Abstract
Human respiratory syncytial virus (RSV) is a severe threat to children and a main cause of acute lower respiratory tract infections. Nevertheless, the intra-host evolution and inter-regional diffusion of RSV are little known. In this study, we performed a systematic surveillance in hospitalized children in Hubei during 2020–2021, in which 106 RSV-positive samples were detected both clinically and by metagenomic next generation sequencing (mNGS). RSV-A and RSV-B groups co-circulated during surveillance with RSV-B being predominant. About 46 high-quality genomes were used for further analyses. A total of 163 intra-host nucleotide variation (iSNV) sites distributed in 34 samples were detected, and glycoprotein (G) gene was the most enriched gene for iSNVs, with non-synonymous substitutions more than synonymous substitutions. Evolutionary dynamic analysis showed that the evolutionary rates of G and NS2 genes were higher, and the population size of RSV groups changed over time. We also found evidences of inter-regional diffusion from Europe and Oceania to Hubei for RSV-A and RSV-B, respectively. This study highlighted the intra-host and inter-host evolution of RSV, and provided some evidences for understanding the evolution of RSV.
Keywords: Respiratory syncytial virus (RSV), Intra-host single nucleotide variation (iSNV), Evolutionary dynamic, Inter-regional diffusion
Highlights
-
•
RSV-A and RSV-B groups co-circulated during surveillance with RSV-B dominant.
-
•
G and F genes are under positive selection pressure within the host.
-
•
G and NS2 genes are undergoing rapid substitution.
-
•
The population size of RSV groups changed over time.
-
•
RSV-A and RSV-B in Hubei were likely imported from Europe and Oceania respectively.
1. Introduction
Respiratory syncytial virus (RSV) belongs to the genus Orthopneumovirus within the family Pneumoviridae (Rima et al., 2017), and is a severe threat to infants and young children worldwide. RSV was estimated to be associated with 33.0 million lower respiratory tract infection cases in children younger than 5 years in 2019, causing 3.6 million hospitalizations and 101 thousand deaths (Li et al., 2022). The health and economic burden of RSV infection in young children exceeded that of influenza (Di Giallonardo et al., 2018), and has now raised concerns by the World Health Organization (Graham, 2017). However, there are currently no licensed RSV vaccines, only some in the process of clinical trials. Notably, RSV reinfection in children can be as frequent as more than 30% under three years old, which is a severe challenge for the vaccine development (Kurzweil et al., 2013; Kutsaya et al., 2016).
The genome of RSV is a non-segmented, single-stranded, negative-sense RNA, approximately 15 kb long. It consists of 10 genes in the following order from 3′ to 5′ terminal: nonstructural protein 1 (NS1) gene - nonstructural protein 2 (NS2) gene - nucleoprotein (N) gene - phosphoprotein (P) gene - matrix (M) gene - short hydrophobic protein (SH) gene - glycoprotein (G) gene - fusion protein (F) gene -transcription processivity factor (M2) gene - large polymerase complex (L) gene (Collins et al., 1986, 2013). Each gene translates into one corresponding protein except M2 gene, which has two overlapping open reading frame (ORF) that encodes two proteins (Collins and Melero, 2011). The virion of RSV is enveloped by lipid and three surface glycoproteins, G, F and SH, and the M protein was located on the inner surface of the envelope, enclosing ribonucleoprotein composed of genome RNA, N, P and L (Pangesti et al., 2018).
RSV has only one serotype and can be divided into two antigenic groups: RSV-A and RSV-B mainly based on the epitope of glycoprotein (Anderson et al., 1985; Mufson et al., 1985). These two groups diverged from the common ancestor phylogenetically about 350 years ago (Zlateva et al., 2005). Both groups can be further subdivided into many genotypes but a consensus on genotype allocation criteria has not been established yet (Goya et al., 2020). Two classification approaches were usually adopted: bootstrap values > 78% and nucleotide similarity > 96% (Peret et al., 1998), or bootstrap values > 70% and pairwise distance < 0.07 (Venter et al., 2001). According to such criteria, RSV-A is generally classified into 13 genotypes (GA1-GA7, NA1-4, ON1 and SAA1) and RSV-B is generally classified into 20 genotypes (GB1-4, SAB1-4, URU1-2 and BA1-10) (Goya et al., 2020; Martinelli et al., 2014; Pangesti et al., 2018). Some genotypes were reported to be associated with clinical features, nevertheless relevant studies were limited and discordant (Midulla et al., 2019).
RSV infection is not a notifiable disease in most countries, with the surveillance being usually combined with other respiratory diseases, and a global systematic surveillance network for RSV infection has not been developed (Pangesti et al., 2018). Currently, the Global Initiative on Sharing All Influenza Data (GISAID, https://www.gisaid.org/) is constructing a global surveillance of RVS to further investigate and elucidate the transmission and genetic backgrounds of RSV. Local surveillances were carried out in specific geographic regions including Africa, the Netherlands, China, Australia, Italy, Argentina and Kenya, and these studies focusing on RSV evolution highlighted the distribution of predominant RSV variants and co-circulation of local endemic sub-lineages (Agoti et al., 2015; Brandenburg et al., 2000; Chen et al., 2021; Di Giallonardo et al., 2018; Martinelli et al., 2014; Trento et al., 2010). However, RSV intra-host genetic diversity and evolution during infection was little known (Grad et al., 2014; Ha Do et al., 2015). In this study, we collected more than 700 samples in hospitalized children in Hubei, China during September 2020 to April 2021. A total of 46 high-quality RSV genome were obtained by mNGS, and intra-host variation, recombination, evolutionary dynamic and inter-regional diffusion analyses were performed to reveal the evolution of RSV. The findings highlighted the intra-host and inter-host evolution of RSV, and the importance of surveillance of the virus.
2. Materials and methods
2.1. Patients and specimens
The study was performed from September 30, 2020 to April 14, 2021 at Wuhan Children's Hospital, a tertiary care hospital in Wuhan, China. Research subjects included hospitalized children with cough and other typical respiratory symptoms without severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infected. Samples were collected using oropharyngeal swabs and stored at −80 °C for subsequent treatment.
2.2. Nucleic acid extraction and amplification
Total nucleic acids were extracted using the BeaverBeads™ Viral DNA/RNA Kit 70406-32FU (Beaverbio, Suzhou, China) on TGuide S32 Automated Extraction (Tiangen, Beijing, China) following the manufacturer's instructions. The RNA library was prepared using TruePrep® RNA Library Prep Kit for Illumina TR503 (Vazyme, Nanjing, China) according to the manufacturer's instructions with several adjustment (Lu et al., 2021). For each library, 8 μL of total nucleic acids extracted from each sample was used without rRNA removal. During reverse transcription, the 1 μL of Oligo(dT)20VN primer was replaced by 2 μL of Oligo(dT)20VN primer and random hexamers at a 1:1 ratio, and the reaction time under 37 °C was increased to 45 min. Then, Tn5 VR10 (Vazyme, Nanjing, China) was applied for nucleic acids fragmentation. During the PCR process, N5 and N7 PCR primers with a final concentration of 0.2 μmol/L were used. After 18 PCR cycles, the library was purified twice using 1× Agencourt AMPure XP beads (Beckman Coulter, Brea, USA) and eluted in 12 μL of nuclease-free water. Concentration of the resulting library was determined by Qubit 3.0 fluorometer with the Qubit dsDNA HS Assay kit (ThermoFisher Scientific, Waltham, USA) and the size distribution of the library was evaluated by Agilent 2100 Bioanalyzer (Agilent, Palo Alto, USA).
2.3. Sequencing and raw reads preprocessing
Sequencing was carried out on the MGISEQ-T7 platform with 150 bp of paired-end raw reads generated, after converting sequencing library style to T7 library. In-house python and Perl scripts were used for quality control and adaptor trimming of raw reads with the quality threshold of Q20 and the minimal length of read 20 nt.
2.4. Taxonomic classification
Taxonomic classification of clean reads was carried out by Kraken2 (version 2.0.9-beta) with nucleotide custom database downloaded through the script “kraken2-build --download-library nt” (Wood et al., 2019). Report files were uploaded to Pivian (https://fbreitwieser.shinyapps.io/pavian/) where the composition rates of reads were calculated and displayed by the percentage bar chart.
2.5. Assembly of virus genome
To confirm the antigen group of RSV and select the reference sequence for genome mapping, de novo assembling of clean reads was performed in MEGAHIT (version 1.2.9) (Li et al., 2015). Contigs in results were searched against the RSV database downloaded from NCBI Virus Database (https://www.ncbi.nlm.nih.gov/labs/virus/) by BLAST 2.11.0+. Genome UU01A-0176-V02 (Genbank accession number: MZ516039) and UU02-0005-V01 (Genbank accession number: MZ516025) were chosen as the reference genome for RSV-A and RSV-B, respectively. Clean reads were aligned to the reference genomes using Samtools (version 1.10) and Bcftools (version 1.11), with the coverage and sequencing depth calculated.
2.6. Intra-host single nucleotide variations detection
Mapping results were subjected to call iSNVs using the tool bam-readcount. Sites with a sequencing depth ≥100 and a minor allele frequency ≥5% were selected as candidate samples for iSNV calling (Ni et al., 2016). Sites with insertion or deletion in coding regions were excluded to avoid possible influence in codon position and translation.
2.7. Evolutionary rate estimation and Bayesian skyline plot
All full-length RSV genomes with clear sampling date were downloaded from GISAID to form the local BLAST database. To generate the datasets, sequences of RSV-A and RSV-B in this study were queried against the local database by the BLASTN program with default parameters and an e-value cutoff of 1 × 10−10. For the two groups, sequences of the top 100 hits were combined with sequences in this study to form two datasets, RSV-A (n = 119) and RSV-B (n = 238), which were then aligned by MAFFT with default parameters. Bayesian Markov Chain Monte Carlo (MCMC) approach was implemented by BEAST software (version 1.10.4) to estimate the substitution rates of full genome and each coding sequencing precisely. For all runs, the GTR Gamma model with Gamma value 4 was used for nucleotide substitution model, along with Relaxed Clock Log Normal and Coalescent Constant Size. The MCMC analyses were carried out using 500 million chain length with sampling every 50 thousand chain length. Tracer software was used to calculate mean evolutionary rate with 10% burn-in.
To clarify the effective population size of the prevalent RSV variants in this study, Bayesian skyline plot (BSP) analyses were conducted for full-length datasets in this part with the tree prior Coalescent Bayesian Skyline and remaining parameters the same. Tracer software was used to generate the BSP plot with 10% burn-in, mean root height and other parameters default.
2.8. Detection of recombination
All full-length RSV genomes with clear sampling date were downloaded from GISAID, and combined with sequences in this study to form two datasets, RSV-A (n = 1130) and RSV-B (n = 1225). Alignments were performed in MAFFT (v7.475). Each alignment was subjected to RDP4 (version 4.101.0.0) to detect recombination under method RDP (Martin and Rybicki, 2000), GENECONV (Padidam et al., 1999), Bootscan (Salminen et al., 1995), Maxchi (Smith, 1992), Chimaera (Posada and Crandall, 2001), 3 Seq (Boni et al., 2007) and SiScan (Gibbs et al., 2000) with default parameters. Datasets used in evolutionary rate estimation were also analyzed in RDP4 for recombination detection. The recombination events detected in RDP4 were further confirmed in SimPlot (version 3.5.1) with window size 200 nucleotides, step size 20 nucleotide and other parameters default.
2.9. Discrete phylogeographic analysis
All RSV sequences with clear sampling date and location were downloaded from GISAID to form the local BLAST database. To generate the datasets for discrete phylogeographic analysis, G gene sequences of RSV-A and RSV-B in this study were queried against the local database with the same steps and parameters as mentioned above. Two datasets were obtained: RSV-A (n = 179) and RSV-B (n = 299). For each datasets, sequences from public database in each dataset were divided into six geographical regions: Africa (AF), Asia (AS), Europe (EU), North America (NA), Oceania (OA), South America (SA) and geographical region of sequences in this study was China/Hubei (CN/HB). To estimate the diffusion rates among geographical regions, the asymmetric substitution model with Bayesian stochastic search variable selection (BSSVS) option was used in BEAST to infer asymmetric diffusion rates between any pairwise location states with other parameters the same as mentioned above. Treeannotator (version 1.10.4) was used to generate a maximum clade credibility (MCC) tree with mean bode heights and 10% burn-in. The same analysis were also carried out on the whole genome datasets used in evolutionary rate estimation.
The actual non-zero rates and mean indicators for all diffusions could be obtained in result log files with 10% burn-in, and the Bayes factor (BF) was calculated in SpreaD3 (version 0.9.6). Significant diffusion pathways were defined as satisfying both BF > 3 and the mean indicator > 0.5. Previous studies defined the degree of rate support as follows: BF ≥ 1000 indicates decisive, 100 ≤ BF ≤ 1000 indicates very strong support, 10 ≤ BF ≤ 100 indicates strong support and 3 ≤ BF ≤ 10 indicates supported. Cytoscape (version 3.7.0) was used to present the diffusion results.
2.10. Statistical analysis
Linear regression between the number of iSNVs in one sample and the average sequencing depth was carried out in python (version 3.6) using the statsmodels (version 0.12.0) package and visualized by the matplotlib (version 2.1.2) package.
3. Results
3.1. Epidemiology of RSV
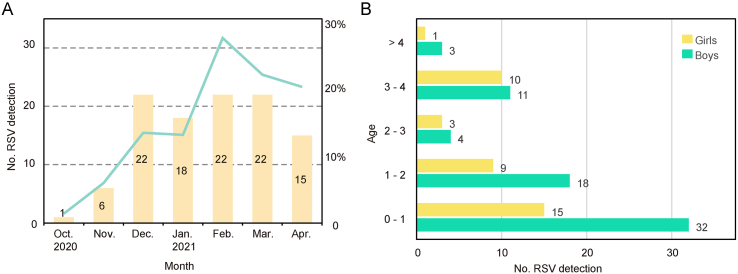
A total of 744 samples were collected and sequenced throughout the duration of this study, among which 106 (14.3%) RSV-positive samples were both diagnosed clinically and detected by mNGS (Supplementary Table S1). The positive rate was lower in October 2020 (1/72, 1.4%) and November 2020 (6/102, 5.9%), and peaked (22/81, 27.2%) in February 2021 (Fig. 1A). Among all 106 cases, 68 (64.2%) were boys and 38 (35.8%) were girls with a boy/girl ratio of 1.8:1 (Fig. 1B). The ages of patients ranged from 1 month to 6 years with a median of 13 months. 76.41% (81/106) of patients were under three years old, showing an obvious pattern that younger children were more susceptible to RSV infection. Patients under one year of age showed the highest detection rate of 21.6%. The detection rates of the group “0–1”, “1–2”, “2–3”, “3–4” and “> 4” were 21.6%, 16.7%, 7.8%, 16.5%, and 2.7%, respectively. The chi-square test showed significant differences among age groups, but the positive rate did not strictly decrease with age. In all age groups, the number of RSV-positive samples of boys exceeded that of girls. No RSV-positive patients died during the study period.
Fig. 1.
Prevalence of RSV in Hubei during 2020–2021. A The number of RSV detections. Green line represents positive rate and yellow bars show the number of positive samples. B Age and gender of patients. Labels around the bar represent the number of patients.
3.2. Taxonomic classification and quality of RSV genome
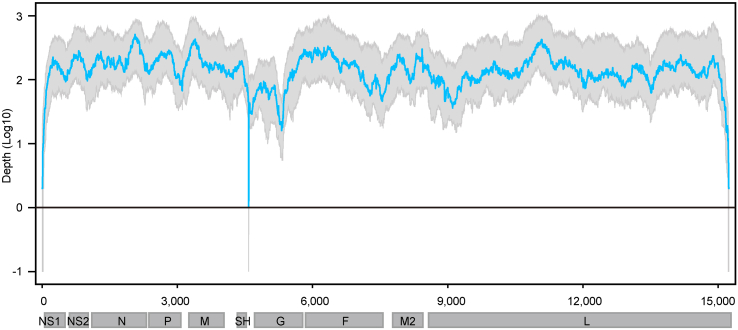
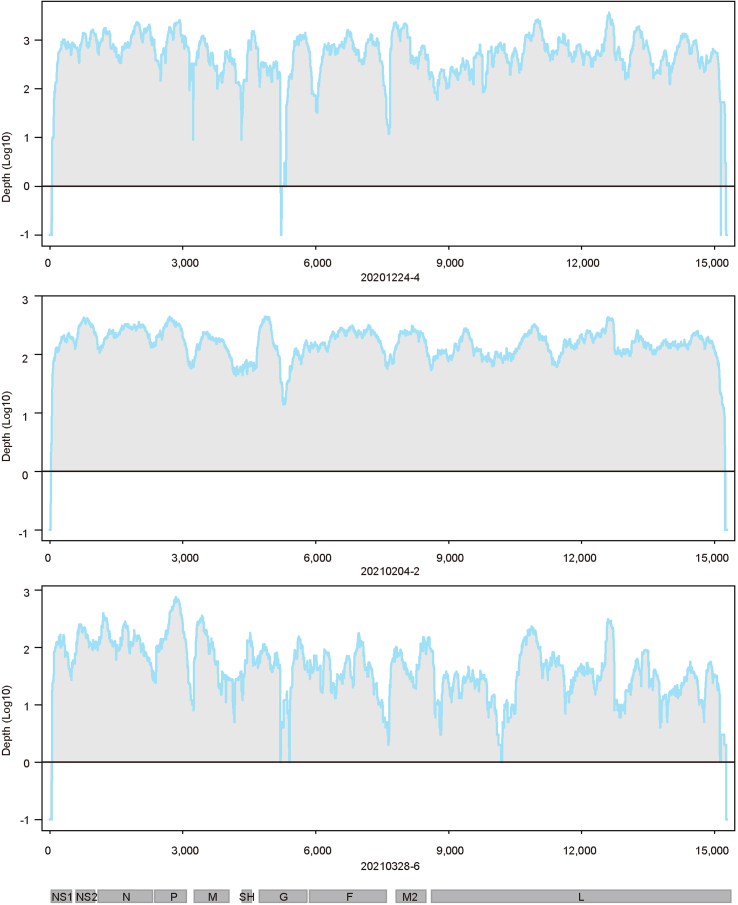
Taxonomic classification of clean reads revealed that host and bacterial reads occupied the majority (>50%, ranging from 52.6% to 93.6% with an average of 76.9%) in all RSV-positive samples (Supplementary Fig. S1). Viral reads had an average ratio of 0.4% (ranging from 0.01% to 9.46% with a median of 0.08%). Nearly the whole genome sequence (coverage > 99%) was able to be acquired from 46 samples, among which 15 genomes were completely covered (Supplementary Table S1). The average sequencing depth of these samples ranged from 25× to 7629× . Only 3 of 46 RSV belonged to RSV-A while others belonged to RSV-B. The average and median depth of no loci in RSV-B genomes were zero (Fig. 2), and the sequencing depth of sites in three RSV-A genomes was also greater than zero except some end sites and several internal sites (Supplementary Fig. S2). These 46 genomes were submitted to the GISAID RSV database (EPI numbers are provided in Supplementary Table S1) and used for further analyses in this study.
Fig. 2.
Genome coverage of RSV-B. Genome coverage of sequenced samples across the RSV-B genome. The x-axis represents the viral genome position, and the y-axis represents the log 10 depth of each site. Sites with zero depth are labeled by −1. Blue Line represent the median sequencing depth, and grey region represents lower quarter to upper quarter percentile of sequencing depth.
3.3. Distribution of iSNVs along the RSV genome
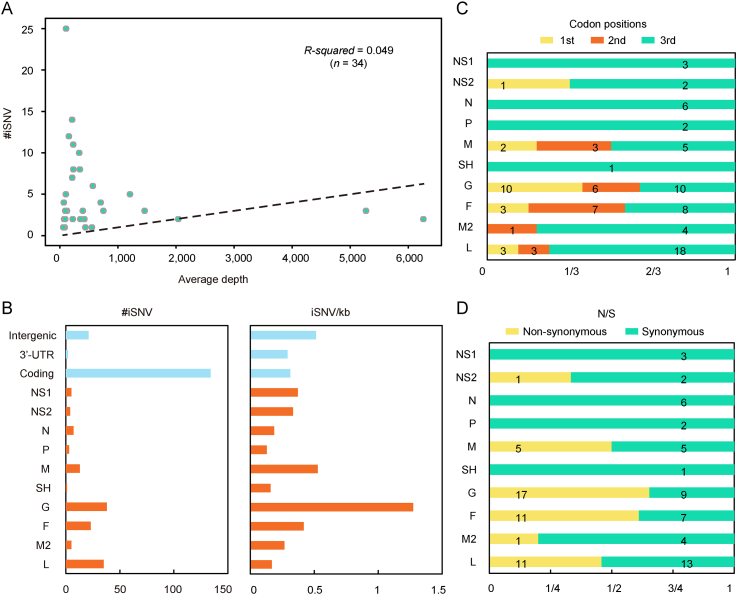
To reveal the viral-host interactions, iSNV analysis was performed. A total of 163 iSNV sites distributed in 34 samples were detected, of which 6 sites were in 2 RSV-A samples while 157 sites were in 32 RSV-B samples. The result of linear regression between the number of iSNVs and the mean sequence depth showed that they were unrelated (R-square = 0.049) (Fig. 3A), suggesting that the number of iSNVs was not affected by the NGS data volume and the results could be used to measure differences in samples (Ni et al., 2016). The iSNV sites in RSV-B samples were used for further analyses.
Fig. 3.
RSV iSNV distributions along the viral genome. A #iSNV versus mean sequencing depth for each site over RSV genomes of patients. Dashed line shows the linear regression. B Total iSNV numbers (left panel) and sum of #iSNV/kb values (right panel) for RSV-B in the study along genomic regions and ORF regions. C, D Distribution of iSNVs at codon positions (C) and non-synonymous (N)/synonymous (S) iSNVs (D) of ORFs.
At the population level, 85.4% (134/157) of iSNVs accumulated in the coding regions (Fig. 3B), which covered about 90% of the entire genome. When normalized by length, the intergenic region was the most iSNV-enriched region with 0.51 iSNV/kb, while the coding region and 3′-UTR showed similar normalized values (Fig. 3B). For each ORF, G gene was the most iSNV-enriched gene before and after normalization (Fig. 3B). A previous study pointed that under neutral selection, the iSNVs should distribute equally in each codon position and the non-synonymous/synonymous (N/S) ratio tends to be similar in all genes (Ni et al., 2016). Here we investigated the iSNVs distribution in codon positions and N/S ration of genes. To exclude the effect of frame shift mutation, samples with insertion or deletion in coding regions were excluded. As expected, most iSNVs (59/98, 60.20%) accumulated in the third codon position, while the numbers of non-synonymous and synonymous mutations were nearly equal (N/S ration 46/52) (Fig. 3C and D). Higher N/S ratio was observed in G and F gene (1.89 and 1.57, respectively), and N/S ratio was no more than one in other genes.
3.4. Recombination and evolutionary dynamic of RSV
Recombination analysis was performed to ensure that viruses for evolutionary dynamic analyses originated from a common ancestor and no recombination events occurred. None of the genomes used for evolutionary dynamic analysis was recombinant according to the RDP4 of the combined datasets including RSVs in this study and those downloaded from GISAID (Supplementary Table S2). To elaborate on this, recombination analysis of datasets used in the evolutionary analysis was also performed in RDP4, and there were possible recombination events detected in three genomes (Supplementary Table S3). However, no significant recombination signal was observed by SimPlot in all three genomes (Supplementary Fig. S3).
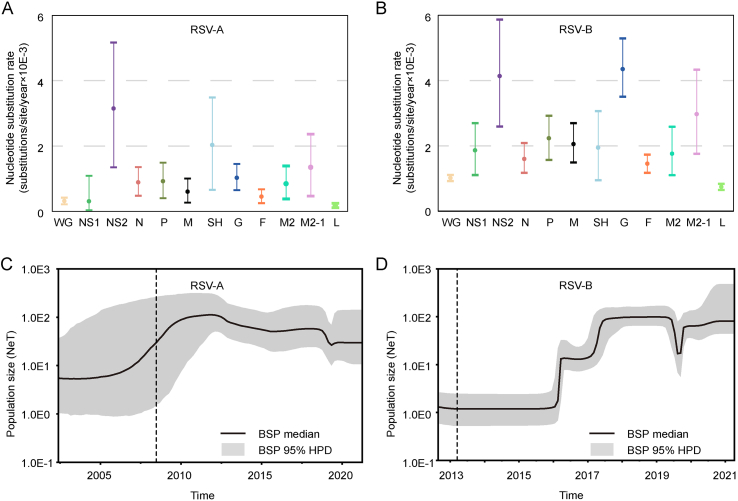
To estimate the evolutionary rates of RSV and explore the temporal relationships of RSVs in this study and related viruses, Bayesian dating and BSP analyses were carried out using BEAST v1.10.4. The evolutionary rates of whole genomes were 3.17 × 10−4 (95% HPD: 2.16 × 10−4 –4.21 × 10−4) and 1.01 × 10−3 (95% HPD: 9.18 × 10−4 –1.11 × 10−3) nucleotide substitutions/site/year for RSV-A and RSV-B, respectively (Fig. 4A and B). Likewise, the mean evolutionary rates of almost all genes in RSV-B were higher than that in RSV-A, except the SH gene. In both RSV groups, the mean evolutionary rates of L genes were the lowest, lower than the whole genomes, and NS2 genes evolved fast with the evolutionary rate being the highest in RSV-A and the second highest in RSV-B. Surface protein G gene in RSV-B showed divergent evolutionary rate compared with F gene and SH gene in RSV-B and G gene in RSV-A, with 3 times, 2.24 times and 4.22 times, respectively (Fig. 4A and B).
Fig. 4.
Evolutionary dynamics of related RSV viruses. A, B Evolutionary rates (substitutions per site per year × 10−3) and 95% HPD values of whole genome and each ORF for related RSV-A (A) and RSV-B (B) viruses. C, D Bayesian skyline plot (BSP) of the whole genome of related RSV-A (C) and RSV-B (D) showing changes in population size. The thick solid black line is the median estimate, and the grey area is the 95% HPD of the genetic diversity estimates. The vertical dash line represents upper 95% HPD of the root.
BSP analysis revealed the effective population sizes and the root dates of the most recent common ancestor of a virus. Analyses of RSV in the present study and related strains with close relatives showed that RSV-A emerged in May 2002 (95% HPD: Feb. 1995–Jun. 2008), underwent a rapid growth and peaked around 2012 (Fig. 4C). Then, the population size decreased slowly and remained stable in 2019. It was estimated that RSV-B first appeared in August 2012 (95% HPD: Jan. 2012–Mar. 2013), and did not change in population size until 2016 (Fig. 4D). After two outbreaks in early 2016 and 2017, the virus population remained at a high level for about three years. At the end of 2019, the population size experienced a fluctuation, where it first decreased then returned to the previous level quickly. It showed that the current population size of both RSV subtypes is at a relatively high level at present, which is likely to persist.
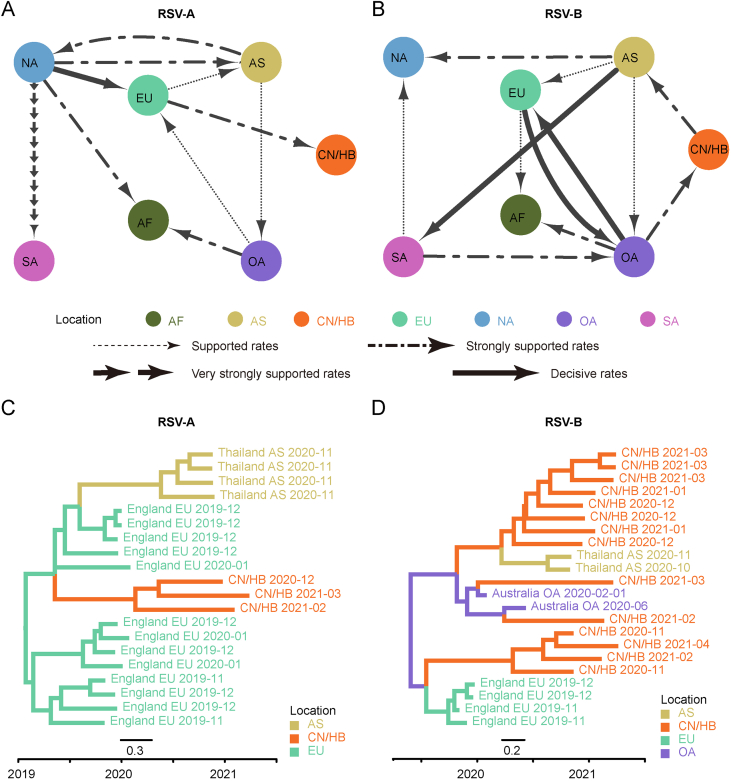
3.5. Inter-regional gene flow of RSV
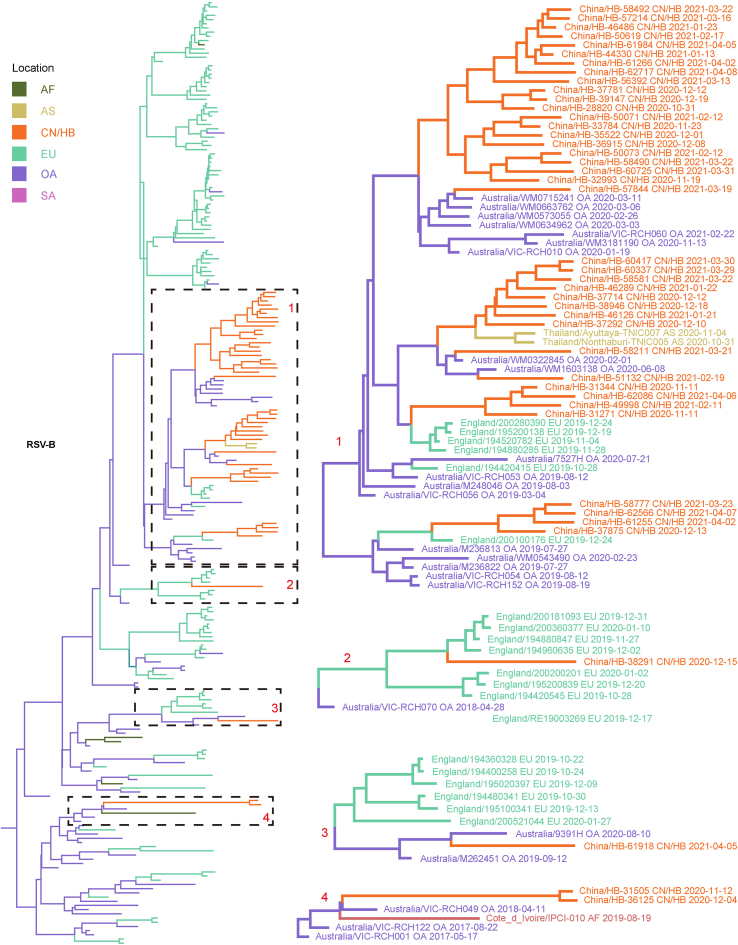
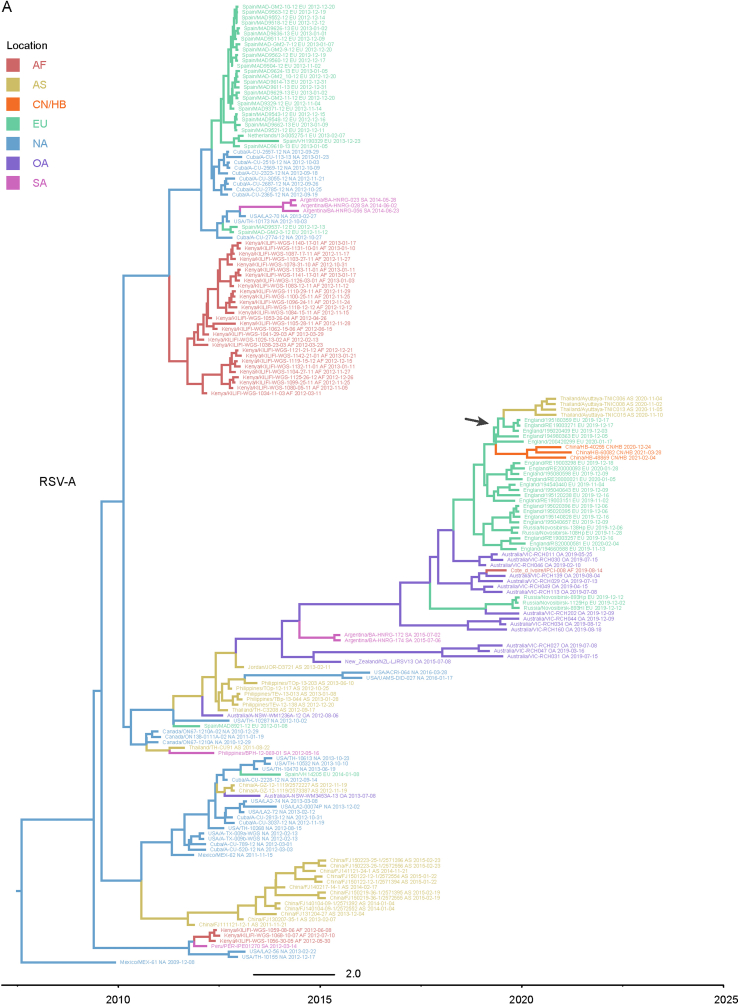
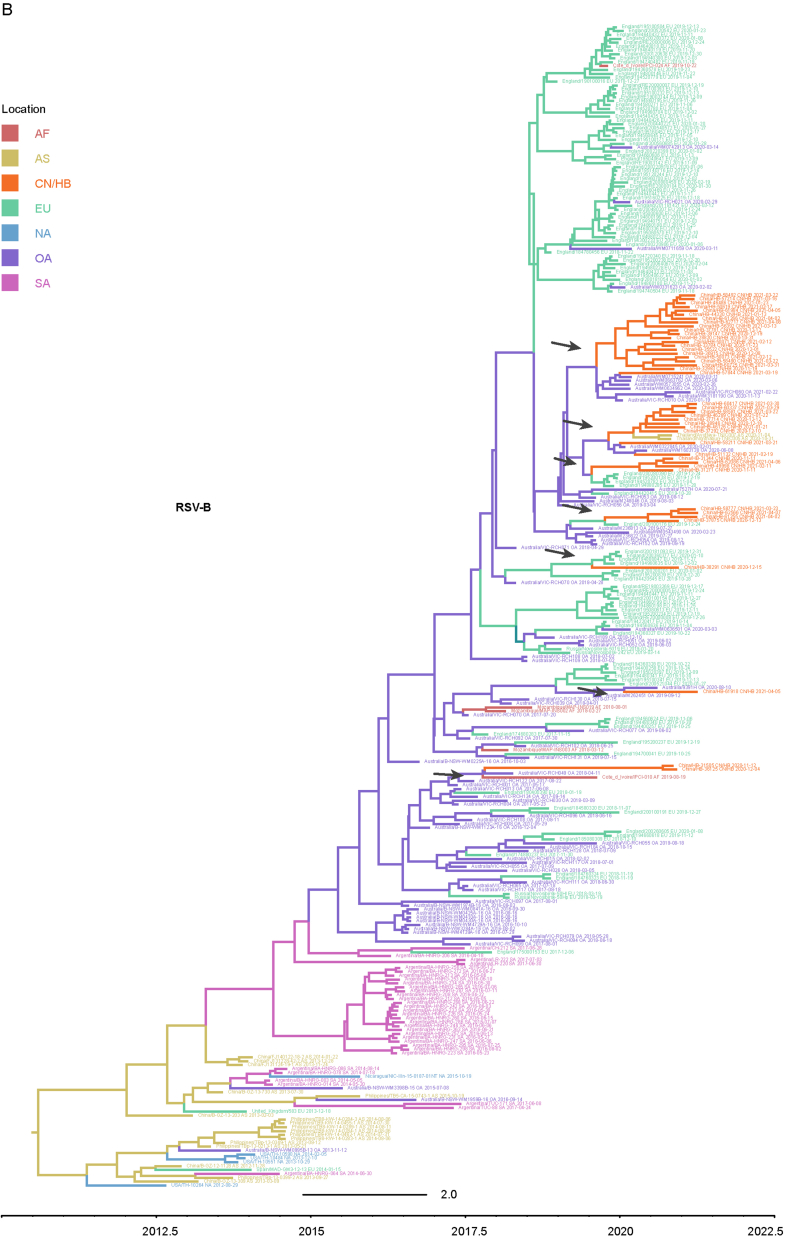
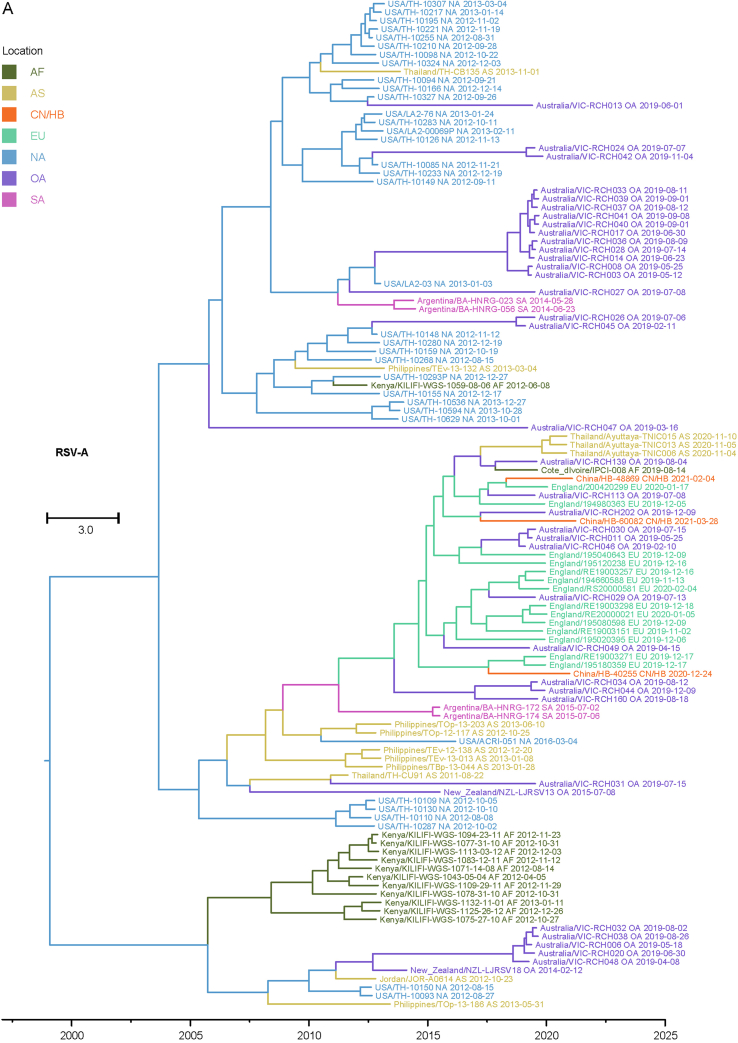
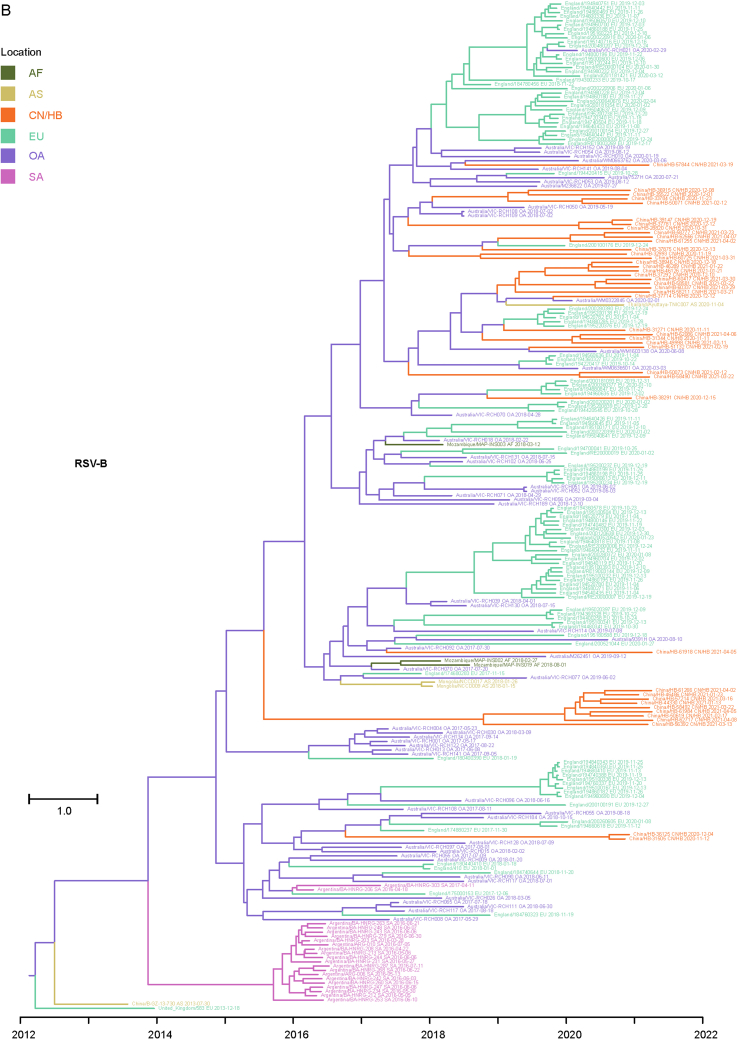
Considering that the number of whole genome sequences in public database was fewer than G gene of RSV (there may be missing samples at some key nodes in the transmission chain), G gene sequences of RSV obtained in this study and the most related viruses were used to infer inter-regional gene flow. RSV-A in this study were clustered with viruses isolated from Europe (Supplementary Fig. S4A), and RSV-B in this study were clustered with viruses isolated from Oceania and Europe (Supplementary Fig. S4B). Viruses in this study from both groups showed rather distant phylogenetic relationships with viruses prevailing previously in Asia including China. It could be observed that location transitions of related viruses from other regions to China/Hubei have taken place at least four times for RSV-B (Fig. 5).
Fig. 5.
Maximum clade credibility tree of RSV-B G gene. Red numbers represent potential inter-regional diffusion events. Colors of branches and labels represent geographical region: AF, Africa; AS, Asia; EU, Europe; NA, North America; OA, Oceania; SA, South America; CN/HB, China/Hubei (this study).
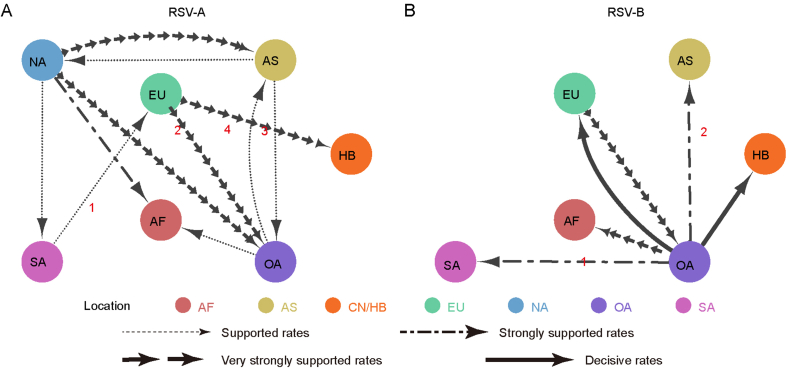
The BSSVS method was applied to provide statistical support for spatial diffusion. The results showed that no diffusion from Asia to China/Hubei was detected (Fig. 6A and B). RSV-A in this study might come from Europe with strongly supported rates (Fig. 6A), and the inter-regional diffusion event was likely to occur in the first half of 2019 from England (Fig. 6C). As for RSV-B group, the diffusion from Oceania to China/Hubei with strongly supported rates could be observed (Fig. 6B), which could have occurred more than one time (Fig. 5), including one diffusion event in the end of 2019 (Fig. 6D). Diffusion of RSV-B in this study to Asia was observed with strongly supported rates (Fig. 6B). Combining with the result of phylogenetic tree, it was highly probable that viruses isolated from Thailand had the same ancestor with viruses in this study, as they both originated in Australia (Fig. 6D).
Fig. 6.
Inter-regional diffusion of RSV G genes. A–B Spatial diffusion pathways of RSV-A (A) and RSV-B (B). Arrows show the direction of spatial diffusion. Node color and annotation represents the geographical locations: AF, Africa; AS, Asia; EU, Europe; NA, North America; OA, Oceania; SA, South America; CN/HB, China/Hubei (this study). Line width and type represent statistically supported migration rates with a mean indicator of >0.5: decisive rates with BF ≥ 1,000, very strongly supported rates with 100 ≤ BF < 1,000, strongly supported rates with 10 ≤ BF < 100 and supported rates with 3 ≤ BF < 10. C–D Partial maximum clade credibility tree of G gene of RSV-A (C) and RSV-B (D) virus. Colors of branches and labels represent geographical region as mentioned above.
The inter-regional diffusion analyses were also carried out on the whole genome datasets to assess the possible impact of insufficient data on the results. The analysis results centered on the RSV strains in this study are basically the same between the two datasets, RSV-A and RSV-B have close phylogenetic relationships with strains in Europe and Oceania, and far away from strains in other regions of Asia. The statistical results of inter-regional diffusion also support that RSV-A was spread from Europe and RSV-B was spread from Oceania (Fig. 6). While the results are quite different for the related RSV strains. For RSV-A, three additional diffusions and one diffusion with reversed direction were obtained based on whole genome datasets compared with those based on G genes (Fig. 6A, Supplementary Fig. S5A; Supplementary Fig. S6A). As for RSV-B, a key node (North America) and five diffusion disappeared and two diffusions with reversed direction in the analysis on whole genomes (Fig. 6B, Supplementary Fig. S5B; Supplementary Fig. S6B). These divergences indicated that the lack of some surveillance data would lead to disparate or even opposite results.
4. Discussion
RSV is the most frequently identified viral pathogen in children with acute lower respiratory tract infection (ALRTI), and is one of the major causes of morbidity and mortality in children younger than five years (Li et al., 2022). In this study, we investigated RSV infection in hospitalized children in Hubei, China during 2020–2021. A total of 744 samples were collected and 106 were RSV-positive. The detection rate of 14.25% was lower than those reported in previous studies focusing on RSV surveillance in hospitalized children, which could reach 30% or even higher (Chen et al., 2021; Rha et al., 2020; Yu et al., 2019; Zhang et al., 2010). One reason for this might be that the preventive measures taken against COVID-19 changed the pathogen spectrum in children hospitalized with ALRTI (Lin et al., 2021). Previous studies pointed out RSV-A and RSV-B groups co-circulated during specific epidemics with one group predominating (Sullender, 2000). Surveillance on RSV in China previously showed that RSV-A was dominant in most years (Yu et al., 2019; Zhang et al., 2010), while it was observed in this study that RSV-B was predominant compared with group RSV-A, reflecting the dynamic of genotype shift of viruses.
About 46 high-quality genomes were attained in this study, which occupied about 1/6 (46/293) of data from China uploaded to GISAID RSV database since 2015. These data also represent the first surveillance of RSV in China after COVID-19 outbreak worldwide. We did not subdivided strains into genotypes further as there is no universal criteria. It has been pointed that a universal standard for the assignment of RSV genotypes is essential to explore the correlation between genotypes and disease severity or geographical or temporal constraints of the virus cycle (Goya et al., 2020).
Intra-host variation is an inherent characteristic of RNA viruses, due to the lack of a proofreading mechanism and the high replication rate of RNA-dependent RNA polymerase, causing the original virus to become a group of variants (Andino and Domingo, 2015). The dynamics of the virus variant population have been highly applied in studying the biological features of human pathogens, including RSV (Grad et al., 2014; Ha Do et al., 2015). The criteria for iSNV site selection could affect the results because there would be many sites with low-frequency mutations if the threshold was too low, making the analysis meaningless. On the other hand, if the threshold is too high, some sites would be missed. In this study, we performed iSNV analysis referring to a previous study (Ni et al., 2016). We identified a total of 163 iSNV sites distributed in 34 samples, and the G gene was the most iSNV-enriched gene with the highest iSNV/kb. The results were in accordance with the current understanding that the G gene is the most variable region in RSV genome because it encodes a surface glycoprotein with cell receptor sites and neutralizing antibody epitopes (Escribano-Romero et al., 2004). From the point of codon position and substitution type, iSNV sites mainly distributed in the third position of each codon and caused synonymous mutations in amino acids (Fig. 3C and D). The ratio of non-synonymous substitution/synonymous substitution of G and F genes is 1.89 and 1.57 respectively, which is greater than 1. According to a previous study (Ni et al., 2016), means that these two genes are under positive selection pressure within the host, and evolution driven by the immune pressure played an important role in this process (Ha Do et al., 2015). Phase III clinical trial of Nirsevimab (Hammitt et al., 2022) has successfully proved that it can protect healthy infants from RSV. The mechanism of its protective effect is to prevent virus infection from fusing with host cells and entering cells by combining with specific regions of RSV F protein. At present, F gene is under positive selection pressure, which is likely to produce mutations that escape Nirsevimab immune protection, thus making its protection ineffective. Therefore, it is more important to monitor the evolution of RSV genome.
Recombination is an important way for virus evolution, and the genome of recombinant contains nucleic acid fragments from different parental viruses. However, recombination in RSV was rarely reported except for one mixed infection study in vitro (Spann et al., 2003), and the co-infection caused by different RSV genotypes was also rarely reported (Yu et al., 2021). In this study, we carried out recombination analysis to exclude its effect on the calculation of evolutionary rates, and no significant recombination signal was discovered (Supplementary Tables S2 and S3). The evolutionary rates of whole genomes were 3.17 × 10−4 and 1.01 × 10−3 nucleotide substitutions/site/year for RSV-A and RSV-B, respectively. Both rates were lower than that reported by previous studies (Pretorius et al., 2013; Yu et al., 2021; Zlateva et al., 2005), indicating a potential sampling bias. Previous studies have reported that the evolutionary rates were different between two groups, particularly that the evolutionary rate of G gene was significantly higher in RSV-B than that in RSV-A (Schobel et al., 2016; Tan et al., 2013). In this study, we observed similar result that the rates in RSV-B were over four times as many as that in RSV-A. In addition, it has been noticed that the evolutionary rate of G gene was significantly higher than that of F gene in RSV-B, but not as high as that documented in previous studies where the evolutionary rate of G genes was ten times higher (Kimura et al., 2016, 2017). Another noteworthy result was that the evolutionary rates of NS2 gene in both RSV groups were high (Fig. 4A and B). It was reported that the nonstructural proteins (NS1 and NS2) only existed in RSV and not in other viruses belonging to the subfamily Pneumovirinae (Pangesti et al., 2018). Previous studies showed that NS1 and NS2 might antagonize against interferon production and signaling to escape from host immune responses both in vitro and in animal models (Lo et al., 2005; Ramaswamy et al., 2006; Spann et al., 2003; Swedan et al., 2009). It seems that the NS proteins could also inhibit cell apoptosis and facilitate and regulate viral replication (Bitko et al., 2007; Spann et al., 2004).
Analyzing whole-genome data is helpful to understand the evolutionary processes of RSV more comprehensively (Agoti et al., 2015), and we followed this in iSNV analysis and evolutionary dynamic analysis. However, only G gene sequences were available in many local studies, so we performed inter-regional diffusion analysis using G genes rather than the whole genomes to prevent missing some key information. According to phylogenetic analysis, we observed that the prevalence of RSV was a combination of global transmission and local prevalence, and viruses detected throughout the duration of this study might have undergone several inter-regional diffusions (Fig. 5). Statistical analysis showed that RSV-A viruses in this study might come from Europe with strongly supported rates (Fig. 6A), and RSV-B viruses might originate from Oceania with strongly supported rates (Fig. 6B). However, lacking data on other cities in China may lead to the neglect of some key geographical transmission nodes. It is well likely that RSV first seeded from Europe or Australia to a populated city in China and then spread to other locations. At the same time, it is unclear whether any surveillance data on key transmission nodes overseas were missing on account of limited global data. The disparate results obtained from regional diffusion based on all current G gene data and whole genome data underscores the urgent need for global surveillance for whole genomes of RSV. Globalization has facilitated the spread of numerous infectious pathogens worldwide, raising serious concerns for public health (Smith et al., 2007). A recent epidemiological and genomic monitoring of RSV in Australia showed that RSV broke out non-seasonally after the downgrade of COVID-19 prevention and control (Eden et al., 2022), which also reminded us of the importance of monitoring other respiratory pathogens after the adjustment of current epidemic prevention and control measures. It is urgent to establish a global detection network and strengthen customs inspection of other infectious pathogens including RSV but not only SARS-CoV-2, so as to provide information for disease control and prevention. At the same time, clinical information on infected patients should be collected and more comprehensive data should be used to detect genomic characteristics of the virus that are statistically related to age/sex preference of infection and disease severity. And a global genomic surveillance network for RSV could help us understand the transmission of the virus worldwide, predict the effectiveness of preventive treatment, and support countermeasure development for all circulating virus variants.
5. Conclusions
In summary, we performed a systematic surveillance in hospitalized children in Hubei during 2020–2021, and found that RSV was still a severe threat to children's health. RSV-A and RSV-B co-circulated during surveillance with RSV-B being predominant. Intra-host variations revealed that G and F genes of RSV-B were under persistent interaction with the host immune system and positive selection pressure. Evolutionary dynamic analysis showed that the evolutionary rates of G and NS2 genes were higher, and the population size of RSV groups changed over time. We also found evidences of inter-regional diffusion of RSV. This study highlighted the intra-host and inter-host evolution of RSV, which would contribute to the control of RSV, and warns of the potential for monoclonal resistance. However, due to the limitations of the current data set, it is difficult for us to obtain more detailed genome sequences domestically and globally, and some key propagation nodes may be missing from statistical results such as geographical diffusion speculation. Therefore, we emphasized that strengthening surveillance by sampling widely, especially during suspected outbreaks, is essential to improve our understanding of the ecology and evolution of RSV.
Data availability
All data generated or analyzed in the current study are included in this published article and its supplementary materials.
Ethics statement
Studies involving human participants were reviewed and approved by Medical Ethics Committee of Wuhan Children's Hospital (Wuhan Maternal and Children Healthcare Hospital) (the ethical approval number: 2021R005-E01). Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author contributions
Yi Yan, Decheng Wang: conceptualization, data curation, formal analysis, investigation, methodology, software, validation, visualization, writing–original draft; Ying Li: conceptualization, data curation, formal analysis, investigation, resources, validation, writing–original draft, writing–review & editing; Zhiyong Wu: data curation, formal analysis, investigation, validation, software; Haizhou Liu: conceptualization, project administration, software, supervision; Yue Shi: formal analysis, investigation, writing–review & editing; Xiaoxia Lu: conceptualization, funding acquisition, project administration, resources, supervision, writing–review & editing; Di Liu: conceptualization, funding acquisition, methodology, project administration, resources, supervision, visualization, writing–review & editing. All the authors read and approved the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest. Di Liu is an editorial board member for Virologica Sinica and was not involved in the editorial review or the decision to publish this article.
Acknowledgement
This study was supported by National Key Research and Development Program of China (2018YFC1603803), National Natural Science Foundation of China (31970548), Knowledge Innovation Program of Wuhan-Basi Research (2022020801010519), Health Commission of Hubei Province (WJ 2021M262) and Natural Science Fund of Hubei Province (2021CFA012). We thank data submitters of RSV in GISAID and GenBank database.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2023.05.001.
Contributor Information
Xiaoxia Lu, Email: lusi74@163.com.
Di Liu, Email: liud@wh.iov.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
figs7.
figs8.
References
- Agoti C.N., Otieno J.R., Munywoki P.K., Mwihuri A.G., Cane P.A., Nokes D.J., Kellam P., Cotten M. Local evolutionary patterns of human respiratory syncytial virus derived from whole-genome sequencing. J. Virol. 2015;89:3444–3454. doi: 10.1128/JVI.03391-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L.J., Hierholzer J.C., Tsou C., Michael Hendry R., Fernie B.F., Stone Y., McIntosh K. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J. Infect. Dis. 1985;151:626–633. doi: 10.1093/infdis/151.4.626. [DOI] [PubMed] [Google Scholar]
- Andino R., Domingo E. Viral quasispecies. Virology. 2015;479–480:46–51. doi: 10.1016/j.virol.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitko V., Shulyayeva O., Mazumder B., Musiyenko A., Ramaswamy M., Look D.C., Barik S. Nonstructural proteins of respiratory syncytial virus suppress premature apoptosis by an NF-κB-Dependent, interferon-independent mechanism and facilitate virus growth. J. Virol. 2007;81:1786–1795. doi: 10.1128/JVI.01420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Posada D., Feldman M.W. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics. 2007;176:1035–1047. doi: 10.1534/genetics.106.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg A.H., Van Beek R., Moll H.A., Osterhaus A.D.M.E., Claas E.C.J. G protein variation in respiratory syncytial virus group A does not correlate with clinical severity. J. Clin. Microbiol. 2000;38:3849–3852. doi: 10.1128/jcm.38.10.3849-3852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhu Y., Wang W., Li C., An S., Lu G., Jin R., Xu B., Zhou Y., Chen A., Li L., Zhang M., Xie Z. A multi-center study on molecular epidemiology of human respiratory syncytial virus from children with acute lower respiratory tract infections in the mainland of China between 2015 and 2019. Virol. Sin. 2021;36:1475–1483. doi: 10.1007/s12250-021-00430-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P.L., Dickens L.E., Buckler-White A., Olmsted R.A., Spriggs M.K., Camargo E., Coelingh K.V. Nucleotide sequences for the gene junctions of human respiratory syncytial virus reveal distinctive features of intergenic structure and gene order. Proc. Natl. Acad. Sci. U. S. A. 1986;83:4594–4598. doi: 10.1073/pnas.83.13.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P.L., Fearns R., Graham B.S. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr. Top. Microbiol. Immunol. 2013;372:3–38. doi: 10.1007/978-3-642-38919-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P.L., Melero J.A. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res. 2011;162:80–99. doi: 10.1016/j.virusres.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giallonardo F., Kok J., Fernandez M., Carter I., Geoghegan J.L., Dwyer D.E., Holmes E.C., Eden J.S. Evolution of human respiratory syncytial virus (RSV) over multiple seasons in New South Wales, Australia. Viruses. 2018;10:1–13. doi: 10.3390/v10090476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden J., Sikazwe C., Xie R., Deng Y., Sullivan S.G., Michie A., Levy A., Cutmore E., Blyth C.C., Britton P.N., Crawford N., Dong X., Dwyer D.E., Edwards K.M., Horsburgh B.A., Foley D., Kennedy K., Minney-smith C., Speers D., Tulloch R.L., Holmes E.C., Dhanasekaran V., Smith D.W., Kok J., Barr I.G., Group, the A.R. study Off-season RSV epidemics in Australia after easing of COVID-19 restrictions. Nat. Commun. 2022;12:1–9. doi: 10.1038/s41467-022-30485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Romero E., Rawling J., García-Barreno B., Melero J.A. The soluble form of human respiratory syncytial virus attachment protein differs from the membrane-bound form in its oligomeric state but is still capable of binding to cell surface proteoglycans. J. Virol. 2004;78:3524–3532. doi: 10.1128/JVI.78.7.3524-3532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs M.J., Armstrong J.S., Gibbs A.J. Sister-scanning: a Monte Carlo procedure for assessing signals in rebombinant sequences. Bioinformatics. 2000;16:573–582. doi: 10.1093/bioinformatics/16.7.573. [DOI] [PubMed] [Google Scholar]
- Goya S., Galiano M., Nauwelaers I., Trento A., Openshaw P.J., Mistchenko A.S., Zambon M., Viegas M. Toward unified molecular surveillance of RSV: a proposal for genotype definition. Influenza Other Respir. Viruses. 2020;14:274–285. doi: 10.1111/irv.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad Y.H., Newman R., Zody M., Yang X., Murphy R., Qu J., Malboeuf C.M., Levin J.Z., Lipsitch M., DeVincenzo J. Within-host whole-genome deep sequencing and diversity analysis of human respiratory syncytial virus infection reveals dynamics of genomic diversity in the absence and presence of immune pressure. J. Virol. 2014;88:7286–7293. doi: 10.1128/JVI.00038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham B.S. Vaccine development for respiratory syncytial virus. Curr Opin Virol. 2017;23:107–112. doi: 10.1016/j.coviro.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha Do L.A., Wilm A., Van Doorn H.R., Lam H.M., Sim S., Sukumaran R., Tran A.T., Nguyen B.H., Tran T.T.L., Tran Q.H., Vo Q.B., Tran Dac N.A., Trinh H.N., Nguyen T.T.H., Le Binh B.T., Le K., Nguyen M.T., Thai Q.T., Vo T.V., Minh Ngo N.Q., Dang T.K.H., Cao N.H., Van Tran T., Ho L.V., Farrar J., De Jong M., Chen S., Nagarajan N., Bryant J.E., Hibberd M.L. Direct whole-genome deep-sequencing of human respiratory syncytial virus A and B from Vietnamese children identifies distinct patterns of inter- and intra-host evolution. J. Gen. Virol. 2015;96:3470–3483. doi: 10.1099/jgv.0.000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammitt L.L., Dagan R., Yuan Y., Baca Cots M., Bosheva M., Madhi S.A., Muller W.J., Zar H.J., Brooks D., Grenham A., Wählby Hamrén U., Mankad V.S., Ren P., Takas T., Abram M.E., Leach A., Griffin M.P., Villafana T. Nirsevimab for prevention of RSV in healthy late-preterm and term infants. N. Engl. J. Med. 2022;386:837–846. doi: 10.1056/NEJMoa2110275. [DOI] [PubMed] [Google Scholar]
- Kimura H., Nagasawa K., Kimura R., Tsukagoshi H., Matsushima Y., Fujita K., Hirano E., Ishiwada N., Misaki T., Oishi K., Kuroda M., Ryo A. Molecular evolution of the fusion protein (F) gene in human respiratory syncytial virus subgroup B. Infect. Genet. Evol. 2017;52:1–9. doi: 10.1016/j.meegid.2017.04.015. [DOI] [PubMed] [Google Scholar]
- Kimura H., Nagasawa K., Tsukagoshi H., Matsushima Y., Fujita K., Yoshida L.M., Tanaka R., Ishii H., Shimojo N., Kuroda M., Ryo A. Molecular evolution of the fusion protein gene in human respiratory syncytial virus subgroup A. Infect. Genet. Evol. 2016;43:398–406. doi: 10.1016/j.meegid.2016.06.019. [DOI] [PubMed] [Google Scholar]
- Kurzweil V., Tang R., Galinski M., Wang K., Zuo F., Cherukuri A., Gasser R.A., Malkin E., Sifakis F., Mendel D.B., Esser M.T. Translational sciences approach to RSV vaccine development. Expert Rev. Vaccines. 2013;12:1047–1060. doi: 10.1586/14760584.2013.824706. [DOI] [PubMed] [Google Scholar]
- Kutsaya A., Teros-Jaakkola T., Kakkola L., Toivonen L., Peltola V., Waris M., Julkunen I. Prospective clinical and serological follow-up in early childhood reveals a high rate of subclinical RSV infection and a relatively high reinfection rate within the first 3 years of life. Epidemiol. Infect. 2016;144:1622–1633. doi: 10.1017/S0950268815003143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C.M., Luo R., Sadakane K., Lam T.W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Li Y., Wang X., Blau D.M., Caballero M.T., Feikin D.R., Gill C.J., Madhi S.A., Omer S.B., Simões E.A.F., Campbell H., Pariente A.B., Bardach D., Bassat Q., Casalegno J.-S., Chakhunashvili G., Crawford N., Danilenko D., Do L.A.H., Echavarria M., Gentile A., Gordon A., Heikkinen T., Huang Q.S., Jullien S., Krishnan A., Lopez E.L., Markić J., Mira-Iglesias A., Moore H.C., Moyes J., Mwananyanda L., Nokes D.J., Noordeen F., Obodai E., Palani N., Romero C., Salimi V., Satav A., Seo E., Shchomak Z., Singleton R., Stolyarov K., Stoszek S.K., Gottberg A. von, Wurzel D., Yoshida L.-M., Yung C.F., Zar H.J., Harish R.V.G.E.N., Nair H., Investigators R. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: a systematic analysis. Lancet. 2022;399:2047–2064. doi: 10.1016/S0140-6736(22)00478-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.X., Lian H. Bin, Lin G.Y., Zhang D.G., Cai X.Y., Cai Z.W., Wen F.Q. Pathogen spectrum changes of respiratory tract infections in children in Chaoshan area under the influence of COVID-19. Epidemiol. Infect. 2021;149:E170. [Google Scholar]
- Lo M.S., Brazas R.M., Holtzman M.J. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat 2 expression and alpha/beta interferon responsiveness. J. Virol. 2005;79:9315–9319. doi: 10.1128/JVI.79.14.9315-9319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Yan Y., Dong L., Han L., Liu Y., Yu J., Chen J., Yi D., Zhang M., Deng X., Wang C., Wang R., Wang D., Wei H., Liu D., Yi C. Integrated characterization of SARS-CoV-2 genome, microbiome, antibiotic resistance and host response from single throat swabs. Cell Discov. 2021;7:1–10. doi: 10.1038/s41421-021-00248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.P., Rybicki E. RDP: detection ofrecombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- Martinelli M., Frati E.R., Zappa A., Ebranati E., Bianchi S., Pariani E., Amendola A., Zehender G., Tanzi E. Phylogeny and population dynamics of respiratory syncytial virus (RSV) A and B. Virus Res. 2014;189:293–302. doi: 10.1016/j.virusres.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Midulla F., Nenna R., Scagnolari C., Petrarca L., Frassanito A., Viscido A., Arima S., Antonelli G., Pierangeli A. How respiratory syncytial virus genotypes influence the clinical course in infants hospitalized for bronchiolitis. J. Infect. Dis. 2019;219:526–534. doi: 10.1093/infdis/jiy496. [DOI] [PubMed] [Google Scholar]
- Mufson M.A., Orvell C., Rafnar B., Norrby E. Two distinct subtypes of human respiratory syncytial virus. J. Gen. Virol. 1985;66:2111–2124. doi: 10.1099/0022-1317-66-10-2111. [DOI] [PubMed] [Google Scholar]
- Ni M., Chen C., Qian J., Xiao H.X., Shi W.F., Luo Y., Wang H.Y., Li Z., Wu J., Xu P.S., Chen S.H., Wong G., Bi Y., Xia Z.P., Li W., Lu H.J., Ma J., Tong Y.G., Zeng H., Wang S.Q., Gao G.F., Bo X.C., Liu D. Intra-host dynamics of Ebola virus during 2014. Nat. Microbiol. 2016;1:1–9. doi: 10.1038/nmicrobiol.2016.151. [DOI] [PubMed] [Google Scholar]
- Padidam M., Sawyer S., Fauquet C.M. Possible emergence of new geminiviruses by frequent recombination. Virology. 1999;265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- Pangesti K.N.A., Abd El Ghany M., Walsh M.G., Kesson A.M., Hill-Cawthorne G.A. Molecular epidemiology of respiratory syncytial virus. Rev. Med. Virol. 2018;28:1–11. doi: 10.1002/rmv.1968. [DOI] [PubMed] [Google Scholar]
- Peret T.C.T., Hall C.B., Schnabel K.C., Golub J.A., Anderson L.J. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J. Gen. Virol. 1998;79:2221–2229. doi: 10.1099/0022-1317-79-9-2221. [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. U. S. A. 2001;19:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretorius M.A., Van Niekerk S., Tempia S., Moyes J., Cohen C., Madhi S.A., Venter M. Replacement and positive evolution of subtype A and B respiratory syncytial virus G-protein genotypes from 1997-2012 in South Africa. J. Infect. Dis. 2013;208:227–237. doi: 10.1093/infdis/jit477. [DOI] [PubMed] [Google Scholar]
- Ramaswamy M., Shi L., Varga S.M., Barik S., Behlke M.A., Look D.C. Respiratory syncytial virus nonstructural protein 2 specifically inhibits type I interferon signal transduction. Virology. 2006;344:328–339. doi: 10.1016/j.virol.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Rha B., Curns A.T., Lively J.Y., Campbell A.P., Englund J.A., Boom J.A., Azimi P.H., Weinberg G.A., Staat M.A., Selvarangan R., Halasa N.B., McNeal M.M., Klein E.J., Harrison C.J., Williams J.V., Szilagyi P.G., Singer M.N., Sahni L.C., Figueroa-Downing D., McDaniel D., Prill M.M., Whitaker B.L., Stewart L.S., Schuster J.E., Pahud B.A., Weddle G., Avadhanula V., Munoz F.M., Piedra P.A., Payne D.C., Langley G., Gerber S.I. Respiratory syncytial virus – associated hospitalizations among young children: 2015–2016. Pediatrics. 2020;146:1–10. doi: 10.1542/peds.2019-3611. [DOI] [PubMed] [Google Scholar]
- Rima B., Collins P., Easton A., Fouchier R., Kurath G., Lamb R.A., Lee B., Maisner A., Rota P., Wang L. ICTV virus taxonomy profile: Pneumoviridae. J. Gen. Virol. 2017;98:2912–2913. doi: 10.1099/jgv.0.000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen M.O., Carr J.K., Burke D.S., Mccutchan F.E. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retrovir. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- Schobel S.A., Stucker K.M., Moore M.L., Anderson L.J., Larkin E.K., Shankar J., Bera J., Puri V., Shilts M.H., Rosas-Salazar C., Halpin R.A., Fedorova N., Shrivastava S., Stockwell T.B., Peebles R.S., Hartert T.V., Das S.R. Respiratory Syncytial Virus whole-genome sequencing identifies convergent evolution of sequence duplication in the C-terminus of the G gene. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- Smith K.F., Sax D.F., Gaines S.D., Guernier V., Guégan J.F. Globalization of human infectious disease. Ecology. 2007;88:1903–1910. doi: 10.1890/06-1052.1. [DOI] [PubMed] [Google Scholar]
- Spann K.M., Collins P.L., Teng M.N. Genetic recombination during coinfection of two mutants of human respiratory syncytial virus. J. Virol. 2003;77:11201–11211. doi: 10.1128/JVI.77.20.11201-11211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann K.M., Tran K.-C., Chi B., Rabin R.L., Collins P.L. Suppression of the induction of alpha, beta, and Gamma interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 2004;78:4363–4369. doi: 10.1128/JVI.78.8.4363-4369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullender W.M. Respiratory syncytial virus genetic and antigenic diversity. Clin. Microbiol. Rev. 2000;13:1–15. doi: 10.1128/cmr.13.1.1-15.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedan S., Musiyenko A., Barik S. Respiratory syncytial virus nonstructural proteins decrease levels of multiple members of the cellular interferon pathways. J. Virol. 2009;83:9682–9693. doi: 10.1128/JVI.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Coenjaerts F.E.J., Houspie L., Viveen M.C., van Bleek G.M., Wiertz E.J.H.J., Martin D.P., Lemey P. The comparative genomics of human respiratory syncytial virus subgroups A and B: genetic variability and molecular evolutionary dynamics. J. Virol. 2013;87:8213–8226. doi: 10.1128/JVI.03278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trento A., Casas I., Calderón A., Garcia-Garcia M.L., Calvo C., Perez-Breña P., Melero J.A. Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60-nucleotide duplication in the G protein gene. J. Virol. 2010;84:7500–7512. doi: 10.1128/JVI.00345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter M., Madhi S.A., Tiemessen C.T., Schoub B.D. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J. Gen. Virol. 2001;82:2117–2124. doi: 10.1099/0022-1317-82-9-2117. [DOI] [PubMed] [Google Scholar]
- Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:1–13. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Liu C., Xiao Y., Xiang Z., Zhou H., Chen L., Shen K., Xie Z., Ren L., Wang J. Respiratory syncytial virus seasonality, beijing, China, 2007–2015. Emerg. Infect. Dis. 2019;25:1127–1135. doi: 10.3201/eid2506.180532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.M., Fu Y.H., Peng X.L., Zheng Y.P., He J.S. Genetic diversity and molecular evolution of human respiratory syncytial virus A and B. Sci. Rep. 2021;11:1–11. doi: 10.1038/s41598-021-92435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.Y., Du L.N., Chen X., Zhao Y., Liu E.M., Yang X.Q., Zhao X.D. Genetic variability of respiratory syncytial viruses (RSV) Prevalent in southwestern China from 2006 to 2009: emergence of subgroup B and A RSV as dominant strains. J. Clin. Microbiol. 2010;48:1201–1207. doi: 10.1128/JCM.02258-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlateva K.T., Lemey P., Moës E., Vandamme A.-M., Van Ranst M. Genetic variability and molecular evolution of the human respiratory syncytial virus subgroup B attachment G protein. J. Virol. 2005;79:9157–9167. doi: 10.1128/JVI.79.14.9157-9167.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed in the current study are included in this published article and its supplementary materials.