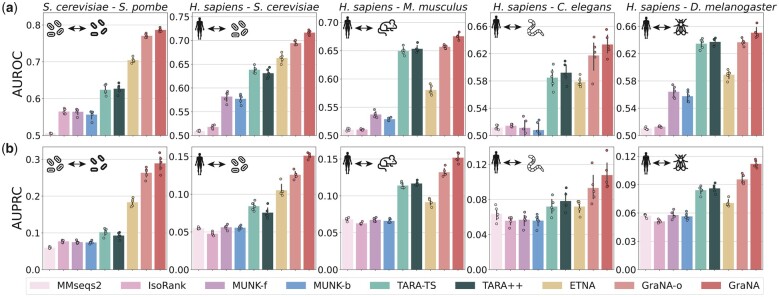
Figure 2.
Performances of NA prediction. GraNA and other baselines were evaluated for aligning functionally similar proteins across five pairs of species, using (a) AUROC and (b) AUPRC as metrics. GraNA-o is a variant of GraNA that only uses orthologs as anchor links whereas GraNA refers to the full model that uses both orthologs and sequence similarity as anchor links. The default E-value cutoff is used for MMseqs2. As MUNK is not a bidirectional NA method, the performances of its forward and backward predictions were shown separately as MUNK-f and MUNK-b. Performances were evaluated using five independent train/test data splits. Raw AUROC and AUPRC scores are provided in Supplementary Tables S4 and S5.