Abstract
Purpose of review
We seek to determine recent advances in kidney pathophysiology that have been enabled or enhanced by artificial intelligence. We describe some of the challenges in the field as well as future directions.
Recent findings
We first provide an overview of artificial intelligence terminologies and methodologies. We then describe the use of artificial intelligence in kidney diseases to discover risk factors from clinical data for disease progression, annotate whole slide imaging and decipher multiomics data. We delineate key examples of risk stratification and prognostication in acute kidney injury (AKI) and chronic kidney disease (CKD). We contextualize these applications in kidney disease oncology, one of the subfields to benefit demonstrably from artificial intelligence using all if these approaches. We conclude by elucidating technical challenges and ethical considerations and briefly considering future directions.
Summary
The integration of clinical data, patient derived data, histology and proteomics and genomics can enhance the work of clinicians in providing more accurate diagnoses and elevating understanding of disease progression. Implementation research needs to be performed to translate these algorithms to the clinical setting.
Keywords: artificial intelligence, diagnosis, machine learning, nephrology, prognosis
INTRODUCTION
Precision medicine is an emerging focus in kidney disease that seeks to integrate large multiscale datasets to understand and tailor the treatment of disease for individual patients. Artificial intelligence, the implementation of computers to solve problems with nominal human intervention [1], has the potential for improved risk prediction and prognostication.
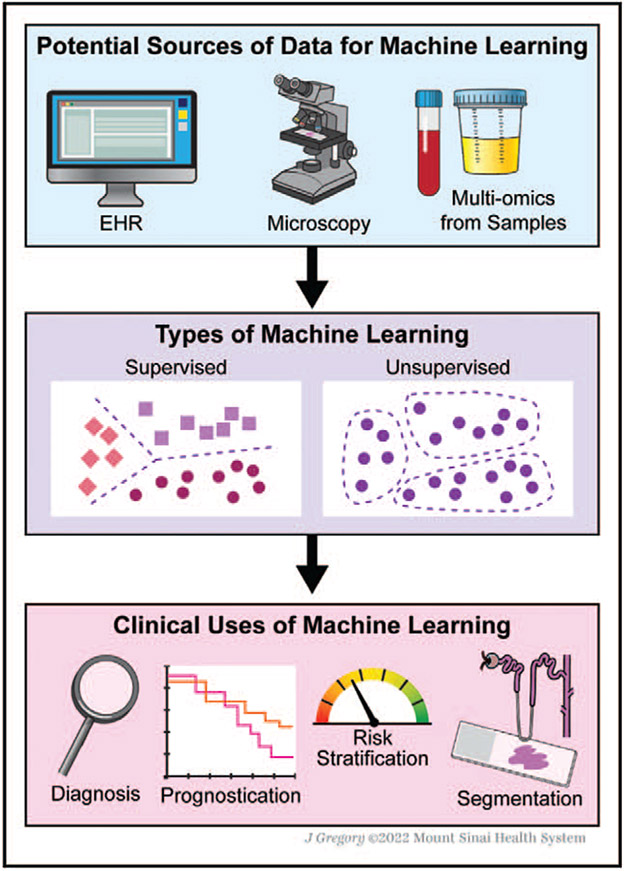
Several advances in technology have allowed for the increased usage of artificial intelligence. The near universal usage of electronic health records (EHRs) allows for incorporation of longitudinal patient data into models. The deployment of whole slide imaging (WSI) technologies, which converts glass slides to digital images, have enabled the application of artificial intelligence to improve diagnostic accuracy and prognosis [2]. Lastly, improvements in sequencing platforms have allowed for genomic and proteomic analysis to further enhance prediction and classification of kidney disease and progression risk. We summarize sources of data and their clinical uses in Fig. 1.
FIGURE 1.
Data sources and clinical applications of machine learning in nephrology.
In this review, we will briefly provide an overview of artificial intelligence and potential sources of data. We then discuss examples of machine learning using various data sources and end with discussions of challenges, ethical considerations and potential future directions.
DEFINITIONS IN ARTIFICIAL INTELLIGENCE
Machine learning is a subset of artificial intelligence that seeks to analyse data by using algorithms that can adapt and improve with repeated data exposure [3]. There are two major subtypes of machine learning: supervised and unsupervised learning. In supervised learning, algorithms are trained on labelled datasets to create models relating input with the outcome and then tasked to predict or classify outcomes in new unlabelled data. In unsupervised learning, algorithms are tasked to identify structures, such as clusters, in unlabelled data [4]. Deep learning is a subset of machine learning that relies on neural networks to identify patterns in large-scale datasets. The artificial neuron serves as the functional unit that receives an input, transforms the information and outputs a result, often to the subsequent neuron [5]. These neurons are organized into layers, and the collation of these layers is termed ‘deep’, leading to the eponymous term deep learning [6].
DATA SOURCES FOR ARTIFICIAL INTELLIGENCE
We provide an overview of potential sources of data, including clinical as well as imaging and -omics data, for machine learning studies.
Patient EHRs amass clinical and laboratory data over a longitudinal time course, providing a source of natural history about disease and its response to treatment for prediction modelling [7]. As each institution’s EHR is unique, care must be used when combining data from several institutions [8]. The Observational Medical Outcomes Partnership (OMOP) common data model provides one potential method for combining data from disparate sources [9]. Several rich databases are available for research (Table 1) [10-21].
Table 1.
Publicly accessible databases for nephrology research.a
| Name | Type of dataset available |
Link | Cohort summary |
|---|---|---|---|
| Chronic Renal Insufficiency Cohort (CRIC) [10] | Clinical, proteomics, transcriptomics | http://www.cristudy.org/Chronic-Kidney-Disease/Chronic-Renal-Insufficiency-Cohort-Study/ | 5499 patients with chronic kidney disease in the United States |
| Cure GlomeruloNephropathies (CureGN) [11] | Clinical, imaging | https://curegn-org.webflow.io/ | 2400 adults and children with minimal change disease (MCD), focal segmental glomerulosclerosis (FSGS), membranous nephropathy (MN) or immunoglobulin A nephropathy/vasculitis (IgAN/IgAV) from 70 centres from the US, Canada, Italy and Poland |
| European Rare Kidney Disease Network (ERK-Net) [12] | Clinical | https://www.erknet.org/ | >70000 patients from 72 nephrology centres in 24 European countries |
| Human Heredity and Health in Africa (H3 Africa) [13] | Clinical, imaging, genomics | https://h3africa.org/index.php/h3africa-kidney-disease-research-network/ | 8000 patients with kidney disease and unaffected controls to determine whether certain genes are uniquely associated with kidney diseases in Africans |
| Human Kidney and Urine Proteome Project (HKUPP) [14] | Proteomics | http://www.hkupp.org/ | International collaboration on human kidney and urine proteomics |
| Japanese Chronic Kidney Disease Database (J-CKD-DB) [15] | Clinical | http://j-ckd-db.jp/ | >100000 patients from 21 university hospitals, with planned linkage to J-RBR in late 2020. |
| Japan Renal Biopsy Registry (J-RBR) [16] | Clinical, imaging | https://jsn.or.jp/en/greeting/ | >40000 patients who underwent renal biopsy |
| Kidney Precision Medicine Project (KPMP) [17] | Clinical, transcriptomics | https://atlas.kpmp.org/ | 2835 total samples from patients with AKI, CKD and control. |
| National Registry of Rare Kidney Diseases (RaDaR) [18] | Clinical | https://ukkidney.org/audit-research/data-permissions/data/radar-database | 26913 patients from 104 sites in the UK |
| Nephroseq [19] | Clinical, transcriptomics | https://www.nephroseq.org/resource/login.html | 26 datasets with 1989 samples with analytic and visualization tools available on the platform |
| NEPhrotic syndrome sTUdy NEtwork (NEPTUNE) [20] | Clinical, imaging, genomics | https://www.neptune-study.org/ | 450 adults and children with minimal change disease, focal segmental glomerulosclerosis, and membranous nephropathy |
| Renal Gene Expression Database [21] | Transcriptomics | http://rged.wall-eva.net/ | 5253 tissue samples and 101 cell lines from NCBI Gene Expression Omnibus |
Approvals may be necessary for access to some databases.
An additional potential source of data is the kidney biopsy [22]. Manual annotation and assessment of whole slide images (WSI) require time and specialist expertise. Automating data extraction and evaluation can reduce variability, facilitate clinical workflow and augment histologic accuracy [23]. Several national and international databases have compiled biopsy samples, alongside clinical, genetic and biomarker data [24].
APPLICATION OF MACHINE LEARNING TO NEPHROLOGY
Advancements in high-throughput technologies have enabled massive collection of biological data, providing more comprehensive information about molecular mechanisms of disease development [5,25]. Parsing meaningful insights necessitates the use of machine learning methods to process these immense datasets, and we review notable examples in the following section.
Acute kidney injury
Acute kidney injury (AKI) is common in hospitalized patients. Artificial intelligence in AKI research has focused on risk prediction, identification and automated notifications. Most models are developed in single centres or a single healthcare system and do not have an external validation cohort, which is necessary to assess the performance and generalizability of models. One study that performed external validation tested a gradient boosted machine model for prediction of AKI stage 2 AKI within 48 h of an observation [26]. Their model had similar discrimination across the three sites used for internal validation and external validation.
Although a majority of studies generate predictions for a single time point, one study used data from the U.S. Department of Veterans Affairs to develop a machine learning model for continuous prediction of AKI [27]. Researchers grouped time-varying features into 6-h time windows to predict risk of AKI at eight-time windows. A major limitation of this study is that more than 90% of the cohort was male, and given differences in AKI incidence between men and women, it is unclear whether this model would generalize to a different population. In addition, urine output was not included as a feature in this model.
Chronic kidney disease progression
Given that patients with chronic kidney disease (CKD) have varying trajectories, it is of interest to identify patients at a high risk or progression. The most commonly used risk prediction tool for CKD progression is the kidney failure risk equation (KFRE) [28,29]. Several studies have attempted to expand on CKD prediction using machine learning. Most recently, Ventrella et al. [30] applied several machine learning techniques to estimate time to when dialysis treatment will be necessary. In their study, they used text mining, several machine learning algorithms, and built models for classification (within 1 year or not) and regression (predict actual number of months to need for dialysis). Overall, the best performing model was the ensemble approach for classification inclusive of features extracted from textual reports. Key limitations include lack of external validation, and the cohort was exclusively white, which limits generalizability.
In a separate study, Chan et al. [31] used data from two biobanks to develop a random forest model to predict risk of a composite kidney end point in patients with CKD and type 2 diabetes mellitus. Their study included three plasma biomarkers and the study population was diverse with nearly one-third African–American. The model outperformed both the clinical model and the KDIGO risk strata. Major limitations to this study were the large amount of missing data for urine results and that the cohort was derived from patients in the Northeastern USA. Lastly, others have unsupervised clustering to identify CKD subgroups with different clinical end points of CKD progression, cardiovascular disease and death [32].
Histopathology
One of the most frequent use of cases of machine learning for histopathologic analysis is to extract the glomeruli and ascertain key histologic findings [33,34]. Manual assessment of glomerular sclerosis, a primary manifestation in a spectrum of kidney diseases and an important component of disease staging, requires expertise that may be lacking in resource-limited settings and introduces intrareader and inter-reader variability in interpretations. As such, many studies have developed machine learning approaches to segment glomeruli derived from biopsy samples and quantify amount of sclerosis [23,34-39]. Kolachalama et al. [40] trained a convolutional neural network (CNN) to correlate renal fibrosis from biopsy samples to clinical phenotypes at the time of biopsy. This approach outperformed a model based on pathologist-estimated fibrosis scores, yet the model’s accuracy would be better ascertained with external validation and incorporation of treatment impact in predicting survival.
In addition to segment identification, artificial intelligence can be implemented to enhance the visual resolution of WSI. Recently, de Haan et al. [41◾] developed a CNN model to transform H&E-stained kidney biopsy samples to computationally generated special stains such as Masson’s Trichrome, periodic acid-Schiff and Jones silver stain. As H&E-stained biopsies are often available well in advance of those prepared by special stains, this can alleviate wait time, which is especially useful for medical conditions in which rapid diagnosis and treatment can significantly improve outcomes, such as rapidly progressive glomerulonephritis. The samples included in this study were stained at a few institutions and imaged by microscopes from the same vendor and model, necessitating future research to generalize results to other facilities.
Genomics and proteomics
Machine learning studies using genetic information have enabled novel molecular phenotyping that increases precision of disease classification that can supplement and even supplant conventional histological diagnosis [42-44]. Using microarray data from 1208 kidney transplant biopsies, Reeve et al. [44] first adopted supervised learning methods to detect molecular features of various types of rejection in kidney transplant, which were quantified in cross-validated classifier scores. They found that late-stage antibody-mediated rejection was associated with the lowest graft survival, which was better predicted using this pipeline rather than histologic diagnosis.
Meanwhile, proteomic analysis of blood and urine samples can be used for diagnosing kidney disease without invasive kidney biopsies [45-47]. One study applied machine learning to mass spectrometry on urinary proteomics to compare four machine learning models for the diagnosis of IgA nephropathy (IgAN), membranous nephropathy and diabetic kidney disease [48]. The best performing model, XGBoost, demonstrated the highest accuracy of 96%, yet increasing sample size and including clinical data in their model could elevate diagnostic power.
Onco-nephrology
Onco-nephrology [49] converges histopathology and large-scale -omics to diagnose and prognosticate cancer of the kidneys.
Machine learning studies in histopathology have strived to distinguish RCC from normal tissue and discover biomarkers that inform diagnosis and prognosis of patients with RCC [50-53]. Using WSI, Tabibu et al. [54] implemented CNN models that classified RCC as clear cell, papillary or chromophobe; identified high probability tumour areas using nuclear and tumour shape; and associated these findings with patient survival. They employed a directed acyclic graph-support vector machine to create multiple binary classification tasks out of the multiclass classification task, which improved model performance and better handled the unequal distribution of RCC subtypes. Furthermore, the combination of proteomics and histology imaging datasets from clear cell RCC patients by Azuaje et al. [55] revealed correlations between select diagnostic proteins and predictions generated by a histology-based classification model, which serves as a proof of concept that demonstrates the possibility of elucidating molecular mechanisms through histopathological analysis.
Genomic studies have similarly sought to classify clear cell RCC and predict prognosis, distinguish tumour stage, classify cancer subtypes and even ascertain the methylation profile of RCC [56-58]. Ali et al. [59] implemented machine learning to classify five kidney cancer subtypes using miRNA data, identifying 35 miRNAs that distinctly contribute to diagnosis. Neighbourhood component analysis was used to distinguish features from miRNAs, and Long Short-Term Memory, a Recurrent Neural Network (a specialized deep learning technique), was implemented to classify miRNA samples into cancer subtypes. Further wet laboratory and clinical evaluations will be necessary to evaluate the utility of these miRNA to renal cancer classification.
Lastly, big data derived from radiologic images have emerged as a cutting-edge application of artificial intelligence in nephropathology. Uhm et al. [60◾◾] developed an integrated framework for the detection and differential diagnosis of five major histologic subtypes of benign and malignant renal tumours using computed tomographic (CT) data from patients with nephrectomies for renal tumours. Their model achieved similar or superior diagnostic performance in comparison to radiologists. These studies underscore the promise of using radiology as another noninvasive modality by which we profile renal cell carcinoma.
CHALLENGES
Machine learning algorithms work best when developed in large, diverse and representative cohorts, yet this is often limited by the abilities to share data across institutions. To address this need, some health systems have de-identified their data and made them freely and publicly available [61], and others have created programmes that collect data nationally [10,11,17,20,21]. Another approach is federated learning, which allows for the training of a prediction model, although all data remain at their respective institutions. This has previously been demonstrated to have better performance compared with models developed at individual sites and then pooled [62].
Although many models exist in the literature, few of them get implemented into clinical practice. One contributing reason may be related to lack of understanding of the models and what are drivers of the models. Providers are unlikely to trust models that do not provide information regarding which features are contributing to these predictions. Models such as deep learning models are particularly difficult to interpret, as they are considered black boxes. In addition, the nephrology and medicine workforce need to integrate informatics and artificial intelligence into their training curriculum to build an understanding of how these tools can improve clinical care [63].
ETHICAL CONSIDERATIONS
Bias in machine learning models refers to the model providing results that are systematically prejudiced due to faulty assumptions. First, models that are trained in one population will perform well in that population but poorly in other populations. In addition, feature selection during the data collection or data cleaning phase can introduce unintended bias: for example, during a medical visit, a person of a certain background or appearance may be more likely to be asked regarding social determinants of health. Therefore, missing data in EHR may not always be missing at random and can lead to bias in models. To mitigate bias, researchers need to ensure that models are developed in representative populations and ensure careful selection of data features. Use of tools such as the Prediction model Risk of Bias Assessment Tool (PROBAST) by researchers can help identify bias in the models [64].
Given the growing use of machine learning in medicine, concerns have grown around privacy and confidentiality of patient data. Patients may not be aware of the secondary use of their EHR data and are not able to opt out of these practices. Although some countries have started to enact laws to address privacy concerns and how machine learning algorithms can be used, this is an area that requires additional work [65]. These data are now often stored in cloud-based databases and protections must be made to ensure that these data do are not obtained by malicious parties [66].
CONCLUSION
Studies discussed in this review exemplify how artificial intelligence can augment diagnostic and prognostic capabilities in nephropathology, presenting a tremendous area of growth in nephrology, as the field lags behind other organ-based research areas in artificial intelligence research [67]. Although in nascent stages of development, leveraging prediction capabilities of artificial intelligence can contribute directly to clinical decision planning to monitor patient status [68] and recommend personalized treatment [69]. Nonetheless, these methodologies will require a multidisciplinary approach to actualize translation.
We believe that artificial intelligence will become an integral asset, not substitute, for clinicians. We foresee artificial intelligence serving as a critical tool by which to complete repetitive, routine tasks while providing bandwidth to clinicians to perform more complex activities [70]. For this reason, we emphasize the need for universal education of pathologists in training programmes to appreciate and apply machine learning algorithms [70]. In addition, deployment of artificial intelligence pipelines is an expensive investment [70]. To persuade healthcare systems to adopt machine learning in the future, implementation research needs to focus on demonstrating value-based care to garner funding. This funding will be especially critical for assessing algorithms in clinical trials that study not only ethnically diverse cohorts [71] but also clinical consequences as a result of implementation that may require recalibration of algorithms [72].
With the ever-growing accumulation of biomedical data, machine learning will continue to deliver exciting advances to nephrology, yet it remains important to distinguish this hope from hype in acknowledging the limitations of machine learning [3]. By using artificial intelligence with awareness of its implementation challenges and observation of its ethical considerations, the synergy of clinical data, histopathology, genomics and proteomics promises a precise and more individualized system of healthcare for everyone.
KEY POINTS.
Several advances in technology have catalyzed the application of artificial intelligence to nephropathology research, such as near universal usage of electronic health records, whole slide imaging technologies, and genomic and proteomic sequencing platforms.
Applying machine learning techniques to large-scale data sets enhances image analysis, such as glomeruli segmentation, as well as various forms of prediction, such as diagnosis, prognostication and risk stratification of kidney diseases.
Although using artificial intelligence comes with its biases, ethical considerations and implementation challenges, synergizing analysis of clinical, histopathological and -omics data through machine learning promises a more individualized system of healthcare.
Acknowledgements
The authors thank Jill Gregory of the Mount Sinai Illustrations Department for her assistance in creating Fig. 1.
Conflicts of interest
G.N.N. has received consulting fees from AstraZeneca, Reata, BioVie and GLG Consulting; has received financial compensation as a scientific board member and advisor to RenalytixAI; and owns equity in RenalytixAI and Pensieve Health as a cofounder. L.C. has received consulting fees from Vifor Pharma, honorarium from Fresenius Medical Care, and is supported in part by K23DK124645. J.J. has no conflicts of interest.
Footnotes
Financial support and sponsorship
None.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
◾ of special interest
◾◾ of outstanding interest
- 1.Hamet P, Tremblay J. Artificial intelligence in medicine. Metabolism 2017;69:S36–S40. [DOI] [PubMed] [Google Scholar]
- 2.Huo Y, Deng R, Liu Q, et al. AI applications in renal pathology. Kidney Int 2021; 99:1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan L, Vaid A, Nadkarni GN. Applications of machine learning methods in kidney disease: hope or hype? Curr Opin Nephrol Hypertens 2020; 29:319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sealfon RSG, Mariani LH, Kretzler M, Troyanskaya OG. Machine learning, the kidney, and genotype–phenotype analysis. Kidney Int 2020; 97:1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martorell-Marugán J, Tabik S, Benhammou Y, et al. Deep learning in omics data analysis and precision medicine. Exon Pub 2019; 37–53. [PubMed] [Google Scholar]
- 6.Somani S, Russak AJ, Richter F, et al. Deep learning and the electrocardiogram: review of the current state-of-the-art. EP Europace 2021; 23:1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nadkarni GN, Coca SG, Wyatt CM. Big data in nephrology: promises and pitfalls. Kidney Int 2016; 90:240–241. [DOI] [PubMed] [Google Scholar]
- 8.Phelan M, Bhavsar NA, Goldstein BA. Illustrating informed presence bias in electronic health records data: how patient interactions with a health system can impact inference. EGEMS (Wash DC) 2017; 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.OMOP Common Data Model [Internet]. Observational Health Data Sciences and Informatics [updated 2022; cited 2022 March 20]. Available from: https://www.ohdsi.org/data-standardization/the-common-data-model/.
- 10.Denker M, Boyle S, Anderson AH, et al. Chronic Renal Insufficiency Cohort Study (CRIC): overview and summary of selected findings. Clin J Am Soc Nephrol 2015; 10:2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillespie BW, Laurin L-P, Zinsser D, et al. Improving data quality in observational research studies: report of the Cure Glomerulonephropathy (CureGN) network. Contemp Clin Trials Commun 2021; 22:100749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassanese G, Wlodkowski T, Servais A, et al. The European Rare Kidney Disease Registry (ERKReg): objectives, design and initial results. Orphanet J Rare Dis 2021; 16:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osafo C, Raji YR, Burke D, et al. Human Heredity and Health (H3) in Africa Kidney Disease Research Network: a focus on methods in sub-Saharan Africa. Clin J Am Soc Nephrol 2015; 10:2279–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto T, Langham RG, Ronco P, et al. Towards standard protocols and guidelines for urine proteomics: a report on the Human Kidney and Urine Proteome Project (HKUPP) symposium and workshop, 6 October 2007, Seoul, Korea and 1 November 2007, San Francisco, CA, USA. Proteomics 2008; 8:2156–2159. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa N, Sofue T, Kanda E, et al. J-CKD-DB: a nationwide multicentre electronic health record-based chronic kidney disease database in Japan. Sci Rep 2020; 10:7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugiyama H, Yokoyama H, Sato H, et al. Japan Renal Biopsy Registry: the first nationwide, web-based, and prospective registry system of renal biopsies in Japan. Clin Exp Nephrol 2011; 15:493–503. [DOI] [PubMed] [Google Scholar]
- 17.de Boer IH, Alpers CE, Azeloglu EU, et al. Rationale and design of the Kidney Precision Medicine Project. Kidney Int 2021; 99:498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerecuk L, Kokocinska M, Parkes S, et al. G33 (P) Overview of rare renal diseases at a paediatric renal centre through the national registry of rare kidney diseases (radar) in the United Kingdom. Archives of Disease in Childhood 2018; 103:A13–A14. [Google Scholar]
- 19.Nephroseq.org [Internet]. The University of Michigan [updated 2022; cited 2022 March 20]. Available from: https://nephroseq.org/resource/login.html.
- 20.Gadegbeku CA, Gipson DS, Holzman LB, et al. Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nephropathy by a multidisciplinary approach. Kidney Int 2013; 83:749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q, Yang B, Chen X, et al. Renal Gene Expression Database (RGED): a relational database of gene expression profiles in kidney disease. Database 2014; 2014:bau092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunningham A, Benediktsson H, Muruve DA, et al. Trends in biopsy-based diagnosis of kidney disease: a population study. Can J Kidney Health Dis 2018; 5:2054358118799690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ginley B, Lutnick B, Jen K-Y, et al. Computational segmentation and classification of diabetic glomerulosclerosis. J Am Soc Nephrol 2019; 30:1953–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barisoni L, Lafata KJ, Hewitt SM, et al. Digital pathology and computational image analysis in nephropathology. Nat Rev Nephrol 2020; 16:669–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubin RF, Rhee EP. Proteomics and metabolomics in kidney disease, including insights into etiology, treatment, and prevention. Clin J Am Soc Nephrol 2020; 15:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Churpek MM, Carey KA, Edelson DP, et al. Internal and external validation of a machine learning risk score for acute kidney injury. JAMA Netw Open 2020; 3:e2012892–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomasev N, Glorot X, Rae JW, et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature 2019; 572:116–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tangri N, Grams ME, Levey AS, et al. Multinational assessment of accuracy of equations for predicting risk of kidney failure: a meta-analysis. JAMA 2016; 315:164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tangri N, Stevens LA, Griffith J, et al. A predictive model for progression of chronic kidney disease to kidney failure. JAMA 2011; 305:1553–1559. [DOI] [PubMed] [Google Scholar]
- 30.Ventrella P, Delgrossi G, Ferrario G, et al. Supervised machine learning for the assessment of chronic kidney disease advancement. Comput Meth Programs Biomed 2021; 209:106329. [DOI] [PubMed] [Google Scholar]
- 31.Chan L, Nadkarni GN, Fleming F, et al. Derivation and validation of a machine learning risk score using biomarker and electronic patient data to predict progression of diabetic kidney disease. Diabetologia 2021; 64:1504–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng Z, Waikar SS, Schmidt IM, et al. Subtyping CKD patients by consensus clustering: the chronic renal insufficiency cohort (CRIC) study. J Am Soc Nephrol 2021; 32:639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Bel T, Hermsen M, Smeets B, et al. Automatic segmentation of histopathological slides of renal tissue using deep learning. In Medical Imaging 2018: Digital Pathology 2018; 10581:1058112. [Google Scholar]
- 34.Jiang L, Chen W, Dong B, et al. A deep learning-based approach for glomeruli instance segmentation from multistained renal biopsy pathologic images. Am J Pathol 2021; 191:1431–1441. [DOI] [PubMed] [Google Scholar]
- 35.Bueno G, Fernandez-Carrobles MM, Gonzalez-Lopez L, Deniz O. Glomerulosclerosis identification in whole slide images using semantic segmentation. Comput Meth Programs Biomed 2020; 184:105273. [DOI] [PubMed] [Google Scholar]
- 36.Marsh JN, Matlock MK, Kudose S, et al. Deep learning global glomerulosclerosis in transplant kidney frozen sections. IEEE Trans Med Imaging 2018; 37:2718–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kannan S, Morgan LA, Liang B, et al. Segmentation of glomeruli within trichrome images using deep learning. Kidney Int Rep 2019; 4:955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uchino E, Suzuki K, Sato N, et al. Classification of glomerular pathological findings using deep learning and nephrologist: AI collective intelligence approach. Int J Med Inform 2020; 141:104231. [DOI] [PubMed] [Google Scholar]
- 39.Hermsen M, de Bel T, den Boer M, et al. Deep learning–based histopathologic assessment of kidney tissue. J Am Soc Nephrol 2019; 30:1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolachalama VB, Singh P, Lin CQ, et al. Association of pathological fibrosis with renal survival using deep neural networks. Kidney Int Rep 2018; 3:464–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.◾. de Haan K, Zhang Y, Zuckerman JE, et al. Deep learning-based transformation of H&E stained tissues into special stains. Nat Commun 2021; 12:4884. This study developed a supervised learning method by which to computationally transform H&E to special stains, which may have important consequences in improving accuracy and speed of preliminary diagnosis.
- 42.Christians U, Klawitter J, Klepacki J, Klawitter J. Chapter Four: the role of proteomics in the study of kidney diseases and in the development of diagnostic tools. In: Edelstein CL, editor. Biomarkers of kidney disease (Second Edition). Academic Press; 2017. pp. 119–223. [Google Scholar]
- 43.Liu P, Lassén E, Nair V, et al. Transcriptomic and proteomic profiling provides insight into mesangial cell function in IgA nephropathy. J Am Soc Nephrol 2017; 28:2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reeve J, Böhmig GA, Eskandary F, et al. Assessing rejection-related disease in kidney transplant biopsies based on archetypal analysis of molecular phenotypes. JCI Insight 2017; 2:e94197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glazyrin YE, Veprintsev DV, Ler IA, et al. Proteomics-based machine learning approach as an alternative to conventional biomarkers for differential diagnosis of chronic kidney diseases. Int J Mol Sci 2020; 21:4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang F, Liu P, Zhao Y, et al. Diagnosis of T-cell-mediated kidney rejection by biopsy-based proteomics and machine learning. medRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruschi M, Granata S, Santucci L, et al. Proteomic analysis of urinary microvesicles and exosomes in medullary sponge kidney disease and autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol 2019; 14:834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao W, Zhang Y, Li X, et al. KDClassifier: a urinary proteomic spectra analysis tool based on machine learning for the classification of kidney diseases. Aging Pathobiol Therap 2021; 3:63–72. [Google Scholar]
- 49.Rosner MH, Jhaveri KD, McMahon BA, Perazella MA. Onconephrology: the intersections between the kidney and cancer. CA Cancer J Clin 2021; 71:47–77. [DOI] [PubMed] [Google Scholar]
- 50.Safarpoor A, Shafiei S, Gonzalez R, et al. Renal cell carcinoma whole-slide image classification and search using deep learning. 2021. [Google Scholar]
- 51.Chen S, Zhang N, Jiang L, et al. Clinical use of a machine learning histopathological image signature in diagnosis and survival prediction of clear cell renal cell carcinoma. Int J Cancer 2021; 148:780–790. [DOI] [PubMed] [Google Scholar]
- 52.Ing N, Huang F, Conley A, et al. A novel machine learning approach reveals latent vascular phenotypes predictive of renal cancer outcome. Sci Rep 2017; 7:13190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fenstermaker M, Tomlins SA, Singh K, et al. Development and validation of a deep-learning model to assist with renal cell carcinoma histopathologic interpretation. Urology 2020; 144:152–157. [DOI] [PubMed] [Google Scholar]
- 54.Tabibu S, Vinod PK, Jawahar CV. Pan-renal cell carcinoma classification and survival prediction from histopathology images using deep learning. Sci Rep 2019; 9:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azuaje F, Kim S-Y, Perez Hernandez D, Dittmar G. Connecting histopathology imaging and proteomics in kidney cancer through machine learning. J Clin Med 2019; 8:1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jagga Z, Gupta D. Classification models for clear cell renal carcinoma stage progression, based on tumor RNAseq expression trained supervised machine learning algorithms. BMC Proc 2014; 8:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang W, Cao Y, Ma X. Novel prognostic prediction model constructed through machine learning on the basis of methylation-driven genes in kidney renal clear cell carcinoma. Biosci Rep 2020; 40:BSR20201604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shon HS, Batbaatar E, Kim KO, et al. Classification of kidney cancer data using cost-sensitive hybrid deep learning approach. Symmetry 2020; 12:154. [Google Scholar]
- 59.Muhamed Ali A, Zhuang H, Ibrahim A, et al. A machine learning approach for the classification of kidney cancer subtypes using miRNA genome data. Appl Sci 2018; 8:2422. [Google Scholar]
- 60.◾◾. Uhm K-H, Jung S-W, Choi MH, et al. Deep learning for end-to-end kidney cancer diagnosis on multiphase abdominal computed tomography. npj PrecOncol 2021; 5:54. This study created an end-to-end deep learning model for classification of five major histological subtypes of renal tumours on multiphase CT, which could have implications in enabling radiologists to noninvasively diagnose kidney cancer patients.
- 61.Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaid A, Jaladanki SK, Xu J, et al. Federated learning of electronic health records to improve mortality prediction in hospitalized patients with COVID-19: machine learning approach. JMIR Med Inform 2021; 9:e24207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rajpurkar P, Chen E, Banerjee O, Topol EJ. AI in health and medicine. Nat Med 2022; 28:31–38. [DOI] [PubMed] [Google Scholar]
- 64.Moons KG, Wolff RF, Riley RD, et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med 2019; 170:W1–W33. [DOI] [PubMed] [Google Scholar]
- 65.Lo Piano S Ethical principles in machine learning and artificial intelligence: cases from the field and possible ways forward. Human Soc Sci Commun 2020; 7:1–7. [Google Scholar]
- 66.De Cristofaro E An overview of privacy in machine learning. arXiv 2020:200508679. [Google Scholar]
- 67.Verma A, Chitalia VC, Waikar SS, Kolachalama VB. Machine learning applications in nephrology: a bibliometric analysis comparing kidney studies to other medicine subspecialities. Kidney Med 2021; 3:762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Improta G, Mazzella V, Vecchione D, et al. Fuzzy logic–based clinical decision support system for the evaluation of renal function in post-transplant patients. J Eval Clin Pract 2020; 26:1224–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barbieri C, Molina M, Ponce P, et al. An international observational study suggests that artificial intelligence for clinical decision support optimizes anemia management in hemodialysis patients. Kidney Int 2016; 90:422–429. [DOI] [PubMed] [Google Scholar]
- 70.Ahmad Z, Rahim S, Zubair M, Abdul-Ghafar J. Artificial intelligence (AI) in medicine, current applications and future role with special emphasis on its potential and promise in pathology: present and future impact, obstacles including costs and acceptance among pathologists, practical and philosophical considerations. A comprehensive review. Diagn Pathol 2021; 16:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Floege J, Mak RH, Molitoris BA, et al. Nephrology research: the past, present and future. Nat Rev Nephrol 2015; 11:677–687. [DOI] [PubMed] [Google Scholar]
- 72.Dahlin E Mind the gap! On the future of AI research. Human Soc Sci Commun 2021; 8:71. [Google Scholar]