Abstract
Glaucoma is the leading cause of irreversible blindness worldwide. In the pathogenesis of glaucoma, activated microglia can lead to retinal ganglion cells (RGCs) apoptosis and death, however, the molecular mechanisms remain largely unknown. We demonstrate that phospholipid scramblase 1 (PLSCR1) is a key regulator promoting RGCs apoptosis and their clearance by microglia. As evidenced in retinal progenitor cells and RGCs of the acute ocular hypertension (AOH) mouse model, overexpressed PLSCR1 induced its translocation from the nucleus to the cytoplasm and cytomembrane, as well as elevated phosphatidylserine exposure and reactive oxygen species generation with subsequent RGCs apoptosis and death. These damages were effectively attenuated by PLSCR1 inhibition. In the AOH model, PLSCR1 led to an increase in M1 type microglia activation and retinal neuroinflammation. Upregulation of PLSCR1 resulted in strongly elevated phagocytosis of apoptotic RGCs by activated microglia. Taken together, our study provides important insights linking activated microglia to RGCs death in the glaucoma pathogenesis and other RGC-related neurodegenerative diseases.
Keywords: Apoptosis, Glaucoma, Phagocytosis, PLSCR1, Retinal ganglion cells
Abbreviations
- AOH
acute ocular hypertension
- CNS
central nervous system
- DAPI
4′,6-diamidino-2-phenylindole
- DCFH-DA
2′,7′-dichlorodihydrofluorescein diacetate
- DHE
dihydroethidium
- GCL
ganglion cell layer
- H&E
hematoxylin and eosin
- hiPSCs
human induced pluripotent stem cells
- IBA1
ionized calcium-binding adaptor molecule 1
- INL
inner nuclear layer
- IOP
intraocular pressure
- IPL
inner plexiform layer
- OGDR
oxygen and glucose deprivation/reoxygenation
- ONC
optic nerve crush
- ONL
outer nuclear layer
- OPL
outer plexiform layer
- PFA
paraformaldehyde
- PI
propidium iodide
- PLSCR1
phospholipid scramblase 1
- PS
phosphatidylserine
- pSIVA
polarity sensitive indicator of viability and apoptosis
- qPCR
quantitative polymerase chain reaction
- RGCs
retinal ganglion cells
- ROS
reactive oxygen species
- RPCs
retinal progenitor cells
- RT
room temperature
- RT-PCR
real-time reverse transcription-polymerase chain reaction
- siRNA
small interfering RNA
- TB
toluidine blue
- TG
transgenic
- TUNEL
terminal deoxynucleotidyl transferase biotin-UTP nick end labeling
- WT
wild-type
Background
Glaucoma, the leading cause of irreversible blindness worldwide, is characterized by progressive retinal ganglion cells (RGCs) degeneration and death and impairment of visual function.1, 2, 3 The pathogenesis of RGCs degeneration and death is complex. The pathophysiology of glaucomatous damage is multifactorial and not completely understood. Activation of retinal microglia has been found to promote RGCs apoptosis and death in the pathogenesis of glaucoma.4,5 However, the underlying molecular mechanisms are still largely unknown.
Microglia are resident immune cells in the human and rodent retina.6 In physiological conditions, microglia are located in the ganglion cell layer (GCL), inner plexiform layer (IPL), and outer plexiform layer (OPL) with ramified morphology.4 In inherited photoreceptor degeneration, activated microglia could facilitate rod death via phagocytosis and secretion of IL-1β.7 In the glaucomatous retina, activated microglia could progressively migrate to GCL, which is thought to be deleterious in the process of retinal degeneration.4
Recent studies indicate that lipid metabolism and turnover play a critical role in the pathogenesis of glaucoma.8,9 Phospholipid scramblase 1 (PLSCR1), associated with lipid metabolism is a calcium-dependent type II single-pass transmembrane protein. Activation of PLSCR1 promotes phosphatidylserine (PS) to expose from the inner leaflet to the outer leaflet of plasma membrane, which is one of the characteristics of early apoptosis and an “eat-me” signal for microglia to recognize and engulf stressed cells.10,11 PLSCR1 might be associated with neuron death, as increased PLSCR1-immunoreactive neurons were observed in the human hippocampus after cerebral ischemia.12 Though downregulated expression of PLSCR1 could inhibit microglial activation for the clearance of virus transduced-astrocytes in the central nervous system (CNS),13 it has not been investigated whether PLSCR1 is directly involved in the microglia activation and neuron death.
In this study, we explored the connection between PLSCR1 and retinal microglia, and revealed their relationships with RGCs death. Our study points out that PLSCR1 is a pivotal regulator promoting RGCs apoptosis and their clearance by activated retinal microglia. Our findings provide novel insights into the mechanisms of microglial activation in glaucoma pathogenesis, which might contribute to potential therapeutic interventions for glaucoma and other RGC-related neurodegenerative diseases.
Materials and methods
Reagents and resources
The detailed information on reagents and resources, including antibodies, chemicals, sequences, and other materials related to our study are listed in the Supplementary Information.
Retinal progenitor cells (RPCs) and cell culture
Human induced pluripotent stem cells (hiPSCs) were purchased from Saibei company (Beijing, China). The procedure of hiPSCs differentiation into RPCs was modified as previously described.14, 15 Briefly, hiPSCs were cultured using retinal induced medium (RIM) in a 12-well culture plate for five days. RIM was change every day. From the sixth day, hiPSCs were cultured using the retinal differentiation medium (RDM) for seven days. From day 12, the cells were transplanted to the retinal progenitor cell differentiation medium (RPCDM). The RPC-like cells were mechanically enriched by scraping out with non-RPC morphology. Enriched RPCs were cultured in retinal progenitor cell medium (RPCM) for three days. Then the RPCs were cultured in Dulbecco's modified Eagle's medium/nutrient mixture F-12 (DMEM/F12) supplemented with 10% fetal bovine serum (FBS) and maintained at 37 °C in an incubator at 5% CO2. The hiPSCs exhibited typical clonal morphology and were characterized by traditional pluripotent stem cell markers OCT4 and Tra-1-60, while RPCs differentiated from hiPSCs were characterized by Pax6, Nestin, CHX10, and LHX2. Flow cytometry was applied to validate differentiation efficiency from hiPSCs to RPCs (Fig. S1). The detailed composition of the cell culture medium was listed in Supplementary information.
Adenoviral transfection and RNA interference
For the adenoviral transfection, the adenovirus packaging expressing PLSCR1 (pAd-PLSCR1) and the control (pAd-NC) vector were obtained from Vigene Biosciences Company (Shandong, China). All the plasmids were verified by DNA sequencing. The adenoviruses were added into the RPC culture medium at a multiplicity of infection (MOI) of 30.
For the RNA interference, effective small interfering RNA (siRNA) targeting human PLSCR1 (si-PLSCR1) and a negative control scrambled siRNA (si-NC) were purchased from GenePharma Company (Shanghai, China) and transfected using Lipofectamine® RNAiMax (Invitrogen, USA) according to the manufacturer's instructions. The transfection was conducted 24 h prior to oxygen and glucose deprivation/reoxygenation (OGDR) treatment. The expression levels of PLSCR1 were verified by Western blot. The sequences used were listed in the Supplementary Information.
OGDR model
To establish the OGDR model, the culture medium of RPCs was replaced with glucose-free DMEM (Gibco) after washing cells twice with PBS. Then, the cells were placed in a 5% CO2 and 95% N2 atmospheric incubator chamber under hypoxic conditions for 3 h at 37 °C. After that, RPCs were cultured in the normal medium again and maintained in a normoxic (5% CO2 and 95% air) atmospheric incubator for 24 h. Control groups were cultured in the normal medium in a normoxic atmospheric incubator for the same duration.
Mice
All the animal procedures were approved by the Animal Care and Ethics Committee of Zhongshan Ophthalmic Center, Sun Yat-Sen University (Guangzhou, China), and all the Use of Animals were performed in accordance with the Association for Research in Vision and Ophthalmology (ARVO) statement. C57BL/6J wild-type (WT) mice and transgenic PLSCR1 (TG-PLSCR1) mice produced on the C57BL/6J background were generated from Gempharmatech Co., Ltd (Jiangsu, China). Two mouse lines showed PLSCR1 overexpression in the retina identified by immunostaining and Western blot were selected to further breeding. Mice aged from 4 to 6 weeks were examined in this study. In all procedures, mice were weighed and anesthetized by intraperitoneal injection of 1% pentobarbital sodium (50 mg/kg, Sigma, USA) and topically anesthetized with 0.5% proparacaine hydrochloride eye drops (Alcon, USA). Their pupils were dilated with topical administration of tropicamide phenylephrine eye drops (Santen, Japan). Animals were maintained on a 12-h light–dark cycle and housed in the Animal Laboratory of Zhongshan Ophthalmic Center.
Acute ocular hypertension (AOH) mouse model
The procedure of AOH model was carried out as our previous study described.3 To establish the AOH model, the WT mice and TG-PLSCR1 mice were anesthetized and their pupils were dilated. The anterior chamber of the right eye was cannulated with a 30-gauge infusion needle connected to a 150-mL bottle of normal saline solution, which was elevated to the height of 150 cm to maintain an IOP of 110 mmHg for 60 min. The left eye without AOH was served as the control group. After the procedure, tobramycin ointment (Alcon, USA) was applied to the eye surface for preventing postoperative infection. Eyes without cataracts, iris injury/bleeding, anterior chamber leakage, or infections were collected for further study.
Optic nerve crush (ONC) injury mouse model
The ONC surgery was performed as previously described.16 After general and topical anesthetization, a small incision was made in the superior-external conjunctiva and orbital muscles were gently put aside with fine forceps to expose the optic nerve. The optic nerve was clamped with self-clamping forceps at about 1 mm behind the eye globe for 5 s. The left eye without crushing served as control. Eyes were harvested on the fifth day after treatment.
Western blot analysis
Western blot analysis was performed as previously described.17 After extraction from RPCs or mouse retinas, the concentrations of the proteins were measured. Equal amounts of proteins were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis resolving gel and 5% stacking gel and transferred onto polyvinylidene fluoride (PVDF) membrane. PVDF membrane was blocked with 5% skim milk in Tris-buffered saline plus 0.1% Tween-20 (TBST) for 1 h at room temperature (RT), and then incubated with primary antibodies (rabbit anti-PLSCR1, 1:1000, Proteintech; rabbit anti-Tubulin,1:1000, Abcam) at 4°C overnight. Then the membranes were incubated with secondary antibodies (HRP-goat anti-rabbit IgG, 1:2000) for 2 h at RT. Proteins signals were developed with SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermos Fisher Scientific) and imaged using a chemiluminescence system (Bio-Rad Laboratories, USA).
Flow cytometry
Cell apoptosis and reactive oxygen species (ROS) were detected by flow cytometric analysis using Annexin V-FITC/PI and 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA), respectively. Approximately 1 × 106 RPCs cultured in a 6-well plate were transfected with adenoviral or siRNA for 48–72 h. The cells were digested by 0.25% trypsin, washed twice with PBS, and then incubated with Annexin V-FITC/PI (Annexin V-FITC Apoptosis Detection Kit, BD Biosciences, USA) or 10 μM DCFH-DA (Reactive Oxygen Species Assay Kit, Beyotime, China) for 20 min. All groups in the experiment were repeated in triplicate. Cell apoptosis and intracellular ROS level were detected by a flow cytometer (BD Biosciences, USA). Data were analyzed by the BD FACSDiva 8.0.1.
Immunofluorescence staining
In cells: Cultured iPSCs and iPSC-RPCs were washed with PBS and fixed with 4% paraformaldehyde (PFA) for 15 min, then washed twice with PBS. Then the cells were incubated in 0.1M PBS containing 3% bovine serum albumin (BSA) and 0.5% Triton X-100 at RT for 1 h. Followed by incubation with primary antibodies (rabbit anti-OCT4, 1:400; mouse anti-Tra-1-60, 1:200; rabbit anti-Pax6, 1:400; mouse anti-CHX10, 1:400; mouse anti-LHX2, 1:400; rabbit anti-nestin, 1:400; rabbit anti-PLSCR1, 1:200) overnight at 4°C, cells were washed with PBS, and incubated with secondary antibodies (1:500) and DAPI (1:5000). Images were captured with a confocal scanning microscope LSM800 (Carl Zeiss, Germany).
In tissues: The mice were overdosed with anesthesia and transcardially perfused with 4% PFA and PBS. The eyes were enucleated and immersed in 4% PFA for 40 min at RT. For frozen cryosections, the eyecups were dehydrated in 10% sucrose, followed by 20% and 30% sucrose, embedded in OCT compounds, frozen, and cut as a 10-μm-thick section. For retinal flat mounts, the retinas were mounted and dissected into a four-leaf clover shape. The superior leaves were labeled by the preservation of the surrounding retinal pigment epithelium and dissected into the largest piece for orientation. The tissues were incubated with primary antibodies (rabbit anti-PLSCR1, 1:200; rabbit anti-RBPMS, 1:300; mouse anti-Brn3a, 1:500; goat anti-IBA1, 1:100; rat anti-CD68, 1:300) overnight (cryosections) or for 72 h (retinal flat mounts) at 4 °C. After washing in PBS, tissues were incubated with secondary antibodies (1:500) for 2 h at RT and counterstained with DAPI (1:2000). Six images were captured at 300 μm from the optic nerve head of each cryosection, while three images were captured from central to peripheral regions in four quadrants of each retina (Fig. 4C). The whole retinal mount and representative images of RBPMS labeled RGCs were acquired by a Zeiss Axio Observer Inverted Microscope (TissueGnostics, Austria) running the TissueFAXS 7.0 software with a 20× objective. Orientation was indicated with S (superior), I (inferior), N (nasal), and T (temporal) axes (Fig. 4D).
Figure 4.
Overexpression of PLSCR1 aggravates RGCs damage and death after AOH. (A) H&E staining shows the thickness of the retina in different groups. Scale bars, 20 μm. (B) Statistic analysis shows the thickness of different layers of the retina significantly decrease in AOH-treated TG-PLSCR1 mice. (C) Indicative map demonstrates that three images (central, middle, and peripheral) were captured in every quadrant of the whole mount retina. A total of 12 fields were assessed for each retina. (D) Immunofluorescence images show that the number of RBPMS labeled RGCs (red) decreased in the AOH-treated mice (5 days). The change is more pronounced in TG-PLSCR1 mice. The upper row shows the density distribution of RGCs in the whole mount retina (Scale bars, 1 mm). The lower row exhibits the magnified micrographs from the middle region in the superior quadrant of the corresponding retinas (white box) (Scale bars, 50 μm). (E) Statistic analysis shows the average survival RGCs numbers from 12 fields per retina. (F) Toluidine blue staining images of optic nerve transverse section demonstrate axon damage in the AOH-treated mice, manifesting reduced axon density, myelin disruption, and fields with gliosis (Scale bars, 5 μm). (G) Statistic analysis of axon damage grade shows TG-PLSCR1 mice have much severer axon damage than WT mice. (B, E, G) Two-tailed Student's t-test (B and G: n = 4; E: n = 5 for each experiment). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and NS indicates difference not significant. Data are mean ± SD. RNFL, retinal nerve fiber layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer. S, superior; I, inferior; N, nasal; T, temporal.
Phosphatidylserine (PS) exposure assay
In vitro, we performed polarity-sensitive indicator of variability and apoptosis (pSIVA-IANBD, Novus Biologicals, USA) in the calcium-dependent states, which can bound to apoptosis cells and be detected by fluorescence. Before commencement of the experiments, cells were washed twice with desktop fluid and supplemented with 2 mM CaCl2 in desktop fluid warmed to 37 °C for 10 min. pSIVA and propidium iodide (PI) were added and incubated in a dark condition at 37 °C for 5 min according to the manufacturer's instructions. The PS on the extracellular face of the plasma membrane was binding by pSIVA, and the damage or necrotic cells were detected by PI. Hoechst (Invitrogen, Thermo Fisher Scientific, USA) reagent was added to stain cell nuclear. We then track the progression and timing of PLSCR1 effect on RPCs apoptosis by live-cell imaging. To visualize PS exposure in vivo, mice were anesthetized and injected 1.5 μL pSIVA solution into the vitreous one day after AOH. The eyes were collected 2 h after pSIVA injection and fixed with 4% PFA for 20 min. The retinal flat mounts were immediately observed by a confocal scanning microscope.
Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL)
To perform TUNEL staining of the retinal cryosections, the cryosections were dried and permeabilized with 0.1% Triton-X100 in PBS for 30 min at RT. After PBS rinsing, the sections were incubated in dark places with TUNEL (In situ Cell Death Detection Kit, Roche Life Science, Switzerland) reaction mixture for 1 h at 37 °C and subsequently incubated with DAPI for 10 min. For the retinal flat mounts, a 30 μl mixture per retina was used for incubation for 2 h at RT in dark places before secondary antibodies incubation. The images were captured with a confocal microscope as above described.
Dihydroethidium (DHE) staining
The 10-μM thick retinal cryosections from different experimental groups were taken to and then covered with a 10 μM DHE solution (Invitrogen). The slides were incubated in a light-protected humidified incubator at 37 °C for 30 min. Sections were mounted with Fluoromount-G and covered with coverslips. Images were taken using an LSM800 confocal scanning microscope.
Histological assessment
The procedure of histologic sections was prepared as described previously.18 Briefly, animals were sacrificed on the fifth day after AOH. A suture was placed on the edge of the inferior conjunctiva to identify the inferior portion of the eye. Eyes were enucleated and fixed in FAS eye fixation solution (Servicebio Technology, China) overnight at RT. After dehydrated in an ethanol series, eyeballs were embedded in paraffin, cut in 4-μm sections through the suture and at the point of the optic nerve head, and mounted on glass slides. Histology sections were stained with hematoxylin and eosin (H&E). The microscopic image of each section was captured at 1 mm on both sides from the optic disc and the thickness of the whole and each layer of the retina were measured using Image J software.
The axon damage of RGCs was evaluated by toluidine blue (TB) staining on the seventh day post-AOH. After 4% PFA perfusion, the mouse optic nerves were cut approximately 1 mm behind the globe, fixed by 2.5% glutaraldehyde for 2 h at RT, and transferred to 4 °C overnight. Followed by washing with phosphate buffer, optic nerves were placed in 1% OsO4 for 2 h at RT and dehydrated with a series of ethanol and isoamyl acetate. The nerves were then embedded in epoxy medium, cut sections at 1 μm with an ultramicrotome, and enhanced with osmium tetroxide-induced myelin staining using 1% TB. Optic nerve was observed under light microscopy and the axon damage was assessed using a semiquantitative optic nerve grading scheme in accordance with previous studies.19,20
Image analysis
The number of interested cells (RGCs, microglia, and apoptosis cells) from each field was counted using Image J software (LOCI, University of Wisconsin, USA) to obtain the average quantification. Areas of immunopositivity (pSIVA, DHE, and CD68) were derived by thresholding images captured under uniform imaging conditions. The unit of each visual field used in statistics was under 20 × objective of an LSM800 confocal scanning microscope (319.45 × 319.45 μm2 per field).
Quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) and quantitative PCR (qPCR)
Total RNA was extracted from retina samples using TRIzol™ reagent, and the final concentration was quantified with a Nanodrop spectrophotometer (ND-1000; NanoDrop Technologies, USA). Next, cDNA was synthesized with a qPCR RT Kit (TOYOBO, Japan). The cDNA was then diluted with SYBR Green Supermix (Bio-Rad) and was analyzed by qPCR for changes in gene expression. GAPDH mRNA was used as an internal control. The primer sequences are listed in Supplementary Information. Each cDNA sample was run in triplicate, superimposed on a standard curve to determine absolute transcript quantities. The relative mRNA expression levels were calculated with the 2−ΔΔCt method as in previous research.21 Data were analyzed using Bio-Rad CFX manager software.
RNA-seq analysis
The RNA-seq analysis was conducted by Berry Genomics Corporation (Beijing, China) as our previous study.22 RNA was extracted from the retinas of WT and TG-PLSCR1 mice with AOH treatment. A total amount of 1 μg RNA per sample was used as input material. Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer's recommendations, and index codes were added to attribute sequences to each sample. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia). Then the library preparations were sequenced on an Illumina NovaSeq platform and 150 bp paired-end reads were generated. Clean data with high quality after processed raw data were aligned with TopHat (v2.0.11) to the mouse genome (GRCm38/mm10). HTSeq v0.6.1 was used to count the reads numbers mapped to each gene. Ingenuity pathway analysis (IPA) (Qiagen Inc., Hilden, Germany) software was applied to analyze each sample's expression values and detect significant differences in gene transcript expression between groups. Sequences have been deposited in the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The GEO accession is GSE186750.
Statistical analysis
SPSS software (version 21.0; IBM SPSS Inc., USA) and GraphPad Prism (version 9.0; GraphPad Inc., USA) were used for statistical analysis and graphics. The summarized data were expressed as mean ± SD obtained from at least three independent experiments. The differences between different groups were calculated by the two-tailed Student's t-test. NS indicates P ≥ 0.05, ∗indicates P < 0.05, indicates P < 0.01, and indicates P < 0.001.
Results
Overexpressed PLSCR1 leads to its translocation in RPCs
To verify the PLSCR1 expression in RPCs, we evaluated the protein expression level of PLSCR1 in the blank group, pAd-NC group, and pAd-PLSCR1 group by Western blot. Compared with the blank group, the expression of PLSCR1 was significantly increased in the pAd-NC and pAd-PLSCR1 groups, especially in the pAd-PLSCR1 group (Fig. 1A, B). Then we performed immunostaining of PLSCR1 to investigate its subcellular location in RPCs. In the blank and pAd-NC group, PLSCR1 was expressed in the nucleus, cytoplasm, and cytomembrane of RPCs; whereas in the pAd-PLSCR1 group, PLSCR1 was not expressed in the nucleus but mainly localized in the cytoplasm and cytomembrane (Fig. 1C). These findings show that a much higher expression of PLSCR1 would trigger its translocation from the nucleus to the cytoplasm and cytomembrane in RPCs.
Figure 1.
Overexpressed PLSCR1 leads to its translocation in RPCs. (A) Western blot shows the protein expression of PLSCR1 in RPCs without pAd treatment (Blank), and RPCs treated with pAd-NC and pAd-PLSCR1. (B) Statistic analysis of the relative protein expression of PLSCR1 normalized to β-Tubulin in RPCs demonstrates the expression increases in pAd-NC and pAd-PLSCR1 groups. Two-tailed Student's t-test (n = 3 for each experiment). ∗P < 0.05, ∗∗∗P < 0.001. Data are mean ± SD. (C) Immunofluorescence shows that PLSCR1 locates in the nucleus, cytoplasm, and cytomembrane in RPCs without pAd treatment and RPCs infected with pAd-NC, while it is translocated from the nucleus to the cytoplasm and cytomembrane with enhanced immunofluorescence in RPCs infected with pAd-PLSCR1. Scale bars, 20 μm.
PLSCR1 regulates PS exposure, cell apoptosis, and ROS generation in RPCs
As PLSCR1 can process PS exposure as an immune response to viral infection,23,24 we use pSIVA, a fixable green fluorescent polarity sensitive indicator of viability and apoptosis, to bind exposure PS and PI to probe death cells to evaluate the effect of PLSCR1 on RPCs function. In the blank group, few RPC was detected with pSIVA/PI fluorescence signal. The pSIVA/PI signals were significantly augmented in the virus-infected groups, of which the pAd-PLSCR1 RPCs demonstrated more striking fluorescence signals (Fig. 2A).
Figure 2.
Overexpressed PLSCR1 promotes phosphatidylserine (PS) exposure, cell apoptosis, and reactive oxygen species (ROS) generation in RPCs. (A) Live-cell imaging using the polarity sensitive indicator of viability and apoptosis (pSIVA) and propidium iodide (PI) shows the PS exposure and cell death increases in RPCs treated with pAd-NC and pAd-PLSCR1. Scale bars, 100 μm. (B) Apoptosis of RPCs (Blank), and RPCs treated with pAd-NC or pAd-PLSCR1 were evaluated by Annexin V-FITC and PI by flow cytometry. (C) Quantification analysis of the early and late apoptotic rate in RPCs (Blank), and RPCs treated with pAd-NC or pAd-PLSCR1. (D) The production of ROS using 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) is detected by flow cytometry. (E) Statistic analysis of the mean intensity of DCFH-DA fluorescence value in different groups shows PLSCR1 significantly increased the ROS generation in RPCs. (A, B, D) RPCs are infected with pAd-NC and pAd-PLSCR1 for 48 h. (C, E) Statistic analysis shows two-tailed Student's t-test (n = 3 for each experiment). ∗P < 0.05, ∗∗P < 0.01. Data are mean ± SD.
We then used flow cytometry of Annexin V-FITC/PI to detect the rate of cell apoptosis and death on RPCs. Consistently, RPCs treated with pAd-PSLCR1 had the highest degree of cell apoptosis among the three groups (Fig. 2B). The early and late apoptotic ratio in the blank group was 6.7% and 12.6% in the pAd-NC treated group, while the percentage of apoptotic cells increased to 19.2% in the pAd-PLSCR1 group (Fig. 2C). For further study, we detected the ROS generation by flow cytometry using DCFH-DA. Compared with the blank group, cytosolic ROS production was significantly elevated in the pAd-NC and pAd-PSLCR1 groups. Among them, the highest cytosolic ROS level was observed in the pAd-PSLCR1 treated RPCs (Fig. 2D, E).
Using RNA interference to knock down PLSCR1 expression in RPCs (Fig. S2), we then investigated whether PLSCR1 inhibition could ameliorate RPCs damage under OGDR conditions. As an in vitro model mimicking AOH, OGDR treatment could enhance pSIVA/PI fluorescence signal, cell apoptosis rate, and ROS accumulation in RPCs. PLSCR1 inhibition by si-PLSCR1 substantially alleviated damage and death of RPCs (Fig. S3). From the above results, we demonstrate that PLSCR1 regulates PS exposure, cell apoptosis, and ROS generation in RPCs.
Upregulated PLSCR1 contributes to its translocation in the retina and optic nerve of TG-PLSCR1 mice
To explore the roles of PLSCR1 in retinal degeneration, TG-PLSCR1 mice were generated using a cDNA coding human PLSCR1 gene transferred into mouse embryos. Increased expression of PLSCR1 was verified by Western blot in the TG-PLSCR1 mice retina (Fig. 3A, B). Then, we use immunostaining of PLSCR1 to examine its distribution in mice retinas. In the retina of WTmice, PLSCR1 was mainly expressed in the GCL and inner nuclear layer (INL). However, in the retina of TG-PLSCR1 mice, PLSCR1 was not only expressed in the GCL and INL, but also expressed in the retinal nerve fiber layer (RNFL), IPL, OPL, and outer nuclear layer (ONL). Also, in the retina and optic nerve of WT mice, PLSCR1 was mainly expressed in the nucleus. Whereas in the retina and optic nerve of TG-PLSCR1 mice, PLSCR1 was expressed in the nucleus and cytoplasm (Fig. 3C).
Figure 3.
Upregulated PLSCR1 contributes to its translocation in the retina and optic nerve of TG-PLSCR1 mice. (A) Western blot results show the PLSCR1 protein expression elevates in the retina of TG-PLSCR1 mice compared with WT. (B) Statistic analysis of PLSCR1 normalized expression level. Two-tailed Student's t-test (n = 3 for each experiment). ∗∗∗P < 0.001. Data are mean ± SD. (C) Immunofluorescence images of PLSCR1 in the retina and optic nerve of WT and TG-PLSCR1 showed different expression and localization. Scale bars, 50 μm. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; ON, optic nerve.
We also examined the distribution of PLSCR1 in the brain and liver tissues from WT and TG-PLSCR1 mice. In WT mice, PLSCR1 was located in the cell nucleus of brain and liver. In TG-PLSCR1 mice, PLSCR1 was expressed in both nucleus and cytoplasm in brain cells, whereas it was translocated to cytoplasm and cytomembrane in liver cells (Fig. S4). These findings demonstrate that overexpressed PLSCR1 contributes to its translocation at the subcellular level in tissue.
Overexpression of PLSCR1 aggravates RGCs damage and death after acute ocular hypertension (AOH)
As the expression of PLSCR1 is elevated in neurons in the cerebral ischemia condition,12 we thus investigated whether PLSCR1 was involved in the glaucomatous damage by detecting its transcriptional expression in the retinas of AOH and ONC model. Our findings revealed that the mRNA level of PLSCR1 significantly increased after treatment (Fig. S5), suggesting that PLSCR1 might be a potential manipulator of retinal injury and neuron death.
To identify the effects of PLSCR1 in the glaucomatous retina, we performed AOH treatment in both WT and TG-PLSCR1 mice to assess the pathological change. The thickness of each layer of the retina, number of RGCs, and axon damage were evaluated. Before AOH treatment, the morphology of the retina and optic nerve exhibited no difference between WT and TG-PLSCR1 mice (Fig. 4A, F). However, compared with WT mice, TG-PLSCR1 mice showed the whole retinal thickness dramatically decreased after AOH, especially in the RNFL, IPL, and INL (Fig. 4A, B). Also, the number of RBPMS labeled RGCs markedly decreased in TG-PLSCR1 mice (Fig. 4D, E). TB staining of optic nerve showed TG-PLSCR1 mice had much severer axon damage of RGCs after AOH, assessed by semiquantitative grading (Fig. 4G).
Altogether, these findings show that PLSCR1 exerts deleterious effects on the glaucomatous retina and optic nerve.
Elevated PLSCR1 facilitates PS exposure, cell apoptosis, and ROS production in RGCs after AOH treatment
Taken overexpressed PLSCR1 promotes PS exposure, apoptosis, and cytosolic ROS in RPCs, we explored whether PLSCR1 overexpression had a similar effect in RGCs of AOH-treated retina. Using pSIVA to label exposed PS, we demonstrated an increased pSIVA signal in GCL one day after AOH, and TG-PLSCR1 mice show a higher level of pSIVA-immunopositivity than WT mice (Fig. 5A, D). Then we used TUNEL staining and DHE to detect cytosolic apoptosis, and ROS in the AOH retinas, respectively. The results showed a significant increase in the number of TUNEL labeled apoptotic cells in the retinas one day after AOH treatment (Fig. 5C). More apoptotic cells were observed in the GCL and INL of TG-PLSCR1 mice than those in the WT mice (Fig. 5C, F). ROS production of the retinas was also assessed on the first days after AOH. Notably, the retinas of TG-PLSCR1 mice with AOH treatment exhibited the highest ROS level (Fig. 5B, E). Our findings indicate overexpression of PLSCR1 aggravates PS exposure, cell apoptosis, and ROS level of RGCs in glaucomatous damage.
Figure 5.
Elevated PLSCR1 facilitates PS exposure, cell apoptosis, and higher ROS production after AOH treatment. (A–C) Immunofluorescence images of annexin-based fluorescent indicator polarity sensitive indicator of viability and apoptosis (pSIVA) in the ganglion cell layer (A), dihydroethidium (DHE) (B), and TUNEL staining (C) shows that PS exposure, ROS level, and retinal cell apoptosis markedly increased in TG-PLSCR1 retinas after AOH. Scale bars, 50 μm. (D–F) Statistic analysis of pSIVA immunopositivity (D), ROS level (E), and TUNEL staining cell number (F) shows two-tailed Student's t-test (n = 3 for each experiment). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and NS indicates difference not significant. Data are mean ± SD.
Overexpressed PLSCR1 promotes activated microglia with increased phagocytosis, M1 polarization, and pro-inflammatory cytokines secretion
On the third day after AOH, we observed the retinal microglia infiltrating the GCL had morphology changed, from ramified shape to ameboid or rod shape (Fig. 6A). Then we performed immunostaining of IBA1 and CD68 to evaluate the number and phagocytic function of microglia, respectively. Our results showed that the number of microglia had no difference between WT and TG-PLSCR1 groups before treatment, yet the TG-PLSCR1 retinas had more microglia compared with the WT retinas after AOH (Fig. 6A, B). CD68 is a lysosome-associated membrane protein and scavenger receptor, and a marker for M1 type microglia.25,26 In both WT and TG-PLSCR1 retinas, the expression of CD68 increased markedly in the GCL co-localized with Iba1 three days after AOH and the AOH-treated TG-PLSCR1 group exhibited a much higher immunopositivity level (Fig. 6A, C).
Figure 6.
Overexpressed PLSCR1 promotes activated microglia with increased phagocytosis, M1 polarization, and pro-inflammatory cytokines secretion. (A) Immunofluorescence images of retinal flat mounts show microglia infiltrating the ganglion cell layer (GCL) with ameboid morphology and upregulation of the phagocytic molecule marker CD68 (green) on the third day after AOH. The immunostaining of CD68 (green) is co-localized within IBA1 labeled microglia (red). Scale bar, 50 μm. (B) Statistic analysis shows the number of IBA1 labeled microglia increases after treatment and more activated microglia in the GCL of TG-PLSCR1 mice compared with WT mice. (C) Statistic analysis shows that the immunopositivity of CD68 is significantly enhanced, demonstrating that microglial activation is much more pronounced in the TG-PLSCR1 mice. (D, E) The mRNA of WT and TG-PLSCR1 mice with AOH treatment are isolated from the mouse retinas. qPCR data show higher mRNA levels of M1 type microglia markers (TNF-α, iNOS, CD86, CCL2, CXCL10, IL-1β, and IL-6) in TG-PLSCR1 mice compared with WT mice; whereas the mRNA level of M2 type microglia markers (IL-10, YM-1, TGF-β, CD206, and Fizz-1) shows no significant difference between groups. (F) RNA-seq analysis identifies significant upregulated microglia-mediated inflammation gene transcripts suggesting a higher level of microglial activation and microglial inflammatory response in TG-PLSCR1 retinas compared with controls. (G) qPCR analysis verifies the genes expression in RNA-seq data. (B–E, G) Two-tailed Student's t-test (B and C: n = 4; D, E, and G: n = 6 for each experiment). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 and NS indicates difference not significant. Data are mean ± SD.
To assess M1 and M2 microglia polarization, we used a qPCR screen to determine gene expression changes in the retina of AOH-treated WT and TG-PLSCR1 mice. The mRNA level of M1 type microglia markers, such as TNF-α, iNOS, and CD86, were distinctly upregulated in the AOH-treated TG-PLSCR1 group, but the mRNA levels of CCL-3 and CCL-5 showed no significant difference (Fig. 6D). The mRNA level of M2 type microglia markers, such as IL-10, YM-1, and CD206, had no significant difference between the two groups (Fig. 6E).
To gain better insight into the pathological neuroinflammation caused by PLSCR1, we performed RNA-seq analysis on retinal tissue from WT and TG-PLSCR1 mice with AOH treatment. As the PLSCR1 pathway has been reported to be involved in microglial activation in neuroinflammation, we investigated the microglia associated molecules and found that the mRNA level of genes in the pathway of immune system process, response to stimulus, cytokine production, response to stress, and inflammatory response were notably upregulated in AOH-treated TG-PLSCR1 mice (Fig. 6F). qPCR validated the gene expression of RNA-seq (Fig. 6G). These data indicate that overexpression of PLSCR1 enhances the inflammatory response in AOH injury by facilitating microglia to exhibit an active state, phagocytic function, and M1 polarization.
PLSCR1 facilitates clearance of apoptotic RGCs by microglia phagocytosis after AOH
According to the above results, the presence of an “eat-me” signal on RGCs and the expression of phagocytic molecules in activated microglia suggests that microglial phagocytosis of RGCs may contribute to RGCs injury and death. To investigate and confirm the interaction between microglia with RGCs, we performed Iba1, TUNEL, and RBPMS immunostaining to label microglia and apoptotic RGCs, respectively. On the third day after AOH when microglia infiltrated the GCL, we observed that Iba1 labeled activated microglia contained DAPI labeled cells that were immunopositive for TUNEL and RBPMS, confirming the microglial engulfment of apoptotic RGCs in both WT and TG-PLSCR1 retinas with AOH treatment (Fig. 7A–C). Assessment of the number of TUNEL- and RBPMS-positive cells, as well as TUNEL-, RBPMS-, and IBA1-positive cells, revealed that TG-PLSCR1 AOH-treated retinas showed significantly more apoptotic RGCs and phagocytosed apoptotic RGCs than WT AOH-treated retinas (Fig. 7D).
Figure 7.
PLSCR1 facilitates clearance of apoptotic RGCs by microglia phagocytosis after AOH. (A) Retinal flat mounts from TG-PLSCR1 mice three days after AOH shows more activated microglia and more apoptotic RGCs than WT AOH-treated mice. Microglia labeled with IBA1 (purple), apoptotic cells labeled with TUNEL (green), and RGCs labeled with RBPMS (red). Arrowhead indicates the apoptotic RGCs phagocytosed by microglia. Scale bar, 20 μm. (B, C) Three-dimensional images reconstruction of the indicated area (white box in A) confirm the apoptotic RGCs inside the soma of microglia. Scale bar, 2 μm. (D) Statistic analysis for TUNEL labeled RGCs with or without microglial phagocytosis in WT and TG-PLSCR1 mice. Two-tailed Student's t-test (n = 3 for each experiment). ∗∗∗P < 0.001. Data are mean ± SD.
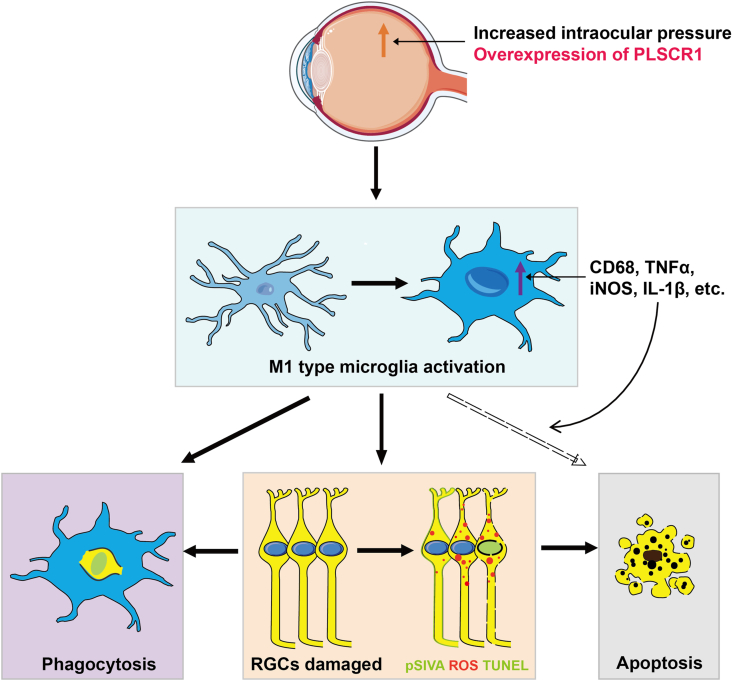
Our results indicate that PLSCR1 promotes RGCs death and clearance by microglial phagocytosis and provide direct evidence of intercellular interaction of activated microglia and apoptotic RGCs in retinal neurodegeneration (Fig. 8).
Figure 8.
Schematic illustrating the activated retinal microglia contribute to RGCs death by phagocytosis and secreting pro-inflammatory cytokines in AOH, and overexpression of PLSCR1 exacerbates this pathological process. In the AOH retina, RGCs become damaged (orange box) marked by exposed phosphatidylserine (PS), TUNEL, and reactive oxygen species (ROS) staining, which induce microglia recruitment. Ramified microglia infiltrate the ganglion cell layer (GCL) three days after AOH, showing ameboid morphology, upregulated phagocytic molecules (CD68), M1 phenotype activation markers (e.g., CD86 and CXCL10), and pro-inflammatory cytokines (e.g., TNF-α and iNOS). The activated microglia, on the one hand, phagocytose a subset of TUNEL labeled RGCs (purple box); on the other hand, additionally influence and potentiate the apoptotic route for RGCs death via pro-inflammatory cytokines secretion, such as TNF-α and IL-1β (grey box). Overexpression of PLSCR1 in the AOH-treated eye increases the PS exposure, apoptosis, and ROS generation of RGCs, and therefore intensifies microglia activation in phagocytosis and pro-inflammatory cytokine production, which aggravates RGCs clearance and death.
Discussion
In the current study, we provide evidence that PLSCR1 is involved in the interactions between microglia and RGCs, the secondary nerve cells of retina. PLSCR1 promotes PS exposure, cell apoptosis, and ROS generation in vitro and in vivo. Upregulation of PLSCR1 in the AOH-treated retina aggravates M1 type microglia activation and phagocytosis of RGCs.
Accumulating studies show that retinal microglia activated and neuroinflammation occurr in the glaucomatous retina, resulting in retinal injury and RGCs death.5,27, 28, 29, 30 Several genes and signaling pathways are found participating in the activation of retinal microglia and RGC pathogenesis, such as CX3CR1, nucleotide-binding leucine-rich repeat-containing receptor (NLR) family, Toll-like receptor (TLR) pathway, and Jak-Stat pathway.28,29,31, 32, 33 However, the direct intercellular interaction between retinal microglia and damaged RGCs is unrevealed.
Phospholipid scramblase activity is involved in the collapse of phospholipid asymmetry at the plasma membrane leading to the externalization of PS, which provides a signal for the recruitment of macrophages or microglia to bind to and engulf the apoptotic cells.34 PLSCR, TMEM16, and XKR family members are specific phospholipid scramblases that contribute to neurodegeneration. PLSCR1 has been found to increase after ischemia injury and modulate microglia-mediated virus infected cell clearance in CNS.12,13 PLSCR3, localized in the mitochondrial membrane, is associated with neuronal vulnerability to brain ischemia.35,36 Deficiency of TMEM16F could relieve the microglial phagocytosis in the pathogenesis of neuropathic pain and cerebral ischemia.37,38 XKR8, a caspase-activated scramblase, is implicated in regulating bipolar cell death and axon clearance.39,40 Also, rhodopsin and other G protein-coupled receptors (GPCRs) are constitutively active as phospholipid scramblases in neurons.41,42 Located on the membrane of photoreceptor disc, rhodopsin is thought to play a role in re-modelling cell membranes and its constitutive activation will lead to retinal degenerations.42,43 The titer of anti-rhodopsin antibodies is found high in normal tension glaucoma patients’ serum.44,45 A Genome-wide association study identifies phospholipid scramblase activity and phospholipid scrambling pathways possibly correlate with high IOP and glaucoma in Westerners.46 Whereas, the specific role of phospholipid scramblase underlying glaucoma pathogenesis remains unidentified.
Taken the essential role of PLSCR1 in the recognition and clearance of stressed cells by microglia in the CNS, we investigate the function and potential mechanism of PLSCR1 in glaucoma pathogenesis and attempt to clarify the intercellular interaction between microglia and stressed RGCs.
In leukemic cells and breast cancer cells, endogenous PLSCR1 is mainly located in the cytoplasm, and it traffics to the nuclear under some conditions.47,48 Different from the distribution in leukemic cells and breast cancer cells, endogenous PLCSR1 is distributed in the whole cell of RPCs, and overexpression of PLSCR1 leads to its translocation from the nucleus to the cytoplasm and cytomembrane. In the retina, endogenous PLSCR1 is located in the cell nucleus of GCL and INL, whereas overexpression leads to its distribution in the cell nucleus and cytoplasm of RNFL, IPL, OPL, and ONL. As the distribution of PLSCR1 in neurons has not been reported previously, our study suggests that PLSCR1 might exhibit different functions with the variation of distribution and expression level after overexpression in neurons.
In many pathological conditions, the intracellular calcium homeostasis is disrupted and subsequently the calcium-mediated signaling cascades activate. PLSCR1 can be activated in the presence of increased cellular calcium and enhances IP3R expression, in turn, influencing intracellular calcium homeostasis.49 Both pAd-NC and pAd-PLSCR1 infection of RPCs can upregulate PLSCR1 expression, PS exposure, cell apoptosis, and ROS generation, while overexpressed PLSCR1 exacerbates these effects. The virus infection of RPCs can be taken as a stimulus, which elevates the expression of PLSCR1 by disrupting intracellular homeostasis, such as the induction of ROS and the activation of apoptotic signals, thus causing PS exposure. Overexpression of PLSCR1 aggravates the intracellular calcium dysregulation and thereby exerts further deleterious effects. Inhibition of PLSCR1 in the OGDR model, nevertheless, could attenuate the RPC damage and apoptosis.
In the mouse retina, AOH and ONC treatment lead to upregulated expression of retinal PLSCR1. Although the distribution of retinal PLSCR1 was distinctly different between WT and TG-PLSCR1 mice, there was no difference in retinal and optic nerve morphology between the two groups before AOH treatment. However, compared with WT mice, the retinas of TG-PLSCR1 mice exhibited more deleterious damage with AOH treatment. In TG-PLSCR1 mice, more PS exposure and TUNEL signals were observed in the GCL on the first day after AOH treatment, and the number of RGCs and the thickness of RNFL and GCL were significantly decreased on the fifth day after AOH treatment. Axon damage was much severer in TG-PLSCR1 mice than controls in the AOH model as well. These findings indicate that PLSCR1 might be served as a stress-responsive gene in glaucomatous neuropathy and overexpression of PLSCR1 will promote the progression of RGC pathogenesis.
On the first day after AOH, the acute hypertension stimulation affected the RGC function and initiated the apoptosis process, presenting as PS exposure and TUNEL signal in the GCL. PS exposure occurred in RGCs can be induced by oxidative stress, such as ROS, which then attracted and activated microglia translocating from the outer layer to the GCL. Then the activated microglia infiltrated the GCL on the third day after AOH with ameboid morphology and upregulation of phagocytic molecules, CD68. Consequently, on the fifth day after AOH, the number of RGCs and the thickness of RNFL and GCL were decreased. TG-PLSCR1 mice showed more serious damage compared with WT mice, implicating that overexpression of PLSCR1 further aggravates the pathological process.
Activated microglia can be divided into two major subtypes, M1 and M2 type.25,50 The markers of M1 polarization are mostly mediators of pro-inflammatory responses whereas M2 markers are considered neuroprotective. It is known that the products of M1 microglia can lead to RGCs death via proinflammatory and oxidative stress pathways in the glaucomatous retina.5,51 Here, M1 types of microglia were significantly increased in the TG-PLSCR1 retinas than WT in the AOH model, whereas there was no difference in the M2 types of microglia. These findings indicate that overexpressed PLSCR1 predominantly promotes retinal microglia polarization towards the M1 phenotype in glaucomatous damage, which can be validated by upregulated expression of microglia-mediated inflammation genes and complement cascade components in the RNA-seq results.
Our three-dimension reconstruction images reveal for the first time the microglia-RGCs interactions during the glaucomatous process. We observe that there is a greater number of infiltrating microglia and apoptotic RGCs in the TG-PLSCR1 AOH-treated retina, and the activated microglia execute the damaged RGCs by phagocytosis its soma. Nonetheless, there are still some apoptotic RGCs that are not phagocytosed by microglia in AOH-treated retinas. We suppose that the rest of apoptotic RGCs without being phagocytosed might be partially caused by the upregulated pro-inflammation cytokines, such as TNF-α and iNOS, secreted from M1 type activated microglia. More M1 type microglial cells are activated by upregulated expression of PLSCR1, which will subsequently trigger more severely retinal damage via phagocytosis and secreting pro-inflammation cytokines.
Conclusions
In summary, we demonstrate that PLSCR1 is a key regulator in promoting RGCs apoptosis and clearance by M1 type microglia, which leads to the retina and optic nerve injury and visual function impairment. Our study points out the connection between PLSCR1 and retinal microglia and their interactions with RGCs degeneration, which will contribute to a better understanding of glaucoma pathogenesis and provide potential therapeutic targets for the treatment of glaucomatous damage or other RGC-related neurodegeneration.
Ethics declaration
All the animals were treated in strict accordance with Animal Research and this study was formally reviewed and approved by the Zhongshan Ophthalmic Center Animal Care and Ethics Committee.
Author contributions
L.Z., L.L., Y.L., Y.Z., and K.Z.: Conceptualization, Supervision, Resources, Writing-review & editing. L.Z., J.L., Q.L. and D.Z.: Investigation, Methodology, Visualization, Writing - original draft. J.L., Q.L., D.Z. M.Z., T.M., B.S., Z.Y., C.L., W.X., L.Z., K.W., X.L., Y.L, F.M., W.L., C.Z., M.L.: Investigation, Data curation, Formal analysis, Software. All authors read and approved the final manuscript.
Conflict of interests
The authors declare that they have no competing interests.
Funding
This work was supported by The National Natural Science Foundation of China (No. 81670894, 81721003, 81570862, 82000915); The National Key Research and Development Program of China (No. 2020YFA0112701); The Pearl River Talents Program-Local Innovative and Research Teams (No. 2017BT01S138); The “100 talents plan” from Sun Yat-sen University; the Open Research Funds of the State Key Laboratory of Ophthalmology (No. 2022KF04); The Guangdong Provincial Key Laboratory of Ophthalmology and Visual Science (No. 2017B030314025); The NSFC/Macao Science and Technology Development Fund (No. 015/2017/AFJ to KZ); the Fundamental Research Funds for the Central Universities, Sun Yat-sen University (No. 22qntd3902).
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgements
We thank the staff of Core Facilities at State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center for technical support. We also gratefully acknowledge the staff of the Laboratory Animal Center at State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.05.036.
Contributor Information
Yizhi Liu, Email: yzliu62@yahoo.com.
Lin Lu, Email: drlulin@126.com.
Ling Zhao, Email: zhaoling6@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Davis B.M., Crawley L., Pahlitzsch M., Javaid F., Cordeiro M.F. Glaucoma: the retina and beyond. Acta Neuropathol. 2016;132(6):807–826. doi: 10.1007/s00401-016-1609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quigley H.A. Glaucoma. Lancet. 2011;377(9774):1367–1377. doi: 10.1016/S0140-6736(10)61423-7. [DOI] [PubMed] [Google Scholar]
- 3.Skowronska-Krawczyk D., Zhao L., Zhu J., et al. P16INK4a upregulation mediated by SIX6 defines retinal ganglion cell pathogenesis in glaucoma. Mol Cell. 2015;59(6):931–940. doi: 10.1016/j.molcel.2015.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silverman S.M., Wong W.T. Microglia in the retina: roles in development, maturity, and disease. Annu Rev Vis Sci. 2018;4:45–77. doi: 10.1146/annurev-vision-091517-034425. [DOI] [PubMed] [Google Scholar]
- 5.Baudouin C., Kolko M., Melik-Parsadaniantz S., Messmer E.M. Inflammation in Glaucoma: from the back to the front of the eye, and beyond. Prog Retin Eye Res. 2021;83 doi: 10.1016/j.preteyeres.2020.100916. [DOI] [PubMed] [Google Scholar]
- 6.Reichenbach A., Bringmann A. Glia of the human retina. Glia. 2020;68(4):768–796. doi: 10.1002/glia.23727. [DOI] [PubMed] [Google Scholar]
- 7.Zhao L., Zabel M.K., Wang X., et al. Microglial phagocytosis of living photoreceptors contributes to inherited retinal degeneration. EMBO Mol Med. 2015;7(9):1179–1197. doi: 10.15252/emmm.201505298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dal Monte M., Cammalleri M., Locri F., et al. Fatty acids dietary supplements exert anti-inflammatory action and limit ganglion cell degeneration in the retina of the EAE mouse model of multiple sclerosis. Nutrients. 2018;10(3):325. doi: 10.3390/nu10030325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Njie-Mbye Y.F., Kulkarni-Chitnis M., Opere C.A., Barrett A., Ohia S.E. Lipid peroxidation: pathophysiological and pharmacological implications in the eye. Front Physiol. 2013;4:366. doi: 10.3389/fphys.2013.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darwich Z., Klymchenko A.S., Kucherak O.A., Richert L., Mély Y. Detection of apoptosis through the lipid order of the outer plasma membrane leaflet. Biochim Biophys Acta. 2012;1818(12):3048–3054. doi: 10.1016/j.bbamem.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 11.Brown G.C., Neher J.J. Microglial phagocytosis of live neurons. Nat Rev Neurosci. 2014;15(4):209–216. doi: 10.1038/nrn3710. [DOI] [PubMed] [Google Scholar]
- 12.Rami A., Sims J., Botez G., Winckler J. Spatial resolution of phospholipid scramblase 1 (PLSCR1), caspase-3 activation and DNA-fragmentation in the human hippocampus after cerebral ischemia. Neurochem Int. 2003;43(1):79–87. doi: 10.1016/s0197-0186(02)00194-8. [DOI] [PubMed] [Google Scholar]
- 13.Tufail Y., Cook D., Fourgeaud L., et al. Phosphatidylserine exposure controls viral innate immune responses by microglia. Neuron. 2017;93(3):574–586. doi: 10.1016/j.neuron.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng F., Chen M., Liu Y., et al. Stage-specific differentiation of iPSCs toward retinal ganglion cell lineage. J Mol Vis. 2016;22:536–547. [PMC free article] [PubMed] [Google Scholar]
- 15.Lamba D.A., Karl M.O., Ware C.B., Reh T.A. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103(34):12769–12774. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X., Liu Y., Jin H., et al. Reactive fibroblasts in response to optic nerve crush injury. Mol Neurobiol. 2021;58(4):1392–1403. doi: 10.1007/s12035-020-02199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu S.T., Sun B.H., Ge J.N., et al. CRLF1-MYH9 interaction regulates proliferation and metastasis of papillary thyroid carcinoma through the ERK/ETV4 axis. Front Endocrinol. 2020;11:535. doi: 10.3389/fendo.2020.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lian Q., Zhao M., Li T., et al. In vivo detecting mouse persistent hyperplastic primary vitreous by Spectralis Optical Coherence Tomography. Exp Eye Res. 2019;181:271–276. doi: 10.1016/j.exer.2019.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Chauhan B.C., Levatte T.L., Garnier K.L., et al. Semiquantitative optic nerve grading scheme for determining axonal loss in experimental optic neuropathy. Invest Ophthalmol Vis Sci. 2006;47(2):634–640. doi: 10.1167/iovs.05-1206. [DOI] [PubMed] [Google Scholar]
- 20.Ebneter A., Casson R.J., Wood J.P., Chidlow G. Microglial activation in the visual pathway in experimental glaucoma: spatiotemporal characterization and correlation with axonal injury. Invest Ophthalmol Vis Sci. 2010;51(12):6448–6460. doi: 10.1167/iovs.10-5284. [DOI] [PubMed] [Google Scholar]
- 21.Yu S.T., Zhong Q., Chen R.H., et al. CRLF1 promotes malignant phenotypes of papillary thyroid carcinoma by activating the MAPK/ERK and PI3K/AKT pathways. Cell Death Dis. 2018;9(3):371. doi: 10.1038/s41419-018-0352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao M., Mei T., Shang B., et al. Defect of LSS disrupts lens development in cataractogenesis. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.788422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang J., Zhu X., Liu J., et al. Inhibition of Hepatitis B virus replication by phospholipid scramblase 1 in vitro and in vivo. Antivir Res. 2012;94(1):9–17. doi: 10.1016/j.antiviral.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Dong B., Zhou Q., Zhao J., et al. Phospholipid scramblase 1 potentiates the antiviral activity of interferon. J Virol. 2004;78(17):8983–8993. doi: 10.1128/JVI.78.17.8983-8993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franco R., Fernández-Suárez D. Alternatively activated microglia and macrophages in the central nervous system. Prog Neurobiol. 2015;131:65–86. doi: 10.1016/j.pneurobio.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Holness C.L., da Silva R.P., Fawcett J., Gordon S., Simmons D.L. Macrosialin, a mouse macrophage-restricted glycoprotein, is a member of the lamp/lgp family. J Biol Chem. 1993;268(13):9661–9666. [PubMed] [Google Scholar]
- 27.Oikawa K., Ver Hoeve J.N., Teixeira L.B.C., et al. Sub-region-specific optic nerve head glial activation in glaucoma. Mol Neurobiol. 2020;57(6):2620–2638. doi: 10.1007/s12035-020-01910-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lozano D.C., Choe T.E., Cepurna W.O., Morrison J.C., Johnson E.C. Early optic nerve head glial proliferation and Jak-Stat pathway activation in chronic experimental glaucoma. Invest Ophthalmol Vis Sci. 2019;60(4):921–932. doi: 10.1167/iovs.18-25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X., Luo C., Cai J., et al. Neurodegenerative and inflammatory pathway components linked to TNF-α/TNFR1 signaling in the glaucomatous human retina. Invest Ophthalmol Vis Sci. 2011;52(11):8442–8454. doi: 10.1167/iovs.11-8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan L., Neufeld A.H. Activated microglia in the human glaucomatous optic nerve head. J Neurosci Res. 2001;64(5):523–532. doi: 10.1002/jnr.1104. [DOI] [PubMed] [Google Scholar]
- 31.Breen K.T., Anderson S.R., Steele M.R., Calkins D.J., Bosco A., Vetter M.L. Loss of fractalkine signaling exacerbates axon transport dysfunction in a chronic model of glaucoma. Front Neurosci. 2016;10:526. doi: 10.3389/fnins.2016.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H., Deng Y., Gan X., et al. NLRP12 collaborates with NLRP3 and NLRC4 to promote pyroptosis inducing ganglion cell death of acute glaucoma. Mol Neurodegener. 2020;15(1):26. doi: 10.1186/s13024-020-00372-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chi W., Li F., Chen H., et al. Caspase-8 promotes NLRP1/NLRP3 inflammasome activation and IL-1β production in acute glaucoma. Proc Natl Acad Sci U S A. 2014;111(30):11181–11186. doi: 10.1073/pnas.1402819111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kodigepalli K.M., Bowers K., Sharp A., Nanjundan M. Roles and regulation of phospholipid scramblases. FEBS Lett. 2015;589(1):3–14. doi: 10.1016/j.febslet.2014.11.036. [DOI] [PubMed] [Google Scholar]
- 35.Dave K.R., Bhattacharya S.K., Saul I., et al. Activation of protein kinase C delta following cerebral ischemia leads to release of cytochrome C from the mitochondria via bad pathway. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0022057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowalczyk J.E., Beresewicz M., Gajkowska B., Zabłocka B. Association of protein kinase C delta and phospholipid scramblase 3 in hippocampal mitochondria correlates with neuronal vulnerability to brain ischemia. Neurochem Int. 2009;55(1–3):157–163. doi: 10.1016/j.neuint.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y., Li H., Li X., et al. TMEM16F aggravates neuronal loss by mediating microglial phagocytosis of neurons in a rat experimental cerebral ischemia and reperfusion model. Front Immunol. 2020;11:1144. doi: 10.3389/fimmu.2020.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Batti L., Sundukova M., Murana E., et al. TMEM16F regulates spinal microglial function in neuropathic pain states. Cell Rep. 2016;15(12):2608–2615. doi: 10.1016/j.celrep.2016.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sapar M.L., Ji H., Wang B., et al. Phosphatidylserine externalization results from and causes neurite degeneration in Drosophila. Cell Rep. 2018;24(9):2273–2286. doi: 10.1016/j.celrep.2018.07.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kautzman A.G., Keeley P.W., Ackley C.R., Leong S., Whitney I.E., Reese B.E. Xkr8 modulates bipolar cell number in the mouse retina. Front Neurosci. 2018;12:876. doi: 10.3389/fnins.2018.00876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li T., Chiou B., Gilman C.K., et al. A splicing isoform of GPR56 mediates microglial synaptic refinement via phosphatidylserine binding. EMBO J. 2020;39(16) doi: 10.15252/embj.2019104136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goren M.A., Morizumi T., Menon I., et al. Constitutive phospholipid scramblase activity of a G protein-coupled receptor. Nat Commun. 2014;5:5115. doi: 10.1038/ncomms6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ernst O.P., Menon A.K. Phospholipid scrambling by rhodopsin. Photochem Photobiol Sci. 2015;14(11):1922–1931. doi: 10.1039/c5pp00195a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romano C., Barrett D.A., Li Z., Pestronk A., Wax M.B. Anti-rhodopsin antibodies in sera from patients with normal-pressure glaucoma. Invest Ophthalmol Vis Sci. 1995;36(10):1968–1975. [PubMed] [Google Scholar]
- 45.Romano C., Li Z., Arendt A., Hargrave P.A., Wax M.B. Epitope mapping of anti-rhodopsin antibodies from patients with normal pressure glaucoma. Invest Ophthalmol Vis Sci. 1999;40(6):1275–1280. [PubMed] [Google Scholar]
- 46.Blue Mountains Eye Study (BMES); Wellcome Trust Case Control Consortium 2 (WTCCC2). Genome-wide association study of intraocular pressure identifies the GLCCI1/ICA1 region as a glaucoma susceptibility locus. Hum Mol Genet. 2013;22(22):4653–4660. doi: 10.1093/hmg/ddt293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang P., Liao R., Chen X., et al. Nuclear translocation of PLSCR1 activates STAT1 signaling in basal-like breast cancer. Theranostics. 2020;10(10):4644–4658. doi: 10.7150/thno.43150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H., Xu J., Zhou Y., et al. PLSCR1/IP3R1/Ca(2+) axis contributes to differentiation of primary AML cells induced by wogonoside. Cell Death Dis. 2017;8(5) doi: 10.1038/cddis.2017.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Q., Ben-Efraim I., Bigcas J.L., Junqueira D., Wiedmer T., Sims P.J. Phospholipid scramblase 1 binds to the promoter region of the inositol 1,4,5-triphosphate receptor type 1 gene to enhance its expression. J Biol Chem. 2005;280(41):35062–35068. doi: 10.1074/jbc.M504821200. [DOI] [PubMed] [Google Scholar]
- 50.Tang Y., Le W. Differential roles of M1 and M2 microglia in neurodegenerative diseases. Mol Neurobiol. 2016;53(2):1181–1194. doi: 10.1007/s12035-014-9070-5. [DOI] [PubMed] [Google Scholar]
- 51.Bosco A., Romero C.O., Breen K.T., et al. Neurodegeneration severity can be predicted from early microglia alterations monitored in vivo in a mouse model of chronic glaucoma. Dis Model Mech. 2015;8(5):443–455. doi: 10.1242/dmm.018788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.