Congenital heart disease (CHD) is the commonest birth defect, affecting approximately 9.4/1 000 live births.1 Atrial Septal Defect (ASD) is one of the commonest CHD clinical phenotypes, which frequently requires treatment either in childhood or adulthood, and can lead to severe complications such as right heart failure and cardiac arrhythmia. Previous genome-wide association studies (GWAS) have identified a region of chromosome 4p16 (Ch4p16) associated with the risk of ASD. The most strongly associated SNPs (rs870142, rs6824295 and rs16835979) lie within a 38.8-kb region of linkage disequilibrium encompassing only the long noncoding RNA STX18-AS1 (also named LOC100507266) (Fig. 1A; Fig. S1a). Associated SNPs are expression quantitative trait locus (eQTLs) for STX18-AS1 in adult ventricular myocardial tissue.2
Figure 1.
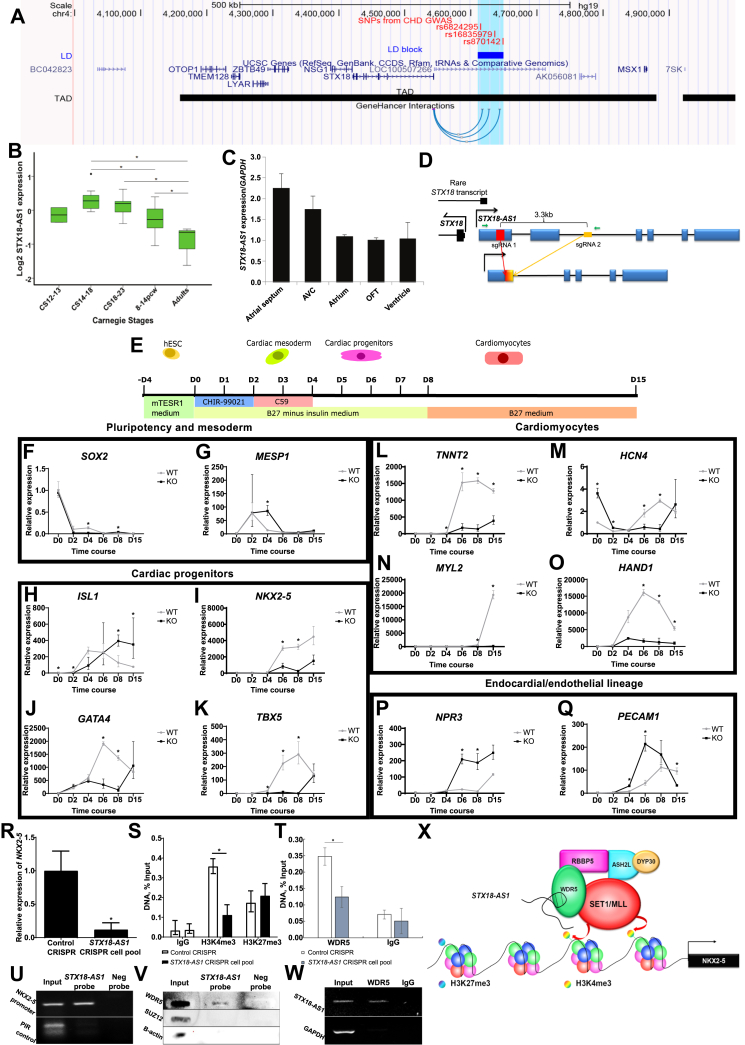
Long non-coding RNA STX18-AS1 regulates in vitro cardiomyocyte differentiation via epigenetic regulation. (A) The relative genomic location of ASD-risk SNPs and STX18-AS1 (LOC100507266) with surrounding genes and chromosome interactions. GWAS identified top risk SNPs for ASD were labeled as red lines with SNP IDs. The linkage disequilibrium block (LD block in blue rectangle spares 38.8 kb) was extracted from the LD map generated with HaploView using data from 1000 Genomes Project (CEU population). Topologically associating domains (TADs) were aligned with data from 3D genome browser; while the GeneHancer Interaction (available as UCSC genome browser track) only shows the regulatory elements within the LD block region using the data from Fishilevich's paper showing the interactions between GeneHancer regulatory elements and genes. (B) Dynamic changes in STX18-AS1 transcription during human heart development. ∗, P < 0.05. 3–5 samples are included for each developmental period of Carnegie Stages (CS). (C)STX18-AS1 transcription in different human heart segments (one sample from CS15). (D) The design of the CRISPR sgRNA pair with the cuts at the first exon and second intron of STX18-AS1. The sgRNAs do not overlap any transcript of STX18. The red rectangle and yellow rectangle represent sgRNA1 and sgRNA2. The gradient colored rectangle indicates the repaired join of two cuts. (E) Time schedule and treatments applied in cardiomyocyte differentiation protocol up to Day 15, and the relevant period of cell stages from hESC to cardiomyocytes. (F, G) The time courses of markers of cell pluripotency (F, SOX2) and cardiac mesoderm (G, MESP1). (H–K) Markers for cardiac progenitors: ISL1 (H), pan-cardiac progenitor marker; NKX2-5(I), GATA4(J), and TBX5 (K), markers for cardiac progenitors and early cardiomyocytes. (L–O) Time courses of markers for cardiomyocytes: TNNT2 (L), pan-cardiomyocyte marker; HCN4 (M), specific marker of atrial cardiomyocytes; MYL2 (N), marker for ventricular cardiomyocytes; HAND1 (O), marker for cardiac mesoderm and ventricular cardiomyocytes. (P, Q) Markers of other lineages: NPR3 (P), marker for endocardial lineage; PECAM1 (Q), marker for endothelial lineage. Data are shown as Mean ± S.E. ∗, P < 0.05. Two-way ANOVA test with Bonferroni adjustment is applied for generating the P values at each time point (n = 3–9). (R)NKX2-5 transcription level was reduced in STX18-AS1 CRISPR cell pool of HepG2. (S, T) Using ChIP, H3K4me3 (S) and WDR5 (T) around the promoter of NKX2-5 was reduced in STX18-AS1 CRISPR cell pool, without changes in H3K27me3. IgG was used as a background control. (U, V) ChIRP-PCR detected the promoter region of NKX2-5 in STX18-AS1 antisense probe pulldown lysate, localizing STX18-AS1 at the NKX2-5 promoter (U). The STX18-AS1 ChIRP probes pulled down WDR5 protein but not SUZ12 and B-actin (background control) detecting with slot blotting (V). A PIR region with rare RNA binding opportunity was used as the background control. (W) RNA immunoprecipitation with anti-WDR5 pulled down STX18-AS1 RNA transcripts, detected with PCR. IgG was used as negative control antibody. (X) The model of STX18-AS1's trans-activing effects on NKX2-5 by interacting with SET1/MLL complex and regulating the histone methylation around the downstream target. Data are shown as Mean ± S.E. ∗, P < 0.05, comparing to Control CRISPR using T-test or Two-way ANOVA (n = 3).
We first confirmed this eQTL association in human atrial tissues and showed it was specific to the myocardium (Fig. S1b–f). GTEx data (https://gtexportal.org/) confirmed that the risk SNPs are not eQTLs of any other genes in this region, including MSX1 and STX18, in cardiac tissues. STX18-AS1, a lowly conserved lncRNA gene without homologues in mouse (Fig. S2) and the only gene interacting with the linkage disequilibrium (LD) block containing the risk SNPs in the GeneHancer regulatory elements database (Fig. 1A), is therefore the strongest regional candidate gene for the ASD association signal.
Based on the most validated transcript ENST00000610009.5 (mapped to GRCh28. p13), we showed the transcription of STX18-AS1 is highly stage-dependent during heart development. Compared to adult tissues, STX18-AS1 was enriched in foetal tissues, including heart, brain, kidney, liver and lung (Fig. S3a). In developing human hearts, the peak expression of STX18-AS1 was identified at Carnegie stage (CS) 14–18 (Fig. 1B), overlapping with the critical period for atrial septation.3 To show the spatial distribution of STX18-AS1 in developing hearts, we conducted whole mount in situ hybridisation with an STX18-AS1 probe on three whole embryonic hearts at CS17-19. In all three hearts, STX18-AS1 expression was detected in AS, outflow tract (OFT), atrioventricular cushion (AVC), and part of the ventricles (Fig. S3b–g). The signals are mainly detected in the developing septum and valves/cushions (Fig. S3h–m). In the single CS15 heart available for RNA extraction, the relative quantity of STX18-AS1 transcripts was found to be higher in the AS than all other segments (Fig. 1C). Thus, levels of STX18-AS1 transcription accompanied Atrial Septal development spatio-temporally in human hearts.
We next investigated STX18-AS1 function in human embryonic stem cell differentiated cardiomyocytes (hESC-CM). Based on the H9 human embryonic stem cell line, we created a clonal STX18-AS1 knockout (KO) line with CRISPR/cas9, targeting the first two exons of STX18-AS1 with paired sgRNAs by removing ∼3.3 kb sequence to stop STX18-AS1 transcription (Fig. 1D). Transcription of the neighboring gene STX18 (one rare transcript of which partially overlaps with STX18-AS1's first exon) was confirmed to be unaffected by the CRISPR design (Fig. S4a). We observed a ∼30% reduction in transcription of another neighboring gene MSX1 in KO cells (Fig. S4b); since MSX1 acts as a “roadblock” to hESC-CM formation, and its deficiency promotes hESC-CM differentiation,4 this would not explain the delayed hESC-CM differentiation from STX18-AS1 KO cells we outline below. Moreover, regards relevance to ASD, loss-of-function mutations in MSX1 in humans cause deficient tooth development without any heart phenotype.5
The STX18-AS1 CRISPR transduction did not change the morphology, proliferation, apoptosis or pluripotency of undifferentiated H9 cells (Fig. S5). Following a monolayer hESC-CM differentiation protocol (Fig. 1E), STX18-AS1 KO cells (Fig. S6) produced no beating colony until Day 10, while control wildtype (WT) cells produced abundant beating CMs at Day 6–8 (Videos S1–3). Throughout differentiation to Day 15, STX18-AS1 KO cells formed beating CM islands of variable sizes and beating rates separated by non-beating cell populations, while the WT CMs formed networks and beat synchronously (Videos S4–5). Cardiomyocytes differentiated from STX18-AS1 deficient hESC cells were less in Cardiac Troponin T positive rate and weak in Cardiac Troponin T signal (Fig. S7).
The following is/are the supplementary data related to this article:
4
5
6
7
8
Supplementary video related to this article can be found at https://doi.org/10.1016/j.gendis.2022.07.010
Time-course data of STX18-AS1 KO hESC-CM differentiation showed an extended peaking time of MESP1 (Day 2–4), delayed activation, and reduced transcription of cardiac progenitor markers (NKX2-5, TBX5, and GATA4), a pan-cardiomyocyte marker (TNNT2), and both ventricular (MYL2 and HAND1) and atrial (HCN4) CM lineage markers at Day 8–15 (Fig. 1F, G and I–O). The increase of ISL1 in KO cells was slower, and prolonged to Day 15, compared to the peak at Day 6 in WT (Fig. 1H). These results suggest that STX18-AS1 KO does not entirely stop the programing of cardiomyocyte differentiation in vitro but prolonged the duration of cardiac progenitor specification and delayed their differentiation into cardiomyocytes. In support of this notion, endocardial/endothelial lineages (NPR3 and PECAM1) increased during the deferred period of forming cardiac progenitors and early cardiomyocytes (Fig. 1P, Q), suggesting potential endocardial/endothelial lineage substitution for CMs during hESC-CM differentiation from STX18-AS1 KO cells.
We next investigated the mechanism of downregulation of the known ASD gene NKX2-5 by STX18-AS1 KO. We detected a 70% reduction of H3K4me3 around NKX2-5, commensurate with the reduced NKX2-5 expression (Fig. 1R, S; Fig. S8a–c), in an STX18-AS1 CRISPR cell pool of HepG2 cells, a cell line chosen owing to its reliably detectable expression of both STX18-AS1 and NKX2-5. In keeping with the reduction of H3K4me3 at NKX2-5, the binding affinity of WDR5, a scaffold protein of the SET1/MLL complex required for the trimethylation of H3K4, as reduced by 50% at the promoter region of NKX2.5 in the STX18-AS1 CRISPR cell pool (Fig. 1T). Using Chromatin isolation by RNA purification (ChIRP) (Fig. S8d–f), STX18-AS1 transcripts were shown to interact both with the promoter sequences of NKX2-5, and with WDR5 (Fig. 1U, V). RNA immunoprecipitation with WDR5 antibody pulled down detectable STX18-AS1 RNA (Fig. 1W), confirming the direct interaction between STX18-AS1 and WRD5 protein. Co-localization of the STX18-AS1 lncRNA and WDR5 protein at the NKX2-5 promoter region suggested a direct epigenetic regulative role of STX18-AS1 on NKX2-5 (Fig. 1X) and potentially other downstream targeted cardiac transcriptional factors.
In summary, we present multiple lines of evidence suggesting that STX18-AS1, a lncRNA at chromosome 4p16, is responsible for the GWAS association with ASD observed in the region. STX18-AS1 has properties consistent with being a critical regulator of multiple core cardiac transcriptional factors promoting cardiac lineage specification, which merit further characterization. As yet, relatively few CHD GWAS studies have been published; these findings demonstrate the feasibility of gene identification and underscore the potential value of larger studies.
Conflict of interests
Authors declare no conflict of interests.
Funding
This work was supported by The University of Manchester-Peking University Health Science Centre Alliance, the China Scholarships Council, and British Heart Foundation Programme Grant RG/15/12/31616. BK holds a British Heart Foundation Personal Chair. Human embryonic/foetal materials were provided by the Joint MRC/Wellcome Trust (grant#MR/R006237/1) Human Developmental Biology Resource (http://hdbr.org).
Acknowledgements
We thank Dr Ruairidh Martin for collection of the samples for eQTL analyses and the data in peripheral blood.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.07.010.
Contributor Information
Yingjuan Liu, Email: yingjuan.liu@manchester.ac.uk.
Bernard D. Keavney, Email: bernard.keavney@manchester.ac.uk.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Liu Y., Chen S., Zühlke L., et al. Global birth prevalence of congenital heart defects 1970-2017: updated systematic review and meta-analysis of 260 studies. Int J Epidemiol. 2019;48(2):455–463. doi: 10.1093/ije/dyz009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordell H.J., Bentham J., Topf A., et al. Genome-wide association study of multiple congenital heart disease phenotypes identifies a susceptibility locus for atrial septal defect at chromosome 4p16. Nat Genet. 2013;45(7):822–824. doi: 10.1038/ng.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faber J.W., Hagoort J., Moorman A.F.M., Christoffels V.M., Jensen B. Quantified growth of the human embryonic heart. Biol Open. 2021;10(2):bio057059. doi: 10.1242/bio.057059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rao J., Pfeiffer M.J., Frank S., et al. Stepwise clearance of repressive roadblocks drives cardiac induction in human ESCs. Cell Stem Cell. 2016;18(3):341–353. doi: 10.1016/j.stem.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Khasawneh R.R., Kist R., Queen R., et al. Msx1 haploinsufficiency modifies the Pax9-deficient cardiovascular phenotype. BMC Dev Biol. 2021;21(1):14. doi: 10.1186/s12861-021-00245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
4
5
6
7
8