Abstract
Melanoma is one of the most commonly diagnosed cancers in the Western world: third in Australia, fifth in the USA and sixth in the European Union. Predicting an individual’s personal risk of developing melanoma may aid them in undertaking effective risk reduction measures. The objective of this study was to use the UK Biobank to predict the 10-year risk of melanoma using a newly developed polygenic risk score (PRS) and an existing clinical risk model. We developed the PRS using a matched case–control training dataset (N = 16 434) in which age and sex were controlled by design. The combined risk score was developed using a cohort development dataset (N = 54 799) and its performance was tested using a cohort testing dataset (N = 54 798). Our PRS comprises 68 single-nucleotide polymorphisms and had an area under the receiver operating characteristic curve of 0.639 [95% confidence interval (CI) = 0.618–0.661]. In the cohort testing data, the hazard ratio per SD of the combined risk score was 1.332 (95% CI = 1.263–1.406). Harrell’s C-index was 0.685 (95% CI = 0.654–0.715). Overall, the standardized incidence ratio was 1.193 (95% CI = 1.067–1.335). By combining a PRS and a clinical risk score, we have developed a risk prediction model that performs well in terms of discrimination and calibration. At an individual level, information on the 10-year risk of melanoma can motivate people to take risk-reduction action. At the population level, risk stratification can allow more effective population-level screening strategies to be implemented.
Keywords: melanoma, polygenic risk scores, risk prediction
Introduction
Currently, adults are identified as being at high risk of melanoma based on a few clinical risk factors including age, ultraviolet (UV) light exposure [1], melanocytic nevus count [2], history of non-melanoma skin cancer [3], skin and hair color [4] and family history of melanoma [5]. While high-risk individuals can be offered appropriate screening and risk reduction options, clinicians often assess these risk factors one at a time, without any way to consider their multiplicative effects. Recent risk prediction models [6–8] have focused on improving screening access to at-risk individuals but one barrier to their implementation in clinical practice is the limited time available during consultations.
Although UV light exposure is a major risk factor for melanoma, there is also a substantial heritable component to melanoma [58%; 95% confidence interval (CI), 43–73%] [9]. A family history of melanoma is a well established risk factor [7,10,11], but there is an excess of familial risk that is due to genetics. A small portion of this genetic risk is due to rare mutations in high-penetrance genes such as CDKN2A and CDK4 [12]; therefore, the vast majority of the total heritability is likely due to polygenic risk. Polygenic risk scores (PRS) are a promising tool to capture risk that is unaccounted for by clinical risk factors for many diseases [13].
In assessing the discriminatory performance of PRS, some studies have included some or all of age, sex and principal components in their models without reporting the performance of the PRS alone [14–16]. This is problematic because these are known risk factors for melanoma and their inclusion in the model confounds the association between the PRS and melanoma, thereby inflating the area under the receiver operating characteristic curve (AUC) above that for PRS alone [17]. Cust et al. (2018) [16], however, did report the increase in AUC from adding the PRS to a clinical model in a study of the general population (Δ = 0.02 in Australian data and Δ = 0.03 in UK data).
Of the studies that reported an AUC with no adjustment for age and sex, Gu et al. (2018) [18] developed a 204-single-nucleotide polymorphism (SNP) PRS that had an AUC of 0.644 in their testing dataset. Steinberg et al. (2022) [19] investigated the performance of several PRSs developed from a meta-analysis [20] and previous studies [16], and found an AUC of 0.656 for the 50-SNP when testing in the UK Biobank data. The paper by Steinberg et al. (2022) [19] is the best comparison with our work (AUC = 0.639).
In this study, we developed a simple risk prediction model comprising our own PRS and an existing clinical risk score [7] to predict the 10-year risk of melanoma. In toto, this model was built with the aim to reduce the clinical burden by considering practice bandwidth and ease of use while improving upon standard clinical risk prediction.
Methods
Ethics approval
The UK Biobank has Research Tissue Bank approval (REC #11/NW/0382) that covers the analysis of data by approved researchers. All participants provided written informed consent to the UK Biobank before data collection began. This research has been conducted using the UK Biobank resource under Application Number 47401.
Data availability
Access to the data used in this study can be obtained by applying directly to the UK Biobank at https://www.ukbiobank.ac.uk/register-apply/. The authors did not receive special access privileges to the data that others would not have. Interested researchers will be able to access the data in the same manner by applying directly to the UK Biobank.
Participants
The UK Biobank [21,22] is a cohort of over 500 000 participants from across the UK who were recruited from 2006 to 2010. A diagnosis of melanoma was ascertained from self-reported data (UK Biobank data field 20001 with code 1003) or from linked cancer registry data. We excluded UK Biobank participants with ages less than 40 years or greater than 69 years. We also excluded non-Caucasian participants, participants who had a diagnosis of melanoma before their baseline assessment date, and participants with less than 6 weeks of follow-up time. We also excluded one of the pairs of individuals with closer than third-degree relatedness and individuals with missing clinical risk factors. Supplementary eTable 1, Supplemental digital content 1, http://links.lww.com/MR/A319 shows the number of individuals excluded after each step of the eligibility criteria. There were 365 326 [196 961 (54%) male and 168 365 (46%) female] individuals after filtering for eligibility, with 2134 incident melanoma cases and 363 192 unaffected participants.
Polygenic risk score training data
We reserved 70% of the data (N = 255 729) for building the PRS. We used this data to create a training subset of cases and controls in which age and sex were controlled by design. First, we divided the cases into groups defined by their quintiles of age: {[40, 52], (52, 59], (59, 62], (62, 66], (66, 69]}. Next, we further divided the cases by gender so that there were 10 groups in total. Then, for each of these 10 groups, we sampled 10 controls per case. By this sampling procedure, we made sure that the case–control ratio was the same across all 10 groups defined by age and sex. The sample size of the PRS training data was 16 434 with 1494 cases and 14 940 controls. The number of cases and controls for each age and sex group for the PRS training data are summarized in Supplementary eTable 2, Supplemental digital content 1, http://links.lww.com/MR/A319.
Combined model development and testing data
We used the remaining 30% of the data (N = 109 597) to perform a cohort analysis to develop the final model comprising the newly developed PRS and the existing clinical model [7]. We limited follow-up to 10 years and we divided the cohort data into halves: development and testing. We used the first half of the cohort data to estimate coefficients for the PRS and the clinical risk score using Cox regression with age as the time axis. In the second half of the cohort data, we assessed the performance of our risk score using Cox regression to estimate the hazard ratio per SD. We computed Harrell’s C-index to assess model discrimination. To examine calibration, we computed the standardized incidence ratio (SIR) of the number of melanoma cases observed in the first 10 years of follow-up compared with the number of cases predicted by the 10-year risk score, overall and by quintile of 10-year risk. Lastly, we refit the model to refine the estimates using the whole cohort data and computed the SIRs of the number of observed melanoma cases compared with the number predicted by population incidence rates, overall and by quintile of 10-year risk.
Polygenic risk score
A PRS is defined as the weighted sum of risk-allele counts of SNPs: , where is the weight for SNP j, Gij is the count (0, 1, 2) of the effect alleles of SNP j for individual i, and p is the number of SNPs in the PRS (see Supplementary eMethods, Supplemental digital content 1, http://links.lww.com/MR/A319). The first step in developing a PRS is to decide which SNPs to use and what effect sizes to assign to them. In this study, we only considered SNPs from the UK Biobank Axiom Array data [22]. We obtained the genome-wide association study effect sizes of SNPs from the summary statistics provided by GenoMEL consortium (the Melanoma Genetics Consortium; http://www.genomel.org), with UK Biobank samples removed. For quality control, we removed SNPs with minor allele frequency less than 10−3, a genotyping rate less than 95% and a Hardy–Weinberg equilibrium P value less than 10−50. We also removed ambiguous SNPs and duplicate variants with the same physical position or refSNP cluster ID number.
To create the PRS, we used the maximum clumping and thresholding method [23]. We selected seven correlation thresholds (0.01, 0.05, 0.10, 0.20, 0.50, 0.80, 0.95), four base clumping window sizes (50, 100, 200, 500; in kb) and 50 P value significance thresholds evenly spaced on a log–log scale. The actual clumping window size used was computed as the base clumping window size divided by the correlation thresholds. The standard clumping and thresholding method was then applied for the combinations of the hyperparameters values to generate 1400 (7 × 4 × 50) risk scores. The best risk score, which maximized the AUC on the PRS training data, was chosen to be our PRS. We used the R package bigsnpr [24] version 1.8.1 to run the maximum clumping and thresholding procedure.
Clinical risk score
The clinical risk score for melanoma was obtained from an Australian-based study [7]. The clinical risk score originally included hair color, nevus density, first-degree family history of melanoma, history of non-melanoma skin cancer and the number of lifetime sunbed sessions. Because nevus density and first-degree family history of melanoma are not available in UK Biobank, we only used the other three risk factors in our study. Hair color was classified as black/brown, light brown, blonde and red. Lifetime sunbed use was classified into three groups: none, 1–10 and >10. Because the UK Biobank only asked about the frequency of solarium or sunlamp use per year instead of lifetime sunbed use, we used a simple conversion to estimate the lifetime sunbed use: frequency greater than six times use per year we converted to the >10 groups for lifetime use; frequency between 1 and 5 per year we converted to the 1–10 group for lifetime use. The clinical risk score is a linear combination of these three risk factors. Table 1 shows the corresponding log-odd ratios (beta coefficients) and the distribution of clinical risk factors in the testing data. The equation for the calculation of the clinical risk score is provided in Supplementary eMethods, Supplemental digital content 1, http://links.lww.com/MR/A319.
Table 1.
Distribution of clinical risk factors in the whole testing data and the beta coefficients
| Risk factor | β coefficient | Case, N = 613 | Control, N = 108 984 |
|---|---|---|---|
| Hair color | |||
| Black/brown | 0 | 205 (33.4%) | 45 954 (42.2%) |
| Light brown | 0.22 | 251 (40.9%) | 45 436 (41.7%) |
| Blonde | 0.91 | 111 (18.1%) | 12 672 (11.6%) |
| Red | 1.46 | 46 (7.5%) | 4922 (4.5%) |
| Lifetime sunbed use | |||
| None | 0 | 558 (91.0%) | 98 681 (90.5%) |
| 1–10 | −0.05 | 40 (6.5%) | 7397 (6.8%) |
| >10 | 0.46 | 15 (2.4%) | 2906 (2.7%) |
| Non-melanoma skin cancer | |||
| No | 0 | 582 (94.9%) | 106 772 (98.0%) |
| Yes | 1.16 | 31 (5.1%) | 2212 (2.0%) |
Ten-year risk score
Our final risk score estimates the 10-year risk of melanoma, accounting for non-melanoma mortality as a competing risk, using the PRS we developed from the case–control training data and the clinical risk score obtained from a previous study [7]. The details for computing the 10-year risk score are provided in Supplementary eMethods, Supplemental digital content 1, http://links.lww.com/MR/A319.
Results
Our PRS comprises 68 SNPs and had an AUC of 0.639 (95% CI = 0.618–0.661). The full list of SNPs is provided in Supplementary eTable 4, Supplemental digital content 1, http://links.lww.com/MR/A319. We have also deposited details of the PRS in PGS Catalog (https://www.pgscatalog.org/) under the accession number PGS003430. The top three SNPs, ranked by odd ratios, were found to be rs149617956, rs1805007 and rs11547464. These SNPs are non-synonymous variants found in moderate-risk genes for melanoma [25]. rs149617956 (E318K) is found in MITF, which was previously found to be associated with nevus count and melanoma development [26,27], and has an odds ratio (OR) of 2.759 (Supplementary eTable 4, Supplemental digital content 1, http://links.lww.com/MR/A319). rs1805007 (R151C) and rs11547464 (R142H) are both found in MC1R that codes for red hair color and have OR of 1.769 and 1.571, respectively (Supplementary eTable 4, Supplemental digital content 1, http://links.lww.com/MR/A319) [28,29]. Finally, a protective SNP, rs55797833 with an OR of 0.610 is located in the 5′UTR of CDKN2A (c.-228A > C) in a putative GRα binding site.
We have tested the 50-SNP PRS from Steinberg et al. (2022) [19] using the same testing data as a comparison to our PRS. We found that the 50-SNP PRS had an AUC of 0.649 (95% CI = 0.627–0.670) which is slightly better than the AUC of our PRS which had an AUC of 0.639 (95% CI = 0.618–0.661). It should be pointed out that our study only considered SNPs from the genotyped data as opposed to imputed data while only 13 SNPs in the 50-SNP PRS can be found in the genotyped data. We found nine of those SNPs were also identified in our PRS. We did not test the 68-SNP PRS from Steinberg et al. (2022) [19] because the meta-analysis used for building that PRS included UK Biobank samples.
Development of combined risk model
Using the first half of the cohort data (307 cases and 54 492 controls), we fitted a Cox regression to estimate coefficients for the PRS and the clinical risk score. The beta coefficients were 0.233 (95% CI = 0.009–0.457; P = 0.042) for the clinical risk score and 0.684 (95% CI = 0.499–0.868; P < 0.001) for the PRS. There was no evidence the proportional hazards assumption was violated for PRS (χ2 = 0.0816, df = 1, P = 0.78), clinical risk score (χ2 = 0.0476, df = 1, P = 0.83) or globally (χ2 = 0.1796, df = 2, P = 0.9). In this dataset, the 10-year risk score had an AUC of 0.645 (95% CI = 0.613–0.676) and the hazard ratio per SD was 1.284 (95% CI = 1.193–1.382).
Performance of combined 10-year risk model
We used the second half of the cohort data (306 cases and 54 492 controls) to test the performance of the combined model. The hazard ratio per SD was 1.332 (95% CI = 1.263–1.406; P < 0.001). Harrell’s C-index for the 10-year risk score was 0.685 (95% CI = 0.654–0.715). As a comparison, Harrell’s C-index was 0.629 (95% CI = 0.596–0.661) for the clinical risk score only and 0.676 (95% CI = 0.645–0.706) for the PRS only. The improvement in Harrell’s C-index for the combined model was 0.056 (P < 0.001) over the clinical risk score alone. In terms of overall calibration of the combined model, the SIR was 1.193 (95% CI = 1.067–1.335) and the model underestimated risk with 306 observed cases versus 256.47 expected cases. When stratified by quintile of risk, the model was well calibrated except for the highest quintile of risk, which underestimated risk, with 140 observed cases and 94.71 expected cases (see Table 2).
Table 2.
Standardized incidence ratios of the number of melanoma cases observed in the first 10 years of follow-up in the second half of the testing data compared with the expected number using 10-year risk
| Observed | Expected | Standardized incidence ratio | 95% confidence interval | |
|---|---|---|---|---|
| Overall | 306 | 256.47 | 1.193 | [1.067–1.335] |
| Quintile of risk | ||||
| 1 | 22 | 23.23 | 0.947 | [0.624–1.439] |
| 2 | 32 | 34.69 | 0.922 | [0.652–1.304] |
| 3 | 54 | 44.95 | 1.201 | [0.920–1.568] |
| 4 | 58 | 58.88 | 0.985 | [0.762–1.274] |
| 5 | 140 | 94.71 | 1.478 | [1.253–1.745] |
Final model
Because the performance in association and discrimination of the model was similar in both halves of the cohort data, we refined the model estimates by re-fitting the model using all of the cohort data and computed the 10-year melanoma risk. The beta coefficients for the final model were 0.319 (95% CI = 0.166–0.471; P < 0.001) for the clinical risk score and 0.741 (95% CI = 0.611–0.871; P < 0.001) for the PRS.
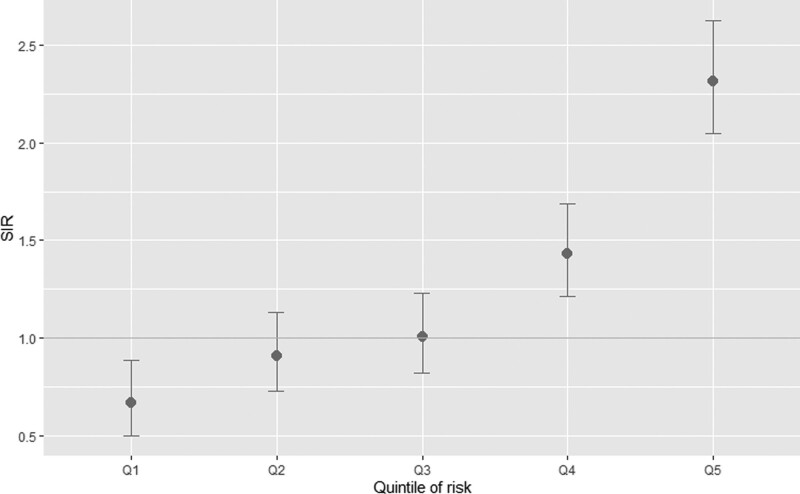
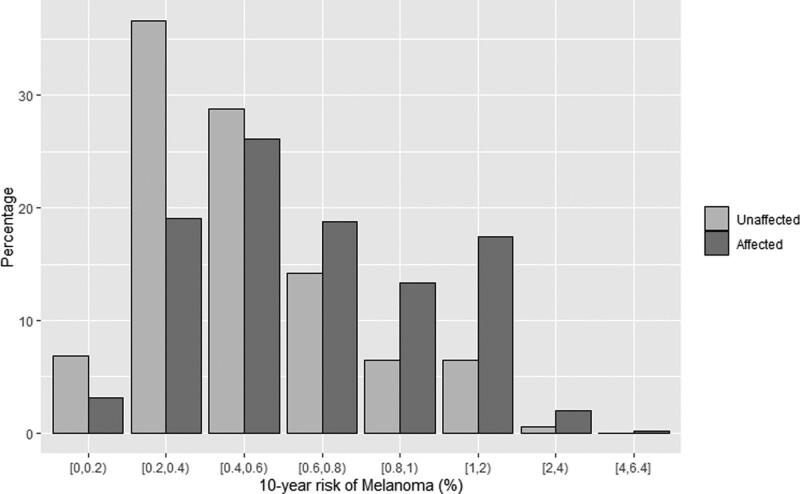
To demonstrate the performance of the model, we computed the SIRs of the number of observed cases compared with the number predicted by age-, sex- and calendar year-specific population incidence rates, overall and by the quintile of 10-year risk (Fig. 1 and Table 3). Overall, the number of observed melanoma cases was higher than the expected number using population incidence rates (613 vs 459.95). Individuals in the top quintile of risk were at 2.3 times the population risk and individuals in the second highest quintile were at 1.4 times the population risk. Individuals in the lowest quintile group were at 0.67 times the population average risk. This is an improvement over the SIR based on clinical risk only, where there is a much smaller difference between the five quintiles (Supplementary eFigure 1, Supplemental digital content 1, http://links.lww.com/MR/A319 and Supplementary eTable 3, Supplemental digital content 1, http://links.lww.com/MR/A319). Figure 2 shows the distribution of the 10-year risk for affected and unaffected individuals.
Fig. 1.
Standardized incidence ratios of the number of observed melanoma cases in the first 10 years of follow-up in the testing data compared with the expected number using population incidence rates by quintile of 10-year risk.
Table 3.
Standardized incidence ratios of the number of melanoma cases observed in the first 10 years of follow-up in the whole testing dataset compared with the expected number using population incidence rates, overall and by quintile of 10-year risk of melanoma
| Observed | Expected | Standardized incidence ratio | 95% confidence interval | |
|---|---|---|---|---|
| Overall | 613 | 459.95 | 1.333 | [1.231–1.443] |
| Quintile of risk | ||||
| 1 | 48 | 72.04 | 0.666 | [0.502–0.884] |
| 2 | 78 | 85.97 | 0.907 | [0.727–1.133] |
| 3 | 95 | 94.32 | 1.007 | [0.824–1.232] |
| 4 | 144 | 100.60 | 1.431 | [1.216–1.685] |
| 5 | 248 | 107.02 | 2.317 | [2.046–2.625] |
Fig. 2.
Distribution of 10-year melanoma risk categorized by different percentage groups.
Discussion
We have developed a model to predict a 10-year risk for melanoma that combines a PRS and a clinical risk score. The risk prediction has good calibration (Table 2) and discrimination (Fig. 1 and Table 3) and can distinguish between individuals at high and low risk. The clinical implications of improved risk prediction are important to increasing screening measures and promoting risk management options for at-risk adults.
No specific screening guidelines exist for melanoma in the USA [30], and limited guidelines exist in Australia, New Zealand, Germany, and the UK for adults at increased risk of developing melanoma. Risk management recommendations for adults at high risk may include full-body skin examination with dermoscopy or photography as well as minimizing UV light exposure [31–34]. Currently, high-risk individuals are identified based on their history of skin cancer, family history of melanoma, skin/hair pigmentation, number of naevi, evidence of skin damage and monogenic carrier susceptibility status.
For the few known high-risk susceptibility genes [35], the same melanoma risk management guidance exists [36]. CDKN2A was the first melanoma susceptibility gene identified; carriers are estimated to have a lifetime risk of 28% (by age 80 years) [37]. Using the combined genetic and clinical risk score, no participants in our study reached a full-lifetime threshold of 28% (equivalent to that of a CDKN2A carrier); however, a quarter of adults with incident melanoma had lifetime risk scores between 4 and 25% (Supplementary eFigure 2, Supplemental digital content 1, http://links.lww.com/MR/A319), which represent two- to 10-fold risk compared to the general population. The SIRs show that participants we identify at increased risk are more likely to develop melanoma compared with the participants identified with standard clinical risk factors.
Incorporating a polygenic component into a clinical risk prediction model can improve the prediction of melanoma risk, with a Harrell’s C-index of 0.685 (95% CI = 0.654–0.715) compared with using hair color, sunbed use and personal history of non-melanoma cancer, which had an AUC of 0.629 (95% CI = 0.596–0.661). This 0.056 increase in AUC is important because it represents a 9% increase in discriminatory performance over the clinical risk score alone.
Importantly, the distribution of adults based on their 10-year risk scores shows the ability of the model to identify a greater number of incident cases with higher 10-year risk scores (and conversely a smaller number of incident cases with lower 10-year risk scores) compared with unaffected participants (Fig. 2). The average participant, aged 40–69 years, had a 10-year risk score of 0.486. We can identify 17.78% of participants who have at least a two-fold increase in risk and 1.29% with a four-fold increase in risk compared to the population.
When we categorize adults by applying a basic two-fold risk threshold, we show the significant clinical value our combined model has over the standard clinical risk factors alone. We classified participants into three risk categories – low, average and high – based on their 10-year risk scores, ≤0.5%, 0.5–1.0% and ≥1.0%, respectively. We are able to better stratify the population utilizing our model (Supplementary eFigure 3a, Supplemental digital content 1, http://links.lww.com/MR/A319) compared with the clinical model alone (Supplementary eFigure 3b, Supplemental digital content 1, http://links.lww.com/MR/A319). Importantly, we identify 10 times as many adults in the high-risk category (≥1% 10-year risk) compared with the clinical model alone; this high-risk category represents adults at twice the population average risk. Conversely, when we identified 60% of the general population at lower-than-average risk (≤0.5% 10-year risk), the SIR for melanoma was well below that of the clinical model alone [0.86 (95% CI = 0.7158–1.0333); 1.1705 (95% CI = 1.0306–1.3295), respectively].
A strength of our study is that we used a matched case–control design to develop the PRS. Many studies include age and sex (and sometimes principal components) as covariates when assessing the discriminatory performance of a PRS. This is misleading because these are known risk factors for melanoma and confound the association between the PRS and melanoma, resulting in an overestimation of the AUC [17].
One limitation of our study is that we did not include nevus density and family history of melanoma in our prediction model because these are not available in the UK Biobank. Nevus count is a very strong predictor of melanoma risk. People with 100 or more nevi over their whole body are found to have seven times the risk compared with people with fewer than 15 nevi [38]. Family history of melanoma is also a strong risk factor. Meta-analyses showed that having a first-degree relative with melanoma increases risk by 2.06 times [39]. Without including these two important risk factors, our risk prediction model has not achieved its full potential. The ethnic breakdown of the UK Biobank is another limitation of the study. These analyses were conducted on people of Northern European ancestry. While melanoma prevalence is much higher in this fair-skinned Caucasian population, phenotypically uncharacteristic (i.e. dark complexion and dark hair) cases of melanoma do occur. When they do occur, they are detected at a later stage and the melanoma-associated mortality is higher. Future studies will address the potential utility of a risk assessment tool that incorporates non-phenotypic factors like polygenic risk [40,41].
Improved stratification of general population adults is possible and could support improved cancer screening recommendations. A total body skin examination is a simple, cost-effective, noninvasive screening tool, yet consensus regarding its implementation is limited by a lack of direct evidence that it reduces melanoma-associated mortality. Given that randomized controlled trials will likely never exist in this space due to sample size, duration and ethical concerns, alternative data-driven metrics must be applied to justify screening recommendations.
Identifying high-risk individuals early can lead to a reduction in late-stage melanoma diagnosis and reduce the substantial medical costs associated with late-stage treatments; however, melanoma prevention efforts are also important for reducing the economic burden. For example, in 2021, the estimated cost for new melanoma cases was $AU 397.9 million in Australia, but nearly half of that was spent on in-situ disease [42]. Consideration of risk stratification tools to address melanoma risk reduction for both prevention and early detection efforts is paramount to lowering the clinical and economic burden of the disease.
Acknowledgements
We wish to thank Mr. Lawrence Whiting for his invaluable expertise in the management of the large data files from the UK Biobank. The analyses undertaken in this study were fully funded by Genetic Technologies Limited. The funders had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Conflicts of interest
C.K.W., G.S.D., N.M.M. and R.A. are employees of Genetic Technologies Limited. E.S. is an employee of Phenogen Science Inc (a subsidiary of Genetic Technologies Limited). Aspects of this manuscript are covered by Provisional Patent Application AU 2022903017, Methods of assessing risk of developing melanoma. C.K.W., G.S.D. and R.A. are named inventors on the patent application, which is assigned to Genetic Technologies Limited.
Supplementary Material
Footnotes
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.melanomaresearch.com.
References
- 1.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Picconi O, Boyle P, et al. Meta-analysis of risk factors for cutaneous melanoma: II. Sun exposure. Eur J Cancer 2005; 41:45–60. [DOI] [PubMed] [Google Scholar]
- 2.Grob JJ, Gouvernet J, Aymar D, Mostaque A, Romano MH, Collet AM, et al. Count of benign melanocytic nevi as a major indicator of risk for nonfamilial nodular and superficial spreading melanoma. Cancer 1990; 66:387–395. [DOI] [PubMed] [Google Scholar]
- 3.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Zanetti R, Masini C, et al. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur J Cancer 2005; 41:2040–2059. [DOI] [PubMed] [Google Scholar]
- 4.Olsen CM, Pandeya N, Thompson BS, Dusingize JC, Green AC, Neale RE, et al.; QSkin Study. Association between phenotypic characteristics and melanoma in a large prospective cohort study. J Invest Dermatol 2019; 139:665–672. [DOI] [PubMed] [Google Scholar]
- 5.Ford D, Bliss JM, Swerdlow AJ, Armstrong BK, Franceschi S, Green A, et al. Risk of cutaneous melanoma associated with a family history of the disease. The International Melanoma Analysis Group (IMAGE). Int J Cancer 1995; 62:377–381. [DOI] [PubMed] [Google Scholar]
- 6.Usher-Smith JA, Emery J, Kassianos AP, Walter FM. Risk prediction models for melanoma: a systematic review. Cancer Epidemiol Biomarkers Prev 2014; 23:1450–1463. [DOI] [PubMed] [Google Scholar]
- 7.Vuong K, Armstrong BK, Weiderpass E, Lund E, Adami HO, Veierod MB, et al.; Australian Melanoma Family Study Investigators. Development and external validation of a melanoma risk prediction model based on self-assessed risk factors. JAMA Dermatol 2016; 152:889–896. [DOI] [PubMed] [Google Scholar]
- 8.Vuong K, Armstrong BK, Drummond M, Hopper JL, Barrett JH, Davies JR, et al. Development and external validation study of a melanoma risk prediction model incorporating clinically assessed naevi and solar lentigines. Br J Dermatol 2020; 182:1262–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mucci LA, Hjelmborg JB, Harris JR, Czene K, Havelick DJ, Scheike T, et al.; Nordic Twin Study of Cancer (NorTwinCan) Collaboration. Familial risk and heritability of cancer among twins in nordic countries. JAMA 2016; 315:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei EX, Li X, Nan H. Having a first-degree relative with melanoma increases lifetime risk of melanoma, squamous cell carcinoma, and basal cell carcinoma. J Am Acad Dermatol 2019; 81:489–499. [DOI] [PubMed] [Google Scholar]
- 11.Frank C, Fallah M, Sundquist J, Hemminki A, Hemminki K. Population landscape of familial cancer. Sci Rep 2015; 5:12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truderung OA, Sagi JC, Semsei AF, Szalai C. Melanoma susceptibility: an update on genetic and epigenetic findings. Int J Mol Epidemiol Genet 2021; 12:71–89. [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med 2020; 12:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potjer TP, van der Grinten TWJ, Lakeman IMM, Bollen SH, Rodríguez-Girondo M, Iles MM, et al. Association between a 46-SNP polygenic risk score and melanoma risk in Dutch patients with familial melanoma. J Med Genet 2021; 58:760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakshi A, Yan M, Riaz M, Polekhina G, Orchard SG, Tiller J, et al. Genomic risk score for melanoma in a prospective study of older individuals. J Natl Cancer Inst 2021; 113:1379–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cust AE, Drummond M, Kanetsky PA, Barrett JH, MacGregor S, Law MH, et al.; Australian Melanoma Family Study Investigators. Assessing the incremental contribution of common genomic variants to melanoma risk prediction in two population-based studies. J Invest Dermatol 2018; 138:2617–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtis D. Clinical relevance of genome-wide polygenic score may be less than claimed. Ann Hum Genet 2019; 83:274–277. [DOI] [PubMed] [Google Scholar]
- 18.Gu F, Chen T-H, Pfeiffer RM, Fargnoli MC, Calista D, Ghiorzo P, et al. Combining common genetic variants and non-genetic risk factors to predict risk of cutaneous melanoma. Hum Mol Genet 2018; 27:4145–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinberg J, Iles MM, Lee JY, Wang X, Law MH, Smit AK, et al. Independent evaluation of melanoma polygenic risk scores in UK and Australian prospective cohorts. Br J Dermatol 2022; 186:823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landi MT, Bishop DT, MacGregor S, Machiela MJ, Stratigos AJ, Ghiorzo P, et al.; GenoMEL Consortium. Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility. Nat Genet 2020; 52:494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015; 12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018; 562:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Privé F, Vilhjálmsson BJ, Aschard H, Blum MGB. Making the most of clumping and thresholding for polygenic scores. Am J Hum Genet 2019; 105:1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prive F, Aschard H, Ziyatdinov A, Blum MGB. Efficient analysis of large-scale genome-wide data with two R packages: bigstatsr and bigsnpr. Bioinformatics 2018; 34:2781–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Y, Hu H, Chen J-S, Hu F, Fowler J, Scheet P, et al. Integrated case-control and somatic-germline interaction analyses of melanoma susceptibility genes. Biochim Biophys Acta Mol Basis Dis 2018; 1864:2247–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature 2011; 480:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potrony M, Puig-Butille JA, Aguilera P, Badenas C, Tell-Marti G, Carrera C, et al. Prevalence of MITF p.E318K in patients with melanoma independent of the presence of CDKN2A causative mutations. JAMA Dermatol 2016; 152:405–412. [DOI] [PubMed] [Google Scholar]
- 28.Raimondi S, Sera F, Gandini S, Iodice S, Caini S, Maisonneuve P, et al. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int J Cancer 2008; 122:2753–2760. [DOI] [PubMed] [Google Scholar]
- 29.Nan H, Xu M, Kraft P, Qureshi AA, Chen C, Guo Q, et al. Genome-wide association study identifies novel alleles associated with risk of cutaneous basal cell carcinoma and squamous cell carcinoma. Hum Mol Genet 2011; 20:3718–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bibbins-Domingo K, Grossman DC, Curry SJ, Davidson KW, Ebell M, Epling JW, et al.; US Preventive Services Task Force. Screening for skin cancer US preventive services task force recommendation statement. JAMA 2016; 316:429–435. [DOI] [PubMed] [Google Scholar]
- 31.Australian Cancer Network Melanoma Guidelines Revision Working Party. Clinical practice guidelines for the management of melanoma in Australia and New Zealand. The Cancer Council Australia and Australian Cancer Network, Sydney and New Zealand Guidelines Group, Wellington; 2008. https://www.health.govt.nz/publication/clinical-practice-guidelines-management-melanoma-australia-and-new-zealand. [Accessed 20 July 2022]. [Google Scholar]
- 32.Marsden JR, Newton-Bishop JA, Burrows L, Cook M, Corrie PG, Cox NH, et al.; British Association of Dermatologists (BAD) Clinical Standards Unit. Revised UK guidelines for the management of cutaneous melanoma 2010. J Plast Reconstr Aesthet Surg 2010; 63:1401–1419. [DOI] [PubMed] [Google Scholar]
- 33.Katalinic A, Eisemann N, Waldmann A. Skin cancer screening in Germany. Documenting melanoma incidence and mortality from 2008 to 2013. Dtsch Arztebl Int 2015; 112:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.German Guideline Program in Oncology (GGPO). Evidence-based guideline on prevention of skin cancer (short version). https://www.leitlinienprogramm-onkologie.de/fileadmin/user_upload/Downloads/Leitlinien/Hautkrebspraeventationsleitlinie_1.1/Short_version_-_Guideline_on_prevention_of_skin_cancer.pdf. [Accessed 20 July 2022].
- 35.Ribero S, Longo C, Glass D, Nathan P, Bataille V. What is new in melanoma genetics and treatment? Dermatology 2016; 232:259–264. [DOI] [PubMed] [Google Scholar]
- 36.National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology (NCCN Guidelines®) Genetic/familial high-risk assessment: breast, ovarian, and pancreatic version 2.2022. [DOI] [PubMed]
- 37.Begg CB, Orlow I, Hummer AJ, Armstrong BK, Kricker A, Marrett LD, et al.; Genes Environment and Melanoma Study Group. Lifetime risk of melanoma in CDKN2A mutation carriers in a population-based sample. J Natl Cancer Inst 2005; 97:1507–1515. [DOI] [PubMed] [Google Scholar]
- 38.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Abeni D, Boyle P, et al. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur J Cancer 2005; 41:28–44. [DOI] [PubMed] [Google Scholar]
- 39.Olsen CM, Carroll HJ, Whiteman DC. Familial melanoma: a meta-analysis and estimates of attributable fraction. Cancer Epidemiol Biomarkers Prev 2010; 19:65–73. [DOI] [PubMed] [Google Scholar]
- 40.Goldenberg A, Vujic I, Sanlorenzo M, Ortiz-Urda S. Melanoma risk perception and prevention behavior among African-Americans: the minority melanoma paradox. Clin Cosmet Investig Dermatol 2015; 8:423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahendraraj K, Sidhu K, Lau CSM, McRoy GJ, Chamberlain RS, Smith FO. Malignant melanoma in African-Americans: a population-based clinical outcomes study involving 1106 African-American patients from the surveillance, epidemiology, and end result (SEER) database (1988-2011). Medicine (Baltimore) 2017; 96:e6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon LG, Leung W, Johns R, McNoe B, Lindsay D, Merollini KMD, et al. Estimated healthcare costs of melanoma and Keratinocyte skin cancers in Australia and aotearoa New Zealand in 2021. Int J Environ Res Public Health 2022; 19:3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Access to the data used in this study can be obtained by applying directly to the UK Biobank at https://www.ukbiobank.ac.uk/register-apply/. The authors did not receive special access privileges to the data that others would not have. Interested researchers will be able to access the data in the same manner by applying directly to the UK Biobank.