Abstract
Cervical cancer is a significant disease affecting women worldwide. Regular cervical examination with gynecologists is important for early detection and treatment planning for women with precancers. Precancer is the direct precursor to cervical cancer. However, there is a scarcity of experts and the experts’ assessments are subject to variations in interpretation. In this scenario, the development of a robust automated cervical image classification system is important to augment the experts’ limitations. Ideally, for such a system the class label prediction will vary according to the cervical inspection objectives. Hence, the labeling criteria may not be the same in the cervical image datasets. Moreover, due to the lack of confirmatory test results and inter-rater labeling variation, many images are left unlabeled. Motivated by these challenges, we propose to develop a pretrained cervix model from heterogeneous and partially labeled cervical image datasets. Self-supervised Learning (SSL) is employed to build the cervical model. Further, considering data-sharing restrictions, we show how federated self-supervised learning (FSSL) can be employed to develop a cervix model without sharing the cervical images. The task-specific classification models are developed by fine-tuning the cervix model. Two partially labeled cervical image datasets labeled with different classification criteria are used in this study. According to our experimental study, the cervix model prepared with dataset-specific SSL boosts classification accuracy by 2.5%↑ than ImageNet pretrained model. The classification accuracy is further boosted by 1.5%↑ when images from both datasets are combined for SSL. We see that in comparison with the dataset-specific cervix model developed with SSL, the FSSL is performing better.
Keywords: Deep Learning, Self-supervised Learning, Federated Learning, Cervix Image Classification
1. Introduction
In the global scenario, cervical cancer is the fourth most common cancer in women. Early detection of pre-cancerous cervical lesions can reduce the premature death of the woman. Hence, it is important to screen the cervix on a regular basis. Among all cervical screening techniques, visual inspection with acetic acid (VIA) is commonly used as it is cheap and available easily. The key limitation of this technique is that it is suffering from inter-and intra-expert variability. Recently growing artificial intelligence and machine learning-based automated image analysis system could address this limitation [6, 10]. Note that hand-crafted feature-based classifiers are known to under performing [12, 7, 5] than the deep learning approaches [8, 9]. Therefore, the researchers prefer to employ deep learning to solve cervical image analysis problems. However, by nature deep learning is data-hungry and needs experts’ involvement for annotating the data [3]. From the current image analysis challenge perspective, cervical image labeling is costly, needs multiple experts’ agreements, and requires multiple diagnostic information. Therefore, to overcome the limitations in developing a robust deep model with limited data, transfer learning [15], i.e. transferring knowledge from natural images becomes a commonly used method. However, the development of domain-specific deep models is the current research focus as it will be providing better image representation.
The conventional approach to overcome data scarcity is uniting data from multiple institutions or organizations. However, conducting such collaboration for centralized learning (CL) has several limitations. First, there might be variations in the visual quality of the images contributed by different organizations. This variation comes from the use of different imaging devices for image acquisition. Moreover, images may not be allowed to share among the collaborated institutions for which federated learning (FL) is an effective solution [14]. However, for federated supervised learning, a special research effort is required to deal with the different class distribution among the institutions [13]. Finally, and most importantly, the cervical image labeling criteria vary and depend on: the availability of other diagnostic results, population under study, treatment planning, severity grading strategy, etc. The variety in the image labeling criteria across datasets makes the task more challenging as it restricts researchers in performing any kind of supervised collaborative (CL or FL) learning.
This paper develops a pretrained cervix model (or cervix model) with self-supervised learning (SSL) using two cervical image datasets that are partially labeled (a part of the dataset is labeled) and labeled with different classification criteria (heterogeneous). Both centralized SSL and federated SSL are experimented. The self-supervised learning is used as the images do not need any expert annotation. Hence, it allows uniting all images from both datasets for cervix model development. There is no way to evaluate the effectiveness of the developed pretrained models. Therefore, a downstream task having labeled data is considered and the pre-trained model is fine-tuned to build a deep model which can be evaluated. In this paper, as a downstream task, we choose to develop two cervical image classification models: (i) classify an image based on the presence of cervical infection and (ii) classify a cervical image based on whether the cervical infection is a precursor of pre-malignancy. Two different datasets are used for these two downstream tasks. Note that, to the best of our knowledge, no cervical image dataset is available in the public domain which can be utilized for automating the visual assessment of acetic acid applied cervix.
In summary, the research presented in [16] and [2] motivate us to do this work. Model genesis presented in [16] proposed to train an encoder-decoder-based deep model to reconstruct original images from synthetically distorted images for obtaining a pretrained model for medical image representation. In [2], visual contrastive learning was employed to learn the natural image representation. In this paper, we hire the concept of contrastive learning from [2] and utilize it for developing pretrained cervical model development. We experiment on both centralized and federated self-supervised learning. Our work is novel in the following two perspectives: (a) it is the first research attempt that focuses on the development of a pretrained cervix model with zero annotation cost; (b) believe to be the first medical image representation research work which consider Federated Self-Supervised Learning (FSSL).
The rest of the paper is organized as follows: Section 2 discusses the background of self-supervised learning, and federated learning and ends with describing how federated self-supervised learning is used for pretrained cervix model development. The experimental protocol is available in Section 3. The analysis of experimental results is presented in Section 4. Finally, Section 5 concludes the paper and mentions the future scope of this work.
2. Methods
2.1. Self-supervised Learning
Self-supervised learning (SSL) is a visual representation learning approach that releases the requirement of expert annotation. Machine learners design a discriminative task directly from the raw images without any expert supervision. The designed task is called the pretext task. The deep model is trained to learn the pretext task which can capture the image semantics (i.e. good initialization weights for related domain’s downstream tasks).
This paper considers contrastive feature learning as the pretext task - i.e. embedding of the images will be well separated and the image and an augmented version will have closer embedding [2]. The SSL model can be trained with the mini-batch of size 2N constructed with N random images and an augmented version of every image. The training loss () between an image and its augmented version is given as:
| (1) |
where is the feature vector of image; is an indicator function evaluating to 1 iff represents transpose operation and is a constant. The loss in a mini-batch is computed across all pairs constructed with an image and its augmented version.
2.2. Federated Self-supervised Learning (FSSL)
Unauthorized access to sensitive data is harmful and is a social threat. Hence, several laws and regulations (GDPR 2018 by the EU, CCPA 2020 in the US, etc) are formed to stop sensitive information sharing (especially medical/banking domain). Therefore due to these legal issues related to data protection collaborated institutions may not be allowed to share raw data among them. In this regard, federated learning (FL) is an effective approach for robust inter-institutional collaborated deep model development from the data distributed in multiple institutions (clients) without sharing the raw data [14].
In this paper, two cervical image datasets are used for developing the cervix model. We assume that two datasets are residing in two different clients and clients are not allowed to share raw data. All images (both labeled and unlabeled) from both datasets are utilized for Federated Self-supervised Learning (FSSL). Two naive federated learning approaches namely, Client-Server Federated Self-supervised Learning (CSFSSL) and Peer-to-Peer Federated Self-supervised Learning (PPFSSL) are considered in this research. In the client-server approach (CSFSSL) there is no direct communication channel among the clients and a central server controls the learning. In this approach, first, the server broadcast a model to both clients, and then independently, at every client, the model is updated based on the loss computed with its images. This weight updating is performed for E-epochs. After that, the clients send the updated models to the server which aggregates the model weights received from both clients. Note that the value of E may vary among the clients. The whole communication round is repeated until the model is fully trained. For simplicity, this paper adopts federated averaging for aggregating the client models. On the other hand, in the Peer-to-Peer approach, no server is present- clients can directly communicate with each other. In this approach, first, a random client is chosen which initialize a model and update weights with SSL by using the available images. The updated model is then sent to another client where SSL is performed and weights are updated further. Thus the communication among the clients takes place circularly. Like CSFSSL, the weight updating in a client is performed for E-epochs and the value of E can be varied among clients. The whole communication round is repeated until the model is fully trained. It is worth mentioning that in both cases clients are not sharing the images however the potential of all images available in both clients are utilized for cervix model building.
2.3. Proposed Approach: Cervical Model Development
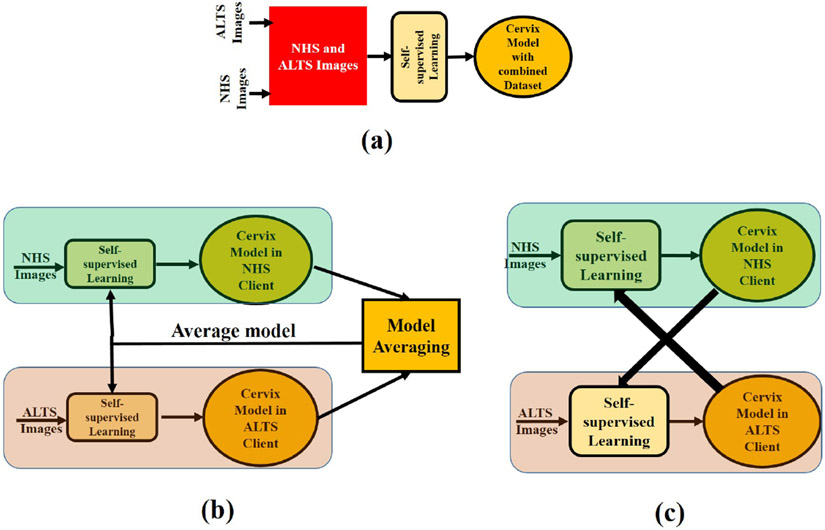
This paper considers two different FSSL approaches presented in Sec 2.2 for cervix model development. Finally, the performance of the FSSL model is compared with centralized SSL (CSSL). As a downstream task, client-specific classification model building is considered. The pictorial representation of the learning frameworks for the CSSL, CSFSSL, and PPFSSL are shown in Fig 1 (a), Fig 1 (b) and Fig 1 (c) respectively. The SSL algorithm discussed in Sec 2.1 is used for model building. The augmented version of an image is obtained with rotation, horizontal and vertical flipping, random shift, random zoom, gamma changing, and brightness changing.
Fig. 1:
Block diagram of the SSL training system- (a) Centralized SSL (CSSL) (b) Client-Server FSSL (CSFSSL) and (c) Peer-to-Peer FSSL (PPFSSL).
3. Experimental Protocol
3.1. Dataset Description
The datasets used in this research come from two distinct cohort studies for cervical examination (NHS [1] and ALTS [11]). These studies are conducted by the researchers at the National Cancer Institute (NCI) of the US National Institutes of Health (NIH). NCI shared a subset of acetic acid-applied cervix images for our research. The images generated during the NHS study are referred to as NHS dataset and the images generated during the ALTS study are referred to as ALTS dataset. Only a subset of the images for both datasets is labeled. The labeling of NHS images performed based on the presence of cervical infection and the labeling of ALTS images performed based on the criteria of whether the cervical infection occurred in the images is a precursor of pre-malignancy. Multiple screening and diagnostic information like visual assessment, HPV, cytology, histopathology, colposcopy, etc are analyzed for labeling the images. For experimental evaluation, both datasets are splitted at the women’s level into three disjoint subsets - train, validation, and test. Table 1 represents the subset-wise number of patients, labeled images, and total available images for the datasets.
Table 1:
Data set splits. For NHS case refers presence of cervical infection and for ALTS case refers cervical infection occurred in a image is a precursors of pre-malignancy.
| Split | Class | Patients | Labeled Images | Total Images | |||
|---|---|---|---|---|---|---|---|
| NHS | ALTS | NHS | ALTS | NHS | ALTS | ||
| Train | Case | 91 | 124 | 182 | 248 | 2029 | 3145 |
| Control | 181 | 242 | 361 | 481 | |||
| Valid | Case | 22 | 31 | 44 | 62 | 520 | 791 |
| Control | 45 | 60 | 90 | 120 | |||
| Test | Case | 25 | 34 | 49 | 68 | ||
| Control | 50 | 65 | 99 | 130 | |||
3.2. Network Architecture
In this paper, ResNet-50 a widely used state-of-the-art deep model is used as the backbone. During SSL, the final classification layer is replaced with a dense layer with a ReLU activation. The number of neurons in the dense layer is varied among [64, 128, 256] and our experimental outcome shows that performances are very closer. Hence, to reduce the computational burden we set the number of the neuron to 64. For the downstream classification task, this dense layer (along with the activation layer) is replaced with a single output neuron with sigmoid activation. The predicted case probability is obtained from the sigmoid layer. Note that full fine-tuning is employed for the dataset-specific downstream classification tasks.
3.3. Competing Methods
This paper compares the performance of cervical image classification for the following six different varying initialization approaches: (i) Random: Random network weights initialization, (ii) ImageNet: Network initialization with pretrained ImageNet model, (iii) Self-supervised Learning (SSL): Network initialization with SSL with the available images in a client. (iv) Centralized Self-supervised Learning (CSSL): Network initialization with SSL with the available images in both clients, (v) Client-Server Federated Self-supervised Learning (CSFSSL): Network initialization with the Client-Server Federated Self-supervised Learning, and (vi) Peer-to-Peer Federated Self-supervised Learning (PPFSSL): Network initialization with the circular SSL. Note that in Random no knowledge is transferred, in ImageNet the knowledge is transferred from a different domain (natural image) than cervical images, and knowledge is transferred from the same domain for the rest of the approaches.
3.4. Parameter Settings
The optimal hyper-parameters for both SSL training and downstream classification model training are chosen empirically. The validation loss is analyzed for parameter selection. SSL uses (See Eq 1), learning rate 0.01, momentum = 0.9, weight decay = 1e – 5, and trained for 50 epochs. The downstream classification models are trained with batch size =4, learning rate 0.001, momentum = 0.09, weight decay = 1e – 6, and trained for 50 epochs. Random shuffling of images is done for both SSL and downstream classification model training. Reverse class weighting is employed to tackle the class imbalance issue during classification model training.
3.5. Implementation
The networks are implemented with Keras [4] a popularly used deep learning toolkit. Regarding computing hardware, two (2) GeForce RTX 2080 Ti GPUs installed with an Intel(R) Xeon(R) Gold 5218 CPU (@ 2.30GHz) is used for training. Note that the federated learning algorithms are logically implemented in the same computing resources.
3.6. Evaluation metrics
As there is no known evaluation metric for evaluating the quality of the developed pretrained model, we evaluate only the dataset-specific downstream class label prediction performances. We constraint the learning approach for the downstream classification task and vary the network initialization (see Sec 3.3). The following four commonly known matrices are computed for performance comparison: (1) Accuracy (ACC), (2) Recall, (3) Precision, (4) F1-Score.
4. Experimental Results and Discussion
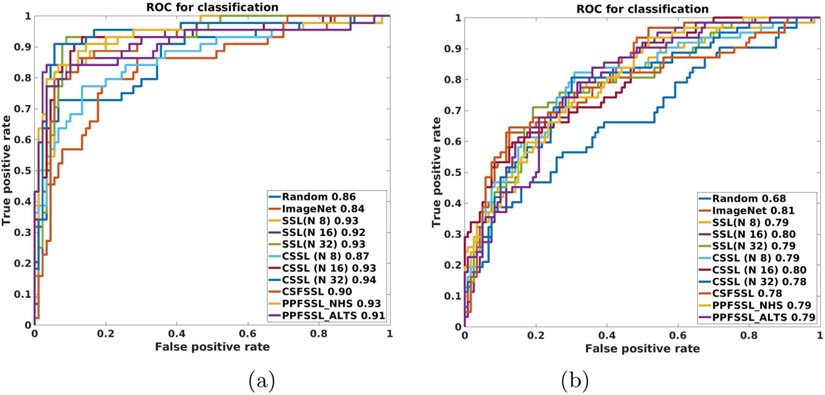
Fig 2 contains the Receiver Operating Curves (ROC) for all downstream classification tasks developed with different model initialization approaches. The quantitative performance (used matrices are described in Sec 3.3) for the same is shown in Table 2. Table 2 shows that for both datasets ImageNet initialization improves the accuracy than random initialization but is unable to improve the recalls. The pretrained network built with SSL is better than transferring knowledge from the ImageNet model (built with natural images). In case of cervical model development with the NHS image dataset, the NHS classification model provides the best accuracy, recall, and F1_Score when the SSL approach with N = 16 is used and the best precision is received when N = 8 is used. In case of cervical model development with the ALTS image dataset, ALTS classification model provides the best accuracy and precision when the SSL approach with N = 16 is used and the best recall and F1_Score is received when N = 8 is used. Our experimental results show that pretrained model developed with SSL makes a noticeable performance improvement over the ImageNet pretrained model which justifies the importance of SSL-based cervical model development. We find that the cervix model initialized with CSSL improves the performance than SSL-based initialization for both downstream tasks which advocates the importance of uniting images from both datasets. For CSSL-based initialization when N = 8, for most cases, the best classification performance are received for both datasets. Therefore, for developing the cervix model with Federated Self-Supervised Learning (FSSL) we set N = 8. For CSFSSL based cervix model development, The value of E (iteration during local model updating) is varied among 1, 5, 10 and according to our experiment E = 1 provides best performance. The PPFSSL-based cervix model development is done with two different approaches. The approaches vary based on the starting client. If the SSL learning starts with the NHS client we term this approach as PPFSSL_NHS and if the SSL learning starts with the ALTS client we term this approach PPFSSL_ALTS. For experimental similarity and comparison, we consider E = 1 for both PPFSSL_NHS and PPFSSL_ALTS. The experimental results show that the development of the cervix model with both PPFSSL and CSFSSL produces comparative classification performance. From Table 2, it is evident that in general, federated SSL performs better than SSL with single client images. Hence, our experimental results support the usefulness of FSSL to overcome data sharing constraints and utilize data from multiple clients to develop a better cervix model.
Fig. 2:
Receiver Operating Curve (ROC): (a) NHS and (b) ALTS. The numeric values represent the AUC values for the classifiers with considered initialization.
Table 2:
Performance evaluation
| Initialization Method |
NHS | ALTS | ||||||
|---|---|---|---|---|---|---|---|---|
| ACC | Recall | Precision | F1_Score | ACC | Recall | Precision | F1_Score | |
| Random | 79.73 | 0.5306 | 0.7879 | 0.6341 | 76.77 | 0.5735 | 0.6964 | 0.6290 |
| ImageNet | 80.41 | 0.4898 | 0.8571 | 0.6234 | 77.78 | 0.5588 | 0.7308 | 0.6333 |
| SSL (N 8) | 83.11 | 0.6327 | 0.8158 | 0.7126 | 79.80 | 0.7206 | 0.7000 | 0.7101 |
| SSL (N 16) | 83.11 | 0.6939 | 0.7727 | 0.7312 | 80.30 | 0.6029 | 0.7736 | 0.6777 |
| SSL (N 32) | 83.11 | 0.6531 | 0.8000 | 0.7191 | 79.29 | 0.6176 | 0.7368 | 0.6720 |
| CSSL (N 8) | 86.49 | 0.7143 | 0.8537 | 0.7778 | 81.82 | 0.7794 | 0.7162 | 0.7465 |
| CSSL (N 16) | 85.81 | 0.7551 | 0.8043 | 0.7789 | 81.31 | 0.6324 | 0.7818 | 0.6992 |
| CSSL (N 32) | 85.14 | 0.7959 | 0.7647 | 0.7800 | 81.31 | 0.6471 | 0.7719 | 0.7040 |
| CSFSSL | 84.46 | 0.6735 | 0.8250 | 0.7416 | 79.80 | 0.6912 | 0.7121 | 0.7015 |
| PPFSSL_NHS | 84.46 | 0.7347 | 0.7826 | 0.7579 | 80.81 | 0.6618 | 0.7500 | 0.7031 |
| PPFSSL_ALTS | 84.46 | 0.7755 | 0.7600 | 0.7677 | 80.30 | 0.6618 | 0.7377 | 0.6977 |
5. Conclusion and Scope of Future Work
In this paper, we highlighted the cervical image analysis challenges owing to labeling scarcity, and variability. Finally, we propose an innovative federated self-supervised learning approach to tackle this. The experimental outcome justifies that the self-supervised learning approach is efficient to cope with the label scarcity and the labeling heterogeneity. The use of Federated Self-Supervised Learning illuminates to tackle data privacy. We believe that the cervix model development presented in this paper is the inaugural attempt in light of a domain-specific task agnostic pre-trained model construction.
The immediate future scope of this work will include the development of an improved cervix model combining multi-source image datasets that are unlabelled, have labeling heterogeneity, and have variation in pixel properties due to imaging devices. The experiment on federated learning is shown in a synthetic environment, however, actual deployment of the proposed federated learning algorithm for uniting multiple institutions is the next future challenge. The proposed concept is generic and can be hired by other medical image analysis tasks.
Acknowledgement
The Intramural Research Program of the National Library of Medicine, a part of the National Institutes of Health (NIH) supports this research. The authors of this paper want to thank Dr. Mark Schiffman of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, and his colleagues for sharing the cervical images with us.
References
- 1.Bratti MC, Rodríguez AC, Schiffman M, Hildesheim A, Morales J, Alfaro M, Guillén D, Hutchinson M, Sherman ME, Eklund C, Schussler J, Buckland J, A Morera L, Cárdenas F, Barrantes M, Pérez E, Cox TJ, Burk R, Herrero R: Description of a seven-year prospective study of human papillomavirus infection and cervical neoplasia among 10000 women in guanacaste, costa rica. Pan American journal of public health 2(15), 75–89 (Feb 2004) [DOI] [PubMed] [Google Scholar]
- 2.Chen T, Kornblith S, Norouzi M, Hinton G: A simple framework for contrastive learning of visual representations. In: H.D., Singh A (eds.) Proceedings of the 37th International Conference on Machine Learning. Proceedings of Machine Learning Research, vol. 119, pp. 1597–1607. PMLR, Virtual (13–18 Jul 2020) [Google Scholar]
- 3.Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, Ferrero E, Agapow PM, Zietz M, Hoffman MM, Xie W, Rosen GL, Lengerich BJ, Israeli J, Lanchantin J, Woloszynek S, Carpenter AE, Shrikumar A, Xu J, Cofer EM, Lavender CA, Turaga SC, Alexandari AM, Lu Z, Harris DJ, DeCaprio D, Qi Y, Kundaje A, Peng Y, Wiley LK, Segler MHS, Boca SM, Swamidass SJ, Huang A, Gitter A, Greene CS: Opportunities and obstacles for deep learning in biology and medicine. Journal of the Royal Society, Interface (April, 2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chollet F, et al. : Keras. https://keras.io (2015) [Google Scholar]
- 5.Fernandes K, Cardoso JS, Fernandes J: Automated methods for the decision support of cervical cancer screening using digital colposcopies. IEEE Access 6, 33910–33927 (2018) [Google Scholar]
- 6.Hu L, Bell D, Antani S, Xue Z, Yu K, Horning MP, Gachuhi N, Wilson B, Jaiswal MS, Befano B, Long LR, Herrero R, Einstein MH, Burk RD, Demarco M, Gage JC, Rodriguez AC, Wentzensen N, Schiffman M: An Observational Study of Deep Learning and Automated Evaluation of Cervical Images for Cancer Screening. JNCI: Journal of the National Cancer Institute 111(9), 923–932 (01 2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim E, Huang X: A Data Driven Approach to Cervigram Image Analysis and Classification, pp. 1–13. Springer Netherlands, Dordrecht: (2013) [Google Scholar]
- 8.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JA, van Ginneken B, Sánchez CI: A survey on deep learning in medical image analysis. Medical Image Analysis 42, 60–88 (2017) [DOI] [PubMed] [Google Scholar]
- 9.Pal A, Chaturvedi A, Chandra A, Chatterjee R, Senapati S, Frangi AF, Garain U: Micaps: Multi-instance capsule network for machine inspection of munro’s microabscess. Computers in Biology and Medicine 140, 105071 (2022) [DOI] [PubMed] [Google Scholar]
- 10.Pal A, Xue Z, Befano B, Rodriguez AC, Long LR, Schiffman M, Antani S: Deep metric learning for cervical image classification. IEEE Access 9, 53266–53275 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schiffman M, Adrianza ME: Ascus-lsil triage study. design, methods and characteristics of trial participants. Acta Cytol 44(5), 726–742 (Sept-Oct 2000) [DOI] [PubMed] [Google Scholar]
- 12.Srinivasan Y, Nutter B, Mitra S, Phillips B, Sinzinger E: Classification of cervix lesions using filter bank-based texture mode. In: 19th IEEE Symposium on Computer-Based Medical Systems (CBMS’06). pp. 832–840 (2006) [Google Scholar]
- 13.Yang M, Wong A, Zhu H, Wang H, Qian H: Federated learning with class imbalance reduction (2020) [Google Scholar]
- 14.Yang Q, Liu Y, Cheng Y, Kang Y, Chen T, Yu H: (2019) [Google Scholar]
- 15.Yang Q, Zhang Y, Dai W, Pan SJ: Transfer Learning. Cambridge University Press; (2020) [Google Scholar]
- 16.Zhou Z, Sodha V, Pang J, Gotway MB, Liang J: Models genesis. Medical Image Analysis 67, 101840 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]