaciens strain EHA105 carrying the PlAMV-GFP or PlAMV-GFP-ΔTGBp2 vector was infiltrated into N. benthamiana. The systemically infected leaves of PlAMV-GFP or the inoculated leaves of PlAMV-GFP-ΔTGBp2 were ground in 0.1 M phosphate buffer (pH 7.0), to which carborundum was then added. Rosette leaves of 4- or 5-week-old A. thaliana plants were rub inoculated with the preparation.
Fluorescence microscopy.
Fluorescence images of the inoculated leaves and infection foci were obtained using an M165 FC fluorescence microscope, a DFC310 FX camera, and LAS software version 4.4.0 (Leica Microsystems, Wetzlar, Germany).
qRT-PCR analysis.
Total RNA was extracted from the inoculated leaves and treated with DNase using an ISOSPIN plant RNA kit (Nippon Gene, Japan). The RNA was then reverse transcribed using a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific, Waltham, MA, USA). The amounts of viral RNA, ACT2, and PR1 were quantified by qRT-PCR using a thermal cycler Dice real-time system (TaKaRa, Shiga, Japan) with TB green premix Ex Taq II (TaKaRa). ACT2 served as an internal control. For the qRT-PCR of viral RNA, the primers were designed for the RdRp coding region, such that only genomic RNA, not subgenomic RNA, was quantified. The sequences of the primers used in the qRT-PCR (PlRep-F3 and PlRep-R3 for viral RNA, Actin2_F and Actin2_R for ACT2, and PR-1_qF and PR-1_qR for PR1) are listed in Table 1. The statistical analysis was based on the Steel-Dwass test.
Y2H assay.
The Y2H assay was performed using the Matchmaker GAL4 two-hybrid system 3 (Clontech). The yeast AH109 strain was cotransformed with pGADT7-EXA1WT or pGADT7-EXA14ebm and pGBKT7-eIF4E1, pGBKT7-eIFiso4E, pGBKT7-nCBP, or pGBKT7. Successful cotransformants were selected on synthetically defined (SD) medium lacking Leu/Trp (–LW). To evaluate the protein interaction, cotransformants were cultured on –LW and on SD medium lacking Leu/Trp/His and containing 20 mM 3-amino-1,2,4-triazole (Sigma-Aldrich, St. Louis, MO, USA) (−LWH+3AT). The plates were incubated at 30°C for 4 days.
Plant total protein extraction and co-IP assay.
For the detection of EXA1 protein, equal amounts of rosette leaves were ground in liquid nitrogen and suspended in SDS-PAGE sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 5% 3-mercapto-1,2-propanediol, 10% glycerol, and 0.0025% bromophenol blue) with 8 M urea. The extract was centrifuged at 1,000 × g for 10 min to remove debris. The supernatant was incubated at 95°C for 5 min and analyzed by Western blotting. For the detection of eIF4E family proteins, equal amounts of rosette leaves were ground in liquid nitrogen and suspended in SDS-PAGE sample buffer. The extract was centrifuged at 15,000 × g for 10 min. The supernatant was incubated at 95°C for 5 min and analyzed by Western blotting.
For the co-IP assay, 3FLAG-tagged proteins, 3myc-tagged proteins, and P19 were transiently coexpressed by agroinfiltration in N. benthamiana. The agroinfiltrated leaves were ground in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% 3-mercapto-1,2-propanediol, 20% glycerol, and 1 tablet of complete mini protease inhibitor cocktail [Roche] per 10 mL) and centrifuged twice at 12,000 × g for 10 min at 4°C to remove debris. The supernatant was mixed with SDS-PAGE loading buffer, and the proteins in the sample were denatured by heat shock at 95°C for 5 min (input). An aliquot was then mixed with anti-FLAG M2 magnetic beads (M8823, Sigma-Aldrich) for at least 2 h at 4°C. The beads were washed six times with wash buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 20% glycerol) and mixed with SDS-PAGE loading buffer. Immunoprecipitated protein was eluted by heat shock at 95°C for 5 min. Input and immunoprecipitated protein samples were analyzed by Western blotting.
Western blotting.
Protein samples were separated by SDS-PAGE and blotted onto a polyvinylidene difluoride (PVDF) membranes. EXA1 and nCBP proteins were detected using anti-EXA1 (22) and anti-nCBP (18) antibodies as previously described. eIF4E1 and eIFiso4E proteins were detected using anti-eIF4E1 (40) and anti-eIFiso4E (42) antibodies, which were kindly provided by Karen Browning and Laura Mayberry (University of Texas at Austin, USA). The 3FLAG- and 3myc-tagged proteins were detected using anti-FLAG (clone M2; Sigma-Aldrich) and anti-Myc (clone 4A6; Millipore, Billerica, MA, USA) antibodies. The membrane was stained with Coomassie brilliant blue (CBB) for the loading control.
Plant transformation.
A. tumefaciens strain EHA105 was transformed with pFAST02-NahG, pFAST01-EXA1pro:EXA1WT, or pFAST01-EXA1pro:EXA14ebm. A. thaliana plants were transformed using the floral-dip method, as described previously (66). T3 homozygous transformants were used.
ACKNOWLEDGMENTS
We thank Kohki Yoshimoto, Shigeyuki Betsuyaku, Jane Parker, Karen Browning, and Laura Mayberry for providing materials.
This research was supported by funds from the Japan Society for the Promotion of Science (no. 17H03770, 20J23036, 21H04722, and 22K19168).
Footnotes
Supplemental material is available online only.
Contributor Information
Yasuyuki Yamaji, Email: ayyamaji@g.ecc.u-tokyo.ac.jp.
W. Allen Miller, Iowa State University
REFERENCES
- 1.Hashimoto M, Maejima K, Yamaji Y, Namba S. 2021. Plant resistance to viruses: natural resistance associated with recessive genes, p 69–80. In Encyclopedia of virology. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 2.Browning KS, Bailey-Serres J. 2015. Mechanism of cytoplasmic mRNA translation. Arabidopsis Book 13:e0176. doi: 10.1199/tab.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joshi B, Lee K, Maeder DL, Jagus R. 2005. Phylogenetic analysis of eIF4E-family members. BMC Evol Biol 5:48. doi: 10.1186/1471-2148-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayberry LK, Allen ML, Nitka KR, Campbell L, Murphy PA, Browning KS. 2011. Plant cap-binding complexes eukaryotic initiation factors eIF4F and eIFISO4F. J Biol Chem 286:42566–42574. doi: 10.1074/jbc.M111.280099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lellis AD, Kasschau KD, Whitham SA, Carrington JC. 2002. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr Biol 12:1046–1051. doi: 10.1016/s0960-9822(02)00898-9. [DOI] [PubMed] [Google Scholar]
- 6.Stein N, Perovic D, Kumlehn J, Pellio B, Stracke S, Streng S, Ordon F, Graner A. 2005. The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J 42:912–922. doi: 10.1111/j.1365-313X.2005.02424.x. [DOI] [PubMed] [Google Scholar]
- 7.Kanyuka K, Druka A, Caldwell DG, Tymon A, McCallum N, Waugh R, Adams MJ. 2005. Evidence that the recessive bymovirus resistance locus rym4 in barley corresponds to the eukaryotic translation initiation factor 4E gene. Mol Plant Pathol 6:449–458. doi: 10.1111/j.1364-3703.2005.00294.x. [DOI] [PubMed] [Google Scholar]
- 8.Reinbold C, Lacombe S, Ziegler-Graff V, Scheidecker D, Wiss L, Beuve M, Caranta C, Brault V. 2013. Closely related poleroviruses depend on distinct translation initiation factors to infect Arabidopsis thaliana. Mol Plant Microbe Interact 26:257–265. doi: 10.1094/MPMI-07-12-0174-R. [DOI] [PubMed] [Google Scholar]
- 9.Nieto C, Morales M, Orjeda G, Clepet C, Monfort A, Sturbois B, Puigdomènech P, Pitrat M, Caboche M, Dogimont C, Garcia-Mas J, Aranda MA, Bendahmane A. 2006. An eIF4E allele confers resistance to an uncapped and non-polyadenylated RNA virus in melon. Plant J 48:452–462. doi: 10.1111/j.1365-313X.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- 10.Udagawa H, Koga K, Shinjo A, Kitashiba H, Takakura Y. 2020. Reduced susceptibility to a tobacco bushy top virus Malawi isolate by loss of function in host eIF(iso)4E genes. Breed Sci 70:313–320. doi: 10.1270/jsbbs.19135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, Ishikawa M. 2004. The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J Virol 78:6102–6111. doi: 10.1128/JVI.78.12.6102-6111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robaglia C, Caranta C. 2006. Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci 11:40–45. doi: 10.1016/j.tplants.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Wang A, Krishnaswamy S. 2012. Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol Plant Pathol 13:795–803. doi: 10.1111/j.1364-3703.2012.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Truniger V, Nieto C, González-Ibeas D, Aranda M. 2008. Mechanism of plant eIF4E-mediated resistance against a Carmovirus (Tombusviridae): cap-independent translation of a viral RNA controlled in cis by an (a)virulence determinant. Plant J 56:716–727. doi: 10.1111/j.1365-313X.2008.03630.x. [DOI] [PubMed] [Google Scholar]
- 15.Gao Z, Johansen E, Eyers S, Thomas CL, Noel Ellis TH, Maule AJ. 2004. The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J 40:376–385. doi: 10.1111/j.1365-313X.2004.02215.x. [DOI] [PubMed] [Google Scholar]
- 16.Contreras-Paredes CA, Silva-Rosales L, Daròs J-A, Alejandri-Ramírez ND, Dinkova TD. 2013. The absence of eukaryotic initiation factor eIF(iso)4E affects the systemic spread of a tobacco etch virus isolate in Arabidopsis thaliana. Mol Plant Microbe Interact 26:461–470. doi: 10.1094/MPMI-09-12-0225-R. [DOI] [PubMed] [Google Scholar]
- 17.Ruud KA, Kuhlow C, Goss DJ, Browning KS. 1998. Identification and characterization of a novel cap-binding protein from Arabidopsis thaliana. J Biol Chem 273:10325–10330. doi: 10.1074/jbc.273.17.10325. [DOI] [PubMed] [Google Scholar]
- 18.Keima T, Hagiwara-Komoda Y, Hashimoto M, Neriya Y, Koinuma H, Iwabuchi N, Nishida S, Yamaji Y, Namba S. 2017. Deficiency of the eIF4E isoform nCBP limits the cell-to-cell movement of a plant virus encoding triple-gene-block proteins in Arabidopsis thaliana. Sci Rep 7:39678. doi: 10.1038/srep39678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez MA, Lin ZD, Moll T, Chauhan RD, Hayden L, Renninger K, Beyene G, Taylor NJ, Carrington JC, Staskawicz BJ, Bart RS. 2019. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol J 17:421–434. doi: 10.1111/pbi.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanssen IM, Thomma BPHJ. 2010. Pepino mosaic virus: a successful pathogen that rapidly evolved from emerging to endemic in tomato crops. Mol Plant Pathol 11:179–189. doi: 10.1111/j.1364-3703.2009.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koh KW, Lu H-C, Chan M-T. 2014. Virus resistance in orchids. Plant Sci 228:26–38. doi: 10.1016/j.plantsci.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto M, Neriya Y, Keima T, Iwabuchi N, Koinuma H, Hagiwara-Komoda Y, Ishikawa K, Himeno M, Maejima K, Yamaji Y, Namba S. 2016. EXA1, a GYF domain protein, is responsible for loss-of-susceptibility to plantago asiatica mosaic virus in Arabidopsis thaliana. Plant J 88:120–131. doi: 10.1111/tpj.13265. [DOI] [PubMed] [Google Scholar]
- 23.Yusa A, Neriya Y, Hashimoto M, Yoshida T, Fujimoto Y, Hosoe N, Keima T, Tokumaru K, Maejima K, Netsu O, Yamaji Y, Namba S. 2019. Functional conservation of EXA1 among diverse plant species for the infection by a family of plant viruses. Sci Rep 9:5958. doi: 10.1038/s41598-019-42400-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsui H, Nomura Y, Egusa M, Hamada T, Hyon G-S, Kaminaka H, Watanabe Y, Ueda T, Trujillo M, Shirasu K, Nakagami H. 2017. The GYF domain protein PSIG1 dampens the induction of cell death during plant-pathogen interactions. PLoS Genet 13:e1007037. doi: 10.1371/journal.pgen.1007037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Z, Huang S, Zhang X, Wu D, Xia S, Li X. 2017. Regulation of plant immune receptor accumulation through translational repression by a glycine-tyrosine-phenylalanine (GYF) domain protein. eLife 6:e23684. doi: 10.7554/eLife.23684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zvereva A, Pooggin M. 2012. Silencing and innate immunity in plant defense against viral and non-viral pathogens. Viruses 4:2578–2597. doi: 10.3390/v4112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bendahmane A, Köhn BA, Dedi C, Baulcombe DC. 1995. The coat protein of potato virus X is a strain-specific elicitor of Rx1-mediated virus resistance in potato. Plant J 8:933–941. doi: 10.1046/j.1365-313x.1995.8060933.x. [DOI] [PubMed] [Google Scholar]
- 28.Matsuo Y, Novianti F, Takehara M, Fukuhara T, Arie T, Komatsu K. 2019. Acibenzolar-S-methyl restricts infection of Nicotiana benthamiana by plantago asiatica mosaic virus at two distinct stages. Mol Plant Microbe Interact 32:1475–1486. doi: 10.1094/MPMI-03-19-0087-R. [DOI] [PubMed] [Google Scholar]
- 29.Kofler MM, Freund C. 2006. The GYF domain. FEBS J 273:245–256. doi: 10.1111/j.1742-4658.2005.05078.x. [DOI] [PubMed] [Google Scholar]
- 30.Mader S, Lee H, Pause A, Sonenberg N. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol 15:4990–4997. doi: 10.1128/MCB.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N. 2005. A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by bicoid and the eIF4E cognate 4EHP. Cell 121:411–423. doi: 10.1016/j.cell.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 32.Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, Drummond EP, Ausubel FM. 2000. Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J 24:205–218. doi: 10.1046/j.1365-313x.2000.00870.x. [DOI] [PubMed] [Google Scholar]
- 33.Rogers EE, Ausubel FM. 1997. Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9:305–316. doi: 10.2307/3870484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nawrath C, Métraux J-P. 1999. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, Ryals J. 1994. A central role of salicylic acid in plant disease resistance. Science 266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- 36.Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J. 1995. Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol Plant Microbe Interact 8:863–870. doi: 10.1094/mpmi-8-0863. [DOI] [PubMed] [Google Scholar]
- 37.Wiermer M, Feys BJ, Parker JE. 2005. Plant immunity: the EDS1 regulatory node. Curr Opin Plant Biol 8:383–389. doi: 10.1016/j.pbi.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 38.Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. 2006. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. Plant Cell 18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alazem M, Lin K-Y, Lin N-S. 2014. The abscisic acid pathway has multifaceted effects on the accumulation of bamboo mosaic virus. Mol Plant Microbe Interact 27:177–189. doi: 10.1094/MPMI-08-13-0216-R. [DOI] [PubMed] [Google Scholar]
- 40.Patrick RM, Mayberry LK, Choy G, Woodard LE, Liu JS, White A, Mullen RA, Tanavin TM, Latz CA, Browning KS. 2014. Two Arabidopsis loci encode novel eukaryotic initiation factor 4E isoforms that are functionally distinct from the conserved plant eukaryotic initiation factor 4E. Plant Physiol 164:1820–1830. doi: 10.1104/pp.113.227785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Callot C, Gallois J-L. 2014. Pyramiding resistances based on translation initiation factors in Arabidopsis is impaired by male gametophyte lethality. Plant Signal Behav 9:e27940. doi: 10.4161/psb.27940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duprat A, Caranta C, Revers F, Menand B, Browning KS, Robaglia C. 2002. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J 32:927–934. doi: 10.1046/j.1365-313x.2002.01481.x. [DOI] [PubMed] [Google Scholar]
- 43.Zafirov D, Giovinazzo N, Bastet A, Gallois JL. 2021. When a knockout is an Achilles’ heel: resistance to one potyvirus species triggers hypersusceptibility to another one in Arabidopsis thaliana. Mol Plant Pathol 22:334–347. doi: 10.1111/mpp.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshida T, Shiraishi T, Hagiwara-Komoda Y, Komatsu K, Maejima K, Okano Y, Fujimoto Y, Yusa A, Yamaji Y, Namba S. 2019. The plant noncanonical antiviral resistance protein JAX1 inhibits potexviral replication by targeting the viral RNA-dependent RNA polymerase. J Virol 93:e01506-18. doi: 10.1128/JVI.01506-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu L, Yang L. 2019. Time to fight: molecular mechanisms of age-related resistance. Phytopathology 109:1500–1508. doi: 10.1094/PHYTO-11-18-0443-RVW. [DOI] [PubMed] [Google Scholar]
- 46.Toribio R, Muñoz A, Castro-Sanz AB, Merchante C, Castellano MM. 2019. A novel eIF4E-interacting protein that forms non-canonical translation initiation complexes. Nat Plants 5:1283–1296. doi: 10.1038/s41477-019-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bush MS, Hutchins AP, Jones AME, Naldrett MJ, Jarmolowski A, Lloyd CW, Doonan JH. 2009. Selective recruitment of proteins to 5′ cap complexes during the growth cycle in Arabidopsis. Plant J 59:400–412. doi: 10.1111/j.1365-313X.2009.03882.x. [DOI] [PubMed] [Google Scholar]
- 48.Cheng S, Gallie DR. 2013. Eukaryotic initiation factor 4B and the poly(A)-binding protein bind eIF4G competitively. Translation (Austin) 1:e24038. doi: 10.4161/trla.24038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng S, Gallie DR. 2010. Competitive and noncompetitive binding of eIF4B, eIF4A, and the poly(A) binding protein to wheat translation initiation factor eIFiso4G. Biochemistry 49:8251–8265. doi: 10.1021/bi1008529. [DOI] [PubMed] [Google Scholar]
- 50.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of elF4G. Mol Cell 3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- 51.Morita M, Ler LW, Fabian MR, Siddiqui N, Mullin M, Henderson VC, Alain T, Fonseca BD, Karashchuk G, Bennett CF, Kabuta T, Higashi S, Larsson O, Topisirovic I, Smith RJ, Gingras A-C, Sonenberg N. 2012. A Novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol Cell Biol 32:3585–3593. doi: 10.1128/MCB.00455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hickey KL, Dickson K, Cogan JZ, Replogle JM, Schoof M, D'Orazio KN, Sinha NK, Hussmann JA, Jost M, Frost A, Green R, Weissman JS, Kostova KK. 2020. GIGYF2 and 4EHP inhibit translation initiation of defective messenger RNAs to assist ribosome-associated quality control. Mol Cell 79:950–962.e6. doi: 10.1016/j.molcel.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zinshteyn B, Sinha NK, Enam SU, Koleske B, Green R. 2021. Translational repression of NMD targets by GIGYF2 and EIF4E2. PLoS Genet 17:e1009813. doi: 10.1371/journal.pgen.1009813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber R, Chung M-Y, Keskeny C, Zinnall U, Landthaler M, Valkov E, Izaurralde E, Igreja C. 2020. 4EHP and GIGYF1/2 mediate translation-coupled messenger RNA decay. Cell Rep 33:108262. doi: 10.1016/j.celrep.2020.108262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X, Clarke JD, Zhang Y, Dong X. 2001. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact 14:1131–1139. doi: 10.1094/MPMI.2001.14.10.1131. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Goritschnig S, Dong X, Li X. 2003. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15:2636–2646. doi: 10.1105/tpc.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huisman MJ, Linthorst HJM, Bol JF, Cornelissen BJC. 1988. The complete nucleotide sequence of potato virus X and its homologies at the amino acid level with various plus-stranded RNA viruses. J Gen Virol 69:1789–1798. doi: 10.1099/0022-1317-69-8-1789. [DOI] [PubMed] [Google Scholar]
- 58.Sonenberg N, Shatkin AJ, Ricciardi RP, Rubin M, Goodman RM. 1978. Analysis of terminal structures of RNA from potato virus X. Nucleic Acids Res 5:2501–2512. doi: 10.1093/nar/5.7.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujimoto Y, Keima T, Hashimoto M, Hagiwara-Komoda Y, Hosoe N, Nishida S, Nijo T, Oshima K, Verchot J, Namba S, Yamaji Y. 2022. Short 5′ untranslated region enables optimal translation of plant virus tricistronic RNA via leaky scanning. J Virol 96:e0214421. doi: 10.1128/jvi.02144-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gamarnik AV, Andino R. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev 12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bastet A, Lederer B, Giovinazzo N, Arnoux X, German-Retana S, Reinbold C, Brault V, Garcia D, Djennane S, Gersch S, Lemaire O, Robaglia C, Gallois J-L. 2018. Trans-species synthetic gene design allows resistance pyramiding and broad-spectrum engineering of virus resistance in plants. Plant Biotechnol J 16:1569–1581. doi: 10.1111/pbi.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shimada TL, Shimada T, Hara-Nishimura I. 2010. A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J 61:519–528. doi: 10.1111/j.1365-313X.2009.04060.x. [DOI] [PubMed] [Google Scholar]
- 63.Himeno M, Maejima K, Komatsu K, Ozeki J, Hashimoto M, Kagiwada S, Yamaji Y, Namba S. 2010. Significantly low level of small RNA accumulation derived from an encapsidated mycovirus with dsRNA genome. Virology 396:69–75. doi: 10.1016/j.virol.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 64.Okano Y, Senshu H, Hashimoto M, Neriya Y, Netsu O, Minato N, Yoshida T, Maejima K, Oshima K, Komatsu K, Yamaji Y, Namba S. 2014. In planta recognition of a double-stranded RNA synthesis protein complex by a potexviral RNA silencing suppressor. Plant Cell 26:2168–2183. doi: 10.1105/tpc.113.120535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Senshu H, Ozeki J, Komatsu K, Hashimoto M, Hatada K, Aoyama M, Kagiwada S, Yamaji Y, Namba S. 2009. Variability in the level of RNA silencing suppression caused by triple gene block protein 1 (TGBp1) from various potexviruses during infection. J Gen Virol 90:1014–1024. doi: 10.1099/vir.0.008243-0. [DOI] [PubMed] [Google Scholar]
- 66.Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S3. Download jvi.00221-23-s0001.pdf, PDF file, 4.1 MB (4.1MB, pdf)