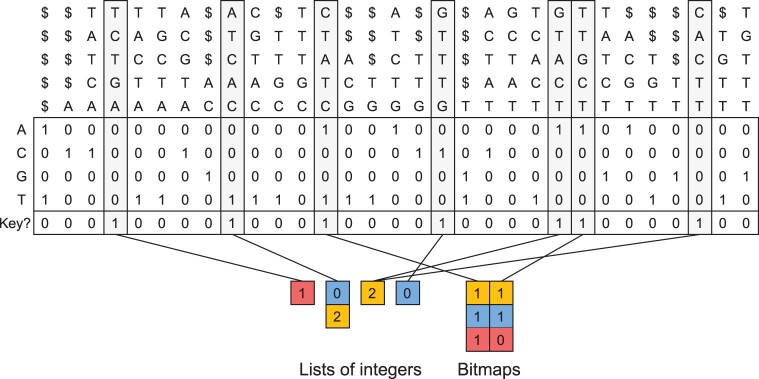
Figure 3.
The Matrix-SBWT k-mer index and the mapping to color sets. This figure continues from the example in Fig. 2. The columns of the SBWT matrix represent the k-mers of the input data, with technical dummy prefixes containing dollar-symbols added to the k-mers ending in the first k positions of the input sequences. The k-mers are shown vertically at the top (for illustration purposes only—they are not explicitly stored), and the SBWT matrix is the binary matrix in the middle with 4 rows. Each row corresponds to a character of the alphabet, and a 1-bit at cell (i, j) indicates that the jth k-mer has a different (k-1)-suffix from the previous k-mer, and has an outgoing edge such that the last character of the edge -mer is the ith character of the alphabet. See Alanko et al. (2022) for a more in-depth explanation. The columns shaded in gray are the key k-mers, which are also marked in the bit vector below the SBWT matrix. The key k-mers are associated with the color sets at the bottom. The sparse sets are encoded as lists of integers, whereas the dense sets are encoded as bit maps. The mapping from key k-mers to the color sets, that is represented by lines in the figure, is implemented by marking with another bit vector (not pictured) whether the set is sparse or dense, and using a bit vector rank query to find the index of the set within the color sets of its type (sparse or dense). Color sets of a single type are stored in concatenated form, with pointers to the starts of the sets.