Abstract
BACKGROUND:
There is an unmet need for fully automated image prescription of the liver to enable efficient, reproducible MRI.
PURPOSE:
To develop and evaluate AI-based liver image prescription.
STUDY TYPE:
prospective.
POPULATION:
570 female/469 male patients(age:56±17yrs) with 72%/8%/20% assigned randomly for training/validation/testing; 2 female/4male healthy volunteers(age:31±6yrs).
FIELD STRENGTH/SEQUENCE:
1.5T, 3.0T; spin echo, gradient echo, bSSFP.
ASSESSMENT:
A total of 1,039 three-plane localizer acquisitions (26,929 slices) from consecutive clinical liver MRI examinations were retrieved retrospectively and annotated by six radiologists. The localizer images and manual annotations were used to train an object-detection convolutional neural network (YOLOv3) to detect multiple object classes (liver, torso, arms) across localizer image orientations and to output corresponding 2D bounding boxes. Whole-liver image prescription in standard orientations was obtained based on these bounding boxes. 2D detection performance was evaluated on test datasets by calculating intersection over union (IoU) between manual and automated labeling. 3D prescription accuracy was calculated by measuring the boundary mismatch in each dimension and percentage of manual volume covered by AI prescription. The automated prescription was implemented on a 3T MR system and evaluated prospectively on healthy volunteers.
STATISTICAL TESTS:
Paired t-tests (threshold=0.05) were conducted to evaluate significance of performance difference between trained networks.
RESULTS:
In 208 testing datasets, the proposed method with full network had excellent agreement with manual annotations, with median IoU>0.91 (interquartile range<0.09) across all seven classes. The automated 3D prescription was accurate, with shifts<2.3cm in superior/inferior dimension for 3D axial prescription for 99.5% of test datasets, comparable to radiologists’ inter-reader reproducibility. The full network had significantly superior performance than the tiny network for 3D axial prescription in patients. Automated prescription performed well across SSFSE, GRE, and bSSFP sequences in the prospective study.
DATA CONCLUSION:
AI-based automated liver image prescription demonstrated promising performance across the patients, pathologies, and field strengths studied.
Keywords: liver, automated scan prescription, AI, deep learning, image prescription
INTRODUCTION
In abdominal MRI, patient experience and cost correlate closely with exam time(1). Recent technical advances have the potential to enable MRI of the abdomen with enhanced efficiency. For example, abbreviated MRI protocols have demonstrated excellent performance for the detection of abdominal lesions as well as diffuse liver disease(1, 2). Further, motion-robust free-breathing quantitative MRI methods are being developed to enable quantification of various imaging biomarkers without the need for breathholds(3–5). These developments may enable fully automated, single button-push focused exams of the liver with multiple advantages in terms of patient throughput, imaging volume, staff training and time, accessibility, and clinical workflow. However, single button-push exams require fully automated prescription of the liver. For these reasons, the current manual image prescription remains a bottleneck for rapid, high-value MRI of the abdomen. Automated prescription may reduce non-value-added preparatory time and improve the reproducibility of image prescription within and across sites(6, 7).
Previous efforts towards automated scan prescription in the liver have relied on traditional image processing tools, such as active shape models, to detect the upper and lower edges of the liver(8, 9). In Goto et al, the upper edge of the liver was detected automatically using an active shape model, and the lower edge was detected using a probabilistic estimate based on automatically-derived statistics of the signal within the liver(8). Although promising, this previous method requires a specialized volumetric, breath-held localizer scan, is limited to prescription of axial series, has been tested in limited numbers of healthy volunteers (n = 24) and simulated deformed livers (n = 7).
Artificial intelligence (AI) methods have enabled automated image prescription in the brain, spine, heart, and kidney, with the potential to improve workflow, efficiency, and prescription accuracy and consistency(6, 10–16). AI has been used to address the persistent challenge of accurately localizing intervertebral discs (IVD) during MR examinations of the lumbar spine in a consistent and time-efficient manner(12). Furthermore, a cardiac MRI prescription method has recently been implemented, which substantially reduces examination time by automating real-time localization(11, 14). However, automated prescription of the liver remains an important unmet need.
Therefore, the purpose of this study was to develop and implement an AI-based fully automated prescription method for liver MRI.
MATERIALS AND METHODS
The overall workflow of the development and validation of the proposed automated prescription method is shown in Figure 1. In an IRB-approved study, the retrospective study was conducted with a waiver of informed consent and the prospective study was performed with informed consent with IRB approval.
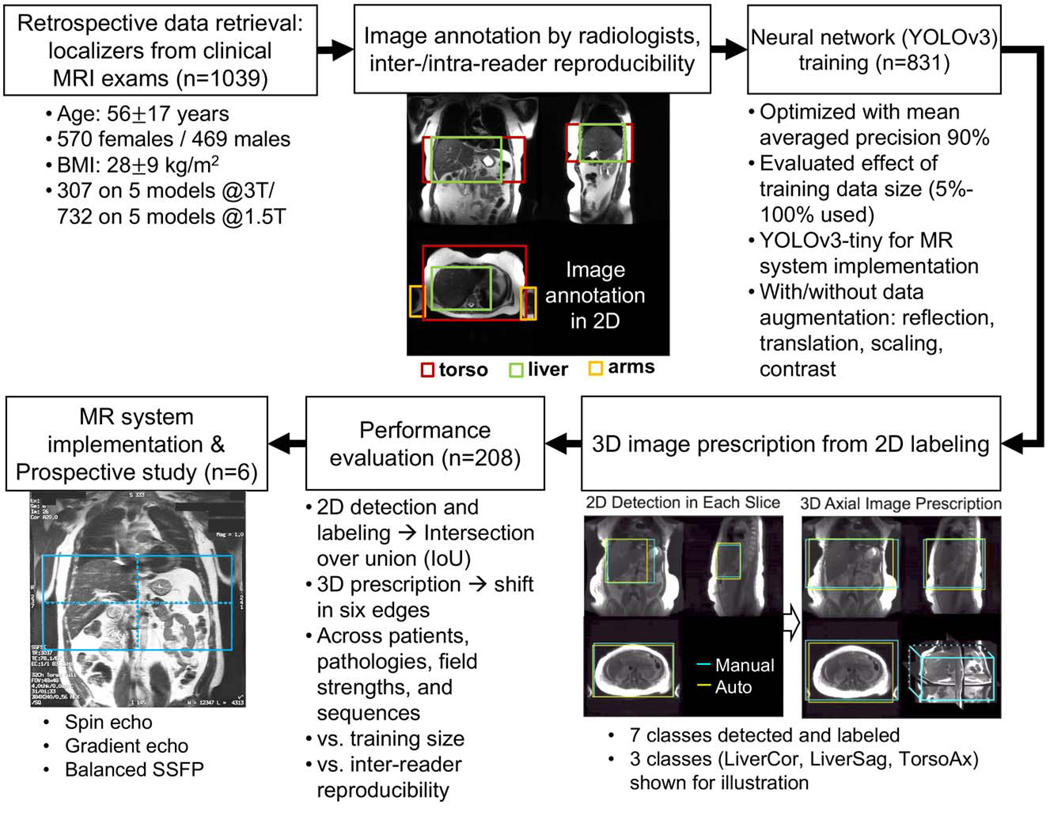
Figure 1.
Flow-chart summary for data retrieval, annotation, training, prescription, evaluation, and scanner implementation. Manual labeling consisted of 7 localization regions (LiverAx, TorsoAx, ArmsAx, LiverCor, TorsoCor, LiverSag, TorsoSag) that was evaluated by assessment of inter-reader reproducibility. A convolutional neural network (CNN) for object detection was trained with 80% of the datasets. The minimum 3D box needed to cover the appropriate labeled 2D boxes was used to obtain the final 3D automated prescription. Evaluation of 2D and 3D boxes was performed on the remaining 20% datasets across patients, pathologies, and acquisition settings. We successfully implemented the method on a clinical MR system and conducted a prospective study with 6 healthy volunteers across acquisition sequences. LiverAx/TorsoAx/ArmsAx: liver/torso/arms in the axial view; LiverCor/TorsoCor: liver/torso in the coronal view; LiverSag/TorsoSag: liver/torso in the sagittal view.
Data and image processing:
In this IRB-approved study, consecutive clinical abdominal MRI exams acquired between 9/23/2008 and 12/17/2019 at the University of Wisconsin Hospitals and Clinics were retrieved retrospectively, with a waiver of informed consent. The clinical MRI examinations were performed to confirm, exclude, or follow-up a wide variety of abdominal disorders. Patients with their BMI recorded within 30 days of one of their scans were included. Only one exam was included for any one patient in this study; the first exam of the patient was included in case of multiple eligible exams. The included exams were chosen randomly for manual annotation and model training and testing. The scanner models used for the exams were Discovery MR750, Discovery MR750w, SIGNA Architect, SIGNA PET/MR and SIGNA Premier at 3T, and Optima MR450w, SIGNA, SIGNA Artist, SIGNA HDx and SIGNA HDxt at 1.5T. The scanners used in this study were all manufactured by GE Healthcare (Waukesha, WI). At the institution of this study, all abdominal MRI examinations start with a three-plane localizer series based on spin-echo or gradient-echo acquisitions. From each exam, the (first) localizer series was retrieved for annotation, curation, and analysis. This series includes multiple abdominal images (21–30 total) in the axial, coronal, and sagittal orientations to allow for prescription of subsequent MR sequences. For manual labeling and automated inference, each individual localizer image slice was read in MATLAB (2021a, Mathworks, Natick, MA). These acquired localizer images typically have spatially heterogeneous intensity due to the receive coil sensitivity profiles, where the subcutaneous fat and other areas near the receive coils are much brighter than the liver. To increase contrast around the liver, the intensity values of each image were normalized to 0 to 1 and values between 0 and 0.2 were mapped to values between 0 and 1 (with values larger than 0.2 saturated to 1). Each resulting normalized image was written as a .png file as the input format to the labeling software and neural network(17). A 80–20 split was used for AI model training and testing in accordance with common practice and to provide sufficient data for training while reserving a meaningful subset for testing. Out of the training set, the ratio between data used for parameter training and for validation of training performance was 9:1. Thus, the data partitioning across training/validation/testing was 72%−8%−20% of the total dataset.
Manual image annotation and intra- and inter-reader reproducibility:
The Radiological Society of North America (RSNA) Medical Imaging Resource Community (MIRC) Clinical Trials Processor (CTP) was used to perform image de-identification. Performance of manual image annotation of all three-plane localizer datasets was distributed among six board-certified abdominal radiologists (J.S., N.P., T.H.O., E.M.L., M.I., and S.B.R.) on the Python platform LabelImg(18, 19). Seven classes of labels were annotated using bounding boxes that span the liver, torso, and arms in specific image orientations (axial: liver, torso, and arms; coronal: liver and torso; sagittal: liver and torso; Figure 1) to enable liver image prescription in any orthogonal orientation. The torso and the arms are needed for coronal prescription to avoid phase wrap in the R/L dimension, while the torso is needed for sagittal prescription in the A/P and S/I dimensions. Arms and torso were annotated at the level of the liver in the superior/inferior dimension. In addition, 50 exams were randomly selected for intra- and inter-reader reproducibility studies. One radiologist (J.S.) labeled the same dataset twice after a month-long delay to assess intra-reader reproducibility. Two radiologists (J.S. and N.P.) labeled the same datasets to assess inter-reader reproducibility.
Training:
A deep convolutional neural network (CNN) based on a You Only Look Once (YOLOv3) architecture was trained for detection, localization, and classification of the aforementioned bounding boxes(20, 21). The network input includes a single image from the three-plane localizer acquisition. The YOLOv3 feature extractor, called Darknet-53 includes 106 layers, and contains skip connections and 3 prediction heads, each processing the image at a different down-sampling rate of 32, 16 and 8, respectively. Detection can thus be achieved at three different scales. The network detects each of the 7 classes of labeled objects described above with a confidence level (0–100%), and outputs the coordinates of the corresponding bounding boxes. Training and validation involved 72% and 8% of all patient datasets respectively (randomly chosen). Different training dataset sizes (5%−100% of all training datasets) were used to evaluate the effect of training size on network performance using 5-fold cross validation. Reflection (horizontally and/or vertically) was used to augment the training set by four-fold.
Additionally, a shallower network based on a lightweight variant of YOLOv3, YOLOv3-tiny, was trained to enable real-time scanner implementation(22). The tiny network includes only 15 layers, and is able to predict bounding boxes 3x faster when implemented on a scanner with CPUs and no GPUs. This shallower network was trained using data augmentation methods: reflections, translation, scaling, and contrast modifications.
Termination of training for both kinds of networks relied on mean average precision (mAP), a common metric to evaluate performance of object localization in multi-class datasets(23).
3D image prescription:
The trained network predicts each bounding box from each test image with a confidence level (0–100%) on its classification label. For each image, the bounding box with the highest confidence level was extracted for each of the liver- and torso-labeled boxes while the top 2 arms-labeled boxes with the highest confidence level were extracted for the arms. Based on the extracted bounding boxes for each image (from either the manual or automated annotations), image prescription for whole-liver acquisition in each orientation was calculated as the minimum 3D bounding box needed to cover all the labeled 2D bounding boxes in the required volume (Supplementary Video S1). Different illustrative examples of 3D prescription along different orientations were considered in this work: 3D axial prescription covers the torso in the anterior/posterior (A/P) and right/left (R/L) dimensions and the liver in superior/inferior (S/I); 3D coronal prescription covers the torso and the arms in the R/L dimension and at the level of the liver in S/I and the liver in A/P; 3D sagittal prescription covers the torso in the A/P and S/I dimensions and the liver in R/L. 3D prescriptions from manual annotation were calculated in a similar fashion.
Evaluation:
The performance of automated 2D object detection for each of the seven classes of labels was measured by mis-classification rate and intersection over union (IoU, the area of the intersection between the manual and automated bounding boxes divided by the area of their union), as intermediate evaluation metrics. To evaluate the performance of the subsequent automated 3D prescription, the mismatch with the manual prescription for each of the six edges of the 3D prescribed bounding box (right, left, anterior, posterior, superior, and inferior) was calculated on the remaining 20% of all patients (randomly chosen). The localizer images, where the S/I shift between manual labeling and AI predictions in 3D axial prescription was larger than 2 cm, were analyzed by a radiologist (J.S.) for subjective evaluation of the causes of errors. Furthermore, the percentage of the 3D volume from manual labeling that was covered by the AI-based prescription was reported as the overlap between AI and manual labeling. The performance of the proposed method was evaluated across patients, pathologies, field strengths and sequences with varying training sizes, full and tiny architectures, with and without data augmentation schemes, and against inter-reader reproducibility results. In this work, the ability to detect the liver’s S/I edges was given particular focus because the S/I edges are expected to be essential to detect for automated prescription of subsequent axial series (which are common in liver imaging).
Online implementation and prospective study:
The automated prescription was implemented for axial prescription on a clinical 3T MR system (MR750, GE Healthcare, Waukesha, WI), and evaluated prospectively on healthy volunteers using different localizer pulse sequences (single-shot fast spin-echo, SSFSE; gradient-echo, GRE; balanced steady-state free-precession, bSSFP) and acquisition parameters. To run the neural networks for prediction during the scan, the trained network weights were converted into TensorFlow, and Python scripts for image pre-processing were integrated into a GE Healthcare investigational automated workflow testing platform(24). Using this platform, after selecting the localizer and the organ to prescribe to, with a single button push DICOM images were passed to the platform and processed (image intensity normalized from the highest intensity pixel and scaled to 0–255), the coordinates of the prescribed field of view were generated and returned, and the scan prescription was updated. In this implementation, the in-plane field-of-view size remained fixed as determined by the user, while the center of FOV and the prescribed volume and the number of axial slices were determined automatically. Once the automated prescriptions were determined, operators (R.G. and C.J.B.) at the console assessed whether the automated prescriptions needed modification in order to proceed to the subsequent scans. This assessment by the operators was recorded for each exam.
Statistical analysis
In the testing datasets from the retrospective study in patients, the performance of the full network trained without any augmentation in various sub-cohort across age, sex, BMI, pathology, acquisition field strength and localizer pulse sequence was measured by the overlap between AI and manual labeling for 3D liver detection and axial prescription. In the same dataset, the performance of the networks with various training settings (full vs. tiny, with/without augmentation) was measured by the mis-classification rate and IoU in 2D detection from individual slices, and the overlap between manual labeling and AI prediction in 3D liver detection and axial, coronal, and sagittal prescriptions. Paired t-tests were conducted to evaluate the statistical significance of performance difference between the trained networks with a threshold of 0.05. Further, the median and inter-quartile range (IQR) of the same performance metrics for the trained networks was compared against the inter- and intra-reader reproducibility of the two radiologists.
In the prospective study in healthy volunteers, the performance of both the full and tiny networks trained without any augmentation was measured by the overlap between manual labeling and AI prediction in 3D liver detection and axial prescriptions across scans acquired with SSFSE, GRE, and bSSFP sequences with various parameters.
RESULTS
Demo code and the YOLOv3 weights obtained after training for this work are shared in the following Zenodo repository: https://doi.org/10.5281/zenodo.7391054.
Overall, 7820 GA MRI examinations were identified, performed in 5351 patients (3022 women and 2329 men). Of the 5351 patients identified, 2285 had their BMI recorded within 30 days of one of their scans. Out of the 2285 exams with patient BMI recorded within 30 days of the scan, 1039 were used for manual annotation and model training and testing. A total of 1,039 exams from 1,039 patients (age 56±17 years, range 0–101 years, 570 females/469 males, BMI 28±9 kg/m2) were collected. Table 1 summarizes the distribution of the patients included in terms of age, sex, BMI, pathology, acquisition field strength and sequence. The localizer acquisitions were based on SSFSE (1,015 exams, 98%) and GRE (24 exams, 2%). Training and validation involved 21,274 localizer slices from 748 patients and 1,850 slices from 83 patients (randomly chosen from all patient datasets). With full augmentation, 2,537,390/268,320 images were used for training/validation of the tiny network. The performance of the proposed method was evaluated on 5,655 localizer slices from the remaining 208 patients. Figure 1 summarizes the data used in this study.
Table 1.
Distribution of patient datasets and testing results across age, sex, BMI, pathology, acquisition field strength and localizer pulse sequence. Overlap between AI and manual labeling for 3D liver detection and axial prescription was high (>91%) across all categories. 3D axial prescription had higher levels of overlap between AI and manual methods than 3D liver detection across all categories. While AI-based liver detection faced challenges in iron overload (signal dropouts), resection and pleural effusion (unusual anatomy), 3D axial prescription maintained high overlap (>95%) between AI and manual labeling across all pathologies. Overlap: percentage of 3D volume from manual labeling covered by AI prescription.
| Sub-cohorts | Number of patients | 3D Liver Detection overlap between AI and manual (%) | 3D Axial Prescription overlap between AI and manual (%) | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Overall | Training | Testing | Median | IQR | Median | IQR | ||
| Age | <21 | 29 | 18 | 11 | 96.95 | 5.29 | 99.53 | 1.50 |
| 21–45 | 245 | 197 | 48 | 95.17 | 6.97 | 97.55 | 4.85 | |
| >45 | 765 | 616 | 149 | 97.04 | 8.53 | 98.45 | 3.14 | |
|
| ||||||||
| Sex | Male | 469 | 373 | 96 | 97.29 | 8.89 | 99.31 | 2.68 |
| Female | 570 | 458 | 112 | 95.95 | 7.15 | 97.68 | 3.58 | |
|
| ||||||||
| BMI | <18.5 | 49 | 36 | 13 | 94.82 | 4.88 | 98.66 | 4.80 |
| 18.5–24.9 | 364 | 295 | 69 | 95.36 | 13.69 | 97.63 | 4.12 | |
| 25.0–29.9 | 319 | 263 | 56 | 97.68 | 6.82 | 99.17 | 2.47 | |
| ≥ 30 | 307 | 237 | 70 | 97.04 | 6.43 | 98.28 | 3.17 | |
|
| ||||||||
| Pathology* | Cirrhosis | 131 | 105 | 26 | 97.60 | 4.79 | 99.72 | 2.37 |
| Iron overload | 71 | 56 | 15 | 93.84 | 7.67 | 100.0 | 2.41 | |
| Metastasis | 353 | 297 | 56 | 96.25 | 9.86 | 98.61 | 3.15 | |
| Steatosis | 312 | 253 | 59 | 96.67 | 6.64 | 98.26 | 4.07 | |
| Ascites | 309 | 246 | 63 | 96.28 | 7.96 | 98.70 | 3.18 | |
| Resection | 39 | 28 | 11 | 91.90 | 6.98 | 97.96 | 3.28 | |
| Pleural effusion | 159 | 125 | 34 | 95.33 | 13.76 | 98.46 | 5.03 | |
|
| ||||||||
| Field strength | 1.5T | 732 | 589 | 143 | 96.28 | 8.37 | 98.38 | 3.82 |
| 3T | 307 | 242 | 65 | 96.50 | 6.60 | 98.30 | 2.67 | |
|
| ||||||||
| Sequence | SSFSE** | 1015 | 809 | 206 | 96.40 | 8.04 | 98.35 | 3.20 |
| GRE | 24 | 22 | 2 | 92.20 | 15.61 | 95.44 | 9.13 | |
|
| ||||||||
| Overall | 1039 | 831 | 208 | 97.62 | 6.51 | 98.48 | 3.00 | |
Pathology information was automatically extracted from medical reports using keyword matching. Actual case counts may be different from reported here.
SSFSE: single-shot fast spin-echo; GRE: gradient-echo
Training of the full network with all training data required ~40 hours (NVIDIA Tesla V100), to reach an mAP of 90% at an IoU threshold of 0.5. With augmentation using reflection, training took ~90 hours. Training of the tiny network with all training data required ~6 hours on the same GPU (~20 hours with reflection, ~150 hours with the full augmentation scheme). For testing, object detection for one entire three-plane localizer (~30 images) on a GPU required ~0.3/0.1 seconds for the full/tiny network.
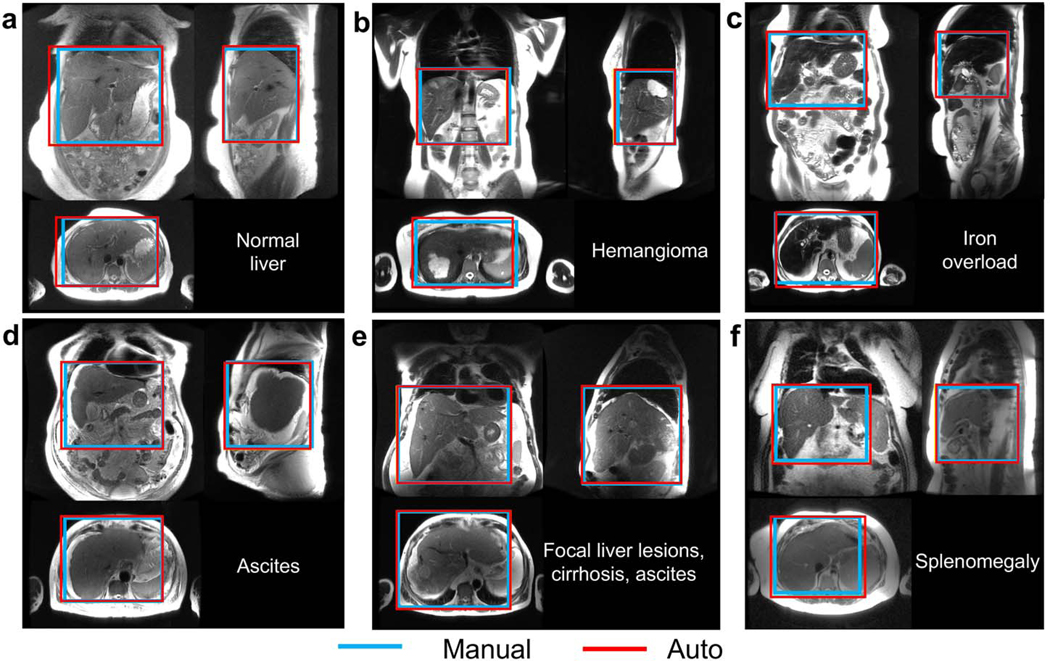
Figure 2 shows representative examples of 3D detection of the liver in a patient with a normal liver, and in five patients with various representative liver pathologies including focal lesions, iron overload, ascites, and cirrhosis.
Figure 2.
3D detection of the liver in all three localizer orientations, with examples of common pathologies. a) In most cases, the liver volume was covered accurately by automated prescription. The automated prescription aligned well with the manual annotation in patients with focal lesions (b), iron overload (c), ascites (d), and/or cirrhosis (e), as well as in a patient with splenomegaly (f), where the spleen abuts the liver and its signal level is similar to that of liver, but the proposed method was still able to identify the liver correctly.
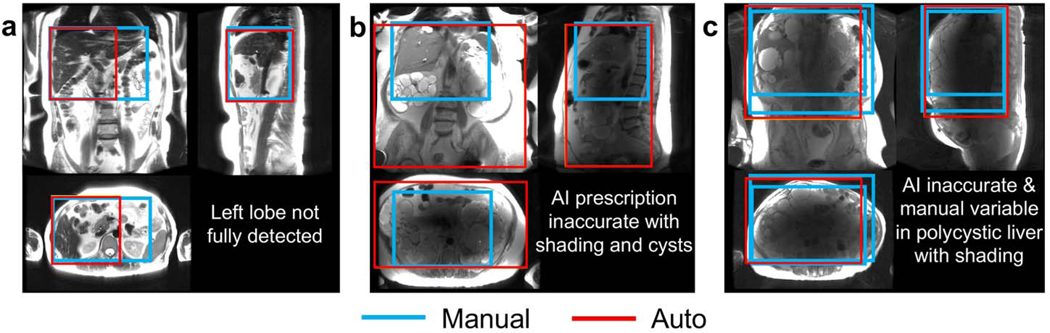
The automated liver detection failed to detect the entirety of the liver with tight margins (2 cm S/I) when localizer slices missed a large portion of the liver and/or provided insufficient signal due to dielectric shading (Figure 3). When the axial localizer images did not cover the liver sufficiently (a), missed prescription of the left lobe was observed due to failed R/L range cross-checking across views. However, 3D axial prescription of this case was entirely adequate (overlap with manual annotation >90%) owing to correct torso detection. Inaccurate automated object detection (b, c) was observed for patients with multiple cysts (b,c), where dielectric shading(25) led to signal dropouts in a large portion of the localizer slices. Inconsistencies in manual annotation between two radiologists and even between repeated annotations by the same radiologist appeared in the more severe case (c).
Figure 3.
Examples of algorithm failures for 3D liver detection. a) Missed coverage in the tip of the lateral segment of the left lobe due to insufficient axial localizer slices. Inaccurate automated object detection was observed for patients with multiple renal cysts (b) or liver cysts (c), or with dielectric shading (b,c) with signal dropouts in the central portion of multiple localizer images. Variation in manual annotation between two radiologists and even between repeated efforts by the same radiologist was observed in severe cases such as (c).
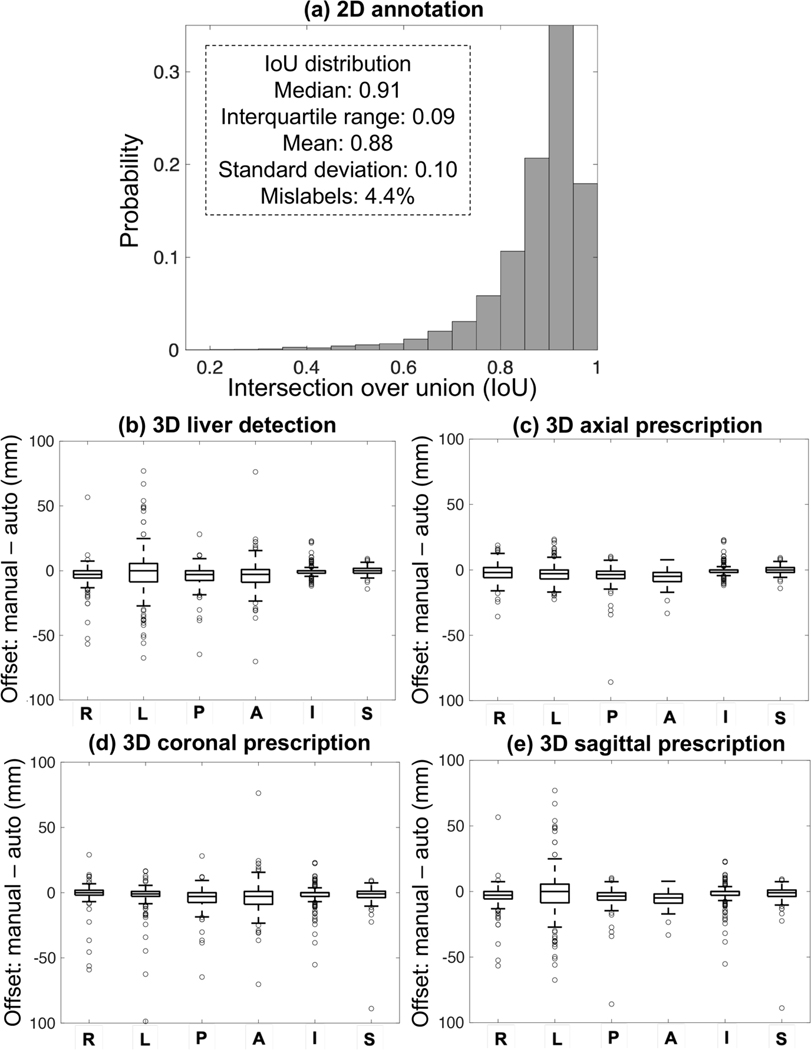
The accuracy of automated 2D and 3D detection and prescription as measured by the mismatch in each dimension is shown in Figure 4. Each of the seven classes had a median IoU>0.91 with interquartile range (IQR) <0.09. The performance of the full YOLOv3 network (without any augmentation) in 3D liver detection and axial, coronal, and sagittal prescription was comparable to that of inter-reader reproducibility. For 3D liver detection, the percentage of manual volume included in the automated prescription had median = 97.6% and IQR = 6.5% while the percentage of manual volume from one radiologist included in the other had median = 97.0% and IQR = 5.0%. The overlap for 3D axial prescription by the full network had median = 98.5% and IQR = 3.0% while the inter-reader reproducibility performance had median = 97.1% and IQR = 1.9%. The maximum S/I shift for 3D axial prescription by the full network was 2.3 cm for 99.5% of the test datasets, while that for the inter-reader reproducibility was 2.4 cm. The overlap for 3D coronal prescription by the full network had median = 98.3% and IQR = 3.8%, while the inter-reader reproducibility performance had median = 97.7% and IQR = 2.2%. The overlap for 3D sagittal prescription by the full network had median =97.9% and IQR = 5.5%, while the inter-reader reproducibility performance had median = 97.3% and IQR = 3.6%. In addition, for the full network the overlap for 3D axial prescription was significantly higher than the overlap for 3D liver detection.
Figure 4.
Accuracy of 2D annotation across all label classes (a), 3D liver detection (b), and 3D image prescription (c-e). IoU histograms for all classes are qualitatively similar, with IoU median >0.91 and interquartile range <0.09. In (b-e), x axis shows the 6 edges: right (R), left (L), posterior (P), anterior (A), inferior (I), and superior (S); y axis shows the difference between automated and manual volumes (0: perfect alignment; negative offset: AI covering more volume; positive offset: missed volume). All boxes are tight around 0. For 3D axial prescription, the shift in the S/I dimension was less than 2.3 cm for 99.5% of the test datasets.
For 2D detection with the full network, IoU histograms for different classes as well as the combined histogram followed similar trends (Supplementary Figure S2). IoU median, IQR, mean, and standard deviation were similar across classes. Liver detection had a lower mis-classification rate than torso detection in all orientations, while 2D detection of the arms in the axial orientation had the highest mis-classification rate. Liver detection in the coronal and sagittal orientations had a mis-classification rate of ~1%.
The performance of the AI-based liver prescription for 3D liver detection and 3D axial prescription across sub-cohorts in age, sex, BMI, pathology, and acquisition field strength and sequence is shown in Table 1. For both 3D liver detection and 3D axial prescription, the overlap between AI and manual labeling was observed across all sub-cohorts was larger than 91%. 3D axial prescription had higher levels of overlap between AI and manual methods than 3D liver detection across all categories. The accuracy of AI-based liver detection was lower in the presence of iron overload (signal dropouts), partial liver resection, anatomical variation, or perihepatic pathologies such as pleural effusion or ascites. 3D axial prescription had an overlap of larger than 95% between AI and manual labeling across all pathologies.
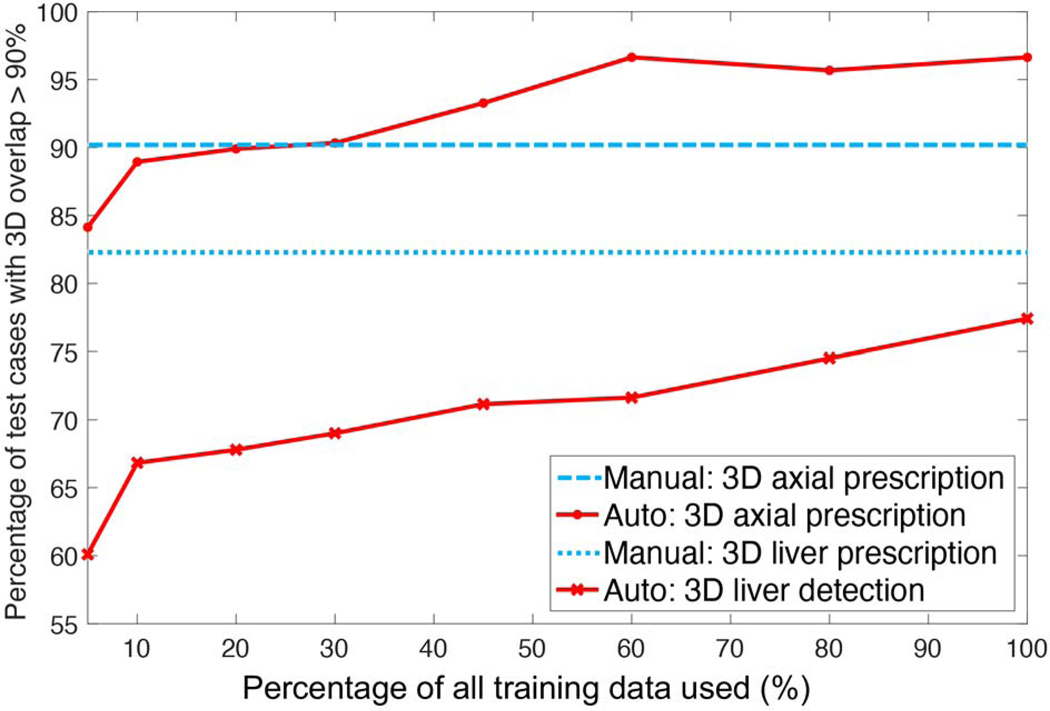
As training size increased, the percentage of test cases with an overlap larger than 90% in 3D between AI and manual prescription increased for 3D liver detection and axial prescription (Figure 5). AI performance for 3D axial prescription plateaued after training with 500 datasets. AI performance for 3D liver detection approached but never reached inter-radiologist reproducibility. Training with ≥ 250 datasets, AI-based 3D axial prescription performed better than (manual) inter-reader reproducibility.
Figure 5.
As training size increased, the percentage of test cases with high overlap (>90%) in 3D between AI and manual prescription increased for 3D liver detection and axial prescription. AI performance for 3D axial prescription plateaued after training with 500 patients’ datasets (60% of training data). AI performance for 3D liver detection approached but never reached radiologists’ inter-reader reproducibility performance. Training with at least 250 datasets (30% of training data), AI-based 3D axial prescription performed better than (manual) inter-reader reproducibility.
Table 2 shows the testing results with various training settings including the tiny and full networks. Overall, the tiny network performance was significantly inferior to the full network for 3D axial prescription. Without any augmentation, the tiny network had an overlap of median = 96.7% and IQR = 6.2%, 95.3%±6.3%, 96.3%±6.1%, 96.5%±5.8% in 3D liver detection, axial, coronal, and sagittal prescriptions, respectively. The maximum S/I shift for 3D axial prescription by the tiny network for 99.5% of the test datasets was 2.3 cm (the same as for the full network). Although augmentation methods reduced the mis-classification rate in individual slices, these augmentation methods did not significantly improve the performance of 3D prescription for either the full (p = 0.23 for 3D axial prescription) or the tiny network (p = 0.26 for 3D axial prescription). Finally, the full network’s mis-classification rate in 2D detection was low and comparable to that of the intra- and inter-reader reproducibility studies (Supplementary Figures S3 and S4).
Table 2.
Testing results with various training settings and inter- and intra-reader reproducibility. The full YOLOv3 network performed better than the tiny network. Although augmentation methods such as reflection and translation/scaling/contrast reduced the mis-classification rate in individual slices, these augmentation methods did not improve the performance of 3D image prescription for either the full or the tiny network. Compared to the performance of the readers (radiologists), the full network’s mis-classification rate in 2D detection was low and on par with the disagreement rate from the intra- and inter-reader reproducibility studies. The full network’s performance in 3D liver detection and axial, coronal, and sagittal prescription was comparable to that of inter-reader reproducibility. Overlap: percentage of 3D volume from manual labeling covered by AI prescription.
| Full network | Tiny network | Reproducibility of manual annotation | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| − | + reflection | − | + reflection | + all augmentation | Inter-reader | Intra-reader | ||
| 2D Annotation IoU between AI and manual (%) | Mis-classify | 4.39 | 3.86 | 9.45 | 9.92 | 6.73 | 6.72 | 4.43 |
| Median | 91.26 | 92.21 | 88.21 | 88.01 | 89.67 | 95.87 | 96.64 | |
| IQR | 8.90 | 8.37 | 10.08 | 10.72 | 10.66 | 4.69 | 5.03 | |
|
| ||||||||
| 3D Liver Detection overlap between AI and manual (%) | Median | 97.62 | 97.66 | 96.65 | 97.17 | 95.17 | 97.02 | 98.91 |
| IQR | 6.51 | 6.52 | 6.16 | 6.49 | 7.49 | 4.96 | 2.46 | |
|
| ||||||||
| 3D Axial Prescription overlap between AI and manual (%) | Median | 98.48 | 98.41 | 95.26 | 95.62 | 96.51 | 97.09 | 99.05 |
| IQR | 3.00 | 2.89 | 6.27 | 5.59 | 5.27 | 1.87 | 1.68 | |
| S/I Shift for ≥99.5% patients | 2.3cm | 2.3cm | 2.3cm | 4.0cm | 4.6cm | 2.4cm | 1.3cm | |
|
| ||||||||
| 3D Coronal Prescription overlap between AI and manual (%) | Median | 98.32 | 98.17 | 96.30 | 96.91 | 95.95 | 97.73 | 98.71 |
| IQR | 3.76 | 3.71 | 6.13 | 5.13 | 5.65 | 2.18 | 2.25 | |
|
| ||||||||
| 3D Sagittal Prescription overlap between AI and manual (%) | Median | 97.89 | 97.90 | 96.53 | 96.92 | 96.55 | 97.31 | 99.53 |
| IQR | 5.50 | 5.37 | 5.79 | 5.92 | 6.79 | 3.64 | 1.29 | |
We successfully implemented the AI-based automated image prescription method with both the full and tiny YOLOv3 networks on a clinical MR system (GE MR750) at the site of this study. Automated image prescription for an entire three-plane localizer dataset on the MRI system CPU required ~10 seconds with the full network and ~3 seconds with the tiny network. For the six volunteers in this study, the online 3D axial prescription was performed by the tiny network to determine the prescription for the subsequent series. In all six volunteers, the whole liver was covered with visually adequate placement. The online AI axial prescriptions were accepted by the operators (R.G. and C.J.B.) at the console without modifications to proceed to the subsequent scans. This allowed for fully automated prescription within 10 seconds (including time for AI prediction and review by the operator) of the previous scan completion.
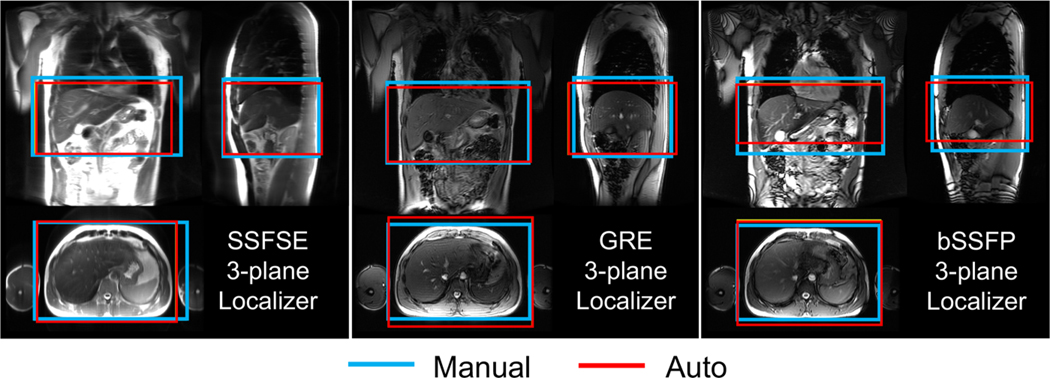
The prospective performance of the AI-based 3D axial prescription in healthy volunteers was similar to that of the retrospective study, using both the tiny network (Table 3) and the full network (Supplementary Table 1). For 3D liver detection, the percentage of manual volume included in the automated prescription by the full network had median = 89.2% and IQR = 14.6%. The overlap for 3D axial prescription by the full network had median = 92.1% and IQR = 13.2%. The 3D liver detection and axial prescription with both networks had overlaps > 80% with all three types of localizer sequences: SSFSE, GRE, and bSSFP, acquired with various parameters (Figure 6). Nevertheless, AI performed best with SSFSE acquisitions (Table 3 and Supplementary Table 1). The performance of both networks had no significant difference (p = 0.06) in 3D axial prescription across the various acquisition settings in the six healthy volunteers.
Table 3.
Testing results for prospective study using the tiny network trained without any augmentation. The online implementation worked well across various localizer sequences for 6 healthy volunteers. Automated prescription demonstrated promising performance (similar to that of the retrospective study) across SSFSE, GRE, and bSSFP sequences with various parameters. The performance of AI-based prescription was best for SSFSE acquisitions. Overlap: percentage of 3D volume from manual labeling covered by AI prescription.
| Sequences | 3D Liver Detection overlap between AI and manual (%) | 3D Axial Prescription overlap between AI and manual (%) | |||
|---|---|---|---|---|---|
|
| |||||
| Median | IQR | Median | IQR | ||
| SSFSE | TE=40ms | 94.74 | 15.77 | 96.39 | 9.54 |
| TE=80ms | 97.36 | 12.24 | 97.65 | 1.58 | |
| TE=120ms | 89.33 | 15.95 | 98.20 | 15.33 | |
|
| |||||
| GRE | Flip angle=20 | 87.26 | 10.52 | 85.01 | 15.79 |
| Flip angle=30 | 86.30 | 9.38 | 94.25 | 6.34 | |
| Flip angle=60 | 79.74 | 26.51 | 87.33 | 13.90 | |
| IR-prep | 85.56 | 19.33 | 84.54 | 14.11 | |
|
| |||||
| bSSFP | In phase | 91.38 | 22.82 | 93.85 | 14.12 |
| Out of phase | 88.44 | 12.56 | 92.09 | 7.94 | |
|
| |||||
| Overall | 89.18 | 14.55 | 92.12 | 13.23 | |
Figure 6.
The AI-based automated image prescription was successfully implemented on a clinical MRI system for prospective scanning. This online implementation performed well across various localizer sequences for 6 healthy volunteers. Automated prescription demonstrated promising performance for 3D axial prescription (similar to that of the retrospective study) across SSFSE, GRE, and bSSFP sequences with various parameters. The performance of AI-based automated prescription was best when using spin-echo (SSFSE) acquisitions.
DISCUSSION
This work proposed an AI-based automated liver image prescription method for abdominal MRI. The proposed method performed well across patients, pathologies, and clinically relevant acquisition settings (spin echo and gradient echo localizers at 1.5T and 3T) in a retrospective study. Further, prospective implementation on a 3T clinical MR system demonstrated rapid and accurate prospective performance in a pilot study with a limited number of healthy volunteers. Importantly, the AI-based automated liver prescription had a performance level comparable to radiologists’ inter-reader reproducibility. This method has the potential to enable automated, efficient, and reproducible image prescription for liver MRI.
The trained networks demonstrated excellent performance for 2D detection of the liver, torso, and the arms from individual localizer slices. However, 2D detection performance was only an intermediate evaluation metric in this study. Thanks to redundancy across multiple slices and orthogonal views, 3D prescription was minimally affected by occasional detection errors in single slices. Importantly, the shift in 3D axial prescription in the testing dataset was less than 2.3 cm in the S/I dimension for 99.5% of the test datasets. This indicates that the addition of narrow safety margins would ensure complete liver coverage in effectively all patients, at the cost of slightly longer acquisitions to cover these safety margins. Finally, although the network was trained primarily with SSFSE localizers, the method performed well with other localizer methods (GRE, bSSFP) with different parameters, suggesting generalizable application across various localizer acquisitions.
There are relevant previous preliminary efforts towards automated scan prescription in the liver using traditional object detection methods(8). The method based on liver-only bounding boxes could only perform axial prescription, while the proposed method has the capability to enable liver image prescription in any orthogonal orientation thanks to the inclusion of torso and arms classes. Importantly, this previous method requires a specialized volumetric, breath-held localizer scan, while the proposed method does not add any additional scan and is based on conventionally acquired localizers. In addition, the previous method has been tested in limited numbers of healthy volunteers (n = 24) and simulated deformed livers (n = 7) while the proposed method has been tested in 208 patients with various pathologies in the liver. The proposed AI-based method may be more robust than the previous method in terms of testing performance. In 24 healthy volunteers, the detection errors of the previous method to detect the upper and lower edges of the liver were 1.87±2.04 mm and −0.90±7.84 mm, respectively(8). The detection errors of the proposed method in 208 patients for the upper and lower edges were 0.31±3.53 mm and −0.68±12.52 mm, respectively. The performance of the proposed method is similar to recent developments in automated organ detection for kidney and spine using YOLOv3-based networks(12, 13). After training with datasets of 14 patients and testing with 41, the reported metrics for 2D detection of kidneys in CT were: average IoU = 0.769 and Dice coefficient = 0.851(13). Detection of intervertebral discs (IVD) in lumbar spine MRI, trained with datasets of 19 patients and tested with 18, obtained a Dice coefficient = 0.839(12). Likely as a result of the larger, diverse training dataset in this study, the IoU and Dice coefficient are both higher (0.878 ± 0.122 and 0.935 ± 0.069). In addition, the object mis-classification rate of the liver of the proposed method (1–4%) is comparable to that in a recent study using a modified YOLOv3 network trained with 5,986 abdominal (axial) CT images (~3%)(21).
This work relied on YOLO, a state-of-the-art and robust object detection system(20). Alternative networks include Single Shot Detector (SSD) and Faster R-CNN(26–28). YOLO and SSD have the advantage of real-time object detection(26). YOLO-based models have been applied to localization of organs with normal activity in 3D PET scans, detection of lung nodules, and detection of the nasal cavity in CT scans, with excellent robustness, speed and accuracy(29–31). YOLOv3 and SSD have shown similar accuracy, but YOLOv3 is about three times faster(13, 20).
The proposed approach presents several important network and workflow design considerations. Among various training settings, the tiny network, with much shallower architecture, showed slightly inferior performance to the full network. However, the 3x speedup of prediction time compared to the full network may make it a practical choice for implementation on current clinical MR systems. Additionally, the current workflow used a single network for all localizer image orientations. Alternatively, one might train a different network for each localizer slice orientation. An important advantage of the current single-network approach is approximately 3x faster prediction for the current MR system implementation, compared to using three networks.
Limitations
Confounding conditions such as multiple cysts, ascites, soft tissue edema, and iron overload, where B1-inhomogeneity leads to dielectric shading artifacts, pose a challenge for automated (and even manual) organ detection in the abdomen(25). Due to the limited number of failure cases with such conditions and artifacts in the testing dataset, the specific cause of detection failures is unknown. Furthermore, the quality of manual and automatic annotation in these cases may be limited by the fixed window/leveling of the input images. Although this did not appear to be a limitation in most cases, dynamic window/leveling may improve the quality of the manual and automatic annotation and provide a more accurate performance assessment. Importantly, this study relied on data from a single center and a single vendor. Finally, the prospective pilot study on a clinical MR system demonstrated promising performance, but only on a small number of healthy subjects.
Training and evaluation of the proposed method using multi-center and multi-vendor data with a wider variety of patient datasets are needed as future work. Evaluation of the model performance for centers that prioritize fast acquisitions and use fewer images per plane for the localizers is needed as future work. More cases with severe dielectric shading and iron overload from other vendors can be collected for training and testing to broaden the applicability of the algorithm. In this study, simple definitions of liver coverage for 3D prescription in axial, coronal, and sagittal orientations were used for illustration. Additional coverage considerations in each orientation can easily be included subsequently. Finally, in this study experienced radiologists (rather than technologists) performed the labeling to train and demonstrate the performance of AI using the best possible manual labeling as the reference. In clinical practice, technologists typically prescribe the field of view for a given patient exam. Future studies aimed at measuring and comparing the inter-reader variability of technologists with AI performance are needed. A previous study showed that a commercial platform (Day optimizing throughput [Dot] engine, Siemens Healthineers, Forchheim, Germany) for automated image prescription contributed significant time savings (2–5 minutes) for whole-body free-breathing exams(32). The effect of the proposed automated liver prescription approach on potential time savings needs to be further examined in future work. Value chain analysis from study acquisition to delivery of actionable information to clinicians and patients may be conducted to determine whether automated prescription improves patient care and workflow efficiency in Radiology departments(33–36).
Conclusion
An AI-based automated liver prescription method developed in this work has promising performance across patients, pathologies, and clinically relevant acquisition settings, well within inter-radiologist reproducibility. Further, this study demonstrated the successful prospective implementation of the method on a clinical MR system. This method has the potential to improve clinical workflow and standardization for MRI of the liver.
Supplementary Material
Grant support:
GE Healthcare and Bracco provide research support to the University of Wisconsin.
Unblinded acknowledgments: We would like to thank Daryn Belden, Wendy Delaney, and Prof. John Garrett from UW Radiology for their assistance with data retrieval, and Dan Rettmann, Lloyd Estkowski, Naeim Bahrami, Ersin Bayram, and Ty Cashen from GE Healthcare for their assistance with implementation of our AI-based liver image prescription on one of the GE scanners at the University of Wisconsin Hospital. We acknowledge support from the NIH (R01-EB031886). We also wish to acknowledge GE Healthcare and Bracco who provide research support to the University of Wisconsin. Dr. Oechtering receives funding from the German Research Foundation (OE 746/1–1). Dr. Reeder is a Romnes Faculty Fellow, and has received an award provided by the University of Wisconsin-Madison Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation.
Footnotes
Data sharing statement: Data generated or analyzed during the study are available from the corresponding author by request [and signing a data use agreement].
References:
- 1.Canellas R, Rosenkrantz AB, Taouli B, et al. Abbreviated MRI Protocols for the Abdomen. RadioGraphics 2019; 39:744–758. [DOI] [PubMed] [Google Scholar]
- 2.Pooler BD, Hernando D, Reeder SB. Clinical Implementation of a Focused MRI Protocol for Hepatic Fat and Iron Quantification. AJR Am J Roentgenol 2019; 213:90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao R, Zhang Y, Wang X, et al. Motion-Robust, High-SNR Liver Fat Quantification Using a 2D Sequential Acquisition with a Variable Flip Angle Approach. Magn Reson Med. 2020;84(4):2004–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang SS, Boyacioglu R, Bolding R, MacAskill C, Chen Y, Griswold MA. Free-Breathing Abdominal Magnetic Resonance Fingerprinting Using a Pilot Tone Navigator. J Magn Reson Imaging JMRI 2021; 54:1138–1151. [DOI] [PubMed] [Google Scholar]
- 5.Wang N, Cao T, Han F, et al. Free-breathing multitasking multi-echo MRI for whole-liver water-specific T1, proton density fat fraction, and R 2 ∗ quantification. Magn Reson Med 2022; 87:120–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth CJ, Boll DT, Wall LK, Merkle EM. Evaluation of MRI acquisition workflow with lean six sigma method: case study of liver and knee examinations. AJR Am J Roentgenol 2010; 195:W150–156. [DOI] [PubMed] [Google Scholar]
- 7.Heye T, Bashir MR. Liver imaging today. Magnetom Flash. 2013;52(2):2013. [Google Scholar]
- 8.Goto T, Kabasawa H. Automated Scan Prescription for MR Imaging of Deformed and Normal Livers. Magn Reson Med Sci 2013; 12:11–20. [DOI] [PubMed] [Google Scholar]
- 9.Goto T, Kabasawa H. Automated Positioning of Scan Planes and Navigator Tracker Locations in MRI Liver Scanning. Med Imaging Technol 2010; 28:245–251. [Google Scholar]
- 10.Massat MB, Bryant M. Thinking inside the box with AI and ML: A new kind of “insider intelligence.” Appl Radiol 2019; 48:34–37. [Google Scholar]
- 11.Barral JK, Overall WR, Nystrom MM, et al. A novel platform for comprehensive CMR examination in a clinically feasible scan time. J Cardiovasc Magn Reson 2014; 16(Suppl 1):W10. [Google Scholar]
- 12.De Goyeneche A, Peterson E, He JJ, Addy NO, Santos J. One-Click Spine MRI. 3rd Med Imaging Meets NeurIPS Workshop Conf Neural Inf Process Syst NeurIPS 2019 Vanc Can. [Google Scholar]
- 13.Lemay A. Kidney Recognition in CT Using YOLOv3. ArXiv191001268 Cs Eess 2019. [Google Scholar]
- 14.Dey D, Slomka PJ, Leeson P, et al. Artificial Intelligence in Cardiovascular Imaging. Curr Treat Options Cardiovasc Med 2019; 21:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itti L, Chang L, Ernst T. Automatic scan prescription for brain MRI. Magn Reson Med 2001; 45:486–494. [DOI] [PubMed] [Google Scholar]
- 16.Lecouvet FE, Claus J, Schmitz P, Denolin V, Bos C, Berg BCV. Clinical evaluation of automated scan prescription of knee MR images. J Magn Reson Imaging 2009; 29:141–145. [DOI] [PubMed] [Google Scholar]
- 17.Redmon J. Darknet: Open Source Neural Networks in C. 2013. [Google Scholar]
- 18.Van Rossum G, Drake FL Jr. Python Reference Manual. Centrum Voor Wiskunde En Informatica Amsterdam; 1995. . [Google Scholar]
- 19.Tzutalin. LabelImg. Git Code 2015; https://github.com/tzutalin/labelImg. [Google Scholar]
- 20.Redmon J, Divvala S, Girshick R, Farhadi A. You Only Look Once: Unified, Real-Time Object Detection. ArXiv150602640 Cs 2016. [Google Scholar]
- 21.Pang S, Ding T, Qiao S, et al. A novel YOLOv3-arch model for identifying cholelithiasis and classifying gallstones on CT images. PloS One 2019; 14:e0217647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adarsh P, Rathi P, Kumar M. YOLO v3-Tiny: Object Detection and Recognition using one stage improved model. In 2020. 6th Int Conf Adv Comput Commun Syst ICACCS; 2020:687–694. [Google Scholar]
- 23.Everingam M, Van Gool L, Williams CKI, Winn J, Zisserman A. The Pascal Visual Object Classes (VOC) Challenge. Int J Comput Vis 88, 303–338 (2010). [Google Scholar]
- 24.Abadi M, Agarwal A, Barham P, et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. 2015. [Google Scholar]
- 25.Huang SY, Seethamraju RT, Patel P, Hahn PF, Kirsch JE, Guimaraes AR. Body MR Imaging: Artifacts, k-Space, and Solutions. RadioGraphics 2015; 35:1439–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Z-Q, Zheng P, Xu S, Wu X. Object Detection with Deep Learning: A Review. ArXiv180705511 Cs 2019. [DOI] [PubMed] [Google Scholar]
- 27.Liu W, Anguelov D, Erhan D, et al. SSD: Single Shot MultiBox Detector. ArXiv151202325 Cs 2016; 9905:21–37. [Google Scholar]
- 28.Ren S, He K, Girshick R, Sun J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. ArXiv150601497 Cs 2016. [DOI] [PubMed] [Google Scholar]
- 29.Afshari S, BenTaieb A, Hamarneh G. Automatic localization of normal active organs in 3D PET scans. Comput Med Imaging Graph Off J Comput Med Imaging Soc 2018; 70:111–118. [DOI] [PubMed] [Google Scholar]
- 30.Ramachandran SS, George J, Skaria S VVV. Using YOLO based deep learning network for real time detection and localization of lung nodules from low dose CT scans. 2018; 10575:105751I. [Google Scholar]
- 31.Laura CO, Hofmann P, Drechsler K, Wesarg S. Automatic Detection of the Nasal Cavities and Paranasal Sinuses Using Deep Neural Networks. In 2019. IEEE 16th Int Symp Biomed Imaging ISBI 2019; 2019:1154–1157. [Google Scholar]
- 32.Koch V, Merklein D, Zangos S, et al. Free-breathing accelerated whole-body MRI using an automated workflow: Comparison with conventional breath-hold sequences. NMR Biomed 2022:e4828. [DOI] [PubMed] [Google Scholar]
- 33.O’Brien JJ, Stormann J, Roche K, et al. Optimizing MRI Logistics: Focused Process Improvements Can Increase Throughput in an Academic Radiology Department. Am J Roentgenol 2017; 208:W38–W44. [DOI] [PubMed] [Google Scholar]
- 34.Recht M, Macari M, Lawson K, et al. Impacting key performance indicators in an academic MR imaging department through process improvement. J Am Coll Radiol JACR 2013; 10:202–206. [DOI] [PubMed] [Google Scholar]
- 35.Tokur S, Lederle K, Terris DD, et al. Process analysis to reduce MRI access time at a German University Hospital. Int J Qual Health Care 2012; 24:95–99. [DOI] [PubMed] [Google Scholar]
- 36.Beker K, Garces-Descovich A, Mangosing J, Cabral-Goncalves I, Hallett D, Mortele KJ. Optimizing MRI Logistics: Prospective Analysis of Performance, Efficiency, and Patient Throughput. Am J Roentgenol 2017; 209:836–844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.