PURPOSE
Acute lymphoblastic leukemia (ALL) is the most prevalent cause of childhood cancer and requires a long course of therapy consisting of three primary phases with interval intensification blocks. Although these phases are necessary to achieve remission, the primary chemotherapeutic agents have potentially serious toxicities, which may lead to delays or discontinuations of therapy. The purpose of this study was to perform a comprehensive pharmacogenomic evaluation of common antileukemic agents and develop a polygenic toxicity risk score predictive of the most common toxicities observed during ALL treatment.
METHODS
This cross-sectional study included 75 patients with pediatric ALL treated between 2012 and 2020 at the University of Florida. Toxicity data were collected within 100 days of initiation of therapy using CTCAE v4.0 for toxicity grading. For pharmacogenomic evaluation, single-nucleotide polymorphisms (SNPs) and genes were selected from previous reports or PharmGKB database. 116 unique SNPs were evaluated for incidence of various toxicities. A multivariable multi-SNP modeling for up to 3-SNP combination was performed to develop a polygenic toxicity risk score of prognostic value.
RESULTS
We identified several SNPs predictive of toxicity phenotypes in univariate analysis. Further multivariable SNP-SNP combination analysis suggest that susceptibility to chemotherapy-induced toxicities is likely multigenic in nature. For 3-SNPscore models, patients with high scores experienced increased risk of GI (P = 2.07E-05, 3 SNPs: TYMS-rs151264360/FPGS-rs1544105/GSTM1-GSTM5-rs3754446), neurologic (P = .0005, 3 SNPs: DCTD-rs6829021/SLC28A3-rs17343066/CTPS1-rs12067645), endocrine (P = 4.77E-08, 3 SNPs: AKR1C3-rs1937840/TYMS-rs2853539/CTH-rs648743), and heme toxicities (P = .053, 3 SNPs: CYP3A5-rs776746/ABCB1-rs4148737/CTPS1-rs12067645).
CONCLUSION
Our results imply that instead of a single-SNP approach, SNP-SNP combinations in multiple genes in drug pathways increases the robustness of prediction of toxicity. These results further provide promising SNP models that can help establish clinically relevant biomarkers allowing for greater individualization of cancer therapy to maximize efficacy and minimize toxicity for each patient.
INTRODUCTION
Acute lymphoblastic leukemia (ALL) is the leading cause of childhood cancer, representing approximately 25% of all cancers in patients younger than 15 years.1 Cure rates have improved from 10% in the 1960s to more than 90% in contemporary clinical trials.2,3 Standard treatment regimens consist of prolonged cytotoxic therapy administered over three primary phases—remission induction, consolidation, and maintenance—with interval intensification blocks.4 This approach, although effective, produces a myriad of chronic health consequences for survivors.5 Interpatient variation in pharmacogenomics can result in unpredictable variability in occurrence of adverse events and toxicity as well as discrepancies in therapeutic efficacy. Dose-limiting toxicities lead to modifications in treatment regimens and dosing schedules. Data have shown these delays and omissions can affect long-term outcomes in patients with pediatric cancer6. Studies in leukemia cohorts have uncovered a need for less toxic approaches without compromising efficacy.
CONTEXT
Key Objective
Patients with acute lymphoblastic leukemia (ALL) are treated with intensive chemotherapy that results in severe toxicities, which can sometimes result in delays or discontinuations of therapy. The objective of this study was to comprehensively evaluate pharmacogenomics of antileukemic agents and establish a polygenic toxicity risk score predictive of the common toxicities observed during ALL treatment.
Knowledge Generated
We took a pharmacological pathway–based pharmacogenomics approach and identified several single-nucleotide polymorphisms (SNPs) associated with individual toxicities. A multi-SNP predictor modeling approach established a 3-SNP combination score with significant association with specific toxicities.
Relevance
Multi-SNP combination approach is more robust and takes into account multiple SNPs predictive of toxicity and thus holds significant clinical relevance. Promising SNP models can help establish clinically relevant biomarkers that can be used preemptively and monitor risk of toxicity and accordingly design interventions to reduce toxicity and improve quality of life in patients with ALL.
Pharmacogenomic biomarkers are an evolving area that may help identify patient-specific factors affecting responses to chemotherapeutic agents. Inherited variation in genes involved in drug metabolism and transport have been described in a multitude of modern drugs.7-11 Genetic polymorphisms can influence the gene expression and/or activity, thereby affecting drug pharmacokinetics and causing interindividual variation in drug levels, which can alter toxicity phenotype and therapeutic efficacy.12 To date, the most understood example of this in pediatric cancer is thiopurine S-methyltransferase (TPMT) and nudix hydrolase 15 (NUDT15) activities.13 First described as early as the 1980s,14 a total of 21 TPMT genetic polymorphisms have since been described and are associated with decreased levels of TPMT enzyme activity and/or thiopurine drug-induced toxicity.15 This knowledge led to standardized practices evaluating TPMT and NUDT15 genetic polymorphisms in patients to tailor dosing before the initiation of purines on the basis of these genetic variations.12,16,17 Numerous efforts have been made to replicate these findings in other key genes.18,19
Herein, we sought to describe single-nucleotide polymorphism (SNP) variants as pharmacogenomic biomarkers predictive of treatment toxicity phenotypes in a cohort of children with ALL. Characterization of such variations could establish clinically relevant predictors, allowing for personalized leukemia therapy tailored toward optimizing drug efficacy while lessening toxicities. The objective of this study was to identify SNPs in target genes associated with drug metabolism or transport that could predict undue toxicity from antileukemic agents in children with ALL.
METHODS
Study Design and Patients
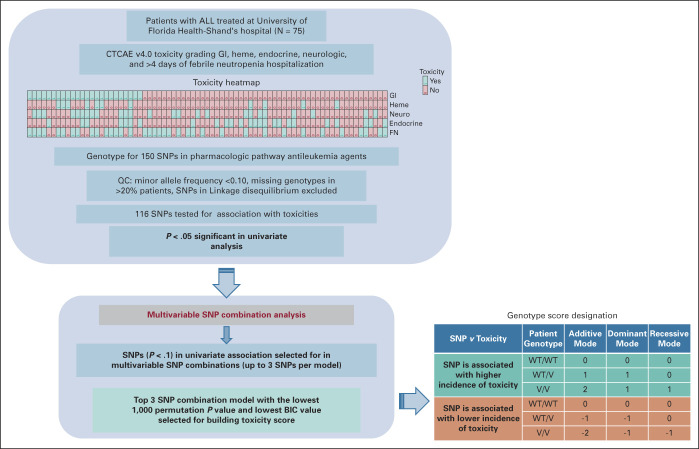
This was a cross-sectional study of subjects treated at a tertiary academic center. Overall study design is shown in Figure 1. Patients age 3 months to 26 years with a diagnosis of de novo or secondary ALL who received induction and consolidation therapy between May 2012 and December 2019 at the University of Florida were eligible for enrollment, regardless of disease risk category, sex, or racial or ethnic background, for enrollment. Study enrollment occurred between February 2019 and May 2020 at the University of Florida. Patients who met eligibility criteria were excluded if they declined participation, or if they were unable to provide an adequate blood specimen. The study protocol was approved by the University of Florida Institutional Review Board (IRB#201802623). All patients provided written informed consent before participating in the study and received treatment according to standard-of-care options at the discretion of their treating physician. To be eligible for assessment, patients were required to have received induction and consolidation chemotherapy at the University of Florida. A total of 75 patients treated between 2012 and 2020 were included in the study.
FIG 1.
Overall study design. In table, 0 = no toxicity and 1 = toxicity as per description in the methods section. FN, Febrile Neutropenia; GI, gastrointestinal; Heme, hematological toxicity; Neuro, neurological toxicity; QC, quality control; SNP, single-nucleotide polymorphism; V, variant; WT, wildtype.
Comprehensive Pharmacogenomic Evaluation
A peripheral blood sample (5-10 mL) for pharmacogenomic testing was collected at a single time point during routine follow-up care. Blood samples were stored in a malignant hematology biorepository for subsequent genomic studies. Genomic DNA was isolated from the samples for further genotyping. 150 SNPs in key candidate genes involved in cellular transport and metabolism of cytarabine, vincristine, methotrexate, daunorubicin/doxorubicin, and mercaptopurine/thioguanine were analyzed. Sequenom genotyping that uses matrix-assisted laser desorption/ionization-time of flight–based chemistry was performed at University of Minnesota, Biomedical Genomics Center. Genes involved in pharmacological chemotherapy agents were obtained from PharmGKB.20 Literature search as well as information from PharmGKB20 was used to select the SNPs. After excluding 27 SNPs with minor-allele frequency (MAF) < 0.1, 1 SNP missing genotypes in >20% of the samples, and 6 SNPs that occurred in linkage disequilibrium (LD) with at least one another SNP, a total of 116 unique SNPs (listed in Appendix Table A1 [Supplementary Table 1]) were included in the study. Toxicities were documented in real time by the primary treatment team according to standard-of-care practices and additionally confirmed by physicians on the study team and included the following: hepatic injury defined by increase in serum total bilirubin >3.0 times the upper normal limit; hematologic impairment including severe neutropenia, thrombocytopenia, and anemia leading to treatment delays; number of hospitalizations for febrile neutropenia (FN); thromboembolic events requiring medical intervention; pancreatitis requiring medical intervention; neurotoxicity defined by detailed neurologic examination and alterations in function; and glucose resistance defined by insulin dependence. Overall, CTCAE v4.0 was used for toxicity grading, and all toxicity events during the first 100 days of therapy that include gastrointestinal (GI), hematologic, neurologic, endocrine toxicities, and prolonged hospitalization (>4 days) because of FN were included in the analysis. Logistic regression models were used to test the association between the 116 SNPs in additive, dominant, and recessive modes of inheritance with all different types of toxicities. Odds ratio (OR) and 95% CI were calculated for each test. P < .05 was considered significant. In this exploratory study, no adjustment for multiple testing was done. For multivariable SNP combination analysis, SNPs with univariate association P < .1 were selected for each toxicity, and then SNP combinations (up to 3 SNPs per model) were tested for association with each toxicity. The combination model with the 1,000 permutation P < .05 and lowest Bayesian information criterion (BIC) value was selected to build a 3-SNP score after considering the mode of inheritance and the direction of association with the toxicity for the individual genotypes. 3-SNP score was generated by summation of genotype scores for the individual SNPs passing the top model. Patients were further classified into three groups: 3SNP_Tox score group of “>0,” “0,” or “<0.” Chi-square and the Cochran-Armitage trend tests were used to test for the association between the toxicity risk score groups and the incidence of each of the evaluated toxicities.
RESULTS
Demographics and Clinical Comparisons
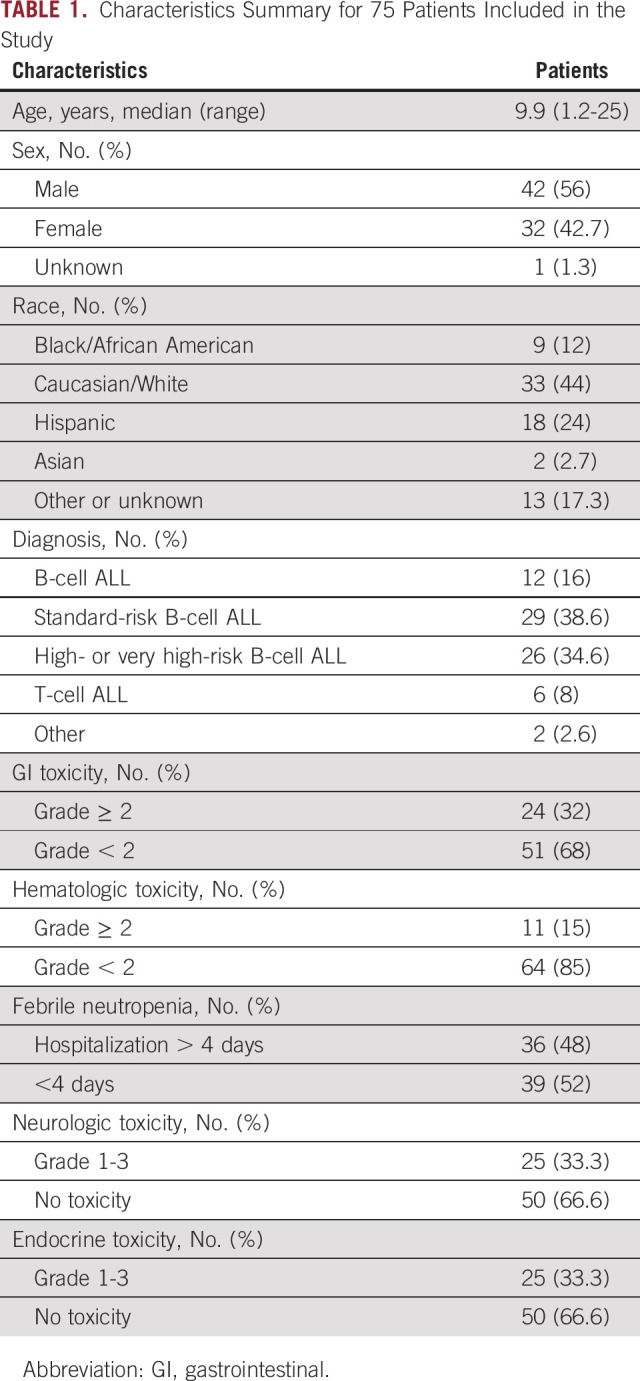
Table 1 summarizes demographics and incidence of toxicities. Twenty-five (33%) patients had neurologic and endocrinologic toxicities (grade 1-3). Twenty-four patients had GI toxicities (grades 2-4), 11 (15%) patients had hematologic toxicities (grade 2-4), and 36 patients were hospitalized at least one time >4 days because of FN.
TABLE 1.
Characteristics Summary for 75 Patients Included in the Study
Univariate Analysis of SNP Association With Toxicity Risk
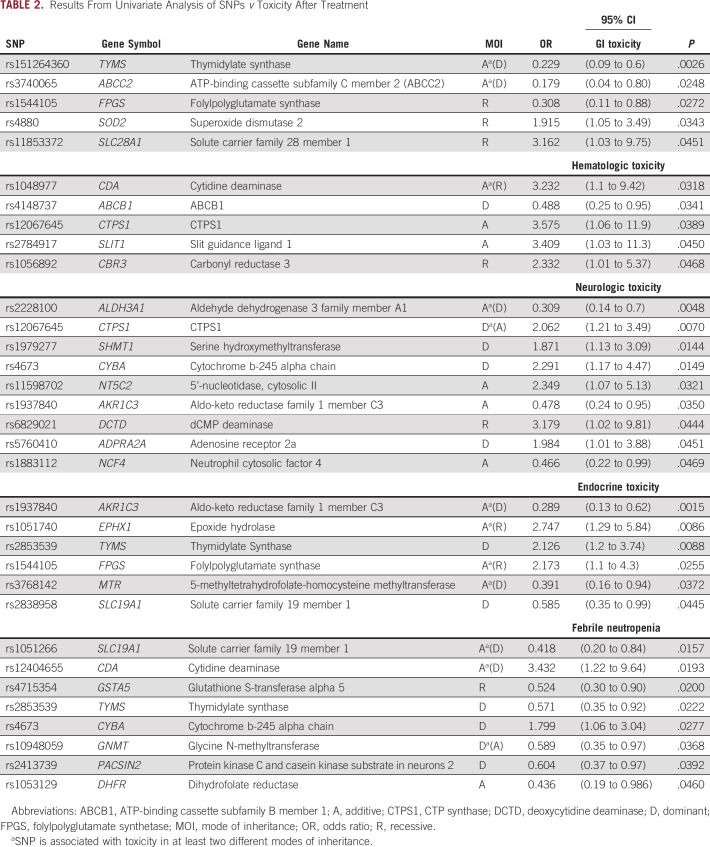
Table 2 provides summary of the univariate analysis. At P < .05, 5 unique SNPs were found significantly associated with GI toxicity, 5 SNPs were associated with hematologic toxicities, 9 SNPs were associated with neurologic toxicity, 6 SNPs were associated with endocrine toxicities, and 8 SNPs were found associated with prolonged hospitalization because of FN. For GI toxicities, variant alleles for SNPs rs151264360 in TYMS, rs3740065 in ABCC2, and rs1544105 in folylpolyglutamate synthetase (FPGS) were found associated with reduced risk of toxicity (OR < 1), while rs11853372 in SLC28A1 and rs4880 in SOD2 were found associated with higher risk of toxicity (OR > 1). For hematologic toxicities, SNPs in CDA (rs1048977), CTPS1 (rs12067645), SLIT1 (rs2784917), and CBR3 (rs1056892) were associated with higher risk of toxicity, while ABCB1 SNP rs4148737 was associated with lower risk of toxicity. Higher risk of neurologic toxicity was associated with variant alleles of rs12067645 (CTPS1), rs1979277 (SHMT1), rs4673 (CYBA), rs11598702 (NT5C2), rs6829021 deoxycytidine deaminase (DCTD), and rs5760410 (ADPRA2A), while rs2228100 (ALDH3A1), rs1883112 (NCF4), and rs1937840 (AKR1C3) were found associated with lower risk of neurologic toxicity. With respect to endocrine toxicity, variant alleles of 3 SNPs (rs1051740 in EPHX1, rs2853539 in TYMs, and rs1544105 in FPGS) were found associated with higher risk of toxicity, while rs1937840 in AKR1C3 and rs2838958 in SLC19A1 were associated with lower risk of toxicity. We also tested length of hospitalization for FN for association with polymorphism in genes important in metabolism of chemotherapy. rs12404655 in CDA and rs4673 in CYBA were associated with higher risk of > 4 days of hospitalization, whereas six other SNPs were predictive of lower risk (rs1051266 in SLC19A1, rs4715354 in GSTA5, rs2853539 in TYMS, rs10948059 in GNMT, rs1053129 in DHFR, and rs2413739 in PACSIN2).
TABLE 2.
Results From Univariate Analysis of SNPs v Toxicity After Treatment
Development of Pharmacogenomics Toxicity Risk Score Models
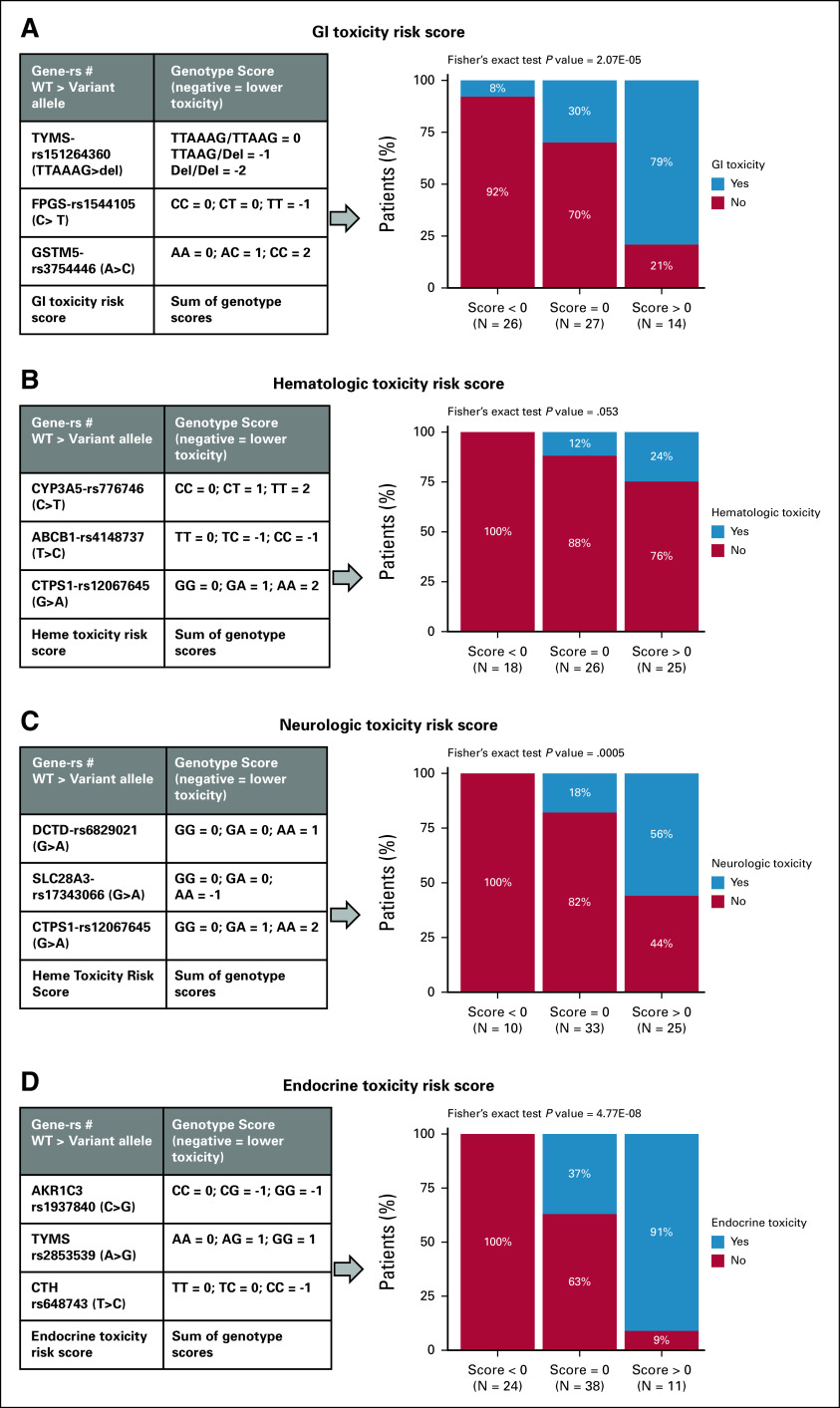
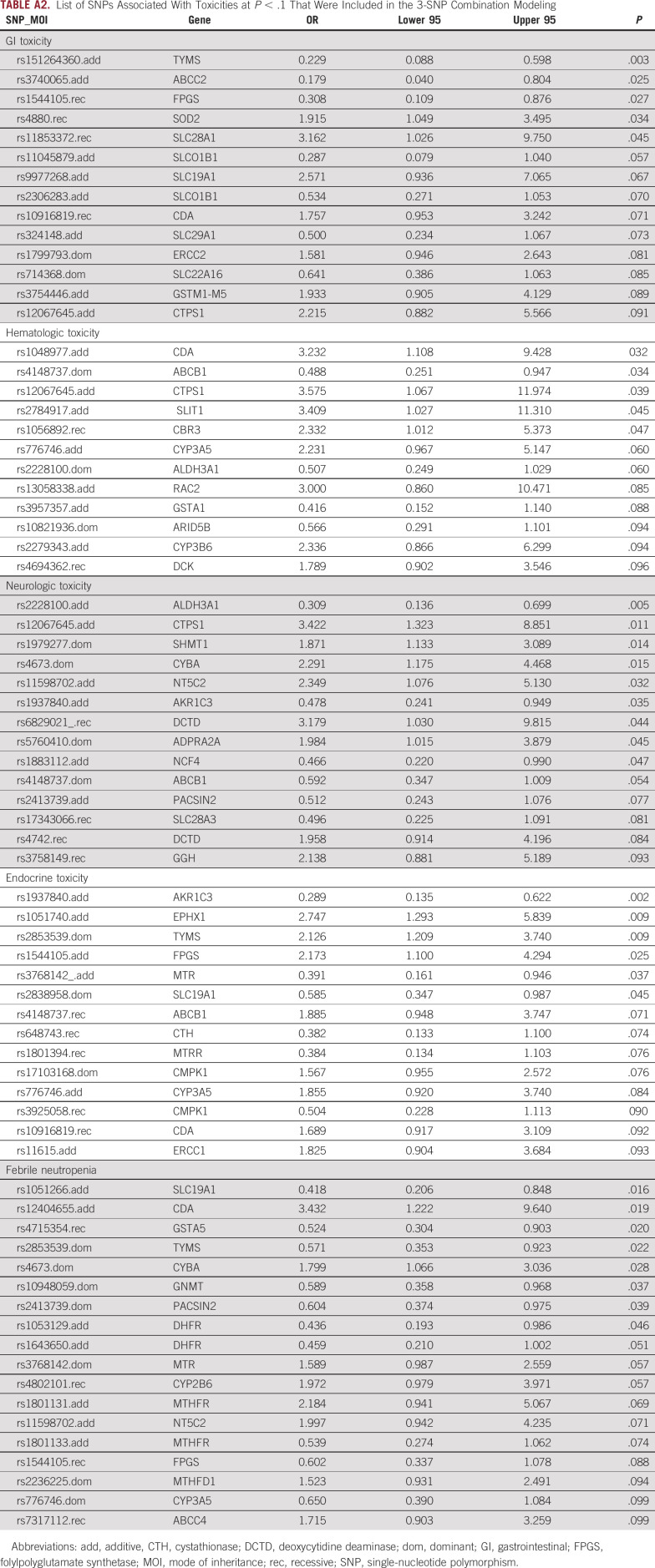
To enhance the clinical utility of the pharmacogenomic discoveries, we performed a multi-SNP predictor modeling to create a pharmacogenomics toxicity risk score as described in Methods (Appendix Table A2 [Supplementary Table 2] provides the list of SNPs included in the modeling as explained above). The best 3-SNP predictor model with lowest BIC and 1,000 permutations test P value of < .05 was selected for each toxicity. Toxicity score was created by adding the genotype scores of the 3 SNPs in the top model with consideration of a direction of association with toxicity (negative for lower toxicity and positive for higher toxicity) as well as mode of inheritance. Overall, higher score meant higher incidence of toxicity.
The top 3-SNP model predictor of GI toxicity included rs151264360 in TYMS (TTAAAG > del), rs1544105 in FPGS (C>T), and rs3754446 GSTM5 (A>C). GI toxicity score was created as shown in Figure 2A. Patients were classified into three groups: score <0, score = 0, and score >0. As shown in the bar plot, patients within >0 score group had higher incidence of GI toxicity compared with those in score <0 or = 0 (GI toxicity incidence: 8% v 30% v 79% in score <0, = 0 or >0, respectively; P = 2.07E-05). The Cochran-Armitage trend test, which tests if the trend of incidence of toxicity increases by increasing score, was also significant for GI toxicity (P = 3.89E-06). For hematologic toxicity, the top model consisted of CYP3A5-rs776746 (C>T), ABCB1-rs4148737 (T>C), and CTPS1-rs12067645 (G>A). Figure 2B shows the genotype scores for SNPs used to create the toxicity score. As shown in the bar plot, none of the patients with score <0 experienced toxicity, whereas for patients with score >0 around 24%, patients experienced significant hematologic toxicity (heme toxicity incidence: 0% v 12% v 24% in score <0, = 0, or >0, respectively; Fisher's exact P = .053; Cochran-Armitage trend test P = .01). All 3 SNPs identified in the model for predicting neurologic toxicities belong to the cytarabine metabolic pathway. Top model included rs6829021 (G>A) in DCTD, rs17343066 (G>A) in uptake transporter SLC28A3, and rs12067645 (G>A) in CTPS1 (Fig 2C). As shown in the bar plot (Figs 2C), none of the patients with score <0 experienced toxicity, while 56% of patients with score >0 experienced significant neurologic toxicity (neurologic toxicity incidence: 0% v 18% v 56% in score <0, = 0 or >0, respectively; Fisher's exact P = .0005; Cochran-Armitage trend test P = 9.27E-05). For endocrine toxicity, top multi-SNP predictor model consisted of AKR1C3-rs1937840(C>G), TYMS-rs2853539(A>G), and cystathionase (CTH)-rs648743(T>C). Again, none of the patients with score <0 experienced any endocrine toxicity compared with 91% of patients with score >0 (endocrine toxicity incidence: 0% v 37% v 91% in score <0, = 0 or >0, respectively; Fisher's exact P = 4.77E-08; Cochran-Armitage trend test P = 5.34E-08; Figure 2D). For FN, the score was not generated as none of the 3 SNP-based model's reached permuted P < .05.
FIG 2.
Toxicity incidence by respective composite 3-SNP score groups within patients with ALL. Top model 3 SNPs that were used to create each toxicity SNP score and bar plot of score group versus incidence of toxicity is shown for (A) GI toxicity, n = 67, (B) hematologic toxicity, n = 71, (C) neurologic toxicity, n = 68, (D) and endocrine toxicity, n = 73. Toxicity risk score was computed just for patients with genotype data available for the 3 SNPs in the model. P values for Fisher's exact test are listed on top of each bar plot. GI, gastrointestinal; SNP, single-nucleotide polymorphism.
DISCUSSION
Although ALL is one of the most successfully treated pediatric malignancies, with the current survival rate >90%, its treatment is related to numerous and sometimes life-threatening toxicities. With the progressive integration of immunotherapy in contemporary clinical trials for leukemia, a greater understanding of toxicities from historical antileukemic agents may allow researchers to tailor future treatment approaches to optimize the balance between standard-of-care chemotherapy and novel agents without sacrificing efficacy. This study focused on the most common chemotherapeutic agents used in the treatment of patients with ALL, and instead of taking a single gene-single drug approach, we performed a comprehensive pharmacogenomics evaluation of key genes implicated in the metabolic pathways of these chemotherapeutic agents. Relationships between SNPs within TPMT and NUDT15 genes and hematologic toxicities have been well established and implemented in standard of care to guide mercaptopurine/thioguanine dosing (Clinical Pharmacogenomics Implementation Consortium Guidelines).17,21 In our cohort, the NUDT15 and TPMT SNPs occurred at a very low frequency. Herein, we focused on common genetic polymorphisms (minor allele frequency of >0.1) within genes relevant in metabolism of chemotherapeutic agents used in ALL. Our focus was on toxicities observed within 100 days of treatment initiation as this time period is critical in obtaining disease remission. The results show a significant association between SNPs in genes of pharmacologic significance to chemotherapeutic agents with toxicities experienced in patients with pediatric ALL.
To enhance the prediction by co-occurrence of multiple variants within a patient, we performed a multi-SNP predictor modeling to identify the most significant 3-SNP combination that is predictive of a particular toxicity incidence. The top 3-SNP model predictive of GI toxicity included (1) rs151264360 is a 6 bp deletion (TTAAAG > del) in thymidylate synthase (TYMS). TYMS catalyzes methylation of dUMP to dTMP and is targeted by methotrexate. This SNP is also referred to as rs11280056 (as a 9 bp deletion), rs34489327, or rs16430 in many publications, and the deletion has been associated with reduced mRNA stability and TYMS expression. Association of this SNP with toxicity and outcome in patients with rheumatoid arthritis receiving methotrexate and patients with cancer treated with methotrexate or other anticancer agents is summarized in the PharmGKB database.20 Its association with reduced toxicity in multiple studies in rheumatoid arthritis22,23 is consistent with our results. (2) rs1544105 (C>T) in FPGS, another gene of relevance to methotrexate. TT genotype of this SNP has been associated with increased response compared with CC and CT genotypes in patients with ALL24,25; however, associations with toxicity has not been reported; and (3) rs3754446 (A>C) maps to GSTM1-GSTM5 locus. GST family of genes is involved in metabolism of wide range of drugs and this SNP has previously associated with outcome in patients with AML.26 For hematologic toxicity, the top 3-SNPs included in the top were the following: (1) rs776746 (C>T, with C allele designated as *3 allele) is the most studied functional SNP in the drug metabolizing enzyme CYP3A5. rs776746 is a splicing SNP, and presence of the C allele (which is more abundant in Caucasian ancestry) results in loss of CYP3A5 expression; (2) rs4148737 (T>C) occurs in a multidrug transporter ABCB1 (also known as PgP1) and has been implicated in efflux of wide range of drugs; and (3) rs12067645 (G>A) is in CTP synthase (CTPS1), which is involved in pyrimidine synthesis and has been associated with cytarabine metabolic pathway. As indicated before, all 3 SNPs in neurologic toxicity mapped to cytarabine metabolic pathway genes and included (1) rs6829021 (G>A) in inactivating enzyme DCTD and (2) rs17343066 (G>A) in uptake transporter SLC28A3. Our group has previously shown this SNP to be associated with intracellular ara-CTP levels in patients with AML.10 SLC28A3 has also been implication in thiopurine and (3) rs12067645 (G>A) in CTPS1 implicated in pyrimidine synthesis. Endocrine toxicity model included 3-SNPs: (1) rs1937840(C>G), in aldoketoreductase 1C3 (AKR1C3), a member of NAD(P)H oxidoreductase. AKR1C3 has been implicated in multiple malignancies including leukemias and is also involved in the metabolism of anthracyclines. This SNP has been associated with increased response to docetaxel and doxorubicin in breast cancer27; (2) rs2853539(A>G) in TYMS, a target of methotrexate. AA genotype for this SNP has been associated with reduced methotrexate response in rheumatoid arthritis previously,25 and (3) rs648743(T>C) in CTH involved in glutathione synthesis, which has previously been associated with sinusoidal obstruction in transplant patients.28 Although FN is one of the life-threatening toxicities and we did identify 8 SNPs predictive of patients receiving >4 days of hospitalization because of FN, none of the multi-SNP predictor model passed the permutated P value threshold of < .05. So, at this time, we did not create a multi-SNP score for this toxicity. One of the reasons for this might be the limited sample size of the study cohort.
Development of the toxicity score by taking direction of association of the SNP with toxicity risks (positive for higher toxicity risk) and mode of inheritance (additive, dominant, or recessive), we propose a pharmacogenomics-based toxicity score for each type of toxicity. Our results show that each described high multi-gene/SNP-based toxicity risk score is significantly associated with a higher incidence of toxicity.
A limitation of the current study was a limited sample size, warranting validation of these findings in a larger cohort of patients with ALL. Additionally, our cohort, although reflective of patients seen at our center, has ethnicity bias with more patients reflective of Caucasian and Hispanic ethnicity. Nonetheless, this approach demonstrates the advantages of multi-SNP prediction modeling compared with single gene-single SNP evaluations and warrants the need to perform similar analysis in other ethnic and racial groups while considering SNPs more prevalent in the population selected. Although preliminary, the results demonstrate the potential use of pharmacogenomic risk scores in individualizing chemotherapy with a goal of reducing toxicities, avoiding toxicity-related omissions and delays in treatment, and designing future trials to incorporate our current knowledge of antileukemic chemotherapy toxicities with novel treatment approaches.
APPENDIX
TABLE A1.
SNPs With Minor Allele Frequency of >0.1 Tested in the Patient's Cohort
TABLE A2.
List of SNPs Associated With Toxicities at P < .1 That Were Included in the 3-SNP Combination Modeling
Abdelrahman H. Elsayed
Patents, Royalties, Other Intellectual Property: I have two patent applications: Patent 1: Title: “Pharmacogenomics score to make decisions on therapy augmentation in AML”; Status: Filed; Number: US Provisional Application No.: 63/233,673 Patent 2: Title: “Methods for Predicting AML Outcome”; Status: Published; Number: PCT/US2020/051961
Biljana Horn
Stock and Other Ownership Interests: My retirement counts contain numerous health companies
Research Funding: Orca Bio
Expert Testimony: multiple
Honoraria: Sobi
Jatinder K. Lamba
Patents, Royalties, Other Intellectual Property: Patent 1: Title: “Pharmacogenomics score to make decisions on therapy augmentation in AML”; Status: Filed; Number: US Provisional Application No.: 63/233,673 Patent 2: Title: “Methods for Predicting AML Outcome”; Status: Published; Number: PCT/US2020/051961 Patent 3: Title: “Development of Novel CD33 Antibodies,” Status: Filed Number: 63/078,686 Patent 4: Title: “CD33-Targeted Cancer Therapy” Status: Nationalized Number: PCT/US2017/026369
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
This manuscript was previously published in abstract form or presented (oral or poster) at the American Society of Hematology (ASH) Annual Meeting, Atlanta, GA, December 11-14, 2021, and the American Society of Clinical Pharmacology and Therapeutics (ASCPT) Meeting, virtual, March 16-18, 2022.
SUPPORT
J.K.L.: R01-CA132946, Live like Bella (9LA04); Larkin: University of Florida Clinical and Translational Science Institute Pilot Grant and Children's Miracle Network Trainee Grant.
AUTHOR CONTRIBUTIONS
Conception and design: Trisha Larkin, Abdelrahman H. Elsayed, Beate Greer, Roya Raffiee, Biljana Horn, Jatinder K. Lamba
Administrative support: Trisha Larkin, Biljana Horn
Provision of study materials or patients: Trisha Larkin, Beate Greer
Collection and assembly of data: Trisha Larkin, Reema Kashif, Roya Raffiee, Beate Greer, Karna Mangrola, Vivek Shastri, Jatinder K. Lamba
Data analysis and interpretation: Trisha Larkin, Abdelrahman H. Elsayed, Roya Raffiee, Beate Greer, Nam Nguyen, Biljana Horn, Jatinder K. Lamba
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Abdelrahman H. Elsayed
Patents, Royalties, Other Intellectual Property: I have two patent applications: Patent 1: Title: “Pharmacogenomics score to make decisions on therapy augmentation in AML”; Status: Filed; Number: US Provisional Application No.: 63/233,673 Patent 2: Title: “Methods for Predicting AML Outcome”; Status: Published; Number: PCT/US2020/051961
Biljana Horn
Stock and Other Ownership Interests: My retirement counts contain numerous health companies
Research Funding: Orca Bio
Expert Testimony: multiple
Honoraria: Sobi
Jatinder K. Lamba
Patents, Royalties, Other Intellectual Property: Patent 1: Title: “Pharmacogenomics score to make decisions on therapy augmentation in AML”; Status: Filed; Number: US Provisional Application No.: 63/233,673 Patent 2: Title: “Methods for Predicting AML Outcome”; Status: Published; Number: PCT/US2020/051961 Patent 3: Title: “Development of Novel CD33 Antibodies,” Status: Filed Number: 63/078,686 Patent 4: Title: “CD33-Targeted Cancer Therapy” Status: Nationalized Number: PCT/US2017/026369
No other potential conflicts of interest were reported.
REFERENCES
- 1.NIH Childhood Acute Lymphoblastic Leukemia Treatment (PDQ)—Health Professional Version National Cancer Institute. https://www.cancer.gov/types/leukemia/hp/child-all-treatment-pdq [PubMed] [Google Scholar]
- 2. Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373:1541–1552. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- 3. Pui CH. Precision medicine in acute lymphoblastic leukemia. Front Med. 2020;14:689–700. doi: 10.1007/s11684-020-0759-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rivera GK, Pinkel D, Simone JV, et al. Treatment of acute lymphoblastic leukemia. 30 years' experience at St Jude Children's Research Hospital. N Engl J Med. 1993;329:1289–1295. doi: 10.1056/NEJM199310283291801. [DOI] [PubMed] [Google Scholar]
- 5. Mulrooney DA, Hyun G, Ness KK, et al. The changing burden of long-term health outcomes in survivors of childhood acute lymphoblastic leukaemia: A retrospective analysis of the St Jude lifetime cohort study. Lancet Haematol. 2019;6:e306–e316. doi: 10.1016/S2352-3026(19)30050-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dulucq S, Laverdiere C, Sinnett D, et al. Pharmacogenetic considerations for acute lymphoblastic leukemia therapies. Expert Opin Drug Metab Toxicol. 2014;10:699–719. doi: 10.1517/17425255.2014.893294. [DOI] [PubMed] [Google Scholar]
- 7. Cairns J, Ingle JN, Dudenkov TM, et al. Pharmacogenomics of aromatase inhibitors in postmenopausal breast cancer and additional mechanisms of anastrozole action. JCI Insight. 2020;5:e137571. doi: 10.1172/jci.insight.137571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang M, Huan G, Wang M. Influence of CYP3A and ABCB1 single nucleotide polymorphisms on the pharmacokinetics/pharmacodynamics of tacrolimus in pediatric patients. Curr Drug Metab. 2018;19:1141–1151. doi: 10.2174/1389200219666180925090228. [DOI] [PubMed] [Google Scholar]
- 9. Bargal SA, Rafiee R, Crews KR, et al. Genome-wide association analysis identifies SNPs predictive of in vitro leukemic cell sensitivity to cytarabine in pediatric AML. Oncotarget. 2018;9:34859–34875. doi: 10.18632/oncotarget.26163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Elsayed AH, Cao X, Crews KR, et al. Comprehensive Ara-C SNP score predicts leukemic cell intracellular ara-CTP levels in pediatric acute myeloid leukemia patients. Pharmacogenomics. 2018;19:1101–1110. doi: 10.2217/pgs-2018-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gbadamosi M, Meshinchi S, Lamba JK. Gemtuzumab ozogamicin for treatment of newly diagnosed CD33-positive acute myeloid leukemia. Future Oncol. 2018;14:3199–3213. doi: 10.2217/fon-2018-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maxwell RR, Cole PD. Pharmacogenetic predictors of treatment-related toxicity among children with acute lymphoblastic leukemia. Curr Hematol Malig Rep. 2017;12:176–186. doi: 10.1007/s11899-017-0376-z. [DOI] [PubMed] [Google Scholar]
- 13. Yang JJ, Landier W, Yang W, et al. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J Clin Oncol. 2015;33:1235–1242. doi: 10.1200/JCO.2014.59.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weinshilboum RM, Sladek SL. Mercaptopurine pharmacogenetics: Monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am J Hum Genet. 1980;32:651–662. [PMC free article] [PubMed] [Google Scholar]
- 15. Salavaggione OE, Wang L, Wiepert M, et al. Thiopurine S-methyltransferase pharmacogenetics: Variant allele functional and comparative genomics. Pharmacogenet Genom. 2005;15:801–815. doi: 10.1097/01.fpc.0000174788.69991.6b. [DOI] [PubMed] [Google Scholar]
- 16. Relling MV, Gardner EE, Sandborn WJ, et al. Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin Pharmacol Ther. 2013;93:324–325. doi: 10.1038/clpt.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Relling MV, Schwab M, Whirl-Carrillo M, et al. Clinical pharmacogenetics implementation consortium guideline for thiopurine dosing based on TPMT and NUDT15 genotypes: 2018 update. Clin Pharmacol Ther. 2019;105:1095–1105. doi: 10.1002/cpt.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang SK, Hong M, Baek J, et al. A common missense variant in NUDT15 confers susceptibility to thiopurine-induced leukopenia. Nat Genet. 2014;46:1017–1020. doi: 10.1038/ng.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zgheib NK, Akra-Ismail M, Aridi C, et al. Genetic polymorphisms in candidate genes predict increased toxicity with methotrexate therapy in Lebanese children with acute lymphoblastic leukemia. Pharmacogenet Genom. 2014;24:387–396. doi: 10.1097/FPC.0000000000000069. [DOI] [PubMed] [Google Scholar]
- 20. Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Relling MV, Ramsey LB. Pharmacogenomics of acute lymphoid leukemia: New insights into treatment toxicity and efficacy. Hematology. 2013;2013:126–130. doi: 10.1182/asheducation-2013.1.126. [DOI] [PubMed] [Google Scholar]
- 22. Chen Y, Zou K, Sun J, et al. Are gene polymorphisms related to treatment outcomes of methotrexate in patients with rheumatoid arthritis? A systematic review and meta-analysis. Pharmacogenomics. 2017;18:175–195. doi: 10.2217/pgs-2016-0158. [DOI] [PubMed] [Google Scholar]
- 23. Ghodke-Puranik Y, Puranik AS, Shintre P, et al. Folate metabolic pathway single nucleotide polymorphisms: A predictive pharmacogenetic marker of methotrexate response in Indian (Asian) patients with rheumatoid arthritis. Pharmacogenomics. 2015;16:2019–2034. doi: 10.2217/pgs.15.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang Z, Tong HF, Li Y, et al. Effect of the polymorphism of folylpolyglutamate synthetase on treatment of high-dose methotrexate in pediatric patients with acute lymphocytic leukemia. Med Sci Monit. 2016;22:4967–4973. doi: 10.12659/MSM.899021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharma S, Das M, Kumar A, et al. Purine biosynthetic pathway genes and methotrexate response in rheumatoid arthritis patients among north Indians. Pharmacogenet Genom. 2009;19:823–828. doi: 10.1097/fpc.0b013e328331b53e. [DOI] [PubMed] [Google Scholar]
- 26. Yee SW, Mefford JA, Singh N, et al. Impact of polymorphisms in drug pathway genes on disease-free survival in adults with acute myeloid leukemia. J Hum Genet. 2013;58:353–361. doi: 10.1038/jhg.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Voon PJ, Yap HL, Ma CYT, et al. Correlation of aldo-ketoreductase (AKR) 1C3 genetic variant with doxorubicin pharmacodynamics in Asian breast cancer patients. Br J Clin Pharmacol. 2013;75:1497–1505. doi: 10.1111/bcp.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huezo-Diaz Curtis P, Uppugunduri CRS, Muthukumaran J, et al. Association of CTH variant with sinusoidal obstruction syndrome in children receiving intravenous busulfan and cyclophosphamide before hematopoietic stem cell transplantation. Pharmacogenomics J. 2018;18:64–69. doi: 10.1038/tpj.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]