Abstract
Lyssaviruses are the causative agents of rabies, a zoonotic, fatal disease that is thought to be ancestral to bats. In the last decade, the detection of bat associated lyssaviruses is increasing also in Europe. Within a retrospective bat associated lyssavirus surveillance study a total of 225 dead bats of 21 bat species were collected in Slovenia between 2012 and 2019 and tested by specific real-time RT-PCR method. The first lyssavirus positive sample in bats in Slovenia was detected using the real-time RT-PCR, the fluorescent antibody test, and next generation sequencing, while the rabies tissue culture inoculation test was unsuccessful due to sample degradation and storage conditions. The nearly complete genome of Divača bat lyssavirus from Slovenia consists of 11,871 nucleotides and reflects the characteristic gene organization known for lyssaviruses, encoding the five viral proteins. Phylogenetic analysis of Divača bat lyssavirus revealed that it belongs to phylogroup I lyssaviruses and is most closely related to Kotalahti bat lyssavirus (KBLV) with 87.20% nucleotide and 99.22% amino acid identity. Together with KBLV, Khujand virus, European bat lyssavirus 2, Bakeloh bat lyssavirus, and Aravan virus, Divača bat lyssavirus was detected in the genus Myotis suggesting its key role in the transmission and maintenance of certain lyssaviruses.
Author summary
Lyssaviruses are the causative agents for rabies, a zoonotic, fatal disease that is thought to be ancestral to bats. The wide geographical distribution and genetic diversity of lyssaviruses detected in bats across Europe indicated the need for lyssavirus surveillance in the Slovenian bat population. Within a retrospective surveillance study on bat-associated lyssavirus in Slovenia, one positive sample was detected. Whole genome sequencing and phylogenetic analyses revealed that the Divača bat lyssavirus genome, from Slovenia, reflects the characteristic gene organization known for lyssaviruses, its classification in phylogroup I, and its close relation to tentative lyssavirus species KBLV. Together with KBLV, KHUV, EBLV 2, BBLV, and ARAV, Divača bat lyssavirus was detected in the genus Myotis suggesting its key role in the transmission and maintenance of certain lyssaviruses. Worldwide increased laboratory surveillance and pan-lyssavirus molecular tools will support the discovery of novel lyssaviruses which could greatly impact recognizing the risk of lyssaviruses to animal and human health.
Introduction
Bats belong to the mammalian order Chiroptera, with more than 1460 species in 21 bat families [1]. They are widely distributed on all continents except Antarctica and play an important role in seed dispersal, pollination, and in organic fertilization through their guano [2]. Bats are a natural reservoir for viruses that are among the deadliest viruses transmitted from wildlife to humans, including rabies, and SARS coronavirus [3]. As reservoir hosts for emerging viral zoonotic diseases, bats also carry viruses from at least 28 different virus families, most of which are likely host-specific and have limited zoonotic potential [4]. The number of viral zoonoses in bats is comparable to the number of viral zoonoses in other mammal or bird orders since variation in the number of viral zoonoses among animal groups arises as a consequence of their species richness [5].
The first described zoonotic disease associated with bats was rabies [6], a zoonotic fatal disease caused by a member of the genus Lyssavirus, within the subfamily Alpharhabdovirinae, family Rhabdoviridae, order Mononegavirales [7]. Lyssaviruses are bullet-shaped enveloped viruses with an approximately 12-kb-long linear single stranded negative-sensed RNA genome encoding nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and RNA-dependent polymerase (L) [7,8]. According to the taxonomic classification, the genus Lyssavirus currently consists of 17 different virus species. According to the most recent ICTV report [9], lyssavirus names are provided here followed by the traditional abbreviations used to identify their isolates: Aravan virus (ARAV), Australian bat lyssavirus (ABLV), Bokeloh bat lyssavirus (BBLV), West Caucasian bat virus (WCBV), Duvenhage virus (DUVV), Taiwan bat lyssavirus (TWBLV), Gannoruwa bat lyssavirus (GBLV), European bat lyssavirus 1 (EBLV-1), European bat lyssavirus 2 (EBLV-2), Ikoma lyssavirus (IKOV), Irkut virus (IRKV), Khujand virus (KHUV), Lagos bat virus (LBV), Lleida bat lyssavirus (LLEBV), Mokola virus (MOKV), rabies virus (RABV), Shimoni bat virus (SHIBV) [9]. Recently, two potentially novel lyssaviruses, Kotalahti bat lyssavirus (KBLV) and Matlo bat lyssavirus (MBLV) have been described [9–12]. Apart from the two virus species MOKV and IKOV within the lyssavirus genus, all lyssaviruses are associated with bats and bats are thought to be the primary ancestral reservoir hosts [13]. In Europe, spillover lyssavirus infections from insectivorous bats involving EBLV-1, EBLV-2, and WCBV have been described in humans [14–17], cats [18,19], sheep [20], and stone martens [21]. All lyssaviruses are thought to be capable of causing central nervous system infection leading to acute progressive encephalomyelitis [13] and death in unvaccinated humans if not treated appropriately [16].
According to phylogenetic and antigenic characteristics, lyssaviruses are divided into two phylogroups [22]. ABLV, ARAV, BBLV, DUVV, EBLV-1, EBLV-2, GBLV, IRKV, KBLV, KHUV, RABV, TWBLV represent phylogroup I, while LBV, MOKV, and SHIBV represent phylogroup II [22–24]. The classification into phylogroups I and II could not be applied to the most divergent lyssaviruses IKOV, LLEBV, MBLV, and WCBV [25]. The highest nucleotide sequence similarity of the glycoprotein gene was described within the phylogroup II with an average of 71.5%, followed by phylogroup I with 70.3% similarity and the group of lyssaviruses not assigned to phylogroups I and II (IKOV, LLEBV, MBLV, WCBV) with 58.2% similarity [25]. According to Fooks et al. [25], rabies vaccines are not effective against the most divergent lyssaviruses (IKOV, LLEBV, MBLV, WCBV) belonging to the group of lyssaviruses not assigned to phylogroups I and II [25].
In Europe, rabies in bats was first reported in Germany in 1954 [26]. With subsequent molecular analyses of rabies virus genomes in European bats, the distinction between classical RABV and EBLV 1 and 2 was described [27]. In European bats, EBLV-1 was detected in Serotine bat (Eptesicus serotinus) and Isabelline serotine bat (Eptesicus isabellinus) [27–29]. EBLV-2 was detected in European bat species Daubenton’s bat (Myotis daubentonii) and Pond bat (Myotis dasycneme) [30], WCBV in Common bent-wing bat (Miniopterus schreibersii) [19,31], LLEBV in Common bent-wing bat (Miniopterus schreibersii) [32], BBLV in Natterer’s bat (Myotis nattereri) [33], and KBLV in Brandt’s bat (Myotis brandtii) [34].
In this manuscript, we describe the discovery and genetic characterisation of a previously unknown bat-associated lyssavirus, Divača bat lyssavirus, in an insectivorous bat species from Slovenia as a result of national retrospective surveillance programme. Based on whole genome sequencing and phylogenetic analysis, Divača bat lyssavirus, together with KBLV, may represent a putative new lyssavirus species in Europe.
Material and methods
Sampling
Between 2012 and 2019, a total of 225 dead bats of 21 bat species from 59 (out of 212) municipalities in Slovenia were collected by bat biologists or volunteers and stored frozen (S1 Table). The collected dead bats were submitted to the National Veterinary Institute, Institute of Microbiology and Parasitology, Virology unit, for lyssavirus diagnosis. Bat biologists provided information on bat species, location (municipality), and year of collection. Bat species were identified using morphological keyes, described in Dietz and Halversen [35]. Additional molecular characterization of the host species from lyssavirus positive bat sample PP-0868/2014, where NGS reads were mapped to cytochrome b and cytochrome c oxidase subunit I [36], confirmed the original assignment classification to Myotis capaccinii.
Brain samples were collected through the foramen occipitale magnum by pipette aspiration. After aspiration of the brain tissue, the cranial cavity was rinsed with RPMI-1640 medium (Thermo Fisher, USA). The collected brain tissue was homogenized in a total volume of 500 μl of RPMI-1640 medium (Thermo Fisher, USA) before long-term storage at < - 60°C.
Permit for capture, disturbance, and temporary taking from the wild and sampling of protected animals was issued by the Agency of the Republic of Slovenia for the Environment No. 35601-35/2010-6.
Diagnostic methods
For molecular detection of lyssaviruses, brain tissue homogenates were subjected to automated RNA extraction using the KingFisher Flex Purification System (Thermo Fisher, USA) and MagMax Core Nucleic Acid Purification Kit (Thermo Fisher, USA) according to the manufacturer’s instructions. The extracted RNA was tested by real time RT-PCR [37,38] to detect lyssaviruses. Real-time RT-PCR with three specific primers LN34forward1, LN34forward2, and LN34reverse, and two probes LN34probe and LN34probeLago targeting highly conserved sequences within the nucleoprotein gene region [37] was performed.
When the lyssavirus-positive sample PP-0868/2014 was detected by real-time RT-PCR, the bat carcass was resampled by opening the cranial cavity to obtain the brain sample, which was subjected to a fluorescent antibody test (FAT) performed as previously described [39].
RTCIT was performed from lyssavirus positive sample PP-0868/2014 as previously described [39]. Three consecutive serial passages were performed to obtain RTCIT results.
Whole genome sequencing and phylogenetic analysis
The lyssavirus positive sample PP-0868/2014 was subjected to next generation sequencing (NGS) to determine the complete genome sequence of lyssavirus using the NGS service of Novogene (Cambridge, UK). From extracted RNA to final data, Novogene’s service included sample preparation, quality control, library construction using the RIP-seq library preparation kit (Illumina, USA), library quality control, sequencing on NovaSeq 6000 (Illumina, USA), using the NovaSeq PE150 kit (Illumina, USA), and data quality control. NGS reads were used for de novo assembly with SPAdes 3.13.0 [40]. Diamond BLASTx [41] and MEGAN 6.11.7 [42] were used for the taxonomic assignment of the assembled contigs. The nucleotide sequence of the assembled lyssavirus genome, named Divača bat lyssavirus (original name of sample PP-0868/2014), was deposited in the GenBank database with accession number OQ428158 and compared with other genomes of 17 lyssavirus species and 2 putative lyssavirus species and annotated using Geneious 20221.1 (Biomatters, New Zealand). The nucleotide and amino acid alignments were constructed using MAFFT [43]. Phylogenetic analysis was performed using IQ-TREE 1.6.12 [44], with ModelFinder [45] determining the best model according to BIC score and 1000 ultrafast bootstrap replicates [46,47] used to test tree reliability.
Results
Virus detection by real-time RT-PCR, FAT, and RTCIT
One sample (original name PP-0868/2014) out of 225 tested samples from 21 different bat species (S1 Table) was found to be lyssavirus positive by the real-time RT-PCR, with a cycle threshold value of 21.82. The sample PP-0868/2014 was from species Long-fingered bat (Myotis capaccinii), which was found dead approximately 700m inside of Škocjan caves in Divača municipality collected in the year 2014. The bat carcass was found in the advance stage of decay and was stored at < - 20°C until brain collection in 2020. The lyssavirus positive brain sample PP-0868/2014 was subjected to further analysis, namely FAT and RTCIT. The FAT slide stained with FITC anti-rabies monoclonal globulin showed positive results with typical lyssavirus staining (S1 Fig). No viral growth was detected on RTCIT for PP-0868/2014 after three consecutive passages.
Whole genome sequencing and phylogenetic analysis
Viral RNA from lyssavirus positive sample PP-0868/2014 was subjected to NGS to determine the complete genome sequence, named Divača bat lyssavirus. NGS yielded 13,048,710 raw reads that were used for de novo assembly. The nearly complete lyssavirus genome sequence with a total length of 11,871 nucleotides (an average sequence depth of 135,333 nucleotides per nucleotide site) was generated, encoding five genes, N, P, M, G, and L in the highly conserved order typical of lyssaviruses. The lengths of the N, P, M, G, and L protein genes were 1356 nt, 894 nt, 609 nt, 1581 nt, and 6384 nt, respectively. Gene lengths and intergenic spacers of Divača bat lyssavirus were found to be similar to all lyssaviruses from phylogroup I.
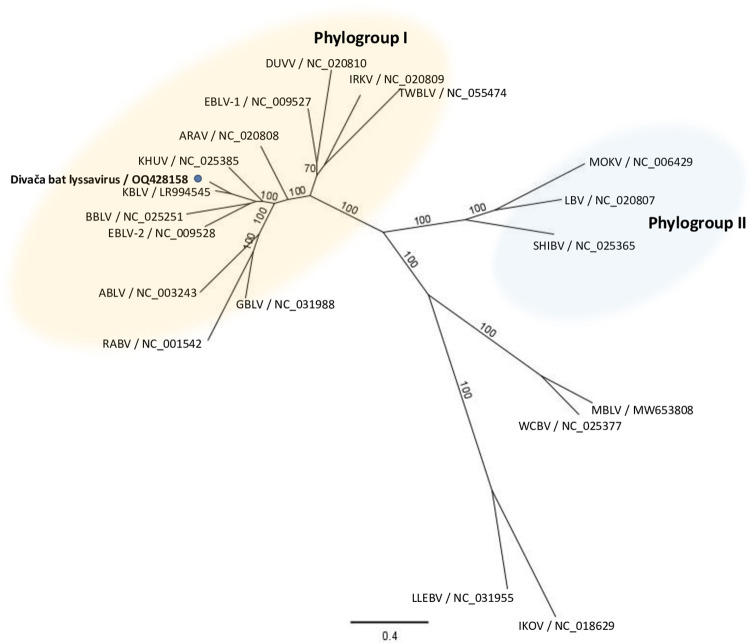
Phylogenetic analysis of the concatenated N+P+M+G+L coding sequences of the assembled Divača bat lyssavirus, 17 lyssavirus species, and 2 putative lyssavirus species (Fig 1) showed that Divača bat lyssavirus clustered with phylogroup I lyssaviruses (Fig 1) and is most closely related to KBLV with 87.20% nucleotide and 99.22% amino acid identity (Table 1). Similar nucleotide and amino acid identities to KBLV were found for the N gene and also for the P, M, G, and L genes. The percent nucleotide and amino acid identities of the N, P, M, G, L, and concatenated N+P+M+G+L genes between Divača bat lyssavirus and other lyssavirus species from GenBank are shown in Table 1.
Fig 1. N+P+M+G+L maximum likelihood phylogenetic tree.
The maximum likelihood phylogeny was constructed with concatenated N+P+M+G+L coding sequences of representative 17 lyssavirus species, 2 tentative lyssavirus species, and Divača bat lyssavirus (indicated with a blue dot) by IQ-TREE using the GTR+F+I+G4 substitution model with 1000 ultrafast bootstrap replicates. Numbers at the nodes indicate ultrafast bootstrap support and the scale bar indicates the number of substitutions per site.
Table 1. Nt and aa % identity of N, P, M, G, L and concatenated N+P+M+G+L genes of 19 different lyssaviruses to Divača bat lyssavirus.
| Lyssavirus species abbrev. | N gene nt/aa % identity to Divača bat lyssavirus | P gene nt/aa % identity to Divača bat lyssavirus | M gene nt/aa % identity to Divača bat lyssavirus | G gene nt/aa % identity to Divača bat lyssavirus | L gene nt/aa % identity to Divača bat lyssavirus | Concatenated N+P+M+G+L genes nt/aa % identity to Divača bat lyssavirus |
|---|---|---|---|---|---|---|
| ARAV | 79.20/ 97.78 |
74.61/ 93.27 |
79.97/ 99.50 |
74.89/ 95.82 |
77.79/ 98.35 |
77.40/ 97.56 |
| ABLV | 76.72/ 97.78 |
70.02/ 89.56 |
77.01/ 98.02 |
70.91/ 92.00 |
75.15/ 96.95 |
74.25/ 95.44 |
| BBLV | 78.69/ 97.78 |
77.29/ 94.28 |
80.13/ 99.01 |
75.94/ 95.42 |
79.14/ 98.64 |
78.47/ 97.67 |
| WCBV | 71.69/ 95.56 |
52.81/ 73.91 |
69.13/ 95.05 |
55.51/ 76.57 |
68.34/ 92.99 |
65.48/ 89.12 |
| DUVV | 76.18/ 96.67 |
65.33/ 88.93 |
77.67/ 98.02 |
72.19/ 93.73 |
73.90/ 97.04 |
73.33/ 95.69 |
| TWBLV | 74.63/ 96.45 |
66.78/ 88.26 |
78.00/ 98.51 |
66.79/ 91.06 |
74.17/ 96.94 |
72.46/ 95.01 |
| GBLV | 77.97/ 98.00 |
71.14/ 88.22 |
76.52/ 99.50 |
74.38/ 94.49 |
77.41/ 98.07 |
76.46/ 96.78 |
| EBLV-1 | 76.99/ 97.12 |
66.89/ 89.60 |
75.37/ 97.03 |
73.71/ 93.51 |
76.05/ 97.41 |
74.99/ 96.09 |
| EBLV-2 | 78.61/ 96.45 |
76.17/ 93.60 |
79.79/ 98.51 |
78.29/ 95.99 |
79.83/ 98.45 |
79.13/ 97.39 |
| IKOV | 68.22/ 91.56 |
49.56/ 73.15 |
68.14/ 94.06 |
52.34/ 74.24 |
65.40/ 90.12 |
62.47/ 86.50 |
| IRKV | 77.29/ 96.90 |
67.78/ 89.93 |
78.33/ 97.52 |
70.10/ 94.27 |
74.94/ 97.27 |
74.11/ 96.12 |
| KHUV | 80.31/ 97.34 |
76.96/ 95.62 |
78.98/ 99.01 |
76.60/ 95.06 |
80.01/ 98.73 |
79.24/ 97.78 |
| LBV | 73.39/ 96.22 |
53.64/ 77.74 |
72.09/ 96.04 |
58.89/ 83.91 |
71.08/ 94.41 |
67.93/ 91.54 |
| LLEBV | 70.07/ 92.89 |
50.44/ 72.73 |
69.46/ 94.55 |
52.66/ 74.90 |
65.52/ 90.55 |
62.66/ 86.97 |
| MOKV | 71.54/ 95.78 |
54.36/ 77.26 |
71.26/ 96.04 |
60.42/ 82.95 |
70.29/ 94.17 |
67.66/ 91.18 |
| RABV | 75.76/ 96.89 |
69.35/ 87.21 |
71.76/ 96.04 |
69.71/ 91.60 |
74.06/ 96.94 |
73.39/ 95.23 |
| SHIBV | 72.88/ 95.11 |
56.35/ 81.33 |
71.76/ 97.03 |
60.48/ 83.52 |
71.19/ 94.22 |
68.54/ 91.57 |
| KBLV | 87.09/ 99.78 |
87.81/ 97.64 |
89.49/ 100.00 |
86.40/ 98.10 |
87.11/ 99.53 |
87.20/ 99.22 |
| MBLV | 72.58/ 95.78 |
52.59/ 71.57 |
68.31/ 95.05 |
54.56/ 75.81 |
68.39/ 92.71 |
65.41/ 88.79 |
Discussion
Several bat lyssavirus surveillance systems have been implemented in Europe and results have shown that passive or retrospective surveillance is better than active surveillance for lyssavirus detection [27,50]. In this study, a retrospective survey was conducted between 2012 and 2019, collecting 225 carcasses from 21 of the 32 bat species living in Slovenia [51]. Lyssavirus was found in one of the two Myotis capaccinii carcasses. Based on a single detection of lyssavirus in a particular bat species, we could not determine whether Myotis capaccinii is the true reservoir host for the Divača bat lyssavirus. Similar results and conclusions were reported by Nokireki et al. [34] who described the putative new lyssavirus species KBLV in Myotis brandtii and also found only one KBLV positive sample. However, the authors of a later study on KBLV [52] suggested that, based on the data that closely related phylogroup I lyssaviruses have also been isolated from bat species of the genus Myotis, Myotis brandtii is probably a reservoir host and not infected by spillover infection from another bat species.
This study is the first to report a lyssavirus detected in Myotis capaccinii, a bat species in which several viruses from the families Astroviridae (3), Coronaviridae (6), Herpesviridae (2), and Paramyxoviridae (1) have been described [53]. Infection of bats with lyssaviruses occurs worldwide, although different virus species occur in different regions, have co-evolved and are therefore linked to specific bat species [54]. In the Americas, only RABV is associated with bats, whereas in Europe, Africa, Asia, and Australia, the other lyssaviruses predominate without RABV being associated with bats [55]. In Europe, numerous bat species have already been identified as infected with lyssaviruses [56]. However, there is no report of a lyssavirus finding for the bat species Myotis capaccinii.
Using NGS, a nearly complete genome sequence of the Divača bat lyssavirus was generated and phylogenetic analysis was performed. According to the ICTV [9], there are several criteria for classifying new species in the genus Lyssavirus. The new species should have a nucleotide identity for the complete N gene of less than 78–80% or a nucleotide identity for the concatenated coding regions of N+P+M+G+L of less than 80%. The Divača bat lyssavirus is most closely related to the putative novel lyssavirus species KBLV with 87.20% and 87.09% nucleotide identity for the concatenated N+P+M+G+L genes and the N gene, respectively, suggesting that Divača bat lyssavirus and KBLV belong to the same putative novel lyssavirus species. Another demarcation criterion of ICTV [9] requires that the new virus should not represent a sister branch to a virus of an established species in the phylogenetic tree of concatenated N+P+M+G+L. Phylogenetic analyses of concatenated N+P+M+G+L revealed that Divača bat lyssavirus represents a sister branch to a putative novel lyssavirus species KBLV, again suggesting that Divača bat lyssavirus and KBLV are members of the same putative novel lyssavirus species. Serological differentiation of a novel lyssavirus species from other lyssavirus species is also one of the species demarcation criteria by ICTV for a novel lyssavirus species, but in our study, Divača bat lyssavirus could not be serologically assessed. The last demarcation criterion of ICTV [9] is that a novel lyssavirus species occupies a distinct ecological niche, as evidenced by host species, pathobiological properties, or geographic range. The Divača bat lyssavirus occupies a specific ecological niche because it originates from a different host than KBLV, which has also been detected in a different geographical location (Fig 2). Taking all four ICTV criteria together to taxonomically classify the novel Divača bat lyssavirus, even though it was found in a new host species almost two thousand kilometres away from the closely related KBLV, we suggest that the Divača bat lyssavirus and KBLV are both members of the same putative novel lyssavirus species.
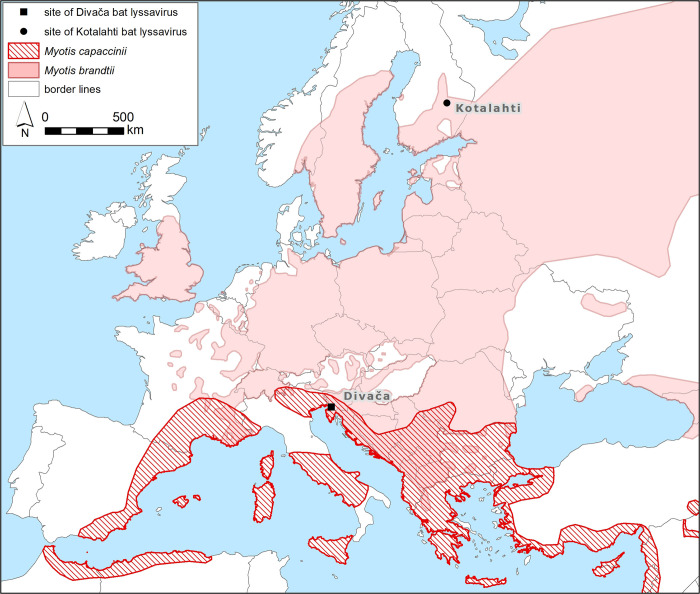
Fig 2. The sites of closely related Divača bat lyssavirus (square) and Kotalahti bat lyssavirus (dot).
The geographical distribution of Myotis capaccinii and Myotis brandtii according to NatureServe and International Union for Conservation of Nature [48,49]. Base layer of the map was made with Natural Earth (https://www.naturalearthdata.com/http//www.naturalearthdata.com/download/10m/cultural/ne_10m_admin_0_sovereignty.zip).
As already aforementioned, Divača bat lyssavirus is most closely related to KBLV. KBLV was detected in Myotis brandtii in Finland in 2017 [34]. In Slovenia, Myotis brandtii is very rare, however, it was found in the vicinity of Škocjan caves [57]. The contact between Myotis capaccinii, species with circum-Mediterranean distribution [48] almost exclusively roosting in caves, and Myotis brandtii, species with wider distribution in Europe [49] primarily roosting in tree holes and cracks, remains unclear as they usually do not share roosts. The geographical distance between Kotalahti in Finland and Divača in Slovenia is quite large, it measures 1886 km (Fig 2). Due to the vast geographical distance and fact that both host species Myotis brandtii and Myotis capaccinii are mostly regional migrants [58], we could speculate that lyssaviruses similar to KBLV and Divača bat lyssavirus could be present in other European countries between Slovenia and Finland, suggesting the need for surveillance programs and the need to increase public awareness.
Despite efforts to isolate viable virus from the sample PP-0868/2014, we were unsuccessful and could not evaluate the vaccine efficacy of conventional rabies vaccines. However, according to the assignment of Divača bat lyssavirus to phylogroup I and high sequence identity with KBLV we could speculate good cross protection by conventional rabies vaccines.
We believe that the complete genome sequence of Divača bat lyssavirus and its phylogenetic relationships to other lyssaviruses identified in our study may be useful for future studies of the lyssavirus genus. For further studies, the isolation from field samples on cell culture will be an important step toward the possibility of the additional analysis of this strain.
In conclusion, Divača bat lyssavirus is most closely related to KBLV, which has been proposed as a separate species within the genus Lyssavirus. Previously published studies on KBLV indicate that it belongs to phylogroup I of the lyssaviruses against which rabies vaccines provide protection. Continued passive surveillance would be required to obtain infectious lyssaviruses for evaluating the efficacy of vaccines relevant to public health. Because Divača bat lyssavirus has the greatest genetic similarity to KBLV, it could be considered to pose a low risk to public health if preventive measures such as vaccination of bat caretakers and prophylactic treatment after human-bat contact are followed.
Supporting information
Lyssavirus positive sample is indicated with blue colour.
(DOCX)
Apple green fluorescence is present in neurons (magnification 20×0.40).
(DOCX)
Data Availability
The nucleotide sequence of the assembled lyssavirus genome, named Divača bat lyssavirus (original name of sample PP-0868/2014), was deposited in the GenBank database with accession number OQ428158.
Funding Statement
This research was funded by the Administration of the Republic of Slovenia for Food Safety, Veterinary, and Plant Protection and by the Slovenian Research Agency (research core funding No. P4-0092). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Simmons N.B. and Cirranello A.L. 2023. Bat Species of the World: A taxonomic and geographic database. Version 1.3. Accessed on 05/09/2023. [Google Scholar]
- 2.Epstein JH. 40 –Emerging Diseases in Bats. In: Miller RE, Lamberski N, Calle PP, editors. Fowler’s Zoo and Wild Animal Medicine Current Therapy, Volume 9: W.B. Saunders; 2019. p. 274–9. [Google Scholar]
- 3.Schountz T, Baker ML, Butler J, Munster V. Immunological Control of Viral Infections in Bats and the Emergence of Viruses Highly Pathogenic to Humans. Frontiers in Immunology. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Letko M, Seifert SN, Olival KJ, Plowright RK, Munster VJ. Bat-borne virus diversity, spillover and emergence. Nature Reviews Microbiology. 2020;18(8):461–71. doi: 10.1038/s41579-020-0394-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mollentze N, Streicker DG. Viral zoonotic risk is homogenous among taxonomic orders of mammalian and avian reservoir hosts. Proceedings of the National Academy of Sciences. 2020;117(17):9423–30. doi: 10.1073/pnas.1919176117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banyard AC, Evans JS, Luo TR, Fooks AR. Lyssaviruses and Bats: Emergence and Zoonotic Threat. Viruses. 2014;6(8):2974–90. doi: 10.3390/v6082974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker PJ, Freitas-Astúa J, Bejerman N, Blasdell KR, Breyta R, Dietzgen RG, et al. ICTV Virus Taxonomy Profile:. J Gen Virol. 2022;103(6). doi: 10.1099/jgv.0.001689 . [DOI] [PubMed] [Google Scholar]
- 8.Walker PJ, Firth C, Widen SG, Blasdell KR, Guzman H, Wood TG, et al. Evolution of Genome Size and Complexity in the Rhabdoviridae. PLOS Pathogens. 2015;11(2):e1004664. doi: 10.1371/journal.ppat.1004664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ICTV https://ictv.global/report/chapter/rhabdoviridae/rhabdoviridae/lyssavirus, accessed on 22nd September 2022.
- 10.Amarasinghe GK, Ayllón MA, Bào Y, Basler CF, Bavari S, Blasdell KR, et al. Taxonomy of the order Mononegavirales: update 2019. Archives of Virology. 2019;164(7):1967–80. doi: 10.1007/s00705-019-04247-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coertse J, Grobler C, Sabeta C, Seamark ECJ, Kearney T, Paweska J, et al. Lyssaviruses in Insectivorous Bats, South Africa, 2003–2018. Emerging Infectious Disease journal. 2020;26(12):3056. doi: 10.3201/eid2612.203592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grobler CS, Coertse J, Markotter W. Complete Genome Sequence of Matlo Bat Lyssavirus. Microbiol Resour Announc. 2021;10(20). Epub 20210520. doi: 10.1128/MRA.00241-21 ; PubMed Central PMCID: PMC8188345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fooks AR, Cliquet F, Finke S, Freuling C, Hemachudha T, Mani RS, et al. Rabies. Nat Rev Dis Primers. 2017;3:17091. Epub 20171130. doi: 10.1038/nrdp.2017.91 . [DOI] [PubMed] [Google Scholar]
- 14.Lumio J, Hillbom M, Roine R, Ketonen L, Haltia M, Valle M, et al. HUMAN RABIES OF BAT ORIGIN IN EUROPE. The Lancet. 1986;327(8477):378. doi: 10.1016/s0140-6736(86)92336-6 [DOI] [PubMed] [Google Scholar]
- 15.Fooks AR, McElhinney LM, Pounder DJ, Finnegan CJ, Mansfield K, Johnson N, et al. Case report: Isolation of a European bat lyssavirus type 2a from a fatal human case of rabies encephalitis. Journal of Medical Virology. 2003;71(2):281–9. doi: 10.1002/jmv.10481 [DOI] [PubMed] [Google Scholar]
- 16.Johnson N, Vos A, Freuling C, Tordo N, Fooks AR, Müller T. Human rabies due to lyssavirus infection of bat origin. Veterinary Microbiology. 2010;142(3):151–9. doi: 10.1016/j.vetmic.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 17.Regnault B, Evrard B, Plu I, Dacheux L, Troadec E, Cozette P, et al. First Case of Lethal Encephalitis in Western Europe Due to European Bat Lyssavirus Type 1. Clinical Infectious Diseases. 2022;74(3):461–6. doi: 10.1093/cid/ciab443 [DOI] [PubMed] [Google Scholar]
- 18.Dacheux L, Larrous F, Mailles A, Boisseleau D, Delmas O, Biron C, et al. European Bat Lyssavirus Transmission among Cats, Europe. Emerging Infectious Disease journal. 2009;15(2):280. doi: 10.3201/eid1502.080637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leopardi S, Barneschi E, Manna G, Zecchin B, Priori P, Drzewnioková P, et al. Spillover of West Caucasian Bat Lyssavirus (WCBV) in a Domestic Cat and Westward Expansion in the Palearctic Region. Viruses. 2021;13(10). doi: 10.3390/v13102064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tjørnehøj K, Fooks AR, Agerholm JS, Rønsholt L. Natural and Experimental Infection of Sheep with European Bat Lyssavirus Type-1 of Danish Bat Origin. Journal of Comparative Pathology. 2006;134(2):190–201. doi: 10.1016/j.jcpa.2005.10.005 [DOI] [PubMed] [Google Scholar]
- 21.Müller T, Cox J, Peter W, Schäfer R, Johnson N, McElhinney LM, et al. Spill-over of European Bat Lyssavirus Type 1 into a Stone Marten (Martes foina) in Germany. Journal of Veterinary Medicine, Series B. 2004;51(2):49–54. doi: 10.1111/j.1439-0450.2003.00725.x [DOI] [PubMed] [Google Scholar]
- 22.Badrane H, Bahloul C, Perrin P, Tordo N. Evidence of Two Lyssavirus Phylogroups with Distinct Pathogenicity and Immunogenicity. Journal of Virology. 2001;75(7):3268–76. doi: 10.1128/JVI.75.7.3268-3276.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunawardena P, Marston D, Ellis R, Wise E, Karawita A, Breed A, et al. Lyssavirus in Indian Flying Foxes, Sri Lanka. Emerging Infectious Disease journal. 2016;22(8):1456. doi: 10.3201/eid2208.151986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuzmin IV, Mayer AE, Niezgoda M, Markotter W, Agwanda B, Breiman RF, et al. Shimoni bat virus, a new representative of the Lyssavirus genus. Virus Research. 2010;149(2):197–210. doi: 10.1016/j.virusres.2010.01.018 [DOI] [PubMed] [Google Scholar]
- 25.Fooks AR, Shipley R, Markotter W, Tordo N, Freuling CM, Müller T, et al. Renewed Public Health Threat from Emerging Lyssaviruses. Viruses. 2021;13(9). doi: 10.3390/v13091769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohr W. Die tollwut. Krankheiten durch Viren: Springer; 1967. p. 536–72. [Google Scholar]
- 27.Schatz J, Fooks AR, McElhinney L, Horton D, Echevarria J, Vázquez-Moron S, et al. Bat Rabies Surveillance in Europe. Zoonoses and Public Health. 2013;60(1):22–34. doi: 10.1111/zph.12002 [DOI] [PubMed] [Google Scholar]
- 28.Vázquez-Morón S, Juste J, Ibáñez C, Ruiz-Villamor E, Avellón A, Vera M, et al. Endemic Circulation of European Bat Lyssavirus Type 1 in Serotine Bats, Spain. Emerging Infectious Disease journal. 2008;14(8):1263. doi: 10.3201/eid1408.080068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller T, Johnson N, Freuling CM, Fooks AR, Selhorst T, Vos A. Epidemiology of bat rabies in Germany. Archives of Virology. 2007;152(2):273–88. doi: 10.1007/s00705-006-0853-5 [DOI] [PubMed] [Google Scholar]
- 30.McElhinney LM, Marston DA, Wise EL, Freuling CM, Bourhy H, Zanoni R, et al. Molecular Epidemiology and Evolution of European Bat Lyssavirus 2. International Journal of Molecular Sciences. 2018;19(1):156. doi: 10.3390/ijms19010156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuzmin IV, Hughes GJ, Botvinkin AD, Orciari LA, Rupprecht CE. Phylogenetic relationships of Irkut and West Caucasian bat viruses within the Lyssavirus genus and suggested quantitative criteria based on the N gene sequence for lyssavirus genotype definition. Virus Res. 2005;111(1):28–43. Epub 20050408. doi: 10.1016/j.virusres.2005.03.008 . [DOI] [PubMed] [Google Scholar]
- 32.Ceballos NA, Morón SV, Berciano J, Nicolás O, López CA, Juste J, et al. Novel Lyssavirus in Bat, Spain. Emerging Infectious Disease journal. 2013;19(5):793. doi: 10.3201/eid1905.121071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freuling CM, Beer M, Conraths FJ, Finke S, Hoffmann B, Keller B, et al. Novel Lyssavirus in Natterer’s Bat, Germany. Emerging Infectious Disease journal. 2011;17(8):1519. doi: 10.3201/eid1708.110201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nokireki T, Tammiranta N, Kokkonen U-M, Kantala T, Gadd T. Tentative novel lyssavirus in a bat in Finland. Transboundary and Emerging Diseases. 2018;65(3):593–6. doi: 10.1111/tbed.12833 [DOI] [PubMed] [Google Scholar]
- 35.Dietz C, von Helversen O. Illustrated Identification Key to the Bats of Europe: Dietz & von Helversen; 2004. [Google Scholar]
- 36.De Benedictis P, Leopardi S, Markotter W, Velasco-Villa A. The Importance of Accurate Host Species Identification in the Framework of Rabies Surveillance, Control and Elimination. Viruses. 2022;14(3). doi: 10.3390/v14030492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wadhwa A, Wilkins K, Gao J, Condori Condori RE, Gigante CM, Zhao H, et al. A Pan-Lyssavirus Taqman Real-Time RT-PCR Assay ort he Detection of Highly Variable Rabies virus and Other Lyssaviruses. PLOS Neglected Tropical Diseases. 2017;11(1):e0005258. doi: 10.1371/journal.pntd.0005258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization, Rupprecht, Charles E, Fooks, Anthony R & Abela-Ridder, Bernadette. (2019). Laboratory techniques in rabies, volume 2, 5th ed. World Health Organization. https://apps.who.int/iris/handle/10665/310837. License: CC BY-NC-SA 3.0 IGO.
- 39.World Health Organization, Rupprecht, Charles E, Fooks, Anthony R & Abela-Ridder, Bernadette. (2018). Laboratory techniques in rabies, volume 1, 5th ed. World Health Organization. https://apps.who.int/iris/handle/10665/310836. License: CC BY-NC-SA 3.0 IGO.
- 40.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. Journal of Computational Biology. 2012;19(5):455–77. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nature Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 42.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome research. 2007;17(3):377–86. Epub 2007/01/25. doi: 10.1101/gr.5969107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution. 2013;30(4):772–80. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Molecular Biology and Evolution. 2015;32(1):268–74. doi: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods. 2017;14(6):587–9. doi: 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast Approximation for Phylogenetic Bootstrap. Molecular Biology and Evolution. 2013;30(5):1188–95. doi: 10.1093/molbev/mst024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Molecular Biology and Evolution. 2018;35(2):518–22. doi: 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.NatureServe and IUCN (International Union for Conservation of Nature) 2007. Myotis capaccinii. The IUCN Red List of Threatened Species. Version 2007. https://www.iucnredlist.org. Downloaded on 03 April 2023. [Google Scholar]
- 49.NatureServe and IUCN (International Union for Conservation of Nature) 2020. Myotis brandtii. The IUCN Red List of Threatened Species. Version 2020. https://www.iucnredlist.org. Downloaded on 03 April 2023. [Google Scholar]
- 50.Klein A, Calvelage S, Schlottau K, Hoffmann B, Eggerbauer E, Müller T, et al. Retrospective Enhanced Bat Lyssavirus Surveillance in Germany between 2018–2020. Viruses. 2021;13(8). doi: 10.3390/v13081538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Presetnik P. First records of the European free-tailed bat Tadarida teniotis (Rafinesque, 1814) in Slovenia. Natura Sloveniae. 2019;21(1): 47–53. [Google Scholar]
- 52.Calvelage S, Tammiranta N, Nokireki T, Gadd T, Eggerbauer E, Zaeck LM, et al. Genetic and Antigenetic Characterization of the Novel Kotalahti Bat Lyssavirus (KBLV). Viruses. 2021;13(1). doi: 10.3390/v13010069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen L, Liu B, Yang J, Jin Q. DbatVir: the database of bat-associated viruses. Database. 2014;2014:bau021. doi: 10.1093/database/bau021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin IV, McCracken GF, Rupprecht CE. Host Phylogeny Constrains Cross-Species Emergence and Establishment of Rabies Virus in Bats. Science. 2010;329(5992):676–9. doi: 10.1126/science.1188836 [DOI] [PubMed] [Google Scholar]
- 55.Vos A, Kaipf I, Denzinger A, Fooks A R, Johnson N, Müller T. European bat lyssaviruses-an ecological enigma. Acta Chiropterologica. 2007;9(1):283–96. [Google Scholar]
- 56.Shipley R, Wright E, Selden D, Wu G, Aegerter J, Fooks AR, et al. Bats and Viruses: Emergence of Novel Lyssaviruses and Association of Bats with Viral Zoonoses in the EU. Tropical Medicine and Infectious Disease. 2019;4(1). doi: 10.3390/tropicalmed4010031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Presetnik P, Zidar S, Gojznikar J, Grgurevič S, Knapič T, Likozar L, et al. A review of Myotis brandtii and Myotis alcathoe records in Slovenia = Pregled nalaza Myotis brandtii i Myotis alcathoe u Sloveniji. Hypsugo. 2021;6(ǂǂ1):28–43. [Google Scholar]
- 58.Hutterer R, Ivanova T, Meyer-Cords CH, Rodrigues L. Bat migration in europe. A review of banding data and literature. Federal Agency for Nature Conser Vation. 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lyssavirus positive sample is indicated with blue colour.
(DOCX)
Apple green fluorescence is present in neurons (magnification 20×0.40).
(DOCX)
Data Availability Statement
The nucleotide sequence of the assembled lyssavirus genome, named Divača bat lyssavirus (original name of sample PP-0868/2014), was deposited in the GenBank database with accession number OQ428158.