Abstract
In recent years, transcriptional roadblocking has emerged as a crucial regulatory mechanism in gene expression, whereby other DNA-bound obstacles can block the progression of transcribing RNA polymerase (RNAP), leading to RNAP pausing and ultimately dissociation from the DNA template. In this review, we discuss the mechanisms by which transcriptional roadblocks can impede RNAP progression, as well as how RNAP can overcome these obstacles to continue transcription. We examine different DNA-binding proteins involved in transcriptional roadblocking and their biophysical properties that determine their effectiveness in blocking RNAP progression. The catalytically dead CRISPR-Cas (dCas) protein is used as an example of an engineered programmable roadblock, and the current literature in understanding the polarity of dCas roadblocking is also discussed. Finally, we delve into a stochastic model of transcriptional roadblocking and highlight the importance of transcription factor binding kinetics and its resistance to dislodgement by an elongating RNAP in determining the strength of a roadblock.
Keywords: RNA polymerase, Transcriptional roadblocking, Transcriptional interference, DNA binding kinetics, CRISPR
Introduction
Gene expression in both prokaryotes and eukaryotes involves sophisticated regulatory mechanisms, with much of this regulation occurring at the transcription level. Transcription is a multistage process that involves the binding of RNA polymerase (RNAP) to a promoter region on the DNA, followed by the unwinding of the double helix. Once the DNA is unwound, RNAP travels along the DNA to synthesize an RNA molecule until it reaches the end of the gene or transcription unit, where it is terminated, and the newly synthesized RNA molecule is released. Transcriptional control takes place at all three stages of this process—initiation, elongation and termination. In this review, we will focus on the regulation that occurs during transcription elongation, where RNAP must navigate various obstacles while traveling along the DNA.
During transcription elongation, RNAP may encounter various obstacles that can impede its progression. These obstacles can include DNA pause sites, which temporarily halt the progression of RNAP, thereby altering the rate of RNA synthesis (Mayer et al. 2017), as well as DNA lesions on the transcribed DNA caused by UV damage or chemical alteration (Gregersen and Svejstrup 2018; Nadon et al. 2022; Strobel et al. 2020). Additionally, RNAP may encounter static or mobile protein roadblocks that hinder its progression. Mobile protein blocks are formed by proteins that move along the same DNA, including other RNA polymerases and DNA polymerases required for DNA metabolism (Helmrich et al. 2011; Le and Wang 2018; Shearwin et al. 2005; Wang et al. 2023). On the other hand, static DNA-bound obstacles are formed by proteins such as transcription factors that bind to specific DNA sites or by non-specific DNA-binding proteins (Choi and Saier 2005; Dole et al. 2004; He and Zalkin 1992; Lewis et al. 2008). These static DNA-bound obstacles can pose a physical barrier that slows down or impedes RNAP progression, thereby affecting gene expression (Epstein et al. 2003; Hao et al. 2014). In the following section, we will discuss the details of these static roadblocks and their effects on transcription elongation.
Transcriptional roadblocking: mechanisms and consequences
The interactions between elongating RNAP and static protein roadblocks have been extensively studied in bacteria using both in vitro and in vivo approaches (Hao et al. 2014; He and Zalkin 1992; Lewis et al. 2008; Voros et al. 2017; Xu et al. 2022). In recent years, similar behaviour has also been observed in eukaryotic systems (Colin et al. 2014; Hedouin et al. 2022; Shukla et al. 2011), highlighting the conserved nature of this transcriptional regulatory mechanism in gene regulation. Several factors have been identified that can affect the outcomes of RNAP-roadblock protein interactions (Fig. 1), including the sequence context of the DNA region where the roadblock is located and the binding kinetics of the roadblock protein (Hao et al. 2014, 2016).
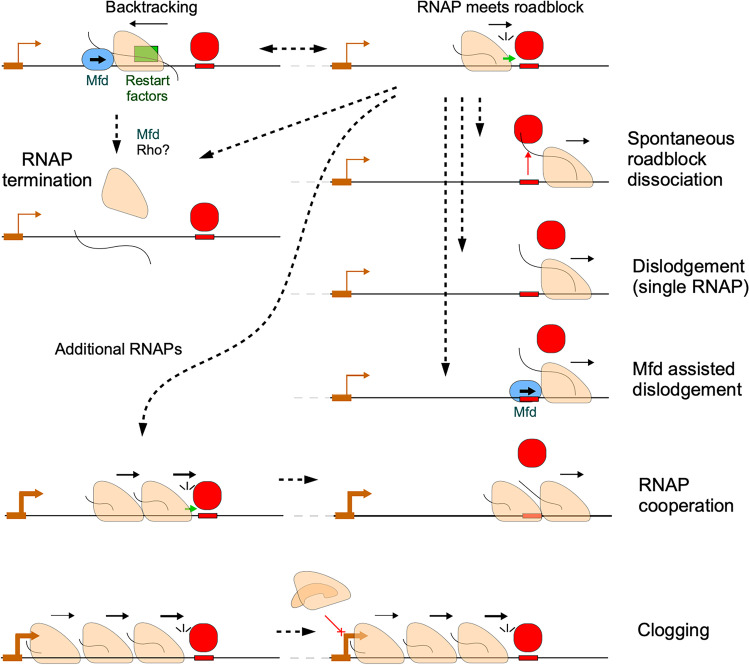
Fig. 1.
Possible outcomes of RNAP-roadblock interactions. When a roadblock protein (red) binds to the DNA and blocks the progression of an RNAP (tan), the RNAP may backtrack along the DNA. However, specialized proteins such as Mfd (blue) or transcript cleavage factors (green) can help reactivate the backtracked RNAP. If the RNAP is not reactivated, it is subject to termination by Mfd or possibly Rho. Alternatively, RNAP may be able to pass the roadblock as a result of spontaneous (unassisted) dissociation of the roadblock protein. RNAP may also actively dislodge the roadblock protein from the DNA, with or without the assistance of Mfd. Multiple RNAP at a roadblock site generally increase the fractional read-through due to RNAP cooperation, which can either increase the roadblock dislodgement or reduce RNAP backtracking and termination. However, when multiple RNAP build up at a roadblock, the promoter site may become obstructed, blocking further initiation from the promoter, in a process known as clogging
When an elongation complex (EC) encounters a roadblock, it may stall and subsequently backtrack along the DNA, causing the 3′ end of the nascent RNA transcript to become displaced from the catalytic site (Fig. 1). This leads to the formation of an inactive RNAP complex that halts further transcription (Nudler 2012; Toulmé et al. 2005). Several mechanisms have evolved to overcome RNAP backtracking (Fig. 1). For example, bacterial transcription factors GreA and GreB can rescue stalled RNAP by cleaving the RNA transcript, generating a new 3′ end at the catalytic site to reinitiate transcription (Abdelkareem et al. 2019; Toulmé et al. 2005; Yuzenkova et al. 2014). Mfd, a bacterial transcription-coupled repair (TCR) factor, can also reactivate a stalled or backtracked RNAP (Borukhov et al. 2005), using its ATP-dependent translocation activity to push the RNAP forward. This allows the 3′ end of the nascent RNA transcript to re-engage at the catalytic site (Park et al. 2002). Once the EC is successfully reactivated, the elongation complex continues transcription, and re-approaches the roadblock (Fig. 1). Several other proteins have also been implicated in preventing EC pausing and backtracking. These include the bacterial transcription factor DksA, which minimises RNAP pausing by reducing nucleotide misincorporation (Roghanian et al. 2015), and the NusG family of transcription factors, which stabilise the minimal transcription bubble (Kang et al. 2018; Turtola and Belogurov 2016). In bacteria, transcription and translation are often coupled, and the ribosome trailing behind the RNAP can exert a "push" force to reactivate transcription of a backtracked polymerase (Nudler 2012; Proshkin et al. 2010). Mathematical modelling suggests that an RNAP trailed by a ribosome is ~ 13 times more resistant to termination than an untrailed RNAP in a head on collision (Hoffmann et al. 2019).
RNAP pausing and backtracking can also occur in eukaryotes as the EC progresses through nucleosomes (Teves et al. 2014) or encounters protein roadblocks (Candelli et al. 2018), and the mechanisms for resolving backtracking are similar to those in bacteria. For example, the eukaryotic NusG family protein Spt4-Spt5 interacts directly with the EC, the upstream DNA, and the nascent transcript near the RNA exit channel to repress RNAP pausing and backtracking (Ehara et al. 2017; Hartzog and Fu 2013; Klein et al. 2011). In addition, RNAP backtracking can also be suppressed by the transcription initiation factor TFIIF, which occurs through transient interactions between TFIIF and RNAP at the pause site, resulting in a conformational change in RNAP (Schweikhard et al. 2014). However, when backtracking is inevitable, mechanisms also exist to resume transcriptional elongation and prevent prolonged RNAP pausing. For instance, the eukaryotic transcriptional factor TFIIS protein can generate a new 3′ end by cleaving the nascent RNA transcript, much like the GreA/GreB proteins in bacteria. This allows the RNA to realign with the active site (Schweikhard et al. 2014). TFIIS can also bind competitively to the backtrack site, displacing and mobilizing RNA, thereby weakening the grip of RNAPII on the backtracked RNA (Cheung and Cramer 2011; Farnung et al. 2022).
One outcome of this cycle of roadblock encounter and backtracking is termination of the RNAP, with the result that downstream transcription is reduced (Fig. 1). We found that Mfd is a major factor in roadblock-induced termination, at least at a strong roadblock, with the absence of Mfd significantly increasing transcription downstream of a LacI roadblock, particularly for weak promoters (Hao et al. 2014). When the EC stalls at the roadblock site for an extended period of time and is unable to overcome the obstacle, Mfd can use its ATPase activity to facilitate the dissociation of RNAP from the DNA, leading to the termination of transcription (Borukhov et al. 2005; Hao et al. 2019; Le et al. 2018). This activity is similar to Mfd’s role in transcription-coupled repair where it removes RNAPs stalled at DNA lesions. Mfd can also remove RNAPs stalled by encounter with a replisome (i.e., a mobile roadblock) that is moving in either the same or opposite direction as the RNAP to mitigate the resultant DNA damage, and genomic instability (Pomerantz and O'Donnell 2010). The bacterial RNA-binding termination factor Rho accounts for approximately 50% of all termination events in E. coli (Cardinale et al. 2008), but it remains unclear whether Rho plays a role in terminating RNAP stalled at roadblocks (Dutta et al. 2008; Peters et al. 2011).
The other outcome for the RNAP is to pass through the roadblock (Fig. 1). To do this, RNAP can simply take advantage of the spontaneous dissociation of the roadblock protein and escape a roadblock site before the protein can rebind to the DNA. Alternatively, RNAP may actively remove the roadblock protein from the DNA, a process we term dislodgment (Hao et al. 2014). Mfd’s ability to push RNAP forward may aid dislodgement (Hao et al. 2014), akin to its ability to assist RNAP in traversing difficult-to-transcribe DNA sequences where RNA secondary structure formation at the RNAP exit channel can lead to RNAP stalling (Ragheb et al. 2021).
When a strong promoter is involved, multiple RNAPs can queue up along the DNA upstream of the roadblock. In general, this results in a larger fraction of RNAPs passing the roadblock site, a process termed cooperation (Epshtein and Nudler 2003; Hao et al. 2014; Jin et al. 2010). The mechanism of cooperation is unclear. Trailing RNAPs can prevent backtracking by assisting the translocation of the leading EC, which should increase the number of roadblock-passing attempts. The presence of multiple RNAPs may also inhibit termination, possibly by blocking access by Mfd. Trailing RNAPs may push the leading RNAP to help it dislodge the roadblock protein. In addition, once a leading RNAP passes the roadblock, the following RNAPs may be able to pass before the roadblock protein rebinds or may even occlude the binding site to inhibit its rebinding.
While a high flux of RNAPs from a strong promoter can increase transcription over a protein roadblock, this effect only occurs up to a certain threshold. At very high rates of promoter firing, transcription can decrease due to a build-up of RNAPs, a process known as clogging (Fig. 1). Clogging occurs when backed-up RNAPs occupy the promoter, blocking transcription initiation and reducing the rate of transcription (Sigurdsson et al. 2010; Washburn et al. 2003; Yuzenkova et al. 2014). This phenomenon is more likely to occur when the roadblock is strong, and when the roadblock site is relatively close to the promoter (Epshtein et al. 2003; Hao et al. 2014).
Natural and engineered protein roadblocks
Only a small number of DNA-binding proteins have been found to be effective roadblocks to transcribing RNAPs. The discovery and study of these proteins has shed light on the important properties of the roadblock proteins that impede the progression of RNAP.
The idea that a static DNA bound complex could block the progression of RNAP downstream of the promoter began with Kassavetis et al. in 1978 who found that RNAP inactivated by rifampicin is unable to initiate a long chain RNA synthesis but retains its ability to bind to promoter sites on the DNA. This then acts as a barrier against other RNAP molecules that have initiated transcription at upstream promoter sites, with the potential to block progression along the DNA (Kassavetis et al. 1978). Similarly, a study by Kingston et al. on RNAP pausing within the E. coli ribosomal RNA operon found that RNAP bound at a downstream promoter, if slow to initiate, can act as a potential barrier to a second RNAP initiated from an upstream promoter (Kingston and Chamberlin 1981).
After the initial demonstrations that an elongating RNAP can be blocked by other DNA occupants that share the same DNA, one of the first roadblock proteins to be extensively studied was the E. coli Lac repressor (LacI). LacI regulates the expression of the lac operon by binding to the operator region of the lac operon, repressing transcription by blocking the binding of RNAP to the promoter. It also serves as a sensor for lactose, binding to it and causing a conformational change that weakens its affinity for the operator region, thus allowing transcription to occur in the presence of lactose (Jacob and Monod 1961).
In 1982, Horowitz et al. investigated the lac and trp operons in bacteria that are expressed in opposite orientations in the same region of DNA. They found that LacI bound at the lacUV5 promoter limited the expression of the trp operon by blocking the RNAP initiated at this operon (Horowitz and Platt 1982). A later study found that LacI, when bound at an operator site, can halt the progression of RNAPs and interrupt transcription (Deuschle et al. 1986). The E. coli LacI repressor was later also shown to be an effective roadblock for eukaryotic RNAPI and II (Deuschle et al. 1990; Kuhn et al. 1990; Reines and Mote 1993). The ability of LacI to block the progression of RNAP was also demonstrated in vitro via single-molecule experiments (Voros et al. 2017; Xu et al. 2022). More recently, our laboratory has used a synthetic biology approach to systematically analyse the LacI roadblocking in vivo (Hao et al. 2014), and determined the key parameters of effective roadblocking (see below).
The discovery of the Lac repressor as a transcriptional roadblock has paved the way for the identification of additional roadblock proteins in recent years. By binding downstream of transcription start sites, these proteins have all been shown to block the progression of elongating RNAP. Examples of such proteins include transcription factors like PurR, GalR and ArsR repressors in E. coli (He and Zalkin 1992; Lewis et al. 2008; Merulla and van der Meer 2015), CcpA and CodY in B. subtilis (Choi and Saier 2005), and various other cellular proteins, such as the dominant growth phase nucleoid protein Fis (Chintakayala et al. 2013), Tus replication terminator protein (Guajardo and Sousa 1999), histone-like nucleoid structuring protein H-NS (Dole et al. 2004), and the SOS response protein LexA (Sancar et al. 1982). In addition, a recent study by Klimuk et al. found that the controller protein C of the bacteria Kpn2I restriction-modification system can also function as a roadblock to RNAP, preventing transcription of its own gene and thus forming a negative feedback loop to control its own expression (Klimuk et al. 2018).
The phenomenon of transcriptional roadblocking is not limited to prokaryotic systems. Next-generation sequencing has shown the widespread occurrence of RNAPII pausing and roadblock termination at sites occupied by transcription factors in both yeast and human cells (Candelli et al. 2018; Mayer et al. 2015). For example, CTCF, a zinc finger binding protein capable of sequence-specific binding to specific DNA sequences known as insulators, was found to pause RNAPII in a mammalian model system (Kornblihtt 2012; Shukla et al. 2011). Similarly, the yeast general regulatory factor Reb1p and repressor activator protein Rap1p have both been shown to block RNAPII progression when bound at their high affinity sites (Colin et al. 2014; Roy and Chanfreau 2018; Yarrington et al. 2012). Notably, the ability of Rap1p to block RNAPII is dependent on its binding orientation, as binding in the inverted orientation can render it susceptible to dislodgement by RNAPII (Yarrington et al. 2012). It has been proposed that the interaction between the C terminus of the Rap1p DNA binding domain and its DNA binding site creates additional contacts, reducing the likelihood of its displacement by RNAP. Finally, the yeast Cbf1 protein was recently shown to serve as a transcriptional roadblock, preventing unscheduled transcription from entering the centromeres and thus safeguarding these critical regions (Hedouin et al. 2022).
Apart from naturally occurring roadblock proteins, several proteins have also been engineered to function as transcriptional roadblocks. One of the first examples is EcoRI Q111, a mutant version of the EcoRI endonuclease that has had its DNA cleavage activity removed via targeted mutagenesis. This modified EcoRI retains its DNA binding site affinity and effectively blocks RNAP progression in vitro (Pavco and Steege 1990). Another example of an engineered roadblock protein is the TALE proteins, which are DNA binding proteins found in Xanthomonas bacteria. TALE proteins recognize specific DNA sequences through repeating units of 33–35 highly conserved amino acids, including two critical amino acids known as repeat variable di-residues (RVDs). By modifying the RVD sequence, TALE proteins have been engineered to bind a lac operator with high affinity, resulting in a more potent roadblock than the Lac repressor (Politz et al. 2013). The catalytically dead CRISPR-Cas (dCas) protein is another example of an engineered roadblock protein, which will be discussed in more detail below.
Programmable roadblocks by catalytically dead CRISPR-Cas proteins
In the last decade, the rapid development of CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR associated) technology, derived from the adaptive immunity system found in a large number of bacteria and archaea (Barrangou et al. 2007; Makarova et al. 2006), has been fundamental in the expansion of genome editing in a variety of organisms, including bacteria, plants, animals, and human cells (Anzalone et al. 2020; Cong et al. 2013; Doudna and Charpentier 2014; Makarova et al. 2015; Mali et al. 2013).
One of the best studied CRISPR-Cas systems is the type-II system, which consists of a single multidomain effector protein known as Cas9. Cas9 can cut the DNA at specific sites designated by a single guide RNA (sgRNA) that is complementary to the target DNA sequence (Gasiunas et al. 2012; Jinek et al. 2012; Wiedenheft et al. 2012). Once the DNA is cut, the natural repair mechanisms of the cell are triggered to generate indel mutations, effectively changing the target DNA sequence. However, for Cas9 to bind and cut DNA, a PAM (Protospacer Adjacent Motif) sequence must be present adjacent to the target DNA sequence (Hille et al. 2018). Different Cas9 proteins have different PAM requirements, and this PAM specificity is crucial for minimizing off-target effects and ensuring precise genome editing. The widely used Cas9 from Streptococcus pyogenes requires an NGG PAM, which restricts the sequence space that is targetable by Cas9. However, structure-guided rational design and directed evolution have been used to significantly broaden the target range of Sp Cas9 (Hu et al. 2018; Kleinstiver et al. 2015; Nishimasu et al. 2018; Walton et al. 2020), thereby enabling genome editing of regions previously inaccessible to Sp Cas9.
While Cas9 is a highly efficient DNA-cutting enzyme, its ability to bind to DNA independently of its nuclease activity has also led to the development of catalytically inactive Cas9, known as dCas9 (Jinek et al. 2012). This mutant version of Cas9 has been used as a versatile molecular scaffold to bring various effectors, such as transcriptional modulators and epigenetic modifiers, to targeted DNA locations (Dominguez et al. 2016; Gilbert et al. 2013). In addition, a number of studies have demonstrated that dCas9 itself can also regulate gene expression in prokaryotes by blocking the progression of RNAPs (Bikard et al. 2013; Peters et al. 2016; Qi et al. 2013; Vigouroux et al. 2018; Widom et al. 2019). Similar to yeast Rap1p protein, the roadblocking activity of dCas9 is dependent on its binding orientation. Effective repression of transcription elongation is observed when targeting the non-template strand, whereas less effective repression is observed when targeting the template strand (Bikard et al. 2013; Hao et al. 2016; Qi et al. 2013). Interestingly, while a DNA-bound dCas9 can also impede the progress of the replisome, this roadblocking activity is not dependent on binding orientation (Doi et al. 2021; Whinn et al. 2019).
The location of dCas9 targeting sites relative to the transcriptional start site is also found to be important for effective roadblocking. Target regions located further from the promoter resulted in less effective roadblocking, possibly due to the extended sequence allowing for multiple RNAPs to build up and use the cooperation effect to ‘push back’ against backtracking or ‘push’ past a bound roadblock. The roadblocking effect is also reduced at a target region very close to the promoter site, but it can still reduce promoter activity either by blocking the promoter site for RNAP initial binding or resulting in a clogging effect from a build-up of RNAP (Bikard et al. 2013).
In addition to RNAP roadblocking, dCas9 can also reduce the expression of a target gene by targeting the promoter sequence to directly block RNAP binding at the promoter. This repression however differs from roadblocking and is not dependent on the orientation of Cas9 binding (Bikard et al. 2013; Qi et al. 2013).
More recent studies of Sp dCas9 have been able to tune dCas9 roadblocking to allow for precise and robust changes in targeted gene expression by controlling complementarity between the guide RNA and the target DNA sequence through the introduction of mismatches in the 20nt sgRNA sequence (Vigouroux et al. 2018). Several studies have found that mutations within the last 6–8 nucleotides at the PAM-distal end of the sgRNA did not have a significant impact on the roadblock activity of dCas9, but mutations within the 7nt at the PAM-proximal end almost completely abolish the dCas9 roadblocking (Fu et al. 2013; Jinek et al. 2012; Widom et al. 2019). It is unclear how much these effects can be explained by reduced occupancy of the site by dCas9 or by increased dislodgement of bound dCas9 by RNAP. Recent structural studies have shown that PAM-distal mismatches can be accommodated either by base skipping and multiple noncanonical base pairing, or by stabilization through reorganization of the RuvC nuclease domain (Bravo et al. 2022; Pacesa et al. 2022). These findings provide new insights for designing better Cas9 variants with increased fidelity.
Other Cas9 orthologs have also demonstrated dCas9 mediated roadblocking, such as Streptococcus thermophilus CRISPR1 and Neisseria meningitidis Cas9 proteins. These orthologs have varying PAM requirements, providing a wider range of potential target sites (Esvelt et al. 2013). More recently, a bioinformatic screen has identified 79 phylogenetically distinct Cas9 orthologs, along with their corresponding gRNA and PAM requirements (Gasiunas et al. 2020). It is likely that many of these Cas9 proteins, when mutated to remove their catalytic activity, will act as roadblocks to elongating RNAPs.
Similar to Cas9, other type-II CRISPR associated single effector proteins from various bacteria are known to be guided to a target site complementary to a region within the RNA guide to cleave a target (Makarova et al. 2015). One such protein is Cas12a (previously known as Cpf1), a single-RNA guided endonuclease from the type-II V CRISPR system that, in contrast to Cas9, requires the presence of an upstream T-rich PAM sequence (Fig. 2) and produces a staggered double stranded break in the DNA (Leenay et al. 2016). A catalytically dead version of Cas12a (dCas12a) has been shown to function as a transcription roadblock (Zetsche et al. 2015) with a strand dependence opposite to that of dCas9 (Fig. 2) (Miao et al. 2019). However, the experiments were conducted using different guide RNAs that target either the template or non-template strand of DNA. It is possible that the differences in roadblocking efficiencies could be due in part to the varying binding affinities of different guide RNA/dCas complexes to DNA. An ideal experiment would be to use the same guide RNA to target the identical DNA sequence in both orientations.
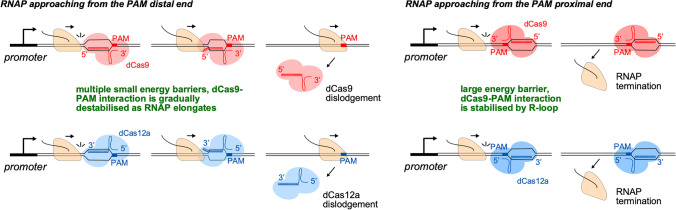
Fig. 2.
Model for the strand-dependent roadblocking by the dCas9 and dCas12a complexes. As RNAP moves forward towards the dCas complex from the PAM-distal end, it weakens the numerous small energetic barriers that form between the guide RNA and DNA. With each step, the dCas-PAM interaction becomes slightly destabilized. As a result, by the time RNAP reaches the PAM end, the final barrier is weak and easily overcome, enabling RNAP to dislodge the dCas complex. Conversely, the dCas-PAM interaction at the PAM-proximal end is fully stabilized by the R-loop, creating a large energetic barrier that RNAP must overcome in a single step
A recent single-molecule study has shed light on the mechanism of orientation-dependent roadblocking by dCas9 and dCas12a (Hall et al. 2022). The DNA-bound dCas complex consists of two distinct regions: an unprotected R-loop-mediated DNA bubble located on the PAM-distal end and tightly clamped DNA located on the PAM-proximal end (Jinek et al. 2014; Stella et al. 2017; Swarts et al. 2017). According to the model of Hall et al., when RNAP approaches the dCas complex from the PAM-distal end, forward translocation forces the re-zipping of the DNA bubble in the dCas complex. This causes the R-loop to collapse and dCas to detach from the DNA, resulting in more permissive transcription from the PAM-distal side. Conversely, when RNAP approaches a bound dCas from the PAM-proximal side, the R-loop in the Cas complex is not readily accessible to the elongating RNAP, resulting in more prohibitive transcription from the PAM-proximal side. In line with this model, a modified sgRNA that can stabilise the R-loop enhances the effectiveness of the dCas9 roadblock when RNAP approaches from the PAM-distal side (Hall et al. 2022). We expect that the positive supercoiling generated ahead of RNAP may also aid R-loop collapse. Importantly, when approaching from the bubble side, each RNAP elongation step need only remove one RNA–DNA basepair. Thus, RNAP encounters a series of small energetic barriers that it can easily overcome one by one. Each step slightly destabilizes the dCas-PAM interaction, so that once RNAP reaches the PAM, the final barrier is very weak. In contrast, when RNAP approaches from the other direction, the dCas-PAM interaction is fully stabilized by the R-loop and may be required to be removed in one step, thus presenting a large energetic barrier (Fig. 2).
What makes a protein a strong or a weak roadblock?
While several DNA-binding proteins have been identified as roadblocks, the majority of DNA binding proteins are not effective transcriptional roadblocks. For example, neither the CI lysogenic repressor nor the Cro repressor of bacteriophage λ bound to their three-operator OR site is a strong roadblock (Hao et al. 2016). Our laboratory has also shown poor roadblocking by the CII activator protein of phage λ (Palmer et al. 2009) and the CI repressor (Shearwin et al. 2002) and CII activator proteins of phage 186 (Hao et al. 2019; Neufing et al. 2001). The inability of the phage CI repressors to block RNAP progression was also shown in a recent in vitro single-molecule study (Lu et al. 2022). This raises questions about what properties of proteins cause them to be strong roadblocks to RNAP.
To understand the key parameters that govern transcriptional roadblocking, we performed systematic experiments combined with modelling to study LacI roadblocking in vivo (Hao et al. 2014). We varied the promoter firing rate (15 strengths from 0.00033 to 0.2 sec−1), the LacI concentration (5 levels from 15 to 254 nM), operator affinity (3 strengths from 0.17 to 3.9 nM), the rate of RNAP termination (wild type and mfd knockout), and promoter–roadblock spacing (3 spacings from 30 to 102 base pairs) and measured the magnitude of roadblocking. To dissect the complex interaction of these factors, we utilized stochastic simulations.
We used a hybrid Gillespie/discrete fixed time-step method. In the absence of RNAP on the DNA, only three possible events can occur: loading of a new RNAP at the promoter (if there is no other RNAP blocking it), and binding or unbinding of LacI, depending on whether the operator is occupied. The next time for each of these processes was determined by the formula ‘– ln(r/k)’, where r is a random number between 0 and 1 and k (s−1) is the rate of the process (Fig. 3). The process with the next shortest time was used to update the system, and the time was incremented by the time associated with that process. This simulation procedure was repeated until an RNAP was loaded on the DNA, at which point the simulation was switched to a fixed time-step mode. In this approach, each step was set to the time taken for RNAP to move forward one base pair (1/40 s), and in each time step, each RNAP attempted to advance 1 bp. All events were assigned a specific rate k (Fig. 3). The occurrence of any particular event, if possible, during the next time step was determined by generating a random number between 0 and 1; if this number was less than 1- e–k, then that event occurred. After all RNAPs were cleared from the DNA, the simulation was switched back to the Gillespie approach.
Fig. 3.
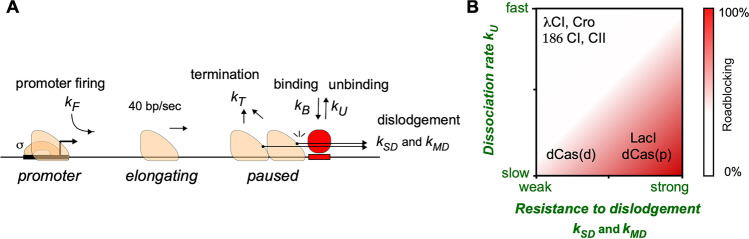
Biophysical determinants of effective transcriptional roadblocking. A) A stochastic model of transcriptional roadblocking indicating the key parameters that govern transcriptional roadblocking. kSD and kMD are the rates of roadblock dislodgement by single or multiple RNAPs, respectively. Figure adapted from Fig. 1C in (Hao et al. 2014). B) The kinetic properties of a DNA-binding protein and its resistance to dislodgement by an elongating RNAP collectively determine its ability to effectively block RNAP progression. λCI, Cro: the CI and Cro repressors from bacteriophage λ; 186 CI, CII: the CI repressor and CII activator of bacteriophage 186; dCas(d): an RNAP approaching dCas complex from a PAM-distal end; dCas(p): an RNAP approaching dCas complex from a PAM-proximal end. Figure adapted from Fig. 2A in (Hao et al. 2017)
The events that are included in the simulation are as follows (Fig. 3A):
Promoter firing: Binding and initiation of transcription by RNAP is simulated as a single-step process with a rate kF.
Binding and unbinding of the roadblock protein: If the operator is unoccupied by either another Lac repressor or elongating RNAPs, the rate of binding of LacI (kB) is influenced by the concentration of LacI and is determined by the product of the binding constant (kon, s−1 M−1) and its concentration. On the other hand, the rate of unbinding (kU, s−1) is dependent on the specific operator sequence.
Elongation: The simulation begins by attempting to move each RNAP forward by 1 bp, starting from the RNAP farthest from the promoter. If an RNAP encounters a roadblock, caused by either LacI or a paused RNAP, it can move forward only if it or the leading RNAP is capable of dislodging LacI. The dislodgement rate by a single RNAP is kSD. If multiple RNAPs are queued in front of a roadblock, a different dislodgement rate (kMD) is applied to account for RNAP cooperation. Importantly, we found that this active dislodgement of the roadblock by RNAP was needed to reproduce the experimental data. A successful dislodgement of LacI was taken to allow all RNAPs queued at the roadblock to advance. Once an RNAP moves past the last position of the operator, a new transcript is counted, and that RNAP is removed from the DNA.
RNAP Termination: Each paused RNAP can be removed from the system with rate kT.
Our modelling highlighted protein kinetics as the key determinant for transcription roadblocking. To be a good roadblock, the protein’s rate of binding must be sufficiently in excess of its unbinding rate to ensure that its fractional occupancy of the roadblock site is high (fractional occupancy = kB/(kB + kU)). However, kB and kU affect roadblocking in different ways. The rate of binding kB has minimal impact on roadblocking unless the promoter is very strong, whereas the rate of unbinding of the roadblock protein kU is critical. If unbinding is rapid, then there is a greater chance that the roadblock protein will spontaneously dissociate before RNAPs stalled at the roadblock are terminated. A low kU implies a higher energy barrier for the protein to leave its site, which should also cause it to be more resistant to active dislodgement by RNAP. Indeed, we found that the fitted rates for dislodgement by single and multiple RNAPs (kSD and kMD) were inversely correlated with kU for the different Lac operators.
Consequently, roadblocking and site occupancy can be adjusted independently to some extent, and proteins with the same fractional occupation of a site, but with distinct kinetics, can have different roadblocking properties. Proteins with slow kinetics (low kB, low kU e.g. LacI), will tend to be strong roadblocks, while proteins with fast binding kinetics (high kB, high kU e.g. λ CI) will tend to be ineffective roadblocks (Fig. 3B).
However, not all proteins with slow binding kinetics are strong roadblocks to elongating RNAP, as demonstrated by dCas9 (Bikard et al. 2013; Hao et al. 2016; Qi et al. 2013)(Fig. 3B). The strong orientation bias for roadblocking cannot be simply explained by binding kinetics because the values of kB and kU for dCas9 are the same, regardless of the direction RNAP approaches from. This could be accounted for in our model by setting different active dislodgement rates for the two directions, thus breaking the correlation between kU and the rates for kSD and kMD. We expect that a more complete model of dCas9 roadblocking would require incorporating multistep unbinding.
Another factor that can affect roadblocking is DNA looping, with weak roadblocks strengthened when participating in long-range interactions on the DNA. For example, a single-molecule study by Voros et al. demonstrated that the LacI repressor can act as a robust roadblock even when bound to a weak lacO2 operator, as long as it participates in a DNA loop to another LacI site (Voros et al. 2017). Similarly, our study on the λ CI repressor found that CI binding in a 3.8 kb loop with the OL binding site improved CI roadblocking at the OR site in vivo (Hao et al. 2016). It is not clear whether these effects can be explained by the expected increased binding rate provided by DNA looping or if the spontaneous or RNAP-assisted unbinding rates are affected. We have recently developed a quantitative in vivo assay to measure the looping efficiencies of various natural and engineered proteins (Hao et al. 2021) that should help understand the mechanism of this effect.
Conclusion and future perspective
With recent advances in high-throughput RNA sequencing techniques, transcriptional roadblocking has increasingly been recognized as an important control mechanism in gene regulation in both bacteria and eukaryotic cells. Roadblock-mediated termination has been shown to prevent uncontrolled gene expression in E. coli (Chintakayala et al. 2013), as well as protect the yeast centromere from off-target transcription (Hedouin et al. 2022). Additionally, roadblock proteins have been suggested to work in tandem with the cellular canonical termination pathways to delimit transcription units and reduce pervasive transcription (Candelli et al. 2018).
In this review, we summarised the mechanisms by which roadblock proteins impede the progression of elongating RNAP and the cellular factors that can facilitate RNAP escape or clearance of roadblocks. We surveyed the literature on factors that influence a DNA-bound protein's effectiveness as a roadblock and identified protein binding dynamics and resistance to RNAP dislodgement as key defining factors for a strong roadblock. Further investigation into the molecular regulation of roadblocking and identification of key attributes for effective roadblocks would significantly enhance our understanding of gene regulation and potentially open new avenues for designing synthetic genetic circuits and therapeutic interventions.
For example, roadblocking regulation has been used to build an arsenic biosensor with ultra-low background and high sensitivity, based on the E. coli arsenite-responsive ArsR repressor (Merulla and van der Meer 2015). Likewise, Chatterjee and colleagues used LacI roadblocking and transcriptional interference (TI) to construct a variety of synthetic two-input logic gates and investigated the factors that influence their behaviour (Bordoy et al. 2019; O'Connor et al. 2021).
Our own research on transcriptional interference between convergent promoters also revealed that transcription factors can indirectly control a target promoter by regulating the interfering promoter (Hao et al. 2016, 2017). This approach is widely used in developmental switches across all kingdoms of life (Dodd et al. 2007; Hongay et al. 2006; Latos et al. 2012) to ensure mutual exclusivity between different developmental programs. However, for this strategy to be effective, the transcription factor must have rapid binding kinetics to maintain site occupancy without blocking RNA polymerase.
Recent discoveries in the use of catalytically dead Cas proteins as programmable roadblocks to RNAP, with the potency of dCas roadblocking that is adjustable through targeting different DNA strands and through guide RNA mismatches, have opened up exciting opportunities to uncover novel aspects of biology. Genome-wide dCas9 knockdown (CRISPRi) identified essential genes in E. coli, including a subset that are highly sensitive to minor expression fluctuations via roadblocking with less effective template targeting (Rousset et al. 2018). Additionally, CRISPRi has been utilized to identify genes required for phage infection by 14 phylogenetically diverse phages (Mutalik et al. 2020; Rousset et al. 2018).
Author contribution
Conceptualization: Nan Hao, Alana Donnelly, Ian B. Dodd, Keith E. Shearwin; writing— original draft preparation: Nan Hao, Alana Donnelly; writing—review and editing: Ian B. Dodd, Keith E. Shearwin; visualization: Nan Hao, Alana Donnelly, Ian B. Dodd; supervision: Keith E. Shearwin.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions The work was funded by an ARC Discovery grant to Keith E. Shearwin [DP160101450].
Data availability
The article type (review article) does not expect raw files.
Declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors (not applicable).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abdelkareem M, Saint-Andre C, Takacs M, Papai G, Crucifix C, Guo X, Ortiz J, Weixlbaumer A (2019) structural basis of transcription: RNA polymerase backtracking and its reactivation. Mol Cell 75(298–309):e294 [DOI] [PMC free article] [PubMed]
- Anzalone AV, Koblan LW, Liu DR (2020) Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38:824–844 [DOI] [PubMed]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 [DOI] [PubMed]
- Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA (2013) Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res 41:7429–7437 [DOI] [PMC free article] [PubMed]
- Bordoy AE, O’Connor NJ, Chatterjee A (2019) Construction of two-input logic gates using Transcriptional Interference. ACS Synth Biol 8:2428–2441 [DOI] [PubMed]
- Borukhov S, Lee J, Laptenko O (2005) Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol Microbiol 55:1315–1324 [DOI] [PubMed]
- Bravo JPK, Liu MS, Hibshman GN, Dangerfield TL, Jung K, McCool RS, Johnson KA, Taylor DW (2022) Structural basis for mismatch surveillance by CRISPR-Cas9. Nature 603:343–347 [DOI] [PMC free article] [PubMed]
- Candelli T, Challal D, Briand JB, Boulay J, Porrua O, Colin J, Libri D (2018) High-resolution transcription maps reveal the widespread impact of roadblock termination in yeast. EMBO J 37:e9749037 [DOI] [PMC free article] [PubMed]
- Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E (2008) Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 320:935–938 [DOI] [PMC free article] [PubMed]
- Cheung AC, Cramer P (2011) Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471:249 [DOI] [PubMed]
- Chintakayala K, Singh SS, Rossiter AE, Shahapure R, Dame RT, Grainger DC (2013) E. coli Fis protein insulates the cbpA gene from uncontrolled transcription. PLoS Genet 9:e1003152 [DOI] [PMC free article] [PubMed]
- Choi S-K, Saier MH (2005) Regulation of sigL expression by the catabolite control protein CcpA involves a roadblock mechanism in Bacillus subtilis: potential connection between carbon and nitrogen metabolism. J Bacteriol 187:6856–6861 [DOI] [PMC free article] [PubMed]
- Colin J, Candelli T, Porrua O, Boulay J, Zhu C, Lacroute F, Steinmetz LM, Libri D (2014) Roadblock termination by reb1p restricts cryptic and readthrough transcription. Mol Cell 56:667–680 [DOI] [PubMed]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823 [DOI] [PMC free article] [PubMed]
- Deuschle U, Gentz R, Bujard H (1986) lac repressor blocks transcribing RNA polymerase and terminates transcription. Proc Natl Acad Sci 83:4134–4137 [DOI] [PMC free article] [PubMed]
- Deuschle U, Hipskind RA, Bujard H (1990) RNA polymerase II transcription blocked by Escherichia coli lac repressor. Science 248:480–483 [DOI] [PubMed]
- Dodd IB, Shearwin KE, Sneppen K (2007) Modelling transcriptional interference and DNA looping in gene regulation. J Mol Biol 369:1200–1213 [DOI] [PubMed]
- Doi G, Okada S, Yasukawa T, Sugiyama Y, Bala S, Miyazaki S, Kang D, Ito T (2021) Catalytically inactive Cas9 impairs DNA replication fork progression to induce focal genomic instability. Nucleic Acids Res 49:954–968 [DOI] [PMC free article] [PubMed]
- Dole S, Nagarajavel V, Schnetz K (2004) The histone-like nucleoid structuring protein H-NS represses the Escherichia coli bgl operon downstream of the promoter. Mol Microbiol 52:589–600 [DOI] [PubMed]
- Dominguez AA, Lim WA, Qi LS (2016) Beyond editing: repurposing CRISPR–Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol 17:5 [DOI] [PMC free article] [PubMed]
- Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096 [DOI] [PubMed]
- Dutta D, Chalissery J, Sen R (2008) Transcription termination factor rho prefers catalytically active elongation complexes for releasing RNA. J Biol Chem 283:20243–20251 [DOI] [PubMed]
- Ehara H, Yokoyama T, Shigematsu H, Yokoyama S, Shirouzu M, Sekine SI (2017) Structure of the complete elongation complex of RNA polymerase II with basal factors. Science 357:921–92 [DOI] [PubMed]
- Epshtein V, Nudler E (2003) Cooperation between RNA polymerase molecules in transcription elongation. Science 300:801–805 [DOI] [PubMed]
- Epshtein V, Toulmé F, Rahmouni AR, Borukhov S, Nudler E (2003) Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J 22:4719–4727 [DOI] [PMC free article] [PubMed]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM (2013) Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods 10:1116 [DOI] [PMC free article] [PubMed]
- Farnung L, Ochmann M, Garg G, Vos SM, Cramer P (2022) Structure of a backtracked hexasomal intermediate of nucleosome transcription. Mol Cell 82(3126–3134):e3127 [DOI] [PubMed]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826 [DOI] [PMC free article] [PubMed]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci 109:E2579–E2586 [DOI] [PMC free article] [PubMed]
- Gasiunas G, Young JK, Karvelis T, Kazlauskas D, Urbaitis T, Jasnauskaite M, Grusyte MM, Paulraj S, Wang PH, Hou Z et al (2020) A catalogue of biochemically diverse CRISPR-Cas9 orthologs. Nat Commun 11:5512 [DOI] [PMC free article] [PubMed]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154:442–451 [DOI] [PMC free article] [PubMed]
- Gregersen LH, Svejstrup JQ (2018) The cellular response to transcription-blocking DNA damage. Trends Biochem Sci 43:327–341 [DOI] [PMC free article] [PubMed]
- Guajardo R, Sousa R (1999) Characterization of the effects of Escherichia coli replication terminator protein (Tus) on transcription reveals dynamic nature of the tus block to transcription complex progression. Nucleic Acids Res 27:2814–2824 [DOI] [PMC free article] [PubMed]
- Hall PM, Inman JT, Fulbright RM, Le TT, Brewer JJ, Lambert G, Darst SA, Wang MD (2022) Polarity of the CRISPR roadblock to transcription. Nat Struct Mol Biol 29:1217–1227 [DOI] [PMC free article] [PubMed]
- Hao N, Krishna S, Ahlgren-Berg A, Cutts EE, Shearwin KE, Dodd IB (2014) Road rules for traffic on DNA—systematic analysis of transcriptional roadblocking in vivo. Nucleic Acids Res 42:8861–8872 [DOI] [PMC free article] [PubMed]
- Hao N, Palmer AC, Ahlgren-Berg A, Shearwin KE, Dodd IB (2016) The role of repressor kinetics in relief of transcriptional interference between convergent promoters. Nucleic Acids Res 44:6625–6638 [DOI] [PMC free article] [PubMed]
- Hao N, Palmer AC, Dodd IB, Shearwin KE (2017) Directing traffic on DNA-How transcription factors relieve or induce transcriptional interference. Transcription 8:120–125 [DOI] [PMC free article] [PubMed]
- Hao N, Crooks MT, Palmer AC, Dodd IB, Shearwin KE (2019) RNA polymerase pausing at a protein roadblock can enhance transcriptional interference by promoter occlusion. FEBS Lett 593:903–917 [DOI] [PMC free article] [PubMed]
- Hao N, Sullivan AE, Shearwin KE, Dodd IB (2021) The loopometer: a quantitative in vivo assay for DNA-looping proteins. Nucleic Acids Res 49:e39 [DOI] [PMC free article] [PubMed]
- Hartzog GA, Fu J (2013) The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta 1829:105–115 [DOI] [PMC free article] [PubMed]
- He B, Zalkin H (1992) Repression of Escherichia coli purB is by a transcriptional roadblock mechanism. J Bacteriol 174:7121–7127 [DOI] [PMC free article] [PubMed]
- Hedouin S, Logsdon GA, Underwood JG, Biggins S (2022) A transcriptional roadblock protects yeast centromeres. Nucleic Acids Res 50:7801–7815 [DOI] [PMC free article] [PubMed]
- Helmrich A, Ballarino M, Tora L (2011) Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell 44:966–977 [DOI] [PubMed]
- Hille F, Richter H, Wong SP, Bratovič M, Ressel S, Charpentier E (2018) The biology of CRISPR-Cas: backward and forward. Cell 172:1239–1259 [DOI] [PubMed]
- Hoffmann SA, Hao N, Shearwin KE, Arndt KM (2019) Characterizing transcriptional interference between converging genes in bacteria. ACS Synth Biol 8:466–473 [DOI] [PubMed]
- Hongay CF, Grisafi PL, Galitski T, Fink GR (2006) Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127:735–745 [DOI] [PubMed]
- Horowitz H, Platt T (1982) Regulation of transcription from tandem and convergent promoters. Nucleic Acids Res 10:5447–5465 [DOI] [PMC free article] [PubMed]
- Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z et al (2018) Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556:57–63 [DOI] [PMC free article] [PubMed]
- Jacob F, Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 3:318–356 [DOI] [PubMed]
- Jin J, Bai L, Johnson DS, Fulbright RM, Kireeva ML, Kashlev M, Wang MD (2010) Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nat Struct Mol Biol 17:745–752 [DOI] [PMC free article] [PubMed]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821 [DOI] [PMC free article] [PubMed]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S et al (2014) Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343:1247997 [DOI] [PMC free article] [PubMed]
- Kang JY, Mooney RA, Nedialkov Y, Saba J, Mishanina TV, Artsimovitch I, Landick R, Darst SA (2018) Structural basis for transcript elongation control by NusG family universal regulators. Cell 173(1650–1662):e1614 [DOI] [PMC free article] [PubMed]
- Kassavetis GA, Kaya KM, Chamberlin MJ (1978) Escherichia coli RNA polymerase-rifampicin complexes bound at promoter sites block RNA chain elongation by Escherichia coli/RNA polymerase and T7-specific RNA polymerase. Biochemistry 17:5798–5804 [DOI] [PubMed]
- Kingston RE, Chamberlin MJ (1981) Pausing and attenuation of in vitro transcription in the rrnB operon of E. coli. Cell 27:523–531 [DOI] [PubMed]
- Klein BJ, Bose D, Baker KJ, Yusoff ZM, Zhang X, Murakami KS (2011) RNA polymerase and transcription elongation factor Spt4/5 complex structure. Proc Natl Acad Sci U S A 108:546–550 [DOI] [PMC free article] [PubMed]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR et al (2015) Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523:481–485 [DOI] [PMC free article] [PubMed]
- Klimuk E, Bogdanova E, Nagornykh M, Rodic A, Djordjevic M, Medvedeva S, Pavlova O, Severinov K (2018) Controller protein of restriction-modification system Kpn2I affects transcription of its gene by acting as a transcription elongation roadblock. Nucleic Acids Res 46:10810–10826 [DOI] [PMC free article] [PubMed]
- Kornblihtt AR (2012) CTCF: from insulators to alternative splicing regulation. Cell Res 22:450 [DOI] [PMC free article] [PubMed]
- Kuhn A, Bartsch I, Grummt I (1990) Specific interaction of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature 344:559 [DOI] [PubMed]
- Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE et al (2012) Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338:1469–1472 [DOI] [PubMed]
- Le TT, Wang MD (2018) Molecular highways-navigating collisions of DNA motor proteins. J Mol Biol 430:4513–4524 [DOI] [PubMed]
- Le TT, Yang Y, Tan C, Suhanovsky MM, Fulbright RM Jr, Inman JT, Li M, Lee J, Perelman S, Roberts JW (2018) Mfd dynamically regulates transcription via a release and catch-up mechanism. Cell 172(344–357):e315 [DOI] [PMC free article] [PubMed]
- Leenay RT, Maksimchuk KR, Slotkowski RA, Agrawal RN, Gomaa AA, Briner AE, Barrangou R, Beisel CL (2016) Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Mol Cell 62:137–147 [DOI] [PMC free article] [PubMed]
- Lewis DE, Komissarova N, Le P, Kashlev M, Adhya S (2008) DNA sequences in gal operon override transcription elongation blocks. J Mol Biol 382:843–858 [DOI] [PMC free article] [PubMed]
- Lu Y, Voros Z, Borjas G, Hendrickson C, Shearwin K, Dunlap D, Finzi L (2022) RNA polymerase efficiently transcribes through DNA-scaffolded, cooperative bacteriophage repressor complexes. FEBS Lett 596:1994–2006 [DOI] [PMC free article] [PubMed]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV (2006) A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1:7 [DOI] [PMC free article] [PubMed]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH (2015) An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol 13:722 [DOI] [PMC free article] [PubMed]
- Mali P, Esvelt KM, Church GM (2013) Cas9 as a versatile tool for engineering biology. Nat Methods 10:957–963 [DOI] [PMC free article] [PubMed]
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS (2015) Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161:541–554 [DOI] [PMC free article] [PubMed]
- Mayer A, Landry HM, Churchman LS (2017) Pause & go: from the discovery of RNA polymerase pausing to its functional implications. Curr Opin Cell Biol 46:72–80 [DOI] [PMC free article] [PubMed]
- Merulla D, van der Meer JR (2015) Regulatable and modulable background expression control in prokaryotic synthetic circuits by auxiliary repressor binding sites. ACS Synth Biol 5:36–45 [DOI] [PubMed]
- Miao C, Zhao H, Qian L, Lou C (2019) Systematically investigating the key features of the DNase deactivated Cpf1 for tunable transcription regulation in prokaryotic cells. Synth Syst Biotechnol 4:1–9 [DOI] [PMC free article] [PubMed]
- Mutalik VK, Adler BA, Rishi HS, Piya D, Zhong C, Koskella B, Kutter EM, Calendar R, Novichkov PS, Price MN et al (2020) High-throughput mapping of the phage resistance landscape in E. coli. PLoS Biol 18:e3000877 [DOI] [PMC free article] [PubMed]
- Nadon JF, Epshtein V, Cameron E, Samatov MR, Vasenko AS, Nudler E, Lafontaine DA (2022) Site-specific photolabile roadblocks for the study of transcription elongation in biologically complex systems. Commun Biol 5:457 [DOI] [PMC free article] [PubMed]
- Neufing PJ, Shearwin KE, Egan JB (2001) Establishing lysogenic transcription in the temperate coliphage 186. J Bacteriol 183:2376–2379 [DOI] [PMC free article] [PubMed]
- Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H et al (2018) Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361:1259–1262 [DOI] [PMC free article] [PubMed]
- Nudler E (2012) RNA polymerase backtracking in gene regulation and genome instability. Cell 149:1438–1445 [DOI] [PMC free article] [PubMed]
- O’Connor NJ, Bordoy AE, Chatterjee A (2021) Engineering transcriptional interference through RNA polymerase processivity control. ACS Synth Biol 10:737–748 [DOI] [PubMed]
- Pacesa M, Lin CH, Clery A, Saha A, Arantes PR, Bargsten K, Irby MJ, Allain FH, Palermo G, Cameron P et al (2022) Structural basis for Cas9 off-target activity. Cell 185(4067–4081):e4021 [DOI] [PMC free article] [PubMed]
- Palmer AC, Ahlgren-Berg A, Egan JB, Dodd IB, Shearwin KE (2009) Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol Cell 34:545–555 [DOI] [PMC free article] [PubMed]
- Park JS, Marr MT, Roberts JW (2002) E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109:757–767 [DOI] [PubMed]
- Pavco PA, Steege D (1990) Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J Biol Chem 265:9960–9969 [PubMed]
- Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CHS, Koo BM, Marta E et al (2016) A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165:1493–1506 [DOI] [PMC free article] [PubMed]
- Peters JM, Vangeloff AD, Landick R (2011) Bacterial transcription terminators: the RNA 3′-end chronicles. J Mol Biol 412:793–813 [DOI] [PMC free article] [PubMed]
- Politz MC, Copeland MF, Pfleger BF (2013) Artificial repressors for controlling gene expression in bacteria. Chem Commun 49:4325–4327 [DOI] [PMC free article] [PubMed]
- Pomerantz RT, O’Donnell M (2010) What happens when replication and transcription complexes collide? Cell Cycle 9:2537–2543 [DOI] [PMC free article] [PubMed]
- Proshkin S, Rahmouni AR, Mironov A, Nudler E (2010) Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328:504–508 [DOI] [PMC free article] [PubMed]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152:1173–1183 [DOI] [PMC free article] [PubMed]
- Ragheb MN, Merrikh C, Browning K, Merrikh H (2021) Mfd regulates RNA polymerase association with hard-to-transcribe regions in vivo, especially those with structured RNAs. Proc Natl Acad Sci USA 5:e2008498118 [DOI] [PMC free article] [PubMed]
- Reines D, Mote J Jr (1993) Elongation factor SII-dependent transcription by RNA polymerase II through a sequence-specific DNA-binding protein. Proc Natl Acad Sci USA 90:1917–1921 [DOI] [PMC free article] [PubMed]
- Roghanian M, Zenkin N, Yuzenkova Y (2015) Bacterial global regulators DksA/ppGpp increase fidelity of transcription. Nucleic Acids Res 43:1529–1536 [DOI] [PMC free article] [PubMed]
- Rousset F, Cui L, Siouve E, Becavin C, Depardieu F, Bikard D (2018) Genome-wide CRISPR-dCas9 screens in E. coli identify essential genes and phage host factors. PLoS Genetics 14:e1007749 [DOI] [PMC free article] [PubMed]
- Roy K, Chanfreau GF (2018) A global function for transcription factors in assisting RNA polymerase II termination. Transcription 9:41–46 [DOI] [PMC free article] [PubMed]
- Sancar A, Sancar GB, Rupp WD, Little JW, Mount DW (1982) LexA protein inhibits transcription of the E. coli uvrA gene in vitro. Nature 298:96 [DOI] [PubMed]
- Schweikhard V, Meng C, Murakami K, Kaplan CD, Kornberg RD, Block SM (2014) Transcription factors TFIIF and TFIIS promote transcript elongation by RNA polymerase II by synergistic and independent mechanisms. Proc Natl Acad Sci 111:6642–6647 [DOI] [PMC free article] [PubMed]
- Shearwin KE, Callen BP, Egan JB (2005) Transcriptional interference–a crash course. Trends Genet 21:339–345 [DOI] [PMC free article] [PubMed]
- Shearwin KE, Dodd IB, Egan JB (2002) The helix-turn-helix motif of the coliphage 186 immunity repressor binds to two distinct recognition sequences. J Biol Chem 277:3186–3194 [DOI] [PubMed]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S (2011) CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479:74–79 [DOI] [PMC free article] [PubMed]
- Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ (2010) Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol Cell 38:202–210 [DOI] [PMC free article] [PubMed]
- Stella S, Alcon P, Montoya G (2017) Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature 546:559–563 [DOI] [PubMed]
- Strobel EJ, Lis JT, Lucks JB (2020) Chemical roadblocking of DNA transcription for nascent RNA display. J Biol Chem 295:6401–6412 [DOI] [PMC free article] [PubMed]
- Swarts DC, van der Oost J, Jinek M (2017) Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol Cell 66(221–233):e224 [DOI] [PMC free article] [PubMed]
- Teves SS, Weber CM, Henikoff S (2014) Transcribing through the nucleosome. Trends Biochem Sci 39:577–586 [DOI] [PubMed]
- Toulmé F, Mosrin-Huaman C, Artsimovitch I, Rahmouni AR (2005) Transcriptional pausing in vivo: a nascent RNA hairpin restricts lateral movements of RNA polymerase in both forward and reverse directions. J Mol Biol 351:39–51 [DOI] [PubMed]
- Turtola M, Belogurov GA (2016) NusG inhibits RNA polymerase backtracking by stabilizing the minimal transcription bubble. Elife 5:e18096 [DOI] [PMC free article] [PubMed]
- Vigouroux A, Oldewurtel E, Cui L, Bikard D, van Teeffelen S (2018) Tuning dCas9's ability to block transcription enables robust, noiseless knockdown of bacterial genes. Mol Syst Biol 14:e7899 [DOI] [PMC free article] [PubMed]
- Voros Z, Yan Y, Kovari DT, Finzi L, Dunlap D (2017) Proteins mediating DNA loops effectively block transcription. Protein Sci 26:1427–1438 [DOI] [PMC free article] [PubMed]
- Walton RT, Christie KA, Whittaker MN, Kleinstiver BP (2020) Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368:290–296 [DOI] [PMC free article] [PubMed]
- Wang L, Watters JW, Ju X, Lu G, Liu S (2023) Head-on and co-directional RNA polymerase collisions orchestrate bidirectional transcription termination. Mol Cell 83(1153–1164):e1154 [DOI] [PMC free article] [PubMed]
- Washburn RS, Wang Y, Gottesman ME (2003) Role of E.coli transcription-repair coupling factor Mfd in Nun-mediated transcription termination. J Mol Biol 329:655–662 [DOI] [PubMed]
- Whinn KS, Kaur G, Lewis JS, Schauer GD, Mueller SH, Jergic S, Maynard H, Gan ZY, Naganbabu M, Bruchez MP et al (2019) Nuclease dead Cas9 is a programmable roadblock for DNA replication. Sci Rep 9:13292 [DOI] [PMC free article] [PubMed]
- Widom JR, Rai V, Rohlman CE, Walter NG (2019) Versatile transcription control based on reversible dCas9 binding. RNA 25:1457–1469 [DOI] [PMC free article] [PubMed]
- Wiedenheft B, Sternberg SH, Doudna JA (2012) RNA-guided genetic silencing systems in bacteria and archaea. Nature 482:331 [DOI] [PubMed]
- Xu W, Yan Y, Artsimovitch I, Dunlap D, Finzi L (2022) Positive supercoiling favors transcription elongation through lac repressor-mediated DNA loops. Nucleic Acids Res 50:2826–2835 [DOI] [PMC free article] [PubMed]
- Yarrington RM, Richardson SM, Lisa Huang CR, Boeke JD (2012) Novel transcript truncating function of Rap1p revealed by synthetic codon-optimized Ty1 retrotransposon. Genetics 190:523–535 [DOI] [PMC free article] [PubMed]
- Yuzenkova Y, Gamba P, Herber M, Attaiech L, Shafeeq S, Kuipers OP, Klumpp S, Zenkin N, Veening JW (2014) Control of transcription elongation by GreA determines rate of gene expression in Streptococcus pneumoniae. Nucleic Acids Res 42:10987–10999 [DOI] [PMC free article] [PubMed]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, Van Der Oost J, Regev A (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The article type (review article) does not expect raw files.