Abstract
TANK-binding kinase 1 (TBK1) is a nodal protein involved in multiple signal transduction pathways. In RNA virus-mediated innate immunity, TBK1 is recruited to the prion-like platform formed by MAVS and subsequently activates the transcription factors IRF3/7 and NF-κB to produce type I interferon (IFN) and proinflammatory cytokines for the signaling cascade. In this study, TRAF7 was identified as a negative regulator of innate immune signaling. TRAF7 interacts with TBK1 and promotes K48-linked polyubiquitination and degradation of TBK1 through its RING domain, impairing the activation of IRF3 and the production of IFN-β. In addition, we found that the conserved cysteine residues at position 131 of TRAF7 are necessary for its function toward TBK1. Knockout of TRAF7 could facilitate the activation of IRF3 and increase the transcript levels of downstream antiviral genes. These data suggest that TRAF7 negatively regulates innate antiviral immunity by promoting the K48-linked ubiquitination of TBK1.
Keywords: TANK-Binding kinase 1 (TBK1), Type I interferon, TRAF7, Ubiquitination, Innate immunity
Highlights
-
•
TRAF7 is a negative regulator of RLR signaling.
-
•
TRAF7 promotes the K48-linked polyubiquitination of TBK1 to inhibit the cellular antiviral response.
-
•
TRAF7's RING domain is necessary for its function in antiviral signaling.
1. Introduction
The innate immune system relies on pattern-recognition receptors (PRRs) to recognize pathogen-associated molecular patterns (Akira et al., 2006; Takeuchi and Akira, 2010) and initiate intracellular and intercellular signaling pathways to remove invading pathogens. Several types of PRRs (such as RIG-I-like receptors (RLRs) (Takeuchi and Akira, 2009), Toll-like receptors (TLRs) (Kawai and Akira, 2008, 2009), NOD-like receptors (NLRs) (Kawai and Akira, 2009), and DNA sensors (Barber, 2011) have been identified to detect invading pathogenic molecules and initiate the type I IFN signaling. Among them, RIG-I recognizes RNA viruses, resulting in conformational changes and exposure of the N-terminal CARD domain (Hu and Shu, 2018). Subsequently, RIG-I interacts with the mitochondrial antiviral protein MAVS (also known as VISA, ISP-I, and CARDIF) via the CARD structural domain (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). Activation of MAVS forms a giant prion-like complex that recruits downstream signaling molecules, including TRAFs, TBK1, and IKK complex (Hou et al., 2011). Next, TBK1 activates transcription factors IRF3/7 and NF-κB to produce type I interferon and proinflammatory cytokines (Tan et al., 2018). Type I interferon further activates the expression of a series of interferon-stimulated genes to initiate adaptive immunity (Ivashkiv and Donlin, 2014).
TBK1, a serine/threonine kinase, involved in multiple signal transductions that belongs to the noncanonical IKK family, similar to IκB kinase ε (IKKε, also known as IKKi). It plays an essential role in regulating the interferon response (Fitzgerald et al., 2003). Multiple pattern recognition receptors (PRRs) activate downstream signaling via TBK1 after virus or bacterial infection, such as TLR3/4-TRIF-TBK1, RIG-I/MDA5-MAVS-TBK1 and cGAS-STING-TBK1 (Tan et al., 2018). Moreover, many studies have shown that TBK1 also plays essential roles in multiple signal transduction pathways. TBK1 can regulate adipose tissue metabolism and inflammation, thus affecting the metabolic balance of glucose and energy (Oral et al., 2017; Zhao et al., 2018). TBK1 phosphorylates and matures optineurin (OPTN) to mediate the xenophagy of damaged mitochondria and pathogens (Pilli et al., 2012; Richter et al., 2016). It has been reported that GSK3β promoted TBK1 self-association and autophosphorylation at Ser172 (Lei et al., 2010). In addition, TBK1 is an essential kinase involved in mitosis, which provides a target for cancer treatment (Pillai et al., 2015). Zhu et al. proposed that TBK1 promotes KRAS-driven tumorigenesis by regulating CCL5 and IL-6 (Zhu et al., 2014). In the non-classical NF-κB signaling pathway, TBK1 promotes the phosphorylation of NIK, and this phosphorylation site, Ser862, is located in the degradation-determining region of NIK. Its phosphorylation promotes the degradation of NIK, which inhibits the activation of the non-classical NF-κB signaling pathway (Cildir et al., 2016). Furthermore, posttranscriptional modifications (including phosphorylation, ubiquitination, acetylation, etc.) of TBK1 are essential for its function. The phosphorylation modification of TBK1 is closely related to its activation level. Phosphorylated TBK1 can activate more TBK1 by trans-auto-activation, thus achieving a cascade amplification of activation (Ma et al., 2012). Acetylation modification of TBK1 also plays a vital role in its activation. Histone deacetylase 9 (HDAC9) can remove the acetylation modification of the lysine residue at position 241 of TBK1, thereby activating TBK1 and its downstream signaling pathway (Li et al., 2016). Besides, ubiquitination is the leading way to regulate the activity of TBK1. DYRK2 could promote the K48-linked ubiquitination and degradation of TBK1 in a kinase-activity-dependent manner (An et al., 2015). NLRP4 has been reported to promote ubiquitination and degradation of TBK1 via the E3 ligase DTX4 (Cui et al., 2012), and Zhang et al. proposed that TRIP promotes ubiquitination and degradation of TBK1 to negatively regulate type I interferon (Zhang et al., 2012). Besides, USP24 promotes the immune evasion of EV71 by restricting the K63-linked polyubiquitination of TBK1(Zang et al., 2023). Recently, an increasing number of teams have conducted in-depth studies on TBK1 function since TBK1 acts as a nodal protein involved in a series of signaling pathways. Although multiple TBK1 regulators have been reported, the other potential mechanisms of TBK1 regulation need to be investigated to maintain intracellular homeostasis.
TRAF7 is a protein that expresses 670 amino acids with E3 ubiquitin ligase activity in its RING domain, capable of auto-ubiquitination in vitro (Bouwmeester et al., 2004). TRAF7 interacts with NEMO and p65 to promote its Lys-29-linked polyubiquitination, leading to targeting these two proteins to lysosomal degradative pathways, reducing the transcriptional activity of NF-κB (Zotti et al., 2011). In addition, TRAF7 cooperates with TRAF6 in the CYLD-mediated inhibition of TLR2-dependent activation signaling of NF-κB (Yoshida et al., 2005). TRAF7 functions in hepatocellular carcinoma and meningioma through its N-terminal interactions and ubiquitin-mediated degradation of KLF4 (He et al., 2020; Dogan et al., 2022). In addition, TRAF7 promotes cell death by promoting K29 ubiquitination of c-FLIP, mediating cells undergoing lysosomal degradation (Scudiero et al., 2012). Wang and Zhang et al. proposed that TRAF7 plays important role in the development of breast cancer and hepatocellular carcinoma by facilitating the K48-linked polyubiquitination of p53 through its RING domain (Wang et al., 2013; Zhang et al., 2021). What's more, TRAF7 possesses SUMO E3 ligase activity. Morita et al. have reported that TRAF7 inhibits c-Myb-induced trans-activation by binding to the DNA binding domain (DBD) of c-Myb through the WD40 repeat domain, which activates SUMO of c-Myb at the Lys-523 and Lys-499 sites (Morita et al., 2005). Wang et al. propose miR126 inhibits apoptosis by reducing TRAF7 expression and ROS formation, suggesting that TRAF7 may be a potential target for preventing and treating vascular diseases such as atherosclerosis (Wang et al., 2015). Our previous studies revealed that TRAF7 potentiated MEKK3-mediated AP1 and CHOP activation (Xu et al., 2004).
TRAF2,3,5,6 are critical signaling molecules in the RLR antiviral signaling pathway. TRAF7, like other TRAFs, contains a RING finger domain (aa 125–160) and an adjacent zinc finger domain (aa 221–287) at its N-terminal end (Zotti et al., 2012). Unlike other TRAFs, TRAF7 contains seven WD40 repeats at its C-terminal. However, it is not clear whether TRAF7 is involved in antiviral signaling. In this study, we found that TRAF7 interacts with TBK1 and promotes K48 ubiquitination of TBK1, negatively regulating the RLR antiviral signaling pathway. Our results provide new insights into the regulation of innate antiviral immunity and expand our understanding of TBK1's post-translational modifications.
2. Materials and methods
2.1. Cell culture and virus
HEK 293, MCF7, and Vero cells (provided by Dr. Hong-Bing Shu, Wuhan University, China) were cultured in Dulbecco's Modified Eagle's Medium, which contains 10% fetal bovine serum and 1% penicillin-streptomycin, and incubated at 37 °C with 5% carbon dioxide. Cells were infected with Sendai virus (SeV) or Vesicular Stomatitis Virus (VSV) for the indicated time. All cell lines were tested for mycoplasma contamination with LookOut® Mycoplasma PCR Detection Kit from Sigma-Aldrich (MO, USA).
2.2. Antibodies, reagents, and plasmids
Mouse monoclonal antibodies (Flag: F3165 and HA: H3663) were purchased from Sigma-Aldrich (MO, USA). Anti-IRF3 (#4302S), anti-P65 (#8242), and anti-phosphorylated P65 (#3033) were purchased from Cell Signaling Technology (MA, USA). Anti-Myc (sc-40), anti-β actin (sc-1616), anti-β tubulin (sc-55529), and anti-LAMB1 (sc-56144) were purchased from Santa Cruz Biotechnology (TX, USA). Anti-TRAF7 (11780-1-AP) and anti-TBK1 (67211-1-Ig) were purchased from Proteintech (Wuhan, China). Anti-TBK1 (#3504) was purchased from Cell Signaling Technology. Alexa Fluor 647-labeled anti-mouse (A0473), Alexa Fluor 488-labeled anti-rabbit (A0423), and HA tag Rabbit Monoclonal Antibody (AF2305) were purchased from Beyotime (Suzhou, China). SeV and VSV were provided by Dr. Hong-Bing Shu (Wuhan University, China). The mammalian expression plasmids used in this study were previously described (Xu et al., 2004; Ling et al., 2018).
2.3. Dual-luciferase reporter assay and transfection
These experiments were consistent with previous descriptions (He et al., 2018; Huang et al., 2022). In brief, HEK 293 cells or TRAF7-deficient cells were plated in 24-well plates and transfected with plasmids containing the IFN-β promoter or ISRE luciferase reporter gene and URL-TK using the calcium phosphate method with a dose of TRAF7 or RIG-I/VISA signaling pathway components. The cells were subsequently analyzed after viral infection for the indicated time.
2.4. Coimmunoprecipitation, western blotting, and native PAGE
These assays were performed as previously described (Chen et al., 2018; Ling et al., 2018). In brief, cells were lysed in NP-40 lysis buffer supplemented with 1 mmol/L EDTA, 150 mmol/L NaCl, 20 mmol/L Tris-base, 1% NP-40, and 1% protease and phosphatase inhibitor cocktail. For immunoprecipitation assays, the lysates were immunoprecipitated with IgG or the appropriate antibodies, and the precipitates were washed two times with lysis buffer containing 0.5 mol/L NaCl. For Western blotting, protein samples were fractionated by SDS-PAGE, and analysis was performed with the indicated antibodies (mouse anti-Flag/HA, 1:4000 dilution, Sigma; rabbit anti-IRF3/P65/p-P65, 1:1000 dilution, CST; mouse anti-Myc/β actin/β tubulin/LAMB1, 1:1000 dilution, Santa Cruz; rabbit anti-TBK1, 1:1000 dilution, CST; rabbit anti-TRAF7, 1:500 dilution, Proteintech). For native PAGE, cell lysates were diluted with 5 × sample buffer, and the samples were performed with a 7.5% acrylamide gel without SDS for analysis.
2.5. CRISPR-Cas9 gene editing, quantitative RT‒PCR and ELISA
The lentil-CRISPR-V2 vector was used to clone double-stranded oligonucleotides of human TRAF7, which were then transfected into HEK 293 cells with psPAX2 and pMD2G. The supernatants were collected after 36 h, and HEK 293 cells were infected with the supernatants. Cells were screened with puromycin (1 μg/mL) for seven days, followed by sorting in 96-well plates to obtain TRAF7-deficient monoclonal cells. The gRNA sequence used for targeting TRAF7 was 5′-GCTACAACCGCTTCTCCGGG-3′. Total RNA extraction and reverse transcription were performed according to the Promega protocols. The qPCR primer sequences were consistent as previously described (Huang et al., 2022). HEK 293 cells and TRAF7-deficient cells were stimulated with viruses for 12 h, and the culture media were collected for measurement of IFN-β by ELISA.
2.6. Immunofluorescence and plaque assays
MCF7 cells were grown on 24-well plates and the mammalian expression vectors HA-TRAF7 and Flag-TBK1 were transfected into cells with Lipofectamine™ 3000 transfection reagent for 36 h. Cells were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 10 min, and blocked with 1% BSA for 60 min. Subsequently, the cells were incubated with mouse anti-Flag and rabbit anti-HA for 60 min at room temperature. Then, the cells were incubated with Alexa Fluor 647-labeled anti-mouse and Alexa Fluor 488-labeled anti-rabbit antibodies. After 60 min, nuclei were stained with 10 μg/mL DAPI at RT for 10 min. Finally, the cells were analyzed with a Leica DMi8 confocal microscope. For plaque assays, HEK 293 cells or TRAF7-deficient cells were plated in 6-well plates, and TRAF7 plasmids were transfected into cells or not for 12 h. Then, the cells were infected with VSV (MOI = 0.2) for 16 h before collecting the supernatant. Viral titers were subsequently calculated by infecting Vero cells with the supernatant.
2.7. Yeast two-hybrid system
Full-length TBK1 protein was used as bait to screen for candidate proteins interacting with TBK1 in a cDNA expression library of 293 cells by a yeast two-hybrid system (Clontech). The screening method was performed as previously described (He et al., 2019).
2.8. Statistical analysis
All statistical analyses were performed with GraphPad Prism 8.0, and are presented as the mean and standard deviation of at least three independent experiments. The Student's t-test was performed to compare the statistical significance of differences between the two groups, and the one-way analysis of variance with Tukey's post hoc analysis was used for multiple comparisons (n ≥ 3). P < 0.05 was considered statistically significant.
3. Results
3.1. TRAF7 interacts with TBK1 specifically
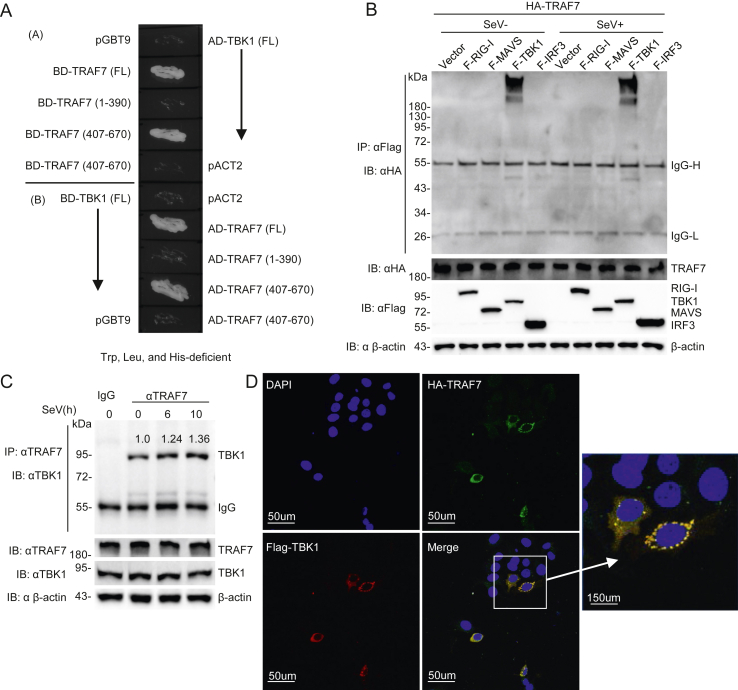
To investigate the regulatory mechanism of TBK1, a yeast two-hybrid assay was performed to screen the HEK 293 cell library using TBK1 as bait. Multiple TBK1-interacting candidate genes were obtained, including TRAF7. To further confirm the interaction of TBK1 with its candidate prey TRAF7, we constructed TRAF7 and its truncations TRAF7 (1–390), TRAF7 (407–670) into the pGBT9 expression vector and pACT2 expression vector, respectively. As shown in Fig. 1A, we co-introduced different pGBT9 and pACT2 expression vectors into yeast strain AH109 and observed whether the transformants could grow on Trp, Leu, and His-deficient plates. The results showed that the full length of TRAF7 and its C-terminal WD40 repeat domain (407–670) interacted with TBK1. Transient transfer and immunoprecipitation results demonstrated that TRAF7 specifically interacts with TBK1, but not with RIG-I, MAVS, and IRF3 in HEK 293 cells (Fig. 1B). The endogenous co-immunoprecipitation assay in HEK 293 cells consistently showed that the interaction between TRAF7 and TBK1 was enhanced upon Sendai virus (SeV) infection (Fig. 1C). Further immunofluorescence experiments in MCF7 cells validated that TRAF7 was colocalized with TBK1 (Fig. 1D). These results indicate that TRAF7 specifically interacts with TBK1 and may play a role in TBK1-mediated antiviral signaling.
Fig. 1.
TRAF7 associates with TBK1. A TRAF7 associated with TBK1 in a yeast two-hybrid system. DNA binding domain fused with TRAF7 (Full length, 1–390, or 407–670) and activation domain fused to TBK1 were co-expressed in AH109 yeast, and DNA binding domain fused with TBK1 and activation domain fused to TRAF7 (Full length, 1–390, or 407–670) were co-expressed in AH109 yeast. B TRAF7 associates with TBK1. HEK 293 cells (∼2 × 105) were transfected with the indicated plasmids. Cells were treated or untreated with SeV (MOI = 1) for 10 h before Co-IP and immunoblot analysis. C Endogenous TRAF7 is associated with TBK1. HEK 293 cells (4 × 106) were infected with SeV (MOI = 1) for indicated times or left untreated before endogenous co-immunoprecipitation and Western blot analysis. D TRAF7 is colocalized with TBK1. MCF7 cells were incubated with anti-HA (green) and anti-Flag (red) antibodies. Nuclei were incubated with DAPI (10 μg/mL). Images were acquired with a Leica DMI8 (The size bar represents 50 μm).
3.2. TRAF7 negatively regulates the RLR-mediated antiviral response
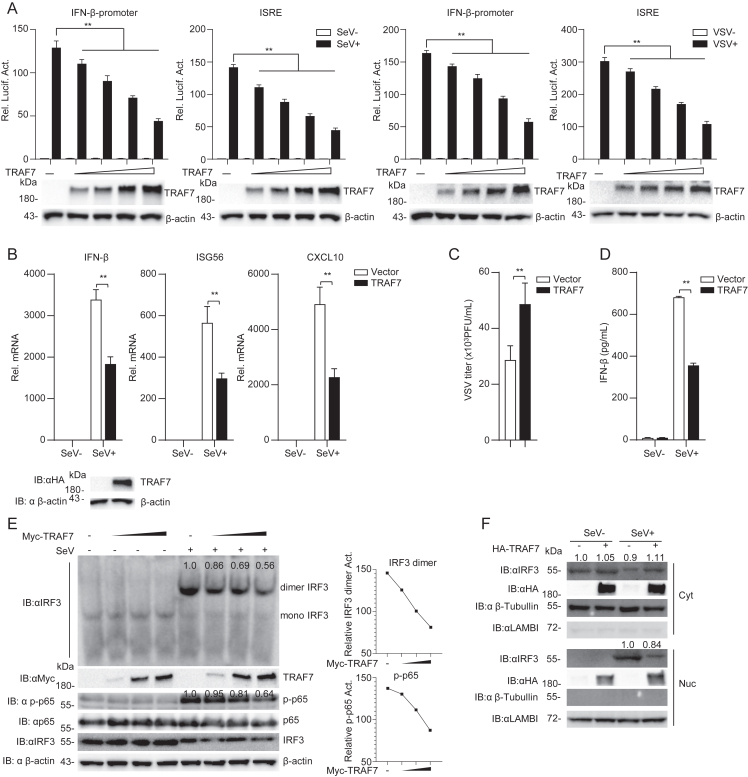
The role of TRAF7 in RNA virus-triggered innate immunity was further investigated. The dual luciferase reporter assays showed that TRAF7 overexpression in HEK 293 cells inhibited SeV and VSV-induced activation of the IFN-β promoter and ISRE in a dose-dependent manner (Fig. 2A). qPCR assays indicated that overexpression of TRAF7 in HEK 293 cells suppressed the transcription level of downstream antiviral genes, including IFN-β, ISG56, and CXCL10, induced by RNA virus (Fig. 2B). Plaque assays showed that TRAF7 overexpression in HEK 293 cells increased VSV titers (Fig. 2C). Furthermore, overexpression of TRAF7 in HEK 293 cells inhibited the production of IFN-β protein (Fig. 2D), the dimerization of IRF3 and the phosphorylation of p65 induced by SeV (Fig. 2E). Further experiments indicated that TRAF7 overexpression suppressed the nuclear translocation of IRF3 (Fig. 2F). These data suggest that TRAF7 is a negative regulator in RLR-mediated signaling.
Fig. 2.
TRAF7 inhibits RLR-mediated innate immunity. A TRAF7 inhibits the activation of the IFN-β promoter and ISRE. HEK 293 cells (1 × 105) were transfected with TRAF7 plasmids, pRL-TK (0.025 μg), and IFN-β promoter (0.025 μg) or ISRE (0.025 μg). After 12 h, the cells were treated with SeV (MOI = 1) for 12 h or left untreated before luciferase analysis. B TRAF7 impairs the transcript levels of downstream antiviral genes and enhances the viral replication ability. HEK 293 cells (2 × 105) were transfected with an empty vector or TRAF7. Cells were treated or untreated with SeV for 12 h before qPCR analysis. C TRAF7 enhances the replication of VSV, HEK 293 cells were transfected with an empty vector or TRAF7 and infected with VSV (MOI = 0.2) for 16 h before collecting the supernatants to infect Vero cells. D TRAF7 reduces the production of IFN-β protein. ELISA of IFN-β in HEK 293 cells transfected for 12 h with TRAF7, followed by infection for 12 h with SeV. E TRAF7 damages the dimerization of IRF3. HEK 293 cells (2 × 105) were transfected with an empty vector or TRAF7. After 12 h, the cells were treated or untreated with SeV for 12 h before immunoblotting analysis. F TRAF7 inhibits the nuclear translocation of IRF3. HEK 293 cells (2 × 106) were transfected with empty vector or TRAF7 plasmids; cells were infected with SeV 0 or 12 h before immunoblot analysis of IRF3 in cytoplasmic (Cyt) and nuclear (Nuc) fractions. (∗∗P < 0.01).
3.3. Knockout of TRAF7 facilitates RNA virus-triggered signaling
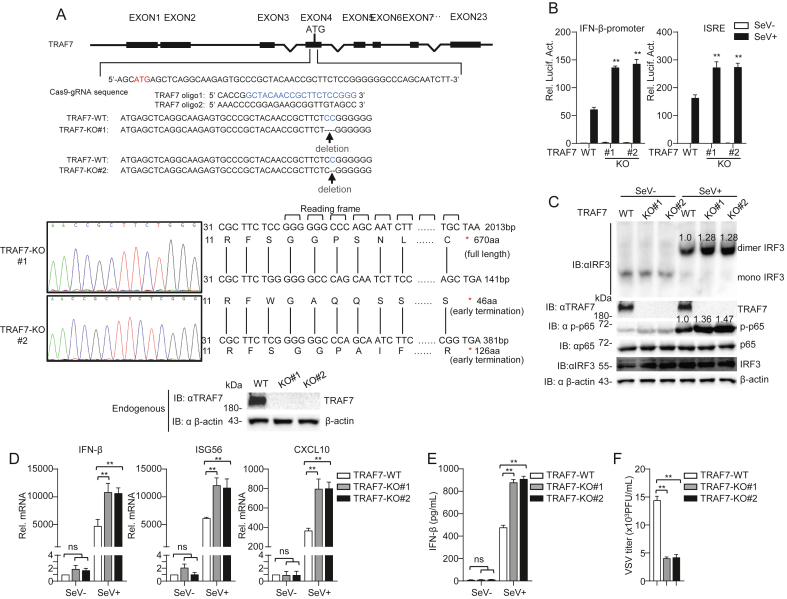
To further explore the function of TRAF7 in RLR-mediated antiviral innate immunity, TRAF7-deficient single clones of HEK 293 cells were obtained using CRISPR/Cas9-mediated genome editing. Sequence analysis revealed TRAF7 genomic base deletions leading to amino acid shift mutations and early termination, and Western blotting experiments showed that endogenous TRAF7 was successfully knockout (Fig. 3A). The dual luciferase reporter assays indicated that the activation of the IFN-β promoter and ISRE was increased in TRAF7-deficient cells (Fig. 3B). In addition, TRAF7 knockout enhanced the dimerization of IRF3 and the phosphorylation of p65 after SeV infection (Fig. 3C). Furthermore, qPCR analysis suggested that the transcription levels of IFN-β, ISG56, and CXCL10 induced by Sendai virus were enhanced in TRAF7-deficient HEK 293 cells (Fig. 3D), and knockout of TRAF7 in HEK 293 cells enhanced the production of IFN-β protein (Fig. 3E). As shown in Fig. 3F, the plaque assay indicated that VSV titers were reduced in TRAF7-deficient HEK 293 cells. These results suggest that TRAF7 deficiency enhances RLR-mediated antiviral signaling.
Fig. 3.
TRAF7 deficiency inhibits RNA virus-mediated signaling. A Knockout efficiencies of TRAF7. A strategy for CRISPR/Cas9-mediated genome editing of TRAF7. The deletions were confirmed by immunoblot analysis and DNA sequence analysis of genomic DNA isolated from TRAF7 wild-type and deletion cell lines. B TRAF7 deficiency facilitates the activation of the IFN-β promoter and ISRE. HEK 293 wild-type and TRAF7-deficient cells (1 × 105) were transfected with pRL-TK (0.025 μg) and IFN-β promoter (0.025 μg) or ISRE (0.025 μg). After 12 h, the cells were treated with SeV (MOI = 1) for 12 h or left untreated before luciferase analysis. C TRAF7 deficiency enhances the dimerization of IRF3. HEK 293 wild-type and TRAF7-deficient cells (2 × 106) were treated with SeV (MOI = 1) for 12 h or untreated before immunoblotting analysis. D Knockout of TRAF7 improves the transcript levels of downstream antiviral genes. HEK 293 wild-type and TRAF7-deficient cells (2 × 106) were treated with SeV (MOI = 1) for 12 h or untreated before qPCR analysis. E Knockout TRAF7 improves the production of IFN-β protein. ELISA of IFN-β in HEK 293 wild-type and TRAF7-deficient cells were infected with SeV for 12 h. F Knockout TRAF7 impairs the ability of viral replication. HEK 293 wild-type and TRAF7-deficient cells were infected or uninfected with VSV (MOI = 0.2) for 16 h before collecting the supernatants to infect Vero cells. (∗∗P < 0.01; ns, no significant difference).
3.4. TRAF7 impairs TBK1-mediated type-I IFN signaling
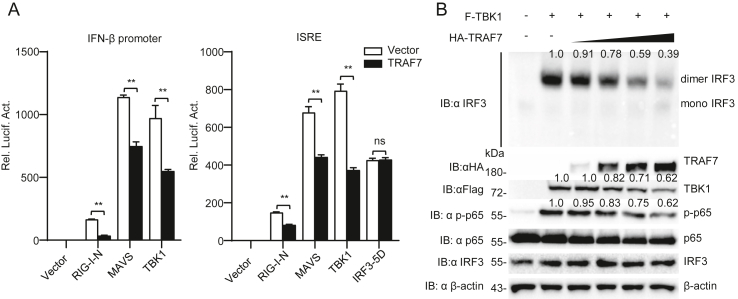
We performed RLR pathway component-mediated reporter gene experiments to further examine the specific site of TRAF7 function in the antiviral signaling pathway. As shown in Fig. 4A, the results indicated that overexpression of TRAF7 impaired the activation of the IFN-β promoter and ISRE mediated by RIG-I-N, VISA, and TBK1. In contrast, it did not affect the activation of ISRE mediated by IRF3-5D. Subsequent experiments showed that overexpression of TRAF7 reduced the dimerization of IRF3 and the phosphorylation of p65 induced by TBK1 (Fig. 4B). These data suggest that TRAF7 negatively regulates RNA virus-triggered innate immunity by targeting TBK1.
Fig. 4.
TRAF7 negatively regulates RLR-mediated signaling by targeting TBK1. A TRAF7 inhibits the activation of IFN-β promoter and ISRE by targeting TBK1. HEK 293 cells (1 × 105) were transfected with the indicated plasmids for 20 h before luciferase analysis. B TRAF7 impairs the TBK1-mediated dimerization of IRF3. HEK 293 cells (2 × 105) were transfected with TRAF7 (dose) and TBK1 for 20 h before immunoblotting analysis. (∗∗P < 0.01; ns, no significant difference).
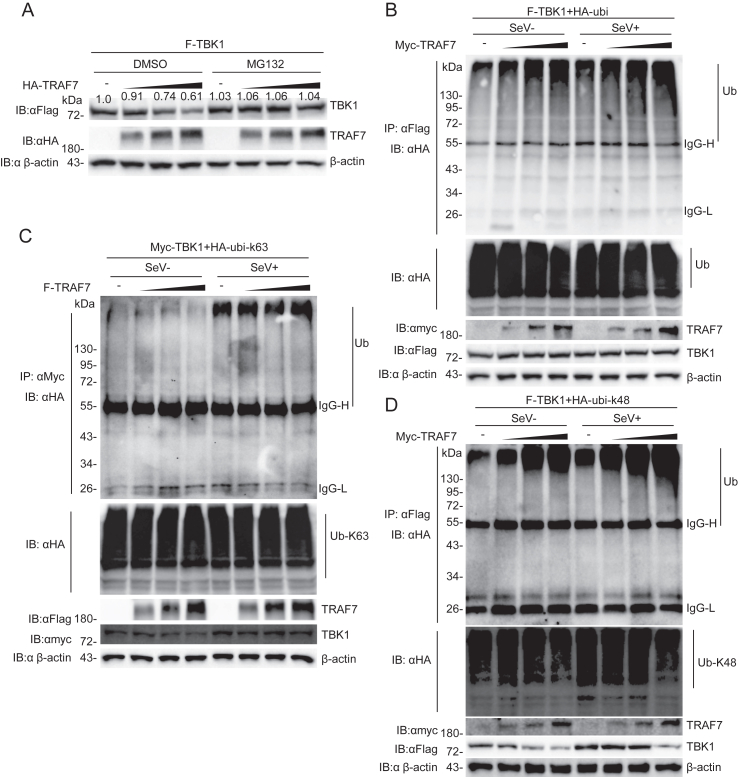
3.5. TRAF7 enhances K48-linked polyubiquitination of TBK1
It has been reported that TRAF7 is a RING-type E3 ubiquitin ligase that has autoubiquitination activity (Xu et al., 2004). As shown in Fig. 4B, TRAF7 inhibited the protein level of TBK1 in a dose-dependent manner. Combined with the ubiquitin ligase activity of TRAF7, we hypothesized that TRAF7 promotes K48 ubiquitination of TBK1 to mediate its degradation. To verify this hypothesis, TRAF7 and TBK1 were cotransfected into HEK 293 cells treated with DMSO or MG132. The results indicated that TRAF7 could enhance the degradation of TBK1 through the proteasome pathway (Fig. 5A). To further investigate the effect of TRAF7 on ubiquitination modifications of TBK1. Coimmunoprecipitation experiments were performed, and the results showed that the overexpression of TRAF7 in HEK 293 cells enhanced K48-linked ubiquitination but had no effect on K63-linked ubiquitination of TBK1 (Fig. 5B–D). These results suggest that TRAF7 promotes K48 ubiquitination of TBK1 to negatively regulate innate immune signaling.
Fig. 5.
TRAF7 promotes the K48-linked ubiquitination of TBK1. A TRAF7 facilitates the proteasomal degradation of TBK1. HEK 293 cells (2 × 105) were transfected with TBK1 and TRAF7 (dose), and the cells were incubated in the presence or absence of MG132 (5 μmol/L) for 10 h. B TRAF7 promotes the ubiquitination of TBK1. HEK 293 cells (2 × 105) were transfected with the indicated plasmids and cells were treated with MG132 (5 μmol/L); after 12 h, the cells were infected or uninfected with SeV (MOI = 1) for 10 h before Co-IP and immunoblotting analysis. C and D TRAF7 promotes the K48-linked ubiquitination of TBK1. HEK 293 cells (2 × 105) were transfected with the indicated plasmids; after 12 h, the cells were infected or uninfected with SeV (MOI = 1) for 10 h before Co-IP and immunoblotting analysis.
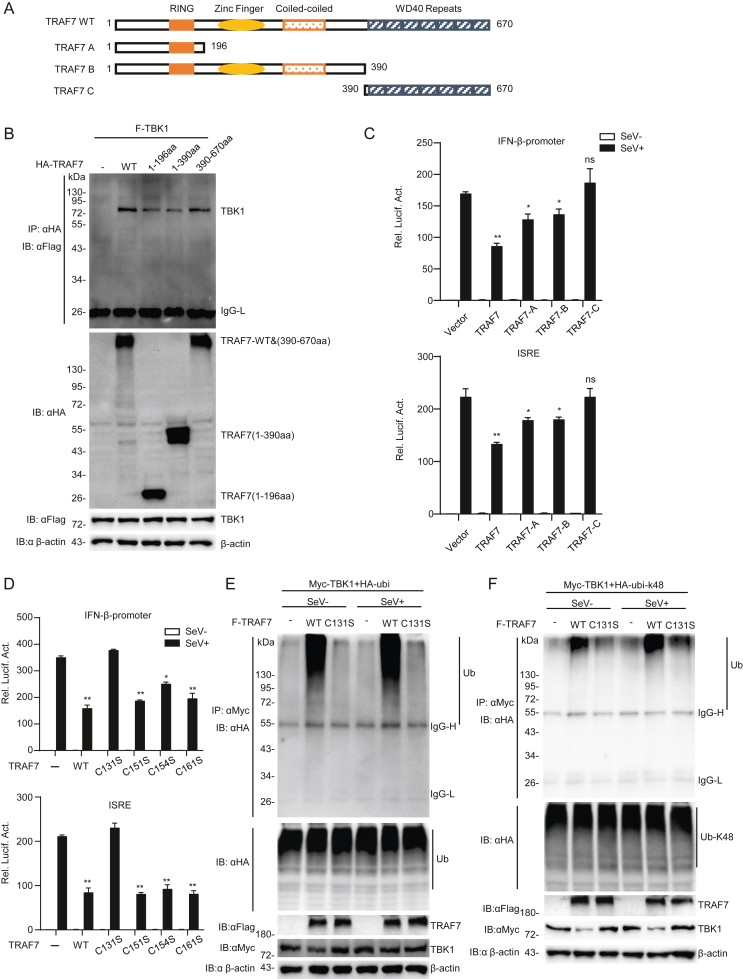
3.6. The RING domain is required for TRAF7 functions in antiviral signaling
To further investigate the domains responsible for TRAF7 inhibiting innate immune responses, a series of deletion mutants of TRAF7 were constructed (Fig. 6A). Then, these TRAF7 mutants were analyzed for TBK1 by co-immunoprecipitation. The results showed that both the RING domain and WD40 repeat domain of TRAF7 were required for its interaction with TBK1 (Fig. 6B). Subsequent reporter gene assays indicated that the RING domain was necessary for TRAF7 to inhibit the activation of the IFN-β-promoter and ISRE after SeV infection (Fig. 6C). To further explore the specific site of TRAF7 function on TBK1 ubiquitination modification, we constructed TRAF7 (C131S, C151S, C154S, C161S), in which the conserved cysteine residues at positions 131, 151, 154, and 161 within the RING domain were replaced with serine independently. Subsequent reporter gene assays indicated that the conserved cysteine residue at position 131 within the RING domain was required for TRAF7 to inhibit the activation of the IFN-β promoter and ISRE after SeV infection (Fig. 6D). We then performed ubiquitination experiments and found that overexpression of TRAF7 (C131S) did not affect WT TBK1 or K48-linked ubiquitination of TBK1 in HEK 293 cells (Fig. 6E and F). These results demonstrated that TRAF7 inhibits innate immune signaling by facilitating the K48-linked ubiquitination of TBK1 through conserved cysteine residues at position 131 in TRAF7's RING domain, leading to TBK1's proteasomal degradation.
Fig. 6.
The RING domain is necessary for TRAF7 functions. A A schematic diagram of wild-type and truncated human TRAF7 mutants. B TRAF7 interacts with TBK1 through its RING domain and WD40 repeat domain. HEK 293 cells (2 × 105) were transfected with TBK1 and TRAF7 deletion mutants for 24 h and treated with MG132 before Co-IP and immunoblotting analysis. C TRAF7 relies on the RING domain to repress the activation of reporter genes. HEK 293 cells (2 × 105) were transfected with the indicated plasmids, and after 12 h, cells were treated or untreated with SeV (MOI = 1) for 12 h before luciferase analysis. D The conserved cysteine residues at position 131 of TRAF7 are necessary for TRAF7's RING domain functions. HEK 293 cells (2 × 105) were transfected with the indicated plasmids, and after 12 h, cells were treated or untreated with SeV (MOI = 1) for 12 h before luciferase analysis. E and F The conserved cysteine residues at position 131 of TRAF7 are necessary for its E3 ligase activity. HEK 293 cells (2 × 105) were transfected with the indicated plasmids; after 12 h, the cells were infected or uninfected with SeV (MOI = 1) for 10 h before Co-IP and immunoblotting analysis. (∗P < 0.05; ∗∗P < 0.01; ns, no significant difference).
4. Discussion
In this study, we demonstrated a novel regulatory mechanism of TRAF7 in innate immunity. Our results showed that TRAF7 overexpression inhibited IFN-β promoter and ISRE activation, IRF3 dimerization and nuclear translocation, and the production of IFN-β protein. Knockout of TRAF7 had the opposite effect, which enhanced the transcript levels of downstream antiviral genes and improved interferon production. In addition, TRAF7, which interacted specifically with TBK1, promoted the K48-linked ubiquitination and degradation of TBK1. These results suggest that TRAF7 negatively regulates RNA virus-mediated antiviral innate immunity by promoting K48-linked ubiquitination of TBK1.
Recently, TBK1 has become a hot spot for research due to its role as a nodal protein in various signaling pathways (Runde et al., 2022). Posttranslational modifications of TBK1 are critical for its function. One of the essential modifications of TBK1 is ubiquitination, but there are still a few known E3 ligases that regulate TBK1. RNF128 facilitates innate immune signaling by promoting the K63-linked ubiquitination of TBK1 (Song et al., 2016). Parkin promotes mitochondrial autophagy by enhancing the K63 ubiquitination of TBK1 (Gao et al., 2021). USP15 inhibits type I interferon production by removing the ubiquitination of TBK1 (Huang et al., 2020). Cui and Zhang et al. proposed that the E3 ligases DTX4 and TRIP negatively regulate type I interferon signaling by facilitating the K48-linked ubiquitination and degradation of TBK1 (Cui et al., 2012; Zhang et al., 2012). Our study revealed that TRAF7 interacts with TBK1 specifically.
We have previously reported that TRAF7 interacts with MEKK3 through the WD40 repeat domain to activate AP-1 signaling (Xu et al., 2004). Bouwmeester et al. reported that the RING domain of TRAF7 has E3 ligase activity (Bouwmeester et al., 2004). Our experiments show that TRAF7 promotes ubiquitination of TBK1 mainly by K48-linked ubiquitination, which is associated with proteasomal degradation, and that TRAF7 weakly promotes ubiquitination of K6, 11, 27, 29 linkages (data not shown), and according to the experimental results in Fig. 6B and C, we speculate that TRAF7 depends on RING domain to inhibit RNA virus-induced innate immune responses, while its WD40 repeat domain interacting with TBK1 may provide support for its function, but is not necessary. Further data suggest that the conserved cysteine residues at position 131 of TRAF7 are necessary for its E3 ligase activity. TRAF7 loses its ability to ubiquitinate TBK1 after cysteine mutation at position 131. However, further experiments are needed to explore the specific site of TBK1 ubiquitination by TRAF7.
We found that TRAF7 was present in both the cytoplasm and nucleus and that the nuclear translocation of TRAF7 increased after SeV infection. This is consistent with what Morita et al. have reported (Morita et al., 2005). Furthermore, we found that overexpression of TRAF7 inhibits the phosphorylation of p65, which is consistent with previous reports (Zotti et al., 2011). TBK1 is known to be involved in metabolic signaling (Xiang et al., 2021), such as adipose metabolism (Beyett et al., 2018), while TRAF7 is involved in multiple tumor signal transduction (Zotti et al., 2017), including meningiomas (Dogan et al., 2022). We propose that TRAF7 may be involved in various metabolic regulatory mechanisms through TBK1, but further studies on the regulatory mechanism of TRAF7 are needed.
5. Conclusions
In summary, our study demonstrates that TRAF7 is a novel E3 ligase that negatively regulates RLR-mediated innate immunity by promoting K48-linked ubiquitination of TBK1 through its RING domain to regulate signal transduction. Those results suggest TRAF7 regulates TBK1 signaling.
Data availability
All the data generated during the current study are included in the manuscript.
Ethics statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author contributions
Liang-Guo Xu: conceptualization, funding acquisition, project administration, resources, and writing - review & editing; Jing-Ping Huang: formal analysis, investigation, software, validation, and writing-original draft; Ya-Xian Yang: investigation and validation; Tian Chen: validation; Dan-Dan Wang: validation; Jing Li: validation.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant Nos. 81971502, 82060298, 31570876).
We sincerely appreciate Dr. Hong-Bing Shu (Wuhan University, Wuhan, China) for providing plasmids and reagents. We thank Jiangxi Normal University, College of Chemistry and Chemical Engineering, for help with the laser confocal microscope analysis.
References
- Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- An T., Li S., Pan W., Tien P., Zhong B., Shu H.B., Wu S. DYRK2 negatively regulates type I interferon induction by promoting TBK1 degradation via Ser527 phosphorylation. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber G.N. Innate immune DNA sensing pathways: STING, AIMII and the regulation of interferon production and inflammatory responses. Curr. Opin. Immunol. 2011;23:10–20. doi: 10.1016/j.coi.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyett T.S., Gan X., Reilly S.M., Chang L., Gomez A.V., Saltiel A.R., Showalter H.D., Tesmer J.J.G. Carboxylic acid derivatives of amlexanox display enhanced potency toward TBK1 and IKKepsilon and reveal mechanisms for selective inhibition. Mol. Pharmacol. 2018;94:1210–1219. doi: 10.1124/mol.118.112185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester T., Bauch A., Ruffner H., Angrand P.O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., Hopf C., Huhse B., Mangano R., Michon A.M., Schirle M., Schlegl J., Schwab M., Stein M.A., Bauer A., Casari G., Drewes G., Gavin A.C., Jackson D.B., Joberty G., Neubauer G., Rick J., Kuster B., Superti-Furga G. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat. Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- Chen T., Wang D., Xie T., Xu L.G. Sec13 is a positive regulator of VISA-mediated antiviral signaling. Virus Gene. 2018;54:514–526. doi: 10.1007/s11262-018-1581-0. [DOI] [PubMed] [Google Scholar]
- Cildir G., Low K.C., Tergaonkar V. Noncanonical NF-kappa B signaling in health and disease. Trends Mol. Med. 2016;22:414–429. doi: 10.1016/j.molmed.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Cui J., Li Y., Zhu L., Liu D., Songyang Z., Wang H.Y., Wang R.F. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat. Immunol. 2012;13:387–395. doi: 10.1038/ni.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan H., Blume C., Patel A., Jungwirth G., Sogerer L., Ratliff M., Ketter R., Herold-Mende C., Jones D.T.W., Wick W., Vollmuth P., Zweckberger K., Reuss D., Von Deimling A., Sahm F. Single-cell DNA sequencing reveals order of mutational acquisition in TRAF7/AKT1 and TRAF7/KLF4 mutant meningiomas. Acta Neuropathol. 2022;144:799–802. doi: 10.1007/s00401-022-02485-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K.A., Mcwhirter S.M., Faia K.L., Rowe D.C., Latz E., Golenbock D.T., Coyle A.J., Liao S.M., Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- Gao B., Yu W., Lv P., Liang X., Sun S., Zhang Y. Parkin overexpression alleviates cardiac aging through facilitating K63-polyubiquitination of TBK1 to facilitate mitophagy. Biochim. Biophys. Acta, Mol. Basis Dis. 2021;1867 doi: 10.1016/j.bbadis.2020.165997. [DOI] [PubMed] [Google Scholar]
- He H., Wu Z., Li S., Chen K., Wang D., Zou H., Chen H., Li Y., Liu Z., Qu C. TRAF7 enhances ubiquitin-degradation of KLF4 to promote hepatocellular carcinoma progression. Cancer Lett. 2020;469:380–389. doi: 10.1016/j.canlet.2019.11.012. [DOI] [PubMed] [Google Scholar]
- He T.S., Chen T., Wang D.D., Xu L.G. HAUS8 regulates RLRVISA antiviral signaling positively by targeting VISA. Mol. Med. Rep. 2018;18:2458–2466. doi: 10.3892/mmr.2018.9171. [DOI] [PubMed] [Google Scholar]
- He T.S., Xie T., Li J., Yang Y.X., Li C.S., Wang W.Y., Cao L.Z., Rao H., Ju C., Xu L.G. THO Complex Subunit 7 Homolog Negatively Regulates Cellular Antiviral Response against RNA Viruses by Targeting TBK1. Viruses-Basel. 2019;11, 158 doi: 10.3390/v11020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou F., Sun L., Zheng H., Skaug B., Jiang Q.-X., Chen Z.J. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M.M., Shu H.B. Cytoplasmic mechanisms of recognition and defense of microbial nucleic acids. Annu. Rev. Cell Dev. Biol. 2018;34:357–379. doi: 10.1146/annurev-cellbio-100617-062903. [DOI] [PubMed] [Google Scholar]
- Huang J.P., Li J., Xiao Y.P., Xu L.G. BAG6 negatively regulates the RLR signaling pathway by targeting VISA/MAVS. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.972184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Liu H., Zhang K., Meng Q., Hu L., Zhang Y., Xiang Z., Li J., Yang Y., Chen Y., Cui S., Tang H., Pei H., Bu Z., Weng C. Ubiquitin-conjugating enzyme 2S enhances viral replication by inhibiting type I IFN production through recruiting USP15 to deubiquitinate TBK1. Cell Rep. 2020;32:108044. doi: 10.1016/j.celrep.2020.108044. [DOI] [PubMed] [Google Scholar]
- Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann. N. Y. Acad. Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009;21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K.J., Takeuchi O., Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Lei C.Q., Zhong B., Zhang Y., Zhang J., Wang S., Shu H.B. Glycogen synthase kinase 3beta regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity. 2010;33:878–889. doi: 10.1016/j.immuni.2010.11.021. [DOI] [PubMed] [Google Scholar]
- Li X., Zhang Q., Ding Y.Y., Liu Y.Q., Zhao D.Z., Zhao K., Shen Q.C., Liu X.G., Zhu X.H., Li N., Cheng Z.Y., Fan G.P., Wang Q.Q., Cao X.T. Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat. Immunol. 2016;17:806. doi: 10.1038/ni.3464. [DOI] [PubMed] [Google Scholar]
- Ling T., Li S.N., Weng G.X., Wang W., Li C., Cao L., Rao H., Shu H.B., Xu L.G. TARBP2 negatively regulates IFN-beta production and innate antiviral response by targeting MAVS. Mol. Immunol. 2018;104:1–10. doi: 10.1016/j.molimm.2018.10.017. [DOI] [PubMed] [Google Scholar]
- Ma X.L., Helgason E., Phung Q.T., Quan C.L., Iyer R.S., Lee M.W., Bowman K.K., Starovasnik M.A., Dueber E.C. Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9378–9383. doi: 10.1073/pnas.1121552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp R. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Morita Y., Kanei-Ishii C., Nomura T., Ishii S. TRAF7 sequesters c-Myb to the cytoplasm by stimulating its sumoylation. Mol. Biol. Cell. 2005;16:5433–5444. doi: 10.1091/mbc.E05-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oral E.A., Reilly S.M., Gomez A.V., Meral R., Butz L., Ajluni N., Chenevert T.L., Korytnaya E., Neidert A.H., Hench R., Rus D., Horowitz J.F., Poirier B., Zhao P., Lehmann K., Jain M., Yu R., Liddle C., Ahmadian M., Downes M., Evans R.M., Saltiel A.R. Inhibition of IKK 3 and TBK1 improves glucose control in a subset of patients with type 2 diabetes. Cell Metabol. 2017;26:157. doi: 10.1016/j.cmet.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai S., Nguyen J., Johnson J., Haura E., Coppola D., Chellappan S. Tank binding kinase 1 is a centrosome-associated kinase necessary for microtubule dynamics and mitosis. Nat. Commun. 2015;6 doi: 10.1038/ncomms10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilli M., Arko-Mensah J., Ponpuak M., Roberts E., Master S., Mandell M.A., Dupont N., Ornatowski W., Jiang S., Bradfute S.B., Bruun J.A., Hansen T.E., Johansen T., Deretic V. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–234. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter B., Sliter D.A., Herhaus L., Stolz A., Wang C., Beli P., Zaffagnini G., Wild P., Martens S., Wagner S.A., Youle R.J., Dikic I. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2016;113:4039–4044. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runde A.P., Mack R., S J.P., Zhang J. The role of TBK1 in cancer pathogenesis and anticancer immunity. J. Exp. Clin. Cancer Res. 2022;41:135. doi: 10.1186/s13046-022-02352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudiero I., Zotti T., Ferravante A., Vessichelli M., Reale C., Masone M.C., Leonardi A., Vito P., Stilo R. Tumor necrosis factor (TNF) receptor-associated factor 7 is required for TNFalpha-induced Jun NH2-terminal kinase activation and promotes cell death by regulating polyubiquitination and lysosomal degradation of c-FLIP protein. J. Biol. Chem. 2012;287:6053–6061. doi: 10.1074/jbc.M111.300137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R.B., Sun L.J., Ea C.K., Chen Z.J.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappa B and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Song G., Liu B., Li Z., Wu H., Wang P., Zhao K., Jiang G., Zhang L., Gao C. E3 ubiquitin ligase RNF128 promotes innate antiviral immunity through K63-linked ubiquitination of TBK1. Nat. Immunol. 2016;17:1342–1351. doi: 10.1038/ni.3588. [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. Innate immunity to virus infection. Immunol. Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tan X., Sun L., Chen J., Chen Z.J. Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol. 2018;72:447–478. doi: 10.1146/annurev-micro-102215-095605. [DOI] [PubMed] [Google Scholar]
- Wang L., Wang L., Zhang S., Qu G., Zhang D., Li S., Liu S. Downregulation of ubiquitin E3 ligase TNF receptor-associated factor 7 leads to stabilization of p53 in breast cancer. Oncol. Rep. 2013;29:283–287. doi: 10.3892/or.2012.2121. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang F., Wu Y., Zuo L., Zhang S., Zhou Q., Wei W., Wang Y., Zhu H. MicroRNA-126 attenuates palmitate-induced apoptosis by targeting TRAF7 in HUVECs. Mol. Cell. Biochem. 2015;399:123–130. doi: 10.1007/s11010-014-2239-4. [DOI] [PubMed] [Google Scholar]
- Xiang S., Song S., Tang H., Smaill J.B., Wang A., Xie H., Lu X. TANK-binding kinase 1 (TBK1): an emerging therapeutic target for drug discovery. Drug Discov. Today. 2021;26:2445–2455. doi: 10.1016/j.drudis.2021.05.016. [DOI] [PubMed] [Google Scholar]
- Xu L.G., Li L.Y., Shu H.B. TRAF7 potentiates MEKK3-induced AP1 and CHOP activation and induces apoptosis. J. Biol. Chem. 2004;279:17278–17282. doi: 10.1074/jbc.C400063200. [DOI] [PubMed] [Google Scholar]
- Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z.H., Shu H.B. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Jono H., Kai H., Li J.D. The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative cross-talk with TRAF6 AND TRAF7. J. Biol. Chem. 2005;280:41111–41121. doi: 10.1074/jbc.M509526200. [DOI] [PubMed] [Google Scholar]
- Zang L., Gu J., Yang X., Yuan Y., Guo H., Zhou W., Ma J., Chen Y., Wu Y., Zheng H., Shi W. Ubiquitin-specific protease 24 promotes EV71 infection by restricting K63-linked polyubiquitination of TBK1. Virol. Sin. 2023;38:75–83. doi: 10.1016/j.virs.2022.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Wang L., Zhao X., Zhao K., Meng H., Zhao W., Gao C. TRAF-interacting protein (TRIP) negatively regulates IFN-beta production and antiviral response by promoting proteasomal degradation of TANK-binding kinase 1. J. Exp. Med. 2012;209:1703–1711. doi: 10.1084/jem.20120024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zhang X., Dong W. TRAF7 contributes to tumor progression by promoting ubiquitin-proteasome mediated degradation of P53 in hepatocellular carcinoma. Cell Death Dis. 2021;7:352. doi: 10.1038/s41420-021-00749-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Wong K.I., Sun X.L., Reilly S.M., Uhm M., Liao Z.J., Skorobogatko Y., Saltiel A.R. TBK1 at the crossroads of inflammation and energy homeostasis in adipose tissue. Cell. 2018;172:731–743. doi: 10.1016/j.cell.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Aref A.R., Cohoon T.J., Barbie T.U., Imamura Y., Yang S., Moody S.E., Shen R.R., Schinzel A.C., Thai T.C., Reibel J.B., Tamayo P., Godfrey J.T., Qian Z.R., Page A.N., Maciag K., Chan E.M., Silkworth W., Labowsky M.T., Rozhansky L., Mesirov J.P., Gillanders W.E., Ogino S., Hacohen N., Gaudet S., Eck M.J., Engelman J.A., Corcoran R.B., Wong K.K., Hahn W.C., Barbie D.A. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov. 2014;4:452–465. doi: 10.1158/2159-8290.CD-13-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotti T., Scudiero I., Vito P., Stilo R. The emerging role of TRAF7 in tumor development. J. Cell. Physiol. 2017;232:1233–1238. doi: 10.1002/jcp.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotti T., Uva A., Ferravante A., Vessichelli M., Scudiero I., Ceccarelli M., Vito P., Stilo R. TRAF7 protein promotes Lys-29-linked polyubiquitination of IkappaB kinase (IKKgamma)/NF-kappaB essential modulator (NEMO) and p65/RelA protein and represses NF-kappaB activation. J. Biol. Chem. 2011;286:22924–22933. doi: 10.1074/jbc.M110.215426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotti T., Vito P., Stilo R. The seventh ring: exploring TRAF7 functions. J. Cell. Physiol. 2012;227:1280–1284. doi: 10.1002/jcp.24011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data generated during the current study are included in the manuscript.