Abstract
Arrested cells/ cellular debris is component left in the zona pellucida after blastocyst hatching. To identify whether expelling arrested cells/cellular debris from blastocysts is a process of human embryo self-correction by eliminating abnormal cells, 21 pairs of trophectoderm (TE) biopsies and the corresponding arrested cells/cellular debris expelled from the blastocysts from July to December 2020 were collected and analyzed using next-generation sequencing (NGS). Then, the NGS results of TE biopsies and the corresponding arrested cells/cellular debris were compared. We identified that 47.6% of blastocysts (10/21) were aneuploidies and mosaicism. A total of 18 groups of arrested cells/cellular debris (85.7%) expelled from blastocysts were abnormal, including nine aneuploid embryos and nine euploid embryos. In the arrested cells/cellular debris, all the chromosomes were affected. In conclusion, mosaicism and aneuploidies are common features of early embryonic development, and the arrested cells/cellular debris expelled from blastocysts provides evidence of early embryonic self-correction.
Keywords: Embryonic self-correction, Arrested cells/cellular debris, Trophectoderm biopsy, Aneuploidy, Mosaicism
Introduction
Mosaic and aneuploid embryos in human are prevalent throughout pre- and post-implantation development [1, 2]. Chromosomal abnormalities in early embryos result from errors during gamete meiosis (most commonly being seen in uniform aneuploidies) and errors during blastomere mitosis (mainly resulting in mosaicism) [3, 4]. When chromosomes fail to separate or separate prematurely into sister chromatids during gamete meiosis, both conditions can result in aneuploidies and then occur in all embryonic cells. That is because when errors occur during meiosis, aneuploid cells are prevalent in human embryos accompanied with cell division after fertilization. Nonetheless, if chromosome separation errors occur in an embryonic cell during mitosis, a mosaic embryo containing diploid and aneuploid cells can be formed consequently [3, 5]. Research has confirmed that the initial stages of human embryonic development are characterized by rapid cell division, which may lead to chromosomal instability resulting in either chromosomal mosaicism or rearrangement [2, 3]. Besides, chromosomal abnormalities in embryos are related to maternal age, embryo morphology, and development speed [6, 7].
Previous studies relied on animal models to investigate chromosomal abnormalities in embryos [8, 9]. However, those models are extremely difficult to provide first-hand evidence for self-correction mechanisms in human embryos. The advancement of in vitro fertilization/intracytoplasmic sperm injection (IVF/ICSI) and cytogenetic techniques for preimplantation genetic testing (PGT) further enhance our understanding of the chromosomal abnormalities in human gametes and preimplantation embryos. Even more intriguing is that several studies reported that human mosaic embryos were capable of producing healthy euploid babies [10–12]. It seems that embryos have self-correction ability to rectify chromosomal errors. Some other studies showed that although during later stages of development, chromosomal abnormalities and mosaicism were still found in blastocysts, data indicated a reduced proportion of aneuploid cells, and even 9.7–40% of aneuploid embryos transformed into euploidies by undergoing complete self-correction [13, 14]. Santos et al. also revealed a marked reduction in the number of abnormal cells in 4, 5, and 8-day embryos over time [15]. Similarly, blastocysts diagnosed with mosaicism when being cultured to day 12 also have been noticed to show euploidy profiles in both ICM and TE-derived lineages, as observed by Popovic et al. [16]. Moreover, euploid human embryonic stem cells can be derived from abnormal embryos, implying that aneuploid cells may be depleted in the presence of euploid cell competition [17]. Thus, a possible hypothesis of a reduced rate of aneuploidies during the blastocyst stage of embryonic development is that an embryo can eliminate its aneuploid cells [18, 19].
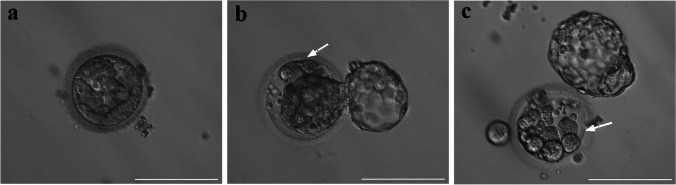
In our daily clinical work, we observed that some cells were eliminated from the blastocysts and left in the zona pellucida after blastocyst hatching (Fig. 1). Because the component of them was unknown, we called them arrested cells/cellular debris. Based on the aforementioned findings, we have hypothesized that this might represent a “correction” mechanism that rescues embryos from mosaicism or aneuploidy by eliminating abnormal cells, and it typically occurs after abnormal cleavage. In the present study, we detected the arrested cells/cellular debris isolated from blastocysts and performed a consistent comparison with the corresponding TE biopsy results to verify the hypothesis of the self-correction mechanism during early embryonic development.
Fig. 1.
Blastocysts with or without expelled arrested cells/cellular debris. a A blastocyst without hatching. b, c Hatching blastocysts with arrested cells/cellular debris left in zona pellucida. White arrows indicated arrested cells/cellular debris
Materials and Methods
Material Source and Ethical permission
In this study, 21 embryos with arrested cells/cellular debris were obtained from 19 patients who accepted PGT at the Department of Reproductive Medicine, Xiangya Hospital, Central South University from July 1st to December 31st, 2020. The indication of PGT included monogenetic disorders, chromosomal rearrangement, repeated implant failure, and unexplained repeated pregnancy loss. The study was approved by the Institutional Review Board of Xiangya Hospital, Central South University. All patients signed informed consent forms and agreed to participate the study.
Embryo Culture and Sampling
Conventional controlled ovarian hyperstimulation was used before ovum pick-up. Then, the collected oocytes were fertilized in vitro by intracytoplasmic sperm injection (ICSI). On days 1, 2, and 3 after fertilization, the embryos were morphologically evaluated and recorded according to Istanbul consensus [20]. On the days 5 or 6, the blastocyst morphology was examined under an inverted microscope to differentiate between TE and ICM. Blastocysts were graded using the Gardner grading system [21], and those with scores of 3BC, 3CB, or better were hatched by using assisted laser hatching for zona pellucida opening. Then, after 4–6 h of blastocyst incubation, 5–8 TE cells were separated using the microlaser-blunt dissection method and aspirated into a biopsy pipette. Besides, a biopsy of the arrested cells/cellular debris remaining in the zona pellucida was also collected and put into a separate polymerase chain reaction tube. Following biopsies, all blastocysts were vitrified using vitrification protocol for personalized embryo transfer in the next cycle after knowing the PGT results.
Sample Processing and Testing
All testing was conducted by Yikon Genomics according to previously described methods [22–24]. Briefly, all TE samples and arrested cells/cellular debris were separately amplified by the multiple annealing and looping-based amplification cycles (MALBAC) single-cell whole-genome amplification kit (Yikon, China). The amplified products were purified using CMPure magnetic beads, and electrophoresis was used to ensure quality control. The target fragmentation was realized using the Covaris M220 DNA Shearing instrument for genomic library construction. NGS testing was carried out using an Illumina HiSeq 2500 system after purification and library quality testing. Sequencing yielded no less than 2 million reads for a single sample. Approximately 20–80% of abnormal cells were classified as mosaicism. One less than 20% of mosaic aneuploid cells was reported as euploidy, and one more than 80% was reported as aneuploidy.
NGS Protocol Validation
Before the initiation of our study, the NGS protocol was validated for accuracy as reported previously [25]. Briefly, Preimplantation Genetic Testing-Aneuploidy kits (semiconductor sequencing) for library construction were tested for accuracy periodically. The total library construction failure rate should not be >3%, the valid data should not be <1Mb, and the genomic coverage should not be <4%. Only if the tested PGT-A met the quality control, they could be used for clinical sample sequencing.
Results
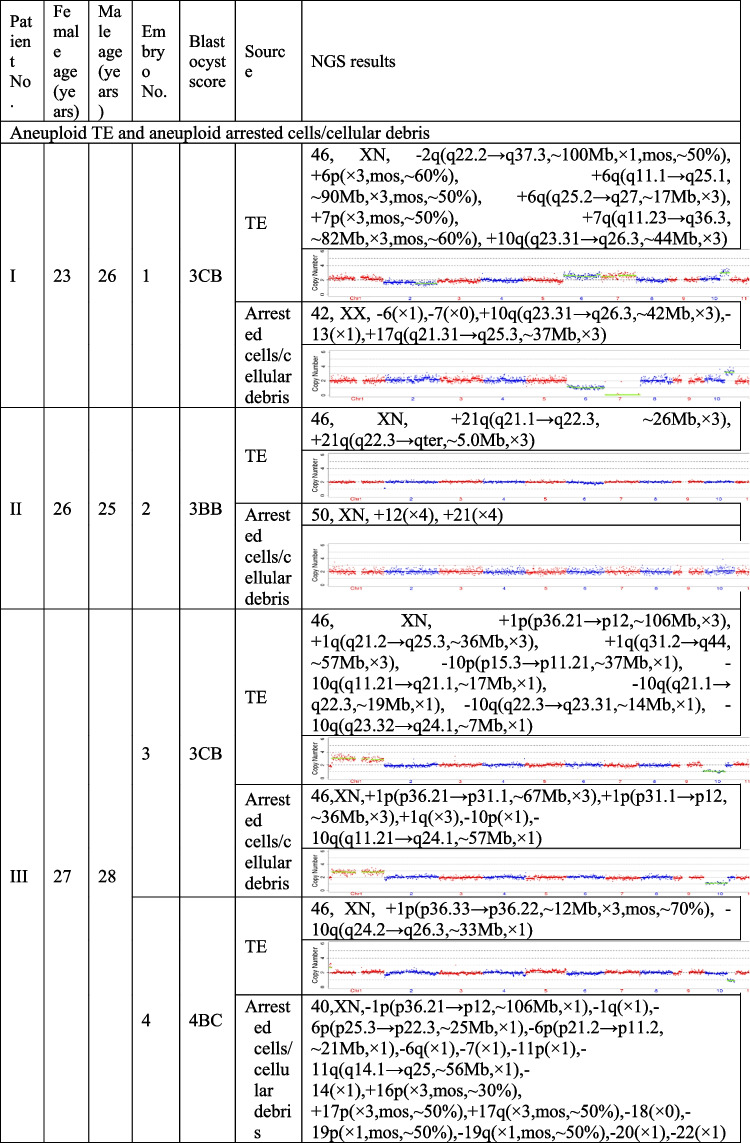
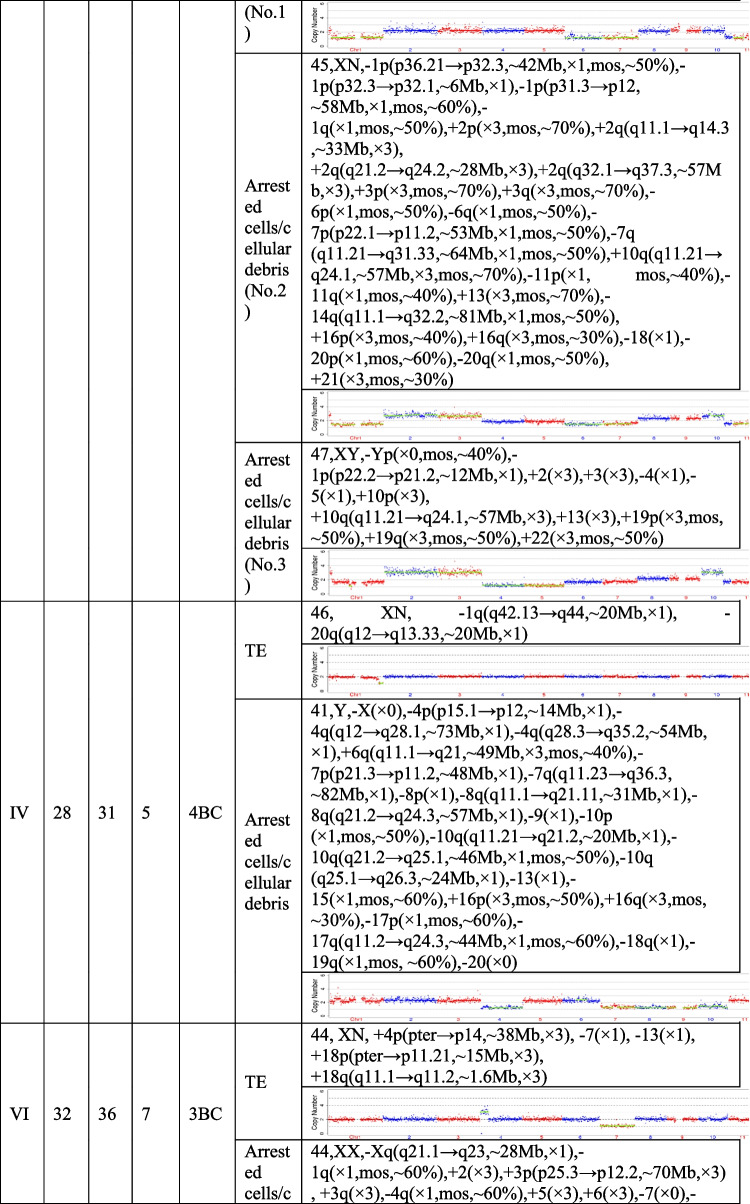
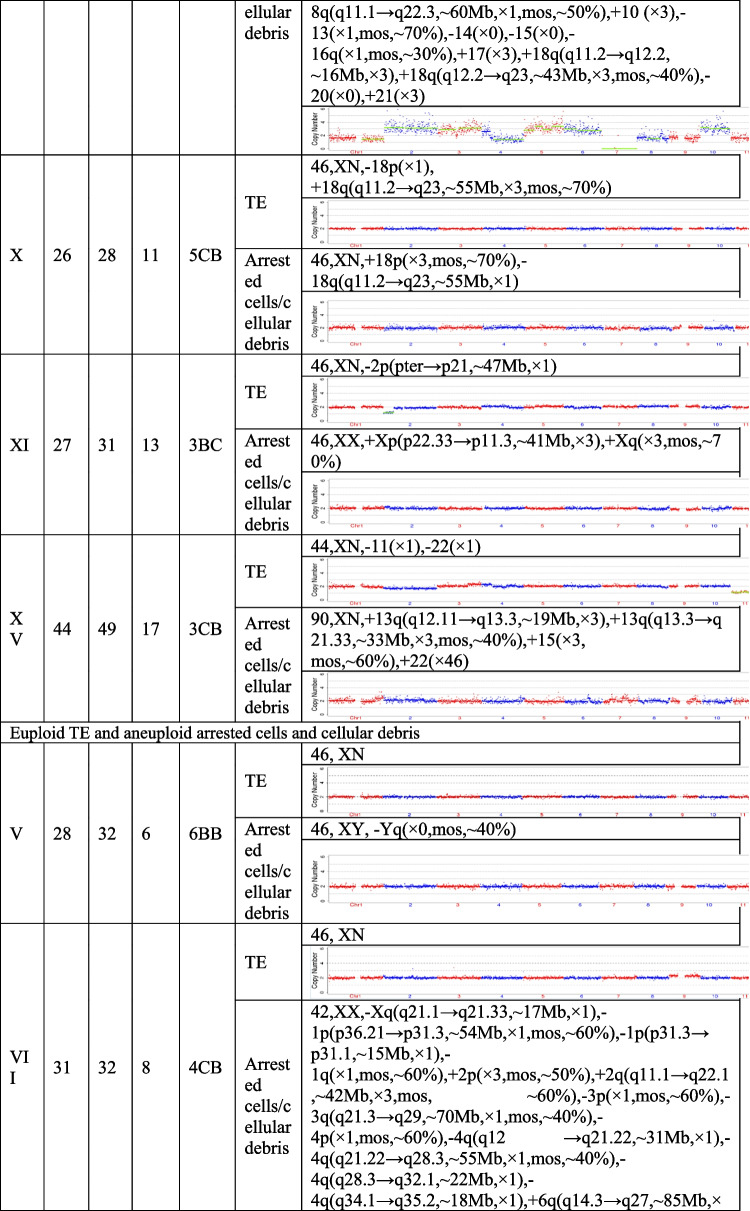
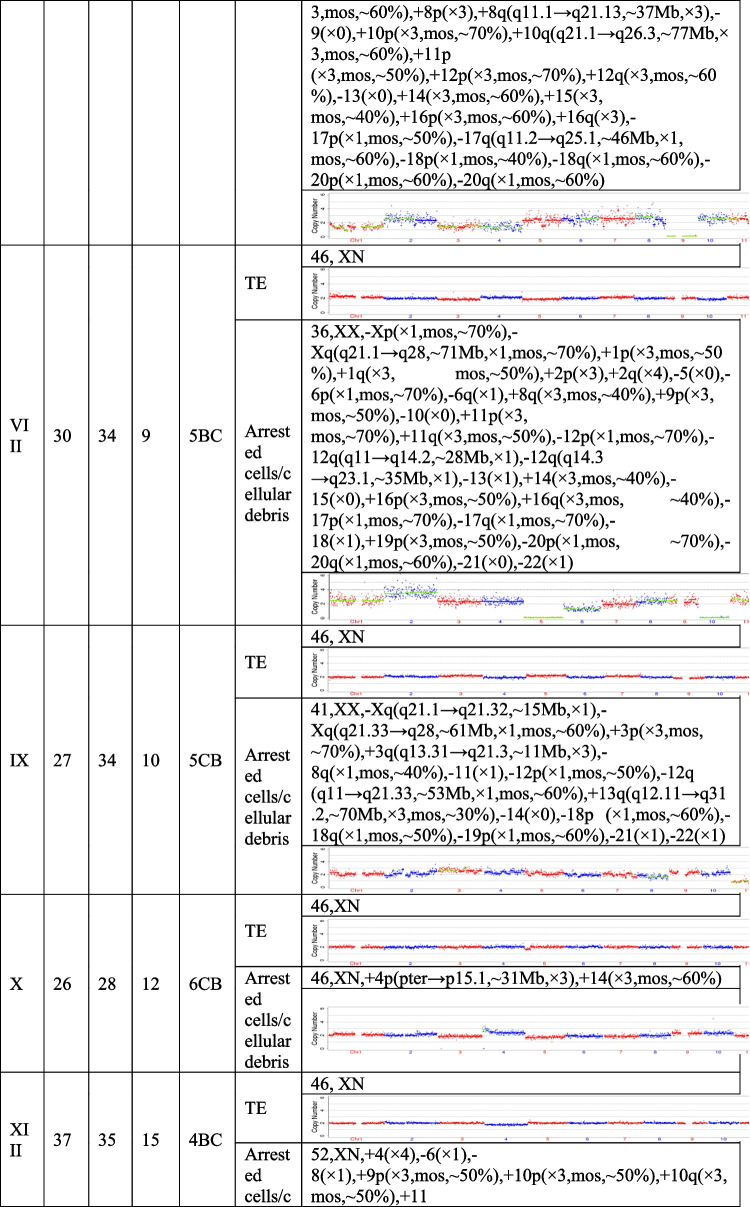
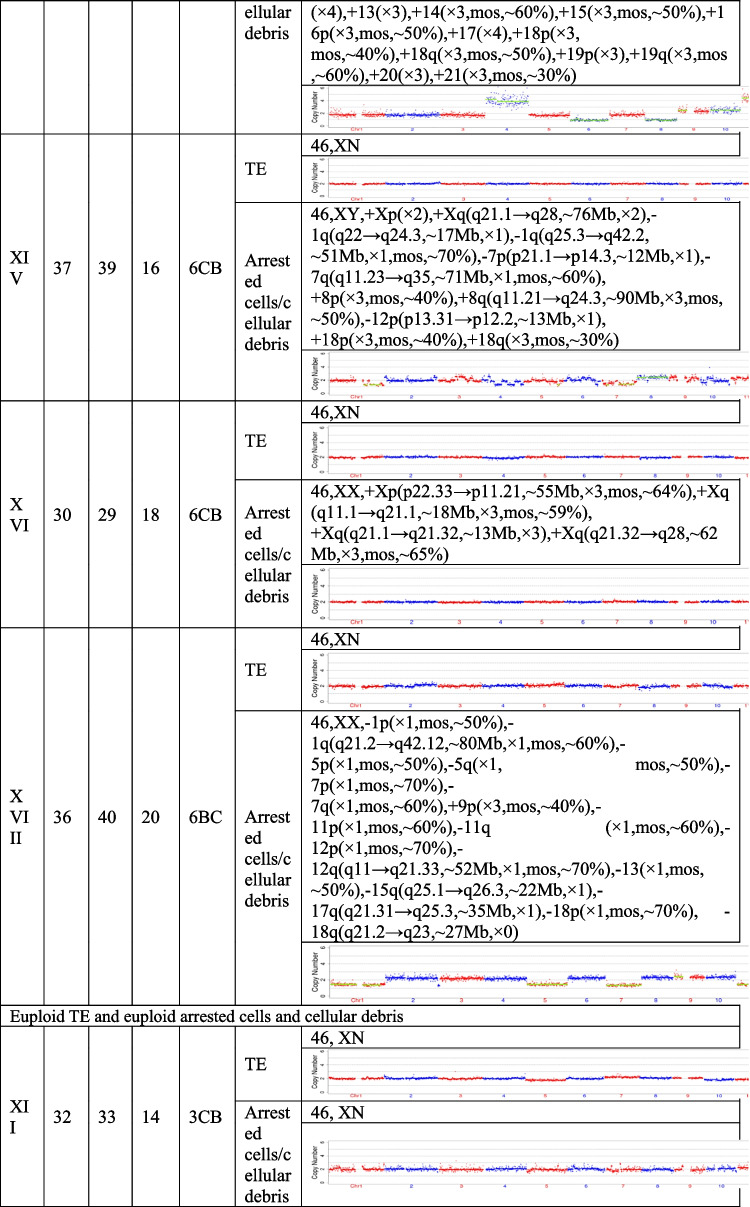
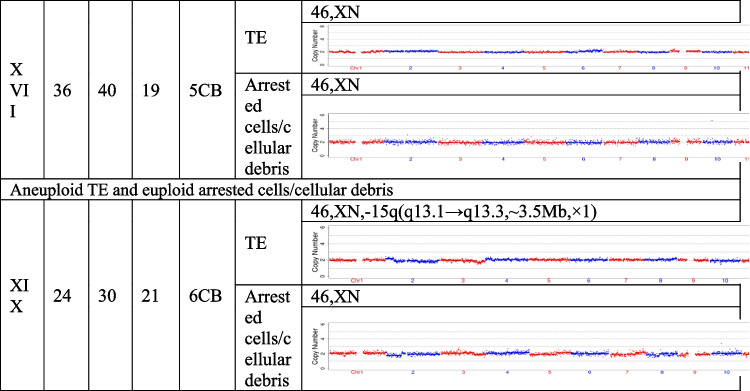
The present study recruited 19 patients (30.59 ± 5.38 years for average female age) who underwent PGT and analyzed a total of 21 pairs of TE biopsies and the arrested cells/cellular debris from their embryos. Couples were from three different groups, including preimplantation genetic testing for chromosomal structural rearrangements (PGT-SR), preimplantation genetic testing for monogenic/single gene defects (PGT-M), and preimplantation genetic testing for aneuploidy (PGT-A). Their ages, embryo morphological grading, and NGS results of their TE biopsies and the corresponding arrested cells/cellular debris are shown in Table 1. In the 21 pairs of TE biopsies and arrested cells/cellular debris, seven pairs were from six couples (27.33 ± 2.69 years for average female age) who accepted PGT-SR, three pairs were from three couples (29.33 ± 1.70 years for average female age) who accepted PGT-M, as well as 11 pairs were from ten couples (32.90 ± 5.89 years for average female age) who accepted PGT-A because of repeated implantation failures or recurrent pregnancy loss.
Table 1.
The clinical information and NGS results of the TE biopsies and the corresponding arrested cells/cellular debris
TE, trophectoderm; NGS, next-generation sequencing; XN = XY or XX.
The TE biopsy results revealed that 47.6% (10/21) of blastocysts were aneuploidies or mosaicism and 52.4% (11/21) of blastocysts were euploidies. In the ten aneuploid blastocysts, nine corresponding arrested cells/cellular debris were abnormal, including seven presenting additional chromosomal rearrangements or increased abnormal chromosomal fragments (No. 1, 2, 4, 5, 7, 13, and 17). Interestingly, the arrested cells/cellular debris of the other one presented euploidy (No. 21). In the 11 euploid blastocysts, nine corresponding arrested cells/cellular debris (81.8%) were aneuploid fragments (No. 6, 8, 9, 10, 12, 15, 16, 18, and 20), and only two pairs of TE biopsies and arrested cells/cellular debris (No. 14 and 19) were found to be both euploid. In total, 18 groups of arrested cells/cellular debris (85.7%) removed from the blastocysts were abnormal.
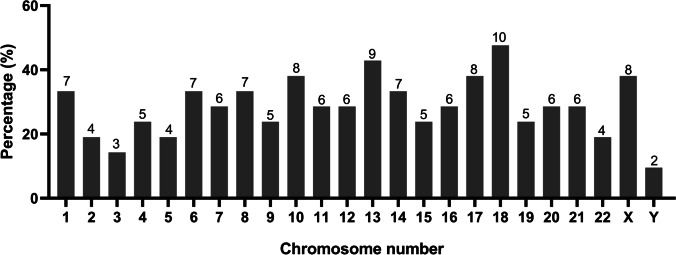
In addition, we calculated the abnormal chromosomal fragments in the 21 groups of arrested cells/cellular debris (the three arrested cells/cell debris in the No. 4 sample were considered as a whole). All the chromosomes were affected. And the abnormal proportion of chromosome 18 was 57.6% (10/21), that of chromosome 13 accounted for 42.9% (9/21), and 38.1% for chromosomes 10, 17 and X (8/21) (Fig. 2).
Fig. 2.
Percentage of affected chromosomes in the 21 groups of arrested cells/cellular debris
Discussion
The present study examined both TE cells and arrested cells/cellular debris expelled from the selected blastocysts. Also, 47.6% (10/21) of the detected blastocysts were aneuploidies or mosaicism. And 18 pairs out of the 21 blastocyst-arrested cells/cellular debris (85.7%) were expelled from the blastocysts with additional chromosomal rearrangements and increased abnormal chromosomal fragments. Among them, nine aneuploid blastocysts and nine euploid blastocysts expelled aneuploid arrested cells/cellular debris. The results confirmed that aneuploid and mosaic embryos were common phenomena during the early embryonic development, and supported the hypothesis that the elimination of chromosomally abnormal cells during early embryonic development might be a self-correction process, allowing to minimize the proportion of abnormal cells in subsequent embryos.
A majority of studies have reported that aneuploidies and mosaic embryos are prevalent during early embryonic development. Munne et al. found that 56% of embryos with the best morphology and development belonged to aneuploidies in patients younger than 35 by detecting more than 6000 embryos, and the proportion was even higher in patients 41 and older [6]. Mertzanidou et al. demonstrated that 71.4% (10/14) of good-quality embryos were mosaic [19]. Our studies examined 21 blastocysts by TE biopsies, and the proportion of aneuploidies and mosaicism was 47.6% (10/21), confirming the physiological phenomenon again.
PGT-A is applied in IVF/ICSI cycles to select euploid embryos, aiming to improve pregnancy outcomes. However, recent studies showed that PGT-A seemed to have no beneficial effects in good-prognosis patients [26, 27]. This is because mosaicism is a physiological feature of early human embryos and had the ability to develop a viable pregnancy [10–12]. Previous evidence also suggests that initial mosaicism or aneuploidy within the embryo may be self-limiting by demonstrating a decrease in the incidence of mosaicism or aneuploidy in vitro over time [13, 15]. However, the underlying mechanisms of normalization of aneuploid cells in human embryos throughout development, including abnormal cell elimination and embryonic self-correction, are confined to a theoretical basis.
Animal studies have suggested that self-correction process involves in cell arrest and apoptosis. Bolton et al. revealed that aneuploid cells in the fetal lineage and placental lineage had different correction ways by using a mouse model of embryo mosaicism caused by a spindle assembly checkpoint inhibitor during the four- to eight-cell division [8]. Aneuploid cells in the fetal lineage were eliminated by apoptosis; nevertheless, those in the placental lineage were eliminated by proliferative defects. Subsequently, their team further showed that aneuploid cells in the fetal lineage were dominantly eliminated in a p53-dependent process involving both autophagy and apoptosis before, during, and after implantation [28]. And mosaic mouse embryos increased cellular proliferation to compensate cell death during implantation stages [28]. Moreover, Daughtry et al. discovered that in rhesus monkey embryos, part of or all abnormal chromosomes as a form of fragments within micronuclei could be lost during cell division, whereas abnormal blastomeres with extensive DNA damage do not progress further into blastocysts [29]. Transcriptomic analyses also have identified that novel candidate genes related to apoptosis regulate early embryo survival and death [30].
In our present study, the comparison of NGS results between TE biopsies and the corresponding arrested cells/cellular debris showed that a total of 18 embryos expelled aneuploid arrested cells/cellular debris, including nine aneuploid blastocysts and nine euploid blastocysts. Especially, seven arrested cells/cellular debris from the aneuploid blastocysts presented additional chromosomal rearrangements or increased abnormal chromosomal fragments. The results directly exhibited strong evidence of the human embryonic self-correction ability. Because of the limited experiment samples, we did not conduct further experiments to investigate mechanism occurred in the process. But according to the animal studies, we can speculate that the arrested cells/cellular debris may be expelled from embryos by cell arrest and apoptosis. In addition, the phenomenon that two euploidies and one aneuploidy expelled euploid arrested cells/cellular debris indicates that the self-correction mechanism in the embryos can be overwhelming sometimes, resulting in expelling some euploid cells. Furthermore, due to the congruency between TE biopsies and arrested cells/cellular debris, these arrested cells/cellular debris are not a suitable source for PGT-A.
Orvieto et al. and Lagalla et al. had analyzed arrested cells/cell debris before [31, 32]. Orvieto et al. called these residues in the zona pellucida as cell debris/fragments and compared them with the corresponding embryos to prove that human embryos had the ability to self- correction. They also mentioned that because of the self-correction mechanism, the efficacy of noninvasive PGT-A (niPGT-A) which analyzed the cell-free DNA in the spent culture media need be suspected. We are in favor of the viewpoint as well. As to Lagalla et al., they analyzed the difference between these excluded cells and TE biopsies and providing the evidence for a potential mechanism of “aneuploidy rescue” of the embryo. However, Orvieto et al.’s research used day 3 discarded human embryos undergoing PGT-M which further cultured until days 5–6. And Lagalla et al. only analyzed excluded cells from irregularly cleaved embryos, not including excluded cells from normally cleaved embryos. Our study used PGT-embryo samples, including normal and abnormal blastocysts. The results may more represent and give evidence for the physiological self-correction phenomenon. Besides, they used array comparative genomic hybridization (aCGH), but our study used NGS technology. As we all know, NGS shows better application than aCGH, with superior sensitivity and higher precision [33]. Furthermore, Orvieto et al.’s study only included 11 pairs of blastocytes and arrested cells/cellular debris, and Lagalla et al. only included 12 pairs. Although it was still small, the sample size of our study was almost twice than theirs to further supplement data in the aspect.
Some inevitable limitations need to be acknowledged and discussed here. First, for ethical reasons, we did not separate ICM and TE for sequencing. Therefore, the source of arrested cells/cellular debris could not be confirmed. Second, it is an observational study, and no experiment data were provided to prove mechanisms because of the limited samples. In the future, studies with a large size are required. The arrested cells/cellular debris samples, ICM and TE biopsies can be separately collected for RNA sequencing to investigate potential self-correction mechanisms of human embryos.
Conclusion
Both mosaic and aneuploid embryos are common during early embryonic development. The residual arrested cells/cellular debris in the zona pellucida of blastocysts provide evidence of self-correction mechanism during early embryonic development.
Acknowledgements
The authors thank the staff in the Department of Reproductive Medicine, Xiangya Hospital, Central South University, without whom the study could not be carried out.
Author Contribution
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Xiaoxia Wang, Jing Zhao, Zhongyuan Yao, Qiuping Xia, Tianli Chang, Jun Zeng, Jiaqi Liu, Yanping Li, and Huimin Zhu. Xiaoxia Wang wrote the first draft of the manuscript. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the Key Project of Research and Development Plan in Hunan Province (2021SK2028).
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethical Approval
The study was approved by Institute Review Board of Department of Reproductive Medicine, Central South University.
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent for Publication
Patients signed informed consent regarding publishing their data and photographs.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author Contribution
Xiaoxia Wang, Jing Zhao and Zhongyuan Yao contributed equally to this work.
References
- 1.Taylor TH, Gitlin SA, Patrick JL, Crain JL, Wilson JM, Griffin DK. The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum Reprod Update. 2014;20:571–581. doi: 10.1093/humupd/dmu016. [DOI] [PubMed] [Google Scholar]
- 2.Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 3.Mantzouratou A, Delhanty JDA. Aneuploidy in the human cleavage stage embryo. Cytogenet Genome Res. 2011;133:141–148. doi: 10.1159/000323794. [DOI] [PubMed] [Google Scholar]
- 4.Fragouli E, Wells D, Delhanty JDA. Chromosome abnormalities in the human oocyte. Cytogenet Genome Res. 2011;133:107–118. doi: 10.1159/000323801. [DOI] [PubMed] [Google Scholar]
- 5.Bielanska M, Tan SL, Ao A. Chromosomal mosaicism throughout human preimplantation development in vitro: incidence, type, and relevance to embryo outcome. Hum Reprod. 2002;17:413–419. doi: 10.1093/humrep/17.2.413. [DOI] [PubMed] [Google Scholar]
- 6.Munne S, Chen S, Colls P, Garrisi J, Zheng X, Cekleniak N, et al. Maternal age, morphology, development and chromosome abnormalities in over 6000 cleavage-stage embryos. Reprod Biomed Online. 2007;14:628–634. doi: 10.1016/s1472-6483(10)61057-7. [DOI] [PubMed] [Google Scholar]
- 7.Fragouli E, Munne S, Wells D. The cytogenetic constitution of human blastocysts: insights from comprehensive chromosome screening strategies. Hum Reprod Update. 2019;25:15–33. doi: 10.1093/humupd/dmy036. [DOI] [PubMed] [Google Scholar]
- 8.Bolton H, Graham SJL, Van der Aa N, Kumar P, Theunis K, Fernandez Gallardo E, et al. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat Commun. 2016;7. 10.1038/ncomms11165 [DOI] [PMC free article] [PubMed]
- 9.Haouzi D, Boumela I, Chebli K, Hamamah S. Global, survival, and apoptotic transcriptome during mouse and human early embryonic development. Biomed Res Int. 2018;2018:1–16. doi: 10.1155/2018/5895628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greco E, Minasi MG, Fiorentino F. Healthy babies after intrauterine transfer of mosaic aneuploid blastocysts. N Engl J Med. 2015;373:2089–2090. doi: 10.1056/NEJMc1500421. [DOI] [PubMed] [Google Scholar]
- 11.Gleicher N, Vidali A, Braverman J, Kushnir VA, Barad DH, Hudson C, et al. Accuracy of preimplantation genetic screening (PGS) is compromised by degree of mosaicism of human embryos. Reprod Biol Endocrin. 2016;14. 10.1186/s12958-016-0193-6 [DOI] [PMC free article] [PubMed]
- 12.Kahraman S, Cetinkaya M, Yuksel B, Yesil M, Pirkevi CC. The birth of a baby with mosaicism resulting from a known mosaic embryo transfer: a case report. Hum Reprod. 2020;35:727–733. doi: 10.1093/humrep/dez309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbash-Hazan S, Frumkin T, Malcov M, Yaron Y, Cohen T, Azem F, et al. Preimplantation aneuploid embryos undergo self-correction in correlation with their developmental potential. Fertil Steril. 2009;92:890–896. doi: 10.1016/j.fertnstert.2008.07.1761. [DOI] [PubMed] [Google Scholar]
- 14.Li M, DeUgarte CM, Surrey M, Danzer H, DeCherney A, Hill DL. Fluorescence in situ hybridization reanalysis of day-6 human blastocysts diagnosed with aneuploidy on day 3. Fertil Steril. 2005;84:1395–1400. doi: 10.1016/j.fertnstert.2005.04.068. [DOI] [PubMed] [Google Scholar]
- 15.Santos MA, Teklenburg G, Macklon NS, Van Opstal D, Schuring-Blom GH, Krijtenburg P, et al. The fate of the mosaic embryo: chromosomal constitution and development of Day 4, 5 and 8 human embryos. Hum Reprod. 2010;25:1916–1926. doi: 10.1093/humrep/deq139. [DOI] [PubMed] [Google Scholar]
- 16.Popovic M, Dheedene A, Christodoulou C, Taelman J, Dhaenens L, Van Nieuwerburgh F, et al. Chromosomal mosaicism in human blastocysts: the ultimate challenge of preimplantation genetic testing? Hum Reprod. 2018;33:1342–1354. doi: 10.1093/humrep/dey106. [DOI] [PubMed] [Google Scholar]
- 17.Lavon N, Narwani K, Golan-Lev T, Buehler N, Hill D, Benvenisty N. Derivation of euploid human embryonic stem cells from aneuploid embryos. Stem Cells. 2008;26:1874–1882. doi: 10.1634/stemcells.2008-0156. [DOI] [PubMed] [Google Scholar]
- 18.Brill A, Torchinsky A, Carp H, Toder V. The role of apoptosis in normal and abnormal embryonic development. J Assist Reprod Gen. 1999;16:512–519. doi: 10.1023/A:1020541019347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mertzanidou A, Wilton L, Cheng J, Spits C, Vanneste E, Moreau Y, et al. Microarray analysis reveals abnormal chromosomal complements in over 70% of 14 normally developing human embryos. Hum Reprod. 2013;28:256–264. doi: 10.1093/humrep/des362. [DOI] [PubMed] [Google Scholar]
- 20.Embryology ASIR. The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod. 2011;26:1270–1283. doi: 10.1093/humrep/der037. [DOI] [PubMed] [Google Scholar]
- 21.Gardner DK, Schoolcraft WB. In vitro culture of human blastocyst. In: Jansen R, Mortimer D (eds.) Towards Reproductive Certainty: Infertility & Genetics Beyond. 1999; Parthenon Press, Carnforth, 377–388.
- 22.Jiao J, Shi B, Sagnelli M, Yang D, Yao Y, Li W, et al. Minimally invasive preimplantation genetic testing using blastocyst culture medium. Hum Reprod. 2019;34:1369–1379. doi: 10.1093/humrep/dez075. [DOI] [PubMed] [Google Scholar]
- 23.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Sci. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi Q, Lu S, Zhou X, Yao F, Hao N, Yin G, et al. Copy number variation sequencing-based prenatal diagnosis using cell-free fetal DNA in amniotic fluid. Prenat Diagn. 2016;36:576–583. doi: 10.1002/pd.4830. [DOI] [PubMed] [Google Scholar]
- 25.Yao Z, Wang X, Zeng J, Zhao J, Xia Q, Zhang L, et al. Chromosomal concordance between babies produced by the preimplantation genetic testing for aneuploidies and trophectoderm biopsies: A prospective observational study. Eur J Obstet Gynecol Reprod Biol. 2023;282:7–11.10.1016/j.ejogrb.2022.12.024. [DOI] [PubMed]
- 26.Yan J, Qin Y, Zhao H, Sun Y, Gong F, Li R, et al. Live birth with or without preimplantation genetic testing for aneuploidy. N Engl J Med. 2021;385:2047–2058. doi: 10.1056/NEJMoa2103613. [DOI] [PubMed] [Google Scholar]
- 27.Munné S, Kaplan B, Frattarelli JL, Child T, Nakhuda G, Shamma FN, et al. Preimplantation genetic testing for aneuploidy versus morphology as selection criteria for single frozen-thawed embryo transfer in good-prognosis patients: a multicenter randomized clinical trial. Fertil Steril. 2019;112:1071–1079. doi: 10.1016/j.fertnstert.2019.07.1346. [DOI] [PubMed] [Google Scholar]
- 28.Singla S, Iwamoto-Stohl LK, Zhu M, Zernicka-Goetz M. Autophagy-mediated apoptosis eliminates aneuploid cells in a mouse model of chromosome mosaicism. Nat Commun. 2020;11. 10.1038/s41467-020-16796-3 [DOI] [PMC free article] [PubMed]
- 29.Daughtry BL, Rosenkrantz JL, Lazar NH, Fei SS, Redmayne N, Torkenczy KA, et al. Single-cell sequencing of primate preimplantation embryos reveals chromosome elimination via cellular fragmentation and blastomere exclusion. Genome Res. 2019;29:367–382. doi: 10.1101/gr.239830.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boumela I, Assou S, Aouacheria A, Haouzi D, Dechaud H, De Vos J, et al. Involvement of BCL2 family members in the regulation of human oocyte and early embryo survival and death: gene expression and beyond. Reprod. 2011;141:549–561. doi: 10.1530/REP-10-0504. [DOI] [PubMed] [Google Scholar]
- 31.Orvieto R, Shimon C, Rienstein S, Jonish-Grossman A, Shani H, Aizer A. Do human embryos have the ability of self-correction? Reprod Biol Endocrin. 2020;18. 10.1186/s12958-020-00650-8 [DOI] [PMC free article] [PubMed]
- 32.Lagalla C, Tarozzi N, Sciajno R, Wells D, Di Santo M, Nadalini M, et al. Embryos with morphokinetic abnormalities may develop into euploid blastocysts. Reprod Biomed Online. 2017;34:137–146. doi: 10.1016/j.rbmo.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 33.Maxwell SM, Colls P, Hodes-Wertz B, McCulloh DH, McCaffrey C, Wells D, et al. Why do euploid embryos miscarry? A case-control study comparing the rate of aneuploidy within presumed euploid embryos that resulted in miscarriage or live birth using next-generation sequencing. Fertil Steril. 2016;106:1414–1419. doi: 10.1016/j.fertnstert.2016.08.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.