PURPOSE
Circulating tumor DNA (ctDNA) has been validated across multiple indications in the adjuvant and surveillance settings. We evaluated whether targeted digital sequencing (TARDIS) may distinguish a partial response (PR) from a complete response (CR) among patients with metastatic renal cell carcinoma (mRCC) receiving immune checkpoint inhibitor (ICI) therapy.
MATERIALS AND METHODS
Eligible patients had mRCC that yielded a PR or CR to ICI therapy. Peripheral blood was obtained at a single time point for ctDNA analysis. TARDIS was used for quantification of average variant allele fractions (VAFs). Our primary objective was to determine the association between VAFs and depth of response (PR v CR). A secondary objective was to determine whether VAFs were associated with disease progression.
RESULTS
Twelve patients were analyzed, nine of whom achieved a PR (75%). Patients received either nivolumab monotherapy (50%) or nivolumab plus ipilimumab (50%). ctDNA analysis incorporated an average of 30 patient-specific mutations (range, 19-35); average coverage depth was 103,342 reads per target. TARDIS quantified a significant difference in VAFs between PR and CR (median, 0.181% [IQR, 0.077%-0.420%] v 0.007% [IQR, 0.0%-0.028%], respectively [P = .014]). Of the 12 patients in the series, six patients demonstrated radiographic progression subsequent to ctDNA assessment. Patients who progressed on subsequent scans had significantly higher ctDNA than those who maintained their response (median, 0.362% [IQR, 0.181%-2.71%] v 0.033% [IQR, 0.007%-0.077%], respectively [P = .026]).
CONCLUSION
In this pilot study, TARDIS accurately differentiated PR from CR among patients with mRCC receiving immunotherapy, and also prospectively identified patients at risk for subsequent progression. Given these findings, we envision subsequent studies that validate these results and investigate the utility of this assay to discern appropriate candidates for discontinuation of immunotherapy.
INTRODUCTION
Immune checkpoint inhibitors (ICIs) have become a cornerstone of frontline therapy for metastatic renal cell carcinoma (mRCC), either with dual ICI therapy or in combination with targeted therapy now representing the standard of care.1-4 An emerging challenge is whether treatment with ICIs can be withdrawn among those patients who achieve a durable response to therapy. The rate of complete response (CR) varies across malignancies with ICI therapy, ranging from 19% to 22% in advanced melanoma and 1% to 3% in advanced non–small-cell lung cancer.5-8 In registrational mRCC trials, frontline ICI-based therapy demonstrated considerably higher rates of partial response (32%) and CR (9%) compared with second-line therapy (24% and 1%, respectively).4,9 Beyond these subsets lies an additional group of patients who achieve a durable response to therapy.
CONTEXT
Key Objective
Previous applications of circulating tumor DNA (ctDNA) assays in patients with renal cell carcinoma (RCC) have yielded low sensitivity and specificity. Targeted digital sequencing (TARDIS) is a novel tumor-informed ctDNA assay capable of using up to 100 baseline mutations. We sought to determine the association between variant allele fractions (VAFs) and depth of response to immunotherapy in patients with metastatic RCC.
Knowledge Generated
TARDIS achieved a significant difference in VAF concentrations between partial responders and complete responders (P = .018). Additionally, TARDIS reached a significant difference in VAF concentrations between those with sustained radiographic response and those who progressed on subsequent imaging (P = .026).
Relevance
To our knowledge, this pilot study represents the first ctDNA assay to effectively discriminate between partial and complete responses to immunotherapy in RCC. These data indicate that TARDIS may be an efficacious platform for detecting molecular residual disease and relapse in metastatic RCC.
Analysis of circulating tumor DNA (ctDNA) is a noninvasive approach to detect and quantify tumor-associated mutations in the plasma of patients with cancer. Current methods for ctDNA analysis can be classified into one of two categories: tumor-agnostic and tumor-informed.10 The former uses standardized, fixed panels that target known somatic mutations without sequencing patient-specific formalin-fixed paraffin-embedded (FFPE) tumor samples to identify alterations. The latter uses next-generation genomic sequencing of patient FFPE tumor tissue to identify patient-specific alterations for the development of bespoke ctDNA panels for subsequent blood surveillance.
One of the major challenges with ctDNA analysis has been achieving sensitivity and quantitative precision in the setting of low ctDNA concentrations, despite limited blood volumes, to effectively prognosticate clinical outcomes.11-14 To this end, we have developed a novel bespoke platform (targeted digital sequencing [TARDIS]) that ascertains up to 100 baseline mutations in patient FFPE tumor tissue to create a personalized assay for blood sample analysis.15 Preliminary results from 33 patients with early and locally advanced breast cancer identified pretreatment ctDNA in 100% of patients at a mean concentration of 0.11%. After neoadjuvant chemotherapy, ctDNA dropped to 0.017% and 0.003% in patients with residual disease versus pathologic CR, respectively.16 In the current study, we hypothesized that if TARDIS is capable of detecting ctDNA in patients with mRCC receiving ICI therapy, then it could discriminate PRs from CRs.
MATERIALS AND METHODS
Patient Selection and Sample Acquisition
Between July 1, 2020, and October 1, 2020, patients diagnosed with mRCC by standard criteria were prospectively identified at a single center using an institutional database.17 Patients were eligible if they achieved a PR or CR to a commercially available ICI (PD-1 and/or CTLA-4 inhibitor). Enrollment was open to patients across all renal cell carcinoma (RCC) histologic subtypes and lines of therapy. Whole blood was collected at a single time point in two 10-mL Cell-Free DNA BCT tubes (Streck, La Vista, NE) from eligible patients. Demographic data were collected for each patient.
The protocol was approved by the institutional scientific review committee, data safety monitoring board, and the institutional review board at the City of Hope Comprehensive Cancer Center. The study conformed to the amended Declaration of Helsinki and the International Conference on Harmonisation Guidelines.
Tumor Genomic Sequencing and Analysis
For whole-genome sequencing (WGS), DNA was extracted from FFPE tumor samples using a truXTRAC Total Kit (Covaris, Inc, Woburn, MA) or from blood as a source of normal DNA using QIAcube automated sample preparation system (QIAGEN USA, Chatsworth, CA). DNA was sheared to a mean size of 200 bp using a model S220 sonicator (Covaris Inc, Woburn, MA) and prepared into libraries using ThruPLEX DNA-Seq Kits (Takara Bio USA, San Jose, CA). Libraries were pooled in equimolar fashion and sequenced at a read length of 2 × 150 bp on an Illumina NovaSeq 6000 instrument using a S4 300-cycle kit (Illumina, San Diego, CA) to a mean coverage of 18× for normal samples (range, 15×-21×) and 26× for tumor samples (range, 14×-43×). Sequencing data processing and variant calling were performed as described.16
Plasma Multiplex TARDIS
TARDIS assay primer pools were designed to target 36 somatic variants per patient, as identified by tumor/normal WGS. After oligonucleotide production, pools were functionally tested using TARDIS assays on sheared control human cell line DNA. Primer pools that passed this quality control were used in the TARDIS assay to probe 2.6-21.1 ng of cell-free DNA isolated from 3.7 to 5.3 mL of matched patient plasma using the MagMAX Cell-Free DNA Isolation Kit (Thermo Fisher Scientific, Waltham, MA) as described.16
TARDIS libraries were sequenced at a read length of 2 × 100 bp on an Illumina NovaSeq 6000 instrument using S1 200-cycle kits. Paired-end sequencing reads were analyzed using the TARDIS data analysis pipeline and aligned to human genome hg19 using BWA-MEM as described.16
Statistical Methods
Patients were treated with an ICI until disease progression or discontinuation because of adverse events, death, or subject/investigator decision. Response to therapy was assessed per clinician's evaluation of computerized tomography of the chest, abdomen, and pelvis. Clinicians were blinded to ctDNA results at the time of radiographic assessments.
Comparison of ctDNA values between groups was made using the Kruskal-Wallis test. Patient characteristics were compared between PR and CR patients with the Fisher exact test or Wilcoxon rank-sum test for discrete and continuous variables, respectively. The significance threshold for type I error was set at 0.05. SASV9.4 software program was used to perform statistical analyses.
RESULTS
Patient Characteristics
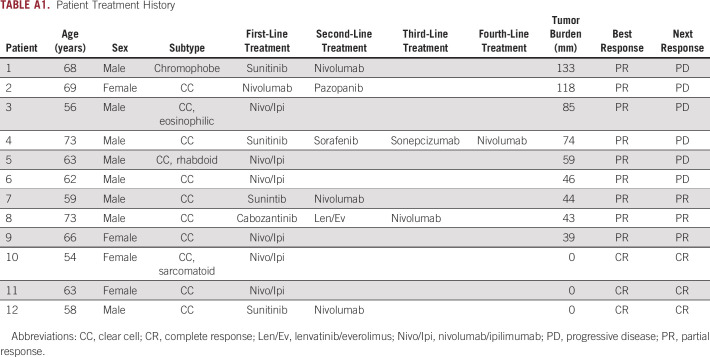
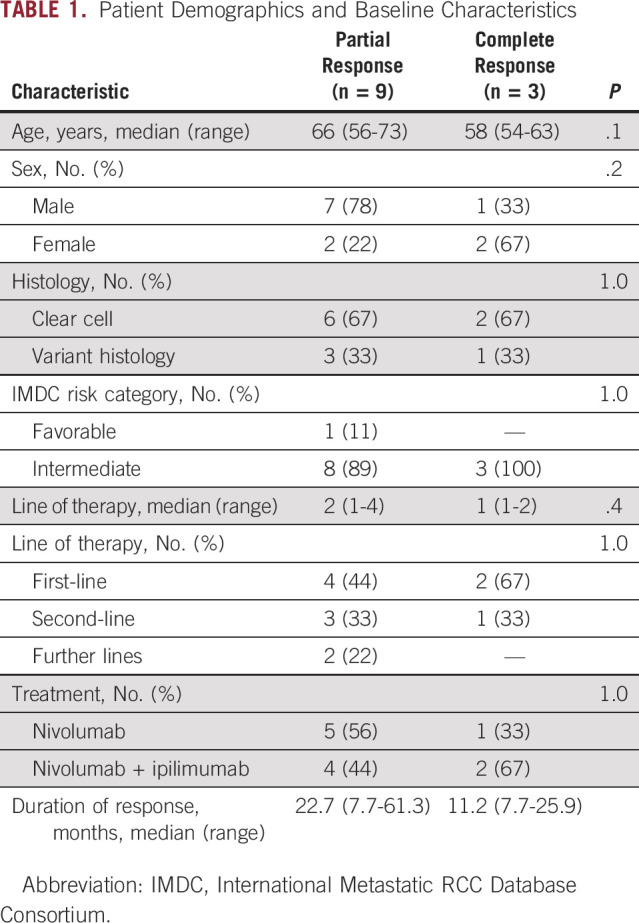
Of 23 patients enrolled, 10 patients did not have sufficient tissue available for WGS and one patient was unable to be evaluated via the TARDIS platform because of suboptimal primer development, leaving 12 patients for ctDNA analysis. Of these 12 patients, 8 (67%) were male and 4 (33%) were female, with a median age of 63 years (range, 54-73). Median lines of therapy received was 1.5 (range, 1-4), median duration of therapy was 24.4 months (range, 7.7-61.3), and median follow-up was 93 days (range, 80-133); no patients were lost to follow-up. Most patients had clear cell histology (92%) and were International Metastatic RCC Database Consortium (IMDC) intermediate risk (92%). All patients had previously received a nephrectomy before initiation of systemic therapy. Treatment was equally distributed between nivolumab plus ipilimumab (50%) and nivolumab monotherapy (50%). Nine patients (75%) achieved a PR, and three patients (25%) achieved a CR. Upon subsequent scans, six patients who previously achieved a PR experienced disease progression. A summary of patient baseline characteristics is presented in Table 1.
TABLE 1.
Patient Demographics and Baseline Characteristics
Tumor Genomic Profiling
Using WGS to identify candidate genes for our bespoke assay, an average of 30 patient-specific mutations (range, 19-35) were identified for the quantification of VAFs. Multiplexed sequencing achieved a mean target coverage of 103,342 reads. We also characterized tumor mutational profiles via whole exome sequencing (WES) as part of routine care. The most frequently altered genes in our series were VHL (67%), SMARCA4 (25%), BAP1 (17%), PBRM1 (17%), SETD2 (17%), and TSC1 (17%). A summary of mutation frequency, mutational signatures, and frequently mutated genes is shown in Figure 1.
FIG 1.
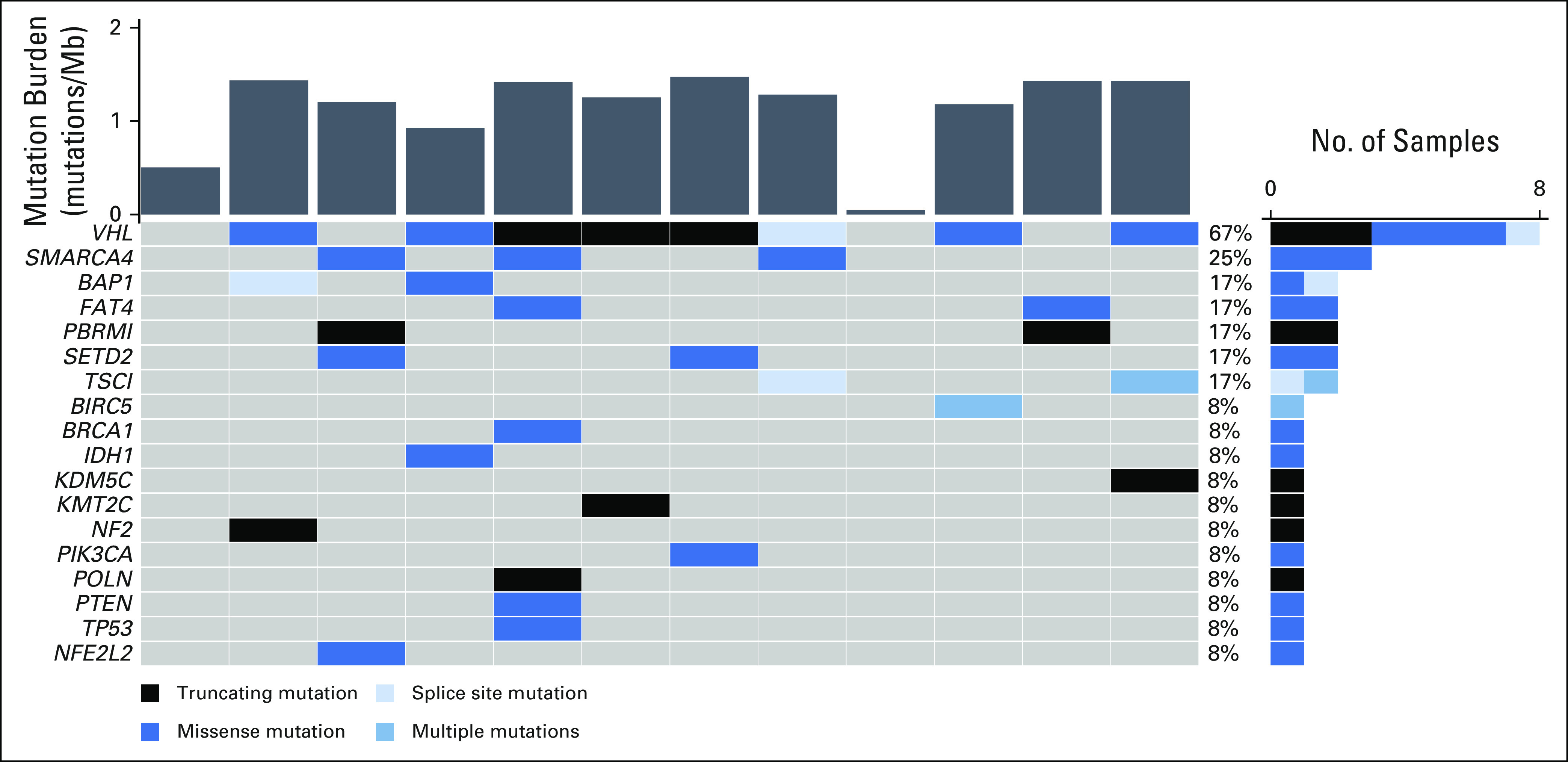
Summary of tumor molecular profiles. Tumor mutational burden (top) along with mutations in frequently mutated genes (bottom) and their relative frequencies (right) for all patients.
ctDNA Detection via Ultrasensitive Multiplex Polymerase Chain Reaction–Based Next-Generation Sequencing
Across our 12 patients, median cell-free DNA yield was 5.1 ng/mL of plasma (range, 2.6-21.1 ng/mL). ctDNA was detected in all but one patient at baseline (median, 0.101%, range, 0%-11.315%). The difference in ctDNA concentration when compared by sex (male v female), age (<65 v ≥65 years), type of therapy (nivolumab v nivolumab plus ipilimumab), or histologic subtype (clear cell v variant) was not statistically significant. Clinical characteristics are depicted in Figure 2.
FIG 2.
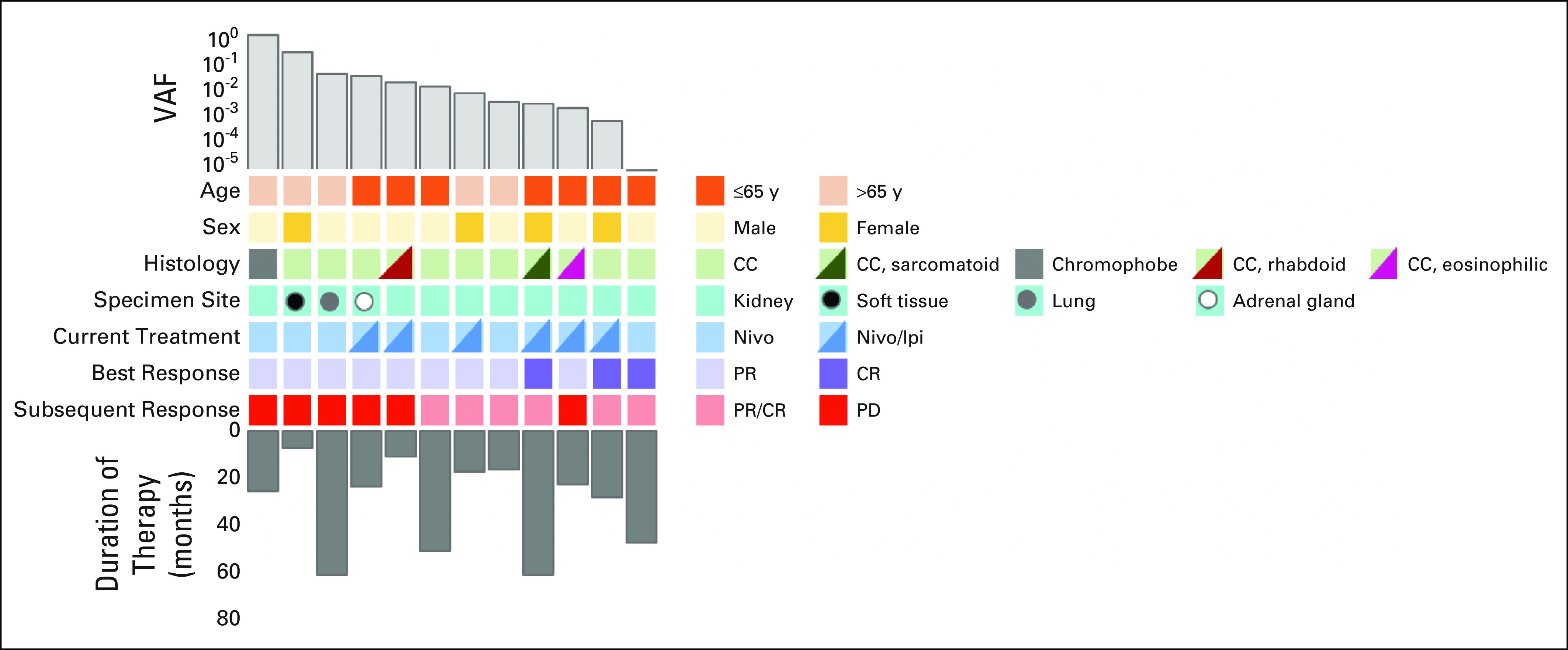
Clinicopathologic and molecular characteristics. Relevant clinical features and VAFs for all patients. CC, clear cell; CR, complete response; Nivo, nivolumab; Nivo/Ipi, nivolumab plus ipilimumab; PD, progressive disease; PR, partial response; VAFs, variant allele fractions.
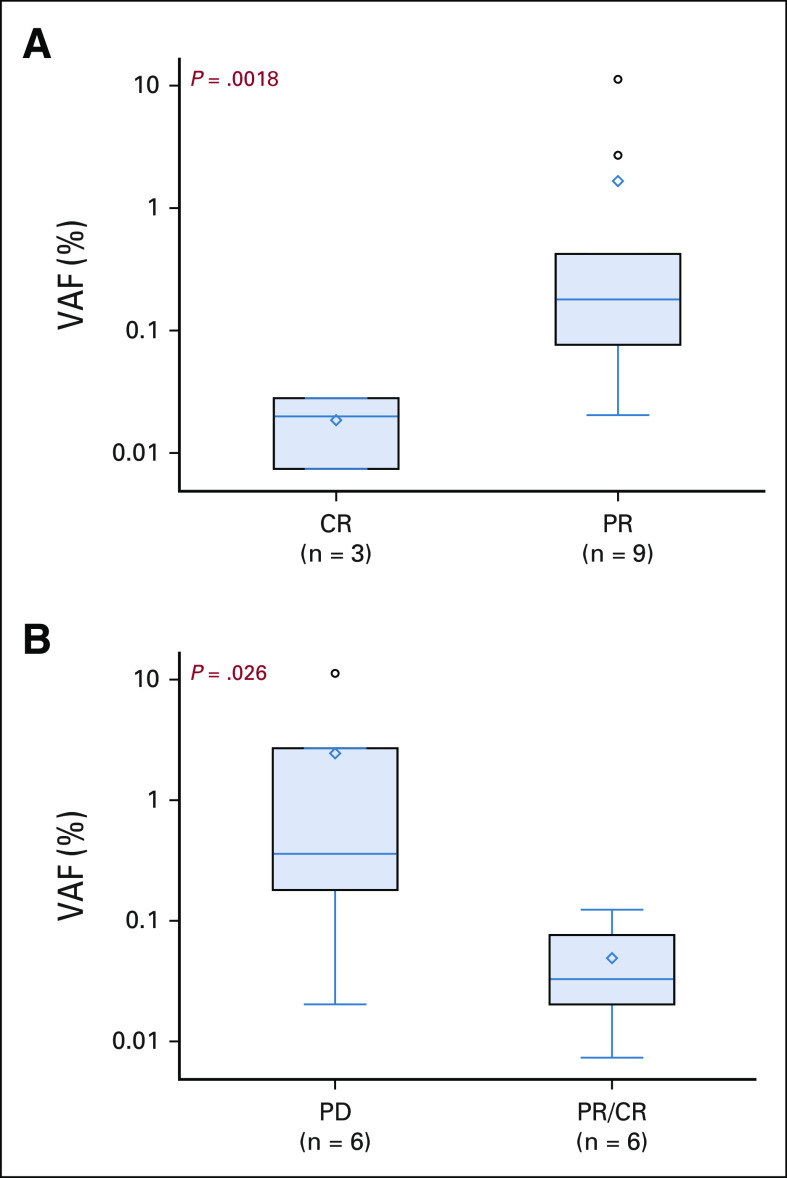
Of the 12 patients in the series, those with a CR had a significantly lower ctDNA concentration than patients with a PR (median, 0.007% [IQR, 0.0%-0.028%] v 0.181% [IQR, 0.077%-0.420%], respectively [P = .014]). Additionally, six patients demonstrated radiographic progression subsequent to ctDNA assessment. Patients who progressed on subsequent scans had significant higher baseline ctDNA than those who maintained their response (median, 0.362% [IQR, 0.181%-2.71%] v 0.033% [IQR, 0.007%-0.077%], respectively [P = .026]). VAF analysis is summarized in Figure 3.
FIG 3.
Association of ctDNA with clinical outcomes. (A) Discrimination of PR and CR using TARDIS. (B) Discrimination of sustained radiographic response versus subsequent progression. CR, complete response; ctDNA, circulating tumor DNA; PD, progressive disease; PR, partial response; TARDIS, targeted digital sequencing.
DISCUSSION
To our knowledge, this pilot study represents the first ctDNA assay to effectively discriminate between PRs and CRs to immunotherapy in mRCC. Our findings suggest that TARDIS may be an effective bespoke platform for monitoring molecular residual disease and relapse in mRCC.
Development of a reliable ctDNA assay for mRCC has historically been a formidable challenge. One prevailing theory is that the low detection rates in RCC—which range from 30% to 40% in older studies using tumor-agnostic assays—are due to low levels of ctDNA shed into the plasma.18-20 However, in the largest assessment of ctDNA in mRCC to date, our group detected genomic alterations in 71.8% of patients,21 commensurate to rates seen in other tumor types.22 Using TARDIS, a targeted tumor-informed approach, yielded higher assay sensitivity and specificity.
Unfortunately, previous efforts using such a tumor-informed approach in mRCC have encountered comparably low yields to their tumor-agnostic counterparts. Using a commercially available bespoke assay guided by tumor WES, Correa et al detected ctDNA in 14 of 34 (41%) patients with RCC of varying stage.23 After definitive surgical resection, 16 of 33 patients deemed ctDNA-negative by the assay subsequently relapsed, corresponding to a negative predictive value of 52%. Similarly, Jang et al24 prospectively applied the aforementioned bespoke assay to five patients with mRCC starting on ICI therapy. These authors reported a concordance rate between ctDNA response and radiographic response of only 60%. One potential explanation for the higher apparent predictive value of TARDIS may lie in our application of WGS rather than WES. By encompassing entire tumor genomes for the development of our bespoke ctDNA panels, we were also able to target noncoding aberrations that would otherwise be overlooked via WES, enabling a greater number of mutations analyzed in plasma DNA.
Our assay builds upon a growing body of evidence that suggests larger quantities of genomic targets may enhance the depth of ctDNA detection. One such commercially available 16-gene assay reached a limit of detection of 0.034% VAF in patients receiving pembrolizumab.25 Another assay, which uses up to 48 tumor-specific variants, reported ctDNA detection at levels as low as 26 parts per million (equivalent to 0.0026% VAF) in patients with postsurgical head and neck squamous cell carcinoma.26 In between lies TARDIS, which in our study incorporated an average of 30 patient-specific mutations and achieved a detection limit of 0.007% VAF. Taken together, these data support the premise that increasing the quantity of patient-specific mutations augments the effective depth of sequencing per sample, improving the sensitivity and quantitative precision for ctDNA analysis.
As previously noted, a modest proportion of patients on ICIs will mount a CR to ICI-based combination therapy, and an even larger subset may have durable PRs. Presumably, patients with a higher ctDNA concentration after initiation of systemic therapy would be candidates for continuation of treatment, and those with a lower ctDNA concentration might be spared further treatment. Although some have proposed using a durable radiographic response as a decision point for treatment discontinuation,27,28 there are obvious limitations with this approach—our study, for instance, highlights the detectability of molecular residual disease even in those patients with a radiographic CR. Additionally, it is challenging to ascertain the presence of active disease in certain sites of metastasis—one prominent example is bone metastases, where a sclerotic reaction can be challenging to differentiate from disease progression.
ctDNA also has potential application in earlier settings. In RCC, the phase III KEYNOTE-564 supports the role of adjuvant pembrolizumab in patients with high-risk localized RCC.29 Presumably, those patients with higher ctDNA burden would have derived greater benefit from therapy. This principle has borne out in muscle-invasive bladder cancer, where a distinct ultrasensitive test has been shown to predict clinical benefit from adjuvant therapy with the ICI atezolizumab using samples collected from a randomized, phase III study (ImVigor010).30 These data have led to the inception of ImVigor011 and TOMBOLA, both of which are biomarker-based randomized trials in muscle-invasive bladder cancer that will allocate treatment with adjuvant atezolizumab on the basis of the presence or absence of ctDNA (NCT04660344 and NCT04138628, respectively).
One of the limitations of this study was its nonrandomized observational design using an institutional database. In an effort to mitigate selection bias, all patients who met the inclusion criteria were included in this study. Another limitation was our high rate (43%) of insufficient tissue for WGS processing, which was primarily because these biopsies were consumed for unrelated research endeavors that predated the study herein. In addition, our sample size was small, lacked survival data, and incorporated one sample at varying time points on therapy; therefore, these data should be viewed as hypothesis-generating. However, we hope to remedy these limitations with a larger prospective validation study of patients with metastatic disease starting on ICI that is currently ongoing at our institution. Blood will be collected at baseline and at consistent time points during therapy. If meaningful distinctions in ctDNA are identified among patients with differing clinical response (eg, CR v PR, as in the current study), we will initiate studies assessing discontinuation of treatment in patients with low ctDNA levels after ICI therapy.
In conclusion, to our knowledge, the data presented herein are the first published report of TARDIS, an ultrasensitive ctDNA assay, for disease surveillance in mRCC. By employing a bespoke approach that uses WGS to identify an average of 30 patient-specific mutations, TARDIS was able to effectively differentiate patients who achieved a PR from those who achieved a CR with immunotherapy. These significant differences seen in ctDNA, if validated in larger series, imply that the assay may play a role in facilitating treatment discontinuation.
APPENDIX 1
TABLE A1.
Patient Treatment History
Alexander Chehrazi-Raffle
Honoraria: OncLive/MJH Life Sciences
Ramya Muddasani
Stock and Other Ownership Interests: AbbVie, Pfizer, Novavax, Cellectar, Forty Seven, Heat Biologics, Advaxis, Curis, Portola Pharmaceuticals, Aveo, Immunomedics, Onconova Therapeutics, NantKwest, NantHealth, Actinium Pharmaceuticals, Mereo BioPharma, DelMar Pharmaceuticals, Agenus, Epizyme, Aprea Therapeutics, Gritstone Bio, Athenex, crispr therapeutics, Vaxart
Nazli Dizman
Consulting or Advisory Role: Vivreon Biosciences
Luis Meza
Honoraria: Ipsen
Travel, Accommodations, Expenses: Ipsen
Tanya Dorff
Consulting or Advisory Role: Bayer, Janssen Oncology, Seattle Genetics, Exelixis, AstraZeneca, Pfizer, Astellas Pharma, Bayer, Sanofi/Aventis
Research Funding: Pfizer (Inst)
Tania Contente-Cuomo
Employment: Translational Genomics Research Institute
Devin Dinwiddie
Research Funding: Exact Sciences (Inst)
Bradon R. McDonald
Patents, Royalties, Other Intellectual Property: Inventor on a patent application licensed to Exact Sciences Corporation related to the TARDIS ctDNA diagnostic assay
Timothy McDaniel
Employment: Delfi Diagnostics
Stock and Other Ownership Interests: Delfi Diagnostics
Research Funding: Exact Sciences
Patents, Royalties, Other Intellectual Property: As an inventor at the Translational Genomics Research Institute, part of City of Hope (TGen), I received payments when my patent was licensed to Exact Sciences, Inc, per TGen's royalty-sharing policy
Travel, Accommodations, Expenses: Delfi Diagnostics
Frederick L. Baehner
Employment: Exact Sciences
Leadership: Exact Sciences
Stock and Other Ownership Interests: Exact Sciences
Muhammed Murtaza
Stock and Other Ownership Interests: PetDx
Consulting or Advisory Role: PetDx, Castle Biosciences
Patents, Royalties, Other Intellectual Property: IP licensed to Exact Sciences, IP licensed to Inivata, Pending patent applications for diagnostic methods
Sumanta K. Pal
This author is an Associate Editor for JCO Precision Oncology. Journal policy recused the author from having any role in the peer review of this manuscript.
Research Funding: Eisai (Inst), Genentech (Inst), Roche (Inst), Exelixis (Inst), Pfizer (Inst), crispr therapeutics (Inst), Allogene Therapeutics (Inst)
Travel, Accommodations, Expenses: crispr therapeutics, Roche
Open Payments Link: https://openpaymentsdata.cms.gov/physician/259259
No other potential conflicts of interest were reported.
SUPPORT
Supported by City of Hope Comprehensive Cancer Center, The Translational Genomics Institute, and Exact Sciences.
A.C.-R. and R.M. contributed equally to this work.
DATA SHARING STATEMENT
Genomic data from tissue and plasma specimens will be deposited at the Translational Genomics Research Institute (TGen) and will be available upon request. The authors defer depositing the participant genomic data in national and international public repositories because of institutional policies and the absence of statements in patient consent forms that would have allowed controlled access distribution and genomic data availability. Deidentified individual participant genomic libraries and clinical data that underlie the results reported in this article are available for transfer on a specific secure server housed at the TGen. Interested investigators can obtain and certify the data transfer agreement (DTA) and submit requests to the principal investigator, S.K.H. Proposals will be vetted by the TGen Data Access Committee. Investigators and institutions who consent to the terms of the DTA form, including but not limited to the use of these data for the purpose of a specific project and only for research purposes, and to protect the confidentiality of the data and limit the possibility of identification of participants in any way whatsoever for the duration of the agreement, will be granted access. TGen will then facilitate the transfer of the requested deidentified data. This mechanism is expected to be via an Aspera High-Speed File Transfer Server but TGen reserves the right to change the specific transfer method at any time, provided appropriate levels of access authorization and control can be maintained.
AUTHOR CONTRIBUTIONS
Conception and design: Alexander Chehrazi-Raffle, Nazli Dizman, Luis Meza, Neal Chawla, Muhammed Murtaza; Sumanta K. Pal
Administrative support: Timothy McDaniel, Jeffrey M. Trent
Provision of study materials or patients: Tania Contente-Cuomo; Devin Dinwiddie
Collection and assembly of data: Alexander Chehrazi-Raffle, Nazli Dizman, JoAnn Hsu, Jasnoor Malhotra, Tania Contente-Cuomo; Devin Dinwiddie, Jeffrey M. Trent, Muhammed Murtaza; Sumanta K. Pal
Data analysis and interpretation: Alexander Chehrazi-Raffle, Ramya Muddasani, Luis Meza, Zeynep B. Zengin, Tanya Dorff, Tania Contente-Cuomo; Devin Dinwiddie, Bradon R. McDonald, Timothy McDaniel, Frederick L. Baehner, Muhammed Murtaza; Sumanta K. Pal
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Alexander Chehrazi-Raffle
Honoraria: OncLive/MJH Life Sciences
Ramya Muddasani
Stock and Other Ownership Interests: AbbVie, Pfizer, Novavax, Cellectar, Forty Seven, Heat Biologics, Advaxis, Curis, Portola Pharmaceuticals, Aveo, Immunomedics, Onconova Therapeutics, NantKwest, NantHealth, Actinium Pharmaceuticals, Mereo BioPharma, DelMar Pharmaceuticals, Agenus, Epizyme, Aprea Therapeutics, Gritstone Bio, Athenex, crispr therapeutics, Vaxart
Nazli Dizman
Consulting or Advisory Role: Vivreon Biosciences
Luis Meza
Honoraria: Ipsen
Travel, Accommodations, Expenses: Ipsen
Tanya Dorff
Consulting or Advisory Role: Bayer, Janssen Oncology, Seattle Genetics, Exelixis, AstraZeneca, Pfizer, Astellas Pharma, Bayer, Sanofi/Aventis
Research Funding: Pfizer (Inst)
Tania Contente-Cuomo
Employment: Translational Genomics Research Institute
Devin Dinwiddie
Research Funding: Exact Sciences (Inst)
Bradon R. McDonald
Patents, Royalties, Other Intellectual Property: Inventor on a patent application licensed to Exact Sciences Corporation related to the TARDIS ctDNA diagnostic assay
Timothy McDaniel
Employment: Delfi Diagnostics
Stock and Other Ownership Interests: Delfi Diagnostics
Research Funding: Exact Sciences
Patents, Royalties, Other Intellectual Property: As an inventor at the Translational Genomics Research Institute, part of City of Hope (TGen), I received payments when my patent was licensed to Exact Sciences, Inc, per TGen's royalty-sharing policy
Travel, Accommodations, Expenses: Delfi Diagnostics
Frederick L. Baehner
Employment: Exact Sciences
Leadership: Exact Sciences
Stock and Other Ownership Interests: Exact Sciences
Muhammed Murtaza
Stock and Other Ownership Interests: PetDx
Consulting or Advisory Role: PetDx, Castle Biosciences
Patents, Royalties, Other Intellectual Property: IP licensed to Exact Sciences, IP licensed to Inivata, Pending patent applications for diagnostic methods
Sumanta K. Pal
This author is an Associate Editor for JCO Precision Oncology. Journal policy recused the author from having any role in the peer review of this manuscript.
Research Funding: Eisai (Inst), Genentech (Inst), Roche (Inst), Exelixis (Inst), Pfizer (Inst), crispr therapeutics (Inst), Allogene Therapeutics (Inst)
Travel, Accommodations, Expenses: crispr therapeutics, Roche
Open Payments Link: https://openpaymentsdata.cms.gov/physician/259259
No other potential conflicts of interest were reported.
REFERENCES
- 1. Motzer R, Alekseev B, Rha S-Y, et al. Lenvatinib plus pembrolizumab or everolimus for advanced renal cell carcinoma. N Engl J Med. 2021;384:1289–1300. doi: 10.1056/NEJMoa2035716. [DOI] [PubMed] [Google Scholar]
- 2. Choueiri TK, Powles T, Burotto M, et al. Nivolumab plus cabozantinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med. 2021;384:829–841. doi: 10.1056/NEJMoa2026982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rini BI, Plimack ER, Stus V, et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med. 2019;380:1116–1127. doi: 10.1056/NEJMoa1816714. [DOI] [PubMed] [Google Scholar]
- 4. Motzer RJ, Tannir NM, McDermott DF, et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N Engl J Med. 2018;378:1277–1290. doi: 10.1056/NEJMoa1712126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garon EB, Hellmann MD, Rizvi NA, et al. Five-year overall survival for patients with advanced non‒small-cell lung cancer treated with pembrolizumab: Results from the phase I KEYNOTE-001 study. J Clin Oncol. 2019;37:2518–2527. doi: 10.1200/JCO.19.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wolchok JD, Chiarion-Sileni V, Gonzalez R, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377:1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non–small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 8. Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2019;381:1535–1546. doi: 10.1056/NEJMoa1910836. [DOI] [PubMed] [Google Scholar]
- 9. Motzer RJ, Escudier B, McDermott DF, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cescon DW, Bratman SV, Chan SM, et al. Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer. 2020;1:276–290. doi: 10.1038/s43018-020-0043-5. [DOI] [PubMed] [Google Scholar]
- 11. Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5:1124. doi: 10.1001/jamaoncol.2019.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abbosh C, Birkbak NJ, Wilson GA, et al. Phylogenetic ctDNA analysis depicts early stage lung cancer evolution. Nature. 2017;545:446–451. doi: 10.1038/nature22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coombes RC, Page K, Salari R, et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin Cancer Res. 2019;25:4255–4263. doi: 10.1158/1078-0432.CCR-18-3663. [DOI] [PubMed] [Google Scholar]
- 14. Christensen E, Birkenkamp-Demtröder K, Sethi H, et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J Clin Oncol. 2019;37:1547–1557. doi: 10.1200/JCO.18.02052. [DOI] [PubMed] [Google Scholar]
- 15. McDonald BR, Contente-Cuomo T, Sammut S-J, et al. Abstract P4-01-21: Multiplexed targeted digital sequencing of circulating tumor DNA to detect minimal residual disease in early and locally advanced breast cancer. Cancer Res. 2019;79:P4-01-21. [Google Scholar]
- 16. McDonald BR, Contente-Cuomo T, Sammut S-J, et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Translational Med. 2019;11:eaax7392. doi: 10.1126/scitranslmed.aax7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amin MB, Greene FL, Edge SB, et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67:93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 18. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamamoto Y, Uemura M, Fujita M, et al. Clinical significance of the mutational landscape and fragmentation of circulating tumor DNA in renal cell carcinoma. Cancer Sci. 2019;110:617–628. doi: 10.1111/cas.13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smith CG, Moser T, Mouliere F, et al. Comprehensive characterization of cell-free tumor DNA in plasma and urine of patients with renal tumors. Genome Med. 2020;12:23. doi: 10.1186/s13073-020-00723-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zengin ZB, Weipert C, Salgia NJ, et al. Complementary role of circulating tumor DNA assessment and tissue genomic profiling in metastatic renal cell carcinoma. Clin Cancer Res. 2021;27:4807–4813. doi: 10.1158/1078-0432.CCR-21-0572. [DOI] [PubMed] [Google Scholar]
- 22. Lanman RB, Mortimer SA, Zill OA, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One. 2015;10:e0140712. doi: 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Correa A, Connolly DC, Balcioglu M, et al. Presence of circulating tumour DNA in surgically resected renal cell carcinoma is associated with advanced disease and poor patient prognosis. Ann Oncol. 2019;30:v32. [Google Scholar]
- 24. Jang A, Jaeger E, Rauterkus G, et al. MP12-02 prospective evaluation of circulating tumor DNA (ctDNA) in detecting early progression on immune checkpoint inhibitors in patients with advanced genitourinary cancers. J Urol. 2022;207:e165. [Google Scholar]
- 25. Bratman SV, Yang SYC, Iafolla MAJ, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer. 2020;1:873–881. doi: 10.1038/s43018-020-0096-5. [DOI] [PubMed] [Google Scholar]
- 26. Flach S, Howarth K, Hackinger S, et al. 884P Personalised circulating cell-free tumour DNA analysis for detection of minimal residual disease and recurrence in patients with head and neck squamous cell carcinoma. Ann Oncol. 2021;32:S795–S796. [Google Scholar]
- 27. Robert C, Marabelle A, Herrscher H, et al. Immunotherapy discontinuation—How, and when? Data from melanoma as a paradigm. Nat Rev Clin Oncol. 2020;17:707–715. doi: 10.1038/s41571-020-0399-6. [DOI] [PubMed] [Google Scholar]
- 28. Zambrana F, Carril-Ajuria L, Gómez de Liaño A, et al. Complete response and renal cell carcinoma in the immunotherapy era: The paradox of good news. Cancer Treat Rev. 2021;99:102239. doi: 10.1016/j.ctrv.2021.102239. [DOI] [PubMed] [Google Scholar]
- 29. Choueiri TK, Tomczak P, Park SH, et al. Pembrolizumab versus placebo as post-nephrectomy adjuvant therapy for patients with renal cell carcinoma: Randomized, double-blind, phase III KEYNOTE-564 study. J Clin Oncol. 2021;39:LBA5. [Google Scholar]
- 30. Powles T, Assaf ZJ, Davarpanah N, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021;595:432–437. doi: 10.1038/s41586-021-03642-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genomic data from tissue and plasma specimens will be deposited at the Translational Genomics Research Institute (TGen) and will be available upon request. The authors defer depositing the participant genomic data in national and international public repositories because of institutional policies and the absence of statements in patient consent forms that would have allowed controlled access distribution and genomic data availability. Deidentified individual participant genomic libraries and clinical data that underlie the results reported in this article are available for transfer on a specific secure server housed at the TGen. Interested investigators can obtain and certify the data transfer agreement (DTA) and submit requests to the principal investigator, S.K.H. Proposals will be vetted by the TGen Data Access Committee. Investigators and institutions who consent to the terms of the DTA form, including but not limited to the use of these data for the purpose of a specific project and only for research purposes, and to protect the confidentiality of the data and limit the possibility of identification of participants in any way whatsoever for the duration of the agreement, will be granted access. TGen will then facilitate the transfer of the requested deidentified data. This mechanism is expected to be via an Aspera High-Speed File Transfer Server but TGen reserves the right to change the specific transfer method at any time, provided appropriate levels of access authorization and control can be maintained.