PURPOSE
NCI-MATCH is a precision medicine trial using genomic testing to allocate patients with advanced malignancies to targeted treatment subprotocols. This report combines two subprotocols evaluating trametinib, a MEK1/2 inhibitor, in patients with Neurofibromatosis 1 (NF1[S1] or GNA11/Q [S2]) altered tumors.
METHODS
Eligible patients had tumors with deleterious inactivating NF1 or GNA11/Q mutations by the customized Oncomine AmpliSeq panel. Prior MEK inhibitor treatment was excluded. Glioblastomas (GBMs) were permitted, including malignancies associated with germline NF1 mutations (S1 only). Trametinib was administered at 2 mg once daily over 28-day cycles until toxicity or disease progression. Primary end point was objective response rate (ORR). Secondary end points included progression-free survival (PFS) at 6 months, PFS, and overall survival. Exploratory analyses included co-occurring genomic alterations and PTEN loss.
RESULTS
Fifty patients were eligible and started therapy: 46 with NF1 mutations (S1) and four with GNA11 mutations (S2). In the NF1 cohort, nonsense single-nucleotide variants were identified in 29 and frameshift deletions in 17 tumors. All in S2 had nonuveal melanoma and GNA11 Q209L variant. Two partial responses (PR) were noted in S1, one patient each with advanced lung cancer and GBM for an ORR of 4.3% (90% CI, 0.8 to 13.1). One patient with melanoma in S2 had a PR (ORR, 25%; 90% CI, 1.3 to 75.1). Prolonged stable disease (SD) was also noted in five patients (four in S1 and one in S2) with additional rare histologies. Adverse events were as previously described with trametinib. Comutations in TP53 and PIK3CA were common.
CONCLUSION
Although these subprotocols did not meet the primary end point for ORR, significant responses or prolonged SD noted in some disease subtypes warrants further investigation.
BACKGROUND
The RAF-MEK-ERK pathway plays a critical role in multiple cellular functions. Activation of this pathway is common in cancer and can result from ligand activation or activating mutations of the upstream receptor tyrosine kinases and RAS, or upregulation or mutations in RAF and MEK. Once activated, ERK1/2 translocates to the nucleus and phosphorylates a number of effector proteins and transcriptional factors that regulate cell proliferation, motility, differentiation, and survival.1,2 Trametinib dimethyl sulfoxide (trametinib) is a potent, allosteric, ATP noncompetitive inhibitor of MEK1/2, and was first approved for clinical use on the basis of activity in BRAF-mutant melanoma.3
CONTEXT
Key Objective
Preclinical data indicate that MEK inhibitors may be effective therapies for tumors harboring Neurofibromatosis 1 (NF1) or GNA11/Q alterations. Therefore, the NCI MATCH precision medicine study included two subprotocols (S1 and S2) testing trametinib, a MEK1/2 inhibitor, in patients with advanced solid tumors.
Knowledge Generated
Although trametinib did not demonstrating meaningful clinical activity by predefined objective response rate in these subprotocols, there were responses noted in one patient each with lung cancer, glioblastoma, and nonuveal melanoma, warranting future studies. No new safety signals were noted.
Relevance
Trametinib should not be recommended for patients with tumors harboring NF1 or GNA11/Q alterations.
Neurofibromatosis 1 (NF1) produces the protein product, neurofibromin. Germline inactivating mutations in NF1 result in Neurofibromatosis type 1, an autosomal dominant familial cancer predisposition syndrome.4 Somatic NF1 mutations or deletions have also been identified in multiple cancer subtypes including, but not limited to, breast adenocarcinoma (2.5%-27.7%), glioblastoma (GBM; 14%-23%), melanoma (12%-30%), lung adenocarcinomas and squamous cell carcinomas (7%-11.8%), and ovarian (12%-34.4%) cancers.5 Neurofibromin is a tumor suppressor that regulates the downstream RAS/RAF/MEK/ERK pathway. Preclinical data demonstrated that tumors with NF1 inactivation are sensitive to MEK inhibitors.6,7 Blockade of MEK led to reduction of neurofibroma in mice harboring NF1 mutations and in longer survival of human-to-murine malignant peripheral nerve sheath tumor xenografts.8 A subset of GBM cell lines with NF1 loss are dependent on the RAF/MEK/ERK pathway for growth, and are sensitive to MEK inhibitors in vitro and in vivo.7 Further supporting evaluation of MEK inhibitors in NF1-mutant tumors, preliminary clinical activity was seen with single-agent MEK inhibition in a phase I pediatric study of patients with progressive and nonprogressive pediatric plexiform neurofibromas, with partial responses (PR) observed in 11 of 18 patients (61%).9
The guanine nucleotide-binding protein G(q) subunit alpha (GNAQ) and guanine nucleotide-binding protein subunit alpha-11 (GNA11) are two closely related large GTPases of the Gαq family.10,11 Mutations in GNAQ or GNA11 have been observed in 1.6%-2% of cancers.12 Mutations in GNAQ or GNA11 eliminate the activity of the heterotrimeric G protein α subunits Gαq and Gα11 GTPase, leading to hyperactivation of signaling downstream of the GNAQ and GNA11, including the RAS/RAF/MEK/ERK pathway.13,14 Uveal melanoma is characterized by functionally active mutations in GNAQ or GNA11 (>80%), and MEK inhibition has been of interest in this disease.15,16 Some nonuveal melanomas also harbor these mutations and appear to have unique clinical and genomic characteristics.17
These data suggest that MEK inhibitors might be new therapeutic candidates for NF1 or GNAQ/11-mutant solid tumors. NCI-MATCH is a precision medicine trial using genomic testing to allocate patients with advanced malignancies to targeted treatment subprotocols. This combined report includes two subprotocols studying trametinib, a MEK1/2 inhibitor, in patients with Neurofibromatosis 1 (NF1 [subprotocol S1] or GNA11/Q [S2]) altered tumors.
METHODS
Patient Eligibility and the Genomic Assay
NCI-MATCH included adult patients with solid tumors, lymphomas, and multiple myeloma whose disease had progressed after at least one line of standard systemic therapy or for whom no standard therapy was available. GBMs were permitted in both cohorts. Malignancies associated with germline NF1 mutations were permitted in S1. Uveal melanoma was excluded from S2, given that several clinical trials were ongoing investigating MEK inhibition in this disease. After informed consent to the master Protocol, patients entered into the screening phase and underwent a fresh tumor biopsy or submitted archived tumor tissue if obtained within 6 months without response to interval treatment. Tumor genomic analysis was performed at one of four central laboratories using a centralized, customized Thermo Fisher Oncomine AmpliSeq NGS panel and selected (PTEN) immunohistochemistry.18,19 After May 11, 2017, patients with prespecified outside genomic assays, performed in Clinical Laboratory Improvement Amendments–accredited and NCI-MATCH–vetted laboratories, were eligible for enrollment; however, only one tumor from S2 had an outside assay and the actionable mutation of interest (aMOI) was confirmed by the Oncomine assay. Patients were assigned to subprotocols using the National Cancer Institute (NCI) Precision Medicine Platform (MATCHBOX) that runs a prospectively defined, NCI-designed informatics rules algorithm, as previously described.20,21 For subprotocol S1, patients whose tumors contained a deleterious, inactivating NF1 mutation were potentially eligible (Appendix Table A1). A function was implemented in MATCHBOX to identify any point mutations creating stop codons leading to premature truncations or any insertions/deletions causing frameshifts, which were predicted to result in a nonfunctional or absent protein. Variants, including missense mutations, were not included for eligibility if there was a lack of adequate evidence that such variants resulted in loss of function in NF1 gene. For subprotocol S2, the actionable mutation list is provided in Appendix Table A1. For both arms, patients were required to meet all eligibility requirements from the master protocol including Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1, absolute neutrophil count of ≥1,500/μL, platelet count ≥100,000/μL, hemoglobin concentration ≥9 g/dL, and adequate kidney and liver function. Except for patients with GBM, others were required to have measurable disease, defined by RECIST version 1.1.22 Patients could not have a prior history of interstitial lung disease, pneumonitis, retinal vein occlusion, left ventricular ejection fraction less than the institutional lower limit of normal, treatment refractory hypertension, or clinically significant ECG changes. Prior treatment with MEK inhibitors was not allowed. The study was approved by the NCI Central Institutional Review Board and is listed in ClinicalTrials.gov (identifier: NCT02465060).
Treatment and Evaluation
Patients were treated with the standard oral dose of trametinib, 2 mg orally once daily fasting with water, continuously, on 28-day cycles until intolerable toxicity or progression. Imaging was performed every two cycles for the first 26 cycles and every three cycles thereafter. Dose reductions were permitted to 1.5 mg and 1.0 mg once daily.
Statistical Methodology and End Point Analysis
The primary end point was objective response rate (ORR), defined as a complete response (CR) or PR, consistent with RECIST version 1.1 criteria for solid tumors.22 For patients with GBM, RANO criteria were used to assess response.23 Both required confirmation at least 4 weeks later. Allowing for a 10% ineligibility rate, the accrual goal was 35 patients for each subprotocol. For subprotocol S1, per the NCI-MATCH master protocol, accrual continued past 35 patients on the basis of protocol criteria allowing up to 6 months of additional accrual and a maximum of 35 additional patients. The proposed design had at least 92% power to distinguish an ORR of 25% from a null of 5% with a one-sided type I error of 1.8%. Further study was considered warranted if >5/31 (16%) patients achieved CR or PR. The null hypothesis is tested at a one-sided significance level of 1.8% for arms accruing more than 31 eligible, treated patients, and at 5% for arms accruing <31 eligible, treated patients. Since S1 and S2 accrued 46 and four eligible and treated patients, respectively, a minimum of seven and two responders are needed to consider trametinib worthy of further study in patients with NF1 or GNA11/Q alterations, respectively. Secondary end points included progression-free survival (PFS) at 6 months, PFS, and overall survival (OS). Exploratory analyses of co-occurring genomic alterations and PTEN loss by immunohistochemistry were also performed.
RESULTS
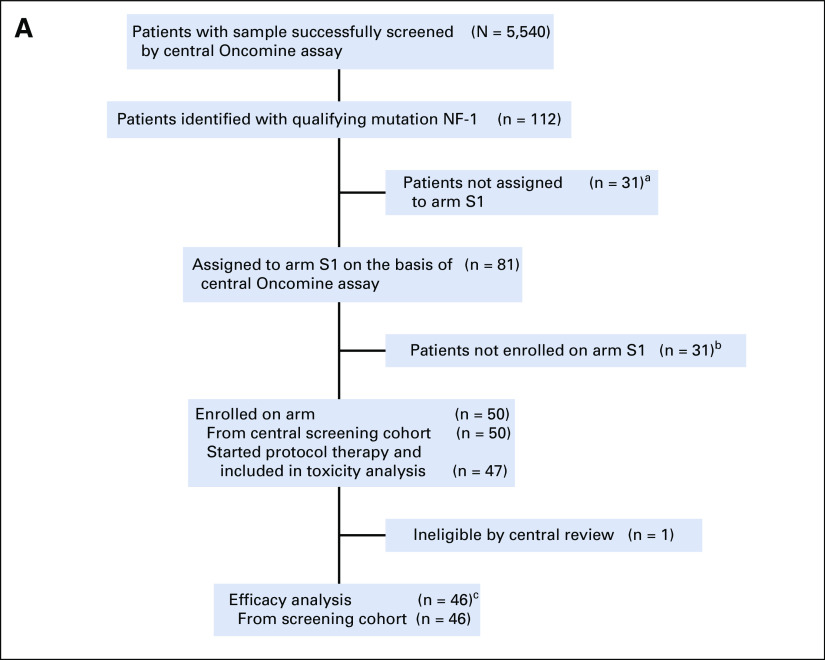
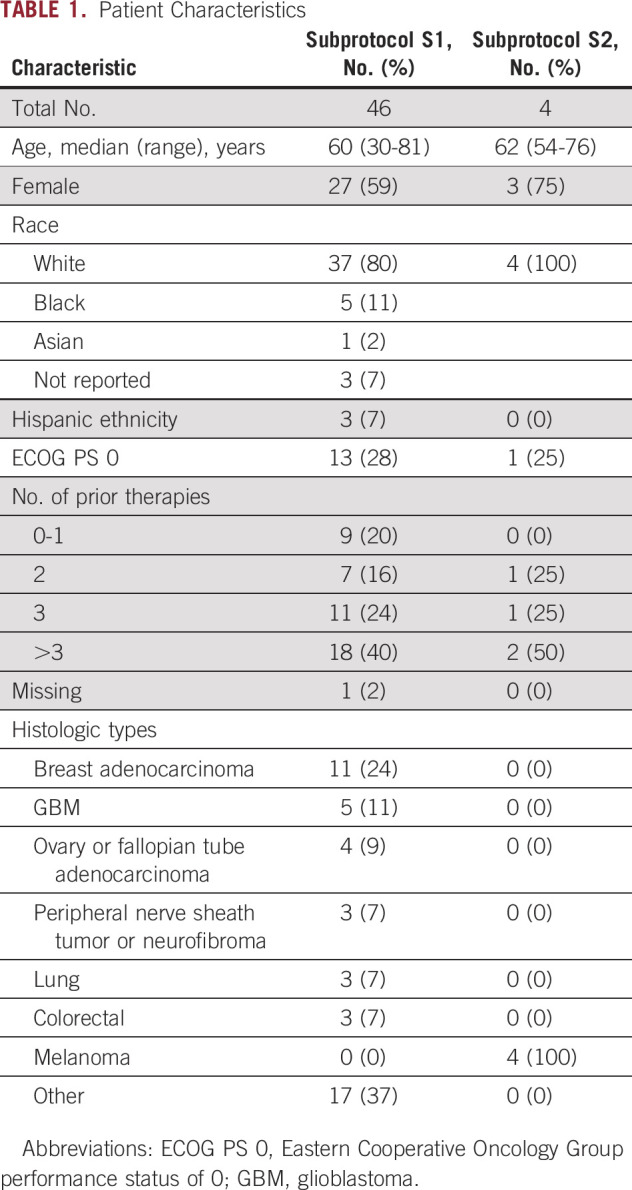
As of arm closures in May and August 2019, 50 and four patients were enrolled in subprotocols S1 and S2, respectively (Fig 1). Subprotocol S2 was closed because of lack of accrual due to the rarity of the mutations outside of uveal melanoma. All eligible patients who received at least one dose of treatment were included in the analysis. Because of ineligibility (one) or never starting treatment (three), data from four patients were not included in S1 efficacy analysis. The first patient was enrolled March 16, 2016, with the last on July 12, 2017, for S1, and July 5, 2016, to January 16, 2019, for S2. Baseline characteristics are described in Table 1. In S1, the primary tumor types enrolled were breast, GBM, lung, colorectal, cancer of the fallopian tube or ovary, and peripheral nerve sheath tumor or neurofibroma. Frameshift deletions (FSD) were noted in 17 (37%) and nonsense single-nucleotide variants (nSNV) in 29 (63%) patients (Appendix Fig A1). Of note, one tumor had both FSD and nSNV, and was classified as FSD. All four patients in S2 had nonuveal melanoma and the same qualifying GNA11 Q209L variant.
FIG 1.
Diagrams for subprotocols (A) S1 and (B) S2. aReasons for not receiving an assignment: study suspended (n = 15), assigned to another arm for which the qualifying variant’s level of evidence was higher (n = 7), before study activation (n = 6), assigned to another arm for which accrual was lower (n = 2), and prior treatment of trametinib (n = 1). bReasons not enrolled after receiving an assignment: active infection and abnormal brain MRI (n = 1), deteriorating performance status (n = 6), eye problems (n = 1), in hospice care (n = 1), inadequate organ/marrow function (n = 2), interstitial lung disease/pneumonitis (n = 2), intervening news/targeted treatment (n = 2), other investigational treatment (n = 1), patient died (n = 6), patient refusal (n = 4), receiving other therapy (n = 2), and unknown (n = 3). cReasons for being nonevaluable: no disease assessment done before death (n = 4), no disease assessment done before NPT (n = 1), baseline scans done outside of specified window (n = 5), withdrew consent (n = 2), different methods of evaluation used (n = 1), no evaluation during treatment (n = 1). dReasons for not receiving an assignment: ineligible histology—uveal melanoma (n = 13). eReasons not enrolled after receiving an assignment: central screening cohort: ineligible histology (n = 1), ineligible histology and inadequate organ/marrow function (n = 1), and unknown (n = 1). NPT, nonprotocol therapy.
TABLE 1.
Patient Characteristics
Subprotocol S1 (NF1-Mutant) Treatment Efficacy
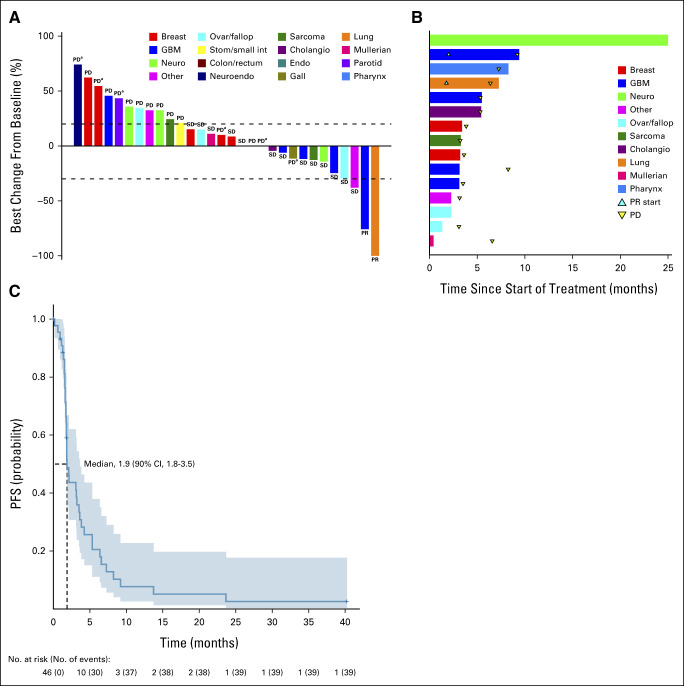
The median number of cycles for patients on S1 was two (range, 1-45). The majority of patients discontinued for disease progression (n = 25; 54%), while 10 of the 46 patients discontinued treatment because of adverse events (22%). In total, 14 patients were not evaluable for response. Patients were nonevaluable because of the following reasons—no disease assessment done before death (n = 4), no disease assessment done before another nonprotocol therapy (NPT; n = 1), baseline scans done outside of specified window (n = 5), withdrawal of consent (n = 2), different methods of evaluation used (n = 1), and no evaluation during treatment (n = 1). One patient remained on study at the time of data analysis. The best confirmed response was PR in two patients (ORR, 4.3%; 90% CI, 0.8 to 13.1), which did not meet the prespecified criteria to consider this agent promising and worthy of further study in NF1-mutant tumors (Figs 2A and 2B). The median PFS for the entire cohort was 1.9 months (Fig 2C) and did not significantly vary by variant type—at 1.8 months for FSD and 3.1 months for nSNV. The estimated 6-month PFS rate was 21% (two-sided 90% CI, 12 to 34) for the whole cohort. By variant type, 6-month PFS was 12% (two-sided 90% CI, 4 to 35) for FSD, and 27% (two-sided 90% CI, 15 to 48) for nSNV. One patient with adenocarcinoma of the lung with a nSNV had a near-CR on the basis of CR of target disease but non-CR in nontarget disease with a PFS of 6.4 months. One patient with GBM and a nSNV achieved a PR after cycle 4 with 75% change on the basis of sum of perpendicular diameters with a PFS of 9.2 months. In addition, four patients experienced prolonged stable disease (SD), one each with GBM (PFS, 8.2 months), squamous cell carcinoma of the pharynx (7.3 months), neurofibroma (40.2 months), and carcinosarcoma of the fallopian tube (6.6 months). A total of five patients with GBM were enrolled; three did not experience clinical benefit as defined by PFS < 6 months. Both patients with GBM who did have clinical benefit had isocitrate dehydrogenase (IDH) wild-type disease and prior temozolomide therapy. In addition, one patient with endometrial cancer who was unevaluable because of baseline scans being out of window had a reported tumor size percent reduction of 39% and a PFS of 23.7 months. Median OS was 10.4 months (90% CI, 7.4 to 13.3) in S1.
FIG 2.
Subprotocol 1 response and Kaplan-Meier curves for PFS. (A) Waterfall plot (N = 30) of best % change from baseline responses. This includes the 32 evaluable patients, excluding two patients who had a best response of PD with new lesions that had no recorded measurements. (B) Treatment duration in the 15 patients who achieved a best response of SD or PR. (C) Kaplan-Meier curve for PFS. aNew lesions. Cholangio, cholangiocarcinoma; endo, endocrine cancer; gall, gallbladder cancer; GBM, glioblastoma; neuro, peripheral nerve sheath tumor or neurofibroma; neuroendo, neuroendocrine carcinoma; ovar/fallop, ovarian/fallopian tube cancer; PD, progressive disease; PFS, progression-free survival; PR, partial response; stom/small int, gastric or small intestine carcinoma; SD, stable disease.
Subprotocol S2 (GNA11) Treatment Efficacy
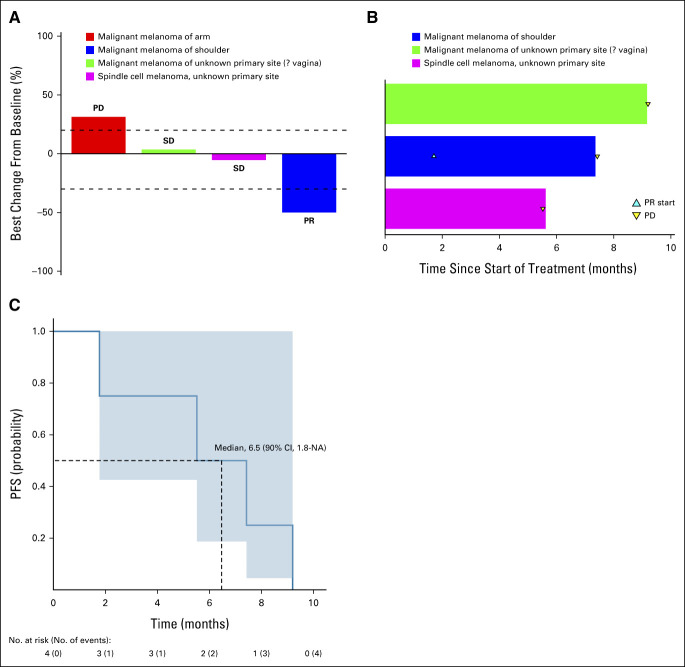
The median number of cycles for the four patients with nonuveal melanoma in this cohort was seven (range, 2-10), with all discontinuing for progressive disease (PD). One patient experienced a PR for an ORR of 25% (90% CI, 1.3 to 75.1; Fig 3A). This patient had a 50% tumor reduction with a PFS of 7.4 months. One patient did experience SD with a PFS of 9.2 months (Fig 3B). In total, this cohort had a 6-month PFS rate of 50% (90% CI, 22 to 100; Fig 3C). Median overall survival was 19.2 months (90% CI, 18.1 to NA) for S2.
FIG 3.
Subprotocol 2 response and Kaplan-Meier curves for PFS. (A) Waterfall plot (N = 4) of best % change from baseline responses. (B) Treatment duration in the three patients who achieved a best response of SD or PR. (C) Kaplan-Meier curve for PFS. NA, not applicable; PD, progressive disease; PFS, progression-free survival; PR, partial response; SD, stable disease.
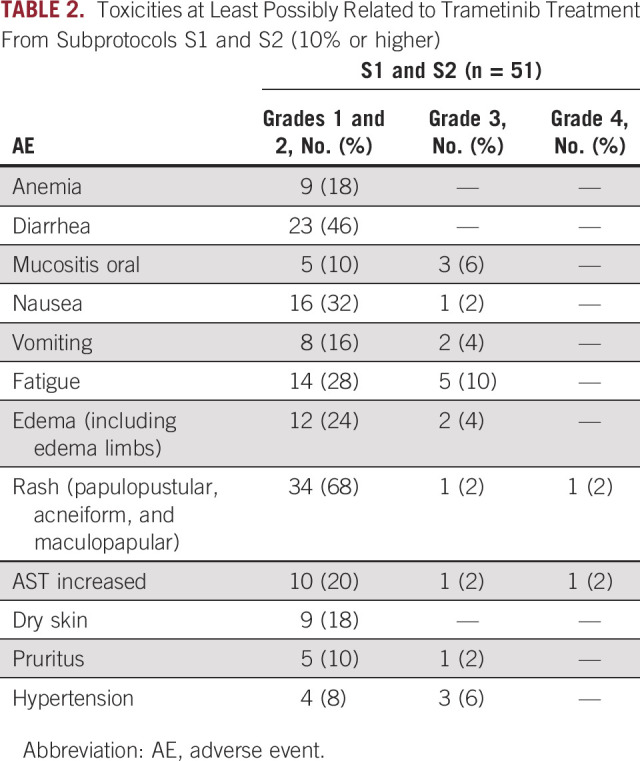
Combined Safety From Subprotocols S1 and S2
All patients who received at least one dose of the study drug were included in the safety analysis (n = 51). No new safety signals were seen with trametinib, with expected side effects of dermatologic and gastrointestinal toxicity, including rash and diarrhea, being the most common (Table 2). There were four cases of grade 5 death recorded during the study therapy in S1, and one death was attributed as possibly related to study drug, with the other three attributed to PD. Three patients (6%) experienced decline in ejection fraction or heart failure, all grade 1-2, a known side effect of MEK inhibitors.
TABLE 2.
Toxicities at Least Possibly Related to Trametinib Treatment From Subprotocols S1 and S2 (10% or higher)
Comutation Data
Comutations were evaluated in the 50 cases (Fig 4). In the S1 cohort (N = 46), TP53 mutations were the most common comutation observed (n = 24; 52%) followed by PIK3CA mutations (n = 13; 28%). No co-occurring mutations were found in the lung cancer with near CR. A BRAF comutation was noted only in one patient with rectal adenocarcinoma. Comutations were uncommon in the S2 cohort. Because of low response rates, formal analysis for response by comutation was not feasible, but no association was noted.
FIG 4.
Co-occurring genomic alterations in subprotocols S1 and S2. Co-occurring genomic alterations using the NCI-MATCH assay color-coded by: left: variant type; top: copy-number variant (pink), SNV (red), indel (purple), and unified gene fusion (magenta) for each eligible patient; and bottom: information regarding histology and best response on treatment: PR, SD, PD, and UE. Cholangio, cholangiocarcinoma; endo, endocrine cancer; gall, gallbladder cancer; GBM, glioblastoma; neuro, peripheral nerve sheath tumor or neurofibroma; neuroendo, neuroendocrine carcinoma; ovar/fallop, ovarian/fallopian tube cancer; PD, progressive disease; PFS, progression-free survival; PR, partial response; SD, stable disease; SNV, single-nucleotide variant; stom/small int, gastric or small intestine carcinoma; UE, unevaluable.
DISCUSSION
Molecularly targeted agents have led to significant advancements in various malignancies, such as HER2-directed therapies in breast cancer,24 epidermal growth factor receptor (EGFR)-targeted agents in lung cancer,25 and more recently, the pan-tumor indication for larotrectinib for solid tumors with NTRK gene fusions.26 Potentially actionable mutations can be identified in low frequencies across multiple tumor types and it remains unknown if targeting these mutations independent of tumor origin will be effective. The NCI-MATCH trial was the first large-scale genomically directed multiarm basket trial in adult malignancies. This report is a combined analysis of two NCI-MATCH subprotocols investigating the MEK inhibitor trametinib in patients with advanced solid tumors with inactivating NF1 mutations (S1 subprotocol) or GNA11 mutations (S2 subprotocol).
Trametinib did not demonstrate a significant ORR in patients with tumors harboring NF1 inactivating mutations (S1). Overall, these results indicate that NF1 mutations alone are not predictive of benefit from MEK inhibitors across heavily pretreated patients with solid tumors. It is possible this disappointing response rate was due to the heavily pretreated cohort, in which responses to molecularly targeted agents may be less frequent.27 Furthermore, it has become clear that there are multiple feedback loops that can lead to resistance to MEK inhibition, including induction of RAS via sprouty-Grb2/SHC/SOS complexes.28 Preclinical studies support combinations with PKC and PI3K inhibitors as these signaling pathways seem to be activated in cell lines with these mutations.29,30 Although co-occurring alterations in resistance pathways such as p53 and PI3 kinase were noted in our study cohort, the study was underpowered to identify specific comutations that mediated resistance to trametinib in these patients with NF1- or GNA11-mutant tumors. None of the patients on these subprotocols had biopsies on progression, which may have helped discern resistance mechanisms. Another potential explanation for the lack of trametinib activity in these subprotocols may be due to the therapeutic window of MEK inhibitors not being wide enough to adequately suppress ERK and lead to cytotoxic results.31
Despite these results, it is notable that two patients (lung adenocarcinoma and GBM) with NF1 mutations did have significant PR with >75% tumor reduction. Furthermore, another patient with GBM in this cohort experienced prolonged SD. Mutations or deletions in the NF1 gene play an important role in the pathogenesis of GBM.32 NF1 is mutated or deregulated in approximately 13% of GBM, and NF1 loss or mutation is observed primarily in the more aggressive mesenchymal subtype.33,34 Previous studies have also indicated a potential role for targeting NF1 in GBM.35,36 One case report also reported potential activity of trametinib in NF1-associated GBM.37 Given the poor prognosis for patients with recurrent IDH wild-type GBM, this observation of activity in NF1-mutant GBM may be worth future study.38
Trametinib is currently approved in combination with dabrafenib for treatment of non–small-cell lung cancer (NSCLC) with BRAF V600E mutations. There is also single-agent activity of trametinib in patients with NSCLC with KRAS mutations.39 It is notable that the patient with NSCLC in our study did not have any of these comutations identified as an alternative explanation for response. In other series, comutations in NF1-mutant lung cancer were only seen in 25% of cases, indicating this may be an important target for this disease.40 Future studies of trametinib alone or in combination with other signaling pathway inhibitors in NF1-mutant lung cancer may be warranted.
The durable activity of MEK inhibitors against neurofibromas has been identified in other studies. One open-label phase II trial of selumetinib in pediatric patients with neurofibromatosis type 1 and symptomatic inoperable plexiform neurofibromas demonstrated 35 of 50 patients (70%) had a confirmed PR with the majority durable for >1 year.41 A phase I/II study of trametinib in a similar patient population showed a 46% response rate.42 In our study, one patient with neurofibroma had a PFS of 40.2 months and remained on study at the time of data cut-off, further supporting future studies of these agents in these low-grade tumors that often cause significant morbidity through pain, disfigurement, and neurologic complications.
The S2 subprotocol was closed because of slow accrual, and all four enrolled patients had nonuveal melanoma. There was a potential signal of benefit in that one of the four patients had a response. The patient with a PR and a PFS of 7.4 months, as well as the patient with SD and a PFS of 9.2 months, is similar to the initial results seen with the single-agent BRAF inhibitor, vemurafenib, which demonstrated a PFS of 5.3 months in BRAF V600E-mutant melanoma. Mutation testing of these tumors led to the discovery of resistance mechanisms to single-agent MEK inhibitor therapy.43 These results suggest that there could be primary or secondary development of resistance to treatment with a single-agent MEK inhibitor, or that additional comutations or other factors could be involved in treatment response. It is noted, however, that MEK inhibitors even in combination with AKT inhibitors have not been successful in GNAQ/GNA11-mutant uveal melanomas,44 although the biology of uveal melanoma is different than that of cutaneous melanoma. Further exploration into whether the spectrum of comutations may be driving these different outcomes is warranted. As anticipated, trametinib was well tolerated with anticipated adverse events of diarrhea and rash leading to infrequent discontinuation. No new safety signals were identified.
In summary, the NCI-MATCH study subprotocol S1 evaluating trametinib in NF1-mutant advanced solid tumors did not meet the primary end point, and subprotocol S2 evaluating trametinib in GNA11-mutant advanced solid tumors had inadequate patient accrual. However, there are potential signals of activity in GBM, lung cancer, and neurofibromas with NF1 mutations, and nonuveal melanoma with GNA11 mutations. These observations could warrant further studies, but are not sufficient to guide treatment recommendations. Future directions include focusing on combinations targeting resistance pathways and studies on selected tumor types.
APPENDIX
FIG A1.
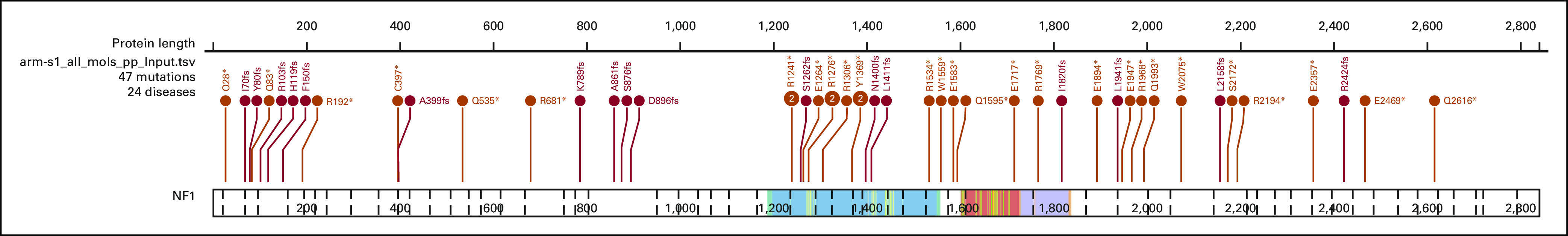
Lollipop plot showing location and sequence of NF1 mutations in S1 cohort.
TABLE A1.
Actionable Mutations for Subprotocol S2: GNA11/GNAQ
Kari B. Wisinski
Honoraria: Pfizer
Consulting or Advisory Role: Sanofi, AstraZeneca, Pfizer, Novartis Institutes for BioMedical Research, Stemline Therapeutics
Research Funding: Novartis (Inst), AstraZeneca (Inst), Pfizer (Inst), Sanofi (Inst), Genentech (Inst), Context Therapeutics (Inst), Oncoceutics (Inst), Zymeworks (Inst), Relay Therapeutics (Inst)
Melissa A. Wilson
Consulting or Advisory Role: Instil Bio
Jason J. Luke
Stock and Other Ownership Interests: Actym Therapeutics, Mavu Pharmaceutical, Pyxis, Alphamab, Tempest Therapeutics, Kanaph Therapeutics, Onc.AI, Arch Oncology, STipe Therapeutics, NeoTX
Consulting or Advisory Role: Bristol Myers Squibb, Merck, EMD Serono, Novartis, Seven Hills Pharma, Janssen, Reflexion Medical, Tempest Therapeutics, Alphamab, AbbVie, Bayer, Incyte, Partner Therapeutics, Synlogic, Werewolf Therapeutics, Ribon Therapeutics, Checkmate Pharmaceuticals, CStone Pharmaceuticals, Nektar, Regeneron, Rubius Therapeutics, Tesaro, Xilio Therapeutics, Xencor, Alnylam, Crown Bioscience, Flame Biosciences, Genentech, Kadmon, KSQ Therapeutics, Immunocore, Inzen Therapeutics, Pfizer, Silicon Therapeutics, TRex Bio, Bright Peak Therapeutics, Onc.AI, STipe Therapeutics, Codiak Biosciences, Day One Therapeutics, Endeavor BioMedicines, Gilead Sciences, Hotspot Therapeutics, SERVIER, STINGthera, Synthekine
Research Funding: Merck (Inst), Bristol Myers Squibb (Inst), Incyte (Inst), Corvus Pharmaceuticals (Inst), AbbVie (Inst), Macrogenics (Inst), Xencor (Inst), Array BioPharma (Inst), Agios (Inst), Astellas Pharma (Inst), EMD Serono (Inst), Immatics (Inst), Kadmon (Inst), Moderna Therapeutics (Inst), Nektar (Inst), Spring Bank (Inst), Trishula Therapeutics (Inst), KAHR Medical (Inst), Fstar (Inst), Genmab (Inst), Ikena Oncology (Inst), Numab (Inst), Replimune (Inst), Rubius Therapeutics (Inst), Synlogic (Inst), Takeda (Inst), Tizona Therapeutics, Inc (Inst), BioNTech (Inst), Scholar Rock (Inst), NextCure (Inst)
Patents, Royalties, Other Intellectual Property: Serial #15/612,657 (Cancer Immunotherapy), Serial #PCT/US18/36052 (Microbiome Biomarkers for Anti-PD-1/PD-L1 Responsiveness: Diagnostic, Prognostic and Therapeutic Uses Thereof)
Travel, Accommodations, Expenses: Bristol Myers Squibb, Array BioPharma, EMD Serono, Janssen, Merck, Novartis, Reflexion Medical, Mersana, Pyxis, Xilio Therapeutics
Hussein A. Tawbi
Consulting or Advisory Role: Novartis, Bristol Myers Squibb, Genentech/Roche, Merck, Eisai, Iovance Biotherapeutics, Karyopharm Therapeutics, Boxer Capital, Jazz Pharmaceuticals, Pfizer, Medicenna
Research Funding: Bristol Myers Squibb (Inst), Novartis (Inst), Merck (Inst), GlaxoSmithKline (Inst), Genentech/Roche (Inst), Celgene (Inst), Dragonfly Therapeutics (Inst), RAPT Therapeutics (Inst)
Fangxin Hong
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Consulting or Advisory Role: Merck Sharp & Dohme
Edith P. Mitchell
Leadership: Corvus Pharmaceuticals
Honoraria: Sanofi, Exelixis
Consulting or Advisory Role: Genentech, Novartis, Merck, Bristol Myers Squib
Speakers' Bureau: Ipsen
Research Funding: Genentech (Inst), Sanofi (Inst)
Robert J. Gray
Research Funding: Agios, Amgen, AstraZeneca, Bristol Myers Squibb, Boehringer Ingelheim, Celgene, Genentech/Roche, Genomic Health, Genzyme, GlaxoSmithKline, Janssen-Ortho, Onyx, Pfizer, Sequenta, Syndax, Novartis, Takeda, AbbVie, Sanofi, Merck Sharp & Dohme
Shuli Li
Employment: Takeda
Stock and Other Ownership Interests: Takeda
P. Mickey Williams
Research Funding: Illumina (Inst)
Patents, Royalties, Other Intellectual Property: I was a co-inventor of the DLBCL cell of origin patent recently filed by the NIH
Stanley R. Hamilton
Research Funding: Minerva Biotechnologies, Intima
Carlos L. Arteaga
Stock and Other Ownership Interests: Provista Diagnostics
Consulting or Advisory Role: Novartis, Lilly, Sanofi, Radius Health, Taiho Pharmaceutical, Puma Biotechnology, Merck, Origimed, Immunomedics, Daiichi Sankyo, Athenex, AstraZeneca, Arvinas
Research Funding: Pfizer, Lilly, Takeda
Other Relationship: Susan G. Komen for the Cure
Uncompensated Relationships: Susan G. Komen for the Cure Foundation
Lyndsay N. Harris
Patents, Royalties, Other Intellectual Property: Philips Healthcare
Peter J. O'Dwyer
Research Funding: Bristol Myers Squibb (Inst), Pfizer (Inst), Novartis (Inst), Genentech (Inst), Mirati Therapeutics (Inst), Celgene (Inst), GlaxoSmithKline (Inst), BBI Healthcare (Inst), Pharmacyclics (Inst), Five Prime Therapeutics (Inst), 47 (Inst), Amgen (Inst), H3 Biomedicine (Inst), Taiho Pharmaceutical (Inst), Array BioPharma (Inst), Lilly/ImClone (Inst), Syndax (Inst), Minneamrita Therapeutics (Inst)
Expert Testimony: Dai-ichi Sankyo
Keith T. Flaherty
Stock and Other Ownership Interests: Clovis Oncology, Loxo, X4 Pharma, Strata Oncology, PIC Therapeutics, Apricity Health, Oncoceutics, FOGPharma, Tvardi Therapeutics, Checkmate Pharmaceuticals, Kinnate Biopharma, Scorpion Therapeutics, ALX Oncology, xCures, Monopteros Therapeutics, Vibliome Therapeutics, Transcode Therapeutics, Soley Therapeutics, Nextech Invest
Consulting or Advisory Role: Novartis, Lilly, Oncoceutics, Tvardi Therapeutics, Takeda, Debiopharm Group, OmRx, Quanta Therapeutics, Immagene
No other potential conflicts of interest were reported.
DISCLAIMER
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
SUPPORT
This study was coordinated by the ECOG-ACRIN Cancer Research Group (Peter J. O'Dwyer, MD, and Mitchell D. Schnall, MD, PhD, Group Co-Chairs) and supported by the National Cancer Institute of the National Institutes of Health under the following award numbers: U10CA180820, U10CA180794, UG1CA233184, UG1CA233329, UG1CA233341, UG1CA233302, UG1CA233180, UG1CA189816, UG1CA233328, U10CA180868, and UG1CA233277, as well as the University of Wisconsin Carbone Cancer Center Support Grant P30 CA014520.
CLINICAL TRIAL INFORMATION
M.A.W. contributed equally to this work.
AUTHOR CONTRIBUTIONS
Conception and design: Kari B. Wisinski, Jason J. Luke, Hussein A. Tawbi, Helen Chen, Robert J. Gray, Lisa M. McShane, Lawrence V. Rubinstein, David Patton, P. Mickey Williams, Stanley R. Hamilton, Barbara A. Conley, Alice P. Chen, Keith T. Flaherty
Financial support: David Patton
Administrative support: Hussein A. Tawbi, Helen Chen, David Patton, Lyndsay N. Harris
Provision of study materials or patients: Kari B. Wisinski, Jason J. Luke, Hussein A. Tawbi, Edith P. Mitchell, David Patton, Robert J. Behrens, Kathryn P. Pennington
Collection and assembly of data: Kari B. Wisinski, Yael Flamand, Jason J. Luke, Edith P. Mitchell, James A. Zwiebel, Robert J. Gray, Shuli Li, Lisa M. McShane, David Patton, P. Mickey Williams, Stanley R. Hamilton, Robert J. Behrens, Lyndsay N. Harris, Alice P. Chen
Data analysis and interpretation: Kari B. Wisinski, Yael Flamand, Melissa A. Wilson, Jason J. Luke, Hussein A. Tawbi, Fangxin Hong, Edith P. Mitchell, Helen Chen, Robert J. Gray, Shuli Li, Lisa M. McShane, David Patton, Stanley R. Hamilton, Kathryn P. Pennington, Carlos L. Arteaga, Lyndsay N. Harris, Alice P. Chen, Keith T. Flaherty
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Kari B. Wisinski
Honoraria: Pfizer
Consulting or Advisory Role: Sanofi, AstraZeneca, Pfizer, Novartis Institutes for BioMedical Research, Stemline Therapeutics
Research Funding: Novartis (Inst), AstraZeneca (Inst), Pfizer (Inst), Sanofi (Inst), Genentech (Inst), Context Therapeutics (Inst), Oncoceutics (Inst), Zymeworks (Inst), Relay Therapeutics (Inst)
Melissa A. Wilson
Consulting or Advisory Role: Instil Bio
Jason J. Luke
Stock and Other Ownership Interests: Actym Therapeutics, Mavu Pharmaceutical, Pyxis, Alphamab, Tempest Therapeutics, Kanaph Therapeutics, Onc.AI, Arch Oncology, STipe Therapeutics, NeoTX
Consulting or Advisory Role: Bristol Myers Squibb, Merck, EMD Serono, Novartis, Seven Hills Pharma, Janssen, Reflexion Medical, Tempest Therapeutics, Alphamab, AbbVie, Bayer, Incyte, Partner Therapeutics, Synlogic, Werewolf Therapeutics, Ribon Therapeutics, Checkmate Pharmaceuticals, CStone Pharmaceuticals, Nektar, Regeneron, Rubius Therapeutics, Tesaro, Xilio Therapeutics, Xencor, Alnylam, Crown Bioscience, Flame Biosciences, Genentech, Kadmon, KSQ Therapeutics, Immunocore, Inzen Therapeutics, Pfizer, Silicon Therapeutics, TRex Bio, Bright Peak Therapeutics, Onc.AI, STipe Therapeutics, Codiak Biosciences, Day One Therapeutics, Endeavor BioMedicines, Gilead Sciences, Hotspot Therapeutics, SERVIER, STINGthera, Synthekine
Research Funding: Merck (Inst), Bristol Myers Squibb (Inst), Incyte (Inst), Corvus Pharmaceuticals (Inst), AbbVie (Inst), Macrogenics (Inst), Xencor (Inst), Array BioPharma (Inst), Agios (Inst), Astellas Pharma (Inst), EMD Serono (Inst), Immatics (Inst), Kadmon (Inst), Moderna Therapeutics (Inst), Nektar (Inst), Spring Bank (Inst), Trishula Therapeutics (Inst), KAHR Medical (Inst), Fstar (Inst), Genmab (Inst), Ikena Oncology (Inst), Numab (Inst), Replimune (Inst), Rubius Therapeutics (Inst), Synlogic (Inst), Takeda (Inst), Tizona Therapeutics, Inc (Inst), BioNTech (Inst), Scholar Rock (Inst), NextCure (Inst)
Patents, Royalties, Other Intellectual Property: Serial #15/612,657 (Cancer Immunotherapy), Serial #PCT/US18/36052 (Microbiome Biomarkers for Anti-PD-1/PD-L1 Responsiveness: Diagnostic, Prognostic and Therapeutic Uses Thereof)
Travel, Accommodations, Expenses: Bristol Myers Squibb, Array BioPharma, EMD Serono, Janssen, Merck, Novartis, Reflexion Medical, Mersana, Pyxis, Xilio Therapeutics
Hussein A. Tawbi
Consulting or Advisory Role: Novartis, Bristol Myers Squibb, Genentech/Roche, Merck, Eisai, Iovance Biotherapeutics, Karyopharm Therapeutics, Boxer Capital, Jazz Pharmaceuticals, Pfizer, Medicenna
Research Funding: Bristol Myers Squibb (Inst), Novartis (Inst), Merck (Inst), GlaxoSmithKline (Inst), Genentech/Roche (Inst), Celgene (Inst), Dragonfly Therapeutics (Inst), RAPT Therapeutics (Inst)
Fangxin Hong
Employment: Pfizer
Stock and Other Ownership Interests: Pfizer
Consulting or Advisory Role: Merck Sharp & Dohme
Edith P. Mitchell
Leadership: Corvus Pharmaceuticals
Honoraria: Sanofi, Exelixis
Consulting or Advisory Role: Genentech, Novartis, Merck, Bristol Myers Squib
Speakers' Bureau: Ipsen
Research Funding: Genentech (Inst), Sanofi (Inst)
Robert J. Gray
Research Funding: Agios, Amgen, AstraZeneca, Bristol Myers Squibb, Boehringer Ingelheim, Celgene, Genentech/Roche, Genomic Health, Genzyme, GlaxoSmithKline, Janssen-Ortho, Onyx, Pfizer, Sequenta, Syndax, Novartis, Takeda, AbbVie, Sanofi, Merck Sharp & Dohme
Shuli Li
Employment: Takeda
Stock and Other Ownership Interests: Takeda
P. Mickey Williams
Research Funding: Illumina (Inst)
Patents, Royalties, Other Intellectual Property: I was a co-inventor of the DLBCL cell of origin patent recently filed by the NIH
Stanley R. Hamilton
Research Funding: Minerva Biotechnologies, Intima
Carlos L. Arteaga
Stock and Other Ownership Interests: Provista Diagnostics
Consulting or Advisory Role: Novartis, Lilly, Sanofi, Radius Health, Taiho Pharmaceutical, Puma Biotechnology, Merck, Origimed, Immunomedics, Daiichi Sankyo, Athenex, AstraZeneca, Arvinas
Research Funding: Pfizer, Lilly, Takeda
Other Relationship: Susan G. Komen for the Cure
Uncompensated Relationships: Susan G. Komen for the Cure Foundation
Lyndsay N. Harris
Patents, Royalties, Other Intellectual Property: Philips Healthcare
Peter J. O'Dwyer
Research Funding: Bristol Myers Squibb (Inst), Pfizer (Inst), Novartis (Inst), Genentech (Inst), Mirati Therapeutics (Inst), Celgene (Inst), GlaxoSmithKline (Inst), BBI Healthcare (Inst), Pharmacyclics (Inst), Five Prime Therapeutics (Inst), 47 (Inst), Amgen (Inst), H3 Biomedicine (Inst), Taiho Pharmaceutical (Inst), Array BioPharma (Inst), Lilly/ImClone (Inst), Syndax (Inst), Minneamrita Therapeutics (Inst)
Expert Testimony: Dai-ichi Sankyo
Keith T. Flaherty
Stock and Other Ownership Interests: Clovis Oncology, Loxo, X4 Pharma, Strata Oncology, PIC Therapeutics, Apricity Health, Oncoceutics, FOGPharma, Tvardi Therapeutics, Checkmate Pharmaceuticals, Kinnate Biopharma, Scorpion Therapeutics, ALX Oncology, xCures, Monopteros Therapeutics, Vibliome Therapeutics, Transcode Therapeutics, Soley Therapeutics, Nextech Invest
Consulting or Advisory Role: Novartis, Lilly, Oncoceutics, Tvardi Therapeutics, Takeda, Debiopharm Group, OmRx, Quanta Therapeutics, Immagene
No other potential conflicts of interest were reported.
REFERENCES
- 1. Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 2. Imperial R, Toor OM, Hussain A, et al. Comprehensive pancancer genomic analysis reveals (RTK)-RAS-RAF-MEK as a key dysregulated pathway in cancer: Its clinical implications. Semin Cancer Biol. 2019;54:14–28. doi: 10.1016/j.semcancer.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 3. Flaherty KT, Robert C, Hersey P, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 4. Patil S, Chamberlain RS. Neoplasms associated with germline and somatic NF1 gene mutations. Oncologist. 2012;17:101–116. doi: 10.1634/theoncologist.2010-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Philpott C, Tovell H, Frayling IM, et al. The NF1 somatic mutational landscape in sporadic human cancers. Hum Genomics. 2017;11:13. doi: 10.1186/s40246-017-0109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Denayer E, de Ravel T, Legius E. Clinical and molecular aspects of RAS related disorders. J Med Genet. 2008;45:695–703. doi: 10.1136/jmg.2007.055772. [DOI] [PubMed] [Google Scholar]
- 7. See WL, Tan IL, Mukherjee J, et al. Sensitivity of glioblastomas to clinically available MEK inhibitors is defined by neurofibromin 1 deficiency. Cancer Res. 2012;72:3350–3359. doi: 10.1158/0008-5472.CAN-12-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jessen WJ, Miller SJ, Jousma E, et al. MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J Clin Invest. 2013;123:340–347. doi: 10.1172/JCI60578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dombi E, Baldwin A, Marcus LJ, et al. Activity of selumetinib in neurofibromatosis type 1-related plexiform neurofibromas. N Engl J Med. 2016;375:2550–2560. doi: 10.1056/NEJMoa1605943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.cBioPortal.org
- 13. Chen X, Wu Q, Depeille P, et al. RasGRP3 mediates MAPK pathway activation in GNAQ mutant uveal melanoma. Cancer Cell. 2017;31:685–696.e6. doi: 10.1016/j.ccell.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Hayre M, Vazquez-Prado J, Kufareva I, et al. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat Rev Cancer. 2013;13:412–424. doi: 10.1038/nrc3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ambrosini G, Pratilas CA, Qin LX, et al. Identification of unique MEK-dependent genes in GNAQ mutant uveal melanoma involved in cell growth, tumor cell invasion, and MEK resistance. Clin Cancer Res. 2012;18:3552–3561. doi: 10.1158/1078-0432.CCR-11-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carvajal RD, Sosman JA, Quevedo JF, et al. Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: A randomized clinical trial. JAMA. 2014;311:2397–2405. doi: 10.1001/jama.2014.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Livingstone E, Zaremba A, Horn S, et al. GNAQ and GNA11 mutant nonuveal melanoma: A subtype distinct from both cutaneous and uveal melanoma. Br J Dermatol. 2020;183:928–939. doi: 10.1111/bjd.18947. [DOI] [PubMed] [Google Scholar]
- 18. Khoury JD, Wang WL, Prieto VG, et al. Validation of immunohistochemical assays for integral biomarkers in the NCI-MATCH EAY131 clinical trial. Clin Cancer Res. 2018;24:521–531. doi: 10.1158/1078-0432.CCR-17-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lih CJ, Harrington RD, Sims DJ, et al. Analytical validation of the next-generation sequencing assay for a nationwide signal-finding clinical trial: Molecular analysis for therapy choice clinical trial. J Mol Diagn. 2017;19:313–327. doi: 10.1016/j.jmoldx.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Conley BA, Flaherty KT. Molecular analysis for therapy choice (NCI-MATCH): A precision medicine signal-seeking trial in oncology. Personalized Med Oncol. 2016;5 https://www.personalizedmedonc.com/publications/pmo/october-2016-vol-5-no-8/molecular-analysis-for-therapy-choice-nci-match/ [Google Scholar]
- 21. Flaherty KT, Gray R, Chen A, et al. The molecular analysis for therapy choice (NCI-MATCH) trial: Lessons for genomic trial design. J Natl Cancer Inst. 2020;112:1021–1029. doi: 10.1093/jnci/djz245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 23. Lin NU, Lee EQ, Aoyama H, et al. Response assessment criteria for brain metastases: Proposal from the RANO group. Lancet Oncol. 2015;16:e270–e278. doi: 10.1016/S1470-2045(15)70057-4. [DOI] [PubMed] [Google Scholar]
- 24. Vogel CL, Cobleigh MA, Tripathy D, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 25. Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N Engl J Med. 2020;382:41–50. doi: 10.1056/NEJMoa1913662. [DOI] [PubMed] [Google Scholar]
- 26. Hong DS, DuBois SG, Kummar S, et al. Larotrectinib in patients with TRK fusion-positive solid tumours: A pooled analysis of three phase 1/2 clinical trials. Lancet Oncol. 2020;21:531–540. doi: 10.1016/S1470-2045(19)30856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seeber A, Chahine G, Nasr F, et al. Treatment according to a comprehensive molecular profiling can lead to a better outcome in heavily pretreated patients with metastatic cancer: Data of a pooled analysis. Cancer J. 2019;25:73–79. doi: 10.1097/PPO.0000000000000358. [DOI] [PubMed] [Google Scholar]
- 28. Lake D, Corrêa SAL, Muller J. Negative feedback regulation of the ERK1/2 MAPK pathway. Cell Mol Life Sci. 2016;73:4397–4413. doi: 10.1007/s00018-016-2297-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen X, Wu Q, Tan L, et al. Combined PKC and MEK inhibition in uveal melanoma with GNAQ and GNA11 mutations. Oncogene. 2014;33:4724–4734. doi: 10.1038/onc.2013.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carvajal RD, Schwartz GK, Tezel T, et al. Metastatic disease from uveal melanoma: Treatment options and future prospects. Br J Ophthalmol. 2017;101:38–44. doi: 10.1136/bjophthalmol-2016-309034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao Y, Adjei AA. The clinical development of MEK inhibitors. Nat Rev Clin Oncol. 2014;11:385–400. doi: 10.1038/nrclinonc.2014.83. [DOI] [PubMed] [Google Scholar]
- 32. Zhu Y, Guignard F, Zhao D, et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Herting CJ, Chen Z, Pitter KL, et al. Genetic driver mutations define the expression signature and microenvironmental composition of high-grade gliomas. Glia. 2017;65:1914–1926. doi: 10.1002/glia.23203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sintupisut N, Liu PL, Yeang CH. An integrative characterization of recurrent molecular aberrations in glioblastoma genomes. Nucleic Acids Res. 2013;41:8803–8821. doi: 10.1093/nar/gkt656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. The Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ameratunga M, McArthur G, Gan H, et al. Prolonged disease control with MEK inhibitor in neurofibromatosis type I-associated glioblastoma. J Clin Pharm Ther. 2016;41:357–359. doi: 10.1111/jcpt.12378. [DOI] [PubMed] [Google Scholar]
- 38. Khan I, Waqas M, Shamim MS. Prognostic significance of IDH 1 mutation in patients with glioblastoma multiforme. J Pak Med Assoc. 2017;67:816–817. [PubMed] [Google Scholar]
- 39. Blumenschein GR, Jr, Smit EF, Planchard D, et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC) Ann Oncol. 2015;26:894–901. doi: 10.1093/annonc/mdv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Redig AJ, Capelletti M, Dahlberg SE, et al. Clinical and molecular characteristics of NF1-mutant lung cancer. Clin Cancer Res. 2016;22:3148–3156. doi: 10.1158/1078-0432.CCR-15-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gross AM, Wolters PL, Dombi E, et al. Selumetinib in children with inoperable plexiform neurofibromas. N Engl J Med. 2020;382:1430–1442. doi: 10.1056/NEJMoa1912735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McCowage GB, Mueller S, Pratilas CA, et al. Trametinib in pediatric patients with neurofibromatosis type 1 (NF-1)–associated plexiform neurofibroma: A phase I/IIa study. J Clin Oncol. 2018;36:10504. [Google Scholar]
- 43. Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shoushtari AN, Kudchadkar RR, Panageas K, et al. A randomized phase 2 study of trametinib with or without GSK2141795 in patients with advanced uveal melanoma. J Clin Oncol. 2016;34:9511. [Google Scholar]