Abstract
An integrative analysis recovered a new species of the Hemiphyllodactylustypus group from a karst formation in Lung Cu Commune, Dong Van District, Ha Giang Province, northeastern Vietnam. Hemiphyllodactyluslungcuensissp. nov. is embedded within clade 6 of the typus group, bearing an uncorrected pairwise sequence divergence of 4.6–20.2% from all other species based on a 1,038 base pair segment of the mitochondrial NADH dehydrogenase subunit 2 gene (ND2). It is diagnosable from other species in clade 6 by statistically significant mean differences in normalized morphometric, meristic, and categorical characters. A multiple factor analysis using the three aforementioned character types recovered its unique, non-overlapping placement in morphospace as statistically significantly different from that of all other species in clade 6. The description of this new Hemiphyllodactylus species contributes to a growing body of literature underscoring the high degree of herpetological diversity and endemism in karst landscapes in Vietnam as well as in the genus Hemiphyllodactylus.
Key words: Genetics, Hemiphyllodactyluslungcuensis sp. nov., integrative approach, karst forest, morphology, Southeast Asia
Introduction
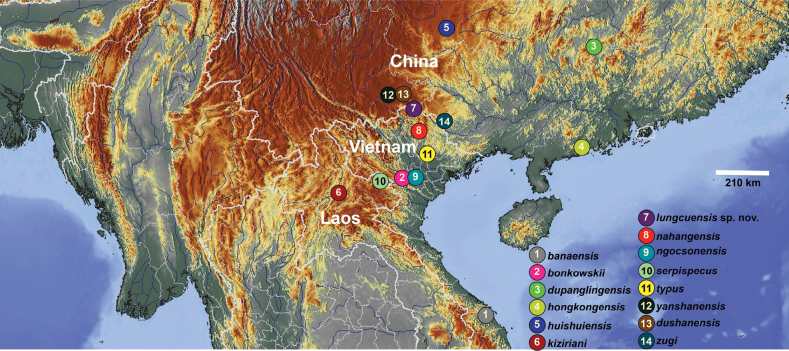
The gekkonid genus Hemiphyllodactylus Bleeker, 1860 consists of 54 species that are widely distributed from southern India and Sri Lanka, across Southeast Asia to the western Pacific (Zug 2010; Grismer et al. 2013, 2018; Agarwal et al. 2019; Agung et al. 2022; Uetz et al. 2022). Despite this broad distribution, their relatively small size, low densities, localized distributions, and cryptic coloration, render them inconspicuous components of the microenvironments they inhabit (Grismer et al. 2020). Bourret (1937) described the first species of Hemiphyllodactylus from Vietnam, namely H.typuschapaensis from Sa Pa, Lao Cai Province. Zug (2010), based on the examination of the holotype of H.typuschapaensis, considered it to be referable to H.yunnanensis Boulenger. Since then, a total of seven species of Hemiphyllodactylus have been recognized in the country, including H.yunnanensis Boulenger, 1903 from northern regions (Zug 2010); H.zugi Nguyen, Lehmann, Le, Duong, Bonkowski and Ziegler 2013 from Cao Bang Province; H.banaensis Ngo, Grismer, Pham and Wood 2014 from Da Nang City; H.bonkowskii and H.ngocsonensis Nguyen, Do, Ngo, Pham, Pham, Le and Ziegler 2020 from Hoa Binh Province; H.nahangensis Do, Pham, Phan, Le, Ziegler and Nguyen 2020 from Tuyen Quang Province; and H.dalatensis Do, Nguyen, Le, Pham, Ziegler and Nguyen 2021 from Lam Dong Province (Fig. 1).
Figure 1.
Distribution of the species of clade 6 of the genus Hemiphyllodactylus.
Based on phylogenetic analyses conducted in previous studies, Hemiphyllodactylus species belonging to clade 6 of the Hemiphyllodactylustypus group have been designated (Agung et al. 2021, 2022). This group includes various species such as H.banaensis, H.bonkowskii, H.nahangensis, H.ngocsonensis, and H.zugi from Vietnam; H.kiziriani Nguyen, Botov, Le, Nophaseud, Zug, Bonkowski, and Ziegler 2014, H.serpispecus Eliades, Phimmachak, Sivongxay, Siler and Stuart 2019 from Laos; and H.dupanglingensis Zhang, Qian, Jiang, Cai, Deng and Yang 2020, H.dushanensis Zhou, Liu and Yang 1981, H.hongkongensis Sung, Lee, Ng, Zhang and Yang 2018, H.huishuiensis Yan, Lin, Guo, Li and Zhou 2016; H.yanshanensis Agung, Chornelia, Grismer, Grismer, Quah, Lu, Tomlinson and Hughes 2022 from southern China.
During herpetological surveys in Lung Cu Commune, Dong Van District, Ha Giang Province, northeastern Vietnam, we collected seven specimens of an unnamed species of Hemiphyllodactylus. The new population differed from the remaining known species of the genus based on morphological and molecular phylogenetic analyses. Therefore, we herein describe it as a new species.
Materials and methods
Species delimitation
The general lineage concept (GLC: de Queiroz 2007) adopted herein proposes that a species constitutes a population of organisms evolving independently from other such populations owing to a lack of, or limited gene flow. By “independently” it is meant that new mutations arising in one species cannot spread readily into another species (Barraclough et al. 2003; de Queiroz 2007). Molecular phylogenies recovered multiple monophyletic mitochondrial lineages of individuals (populations) that were used to develop initial species-level hypotheses, the grouping stage of Hillis (2019). Discrete color pattern data and univariate and multivariate analyses of morphological data were then used to search for characters and morphospatial patterns consistent with the tree-designated species-level hypotheses, the construction of boundaries representing the hypothesis-testing step of Hillis (2019), thus providing independent diagnoses to complement the molecular analyses. In this way, delimiting (phylogeny) and diagnosing (taxonomy) species are not conflated (Frost and Hillis 1990; Frost and Kluge 1994; Hillis 2019).
Sampling
Field surveys were conducted during the month of August in 2017 and 2022 in Lung Cu Commune, Dong Van District, Ha Giang Province, northeastern Vietnam. After having been photographed alive, newly collected individuals, were anaesthetized and euthanized in a closed vessel with a piece of cotton wool containing ethyl acetate (Simmons 2002), fixed in 85% ethanol, thereafter and subsequently transferred to 70% ethanol for permanent storage. Tissue samples were preserved separately in 70% ethanol. Specimens were deposited in the collection of the Vietnam National University of Forestry (VNUF) and the Institute of Ecology and Biological Resources (IEBR), Hanoi, Vietnam.
Morphological data
Terminology of morphological characters follows Zug (2010), Nguyen et al. (2013), and Grismer et al. (2021). Measurements (morphometrics) were taken with a digital caliper to the nearest 0.1 mm on the left side of the body. Character abbreviations and descriptions are as follows: SVL: snout-vent length (from the tip of snout to the vent); TaL: tail length (from the vent to the tip of the tail, original and regenerated); TrunkL: trunk length (from the posterior margin of the forelimb at its insertion point on the body to the anterior margin of the hindlimb insertion point on the body); HeadL: head length (from the posterior margin of the retroarticular process of the lower jaw to the tip of the snout); HeadW: head width (measured at the angle of the jaws); EyeD: eye diameter (the greatest horizontal diameter of the eyeball); SnEye: snout-eye length (from anteriormost margin of the eyeball to the tip of snout); NarEye: nares-eye length (from the anterior margin of the eyeball to the posterior margin of the external nares); SnW: internarial width (measured between the nares across the rostrum); and EarD: ear diameter (maximum diameter of the ear opening).
The following meristic characters were counted using the same dissecting microscope: Chin: chin scales (the number of scales contacting the infralabials and mental from the juncture of the second and third infralabials on the left side to the juncture of the second and third infralabials on the right side); CN: circumnasal scales (the number of scales abutting the external naris, exclusive of the rostral and first supralabial); SnS: the number of scales between the supranasals; SL: supralabial scales (number of enlarged scales bordering the mouth on the upper jaw from the rostral to a point in line with the posterior margin of the orbit); IL: infralabial scales (the number of enlarged scales bordering the mouth on the lower jaw from the mental to a point in line with the posterior margin of the orbit); VS: ventral scales (the number of longitudinal ventral scales at mid-body contained within one eye diameter); DS: dorsal scales (the number of longitudinal dorsal scales at mid-body contained within one eye diameter); lamellae formulae determined as the number of U-shaped, subdigital lamellae (split and single) on the digital pads on digits II–V of the hands and feet (on the left); SL1F: the number of subdigital lamellae wider than long on the first finger; SL1T: the number of subdigital lamellae wider than long on the first toe; the total number of precloacal and femoral pores (i.e. the contiguous or discontinuous rows of femoral and precloacal pore-bearing scales); and CloacS: the number of cloacal spurs.
The categorical color pattern characters evaluated were the presence or absence of dark markings on the dorsum (BodPartn); presence or absence of a dark pre– and/ or postorbital stripe extending onto at least the neck (PosOStrp); the presence or absence of a linear series of white postorbital and dorsolateral spots on the trunk (PosOTrSpt); and the presence or absence of pale–colored, anteriorly projecting arms of the pale–colored postsacral marking (PosSacrlMakg).
Genetic data
We obtained 1,038 base pairs of the NADH dehydrogenase subunit 2 (ND2) gene from 45 specimens from GenBank and five newly sequenced specimens from Lung Cu Commune, Dong Van District, Ha Giang Province, northeastern Vietnam for the phylogenetic analyses (Table 1). Catalog numbers are abbreviated as follows: CSUFT, Central South University of Forestry and Technology, Changsha, China; ITBCZ, Institute of Tropical Biology, Zoological Collection; KIZ: Kunming Institute of Zoology; LSUHC, La Sierra University Herpetology Collection; NJUN, Nanjing Normal University, Nanjing; MVZ, Museum of Vertebrate Zoology; NUOL, National University of Laos; SYS, The Museum of Biology, Sun Yatsen University (SYS), Guangzhou; ZFMK, Zoologisches Forschunginstitut und Museum Alexander Koenig, Bonn, Germany. Hemiphyllodactylusharterti (Werner, 1900) of the H.harterti group was used to root the tree following Grismer et al. (2013). For molecular phylogenetic analyses, we used the protocols of Le et al. (2006) for DNA extraction, amplification, and sequencing. Tissue samples were extracted using DNeasy blood and tissue kit, Qiagen (California, USA). Extracted DNA from the fresh tissue was amplified by PCR mastermix (Fermentas, Canada). A fragment of the mitochondrial gene ND2 (NADH dehydrogenase subunit 2) was amplified using the primer pair ND2f101A (CAACAGAAGCCACAACAAAAT) and HemiR (GAAGAAGAGGCTTGGKAGGCT) (Nguyen et al. 2013). The PCR volume consisted of 21 μl (10 μl of mastermix, 5 μl of water, 2 μl of each primer at 10 pmol/μl and 2 μl of DNA or higher depending on the quantity of DNA in the final extraction solution). The following temperature profile for PCR was used: 95 °C for 5 min to activate the taq; with 40 cycles at 95 °C for 30 s, 50 °C for 45 s, 72 °C for 60 s; and the final extension at 72 °C for 6 min. PCR products were subjected to electrophoresis through a 1% agarose gel (UltraPureTM, Invitrogen). Gels were stained for 10 minutes in 1×TBE buffer at 2 pg/ml of ethidium-bromide and visualized under UV light. Successful amplifications were purified to eliminate PCR components using GeneJETTM PCR Purification kit (Fermentas, Canada). Purified PCR products were sent to 1st Base (Selangor, Malaysia) for sequencing.
Table 1.
Samples used for the phylogenetic analyses.
| Species | Catalog no. | Locality | GenBank no. |
|---|---|---|---|
| Hemiphyllodactylusbanaensis | ITBCZ 2450 | Ba Na-Nui Chua, Vietnam | KF219783 |
| H.bonkowskii | IEBR 4694 | Hang Kia-Pa Co NR, Mai Chau, Hoa Binh, Vietnam | MT415553 |
| IEBR 4695 | Hang Kia-Pa Co NR, Mai Chau, Hoa Binh, Vietnam | MT415554 | |
| H.dupanglingensis | CSUFT 00401 | Dupangling, Hunan, China | MT576070 |
| CSUFT 00405 | Dupangling, Hunan, China | MT576071 | |
| H.hongkongensis | SYS r001728 | Aberdeen Country Park, Hong Kong | MF893330 |
| SYS r001729 | Aberdeen Country Park, Hong Kong | MF893331 | |
| SYS r001730 | Aberdeen Country Park, Hong Kong | MF893332 | |
| SYS r001735 | Aberdeen Country Park, Hong Kong | MF893333 | |
| H.huishuiensis | NJNUh00736 | Huishui, Guizhou, China | KU519708 |
| NJNUh00851 | Huishui, Guizhou, China | KU519707 | |
| NJNUh00852 | Huishui, Guizhou, China | KU519709 | |
| NJNUh00857 | Huishui, Guizhou, China | KU519710 | |
| NJNUh00858 | Huishui, Guizhou, China | KU519711 | |
| H.kiziriani | IEBR A.2014.3 | Luang Prabang, Laos | KJ676800 |
| IEBR A.2014.4 | Luang Prabang, Laos | KJ676801 | |
| IEBR A.2014.5 | Luang Prabang, Laos | KJ676802 | |
| Hemiphyllodactyluslungcuensis sp. nov. | VNUF R.2021.01 | Lung Cu, Dong Van, Ha Giang, Vietnam | OR067852 |
| IEBR R.5151 | Lung Cu, Dong Van, Ha Giang, Vietnam | OR067853 | |
| VNUF R.2021.03 | Lung Cu, Dong Van, Ha Giang, Vietnam | OR067851 | |
| VNUF R.2023.01 | Lung Cu, Dong Van, Ha Giang, Vietnam | OR067854 | |
| VNUF R.2023.02 | Lung Cu, Dong Van, Ha Giang, Vietnam | OR067855 | |
| H.nahangensis | IEBR 4741 | Na Hang, Tuyen Quang, Vietnam | MT711191 |
| IEBR 4742 | Na Hang, Tuyen Quang, Vietnam | MT711192 | |
| IEBR 4743 | Na Hang, Tuyen Quang, Vietnam | MT711193 | |
| H.ngocsonensis | IEBR 4689 | Ngoc Son-Ngo Luong NR, Lac Son, Hoa Binh, Vietnam | MT415551 |
| IEBR 4690 | Ngoc Son-Ngo Luong NR, Lac Son, Hoa Binh, Vietnam | MT415552 | |
| H.serpispecus | NUOL 00476 | Tham Ngou Leium Cave, Viengxay, Houaphan, Laos | MK307996 |
| H.typus | MVZ 226500 | Vinh Phuc, Vietnam | KF219798 |
| H.yanshanensis | KIZ062101 | Yunnan, China | ON676172 |
| KIZ062102 | Yunnan, China | ON676173 | |
| KIZ062090 | Yunnan, China | ON676161 | |
| KIZ062091 | Yunnan, China | ON676162 | |
| KIZ062092 | Yunnan, China | ON676163 | |
| KIZ062093 | Yunnan, China | ON676164 | |
| KIZ062094 | Yunnan, China | ON676165 | |
| KIZ062095 | Yunnan, China | ON676166 | |
| KIZ062096 | Yunnan, China | ON676167 | |
| KIZ062097 | Yunnan, China | ON67618 | |
| KIZ062098 | Yunnan, China | ON676169 | |
| KIZ062099 | Yunnan, China | ON676170 | |
| KIZ062100 | Yunnan, China | ON676171 | |
| H.dushanensis | isolate_N1 | Yunnan, China | FJ971016 |
| isolate_N2 | Yunnan, China | FJ971017 | |
| H.zugi | ZFMK 94782 | Ha Lang, Cao Bang, Vietnam | KF575153 |
| IEBR A.2013.20 | Ha Lang, Cao Bang, Vietnam | KF575151 | |
| IEBR A.2013.21 | Ha Lang, Cao Bang, Vietnam | KF575152 | |
| H.harterti | LSUHC10383 | Bukit Larut, Malaysia | KF219760 |
| LSUHC10384 | Bukit Larut, Malaysia | KF219761 |
Phylogenetic analyses
Maximum likelihood (ML) and Bayesian inference (BI) analyses were used to estimate the phylogenetic relationships among the sampled sequences in the alignment. An ML phylogeny was estimated using the IQ-TREE webserver (Nguyen et al. 2015; Trifinopoulos et al. 2016) preceded by the selection of substitution models using the Bayesian Information Criterion (BIC) in Model Finder (Kalyaanamoorthy et al. 2017), which supported TN+F+G4 as the best fit model of evolution for ND2 codon position 1, TN+F+G4 for position 2 and TIM2+F + G4 for position 3. One thousand bootstrap pseudoreplicates via the ultrafast bootstrap (UFB; Hoang et al. 2018) approximation algorithm were employed and nodes having ML UFB values of 95 and above were considered highly supported (Minh et al. 2013).
The Bayesian inference (BI) analysis was carried out using Mr Bayes 3.2.7. (Ronquist et al. 2012) XSEDE on the CIPRES Science Gateway (Cyberinfrastructure for Phylogenetic Research; Miller et al. 2010), employing the GTR+I+G model of evolution to all partitions. Two independent Markov chain Monte Carlo (MCMC) simulations were performed each with four chains, three hot and one cold. The MCMC simulation was run for 100 million generations, sampled every 10,000 generations, and the first 10% of each run was discarded as burn-in. Convergence and stationarity of all parameters from both runs were checked using Tracer v1.6 (Rambaut et al. 2014) to ensure effective sample sizes (ESS) were above 200. Post-burn-in sampled trees from both runs were combined using the sumt function in MrBayes and a 50% majority-rule consensus tree was constructed. Nodes with Bayesian posterior probabilities (BPP) of 0.95 and above were considered highly supported (Huelsenbeck et al. 2001; Wilcox et al. 2002). Uncorrected pairwise sequence divergences (p-distance) among and within species were calculated in MEGA11 (Kumar et al. 2016) (Table 2).
Table 2.
Uncorrected genetic P-distances (%) in ND2 gene of Hemiphyllodactylus. Intraspecific p-distance are in bold font, n/a = data not applicable.
| banaensis | bonkowskii | dupanglingensis | dushanensis | harterti | hongkongensis | huishuiensis | kiziriani | lungcuensis sp. nov. | nahangensis | ngocsonensis | serpispecus | typus | yanshanensis | zugi | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| banaensis | n/a | ||||||||||||||
| bonkowskii | 13.0 | 0.01 | |||||||||||||
| dupanglingensis | 16.9 | 15.3 | 0.00 | ||||||||||||
| dushanensis | 17.3 | 14.7 | 5.1 | 0.00 | |||||||||||
| harterti | 30.4 | 26.5 | 27.8 | 29.1 | 0.04 | ||||||||||
| hongkongensis | 16.3 | 14.6 | 7.9 | 7.1 | 27.5 | 0.01 | |||||||||
| huishuiensis | 15.7 | 15.4 | 7.1 | 6.4 | 28.9 | 7.9 | 0.01 | ||||||||
| kiziriani | 22.7 | 20.0 | 20.3 | 20.5 | 29.6 | 20.0 | 20.5 | 0.00 | |||||||
| lungcuensis sp. nov. | 15.8 | 13.8 | 6.2 | 4.8 | 27.2 | 7.0 | 5.3 | 20.2 | 0.02 | ||||||
| nahangensis | 15.5 | 13.6 | 5.8 | 5.5 | 25.3 | 7.8 | 6.3 | 20.1 | 4.6 | 0.01 | |||||
| ngocsonensis | 14.5 | 12.8 | 7.6 | 7.9 | 27.0 | 9.2 | 7.8 | 19.3 | 6.6 | 7.0 | 0.02 | ||||
| serpispecus | 12.2 | 7.5 | 16.6 | 16.0 | 25.7 | 16.4 | 15.4 | 21.4 | 14.4 | 13.2 | 12.6 | n/a | |||
| typus | 15.2 | 14.6 | 7.2 | 6.3 | 27.9 | 8.9 | 7.4 | 21.2 | 5.6 | 4.1 | 8.8 | 15.6 | n/a | ||
| yanshanensis | 17.1 | 15.2 | 8.6 | 8.3 | 26.0 | 9.4 | 4.4 | 22.2 | 6.8 | 7.3 | 8.5 | 14.7 | 8.2 | 0.00 | |
| zugi | 16.0 | 14.0 | 4.8 | 5.8 | 27.0 | 7.4 | 7.0 | 19.9 | 6.8 | 6.2 | 7.5 | 15.0 | 7.3 | 8.1 | 0.00 |
Statistical analyses
We compared the Lung Cu population to its closest relatives Hemiphyllodactylushuishuiensis and H.yanshanensis from southern China and to other species in clade 6. Hemiphyllodactylusdushanensis was not evaluated due to the absence of comparative morphological data. Morphological data used for the statistical analyses were obtained from the Lung Cu population (seven specimens) and data from 76 specimens of 12 other species available from previous studies (Ngo et al. 2014; Nguyen et al. 2014; Yan et al. 2016; Sung et al. 2018; Eliades et al. 2019; Nguyen et al. 2020; Do et al. 2020; Zhang et al. 2020; Agung et al. 2022). Raw data for H.typus (four specimens [cat. Nos. CUMZ. R 2013.19; RBWS 19309; JAV bg. R.39; MS92002]) was obtained from the La Sierra University Herpetological Collection, La Sierra University, Riverside, California, USA. Museum Abbreviations: CUMZ - Museum of Zoology, Chulalongkorn University, Bangkok, Thailan; JAV-J.-M. VINSON; MS-Montri Sumontha; RBWS: Royal Belgian Institute of Natural Sciences. The raw morphological data are provided in Tables 3, 4.
Table 3.
Morphometric and categorical data used in the analyses from specimens of Hemiphyllodactylus members within clade 6; m = male; f = female.
| Species | Museum no. | Sex | Morphometric data | Categorical data | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SVL | TrunkL | HeadL | HeadW | SnEye | NarEye | EyeD | SnW | PosOStrp | BodPartn | PosSacrlMakg | PosOTrSpt | |||
| banaensis | ITBCZ 2465 | m | 47.3 | 21.7 | 11.2 | 7.2 | 4.2 | 3.2 | 2.7 | 1.5 | No | Yes | Yes | No |
| ITBCZ 2461 | m | 48.2 | 23.8 | 10.8 | 7.6 | 4.5 | 3.5 | 2.7 | 1.5 | No | Yes | Yes | No | |
| ITBCZ 2450 | f | 48.3 | 23.8 | 10.7 | 7.2 | 4 | 3.3 | 2.7 | 1.6 | No | Yes | Yes | No | |
| ITBCZ 2462 | f | 50.9 | 24.4 | 12 | 7.9 | 4.6 | 3.6 | 2.7 | 1.7 | No | Yes | Yes | No | |
| ITBCZ 2463 | f | 50 | 23.7 | 11.3 | 7.4 | 4.1 | 3.5 | 2.5 | 1.7 | No | Yes | Yes | No | |
| ITBCZ 2464 | f | 51 | 24.1 | 12 | 7.9 | 4.5 | 3.6 | 2.6 | 1.8 | No | Yes | Yes | No | |
| ITBCZ 2466 | f | 50.6 | 24.4 | 11.5 | 7.9 | 5.2 | 3.5 | 2.5 | 1.7 | No | Yes | Yes | No | |
| ITBCZ 2467 | f | 50.1 | 22.3 | 11 | 7.5 | 4.4 | 3.1 | 2.4 | 1.7 | No | Yes | Yes | No | |
| ITBCZ 2468 | f | 45.2 | 20.4 | 10 | 7 | 3.8 | 2.8 | 2.1 | 1.5 | No | Yes | Yes | No | |
| ITBCZ 2469 | f | 49 | 23 | 10.9 | 7.4 | 4.3 | 3.4 | 2.7 | 1.8 | No | Yes | Yes | No | |
| bonkowskii | IEBR 4689 | m | 45.8 | 23.6 | 10.5 | 8.6 | 4.9 | 3.2 | 3.1 | 1.9 | Yes | Yes | Yes | Yes |
| IEBR 4690 | m | 40.7 | 20.9 | 9.6 | 7.9 | 4 | 3.1 | 2.9 | 1.9 | Yes | Yes | Yes | Yes | |
| IEBR 4691 | f | 44.9 | 21.5 | 10.3 | 8.5 | 4.7 | 3.3 | 2.7 | 2.1 | Yes | Yes | Yes | Yes | |
| IEBR 4693 | f | 45.4 | 22.5 | 10.5 | 9.2 | 5.1 | 3.6 | 3.1 | 2.1 | Yes | Yes | Yes | Yes | |
| IEBR 4692 | f | 48 | 26.7 | 11.6 | 9.4 | 5.2 | 3.6 | 3.1 | 2.4 | Yes | Yes | Yes | Yes | |
| IEBR 4749 | f | 36.6 | 17.6 | 8.9 | 7.1 | 4 | 2.9 | 2.3 | 1.7 | Yes | Yes | Yes | Yes | |
| dupanglingensis | CSUFT 00401 | f | 40.8 | 18.7 | 9.8 | 7.5 | 4.5 | 3.3 | 2.5 | 1.8 | Yes | Yes | No | Yes |
| CSUFT 00402 | m | 41 | 19.4 | 11.2 | 7.5 | 4.7 | 3.2 | 2.6 | 1.5 | Yes | Yes | No | Yes | |
| CSUFT 00403 | m | 39.5 | 17.2 | 10.3 | 7.2 | 4.2 | 3 | 2.5 | 1.4 | Yes | Yes | No | Yes | |
| CSUFT 00404 | m | 40.8 | 19.7 | 11.1 | 7.6 | 4.3 | 3.1 | 2.7 | 1.6 | Yes | Yes | No | Yes | |
| CSUFT 00405 | m | 39.4 | 18.1 | 10.5 | 6.8 | 4.3 | 3.2 | 2.5 | 1.7 | Yes | Yes | No | Yes | |
| CSUFT 00406 | m | 44.7 | 22.8 | 10.9 | 7.8 | 4.6 | 3.3 | 2.7 | 1.7 | Yes | Yes | No | Yes | |
| hongkongensis | SYS r001735 | m | 33.6 | 15.8 | 9.3 | 6.9 | 3.1 | 2.4 | 2 | 1.1 | Yes | Yes | No | Yes |
| SYS r001728 | f | 37.5 | 19.4 | 9.5 | 5.2 | 3.2 | 2.6 | 2.2 | 1.2 | Yes | Yes | No | Yes | |
| SYS r001729 | f | 37.4 | 19.3 | 9.9 | 6.4 | 3.7 | 2.5 | 2.3 | 1.1 | Yes | Yes | No | Yes | |
| SYS r001730 | f | 40.8 | 21 | 10.4 | 8 | 3.8 | 2.9 | 2.4 | 1.2 | Yes | Yes | No | Yes | |
| SYS r001731 | f | 42.1 | 21.3 | 10.3 | 7.5 | 3.6 | 3.1 | 2.3 | 1.3 | Yes | Yes | No | Yes | |
| SYS r001732 | f | 43 | 22 | 11.2 | 8.2 | 4 | 3 | 2.5 | 1.4 | No | No | No | Yes | |
| SYS r001733 | f | 38.9 | 20.4 | 10.4 | 8.2 | 3.6 | 3 | 2.3 | 1.3 | Yes | Yes | No | Yes | |
| SYS r001734 | m | 32.3 | 15.6 | 8.7 | 7 | 3.2 | 2.6 | 2.3 | 1.1 | Yes | Yes | No | Yes | |
| huishuiensis | NJNUh00851 | m | 42.6 | 20.5 | 9.5 | 7.8 | 3.9 | 2.8 | 2.7 | 1.2 | Yes | Yes | Yes | No |
| NJNUh00859 | m | 47.4 | 22.4 | 13.9 | 8.5 | 4.4 | 3.1 | 2.7 | 1.4 | Yes | Yes | Yes | No | |
| NJNUh00852 | f | 44.3 | 22.3 | 9.8 | 7.5 | 3.8 | 2.8 | 2.5 | 1.2 | Yes | Yes | Yes | No | |
| NJNUh00854 | f | 51.2 | 25 | 10.8 | 8.8 | 4.8 | 3.6 | 2.7 | 1.5 | Yes | Yes | Yes | No | |
| NJNUh00855 | f | 49.3 | 24.9 | 11.9 | 8.4 | 4.8 | 3.2 | 2.8 | 1.2 | Yes | Yes | Yes | No | |
| NJNUh00856 | f | 36.2 | 17.3 | 8.4 | 7.1 | 3.1 | 2.5 | 2.4 | 1 | Yes | Yes | Yes | No | |
| NJNUh00857 | f | 45.5 | 21.5 | 10.1 | 8 | 4.2 | 3.4 | 2.4 | 1.1 | Yes | Yes | Yes | No | |
| NJNUh00858 | f | 35.7 | 17.4 | 8.4 | 6.3 | 3.7 | 2.5 | 2.3 | 1.1 | Yes | Yes | Yes | No | |
| kiziriani | IEBR A.2014.3 | m | 40.1 | 19.7 | 7.4 | 6.7 | 4.2 | 3.2 | 2.7 | 1.7 | Yes | Yes | Yes | Yes |
| IEBR A.2014.4 | m | 35.1 | 16.4 | 6 | 6.3 | 3.8 | 3 | 2.5 | 1.3 | Yes | Yes | Yes | Yes | |
| VNMN A.2014.1 | m | 37.6 | 18.8 | 6.7 | 6.9 | 3.5 | 2.7 | 2.4 | 1.4 | Yes | Yes | Yes | Yes | |
| ZFMK 95702 | m | 36.2 | 18.8 | 6.3 | 7.2 | 3.8 | 2.8 | 2.7 | 1.3 | Yes | Yes | Yes | Yes | |
| kiziriani | IEBR A.2014.5 | f | 40.8 | 20.7 | 7.1 | 6.8 | 3.9 | 3.1 | 2.85 | 1.4 | Yes | Yes | Yes | Yes |
| NUOL R-2014.1 | f | 36.3 | 16.4 | 6.1 | 6.7 | 3.8 | 2.9 | 2.4 | 1.3 | Yes | Yes | Yes | Yes | |
| NUOL R-2014.2 | f | 40 | 20.1 | 6.3 | 7.4 | 4 | 3 | 2.4 | 1.3 | Yes | Yes | Yes | Yes | |
| VNMN A.2014.2 | f | 40.8 | 19.2 | 7.3 | 6.8 | 4.1 | 3.3 | 2.6 | 1.4 | Yes | Yes | Yes | Yes | |
| ZFMK 95703 | f | 36.7 | 19 | 6.2 | 6.6 | 3.8 | 3 | 2.4 | 1.4 | Yes | Yes | Yes | Yes | |
| ZFMK 95704 | f | 40.1 | 22 | 7.3 | 7.3 | 4.4 | 3.3 | 2.7 | 1.5 | Yes | Yes | Yes | Yes | |
| Hemiphyllodactyluslungcuensis sp. nov. | VNUF R.2021.01 | m | 39.2 | 20.3 | 11 | 8 | 4.3 | 3.2 | 1.8 | 1.5 | Yes | Yes | Yes | Yes |
| IEBR R.5151 | m | 43 | 23 | 11.6 | 8 | 4.5 | 2.9 | 2.1 | 2 | Yes | Yes | Yes | Yes | |
| VNUF R.2021.03 | m | 44.2 | 23.1 | 11.1 | 7 | 5 | 3.6 | 2.4 | 1.7 | Yes | Yes | Yes | Yes | |
| IEBR R.5152 | f | 39.2 | 19 | 8.5 | 7.3 | 4.1 | 3.2 | 2.5 | 1.9 | Yes | Yes | Yes | Yes | |
| VNUF R.2021.08 | f | 43.5 | 22.3 | 10.3 | 8 | 4.2 | 3.3 | 2.1 | 2.1 | Yes | Yes | Yes | Yes | |
| VNUF R.2023.01 | m | 35.3 | 17 | 8.9 | 6.9 | 3.6 | 3.13 | 1.8 | 1.4 | Yes | Yes | Yes | Yes | |
| VNUF R.2023.02 | f | 37.4 | 19.3 | 8.5 | 7 | 3.9 | 3 | 1.8 | 1.4 | Yes | Yes | Yes | Yes | |
| nahangensis | IEBR 4741 | m | 43.6 | 21.3 | 10.7 | 7.4 | 3.8 | 3.1 | 2.5 | 1.3 | No | Yes | Yes | Yes |
| IEBR 4742 | m | 43.5 | 21.3 | 10.2 | 7.1 | 4 | 3.2 | 2.6 | 1.1 | No | Yes | Yes | Yes | |
| IEBR 4743 | f | 41.4 | 20.9 | 10 | 6.9 | 3.5 | 2.8 | 2.7 | 1 | No | Yes | Yes | Yes | |
| ngocsonensis | IEBR 4694 | m | 45.5 | 23.6 | 9.8 | 8.5 | 4.5 | 3.3 | 2.8 | 1.7 | Yes | No | Yes | Yes |
| IEBR 4695 | m | 45.9 | 23.7 | 10.2 | 8.2 | 4.9 | 3.8 | 2.8 | 1.8 | Yes | No | Yes | Yes | |
| IEBR 4696 | f | 46 | 23.1 | 10.5 | 8.6 | 4.6 | 3.3 | 2.7 | 1.7 | Yes | No | Yes | Yes | |
| IEBR 4697 | f | 46.9 | 22.6 | 10.5 | 8.5 | 4.6 | 3.1 | 2.8 | 1.8 | Yes | No | Yes | Yes | |
| serpispecus | NUOL 00476 | m | 41.9 | 21.4 | 10.4 | 7.7 | 4 | 3.3 | 2.4 | 1.5 | Yes | Yes | Yes | Yes |
| typus | JAV bg. R.39 | f | 34.3 | 18 | 8 | 5.3 | 3.3 | 2.5 | 1.7 | 1.2 | Yes | Yes | Yes | Yes |
| MS92002 | f | 34.5 | 20.1 | 9.1 | 5 | 3.5 | 2.6 | 2.1 | 0.7 | Yes | Yes | Yes | Yes | |
| CUMZ R 2003.19 | f | 36.3 | 22.1 | 8.1 | 4.9 | 3.1 | 2.5 | 2 | 1.1 | Yes | Yes | Yes | Yes | |
| RBWS 19309 | f | 22.6 | 12.4 | 6.6 | 4.5 | 2.4 | 1.9 | 1.5 | 0.8 | Yes | Yes | Yes | Yes | |
| yanshanensis | KIZ 062090 | m | 40.0 | 18.9 | 7.3 | 7.7 | 4.1 | 3.2 | 2.3 | 1.5 | Yes | Yes | Yes | No |
| KIZ 062091 | f | 43.9 | 20.1 | 8.2 | 8.4 | 4.2 | 3.2 | 2.7 | 1.5 | Yes | Yes | Yes | No | |
| KIZ 062092 | f | 46.3 | 22.3 | 8.3 | 8.6 | 4.6 | 3.5 | 2.4 | 1.6 | Yes | Yes | Yes | No | |
| KIZ 062093 | f | 38.8 | 18.8 | 8.1 | 8.4 | 4.3 | 3.4 | 2.7 | 1.6 | Yes | Yes | Yes | No | |
| KIZ 062094 | f | 40.8 | 19.8 | 7.3 | 7.7 | 4.1 | 3.1 | 2.3 | 1.4 | Yes | Yes | Yes | No | |
| KIZ 062095 | f | 41.2 | 20.1 | 7.5 | 8.1 | 4.5 | 3.1 | 2.2 | 1.6 | Yes | Yes | Yes | No | |
| KIZ 062096 | f | 42.6 | 19.3 | 7.8 | 8.3 | 4.2 | 3.2 | 2.6 | 1.5 | Yes | Yes | Yes | No | |
| KIZ 062097 | m | 43.5 | 19.7 | 7.4 | 8.6 | 4.1 | 3.1 | 2.3 | 1.5 | Yes | Yes | Yes | No | |
| KIZ 062098 | m | 38.6 | 18.2 | 7.3 | 7.7 | 3.8 | 3.0 | 2.5 | 1.5 | Yes | Yes | Yes | No | |
| KIZ 062099 | m | 34.0 | 17.4 | 6. | 6.4 | 3.5 | 2.4 | 1.7 | 1.3 | Yes | Yes | Yes | No | |
| KIZ 062100 | f | 37.0 | 17.7 | 6.8 | 7.6 | 3.7 | 3.0 | 2.4 | 1.4 | Yes | Yes | Yes | No | |
| KIZ 062101 | f | 43.1 | 19.5 | 8.1 | 8.2 | 4.3 | 3.4 | 2.6 | 1.7 | Yes | Yes | Yes | No | |
| KIZ 062102 | f | 41.1 | 19.9 | 7.5 | 7.8 | 4.3 | 3.4 | 2.3 | 1.5 | Yes | Yes | Yes | No | |
| zugi | IEBR A.2013.20 | m | 44.6 | 22.4 | 10.1 | 7.5 | 4.9 | 3.1 | 2.9 | 1.7 | Yes | Yes | No | Yes |
| ZFMK 94781 | m | 38.3 | 21.4 | 9.2 | 6.9 | 4.1 | 2.9 | 2.6 | 1.5 | Yes | Yes | No | Yes | |
| IEBR A.2013.21 | m | 40.1 | 20.8 | 9 | 7.1 | 4.1 | 2.6 | 2.7 | 1.5 | Yes | Yes | No | Yes | |
Table 4.
Meristic data used in the analyses from specimens of Hemiphyllodactylus members within clade 6; m = male; f = female.
| Species | Museum no. | Sex | Meristic data | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chin | CN | SL | IL | VS | DS | SL1F | SL1T | |||
| banaensis | ITBCZ 2465 | m | 6 | 3 | 11 | 10 | 9 | 18 | 5 | 5 |
| ITBCZ 2461 | m | 6 | 3 | 11 | 9 | 10 | 18 | 5 | 5 | |
| ITBCZ 2450 | f | 6 | 3 | 11 | 10 | 10 | 18 | 5 | 5 | |
| ITBCZ 2462 | f | 7 | 3 | 12 | 11 | 11 | 20 | 5 | 5 | |
| ITBCZ 2463 | f | 6 | 3 | 9 | 10 | 10 | 18 | 5 | 5 | |
| ITBCZ 2464 | f | 6 | 3 | 10 | 10 | 10 | 17 | 5 | 5 | |
| ITBCZ 2466 | f | 6 | 3 | 10 | 10 | 9 | 17 | 5 | 5 | |
| ITBCZ 2467 | f | 6 | 3 | 12 | 11 | 10 | 19 | 5 | 4 | |
| ITBCZ 2468 | f | 6 | 3 | 11 | 11 | 12 | 20 | 5 | 5 | |
| ITBCZ 2469 | f | 7 | 3 | 11 | 11 | 10 | 20 | 5 | 5 | |
| bonkowskii | IEBR 4689 | m | 7 | 3 | 10 | 10 | 13 | 24.5 | 5 | 5 |
| IEBR 4690 | m | 7 | 3 | 9 | 9 | 15 | 25 | 5 | 5 | |
| IEBR 4691 | f | 5 | 3 | 9 | 9 | 15 | 25 | 5 | 5 | |
| IEBR 4693 | f | 7 | 3 | 9.5 | 8 | 15 | 25.5 | 5 | 4 | |
| IEBR 4692 | f | 7 | 3 | 10 | 10.5 | 14 | 25 | 5 | 5 | |
| IEBR 4749 | f | 6 | 3 | 10 | 9 | 14 | 26 | 5 | 5 | |
| dupanglingensis | CSUFT 00401 | f | 10 | 4 | 11 | 12 | 11 | 15 | 5 | 5 |
| CSUFT 00402 | m | 10 | 3 | 11 | 9 | 11 | 15 | 4 | 4 | |
| CSUFT 00403 | m | 9 | 4 | 11 | 9 | 10 | 16 | 5 | 4 | |
| CSUFT 00404 | m | 11 | 3 | 12 | 10 | 10 | 15 | 5 | 5 | |
| CSUFT 00405 | m | 11 | 3 | 11 | 10 | 11 | 14 | 5 | 5 | |
| CSUFT 00406 | m | 11 | 3 | 14 | 9 | 11 | 14 | 5 | 5 | |
| hongkongensis | SYS r001735 | m | 5 | 3 | 10 | 9.5 | 10 | 15 | 5 | 5 |
| SYS r001728 | f | 6 | 3 | 11 | 10 | 10 | 13 | 4 | 5 | |
| SYS r001729 | f | 5 | 3 | 12 | 10 | 9 | 14 | 4 | 5 | |
| SYS r001730 | f | 6 | 3 | 12 | 10 | 9 | 13 | 4 | 5 | |
| SYS r001731 | f | 6 | 3 | 10 | 10 | 9 | 12 | 3 | 5 | |
| SYS r001732 | f | 5 | 4 | 11.5 | 10.5 | 9 | 14 | 4 | 5 | |
| SYS r001733 | f | 6 | 3 | 11 | 10.5 | 10 | 13 | 4 | 5 | |
| SYS r001734 | m | 6 | 3 | 11 | 10.5 | 9 | 14 | 5 | 5 | |
| huishuiensis | NJNUh00851 | m | 10 | 3 | 11 | 11 | 9 | 14 | 3 | 3 |
| NJNUh00859 | m | 8 | 3 | 11 | 10.5 | 9 | 15 | 3 | 3 | |
| NJNUh00852 | f | 10 | 3 | 10 | 10 | 8 | 13 | 3 | 3 | |
| NJNUh00854 | f | 10 | 3 | 9 | 9 | 7 | 13 | 3 | 3 | |
| NJNUh00855 | f | 10 | 3 | 10 | 9 | 8 | 13 | 3 | 3 | |
| NJNUh00856 | f | 10 | 3 | 10 | 10 | 7 | 13 | 3 | 3 | |
| NJNUh00857 | f | 8 | 3 | 10 | 10 | 8 | 14 | 3 | 3 | |
| NJNUh00858 | f | 10 | 3 | 10 | 10 | 9 | 14 | 3 | 3 | |
| kiziriani | IEBR A.2014.3 | m | 9 | 4 | 11 | 10.5 | 13.5 | 21.5 | 5 | 5 |
| IEBR A.2014.4 | m | 9 | 4 | 11 | 9 | 14.5 | 26 | 5 | 5 | |
| VNMN A.2014.1 | m | 7 | 4 | 10 | 10 | 13.5 | 22.5 | 5 | 5 | |
| ZFMK 95702 | m | 6 | 4 | 10.5 | 9 | 14 | 23.5 | 5 | 5 | |
| IEBR A.2014.5 | f | 6 | 4 | 10.5 | 10 | 11.5 | 20.5 | 5 | 5 | |
| kiziriani | NUOL R-2014.1 | f | 9 | 4 | 11 | 10.5 | 13.5 | 23.5 | 5 | 5 |
| NUOL R-2014.2 | f | 6 | 4 | 10 | 10 | 11 | 18 | 5 | 5 | |
| VNMN A.2014.2 | f | 8 | 4 | 11 | 9 | 12 | 24 | 5 | 5 | |
| ZFMK 95703 | f | 8 | 4 | 10 | 10.5 | 13 | 25 | 5 | 5 | |
| ZFMK 95704 | f | 9 | 4 | 10.5 | 9.5 | 12.5 | 23.5 | 5 | 5 | |
| Hemiphyllodactyluslungcuensis sp. nov. | VNUF R.2021.01 | m | 8 | 3 | 12 | 10.5 | 7 | 13 | 4 | 4 |
| IEBR R.5151 | m | 8 | 3 | 11 | 11 | 6 | 13 | 4 | 4 | |
| VNUF R.2021.03 | m | 9 | 3 | 12 | 10 | 7 | 15 | 4 | 4 | |
| IEBR R.5152 | f | 9 | 2 | 11 | 10 | 9 | 16 | 4 | 4 | |
| VNUF R.2021.08 | f | 10 | 3 | 11 | 10 | 7 | 12 | 3 | 4 | |
| VNUF R.2023.01 | m | 10 | 3 | 11.5 | 9.5 | 10 | 17 | 4 | 4 | |
| VNUF R.2023.02 | f | 10 | 3 | 11 | 9.5 | 11 | 16 | 4 | 4 | |
| nahangensis | IEBR 4741 | m | 8 | 3 | 11 | 9.5 | 9 | 22 | 3 | 3 |
| IEBR 4742 | m | 8 | 2.5 | 11.5 | 10.5 | 9 | 18 | 3 | 3 | |
| IEBR 4743 | f | 9 | 3 | 11 | 10.5 | 13 | 23 | 3 | 3 | |
| ngocsonensis | IEBR 4694 | m | 8 | 3 | 10.5 | 10 | 14 | 20.5 | 5 | 5 |
| IEBR 4695 | m | 6 | 3 | 10 | 10 | 13 | 20.5 | 4 | 5 | |
| IEBR 4696 | f | 8 | 3 | 10 | 9 | 15 | 19.5 | 5 | 5 | |
| IEBR 4697 | f | 8 | 3 | 10 | 9.5 | 14 | 19.5 | 5 | 6 | |
| serpispecus | NUOL 00476 | m | 7 | 3 | 11 | 9 | 10 | 26 | 4 | 4 |
| typus | JAV bg. R.39 | f | 10 | 6 | 10.5 | 10 | 9 | 14 | 4 | 4 |
| MS92002 | f | 15 | 5 | 12 | 11 | 12 | 15 | 4 | 4 | |
| CUMZ R 2003.19 | f | 11 | 5 | 11.5 | 11 | 11 | 15 | 4 | 4 | |
| RBWS 19309 | f | 15 | 5 | 11.5 | 11 | 12 | 16 | 4 | 5 | |
| yanshanensis | KIZ 062090 | m | 9 | 6 | 9.5 | 10 | 8 | 14 | 5 | 5 |
| KIZ 062091 | f | 8 | 5 | 11.5 | 11 | 7 | 15 | 5 | 5 | |
| KIZ 062092 | f | 11 | 5 | 11 | 10.5 | 8 | 15 | 5 | 5 | |
| KIZ 062093 | f | 8 | 5 | 10.5 | 10 | 13 | 22 | 5 | 5 | |
| KIZ 062094 | f | 9 | 5 | 12 | 9.5 | 7 | 14 | 5 | 5 | |
| KIZ 062095 | f | 8 | 5 | 11 | 10.5 | 7 | 13 | 5 | 6 | |
| KIZ 062096 | f | 8 | 5 | 11.5 | 11 | 8 | 15 | 4 | 5 | |
| KIZ 062097 | m | 8 | 5 | 10.5 | 10 | 7 | 14 | 4 | 5 | |
| KIZ 062098 | m | 9 | 5 | 10 | 9.5 | 8 | 18 | 5 | 5 | |
| KIZ 062099 | m | 8 | 5 | 11 | 11 | 8 | 15 | 5 | 5 | |
| KIZ 062100 | f | 8 | 5 | 10 | 9.5 | 11 | 20 | 5 | 5 | |
| KIZ 062101 | f | 10 | 5 | 11 | 10 | 8 | 14 | 4 | 5 | |
| KIZ 062102 | f | 10 | 5 | 10.5 | 9.5 | 7 | 11 | 4 | 5 | |
| zugi | IEBR A.2013.20 | m | 9 | 3 | 12.5 | 11 | 15 | 21.5 | 4 | 4.5 |
| ZFMK 94781 | m | 12 | 2 | 11 | 10.5 | 15 | 20.5 | 5 | 5 | |
| IEBR A.2013.21 | m | 9 | 2.5 | 10.5 | 11 | 16 | 21.5 | 5 | 5 | |
All morphological analyses were conducted in R v.4.2.1 (R Core Team, 2021). An ANOVA was conducted on morphometric and meristric characters with statistically similar variances (i.e. p values > 0.05 in a Levene’s test) to search for the presence of statistically significant mean differences (p < 0.05) across the data set. Characters bearing statistically significant differences were subjected to a TukeyHSD test to ascertain which population pairs differed significantly from each other for those particular characters. Boxplots were generated in order to visualize the range, mean, and degree of differences among species bearing statistically different mean values for sets of characters.
Morphometric characters used in the statistical analyses were SVL, TrunkL, HeadL, HeadW, SnEye, NarEye, EyeD, and SnW. Tail metrics were not used due to the high degree incomplete sampling (i.e., regenerated, broken, or missing). To remove potential effects of allometry on morphometric traits (sec. Chan and Grismer 2022), we used the following equation: Xadj= log(X)-β[log(SVL)-log(SVLmean)], where Xadj = adjusted value; X = measured value; β = unstandardized regression coefficient for each population; and SVLmean = overall average SVL of all populations (Thorpe 1975, 1983; Turan 1999; Lleonart et al. 2000, accessible in the R package GroupStruct (available at https://github.com/chankinonn/GroupStruct). The morphometrics of each species were normalized separately and then concatenated into a single data set so as not to conflate potential intra- with interspecific variation (Reist 1986; McCoy et al. 2006) (Table 3). Meristic characters (scale counts) used in statistical analyses were Chin, CN, SL, IL, VS, DS, SL1F, and SL1T (Table 4). Precloacal and femoral pores were excluded from the multivariate analyses due to their absence in females. Categorical characters analyzed were the presence or absence of BodPartn; PosOStrp; PosOTrSpt; and PosSacrlMakg (Table 3).
Morphospatial relationships based on the normalized morphometrics and meristics were visualized using principal component analysis (PCA) from the ADEGENET package in R (Jombart et al. 2010) to determine if their positioning was consistent with the putative species boundaries delimited by the molecular phylogenetic analyses and defined by the univariate analyses (see above). PCA, implemented using the “prcomp()” command in R, is an indiscriminate analysis plotting the overall variation among individuals (i.e. data points) while treating each individual independently (i.e. not coercing data points into pre-defined groups). Subsequent to the PCA, a discriminant analysis of principle components (DAPC) was used to test for corroboration and further discrimination of morphospatial differences among the putative species. DAPCa priori groups the individuals of each predefined population inferred from the phylogeny into separate clusters (i.e. plots of points) bearing the smallest within-group variance that produce linear combinations of centroids having the greatest between-group variance (i.e. linear distance; Jombart et al. 2010). DAPC relies on standardized data from its own PCA as a prior step to ensure that variables analyzed are not correlated and number fewer than the sample size. Principal components with eigenvalues accounting for 90–95% of the variation in the data set were retained for the DAPC analysis according to the criterion of Jombart et al. (2010).
To test and further corroborate the PCA and DAPC analyses, we conducted a multiple factor analysis (MFA) on the above mentioned morphological characters plus the categorical color pattern charactrers for a near total evidence data set (see Tables 5, 6). The MFA was implemented using the mfa () command in the R package FactorMineR (Husson et al. 2017) and visualized using the Factoextra package (Kassambara and Mundt 2017). MFA is a global, unsupervised, multivariate analysis that incorporates qualitative and quantitative data (Pagès 2015), making it possible to analyze different data types simultaneously in a nearly total evidence environment. In an MFA, each individual is described by a different set of variables (i.e. characters) which are structured into different data groups in a global data frame–in this case, quantitative data (i.e. meristics and normalized morphometrics) and categorical data (i.e. color pattern). In the first phase of the analysis, separate multivariate analyses are carried out for each set of variables–principal component analyses (PCA) for the quantitative data sets and a multiple correspondence analysis (MCA) for categorical data. The data sets are then normalized separately by dividing all their elements by the square root of their first eigenvalues. For the second phase of the analysis, the normalized data sets are concatenated into a single matrix for a global PCA. Standardizing the data in this manner prevents one data type from overleveraging another. In other words, the normalization of the data in the first phase prevents data types with the highest number of characters or the greatest amount of variation from outweighing other data types in the second phase. This way, the contribution of each data type to the overall variation in the data set is scaled to define the morphospatial distance between individuals as well as calculating the contribution of each data type and character to the overall variation in the data set (Pagès 2015; Kassambara and Mundt 2017).
Table 5.
Significant p-values from the results of the ANOVA and Turkey HSD analyses comparing all combinations of species pairs. Abbreviations are in the Materials and methods.
| Morphological characters | SVL | TrunkL | HeadL | HeadW | SnEye | NarEye | EyeD | SnW | Chin | CN | SL | VS | DS | SL1F | SL1T |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| lungcuensis sp. nov. vs. banaensis | 0.002 | <0.001 | 0.005 | <0.001 | <0.001 | 0.002 | 0.002 | 0.002 | |||||||
| typus vs. lungcuensis sp. nov. | 0.003 | 0.026 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||||
| lungcuensis sp. nov. vs. kiziriani | 0.004 | 0.00 | <0.001 | <0.001 | <0.001 | <0.001 | 0.00 | 0.00 | |||||||
| ngocsonensis vs. lungcuensis sp. nov. | <0.001 | 0.032 | <0.001 | <0.001 | <0.001 | <0.001 | |||||||||
| typus-lungcuensis | <0.001 | <0.001 | <0.001 | 0.026 | 0.003 | ||||||||||
| yanshanensis vs. lungcuensis sp. nov. | 0.03 | 0.002 | 0.00 | ||||||||||||
| lungcuensis sp. nov. vs. bonkowskii | 0.008 | <0.001 | <0.001 | 0.02 | 0.002 | 0.002 | 0.00 | 0.00 | 0.00 | ||||||
| lungcuensis sp. nov. vs. hongkongensis | <0.001 | <0.001 | <0.001 | ||||||||||||
| lungcuensis sp. nov. vs. huishuiensis | <0.001 | <0.001 | |||||||||||||
| nahangensis vs. lungcuensis sp. nov. | <0.001 | <0.001 | <0.001 | <0.001 | |||||||||||
| zugi c lungcuensis sp. nov. | <0.001 | <0.001 | <0.001 | <0.001 | |||||||||||
| lungcuensis sp. nov. vs. dupanglingensis | 0.027 | ||||||||||||||
| serpispecus vs. lungcuensis sp. nov. | <0.001 | <0.001 |
Table 6.
Summary statistics of morphometric and meristic characters among the Hymiphyllodactylus species in clade 6.
| Species | SVL | TrunkL | HeadL | HeadW | SnEye | NarEye | EyeD | SnW | Chin | CN | SL | IL | VS | DS | SL1F | SL1T |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| banaensis (n = 10) | ||||||||||||||||
| Mean | 1.69 | 1.36 | 1.05 | 0.87 | 0.64 | 0.52 | 0.41 | 0.22 | 6.2 | 3 | 10.8 | 10.3 | 10.1 | 18.5 | 5 | 4.9 |
| SD | 0.017 | 0.014 | 0.012 | 0.009 | 0.027 | 0.021 | 0.031 | 0.019 | 0.422 | 0 | 0.919 | 0.674 | 0.876 | 1.179 | 0 | 0.316 |
| Lower | 1.66 | 1.34 | 1.03 | 0.86 | 0.60 | 0.48 | 0.36 | 0.19 | 6 | 3 | 9 | 9 | 9 | 17 | 5 | 4 |
| Upper | 1.71 | 1.39 | 1.07 | 0.89 | 0.69 | 0.56 | 0.45 | 0.26 | 7 | 3 | 12 | 11 | 12 | 20 | 5 | 5 |
| bonkowskii (n = 6) | ||||||||||||||||
| Mean | 1.64 | 1.34 | 1.01 | 0.93 | 0.67 | 0.52 | 0.45 | 0.31 | 6.5 | 3 | 9.58 | 9.25 | 14.33 | 25.17 | 5 | 4.83 |
| SD | 0.043 | 0.020 | 0.011 | 0.012 | 0.019 | 0.020 | 0.027 | 0.023 | 0.837 | 0 | 0.491 | 0.880 | 0.816 | 0.516 | 0 | 0.408 |
| Lower | 1.56 | 1.32 | 1.00 | 0.91 | 0.63 | 0.48 | 0.42 | 0.27 | 5 | 3 | 9 | 8 | 13 | 24.5 | 5 | 4 |
| Upper | 1.68 | 1.37 | 1.03 | 0.95 | 0.69 | 0.54 | 0.49 | 0.33 | 7 | 3 | 10 | 10.5 | 15 | 26 | 5 | 5 |
| dupanglingensis (n = 6) | ||||||||||||||||
| Mean | 1.61 | 1.28 | 1.03 | 0.87 | 0.65 | 0.50 | 0.41 | 0.21 | 10.33 | 3.33 | 11.67 | 9.83 | 10.67 | 14.83 | 4.83 | 4.67 |
| SD | 0.020 | 0.011 | 0.021 | 0.013 | 0.015 | 0.013 | 0.011 | 0.038 | 0.82 | 0.516 | 1.211 | 1.169 | 0.516 | 0.753 | 0.408 | 0.516 |
| Lower | 1.60 | 1.27 | 0.99 | 0.85 | 0.63 | 0.49 | 0.40 | 0.16 | 9 | 3 | 11 | 9 | 10 | 14 | 4 | 4 |
| Upper | 1.65 | 1.30 | 1.05 | 0.88 | 0.67 | 0.52 | 0.43 | 0.26 | 11 | 4 | 14 | 12 | 11 | 16 | 5 | 5 |
| hongkongensis (n = 8) | ||||||||||||||||
| Mean | 1.58 | 1.29 | 1.00 | 0.85 | 0.55 | 0.44 | 0.36 | 0.08 | 5.63 | 3.13 | 11.06 | 10.13 | 9.38 | 13.5 | 4.13 | 5 |
| SD | 0.044 | 0.012 | 0.012 | 0.061 | 0.022 | 0.024 | 0.021 | 0.022 | 0.518 | 0.35 | 0.776 | 0.353 | 0.518 | 0.926 | 0.641 | 0 |
| Lower | 1.52 | 1.27 | 0.98 | 0.72 | 0.51 | 0.41 | 0.32 | 0.05 | 5 | 3 | 10 | 9.5 | 9 | 12 | 3 | 5 |
| Upper | 1.63 | 1.30 | 1.01 | 0.91 | 0.58 | 0.47 | 0.39 | 0.12 | 6 | 4 | 12 | 10.5 | 10 | 15 | 5 | 5 |
| huishuiensis (n = 8) | ||||||||||||||||
| Mean | 1.64 | 1.33 | 1.01 | 0.89 | 0.61 | 0.47 | 0.41 | 0.08 | 9.5 | 3 | 10.13 | 9.94 | 8.13 | 13.63 | 3 | 3 |
| SD | 0.058 | 0.011 | 0.044 | 0.016 | 0.027 | 0.024 | 0.020 | 0.036 | 0.93 | 0 | 0.641 | 0.678 | 0.835 | 0.744 | 0 | 0 |
| Lower | 1.55 | 1.32 | 0.97 | 0.87 | 0.58 | 0.44 | 0.37 | 0.03 | 8 | 3 | 9 | 9 | 7 | 13 | 3 | 3 |
| Upper | 1.71 | 1.35 | 1.11 | 0.92 | 0.66 | 0.52 | 0.44 | 0.13 | 10 | 3 | 11 | 11 | 9 | 15 | 3 | 3 |
| kiziriani (n = 10) | ||||||||||||||||
| Mean | 1.58 | 1.28 | 0.82 | 0.84 | 0.59 | 0.48 | 0.41 | 0.15 | 7.7 | 4 | 10.55 | 9.8 | 12.9 | 22.8 | 5 | 5 |
| SD | 0.025 | 0.024 | 0.019 | 0.019 | 0.021 | 0.021 | 0.024 | 0.031 | 1.337 | 0 | 0.438 | 0.632 | 1.125 | 2.312 | 0 | 0 |
| Lower | 1.55 | 1.25 | 0.78 | 0.81 | 0.55 | 0.44 | 0.37 | 0.100 | 6 | 4 | 10 | 9 | 11 | 18 | 5 | 5 |
| Upper | 1.61 | 1.32 | 0.85 | 0.87 | 0.63 | 0.51 | 0.45 | 0.22 | 9 | 4 | 11 | 10.5 | 14.5 | 26 | 5 | 5 |
| lungcuensis (n = 7) | ||||||||||||||||
| Mean | 1.60 | 1.31 | 0.10 | 0.87 | 0.63 | 0.50 | 0.31 | 0.23 | 9.14 | 2.86 | 11.36 | 10.07 | 8.14 | 14.57 | 3.86 | 4 |
| SD | 0.037 | 0.013 | 0.039 | 0.026 | 0.022 | 0.028 | 0.049 | 0.046 | 0.900 | 0.378 | 0.476 | 0.535 | 1.864 | 1.902 | 0.378 | 0 |
| Lower | 1.55 | 1.29 | 0.94 | 0.83 | 0.59 | 0.45 | 0.27 | 0.16 | 8 | 2 | 11 | 9.5 | 6 | 12 | 3 | 4 |
| Upper | 1.65 | 1.33 | 1.06 | 0.91 | 0.66 | 0.54 | 0.41 | 0.30 | 10 | 3 | 12 | 11 | 11 | 17 | 4 | 4 |
| nahangensis (n = 3) | ||||||||||||||||
| Mean | 1.63 | 1.33 | 1.01 | 0.85 | 0.58 | 0.48 | 0.42 | 0.05 | 8.33 | 2.83 | 11.17 | 10.17 | 10.33 | 21 | 3 | 3 |
| SD | 0.012 | 4.15E-05 | 0.008 | 0.007 | 0.013 | 0.007 | 0.006 | 0.034 | 0.577 | 0.289 | 0.289 | 0.577 | 2.309 | 2.646 | 0 | 0 |
| Lower | 1.62 | 1.33 | 1.00 | 0.84 | 0.57 | 0.47 | 0.41 | 0.017 | 8 | 2.5 | 11 | 9.5 | 9 | 18 | 3 | 3 |
| Upper | 1.64 | 1.33 | 1.02 | 0.86 | 0.59 | 0.49 | 0.42 | 0.09 | 9 | 3 | 11.5 | 10.5 | 13 | 23 | 3 | 3 |
| ngocsonensis (n = 4) | ||||||||||||||||
| Mean | 1.66 | 1.37 | 1.01 | 0.93 | 0.66 | 0.53 | 0.44 | 0.24 | 7.5 | 3 | 10.13 | 9.63 | 14 | 20 | 4.75 | 5.25 |
| SD | 0.006 | 0.004 | 0.008 | 0.010 | 0.016 | 0.029 | 0.010 | 0.007 | 1 | 0 | 0.25 | 0.479 | 0.816 | 0.577 | 0.5 | 0.5 |
| Lower | 1.66 | 1.36 | 1.00 | 0.91 | 0.65 | 0.50 | 0.43 | 0.23 | 6 | 3 | 10 | 9 | 13 | 19.5 | 4 | 5 |
| Upper | 1.67 | 1.37 | 1.02 | 0.94 | 0.69 | 0.57 | 0.45 | 0.25 | 8 | 3 | 10.5 | 10 | 15 | 20.5 | 5 | 6 |
| serpispecus (n = 1) | ||||||||||||||||
| Mean | 1.62 | 1.33 | 1.01 | 0.88 | 0.59 | 0.51 | 0.38 | 0.17 | 7 | 3 | 11 | 9 | 10 | 26 | 4 | 4 |
| SD | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Lower | 1.62 | 1.33 | 1.01 | 0.88 | 0.59 | 0.51 | 0.38 | 0.17 | 7 | 3 | 11 | 9 | 10 | 26 | 4 | 4 |
| Upper | 1.62 | 1.33 | 1.01 | 0.88 | 0.59 | 0.51 | 0.38 | 0.17 | 7 | 3 | 11 | 9 | 10 | 26 | 4 | 4 |
| typus (n = 4) | ||||||||||||||||
| Mean | 1.50 | 1.26 | 0.90 | 0.69 | 0.49 | 0.38 | 0.26 | -0.03 | 12.75 | 5.25 | 11.36 | 10.75 | 11 | 15 | 4 | 4.25 |
| SD | 0.096 | 0.026 | 0.028 | 0.016 | 0.027 | 0.015 | 0.033 | 0.115 | 2.630 | 0.5 | 0.629 | 0.5 | 1.414 | 0.816 | 0 | 0.5 |
| Lower | 1.35 | 1.22 | 0.88 | 0.68 | 0.46 | 0.36 | 0.21 | -0.19 | 10 | 5 | 10.5 | 10 | 9 | 14 | 4 | 4 |
| Upper | 1.56 | 1.28 | 0.94 | 0.72 | 0.52 | 0.40 | 0.29 | 0.07 | 15 | 6 | 12 | 11 | 12 | 16 | 4 | 5 |
| yanshanensis (n = 13) | ||||||||||||||||
| Mean | 1.61 | 1.29 | 0.87 | 0.90 | 0.62 | 0.50 | 0.38 | 0.18 | 8.77 | 5.077 | 10.77 | 10.15 | 8.23 | 15.38 | 4.69 | 5.08 |
| SD | 0.035 | 0.012 | 0.021 | 0.017 | 0.018 | 0.024 | 0.039 | 0.019 | 1.013 | 0.277 | 0.696 | 0.591 | 1.787 | 2.959 | 0.480 | 0.277 |
| Lower | 1.53 | 1.27 | 0.84 | 0.87 | 0.59 | 0.46 | 0.31 | 0.16 | 8 | 5 | 9.5 | 9.5 | 7 | 11 | 4 | 5 |
| Upper | 1.67 | 1.31 | 0.93 | 0.94 | 0.65 | 0.55 | 0.45 | 0.21 | 11 | 6 | 12 | 11 | 13 | 22 | 5 | 6 |
| zugi (n = 3) | ||||||||||||||||
| Mean | 1.61 | 1.33 | 0.97 | 0.86 | 0.64 | 0.46 | 0.44 | 0.19 | 10 | 2.5 | 11.33 | 10.83 | 15.33 | 21.17 | 4.67 | 4.83 |
| SD | 0.034 | 0.010 | 0.012 | 0.001 | 0.013 | 0.031 | 0.001 | 0.009 | 1.732 | 0.5 | 1.041 | 0.289 | 0.577 | 0.577 | 0.577 | 0.289 |
| Lower | 1.58 | 1.32 | 0.96 | 0.85 | 0.62 | 0.42 | 0.436 | 0.185 | 9 | 2 | 10.5 | 10.5 | 15 | 20.5 | 4 | 4.5 |
| Upper | 1.65 | 1.34 | 0.98 | 0.86 | 0.65 | 0.48 | 0.438 | 0.202 | 12 | 3 | 12.5 | 11 | 16 | 21.5 | 5 | 5 |
Results
Phylogenetic analyses
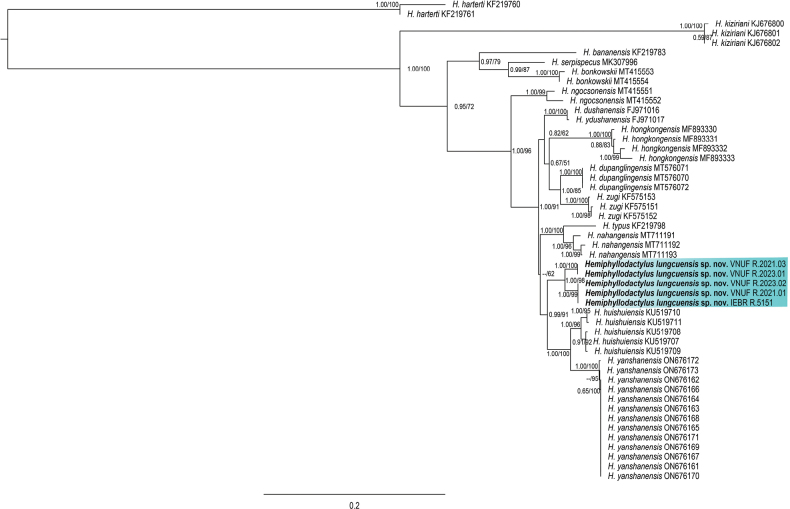
The ML and BI analyses returned well-supported, nearly identical topologies for all the known species of clade 6 (Agung et al. 2022) and were concordant with genus-wide phylogenies from previous analyses (Grismer et al. 2013; Grismer et al. 2020; Agung et al. 2022). The specimens from Lung Cu Commune, Dong Van District, Ha Giang Province, northeastern Vietnam, formed a monophyletic lineage in the typus group (Grismer et al. 2013) embedded within a larger lineage of Indochinese species where it is nested within clade 6. Herein, it was recovered as the well-supported (BI 0.99/ML 91) sister species to a lineage comprised of H.huishuiensis and H.yanshanensis (Fig. 2). The Lung Cu lineage differs from H.huishuiensis and H.yanshanensis by having uncorrected average pairwise sequence divergences of 5.3% and 6.8%, respectively (Table 2). In terms of genetic distance, the Lung Cu population is similar to H.nahangensis and H.dushanensis, but it has a genetic distance of 4.6% and 4.8% from H.nahangensis and H.dushanensis, respectively. Additionally, significant differences of morphometric and meristic traits among them are indicated in the comparisons. As such we hypothesize this population represents a new evolutionary lineage and is described as a new species below.
Figure 2.
Maximum likelihood consensus tree of clade 6 (excepting Hemiphyllodactylusharterti used to root the tree following Grismer et al. 2013) of Hemiphyllodactylus with Bayesian posterior probabilities (BPP) and Ultrafast Bootstrap support (UFB) value coding at the nodes, respectively.
Statistical analyses
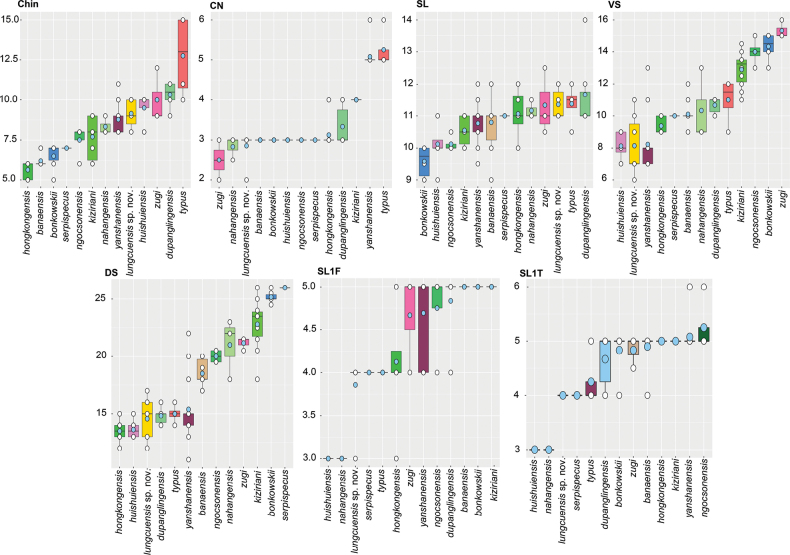
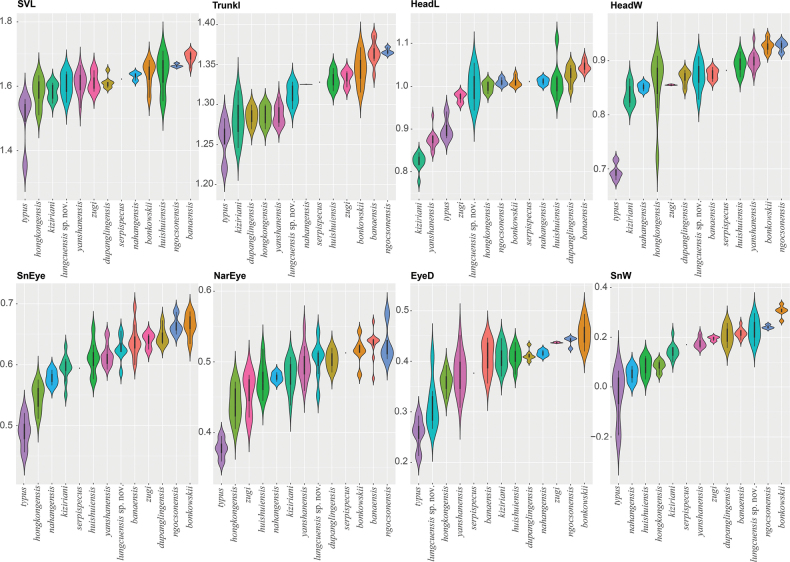
The ANOVA and TukeyHSD post hoc tests of the adjusted morphometric and meristic characters were consistent with the phylogenetic results and their high degree of pairwise genetic distance among the species in recovering a number of statistically significant mean differences between the Lung Cu population and all other species (Tables 5, 6). Variation in all morphometric and metric characters are visualized in Figs 3, 4.
Figure 3.
Boxplot comparisons of significantly different meristic characters between the Lung Cu population and other Hemiphyllodactylus species of clade 6. Pale blue circles are means and the black horizontal bars are medians.
Figure 4.
Violin plots of the significantly different morphometric characters between the Lung Cu population and other Hemiphyllodactylus species of clade 6 overlain with box plots showing the range, frequency, mean (white dot), and 50% quartile (black rectangle) of the size-adjusted morphometric and meristic characters. New species in bold.
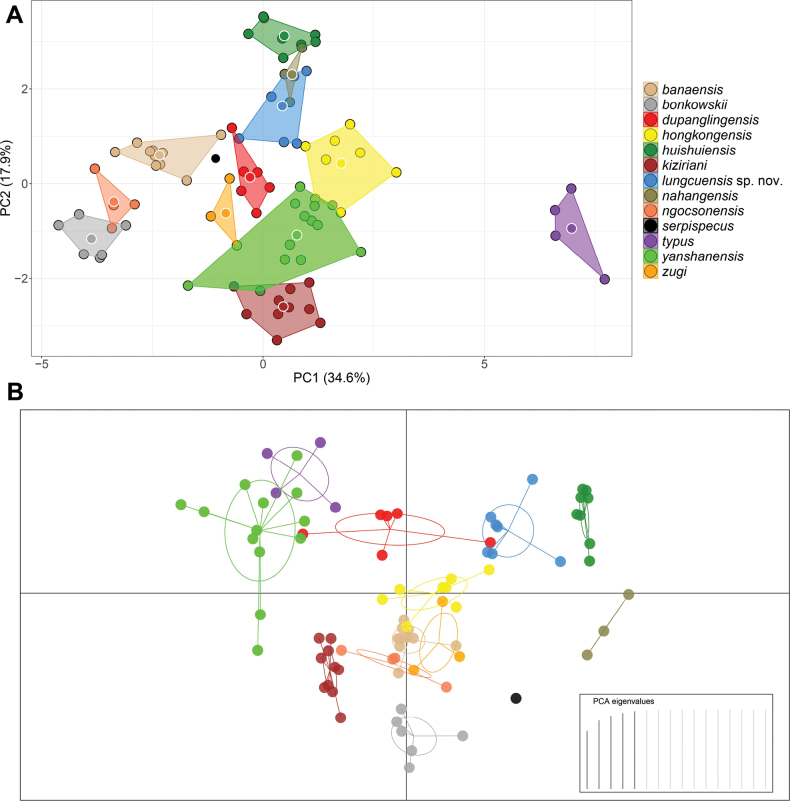
The clustering of species in the PCA of clade 6 showed the first two principal components (PC1 and PC2) recovered 52.5% of the variation in the normalized morphometric and meristic data set (Fig. 5A). PC1 accounted for 34.6% of the variation in the data set and loaded most heavily for trunk length (TrunkL), head width (HeadW), snout-eye length (SnEye), nares-eye length (NarEye), and eye diameter (EyeD). PC2 accounted for an additional 17.9% of the data set and loaded most heavily for head length (HeadL), circumnasal scales (CN), dorsal scales (DS), the number of subdigital lamellae wider than long on the first finger (SL1F), and the number of subdigital lamellae wider than long on the first toe (SL1T) along PC2 (Fig. 5A, Table 7). The PCA recovered Hemiphyllodactyluslungcuensis sp. nov. to be widely separated from most other species and only overlapping with the distantly related H.nahangensis and minimally overlapping with H.dupanglingensis. The new species is well-separated from most other species in the DAPC and only slightly overlapping with the distantly related H.dupanglingensis (Fig. 5B). More importantly, its 66% confidence ellipse does not overlap with that of any other species.
Figure 5.
A principal component analysis (PCA) of Hemiphyllodactylus species of clade 6 showing their morphospatial relationships along the first two principal components B discriminant analysis of principal components (DAPC) based on retention of the first five PCs accounting for 98% of the variation.
Table 7.
Summary statistics of the principal component analysis for Hemiphyllodactylus species.
| PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 | PC8 | |
| Standard deviation | 2.354041057 | 1.693896742 | 1.274819788 | 1.167631895 | 1.035772699 | 0.858583005 | 0.792572769 | 0.700362564 |
| Proportion of Variance | 0.34634 | 0.17933 | 0.10157 | 0.08521 | 0.06705 | 0.04607 | 0.03926 | 0.03066 |
| Cumulative Proportion | 0.34634 | 0.52567 | 0.62725 | 0.71246 | 0.77951 | 0.82558 | 0.86484 | 0.8955 |
| eigen | 5.541509296 | 2.869286174 | 1.625165491 | 1.363364241 | 1.072825085 | 0.737164777 | 0.628171593 | 0.49050772 |
| SVL | -0.275684939 | 0.230671227 | -0.183268856 | 0.035839657 | -0.118110503 | 0.272234498 | -0.353548073 | 0.532231802 |
| TrunkL | -0.308028087 | 0.226577926 | 0.113182 | 0.165672749 | -0.09837754 | 0.21375662 | 0.23148674 | 0.352374067 |
| HeadL | -0.182206073 | 0.401679247 | 0.111064477 | 0.286694098 | -0.082466549 | -0.270566918 | 0.230170931 | -0.024756545 |
| HeadW | -0.316234879 | 0.087646379 | -0.266341502 | -0.227593583 | 0.011593053 | 0.056636999 | -0.034411085 | -0.551718699 |
| SnEye | -0.354053441 | 0.028071822 | -0.174272185 | 0.028737797 | 0.370906143 | -0.005790557 | 0.061744748 | -0.038483117 |
| NarEye | -0.343202425 | 0.013213311 | -0.227705587 | -0.097414199 | 0.248586294 | -0.037810729 | -0.050598906 | 0.110097139 |
| EyeD | -0.316496337 | -0.057054094 | 0.203121932 | -0.054096772 | 0.11278894 | 0.198377934 | -0.488753315 | -0.270887117 |
| SnW | -0.331682586 | -0.118626 | -0.220099451 | 0.078123766 | 0.10976895 | -0.079025168 | 0.348298246 | -0.089036833 |
| Chin | 0.209598984 | 0.041124515 | 0.055504502 | 0.015768954 | 0.762755038 | 0.126936754 | 0.252975725 | 0.142123111 |
| CN | 0.219334589 | -0.306220475 | -0.340275868 | -0.238245722 | 0.106513528 | 0.269505437 | -0.086774889 | 0.202680044 |
| SL | 0.150823615 | 0.072482948 | -0.141526231 | 0.569582987 | 0.281130944 | -0.34518498 | -0.525400396 | -0.03369833 |
| IL | 0.148019724 | 0.047098158 | -0.160448641 | 0.539731081 | -0.078204547 | 0.69478363 | 0.119615798 | -0.305781505 |
| VS | -0.159093977 | -0.32427077 | 0.507208426 | 0.179912633 | 0.107284233 | 0.046759987 | 0.085474597 | -0.042903745 |
| DS | -0.216885816 | -0.316665593 | 0.415917222 | 0.013670345 | 0.048885259 | 0.15087592 | -0.139750976 | 0.074486639 |
| SL1F | -0.160449559 | -0.454450117 | -0.164215408 | 0.230860029 | -0.051722119 | -0.131281578 | 0.073369609 | 0.169471363 |
| SL1T | -0.088298003 | -0.43959919 | -0.276677158 | 0.243587056 | -0.224574637 | -0.154636155 | 0.081186695 | 0.024901385 |
| PC9 | PC10 | PC11 | PC12 | PC13 | PC14 | PC15 | PC16 | |
| Standard deviation | 0.645837646 | 0.528806731 | 0.4949505 | 0.460869402 | 0.422836018 | 0.392344947 | 0.348783481 | 0.252014728 |
| Proportion of Variance | 0.02607 | 0.01748 | 0.01531 | 0.01328 | 0.01117 | 0.00962 | 0.0076 | 0.00397 |
| Cumulative Proportion | 0.92157 | 0.93905 | 0.95436 | 0.96763 | 0.97881 | 0.98843 | 0.99603 | 1 |
| eigen | 0.417106265 | 0.279636559 | 0.244975998 | 0.212400606 | 0.178790298 | 0.153934558 | 0.121649916 | 0.063511423 |
| SVL | -0.007653797 | 0.504865284 | 0.023054857 | 0.048923188 | -0.104515394 | -0.134955096 | 0.216496623 | -0.075553468 |
| TrunkL | -0.155806654 | -0.402668791 | 0.169935171 | -0.50108859 | 0.030208721 | 0.261221952 | -0.105888367 | 0.183446071 |
| HeadL | -0.196823028 | -0.042928589 | 0.308909057 | 0.583413488 | -0.235482569 | 0.083561907 | -0.07755344 | -0.18027682 |
| HeadW | -0.057355934 | 0.018795152 | 0.420271 | -0.230081521 | -0.169918109 | 0.020732474 | 0.445117482 | 0.004175194 |
| SnEye | 0.006655124 | -0.05473374 | 0.125911056 | -0.081337071 | 0.26677457 | -0.615512405 | -0.423767342 | -0.211307545 |
| NarEye | 0.209982569 | -0.475190325 | -0.461292899 | 0.279036274 | -0.29657524 | -0.040045604 | 0.211899966 | 0.218583377 |
| EyeD | -0.467694393 | 0.020586761 | -0.201246365 | 0.150990454 | 0.146331073 | 0.329187066 | -0.274245212 | 0.024651913 |
| SnW | 0.373629734 | 0.470794413 | -0.10280995 | -0.013393282 | 0.031119697 | 0.456870887 | -0.272519277 | 0.133686181 |
| Chin | -0.293197569 | 0.139059923 | -0.038969703 | -0.051313747 | -0.052718431 | 0.18026194 | 0.28186905 | -0.211579231 |
| CN | -0.036235154 | -0.135400518 | 0.485735592 | 0.236559781 | -0.276203918 | 0.132252018 | -0.350233189 | 0.158104094 |
| SL | 0.189523962 | -0.059900251 | 0.171907156 | -0.215076887 | -0.077003357 | 0.119789393 | -0.02904125 | 0.125441842 |
| IL | 0.112573099 | -0.066456621 | -0.117163024 | 0.149019989 | 0.026731134 | -0.082213823 | 0.038834193 | -0.012764391 |
| VS | -0.060117023 | 0.224584718 | 0.102501327 | -0.019925796 | -0.380657113 | -0.333667822 | 0.032368952 | 0.484590421 |
| DS | 0.471295976 | -0.169334813 | 0.207180516 | 0.049104744 | -0.028195629 | 0.15739267 | 0.109826908 | -0.543616511 |
| SL1F | -0.192515088 | -0.071305429 | 0.182854605 | 0.25220064 | 0.565596931 | 0.028793369 | 0.377898255 | 0.17530195 |
| SL1T | -0.364000904 | -0.005236656 | -0.226318873 | -0.228651157 | -0.413553347 | -0.023541078 | -0.058595931 | -0.412884904 |
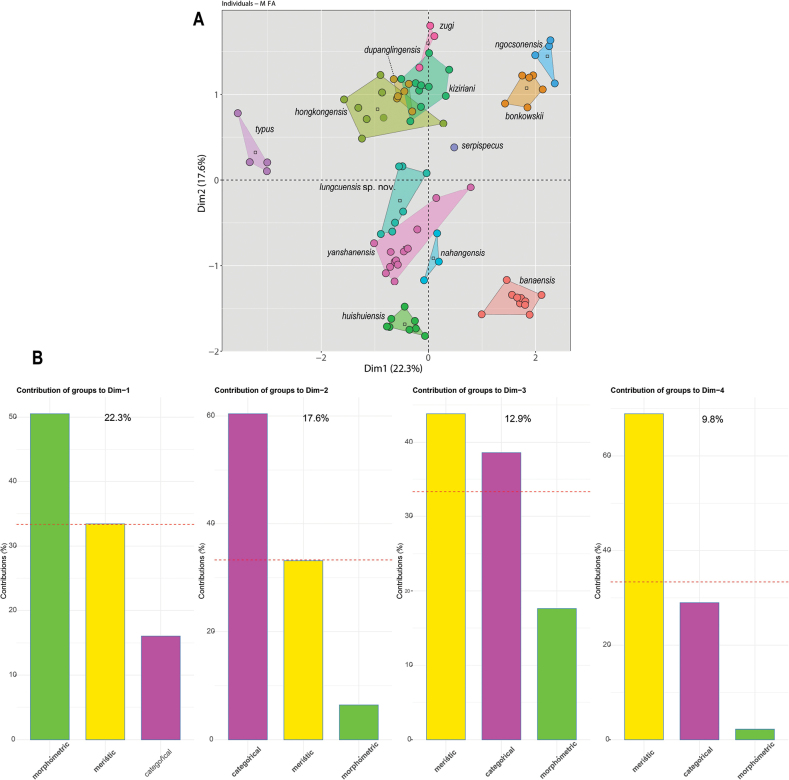
The MFA analysis recovered all species as being separated from one another including Hemiphyllodactyluslungcuensis sp. nov. and H.nahangensis (Fig. 6A). The normalized morphometric data contributed to approximately 50% of the variation along Dim-1 followed by the meristic and categorical data. For Dim-2, the categorical data contributed 60% of the variation followed by meristic and mensural data. Dim-3 showed that meristic data contributed 45% of the variation followed by categorical and mensural data. Finally, Dim-4 showed that meristic data contributed 70% of the variation followed by categorical and mensural data (Fig. 6B).
Figure 6.
AMFA scatter plot showing the morphospatial relationships among the Hemiphyllodactylus species of clade 6 B bar graphs showing the percent contribution of each data type to the overall variation in the data set for the first four dimensions. The dashed red line in the bar graphs indicates the expected average value if the contributions of each data type were equal.
Taxonomy
. Hemiphyllodactylus lungcuensis sp. nov.
64879394-8DB8-5685-9534-C5A76EAF82B1
https://zoobank.org/7E96A770-35D0-4B50-82B5-F8C7A4A817FD
Figs 7 , 8 Verbatim name. Lungcu Slender Gecko
Figure 7.
Dorsal views of Hemiphyllodactyluslungcuensis sp. nov. in life from Ha Giang Province, northeastern Vietnam A adult male holotype VNUF R.2021.01 B adult female paratype VNUF R.2021.08.
Figure 8.
Type series of Hemiphyllodactyluslungcuensis sp. nov. in preservative from Ha Giang Province, northeastern Vietnam. A from right to left: Adult male holotype (VNUF R.2021.01), adult female paratype (IEBR R.5251), adult female paratype (VNUF R.2021.08), adult male paratype (IEBR R.5151), adult male paratype (VNUF R.2021.3). B From right to left: Adult female paratype (VNUF R.2023.02), and adult female paratype (VNUF R.2023.01).
Material examined.
Holotype.VNUF R.2021.01 (Field no. LC17.01), adult male, collected by H.M. Ly on 25 August 2017 from Lung Cu Commune, Dong Van District, Ha Giang Province (23°21'N, 105°19'E, at an elevation of 1391 m a.s.l.). Paratypes.IEBR R.5151 (Field no. LC17.02), VNUF R.2021.03 (Field no. LC17.03), adult males, IEBR R.5152 (Field no. ST17.01), VNUF R.2021.08 (Field no. HG17.08), adult females, collected at the same locality as holotype on 25 August 2017; two adult females, VNUF R.2023.01 (Field no. LC22.01) and VNUF R.2023.02 (Field no. LC22.02), collected at the same locality as the holotype on 5 August 2022.
Diagnosis.
Hemiphyllodactyluslungcuensis sp. nov. can be distinguished from its congeners by a combination of the following characters: a maximum SVL of 44.2 mm; trunk not particularly elongate (AG/SVL ratio 0.52); chin scales 8–10, distinct enlarged postmentals; circumnasal scales two or three; supralabial scales 11 or 12; infralabial scales 10 or 11; ventral scale rows 6–9; dorsal scale rows 12–16; digital lamellae formula 4(3)–4–5(4)–4 (forefoot) and 4–5–5–5 (hindfoot); precloacal pores 17–25 and continuous series in males, 0–22 pitted precloacal scales in females; cloacal spur single in both sexes; enlarged subcaudal scales absent; dorsal surface brown sand color, with irregular dark brown streaks; a distinct dark postorbital stripe extending to at least the base of the neck, uneven dark streaks running along the flanks and ending at the base of tail, a cream-colored V-shaped postsacral mark with anteriorly projecting arms; a pale yellow ventral view of the body and limbs; and unpigmented caecum and gonads.
Description of the holotype.
Adult male, SVL 39.2 mm; tail length (TaL 37.3 mm); body compressed (TrunkL 20.3 mm); head triangular in dorsal profile, depressed, distinct from neck, and head longer than wide (HeadL 11.0 mm, HeadW 8.0 mm); eye large, pupils vertical (EyeD 1.8 mm); snout-eye distance (SnEye 4.3 mm); snout moderate, round in dorsal profile, longer than eye diameter (EyeD/SnEye 0.42); nare-eye distance (NarEye 3.2 mm); internarial distance (SnW 1.5 mm); ear opening oblique (EarD 0.7 mm).
Proportions: EarD/EyeD 0.39, TrunkL/SVL 0.52, HeadL/SVL 0.28, HeadW/SVL 0.20, HeadW/HeadL 0.73, SnEye/HeadL 0.39, NarEye/HeadL 0.29, EyeD/HeadL 0.16, SnW/HeadL 0.14, EyeD/NarEye 0.56, SnW/HeadW 0.19.
Scalation: Rostral large, wider than high, partially divided dorsally, bordered posteriorly by two large supranasals and two internasals (SnS); nares in contact with rostral anteriorly, first supralabial ventrally, supranasal dorsally, and three nasals posteriorly on each side; supralabial scales 12/12 (left/right), infralabial scales 11/10 (left/right), gradually decreasing in size towards angle of jaw; scales on head small, round, largest on rostrum; mental triangular, bordered by first infralabials and posteriorly by two enlarged postmentals, eight chin scales touching internal edge of infralabials and mental between the juncture of the second and third infralabials on each side of the head, anterior pair enlarged; ventral scales smooth and flat, much larger than dorsal scales, subimbricate, in seven rows (contained within one eye diameter); dorsal scales small, granular in 13 rows at midbody on dorsum (contained within one eye diameter), enlarged tubercles absent; fore-limbs relatively short, covered dorsally with granular, subimbricate scales, smaller smooth scales ventrally; 25 pore bearing femoral and precloacal sacles, in a continuous row; terminal two phalanges free, claws absent on first finger and on first toe,large triangular lamellae present on the second to fifth digits of both the forefoot and hindfoot pads, with digital formula 4–4–5–4 (forefoot) and 4–5–5–5 (hindfoot); four on first fingers, four on first toes; cloacal spurs (CloacS) one on each side; semi-regenerated tail; caudal scales flat, not forming distinct caudal segments; caecum and gonads unpigmented.
Coloration in life.
Dorsal surface of head, body, and limbs brown sand color, with irregularly shaped dark-brown streaks; a dark postorbital stripe, distinct, extending to at least base of neck; uneven dark streaks running along the flanks and ending at the base of tail; two dark dots on dorsal surface of tail; dorsal surface of tail has dark bands evenly spaced from base of tail to tip of tail; postsacral mark cream-colored and V-shaped, bearing anteriorly projecting arms; ventral surface of body and limbs pale yellow; subcaudal region pale peach.
Sexual dimorphism and variation.
The scale counts vary among the type series: chin scales 9 or 10; scales between supranasals one or three; supralabials 11; infralabials 10; ventral scale rows seven or nine; 0–22 pitted precloacal scales in females (distinct pore-bearing scales in males). Dorsal surface of the tail has dark bands evenly spaced from the base of the tail to the tip of the tail in females. Subcaudal region of original part of tail of the adult male paratype (IEBR R.5151) is reddish-orange, while the regenerated portion of the tail is uniformly dark grey. For other morphological characters of the paratypes are given in Table 8.
Table 8.
Morphological characters of Hemiphyllodactyluslungcuensis sp. nov. (measurements in mm, * = regenerated or broken, ?= missing data, Min = minimum, Max = maximum, other abbreviations defined in the text).
| VNUF R.2021.01 | IEBR R.5151 | VNUF R.2021.03 | VNUF R.2023.01 | VNUF R.2023.02 | IEBR R.5152 | VNUF R.2021.08 | Min-Max | |
|---|---|---|---|---|---|---|---|---|
| Holotype | Paratype | Paratype | Paratype | Paratype | Paratype | Paratype | ||
| Sex | male | male | male | female | female | female | female | |
| SVL | 39.2 | 43 | 44.2 | 35.3 | 37.4 | 39.2 | 43.5 | 35.3–44.2 |
| TaL | 37.3 | 25* | 42.7 | 30.4* | 27.3* | 4.3* | 34.7 | 34.7–42.7 |
| TrunkL | 20.3 | 23.0 | 23.1 | 17.0 | 19.3 | 19.0 | 22.3 | 17.0–23.1 |
| HeadL | 11.0 | 11.6 | 11.1 | 8.9 | 8.5 | 8.5 | 10.3 | 8.5–11.6 |
| HeadW | 8.0 | 8.0 | 7.0 | 6.9 | 7.0 | 7.3 | 8.0 | 6.9–8.0 |
| EyeD | 1.8 | 2.1 | 2.4 | 1.8 | 1.8 | 2.5 | 2.1 | 1.8–2.5 |
| SnEye | 4.3 | 4.5 | 5.0 | 3.6 | 3.9 | 4.1 | 4.2 | 3.6–5.0 |
| NarEye | 3.2 | 2.9 | 3.6 | 3.1 | 3.0 | 3.2 | 3.3 | 2.9–3.6 |
| SnW | 1.5 | 2.0 | 1.7 | 1.4 | 1.4 | 1.9 | 2.1 | 1.4–2.1 |
| EarD | 0.7 | 0.9 | 0.8 | 0.6 | 0.5 | 0.6 | 0.8 | 0.5–0.9 |
| EarD/EyeD | 0.39 | 0.43 | 0.33 | 0.33 | 0.28 | 0.24 | 0.38 | 0.24–0.43 |
| TrunkL/SVL | 0.52 | 0.53 | 0.52 | 0.48 | 0.52 | 0.48 | 0.51 | 0.48–0.53 |
| HeadL/SVL | 0.28 | 0.27 | 0.25 | 0.25 | 0.23 | 0.22 | 0.24 | 0.22–0.28 |
| HeadW/SVL | 0.20 | 0.19 | 0.16 | 0.20 | 0.19 | 0.19 | 0.18 | 0.16–0.20 |
| HeadW/HeadL | 0.73 | 0.69 | 0.63 | 0.78 | 0.82 | 0.86 | 0.78 | 0.63–0.86 |
| SnEye/HeadL | 0.39 | 0.39 | 0.45 | 0.40 | 0.46 | 0.48 | 0.41 | 0.39–0.48 |
| NarEye/HeadL | 0.29 | 0.25 | 0.32 | 0.35 | 0.35 | 0.38 | 0.32 | 0.25–0.38 |
| EyeD/HeadL | 0.16 | 0.18 | 0.22 | 0.20 | 0.21 | 0.29 | 0.20 | 0.21–0.29 |
| SnW/HeadL | 0.14 | 0.17 | 0.15 | 0.16 | 0.16 | 0.22 | 0.20 | 0.14–0.22 |
| EyeD/NarEye | 0.56 | 0.72 | 0.67 | 0.58 | 0.60 | 0.78 | 0.64 | 0.56–0.78 |
| SnW/HeadW | 0.19 | 0.25 | 0.24 | 0.20 | 0.20 | 0.26 | 0.26 | 0.19–0.26 |
| Chin | 8 | 8 | ? | 10 | 10 | 9 | 10 | 8–10 |
| CN | 3 | 3 | 3 | 3 | 3 | 2 | 3 | 2–3 |
| SnS | 2 | 2 | 2 | 2 | 1 | 1 | 3 | 1–3 |
| SL (left/right) | 12/12 | 11/11 | 12/12 | 12/11 | 11/11 | 11/11 | 11/11 | 11–12 |
| IL (left/right) | 11/10 | 11/11 | 10/10 | 10/9 | 10/9 | 10/10 | 10/10 | 9–11 |
| VS | 7 | 6 | 7 | 10 | 11 | 9 | 7 | 6–11 |
| DS | 13 | 13 | 15 | 17 | 16 | 16 | 12 | 12–17 |
| Lamellae formula of forelimbs II–V (left) | 4454 | * | 44?? | 4454 | 4454 | 4444 | 3454 | 4(3)45(4)4 |
| Lamellae formula of hind limbs II–V (left) | 4555 | 4555 | 4555 | 4555 | 4555 | 4555 | 4555 | 4555 |
| SL1F | 4 | 4 | ? | 4 | 4 | 4 | 3 | 3–4 |
| SL1T | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| Total femoroprecloacal pores | 25 | 17 | 23 | 16 | 0 | 0 | 22 | 17–25 (in males) |
| CloacS on each side | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1–2 |
| Precloacal and femoral pore series separate (1) or continuous (0) | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Subcaudals enlarged, plate-like | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Distribution.
Hemiphyllodactyluslungcuensis sp. nov. is currently known only from the type locality in Lung Cu Commune, Dong Van District, Ha Giang Province, northeastern Vietnam (Fig. 1).
Etymology.
Specific epithet lungcuensis is a toponym in reference to the type locality of the species. For the common names, we suggest Lungcu Slender Gecko (English) and Thạch sùng dẹp lũng cú (Vietnamese).
Natural history.
The species is known from a karstic habitat. The type series were found on karst cliffs between 19:00 and 20:30, approximately 1–1.5 m above the ground. The surrounding habitat was a disturbed karst forest with small hardwood and shrub trees, and was located near residential areas and road systems (Fig. 9). The type locality of the new species is located in an unprotected area, and thus we need to clarify the impact and threats of the localized disturbance so as to put in place effective conservation measures.
Figure 9.
Habitat of Hemiphyllodactyluslungcuensis sp. nov. in Lung Cu Commune, Dong Van District, Ha Giang Province, northeastern Vietnam.
Comparisons.
The molecular analyses indicate that Hemiphyllodactyluslungcuensis sp. nov. is a phylogenetically distinct lineage of the clade 6 that is not embedded within any lineage of the other 13 species of clade 6 (Agung et al. 2022) and bears an uncorrected pairwise sequence divergence of 4.6–20.2% from them (Fig. 1, Table 2). Hemiphyllodactyluslungcuensis sp. nov. is the sister species to a clade composed of H.huishuiensis and H.yanshanensis (Fig. 1) from which it bears an uncorrected pairwise sequence divergence of 5.3% from H.huishuiensis and 6.9% from H.yanshanensis (Table 2). Hemiphyllodactyluslungcuensis sp. nov. differs significantly from H.huishuiensis in mean values of EyeD (0.31 vs. 0.41, p = 0.001) and SnW (0.23 vs. 0.08, p = 0.001) and from H.yanshanensis in mean values of TrunkL (1.31 vs. 1.29, p = 0.03) and EyeD (0.31 vs. 0.38, p = 0.002). In addition, Hemiphyllodactyluslungcuensis sp. nov. differs significantly from H.nahangensis in mean values of EyeD (0.31 vs. 0.42, p = 0.001), SnW (0.23 vs. 0.05, p = 0.001), DS (14.75 vs. 21, p = 0.001), and SL1T (4 vs. 3, p = 0.001) (Tables 6, 7). Statistically significant differences of morphometric and meristic traits between Hemiphyllodactyluslungcuensis sp. nov. and all other species in clade 6 are presented in Tables 6, 7. Due to the unavailability of individual specimens of H.dushanensis for statistical analysis, we obtained integrated data of this species from Do et al. (2020: table 4) for comparison, the new species can be distinguished from H.dushanensis by the presence of dark postorbital stripe, dark dorsolateral stripe, and dark dorsal transverse blotches (vs. absent in H.dushanensis).
Discussion
Phylogenetic analyses indicate that the newly discovered species is closely related to H.nahangensis (with a genetic pairwise distance of 4.6%) found in Vietnam and H.dushanensis (with a genetic pairwise distance of 4.8%) found in China. However, the type locality of the new species is located in Lung Cu Commune, Dong Van District, Ha Giang Province, northeastern Vietnam, which is approximately 100 km away from the type locality of H.nahangensis in Sinh Long Commune, Na Hang District, Tuyen Quang Province, northeastern Vietnam (Do et al. 2020) and approximately 350 km away from the type locality of H.dushanensis in Dushan County, Guizhou Province, China. As such, there is significant geographical separation between the new species and its closest known relatives.
The recognition of Hemiphyllodactyluslungcuensis sp. nov. increases the number of Hemiphyllodactylus in Vietnam to eight. However, the diversity of Hemiphyllodactylus in Vietnam is still likely underestimated, particularly in karst landscapes in northern Vietnam (Do et al. 2020). In comparison, the southern Chinese provinces of Yunnan and Guangxi, which run along the border with Vietnam, have seen the description of three new species of Hemiphyllodactylus from the karst forests in the last two years (Agung et al. 2021, 2022; Uetz et al. 2022). Therefore, further field research is required to uncover the unrealized diversity of Hemiphyllodactylus in the largest karst formation in northern Vietnam.
Supplementary Material
Acknowledgments
We are grateful to the Vietnam National University of Forestry, Hanoi and the Lung Cu Commune People’s Committee, Dong Van District, Ha Giang Province for supporting our field work and issuing permit numbers (05 GT/2017/ĐHLN and 14 GT/2022/ĐHLN). We thank H. M. Ly for his assistance in the field. V.Q. Luu thanks C.V. Tran, K.V. Phung, and D.H. Vu (VNUF, Hanoi) for supporting his work. For the fruitful cooperation within joint research projects, we cordially thank S.V. Nguyen (IEBR, Hanoi), as well as T. Pagel and C. Landsberg (Cologne Zoo). We thank reviewers for providing helpful comments on the manuscript.
Citation
Luu VQ, Nguyen TH, Do HQ, Pham CT, Hoang TT, Nguyen TQ, Le MD, Ziegler T, Grismer JL, Grismer LL (2023) A new species of Hemiphyllodactylus (Squamata, Gekkonidae) from Ha Giang Province, Vietnam. ZooKeys 1167: 353–382. https://doi.org/10.3897/zookeys.1167.103713
Contributor Information
Vinh Quang Luu, Email: qvinhfuv@yahoo.com.au.
L. Lee Grismer, Email: lgrismer@lasierra.edu.
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
This research is funded by the Vietnam Academy of Science and Technology (Grant Number CSCL09.02/22-23). Research of Thuong Huyen Nguyen was funded by the Master, PhD Scholarship Programme of Vingroup Innovation Foundation (VINIF), code VINIF.2022.TS125. Research of Vinh Quang Luu in Herpetology Laboratory, Department of Biology, La Sierra University, U.S. was supported by the Fulbright Program.
Author contributions
All authors contributed to this work. Methodology: L. Lee Grismer, Vinh Quang Luu, Jess L. Grismer; collecting data: Thuong Huyen Nguyen, Quyen Hanh Do, Vinh Quang Luu, Tuoi Thi Hoang; data analysis: Vinh Quang Luu, Tuoi Thi Hoang, L. Lee Grismer, Jesse L. Grismer, Minh Duc Le; writing—original draft preparation, Thuong Huyen Nguyen, Quyen Hanh Do, Vinh Quang Luu, Tuoi Thi Hoang; writing—review and editing: L. Lee Grismer, Minh Duc Le, Cuong The Pham, Thomas Ziegler, Truong Quang Nguyen, Vinh Quang Luu; supervision: L. Lee Grismer, Thomas Ziegler, Truong Quang Nguyen.
Author ORCIDs
Vinh Quang Luu https://orcid.org/0000-0002-0634-1338
Thuong Huyen Nguyen https://orcid.org/0000-0002-4303-525X
Quyen Hanh Do https://orcid.org/0000-0002-9437-4673
Cuong The Pham https://orcid.org/0000-0001-5158-4526
Truong Quang Nguyen https://orcid.org/0000-0002-6601-0880
Minh Duc Le https://orcid.org/0000-0002-2953-2815
Thomas Ziegler https://orcid.org/0000-0002-4797-609X
Jesse L. Grismer https://orcid.org/0000-0002-2542-5430
L. Lee Grismer https://orcid.org/0000-0001-8422-3698
Data availability
All of the data that support the findings of this study are available in the main text or Supplementary Information.
References
- Agarwal I, Khandekar A, Giri VB, Ramakrishnan U, Karanth KP. (2019) The hills are alive with geckos! A radiation of a dozen species on sky islands across peninsular India (Squamata: Gekkonidae, Hemiphyllodactylus) with the description of three new species. Organisms, Diversity & Evolution 19(2): 341–361. 10.1007/s13127-019-00392-5 [DOI] [Google Scholar]
- Agung AP, Grismer LL, Grismer JL, Quah ES, Chornelia A, Lu JM, Hughes LC. (2021) A new species of Hemiphyllodactylus Bleeker (Squamata: Gekkonidae) from Yunnan, China and its phylogenetic relationship to other congeners. Zootaxa 4980(1): 1–27. 10.11646/zootaxa.4980.1.1 [DOI] [PubMed] [Google Scholar]
- Agung AP, Chornelia A, Grismer LL, Grismer JL, Quah ESH, Lu J, Tomlinson KW, Hughes AC. (2022) Description of two new species of Hemiphyllodactylus (Reptilia: Gekkonidae) from karst landscapes in Yunnan, China, highlights complex conservation needs. Zoological Research 43(5): 767–786. 10.24272/j.issn.2095-8137.2022.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough TG, Birky Jr CW, Burt A. (2003) Diversification in sexual and asexual organisms. Evolution; International Journal of Organic Evolution 57: 2166–2172. 10.1554/02-339 [DOI] [PubMed] [Google Scholar]
- Boulenger GA. (1903) Descriptions of new lizards in the collection of the British Museum. The Annals and Magazine of Natural History, Series 7 12(70): 429–435. 10.1080/00222930308678877 [DOI]
- Bourret R. (1937) Lézards et Serpents Reçus au Laboratoire des Sciences Naturelles de l’Univeristé au Cours de l’Année 1937. Descriptions de Deux Espèces et de Deux Variétés Nouvelles. Bulletin Général de l’Instruction Publique, Gouvernment Général de l’Indochiné 4: 57–80. [Google Scholar]
- Chan KO, Grismer LL. (2022) GroupStruct: An R package for allometric size correction. Zootaxa 5124(4): 471–482. 10.11646/zootaxa.5124.4.4 [DOI] [PubMed] [Google Scholar]
- de Queiroz K. (2007) Species concepts and species delimitation. Systematic Biology 56(6): 879–886. 10.1080/10635150701701083 [DOI] [PubMed] [Google Scholar]
- Do QH, Pham CT, Phan TQ, Le MD, Ziegler T, Nguyen TQ. (2020) A new species of Hemiphyllodactylus (Squamata: Gekkonidae) from Tuyen Quang Province, Vietnam. Zootaxa 4821(3): 511–532. 10.11646/zootaxa.4821.3.5 [DOI] [PubMed] [Google Scholar]
- Do QH, Nguyen KV, Le MD, Pham CT, Ziegler T, Nguyen TQ. (2021) A new species of Hemiphyllodactylus Bleeker, 1860 (Squamata: Gekkonidae) from Da Lat Plateau, Vietnam. Zootaxa 5023(1): 093–106. 10.11646/zootaxa.5023.1.5 [DOI] [PubMed] [Google Scholar]
- Eliades SJ, Phimmachak S, Sivongxay N, Siler CD, Stuart BL. (2019) Two new species of Hemiphyllodactylus (Reptilia: Gekkonidae) from Laos. Zootaxa 4577(1): 131–147. 10.11646/zootaxa.4577.1.8 [DOI] [PubMed] [Google Scholar]
- Frost DR, Hillis DM. (1990) Species in concept and practice: Herpetological application. Herpetologica 46: 87–104. [Google Scholar]
- Frost DR, Kluge AG. (1994) A consideration of the epistemology in systematic biology, with special reference to species. Cladistics 10(3): 259–294. 10.1111/j.1096-0031.1994.tb00178.x [DOI] [Google Scholar]
- Grismer LL, Chit MT, Pawangkhanant P, Nazarov RA, Zaw T, Poyarkov NA. (2021) The phylogeny of Hemiphyllodactylus Bleeker, 1860 (Squamata: Gekkonidae) with a description of a new species from the Mangin Range, SagaingRegion, northern Myanmar. Journal of Natural History 54(29–30): 1913–1931. 10.1080/00222933.2020.1833095 [DOI] [Google Scholar]
- Grismer LL, Wood Jr PL, Anuar S, Muin MA, Quah ESH, Mcguire JA, Brown RM, Ngo TV, Pham TH. (2013) Integrative taxonomy uncovers high levels of cryptic species diversity in Hemiphyllodactylus Bleeker, 1860 (Squamata: Gekkonidae) and the description of a new species from Peninsular Malaysia. Zoological Journal of the Linnean Society 169(4): 849–880. 10.1111/zoj.12064 [DOI] [Google Scholar]
- Grismer LL, Wood Jr PL, Zug GR, Thura MK, Grismer MS, Murdoch ML, Quah ESH, Lin A. (2018) Two more new species of Hemiphyllodactylus Bleeker (Squamata: Gekkonidae) from the Shan Hills of eastern Myanmar (Burma). Zootaxa 4483(2): 295–316. 10.11646/zootaxa.4483.2.4 [DOI] [PubMed] [Google Scholar]
- Grismer LL, Wood Jr PL, Quah ESH, Thura MK, Oaks JR, Lin A. (2020) Four new Burmese species of Hemiphyllodactylus Bleeker (Squamata: Gekkonidae) from distantly unrelated parapatric clades from the Shan Plateau and Salween Basin. Zootaxa 4758(1): 45–82. 10.11646/zootaxa.4758.1.2 [DOI] [PubMed] [Google Scholar]
- Hillis DM. (2019) Species delimitation in herpetology. Journal of Herpetology 53(1): 3–12. 10.1670/18-123 [DOI] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. (2018) UFBoot2: Improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35(2): 518–522. 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. (2001) Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294(5550): 2310–2314. 10.1126/science.1065889 [DOI] [PubMed] [Google Scholar]
- Husson F, Josse J, Le S, Mazet J. (2017) FactoMine R: Exploratory Data Analysis and Data Mining. R package version 1.36.
- Jombart T, Devillard S, Balloux F. (2010) Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genetics 11(1): 1–15. 10.1186/1471-2156-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14(6): 587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A, Mundt F. (2017) factoextra: Extract and Visualize the Result of Multivariate Data Analyses. R package version 1.0.5.999.
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular evolutionary genetics analysis version 11.0 for bigger datasets. Molecular Biology and Evolution 33(7): 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le M, Raxworthy CJ, McCord WP, Mertz L. (2006) A molecular phylogeny of tortoises (Testudines: Testudinidae) based on mitochondrial and nuclear genes. Molecular Phylogenetics and Evolution 40(2): 517–531. 10.1016/j.ympev.2006.03.003 [DOI] [PubMed] [Google Scholar]
- Lleonart J, Salat J, Torres GJ. (2000) Removing allometric effects of body size in morphological analysis. Journal of Theoretical Biology 205(1): 85–93. 10.1006/jtbi.2000.2043 [DOI] [PubMed] [Google Scholar]
- McCoy MW, Bolker BM, Osenberg CW, Miner BG, Vonesh JR. (2006) Size correction: Comparing morphological traits among populations and environments. Oecologia 148(4): 547–554. 10.1007/s00442-006-0403-6 [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, Louisiana, 14 November 2010, 1–8. 10.1109/GCE.2010.5676129 10.1109/GCE.2010.5676129 10.1109/GCE.2010.5676129 [DOI]
- Minh Q, Nguyen MAT, von Haeseler A. (2013) Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30(5): 1188–1195. 10.1093/molbev/mst024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo TV, Grismer LL, Pham TH, Wood Jr PL. (2014) A new species of Hemiphyllodactylus Bleeker, 1860 (Squamata: Gekkonidae) from Ba Na-Nui Chua Nature Reserve, Central Vietnam. Zootaxa 3760(4): 539–552. 10.11646/zootaxa.3760.4.3 [DOI] [PubMed] [Google Scholar]
- Nguyen TQ, Botov A, Le MD, Nophaseud L, Zug G, Bonkowski M, Ziegler T. (2014) A new species of Hemiphyllodactylus (Reptilia: Gekkonidae) from northern Laos. Zootaxa 3827(1): 45–46. 10.11646/zootaxa.3827.1.4 [DOI] [PubMed] [Google Scholar]
- Nguyen TQ, Lehmann T, Le MD, Duong HT, Bonkowski M, Ziegler T. (2013) A new species of Hemiphyllodactylus (Reptilia: Gekkonidae) from northern Vietnam. Zootaxa 3736(1): 89–98. 10.11646/zootaxa.3736.1.5 [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TQ, Do QH, Ngo HT, Pham AV, Pham CT, Le MD, Ziegler T. (2020) Two new species of Hemiphyllodactylus (Squamata: Gekkonidae) from Hoa Binh Province, Vietnam. Zootaxa 4801(3): 513–536. 10.11646/zootaxa.4801.3.5 [DOI] [PubMed] [Google Scholar]
- Pagès J. (2015) Multiple Factor Analysis by Example Using R. CRC Press, New York, 272 pp. 10.1201/b17700 [DOI] [Google Scholar]
- R Core Team (2021) R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna. 2018. http://www.R-project.org [Accessed 1 December 2022]
- Rambaut A, Suchard MA, Xie D, Drummond AJ. (2014) Tracer. Version 1.7. http://tree.bio.ed.ac.uk/software/tracer/ [Accessed 15 January 2023]
- Reist JD. (1986) An empirical evaluation of coefficients used in residual and allometric adjustment of size covariation. Canadian Journal of Zoology 64(6): 1363–1368. 10.1139/z86-203 [DOI] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons JE. (2002) Herpetological collecting and collections management. Revised edition. Society for the Study of Amphibians and Reptiles. Herpetological Circular 31: 1–153. [Google Scholar]
- Sung YH, Lee W, Ng H, Zhang Y, Yang J. (2018) A new species of Hemiphyllodactylus (Squamata: Gekkonidae) from Hong Kong. Zootaxa 4392(2): 361–373. 10.11646/zootaxa.4392.2.8 [DOI] [PubMed] [Google Scholar]
- Thorpe RS. (1975) Quantitative handling of characters useful in snake systematics with particular reference to interspecific variation in the Ringed Snake Natrixnatrix (L.). Biological Journal of the Linnean Society. Linnean Society of London 7(1): 27–43. 10.1111/j.1095-8312.1975.tb00732.x [DOI] [Google Scholar]
- Thorpe RS. (1983) A review of the numerical methods for recognizing and analyzing racial differentiation. In: Felsenstein J. (Ed.) Numerical Taxonomy.NATO ASI Series (Series G: Ecological Sciences). Vol. 1. Springer-Verlag, Berlin, 404–423. 10.1007/978-3-642-69024-2_43 [DOI]
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. (2016) W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44(W1): W232–W235. 10.1093/nar/gkw256 [DOI] [PMC free article] [PubMed]
- Turan C. (1999) A note on the examination of morphometric differentiation among fish populations: The Truss System. Turkish Journal of Zoology 23: 259–263. [Google Scholar]
- Uetz P, Freed P, Aguilar R, Reyes F, Hošek J [Eds] (2022) The Reptile Database. http://www.reptile-database.org [Accessed 22 February 2023]
- Werner F. (1900) Beschreibung einiger noch unbekannter neotropischer und indischer Reptilien. Zoologischer Anzeiger 23: 196–198. [Google Scholar]
- Wilcox TP, Zwickl DJ, Heath TA, Hillis DM. (2002) Phylogenetic relationships of the Dwarf Boas and a comparison of Bayesian and bootstrap measures of phylogenetic support. Molecular Phylogenetics and Evolution 25(2): 361–371. 10.1016/S1055-7903(02)00244-0 [DOI] [PubMed] [Google Scholar]
- Yan J, Lin Y, Guo W, Li P, Zhou KY. (2016) A new species of Hemiphyllodactylus Bleeker, 1860 (Squamata: Gekkonidae) from Guizhou, China. Zootaxa 4117(4): 543–554. 10.11646/zootaxa.4117.4.6 [DOI] [PubMed] [Google Scholar]
- Zhang B, Qian TY, Jiang XJ, Cai B, Deng XJ, Yang DD. (2020) A new species of Hemiphyllodactylus Bleeker, 1860 (Reptilia: Squamata) from Hunan Province, China. Asian Herpetological Research 11(3): 183–193. 10.16373/j.cnki.ahr.190066 [DOI] [Google Scholar]
- Zhou KY, Liu YZ, Yang GP . (1981) Three new subspecies of Hemiphyllodactylusyunnanensis (Boulenger) from China (Lacertiformes: Gekkonidae). Acta Zootaxonomica Sinica, 6: 202–209. [in Chinese, English translation by H. Ota (1996) in Smithsonian Herpetological Information Service, 110, 1–8, 1 pl.]. 10.5479/si.23317515.110.1 [DOI]
- Zug GR. (2010) Speciation and dispersal in a low diversity taxon: The slender geckos Hemiphyllodactylus (Reptilia, Gekkonidae). Smithsonian Contributions to Zoology 631(631): 1–70. 10.5479/si.00810282.631 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data that support the findings of this study are available in the main text or Supplementary Information.