Abstract
PURPOSE
Circulating tumor DNA (ctDNA) genotyping on the basis of next-generation sequencing (NGS) may guide targeted therapy for metastatic colorectal cancer (mCRC). However, the validity of NGS-based ctDNA genotyping for RAS/BRAF V600E mutation assessment and the efficacy of anti-EGFR and BRAF-targeted therapies on the basis of ctDNA results remains unclear.
PATIENTS AND METHODS
The performance of NGS-based ctDNA genotyping for RAS/BRAF V600E mutation assessment was compared with that of a validated polymerase chain reaction–based tissue testing in patients with mCRC enrolled in the GOZILA study, a nationwide plasma genotyping study. The primary end points were concordance rate, sensitivity, and specificity. The efficacy of anti-EGFR and BRAF-targeted therapies on the basis of ctDNA were also evaluated.
RESULTS
In 212 eligible patients, the concordance rate, sensitivity, and specificity were 92.9% (95% CI, 88.6 to 96.0), 88.7% (95% CI, 81.1 to 94.0), and 97.2% (95% CI, 92.0 to 99.4) for RAS and 96.2% (95% CI, 92.7 to 98.4), 88.0% (95% CI, 68.8 to 97.5), and 97.3% (95% CI, 93.9 to 99.1) for BRAF V600E, respectively. In patients with a ctDNA fraction of ≥1.0%, sensitivity rose to 97.5% (95% CI, 91.2 to 99.7) and 100% (95% CI, 80.5 to 100.0) for RAS and BRAF V600E mutations, respectively. In addition to a low ctDNA fraction, previous chemotherapy, lung and peritoneal metastases, and interval between dates of tissue and blood collection were associated with discordance. The progression-free survival of anti-EGFR therapy and BRAF-targeted treatment was 12.9 months (95% CI, 8.1 to 18.5) and 3.7 (95% CI, 1.3 to not evaluated) months, respectively, for matched patients with RAS/BRAF V600E results by ctDNA.
CONCLUSION
ctDNA genotyping effectively detected RAS/BRAF mutations, especially with sufficient ctDNA shedding. Clinical outcomes support ctDNA genotyping for determining the use of anti-EGFR and BRAF-targeted therapies in patients with mCRC.
Using a large-scale plasma genomic profiling program (SCRUM-Japan GOZILA), we validated ctDNA genotyping for RAS and BRAF V600E in metastatic colorectal cancer comparing with a validated tissue PCR-based testing.
INTRODUCTION
In metastatic colorectal cancer (mCRC), assessment of a growing number of biomarkers, such as KRAS/NRAS (RAS) and BRAF V600E mutations,1-3 microsatellite instability (MSI) status,4,5 and HER2 amplifications6,7 is required for optimal treatment selection. Hence, multigene next-generation sequencing (NGS) instead of sequential or parallel polymerase chain reaction (PCR)–based testing for each biomarker may be preferred; however, some disadvantages, such as the lengthy time to return results, the cost, invasiveness, and the difficulties related to procedures, limit the use of tissue-based NGS for biomarker testing before the initiation of first-line treatment.
CONTEXT
Key Objective
Using a large-scale plasma genomic profiling program (SCRUM-Japan GOZILA), we aimed to validate the performance of next-generation sequencing–based circulating tumor DNA (ctDNA) genotyping for RAS and BRAF V600E in metastatic colorectal cancer (mCRC) by comparing with a validated tissue polymerase chain reaction–based RAS/BRAF testing.
Knowledge Generated
Our findings demonstrated the concordance rate, sensitivity, and specificity of 92.9%, 88.7%, and 97.2% for RAS and 96.2%, 88.0%, and 97.3% for BRAF V600, respectively. Low ctDNA fraction, previous chemotherapy, lung and peritoneal metastases, and long interval between dates of tissue and blood collection were associated with discordance. Patients with wild-type RAS or BRAF V600E by ctDNA genotyping were likely to have the efficacy of targeted therapies similar to those who received treatment on the basis of tissue testing.
Relevance
Our study supports the use of ctDNA genotyping in the assessment of RAS and BRAF V600E mutations in patients with mCRC.
SCRUM-Japan GOZILA is one of the largest circulating tumor DNA (ctDNA) genomic profiling studies. This study demonstrated that ctDNA NGS had markedly faster turnaround time than tissue NGS (within two weeks),8 supporting the potential of ctDNA NGS for guiding first-line treatment of mCRC. Data from GOZILA also validated ctDNA NGS for the assessment of MSI and HER2 amplification with the concordance of 98.2% and 82.7%, respectively.7,9 Herein, we conducted a validation study by comparing NGS-based ctDNA genotyping for RAS/BRAF with tissue PCR-based RAS/BRAF testing, which has been approved as a companion diagnostic for patients with mCRC in Japan.
PATIENTS AND METHODS
Study Design and Patients
We aimed to compare the performance of ctDNA RAS/BRAF assessment with that of tissue-based RAS/BRAF assessment in the SCRUM-Japan GOZILA study. Patients with mCRC enrolled in GOZILA between August 2018 and February 2020 who had available plasma- and tissue-based RAS/BRAF results before anti-EGFR therapy initiation were included in this study.
GOZILA is a nationwide plasma genomic profiling study involving 31 core cancer institutions in Japan. Patients with metastatic gastrointestinal cancers were eligible for enrollment. All enrolled patients provided written informed consent, and ctDNA genotyping was conducted using Guardant360 CDx (Guardant Health, Inc, Redwood City, CA).10 To avoid ctDNA shedding suppression because of chemotherapy, participants were required to have disease progression during systemic chemotherapy and should not have started subsequent therapy at the time of blood sampling.
This study was conducted in accordance with the Declaration of Helsinki and the Japanese Ethical Guidelines for Medical and Biological Research Involving Human Subjects. All study protocols were approved by the institutional review board of each participating institution and registered at the University Hospital Medical Information Network Clinical Trials Registry (UMIN000046220).
ctDNA Genotyping in the GOZILA Study
NGS analysis of ctDNA was performed using Guardant360 CDx at Guardant Health, a Clinical Laboratory Improvement Amendments–certified, College of American Pathologists–accredited laboratory approved by the US Food and Drug Administration, the Japanese Pharmaceuticals and Medical Devices Agency, and New York State Department of Health, as previously described.11 The assay detects single-nucleotide variations (SNVs), indels, fusions, and copy-number alterations in 74 genes, including the RAS and BRAF mutations reported in RASKET-B, with reportable ranges of ≥0.04, ≥0.02, and ≥0.004% and ≥2.12 copies, respectively, and MSI.
To estimate ctDNA clonality for somatic SNVs, indels, and fusions, relative clonality was defined as the variant allelic fraction (VAF) of the relevant mutation detected in ctDNA divided by the maximum VAF detected in the plasma sample. ctDNA fraction was estimated using the surrogate of maximum VAF. RAS and BRAF V600E mutations were also included in the ctDNA calculation if they had the maximum VAF in plasma. All reported somatic variants, including variants of uncertain significance, were used for calculation of the ctDNA fraction and clonality, whereas putative germline variants were excluded.
Tissue RAS/BRAF Testing
Tissue samples were analyzed using the RASKET-B kit (MBL, Nagoya, Japan), an approved companion diagnostic for cetuximab or panitumumab in mCRC. RASKET-B detects 53 mutations, including those in codon 12 (G12S, G12C, G12R, G12D, G12V, and G12A), codon 13 (G13S, G13C, G13R, G13D, G13V, and G13A), codon 59 (A59T and A59G), codon 61 (Q61K, Q61E, Q61L, Q61P, Q61R, and Q61H), codon 117 (K117N), and codon 146 (A146T, A146P, and A146V) in both KRAS and NRAS as well as BRAF V600E. The assay uses PCR-reverse sequence-specific oligonucleotide and Luminex system.12
End Points
The primary end points were the concordance rate, sensitivity, and specificity between plasma RAS/BRAF genotyping and tissue RAS/BRAF testing. At the planning stage, plasma RAS/BRAF testing was considered effective if the concordance rate, sensitivity, and specificity were ≥85%, ≥80%, and ≥90%, respectively. The threshold values were determined on the basis of previous studies that compared ctDNA-based genotyping with tissue-based genotyping and showed that the concordance rate, sensitivity, and specificity were 82.5%-93.3%, 76.0%-92.6%, and 82.4%-98.2% for RAS mutations and 93.2%-100%, 71.4%-100%, and 97.3%-100% for BRAF V600E mutations, respectively.13-19
Because detection of RAS/BRAF mutations may be less efficient in plasma samples with a low ctDNA fraction (as estimated by the surrogate of maximum VAF), concordance, sensitivity, and specificity were assessed in a patient subset defined by various ctDNA fraction cutoff values (≥0.1%, ≥0.2%, and ≥1.0%) as secondary end points. In addition, the concordance rate, sensitivity, and specificity were also assessed for each codon of KRAS and NRAS, according to the site of metastatic disease, and between patients who had received chemotherapy before plasma collection and those who did not. The best objective response and progression-free survival (PFS) were evaluated in patients without RAS mutation by tissue or ctDNA testing who received anti-EGFR therapy and in patients with BRAF V600E mutations detected by tissue or ctDNA testing who received encorafenib plus cetuximab with or without binimetinib.
Statistical Analysis
All analyses were conducted on all patients. Quantitative data are represented as median and range. The concordance rate between the plasma- and tissue-based tests for RAS/BRAF mutational status and their 95% CIs on the basis of the exact binomial distribution for each condition were estimated. Tumor response was assessed in patients with measurable lesions using the RECIST version 1.1. PFS was measured from the date of therapy initiation to the date of disease progression by investigator judgment or death from any cause. The PFS rate was estimated by the Kaplan-Meier method. All statistical analyses were performed using SAS Release version 9.4 (SAS Institute, Cary, NC).
RESULTS
Patients
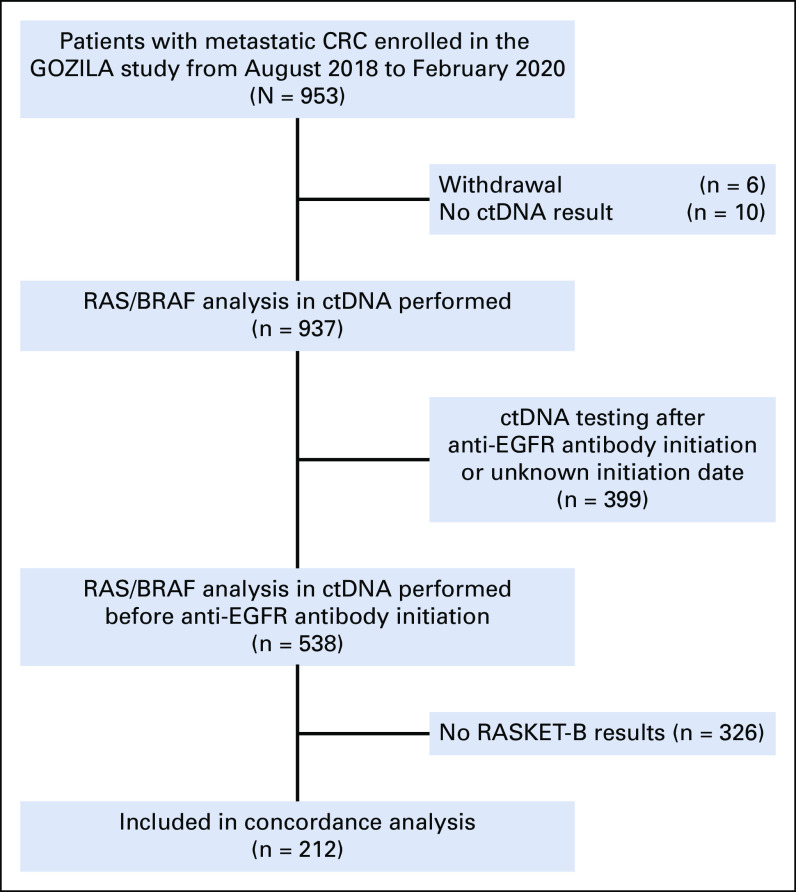
Of the 953 patients with mCRC enrolled in GOZILA between August 2018 and February 2020, 212 underwent both tissue RASKET-B and Guardant360 CDx testing and had not been treated with an anti-EGFR antibody before ctDNA testing (Fig 1). The patients' characteristics are presented in the Data Supplement ([Table S1]). The median age was 61.5 years, and 115 patients (54.2%) were male. In 63.7% of patients, the primary tumor site was at the left colorectum, and the liver was the most common metastatic site (125 patients, 59%). Meanwhile, 146 (68.9%) patients received chemotherapy before ctDNA blood collection. The median time from shipping samples to reporting results was 7 days for both tissue and plasma genotyping.
FIG 1.
Flow diagram of patient selection. Among 953 patients with metastatic CRC enrolled in the GOZILA study from August 2018 to February 2020, 212 patients with RASKET-B and ctDNA results before anti-EGFR therapy were included in this concordance analysis. CRC, colorectal cancer; ctDNA, circulating tumor DNA.
Clinical Validation
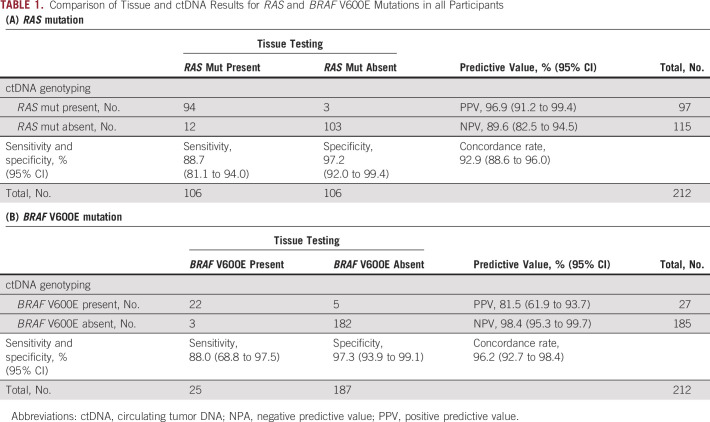
RAS mutations were detected in 106 of 212 (50.0%) patients by tissue testing; ctDNA analysis confirmed RAS mutations in 94 of 106 tissue RAS-mutant patients (88.7%; 95% CI, 81.1 to 94.0) and did not detect RAS mutations in 103 of 106 patients with a tissue RAS wild-type result (97.2%; 95% CI, 92.0 to 99.4; Table 1A). Using the tissue-based kit, BRAF V600E mutations were detected in 25 of 212 patients (11.8%). On ctDNA genotyping, BRAF V600E mutations were detected in 22 of 25 patients with tissue BRAF V600E mutations (88.0%; 95% CI, 68.8 to 97.5) and were not detected in 182 of 187 patients with a tissue BRAF wild-type tumor (97.3%; 95% CI, 93.9 to 99.1; Table 1B). Therefore, there were concordance rates of 92.9% and 96.2%, sensitivities of 88.7% and 88.0%, and specificities of 97.2% and 97.3% for RAS and BRAF V600E mutations, respectively (Tables 1A and 1B).
TABLE 1.
Comparison of Tissue and ctDNA Results for RAS and BRAF V600E Mutations in all Participants
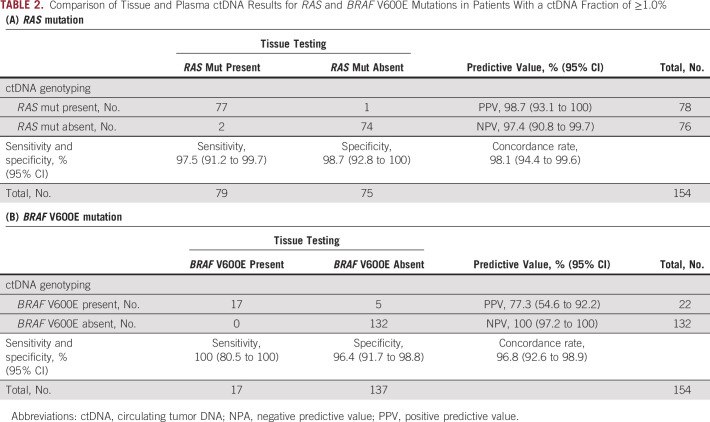
Of all 212 patients, 204 patients had reportable variants in ctDNA, with the remaining eight patients in whom passed quality control, but no variants were detected. In 204 patients with available ctDNA fraction results, we evaluated concordance according to various cutoff values. RAS and BRAF detection by ctDNA genotyping was better for samples with ctDNA fraction of ≥0.1%, ≥0.2% and ≥1.0% (Table 2, Data Supplement [Tables S2 and S3]). In patients with a ctDNA fraction of ≥1.0%, the concordance rate was 98.1% (95% CI, 94.4 to 99.6) and 96.8% (95% CI, 92.6 to 98.9), the sensitivity was 97.5% (95% CI, 91.2 to 99.7) and 100% (95% CI, 80.5 to 100.0), and the specificity was 98.7% (95% CI, 92.8 to 100.0) and 96.4% (95% CI, 91.7 to 98.8) for RAS and BRAF V600E mutations, respectively. In addition, the negative predictive value (NPV) also increased for RAS and BRAF V600E mutations (Tables 2A and 2B). The analysis of sensitivity in correlation with the continuous variable of ctDNA fraction revealed an improvement in the sensitivity of ctDNA testing for RAS and BRAF V600E mutations as the ctDNA fraction cutoff increased (Data Supplement [Figs S1A and S1B]). In addition, the sensitivity and concordance for both RAS and BRAF were better in patients previously untreated or with liver metastases (Data Supplement [Tables S4 and S5]). We evaluated specific KRAS and NRAS mutations and found that most showed a concordance rate of 100% except for some variants, including KRAS G12D, G12A, Q61L, Q61H, and A146T and NRAS Q61E and Q61R (Data Supplement [Table S6]).
TABLE 2.
Comparison of Tissue and Plasma ctDNA Results for RAS and BRAF V600E Mutations in Patients With a ctDNA Fraction of ≥1.0%
We also investigated the characteristics associated with discordance of RAS/BRAF status between tissue and ctDNA testing. The median ctDNA fraction of patients with RAS and BRAF V600E mutations detected on both tissue and ctDNA (tissue+/ctDNA+) was 9.4% and 6.6%, respectively, whereas the median ctDNA fraction among patients with tissue+/ctDNA– was 0.2% and 0.6%, respectively (P = .002 for RAS, P = .117 for BRAF V600E; Fig 2). The median ctDNA relative clonality of RAS mutations was 0.77 in the tissue+/ctDNA+ group and 1.00 in the tissue–/ctDNA+ group (P = .519). For BRAF V600E, the relative clonality was 0.92 in the tissue+/ctDNA+ group and 0.98 in the tissue–/ctDNA+ group (P = .923; Fig 3). There is no significant correlation between the presence of RAS/BRAF mutations in tissues and the median relative clonality of the corresponding mutations in ctDNA.
FIG 2.
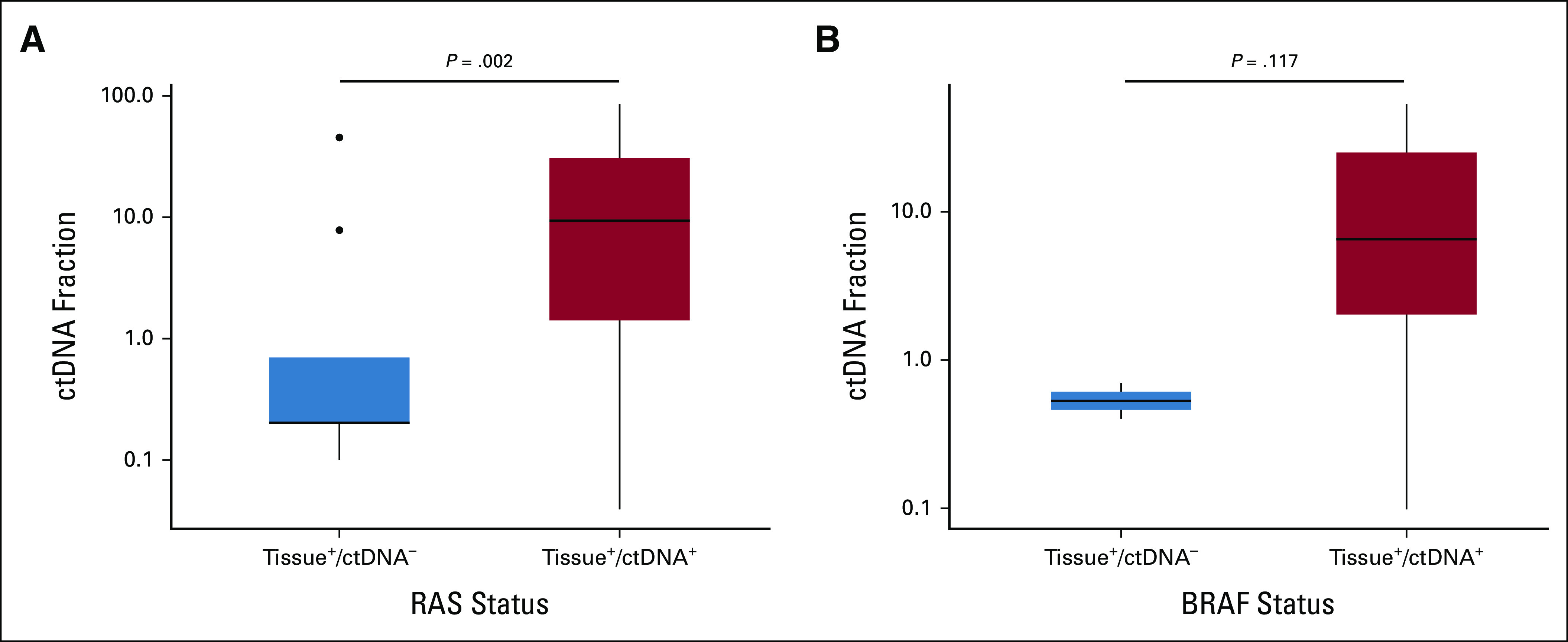
Box plot of ctDNA fraction compared between patients with ctDNA–/tissue+ and ctDNA+/tissue+ for RAS and BRAF V600E mutations. The ctDNA fraction was lower in patients with tissue+/ctDNA– for (A) RAS mutation and (B) BRAF V600E mutation. •Signifies an outlier. ctDNA, circulating tumor DNA.
FIG 3.
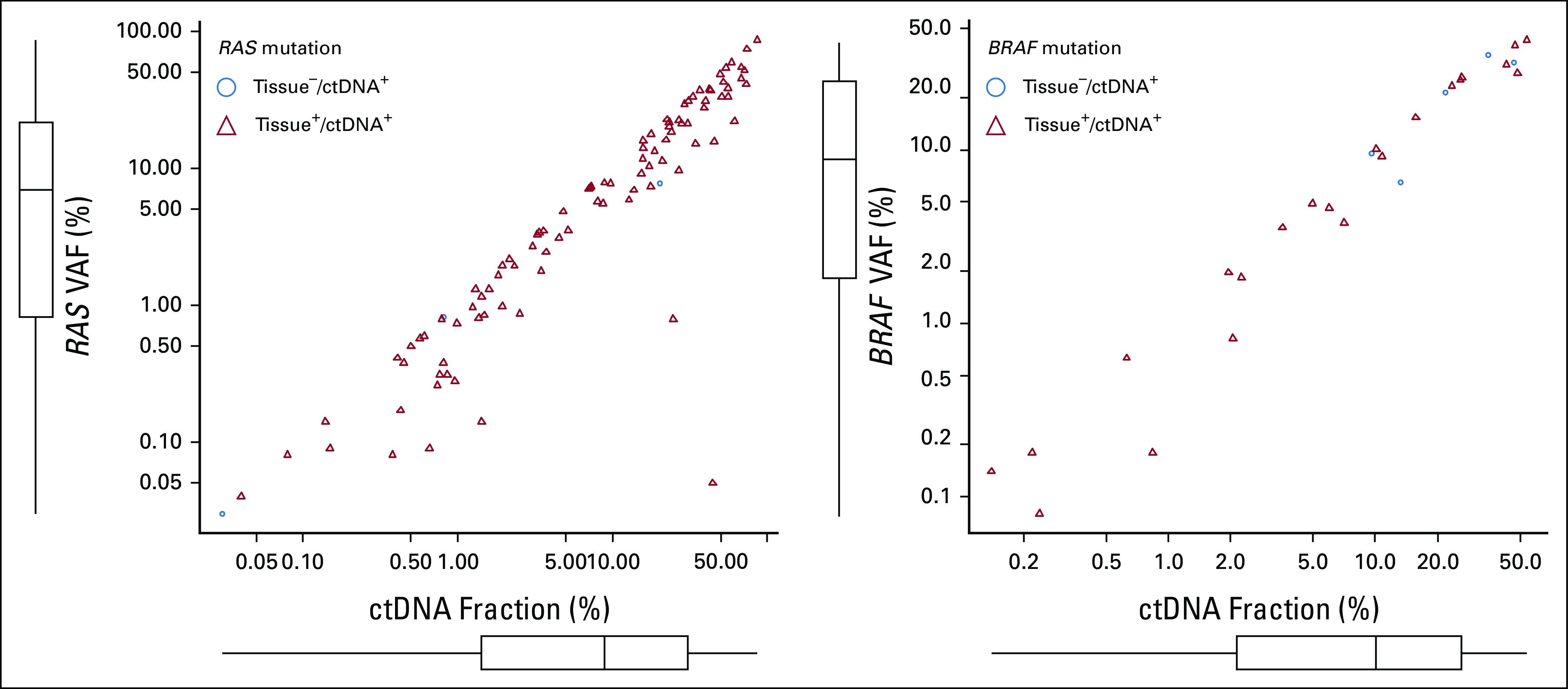
Scatterplot showing correlation between ctDNA fraction and VAF of (A) RAS mutation and (B) BRAF V600E mutation. X and Y axes represent the ctDNA fraction and VAF of RAS and BRAF V600E mutations, respectively. ctDNA, circulating tumor DNA; VAF, variant allelic fraction.
The median interval between the dates of tissue and ctDNA collection was significantly shorter in patients with concordant results for RAS mutation than in those with discordant results (274.5 v 365 days; P = .045). As for BRAF V600E mutation, the interval was also shorter in patients with concordant v discordant results (282.5 v 328 days; P = .372) although the difference was not significant. Indeed, in treatment-naïve patients with blood and tissue collected within 3 months, the concordance rate was 98.0% for both RAS and BRAF mutation, which was better than overall results (Data Supplement [Table S7]).
Efficacy of Targeted Therapies according to ctDNA Genotyping Results
To evaluate the clinical utility of plasma-based RAS/BRAF assessment, we investigated the efficacy of anti-EGFR therapy in patients without ctDNA RAS mutations. The anti-EGFR antibody was given to 22 patients in first-line therapy, all of whom had RAS/BRAF wild-type status according to both tissue and ctDNA testing. All but one of the 22 patients with first-line anti-EGFR therapy received an anti-EGFR antibody combined with chemotherapy. In all 22 patients, the overall response rate and median PFS were 54.5% (95% CI, 32.2 to 75.6) and 12.9 months (95% CI, 8.1 to 18.5), respectively (Data Supplement [Fig S2]). We also investigated the efficacy of anti-EGFR therapy on the basis of wild-type RAS in ctDNA in four patients who did not have tissue results in GOZILA. Of these, treatment of two patients was still ongoing, one with the best of response (BoR) of partial response (PR) and a PFS of 24.4 months and the other with BoR of stable disease (SD) and a PFS of 11.8 months (Data Supplement [Table S8]). In three patients who received anti-EGFR antibody on the basis of wild-type RAS in ctDNA before the return of tissue results, the PFS was more than 8.1 months and PR as BoR was achieved in two patients, one of whom successfully underwent conversion surgery (Data Supplement [Table S9]).
To evaluate the clinical utility of plasma-based BRAF assessment, we investigated the efficacy of encorafenib plus cetuximab with or without binimetinib in patients with ctDNA BRAF V600E. This regimen was given to five patients with BRAF V600E mutation confirmed in both tissue and ctDNA and to one patient with BRAF V600E mutation confirmed by tissue testing alone. The median PFS in six patients with BRAF mutant in tissue-based testing and treated with BRAF-targeted therapy, including five patients with BRAF mutant in ctDNA, was 3.7 months (95% CI, 1.3 to not evaluated (NE); Data Supplement [Fig S2]). Of four patients who received BRAF-targeted treatment on the basis of BRAF V600E in ctDNA and had no tissue results, three patients had PR as BoR with the PFS ranging from 3.6 to 14.2 months (Data Supplement [Table S10]).
DISCUSSION
This validation study of RAS/BRAF mutation assessment by ctDNA genotyping compared with tissue-based testing met its primary end point as shown by the concordance rate, sensitivity, and specificity of 92.9%, 88.7%, and 97.2% and 96.2%, 88.0%, and 97.3% for RAS and BRAF V600E mutations, respectively. For patients without detectable RAS mutations and for those with BRAF V600E mutations detected by ctDNA genotyping, the efficacy of appropriate targeted therapies was suggested to be similar to that observed for patients who received treatment on the basis of tissue testing although further investigation is necessary.
In our study, ctDNA genotyping for RAS and BRAF V600E mutations demonstrated concordance with tissue-based testing. Previously, PCR-based ctDNA testing for point mutations showed the concordance rate, sensitivity, and specificity of 86.4%-93.3%, 82.1%-92.6%, and 90.4%-94.0% and 93.2%-100%, 71.4%-100%, and 97.3%-100% for RAS and BRAF V600E mutations, respectively.13-16 By contrast, NGS-based ctDNA genotyping showed a slightly lower sensitivity compared with PCR-based ctDNA testing, with the concordance rate, sensitivity, and specificity of 82.5%-85.2%, 76.0%-82.8%, and 82.4%-98.2% and 97.3%-98.4%, 76.7%-83.3%, and 98.9%-100% for RAS and BRAF V600E mutations, respectively.17-19 Unlike previous reports of NGS-based ctDNA assays, which were limited by their small sample size and analytical sensitivity of the specific assays used, we successfully demonstrated relevant sensitivity from a sufficiently sized clinical study sample. Our results are qualitatively similar to those from a study comparing the same ctDNA testing platform with the physician's choice of standard tissue testing in 155 patients with untreated mCRC.20 In that study, the sensitivity and specificity for ctDNA NGS were 87.7% and 88.6% for RAS mutations and 100% and 97.6% for BRAF V600E, respectively.
We showed 12 of 106 (11.3%) patients and 3 of 25 (12.0%) patients with tissue+/ctDNA– for RAS and BRAF V600E mutations respectively, which was possibly related to low tumor shedding. Supporting this hypothesis, the sensitivity reached 97.5% and 100% for RAS and BRAF V600E mutations, respectively, in patients with a ctDNA fraction of ≥1.0%. These results were consistent with our previous study on MSI concordance, demonstrating that the sensitivity of MSI in patients with gastrointestinal cancer with a ctDNA fraction of ≥1.0% was 100% compared with overall patients whose sensitivity was 71.4%.9 Subsequently, the NPV for RAS and BRAF V600E mutations also improved in these patients. Furthermore, the sensitivity was higher in patients with liver metastases who had high ctDNA shedding, consistent with previous reports.21,22 These findings suggest that tissue testing may be required when ctDNA genotyping fails to detect RAS and BRAF mutations because of a low ctDNA fraction (eg, <1.0%).
Conversely, some patients had ctDNA+/tissue– results. ctDNA testing results can reflect the molecular characteristics of all tumor cells throughout the body, which may be heterogeneous within and between primary and metastatic sites. Intratumoral or intertumoral spatial heterogeneity of RAS/BRAF in mCRC has been reported.23-25 Previously, we have shown that lower clonality (<30%) results in a lower positive predictive value, which may reflect the spatial heterogeneity of the tumor.8 However, in the present study, clonality in patients with ctDNA+/tissue– profiles was not necessarily low, suggesting a false-negative result from tissue-based testing. In addition to spatial heterogeneity, temporal heterogeneity may contribute to testing discordance. In our study, the median interval between the tissue and ctDNA collection dates was significantly shorter in patients with concordant RAS findings than those with discordance. These findings suggest that changes in the tumor molecular profile might have occurred over time because of either natural tumor progression or exposure to selective pressures caused by chemotherapy.
The high performance of ctDNA testing for detection of RAS and BRAF V600E mutations suggests the potential of ctDNA-guided anti-EGFR and BRAF-targeted therapies although the efficacy for patients who have biomarker positivity only in ctDNA was still unknown since patients who received these treatments on the basis of tissue and ctDNA results were overlapped. Some patients each with RAS and BRAF V600E mutation who received targeted therapy on the basis of only ctDNA results without tissue-based testing achieved a long PFS, but a further validation study in a large number of patients would be needed.
There are several limitations in our study. First, we included patients who had been treated with chemotherapy before blood collection for ctDNA testing, but tumor samples used for tissue testing were collected before chemotherapy. Despite lower sensitivity and concordance for ctDNA collected from such patients, the overall study results exceeded the prespecified threshold for the primary end points. Second, we compared ctDNA-based profiling with PCR-based tissue testing currently used in practice, not with the NGS-based tissue testing. Indeed, the failure of the PCR test is one possible reason for the discordance between ctDNA and tissue tests. However, because no patients with RAS and BRAF V600E mutations detected in ctDNA but not in tissue had undergone an NGS test in our cohort, we could not address this issue. Further comparisons between ctDNA- and tissue-based NGS will be needed in the future. Finally, our study included a small number of patients treated with anti-EGFR therapy and BRAF-targeted treatment after ctDNA genotyping and most of them had consistent tissue-based results. Further investigations are required to clarify the correlation between results of ctDNA genotyping and treatment efficacy.
In conclusion, our findings validated the use of ctDNA genotyping for the detection of RAS and BRAF mutations in patients with mCRC. ctDNA NGS assay demonstrated high concordance rate, sensitivity, and specificity with an approved tissue-based test. Particularly, for patients with tumors that sufficiently shed DNA, the NPV for RAS and BRAF mutations provides confidence for the use of anti-EGFR therapy. As ctDNA can provide accurate, comprehensive, and real-time information that reflects the spatial and temporal heterogeneities of the tumor, it has the potential to guide appropriate therapy for patients with mCRC.
ACKNOWLEDGMENT
I would like to thank patients who had participated in the GOZILA study and their families, GOZILA investigators and site personnel, Translational Research Support Office in National Cancer Center Hospital East, and Guardant Health Inc.
Yoshiaki Nakamura
Honoraria: Chugai Pharma, Guardant Health AMEA, Merck
Research Funding: Taiho Pharmaceutical (Inst), Guardant Health (Inst), Genomedia (Inst), Chugai Pharma (Inst), Seagen (Inst), Roche (Inst)
Tadamichi Denda
Honoraria: Ono Pharmaceutical
Speakers' Bureau: Daiichi-Sankyo, Ono Pharmaceutical
Research Funding: Ono Pharmaceutical (Inst), Amgen (Inst), MSD (Inst), Pfizer (Inst), Bristol Myers Squibb Foundation (Inst)
Takashi Ohta
Honoraria: Bristol Myers Squibb Japan, Chugai Pharma, Teijin Pharma, Takeda, Taiho Pharmaceutical, Eisai, Yakult Honsha
Research Funding: Takeda
Taito Esaki
Honoraria: Lilly, Taiho Pharmaceutical, Daiichi Sankyo, Chugai Pharma, Bristol Myers Squibb Japan (Inst)
Research Funding: Daiichi Sankyo (Inst), MSD (Inst), Novartis (Inst), Ono Pharmaceutical (Inst), Astellas Pharma (Inst), Lilly (Inst), Bayer (Inst), Nihon Kayaku (Inst), Amgen Astellas BioPharma (Inst), Parexel (Inst), IQVIA (Inst), Quintiles (Inst), Eisai (Inst), Pfizer (Inst), Chugai Pharma (Inst), Syneos Health (Inst), Asahi Kasei (Inst), Amgen (Inst), Dainippon Sumitomo (Inst)
Manabu Shiozawa
Speakers' Bureau: Lilly Japan, Merck Serono, Taiho Pharmaceutical, Yakult Honsha, Takeda, Ono Pharmaceutical, Johnson & Johnson, Kaken Pharmaceutical
Kensei Yamaguchi
Consulting or Advisory Role: Bristol Myers Squibb Japan, Daiichi Sankyo
Speakers' Bureau: Chugai Pharma, Bristol Myers Squibb Japan, Takeda, Taiho Pharmaceutical, Lilly, Ono Pharmaceutical, Daiichi Sankyo, Merck
Research Funding: Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Daiichi Sankyo (Inst), Lilly (Inst), Gilead Sciences (Inst), Yakult Honsha (Inst), Chugai Pharma (Inst), Boehringer Ingelheim (Inst), Eisai (Inst), MSD Oncology (Inst), Sanofi (Inst), Bristol Myers Squibb (Inst)
Kentaro Yamazaki
Honoraria: Chugai Pharma, Daiichi Sankyo, Yakult Honsha, Takeda, Bayer, Merck Serono, Taiho Pharmaceutical, Lilly, Sanofi, Ono Pharmaceutical, MSD, Bristol Myers Squibb
Research Funding: Taiho Pharmaceutical (Inst)
Yu Sunakawa
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Takeda, Bayer Yakuhin, Bristol Myers Squibb Japan, Lilly Japan, Merck, Sysmex, MSD K.K, Ono Pharmaceutical, Daiichi Sankyo, Guardant Health, Incyte
Consulting or Advisory Role: Bristol Myers Squibb Japan, MSD K.K, Daiichi Sankyo, Merck
Research Funding: Taiho Pharmaceutical, Takeda, Chugai Pharma, Lilly Japan, Sanofi, Otsuka
Takeshi Kato
Honoraria: Chugai Pharma, Ono Pharmaceutical, Takeda, Lilly Japan, Asahi Kasei
Research Funding: Chugai Pharma
Naohiro Okano
Honoraria: Taiho Pharmaceutical, Bayer Yakuhin, Lilly Japan, Chugai Pharma, Ono Pharmaceutical, Takeda, Eisai, Daiichi Sankyo
Hiroya Taniguchi
Honoraria: Bayer, Sanofi, Takeda, Chugai Pharma, Taiho Pharmaceutical, Lilly Japan, Merck Serono, Yakult Honsha, Medical & Biological Laboratories Co., Ltd, Bristol Myers Squibb Japan, MSD K.K, Novartis, Daiichi Sankyo, Mitsubishi Tanabe Pharma, Nippon Kayaku, Ono Yakuhin
Research Funding: Dainippon Sumitomo Pharma (Inst), Array BioPharma (Inst), MSD Oncology (Inst), Ono Pharmaceutical (Inst), Daiichi Sankyo (Inst), Sysmex (Inst), Novartis (Inst), Takeda (Inst)
Taro Sato
Honoraria: Chugai Pharma, Merck Serono, Bristol Myers Squibb, Takeda, Yakult Honsha, Lilly, Bayer Yakuhin, Ono Pharmaceutical, Merck, Astellas Pharma, Taiho Pharmaceutical, Nihon Kayaku, Daiichi-Sankyo
Consulting or Advisory Role: Bayer Yakuhin, Lilly, Ono Pharmaceutical, Takara Bio, Merck Serono, Nihon Kayaku
Research Funding: Yakult Honsha (Inst), Chugai Pharma (Inst), Ono Pharmaceutical (Inst), Sanofi (Inst), Lilly (Inst), Daiichi Sankyo (Inst), Merck (Inst), Merck Serono (Inst), Gilead Sciences (Inst), Dainippon Sumitomo Pharma (Inst), IQVIA (Inst)
Eiji Oki
Speakers' Bureau: Chugai Pharma, Lilly Japan, Takeda, Ono Pharmaceutical, Bristol Myers Squibb Japan, Taiho Pharmaceutical
Research Funding: Guardant Health
Tomohiro Nishina
Honoraria: Taiho Pharmaceutical, Ono Pharmaceutical, Bristol Myers Squibb Japan
Research Funding: MSD (Inst), Ono Pharmaceutical (Inst), Astellas Pharma (Inst), Daiichi Sankyo/UCB Japan (Inst), Bristol Myers Squibb Japan (Inst), Chugai Pharma (Inst), Taiho Pharmaceutical (Inst), AstraZeneca (Inst)
Yoshito Komatsu
Honoraria: Lilly Japan, Taiho Pharmaceutical, Chugai Pharma, Takeda, Bayer Yakuhin, Bristol Myers Squibb Co, Sanofi/Aventis, Merck, Yakult Honsha, Ono Pharmaceutical, Nipro Corporation, Moroo Co, Asahi Kasei, Mitsubishi Tanabe Pharma, Otsuka, Medical Review Co., Ltd, Daiichi Sankyo
Research Funding: MSD K.K, Taiho Pharmaceutical, Yakult Honsha, Bayer Yakuhin, Daiichi Sankyo Co., Ltd, Ono Pharmaceutical, NanoCarrier, Eisai, Sanofi/Aventis, Sysmex, Shionogi, IQVIA, Parexel International Corporation, Astellas Pharma, Mediscience Planning, Sumitomo Dainippon Pharma Co., Ltd, A2 Healthcare, Incyte, Lilly (Inst), Nipro Corporation (Inst), BeiGene (Inst)
Nobuhisa Matsuhashi
Honoraria: AMCO, Chugai Pharma, Daiichi Sankyo, Lilly Japan, Gunze Medical Limited, Johnson & Johnson, Merck KGaA, Taiho Pharmaceutical, Yakult Honsha
Research Funding: Abbott (Inst), Asahi Kasei (Inst), Chugai Pharma (Inst), Covidien (Inst), Daiichi Sankyo (Inst), Eisai (Inst), Lilly Japan (Inst), EPS Holdings (Inst), EPS Corporation (Inst), Johnson & Johnson (Inst), Kaken Pharmaceutical (Inst), Kyowa Kirin (Inst), MSD K.K (Inst), Nippon Kayaku (Inst), Ono Pharmaceutical (Inst), Otsuka (Inst), Shift Zero (Inst), Taiho Pharmaceutical (Inst), TERUMO (Inst), Tsumura & Co (Inst)
Masahiro Goto
Honoraria: Daiichi Sankyo Company, Limited, Ono Pharmaceutical, Taiho Pharmaceutical, MSD K.K, Takeda, Sumitomo Dainippon Pharma Co., Ltd, Yakult Pharmaceutical, Eli Lilly Japan K.K
Research Funding: Chugai Pharma, Taiho Pharmaceutical, Nippon Kayaku
Hisateru Yasui
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Bristol Myers Squibb Japan, Daiichi Sankyo, Lilly Japan, Yakult Honsha, Bayer Yakuhin, Takeda, Ono Pharmaceutical
Research Funding: MSD (Inst), Ono Pharmaceutical (Inst), Astellas Pharma (Inst), AstraZeneca (Inst), Daiichi Sankyo (Inst)
Toshikazu Moriwaki
Speakers' Bureau: Taiho Pharmaceutical, Chugai Pharma, Yakult Honsha, Takeda, Merck Serono, Lilly Japan, Bayer Yakuhin, Ono Pharmaceutical
Research Funding: Taiho Pharmaceutical (Inst), MSD (Inst), Takeda (Inst), Yakult Honsha (Inst), Asahi Kasei (Inst), Isofol Medical (Inst)
Naoki Takahashi
Honoraria: Ono Pharmaceutical, Bristol Myers Squibb Japan, Taiho Pharmaceutical
Shogen Boku
Speakers' Bureau: Chugai Pharma, Daiichi Sankyo Co. Ltd, Taiho Pharmaceutical, Yakult Honsha
Research Funding: Daiichi Sankyo Co. Ltd (Inst)
Masashi Wakabayashi
Honoraria: Nihon Medi-Physics
Takayuki Yoshino
Honoraria: Chugai Pharma, Merck, Bayer Yakuhin, Ono Pharmaceutical, MSD K.K
Research Funding: MSD (Inst), Daiichi Sankyo Company, Limited (Inst), Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Amgen (Inst), Sanofi (Inst), Pfizer (Inst), Genomedia (Inst), Sysmex (Inst), Nippon Boehringer Ingelheim (Inst), Chugai Pharma (Inst)
No other potential conflicts of interest were reported.
SUPPORT
Supported by SCRUM-Japan Funds, Guardant Health AMEA.
DATA SHARING STATEMENT
To protect the privacy and confidentiality of patients in this study, clinical data and all variant data are not made publicly available in a repository or in the supplementary material of the article, but they will be available upon reasonable request to the corresponding author.
AUTHOR CONTRIBUTIONS
Conception and design: Takeshi Kato, Taro Sato, Eiji Oki, Takayuki Yoshino
Financial support: Takayuki Yoshino
Administrative support: Yoshiaki Nakamura, Nobuhisa Matsuhashi, Takayuki Yoshino
Provision of study materials or patients: Manabu Shiozawa, Kentaro Yamazaki, Takeshi Kato, Naohiro Okano, Taro Sato, Eiji Oki, Tomohiro Nishina, Yoshito Komatsu, Masahiro Goto, Hisateru Yasui, Koushiro Ohtsubo, Naoki Takahashi, Ryuta Mitani, Mihoko Yuasa, Takayuki Yoshino
Collection and assembly of data: Tadamichi Denda, Takashi Ohta, Taito Esaki, Manabu Shiozawa, Kensei Yamaguchi, Kentaro Yamazaki, Yu Sunakawa, Naohiro Okano, Hiroya Taniguchi, Taro Sato, Eiji Oki, Tomohiro Nishina, Yoshito Komatsu, Nobuhisa Matsuhashi, Masahiro Goto, Hisateru Yasui, Koushiro Ohtsubo, Toshikazu Moriwaki, Naoki Takahashi, Yosuke Horita, Ryuta Mitani, Mihoko Yuasa, Takayuki Yoshino
Data analysis and interpretation: Yu Aoki, Yoshiaki Nakamura, Kentaro Yamazaki, Takeshi Kato, Taro Sato, Eiji Oki, Yoshito Komatsu, Shogen Boku, Masashi Wakabayashi, Takashi Ikeno, Takayuki Yoshino
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Yoshiaki Nakamura
Honoraria: Chugai Pharma, Guardant Health AMEA, Merck
Research Funding: Taiho Pharmaceutical (Inst), Guardant Health (Inst), Genomedia (Inst), Chugai Pharma (Inst), Seagen (Inst), Roche (Inst)
Tadamichi Denda
Honoraria: Ono Pharmaceutical
Speakers' Bureau: Daiichi-Sankyo, Ono Pharmaceutical
Research Funding: Ono Pharmaceutical (Inst), Amgen (Inst), MSD (Inst), Pfizer (Inst), Bristol Myers Squibb Foundation (Inst)
Takashi Ohta
Honoraria: Bristol Myers Squibb Japan, Chugai Pharma, Teijin Pharma, Takeda, Taiho Pharmaceutical, Eisai, Yakult Honsha
Research Funding: Takeda
Taito Esaki
Honoraria: Lilly, Taiho Pharmaceutical, Daiichi Sankyo, Chugai Pharma, Bristol Myers Squibb Japan (Inst)
Research Funding: Daiichi Sankyo (Inst), MSD (Inst), Novartis (Inst), Ono Pharmaceutical (Inst), Astellas Pharma (Inst), Lilly (Inst), Bayer (Inst), Nihon Kayaku (Inst), Amgen Astellas BioPharma (Inst), Parexel (Inst), IQVIA (Inst), Quintiles (Inst), Eisai (Inst), Pfizer (Inst), Chugai Pharma (Inst), Syneos Health (Inst), Asahi Kasei (Inst), Amgen (Inst), Dainippon Sumitomo (Inst)
Manabu Shiozawa
Speakers' Bureau: Lilly Japan, Merck Serono, Taiho Pharmaceutical, Yakult Honsha, Takeda, Ono Pharmaceutical, Johnson & Johnson, Kaken Pharmaceutical
Kensei Yamaguchi
Consulting or Advisory Role: Bristol Myers Squibb Japan, Daiichi Sankyo
Speakers' Bureau: Chugai Pharma, Bristol Myers Squibb Japan, Takeda, Taiho Pharmaceutical, Lilly, Ono Pharmaceutical, Daiichi Sankyo, Merck
Research Funding: Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Daiichi Sankyo (Inst), Lilly (Inst), Gilead Sciences (Inst), Yakult Honsha (Inst), Chugai Pharma (Inst), Boehringer Ingelheim (Inst), Eisai (Inst), MSD Oncology (Inst), Sanofi (Inst), Bristol Myers Squibb (Inst)
Kentaro Yamazaki
Honoraria: Chugai Pharma, Daiichi Sankyo, Yakult Honsha, Takeda, Bayer, Merck Serono, Taiho Pharmaceutical, Lilly, Sanofi, Ono Pharmaceutical, MSD, Bristol Myers Squibb
Research Funding: Taiho Pharmaceutical (Inst)
Yu Sunakawa
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Takeda, Bayer Yakuhin, Bristol Myers Squibb Japan, Lilly Japan, Merck, Sysmex, MSD K.K, Ono Pharmaceutical, Daiichi Sankyo, Guardant Health, Incyte
Consulting or Advisory Role: Bristol Myers Squibb Japan, MSD K.K, Daiichi Sankyo, Merck
Research Funding: Taiho Pharmaceutical, Takeda, Chugai Pharma, Lilly Japan, Sanofi, Otsuka
Takeshi Kato
Honoraria: Chugai Pharma, Ono Pharmaceutical, Takeda, Lilly Japan, Asahi Kasei
Research Funding: Chugai Pharma
Naohiro Okano
Honoraria: Taiho Pharmaceutical, Bayer Yakuhin, Lilly Japan, Chugai Pharma, Ono Pharmaceutical, Takeda, Eisai, Daiichi Sankyo
Hiroya Taniguchi
Honoraria: Bayer, Sanofi, Takeda, Chugai Pharma, Taiho Pharmaceutical, Lilly Japan, Merck Serono, Yakult Honsha, Medical & Biological Laboratories Co., Ltd, Bristol Myers Squibb Japan, MSD K.K, Novartis, Daiichi Sankyo, Mitsubishi Tanabe Pharma, Nippon Kayaku, Ono Yakuhin
Research Funding: Dainippon Sumitomo Pharma (Inst), Array BioPharma (Inst), MSD Oncology (Inst), Ono Pharmaceutical (Inst), Daiichi Sankyo (Inst), Sysmex (Inst), Novartis (Inst), Takeda (Inst)
Taro Sato
Honoraria: Chugai Pharma, Merck Serono, Bristol Myers Squibb, Takeda, Yakult Honsha, Lilly, Bayer Yakuhin, Ono Pharmaceutical, Merck, Astellas Pharma, Taiho Pharmaceutical, Nihon Kayaku, Daiichi-Sankyo
Consulting or Advisory Role: Bayer Yakuhin, Lilly, Ono Pharmaceutical, Takara Bio, Merck Serono, Nihon Kayaku
Research Funding: Yakult Honsha (Inst), Chugai Pharma (Inst), Ono Pharmaceutical (Inst), Sanofi (Inst), Lilly (Inst), Daiichi Sankyo (Inst), Merck (Inst), Merck Serono (Inst), Gilead Sciences (Inst), Dainippon Sumitomo Pharma (Inst), IQVIA (Inst)
Eiji Oki
Speakers' Bureau: Chugai Pharma, Lilly Japan, Takeda, Ono Pharmaceutical, Bristol Myers Squibb Japan, Taiho Pharmaceutical
Research Funding: Guardant Health
Tomohiro Nishina
Honoraria: Taiho Pharmaceutical, Ono Pharmaceutical, Bristol Myers Squibb Japan
Research Funding: MSD (Inst), Ono Pharmaceutical (Inst), Astellas Pharma (Inst), Daiichi Sankyo/UCB Japan (Inst), Bristol Myers Squibb Japan (Inst), Chugai Pharma (Inst), Taiho Pharmaceutical (Inst), AstraZeneca (Inst)
Yoshito Komatsu
Honoraria: Lilly Japan, Taiho Pharmaceutical, Chugai Pharma, Takeda, Bayer Yakuhin, Bristol Myers Squibb Co, Sanofi/Aventis, Merck, Yakult Honsha, Ono Pharmaceutical, Nipro Corporation, Moroo Co, Asahi Kasei, Mitsubishi Tanabe Pharma, Otsuka, Medical Review Co., Ltd, Daiichi Sankyo
Research Funding: MSD K.K, Taiho Pharmaceutical, Yakult Honsha, Bayer Yakuhin, Daiichi Sankyo Co., Ltd, Ono Pharmaceutical, NanoCarrier, Eisai, Sanofi/Aventis, Sysmex, Shionogi, IQVIA, Parexel International Corporation, Astellas Pharma, Mediscience Planning, Sumitomo Dainippon Pharma Co., Ltd, A2 Healthcare, Incyte, Lilly (Inst), Nipro Corporation (Inst), BeiGene (Inst)
Nobuhisa Matsuhashi
Honoraria: AMCO, Chugai Pharma, Daiichi Sankyo, Lilly Japan, Gunze Medical Limited, Johnson & Johnson, Merck KGaA, Taiho Pharmaceutical, Yakult Honsha
Research Funding: Abbott (Inst), Asahi Kasei (Inst), Chugai Pharma (Inst), Covidien (Inst), Daiichi Sankyo (Inst), Eisai (Inst), Lilly Japan (Inst), EPS Holdings (Inst), EPS Corporation (Inst), Johnson & Johnson (Inst), Kaken Pharmaceutical (Inst), Kyowa Kirin (Inst), MSD K.K (Inst), Nippon Kayaku (Inst), Ono Pharmaceutical (Inst), Otsuka (Inst), Shift Zero (Inst), Taiho Pharmaceutical (Inst), TERUMO (Inst), Tsumura & Co (Inst)
Masahiro Goto
Honoraria: Daiichi Sankyo Company, Limited, Ono Pharmaceutical, Taiho Pharmaceutical, MSD K.K, Takeda, Sumitomo Dainippon Pharma Co., Ltd, Yakult Pharmaceutical, Eli Lilly Japan K.K
Research Funding: Chugai Pharma, Taiho Pharmaceutical, Nippon Kayaku
Hisateru Yasui
Honoraria: Taiho Pharmaceutical, Chugai Pharma, Bristol Myers Squibb Japan, Daiichi Sankyo, Lilly Japan, Yakult Honsha, Bayer Yakuhin, Takeda, Ono Pharmaceutical
Research Funding: MSD (Inst), Ono Pharmaceutical (Inst), Astellas Pharma (Inst), AstraZeneca (Inst), Daiichi Sankyo (Inst)
Toshikazu Moriwaki
Speakers' Bureau: Taiho Pharmaceutical, Chugai Pharma, Yakult Honsha, Takeda, Merck Serono, Lilly Japan, Bayer Yakuhin, Ono Pharmaceutical
Research Funding: Taiho Pharmaceutical (Inst), MSD (Inst), Takeda (Inst), Yakult Honsha (Inst), Asahi Kasei (Inst), Isofol Medical (Inst)
Naoki Takahashi
Honoraria: Ono Pharmaceutical, Bristol Myers Squibb Japan, Taiho Pharmaceutical
Shogen Boku
Speakers' Bureau: Chugai Pharma, Daiichi Sankyo Co. Ltd, Taiho Pharmaceutical, Yakult Honsha
Research Funding: Daiichi Sankyo Co. Ltd (Inst)
Masashi Wakabayashi
Honoraria: Nihon Medi-Physics
Takayuki Yoshino
Honoraria: Chugai Pharma, Merck, Bayer Yakuhin, Ono Pharmaceutical, MSD K.K
Research Funding: MSD (Inst), Daiichi Sankyo Company, Limited (Inst), Ono Pharmaceutical (Inst), Taiho Pharmaceutical (Inst), Amgen (Inst), Sanofi (Inst), Pfizer (Inst), Genomedia (Inst), Sysmex (Inst), Nippon Boehringer Ingelheim (Inst), Chugai Pharma (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1. Douillard J-Y, Oliner KS, Siena S, et al. Panitumumab–FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 2. Douillard J-Y, Siena S, Cassidy J, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: The PRIME study. J Clin Oncol. 2010;28:4697–4705. doi: 10.1200/JCO.2009.27.4860. [DOI] [PubMed] [Google Scholar]
- 3. Kopetz S, Grothey A, Yaeger R, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E–mutated colorectal cancer. N Engl J Med. 2019;381:1632–1643. doi: 10.1056/NEJMoa1908075. [DOI] [PubMed] [Google Scholar]
- 4. André T, Shiu K-K, Kim TW, et al. Pembrolizumab in microsatellite-instability–high advanced colorectal cancer. N Engl J Med. 2020;383:2207–2218. doi: 10.1056/NEJMoa2017699. [DOI] [PubMed] [Google Scholar]
- 5. Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair–deficient/microsatellite instability–high metastatic colorectal cancer. J Clin Oncol. 2018;36:773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 6. Sartore-Bianchi A, Trusolino L, Martino C, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): A proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17:738–746. doi: 10.1016/S1470-2045(16)00150-9. [DOI] [PubMed] [Google Scholar]
- 7. Nakamura Y, Okamoto W, Kato T, et al. Circulating tumor DNA-guided treatment with pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer: A phase 2 trial. Nat Med. 2021;27:1899–1903. doi: 10.1038/s41591-021-01553-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakamura Y, Taniguchi H, Ikeda M, et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med. 2020;26:1859–1864. doi: 10.1038/s41591-020-1063-5. [DOI] [PubMed] [Google Scholar]
- 9. Nakamura Y, Okamoto W, Denda T, et al. Clinical validity of plasma-based genotyping for microsatellite instability assessment in advanced GI cancers: SCRUM-Japan GOZILA substudy. JCO Precis Oncol. 2022 doi: 10.1200/PO.21.00383. 10.1200/PO.21.00383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.guardant360cdx.com 2023/2/12 21:50:45.pdf [Internet] https://guardant360cdx.com/wp-content/uploads/securepdfs/2023/01/Guardant360-CDx-Technical-Information-US.pdf [Google Scholar]
- 11. Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24:3539–3549. doi: 10.1158/1078-0432.CCR-17-3831. [DOI] [PubMed] [Google Scholar]
- 12. Taniguchi H, Okamoto W, Muro K, et al. Clinical validation of newly developed multiplex kit using Luminex xMAP technology for detecting simultaneous RAS and BRAF mutations in colorectal cancer: Results of the RASKET-B study. Neoplasia. 2018;20:1219–1226. doi: 10.1016/j.neo.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thierry AR, Mouliere F, El Messaoudi S, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med. 2014;20:430–435. doi: 10.1038/nm.3511. [DOI] [PubMed] [Google Scholar]
- 14. Bando H, Kagawa Y, Kato T, et al. A multicentre, prospective study of plasma circulating tumour DNA test for detecting RAS mutation in patients with metastatic colorectal cancer. Br J Cancer. 2019;120:982–986. doi: 10.1038/s41416-019-0457-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. García-Foncillas J, Alba E, Aranda E, et al. Incorporating BEAMing technology as a liquid biopsy into clinical practice for the management of colorectal cancer patients: An expert taskforce review. Ann Oncol. 2017;28:2943–2949. doi: 10.1093/annonc/mdx501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ueda K, Yamada T, Ohta R, et al. BRAF V600E mutations in right-side colon cancer: Heterogeneity detected by liquid biopsy. Eur J Surg Oncol. 2022;48:1375–1383. doi: 10.1016/j.ejso.2022.01.016. [DOI] [PubMed] [Google Scholar]
- 17. Wang F, Huang Y-S, Wu H-X, et al. Genomic temporal heterogeneity of circulating tumour DNA in unresectable metastatic colorectal cancer under first-line treatment. Gut. 2022;71:1340–1349. doi: 10.1136/gutjnl-2021-324852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mas L, Bachet J-B, Taly V, et al. BRAF mutation status in circulating tumor DNA from patients with metastatic colorectal cancer: Extended mutation analysis from the AGEO RASANC study. Cancers. 2019;11:998. doi: 10.3390/cancers11070998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bachet JB, Bouché O, Taieb J, et al. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: The AGEO RASANC prospective multicenter study. Ann Oncol. 2018;29:1211–1219. doi: 10.1093/annonc/mdy061. [DOI] [PubMed] [Google Scholar]
- 20. Benavides M, Alcaide-Garcia J, Torres E, et al. Clinical utility of comprehensive circulating tumor DNA genotyping compared with standard of care tissue testing in patients with newly diagnosed metastatic colorectal cancer. ESMO Open. 2022;7:100481. doi: 10.1016/j.esmoop.2022.100481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bando H, Nakamura Y, Taniguchi H, et al. Effects of metastatic sites on circulating tumor DNA in patients with metastatic colorectal cancer. JCO Precis Oncol. 2022 doi: 10.1200/PO.21.00535. 10.1200/PO.21.00535 [DOI] [PubMed] [Google Scholar]
- 22. Kagawa Y, Elez E, García-Foncillas J, et al. Combined analysis of concordance between liquid and tumor tissue biopsies for RAS mutations in colorectal cancer with a single metastasis site: The METABEAM study. Clin Cancer Res. 2021;27:2515–2522. doi: 10.1158/1078-0432.CCR-20-3677. [DOI] [PubMed] [Google Scholar]
- 23. Yamada T, Iwai T, Takahashi G, et al. Utility of KRAS mutation detection using circulating cell‐free DNA from patients with colorectal cancer. Cancer Sci. 2016;107:936–943. doi: 10.1111/cas.12959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmiegel W, Scott RJ, Dooley S, et al. Blood‐based detection of RAS mutations to guide anti‐EGFR therapy in colorectal cancer patients: Concordance of results from circulating tumor DNA and tissue‐based RAS testing. Mol Oncol. 2017;11:208–219. doi: 10.1002/1878-0261.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grasselli J, Elez E, Caratù G, et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol. 2017;28:1294–1301. doi: 10.1093/annonc/mdx112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
To protect the privacy and confidentiality of patients in this study, clinical data and all variant data are not made publicly available in a repository or in the supplementary material of the article, but they will be available upon reasonable request to the corresponding author.