Abstract
Background
Validated computable eligibility criteria use real-world data and facilitate the conduct of clinical trials. The Genomic Medicine at VA (GenoVA) Study is a pragmatic trial of polygenic risk score testing enrolling patients without known diagnoses of 6 common diseases: atrial fibrillation, coronary artery disease, type 2 diabetes, breast cancer, colorectal cancer, and prostate cancer. We describe the validation of computable disease classifiers as eligibility criteria and their performance in the first 16 months of trial enrollment.
Methods
We identified well-performing published computable classifiers for the 6 target diseases and validated these in the target population using blinded physician review. If needed, classifiers were refined and then underwent a subsequent round of blinded review until true positive and true negative rates ≥80% were achieved. The optimized classifiers were then implemented as pre-screening exclusion criteria; telephone screens enabled an assessment of their real-world negative predictive value (NPV-RW).
Results
Published classifiers for type 2 diabetes and breast and prostate cancer achieved desired performance in blinded chart review without modification; the classifier for atrial fibrillation required two rounds of refinement before achieving desired performance. Among the 1,077 potential participants screened in the first 16 months of enrollment, NPV-RW of the classifiers ranged from 98.4% for coronary artery disease to 99.9% for colorectal cancer. Performance did not differ by gender or race/ethnicity.
Conclusions
Computable disease classifiers can serve as efficient and accurate pre-screening classifiers for clinical trials, although performance will depend on the trial objectives and diseases under study.
INTRODUCTION
Pragmatic clinical trials facilitate experimental studies in large populations while minimizing perturbations to the delivery of usual health care.1 The benefits of embedding trials in routine health care include research cost-efficiency and increased sample sizes, which in turn improve statistical power. Although more strictly protocolized explanatory trials are considered the more rigorous study design to demonstrate efficacy, pragmatic trials are better suited to measuring the real-world clinical impact of an intervention.2
An important tool for enabling the conduct of pragmatic trials is the electronic health record (EHR).3,4 In particular, clinical data stored in the EHR, such as medical diagnoses or treatment history, can be readily used to identify large and representative cohorts of patients who are eligible or ineligible for inclusion in a clinical trial.5 Combinations of structured data such as demographics, prescriptions, and diagnosis codes are easily computable as inclusion and exclusion criteria for a pragmatic trial. However, clinical data in the EHR are collected primarily for clinical care, billing, and administrative purposes. As a result, their secondary use for research may be limited by missingness or insufficient accuracy,6 which can impair trial performance. If computable criteria exclude too many patients who are in fact eligible, the trial might identify an insufficient number of eligible patients. If the criteria include too many patients who are in fact ineligible, the study will need to expend resources to introduce additional screening procedures and increase recruitment efforts to compensate for poor efficiency. Errors in either direction can result in a patient sample unrepresentative of the target population.
The Genomic Medicine at VA (GenoVA) Study (Clinicaltrials.gov identifier NCT04331535) is a pragmatic randomized controlled trial of polygenic risk score testing among adult primary care patients aged 50–70 without a diagnosis of any one of six common diseases.7 Polygenic risk scores have emerged as a new clinical tool with potential utility for improved risk stratification in an era of precision medicine.8 The GenoVA Study enrolls patients from the Veterans Affairs Boston Healthcare System (VA Boston), part of the national Veterans Health Administration (VHA). Conducting pragmatic clinical trials in VHA is facilitated by more than 20 years of EHR and other data from over 20 million US Veterans, but these data are still susceptible to inaccuracies and the missingness that results from a patient population who variably receives healthcare in non-VHA settings.9 The study procedures of the GenoVA Study afforded the opportunity to evaluate the usefulness of computable disease classifiers for use as pre-screening exclusion criteria in a clinical trial. Here, we describe our approach to the development of those classifiers and their real-world performance during the first 16 months of the trial.
METHODS
Setting
Nationally, VHA provides health care and social services for over 9 million US Veterans in a network of almost 1,300 facilities.10 Patients are eligible to receive VHA healthcare by meeting certain criteria, based on military service, service-related disability and income.11 The GenoVA Study is recruiting patients from the three major clinical centers and five outpatient clinics comprising VA Boston, which provides health care to about 61,000 Veterans in Eastern Massachusetts annually.12 The objective of the GenoVA Study is to measure the two-year clinical impact of measuring and reporting polygenic risk scores for six common diseases with established prevention strategies: atrial fibrillation (AFib), coronary artery disease (CAD), type 2 diabetes mellitus (T2D), breast cancer (BrCa), colorectal cancer (CRCa), and prostate cancer (PrCa). The need to identify a sufficiently large eligible patient population while excluding patients with known diagnoses of these conditions at trial baseline motivated the present study.
Data sources
Data for this study derive from three sources: the VHA Corporate Data Warehouse (CDW); the Veterans Health Information Systems and Technology Architecture (VistA), the main EHR system used by VHA providers; and from eligibility telephone screen surveys of potential GenoVA Study participants at baseline. The CDW is a relational database that houses clinical, accounting, and other administrative data since 1999, and is updated nightly from VistA.13,14 The organization of the CDW facilitates queries of structured data such as diagnosis codes and prescriptions.
Identification of validated EHR classifiers
For this pragmatic trial with a target enrollment of 1,076 participants, our goal was to develop an efficient structured data classifier for each of the six target diseases (Figure 1). For each disease, we searched PubMed for publications originating from VHA in or after 2006, the year CDW became active. For each disease, we selected the most recent publication reporting a sensitivity and, when available, positive predictive value (PPV), above 80%. When comparative studies or systematic reviews included several classifiers with comparable performance, we selected the classifier with the fewest components, as we expected these to be the most amenable to subsequent optimization, as well as reasonable computation times for nightly querying. In cases where performance data were not published, we used the most recently published classifier. Suitable validation studies or systematic reviews that included VHA performance characteristics were identified for all diseases except BrCa, for which we instead identified a non-VHA systematic review. Additional details about the classifiers selected from the literature are found in the Supplemental Methods.
Figure 1. Validation and iterative refinement of disease classifier.
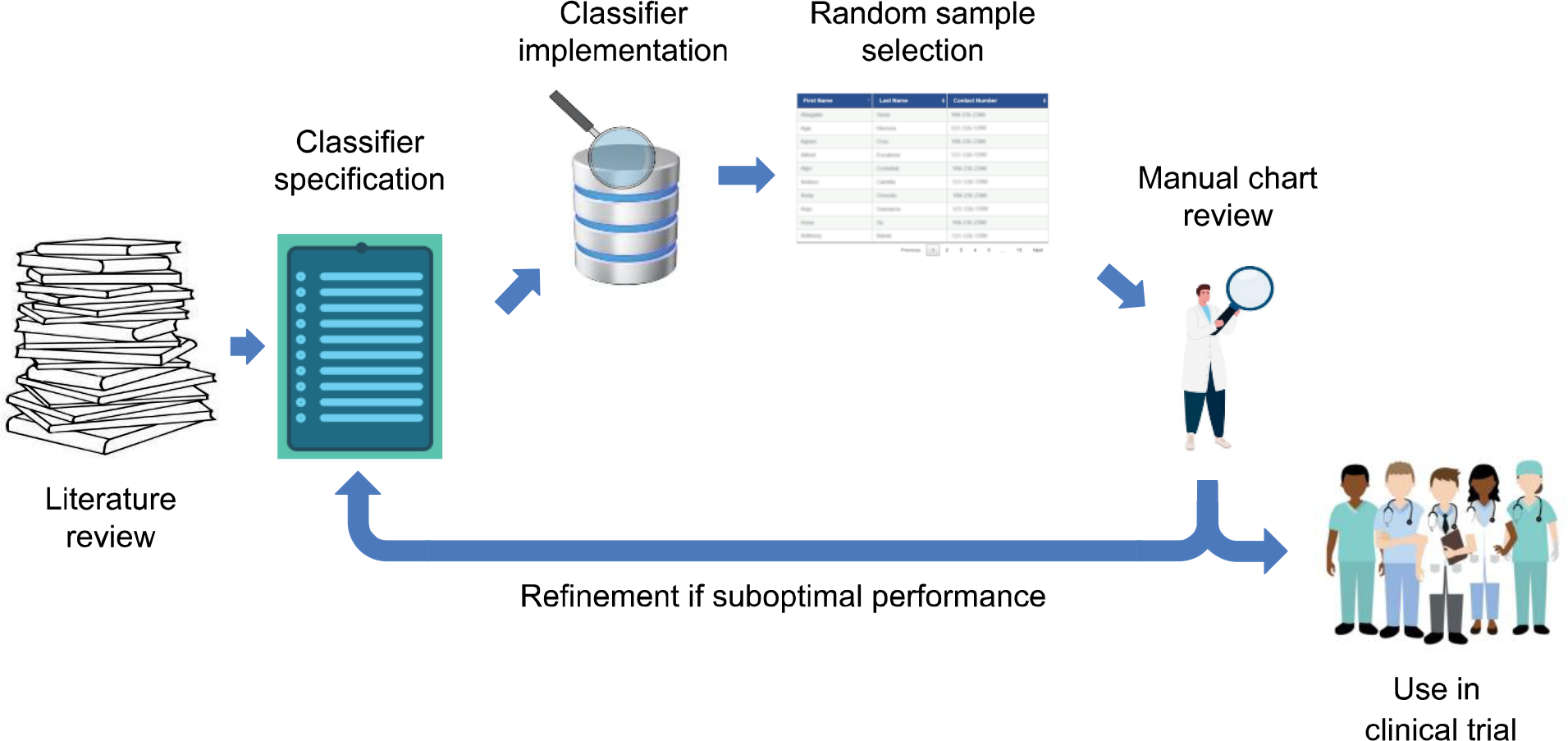
For a given disease, a relevant disease classifier is identified through a literature search, based on criteria such as ease of computability, suitability for the target EHR, and performance. The classifier is implemented in the target EHR and used to draw a random selection of positive and negative cases for manual clinician review. Cycles of classifier refinement and additional clinician review occurs until desired performance is achieved, after which the classifier is implemented for trial recruitment.
Initial application of previously published classifiers
Initial application of the published classifiers to VHA data was straightforward but did require additional decisions and adaptations. We applied International Classification of Diseases (ICD) code classifiers only to outpatient and inpatient diagnosis tables in CDW, and not to the less specific outpatient problem list tables. We applied medication classifiers to both VHA outpatient prescription tables and non-VHA medication list tables in CDW. Most notably, all of the published classifiers except BrCa used ICD, Ninth Revision, Clinical Modification (ICD-9-CM) and Procedure Codes (ICD-9-PCS) instead of the more contemporary ICD-10-CM or ICD-10-PCS codes, to which VHA transitioned in 2015.15 In order to allow the classifier to process both pre-2015 and post-2015 ICD codes, we implemented a semiautomated method to convert ICD-9-CM and ICD-9-PCS codes to their equivalent ICD-10 codes, using conversion tables provided by the Centers for Medicare and Medicaid Services (CMS).16 First, using an automated extraction procedure, written in R 3.5.0,17 we collected all corresponding ICD-10 codes, from both the ICD-9 to ICD-10 conversion table and the ICD-10 to ICD-9 conversion table included in the General Equivalence Mappings (GEMS). Manual review of GEMS tables identified additional relevant ICD-10 codes not mapped through the automated extraction procedure. Our final set of ICD-10 codes was based on the machine-extracted codes, with minor additions and removals as suggested by the manual text search. Although, during design, the addition of post-2015 ICD codes lacked effect on the discriminative characteristics of the classifier, we proceeded with their use, in order to preempt any potential effect on GenoVA recruitment that may be brought about by upcoming transition from VistA to Cerner EHR.
We enhanced the three cancer classifiers by adding inputs from the CDW Oncology tables from the VA Central Cancer Registry, a high-quality dataset containing International Classification of Diseases for Oncology (ICD-O)-encoded diagnoses, abstracted by professional cancer registrars.18 Because the Oncology tables only became available in 2016, literature review identified a VHA classifier using ICD-O-3 codes solely for CRCa. Preliminary chart reviews showed a perfect PPV for CRCa using only the Oncology tables, and so we decided to use Oncology tables for the BrCa and PrCa classifiers as well.
Validation and iterative refinement of the classifiers
In order to measure the performance of each classifier, we compared its performance against manual expert chart reviews. After applying the classifiers to the otherwise eligible population of VA Boston patients (see Real-world performance below), for each disease we randomly selected 10 patient records for which the classifier indicated the presence of a target disease (positive-per-classifier) and 10 records for which the classifier indicated the absence of the target disease (negative-per-classifier). A licensed physician blinded to these classifications reviewed all available clinical data in the corresponding 20 records through VistA to ascertain whether, in his clinical judgment, the patient had any diagnosis of the target disease. After blinded review, the physician was unblinded to classification discrepancies and given the opportunity to reassess and reclassify the discrepant records.
We used this final classification from unblinded clinical chart review (positive-per-review and negative-per-review) as the gold standard classifications. We then assessed the performance of each classifier as follows. A true positive (TP) record was a positive-per-review record that was classified as positive by the classifier. A false negative (FN) record was a positive-per-review record that was classified as negative by the classifier. True negative (TN) and false positive (FP) records were similarly defined. On both passes, Using these definitions, a classifier’s true negative rate (TNR) was defined as TN/(TN+FP), and its true positive rate (TPR) as TP/(TP+FN).
After each round of blinded chart review and unblinded opportunity for reassessment, we revised any classifier whose TPR or TNR was below 80% (see Results). Each time, the refinement strategy was inferred by manual inspection of misclassified records. We performed subsequent rounds of classifier modification and chart review of a new set of 20 records until TPR and TNR were ≥80% for each disease.
Real-world performance
Once optimized, classifiers were put into production for the ongoing GenoVA Study.7 Trial inclusion criteria are age 50 to 70, absence of the six target diseases, VHA health insurance, a primary care provider (PCP) relationship at VA Boston, and at least one clinical care visit or admission at VA Boston in the previous 12 months. We implement the classifiers within a Structured Query Language (SQL) stored procedure (Microsoft SQL Server 13.0, Microsoft SQL Server Management Studio 16.0). The stored procedure queries CDW for patient-PCP relationships from the Primary Care Management Modules and visit-associated stop codes and provider role tables (Supplemental Table 1). An automated scheduled task refreshes the eligibility table nightly, to identify new eligible patients at VA Boston and to remove patients newly diagnosed with one of the exclusionary diseases or who age out of eligibility. Due to these temporal changes, the number of eligible patients identified nightly by the classifier varies by a small, non-zero amount.
GenoVA Study research staff regularly query the eligibility table to send trial recruitment mailings to potentially eligible participants. Mailings are followed by a telephone eligibility screen, during which staff use a phone script to ask whether the patient has ever been diagnosed with any of the 6 target diseases:
Could you tell me whether you’ve even been told by a healthcare provider that you have any of the following conditions?:
Coronary artery disease, such as a heart attack, coronary bypass surgery, or stents in the blood vessels in your heart?
Diabetes?
Atrial fibrillation or an unusual heart rhythm?
Colon cancer or rectal cancer?
Prostate cancer?
Breast cancer?
Study staff follow each positive response with more detailed questions about relevant symptoms, diagnostic tests, medications, and procedures; cases where the research staff is uncertain about diagnosis are escalated to a study physician for chart review and final determination.
The first 16 months of GenoVA Study trial recruitment (June 2020-November 2021) affords the opportunity to assess the performance of our disease classifiers, including counts of what we term real-world true negatives (TN-RW) and false negatives (FN-RW). By extension, we report the classifiers’ real-world negative predictive value (NPV-RW), calculated as the proportion of TN-RW within the set of screened predicted-negative patients (TN-RW + FN-RW). We additionally examined these performance metrics by patient sex and by race/ethnicity, dichotomized as non-Hispanic white and all other, based on administrative data from the CDW.
RESULTS
Optimization of disease classifiers
In April 2020, we identified 20,518 VA Boston patients meeting age, insurance, and PCP relationship criteria. Without additional modification beyond the addition of GEMS-derived codes and VACCR data, described above, all 6 published classifiers yielded a TPR of 100%. TNR ranged from 71% to 91% and was below the optimal threshold of 80% for 2 diseases: AFib and CAD (Table 1). Manual inspection of FP records revealed that a majority were cases in which ICD codes were used for preliminary diagnoses, which were subsequently refuted. For example, ICD code I20.9 for unspecified angina pectoris was used in multiple instances, for patients whose subsequent testing did not confirm a CAD diagnosis. Therefore, during the first round of optimization of the AFib and CAD classifiers, we modified the classifier by requiring two diagnostic codes on two distinct dates.
Table 1.
Performance of classifiers as published and after iterative refinement.
| Metric | AFib | CAD | T2D | BrCa | CRCa | PrCa |
|---|---|---|---|---|---|---|
| Published classifiers | ||||||
| Blinded TPR | 6/6 (100%) | 7/7 (100%) | 9/9 (100%) | 9/9 (100%) | 8/8 (100%) | 9/9 (100%) |
| Blinded TNR | 10/14 (71%) | 10/13 (77%) | 10/11 (91%) | 10/11 (91%) | 10/12 (83%) | 10/11 (91%) |
| Call changes | 0/4 (0%) | 0/3 (0%) | 0/1 (0%) | 0/1 (0%) | 1/2 (50%) | 0/1 (0%) |
| Unblinded TPR | 6/6 (100%) | 7/7 (100%) | 9/9 (100%) | 9/9 (100%) | 9/9 (100%) | 9/9 (100%) |
| Unblinded TNR | 10/14 (71%) | 10/13 (77%) | 10/11 (91%) | 10/11 (91%) | 10/11 (91%) | 10/11 (91%) |
| Modification (Round 1) | ||||||
| Blinded TPR | 6/7 (85%) | 8/8 (100%) | — | — | — | — |
| Blinded TNR | 9/13 (69%) | 10/12 (83%) | — | — | — | — |
| Call changes | 1/5 (20%) | 0/2 (0%) | — | — | — | — |
| Unblinded TPR | 6/6 (100%) | 8/8 (100%) | — | — | — | — |
| Unblinded TNR | 10/14 (71%) | 10/12 (83%) | — | — | — | — |
| Modification (Round 2) | ||||||
| Blinded TPR | 6/6 (100%) | — | — | — | — | — |
| Blinded TNR | 10/14 (71%) | — | — | — | — | — |
| Call changes | 2/4 (50%) | — | — | — | — | — |
| Unblinded TPR | 8/8 (100%) | — | — | — | — | — |
| Unblinded TNR | 10/12 (83%) | — | — | — | — | — |
TP, TN, FP and FN records defined by the performance of the computable classifier against the reference (here, physician chart review, first blinded and then unblinded to computed classification, see Methods). TPR was defined as the ratio TP/(TP+FN). TNR was defined as the ratio TN/(TN+FP). Call changes quantify the number of charts for which the reviewer changed his assessment of the medical record, after being informed about a discrepancy between his blinded classification and the computerized classification.
The first round of refinement significantly optimized the CAD classifier (TNR 83%) but was less effective for the AFib classifier (TNR 71%). Further manual review identified misclassified patients for whom AFib diagnostic codes had been erroneously used during encounters with the anticoagulation clinic. To address this, we further optimized the AFib classifier by excluding diagnostic codes originating from pharmacy staff. This additional modification achieved a TNR of 83% for AFib.
Real-world evaluation of disease classifiers
In November 2021, our data query identified 18,432 VA Boston patients meeting age and primary care relationship criteria for the GenoVA Study, of whom 8,383 (45.5%) were predicted by the optimized classifiers to have at least one of the exclusionary diseases. Of these, 7.2% (1,333/18,432) were classified as having AFib, 16.8% (3,098/18,432) as CAD, 31.2% (5,745/18,432) as T2D, and 0.8% (143/18,432) as CRCa. In addition, 6.2% (1,034/16,690) of men and 7.0% (122/1,742) of women were classified as having PrCa and BrCa, respectively.
The optimized disease classifiers were implemented as pre-screening exclusion criteria for the GenoVA Study trial, which began participant recruitment in June 2020. By November 2021, study staff had sent recruitment letters to 3,950 apparently eligible patients classified as not having been diagnosed with any of the 6 target diseases. Of these, 1,735 were reached by phone, of whom 658 declined study participation and 1,077 completed the telephone eligibility screen. Among these, phone screening identified only 54 (54/1,077, 5.0%) patients who self-reported one or more diagnoses of the target diseases. Fifty-two patients reported only one disease diagnosis and two patients reported two separate disease diagnoses (AFib with T2D and CAD with T2D). By individual disease, misclassifications were observed in as few as 2 cases for CRCa and as many as 20 cases for CAD, corresponding to NPV-RW between 98.4% and 99.7% (Table 2). As shown in Figure 2, lower NPV-RW values were observed for diseases with greater per-classifier prevalences in the target population. This observation is consistent with the general rule that NPV and prevalence are inversely related, with the caveat that, in our case, per-classifier prevalences are a biased estimator of true prevalence. Manual review of the FN records indicated that a high proportion were very recent diagnoses. Classifier performance did not vary appreciably by patient sex or race/ethnicity (Table 2).
Table 2.
Performance of optimized classifiers during first 16 months of implementation in GenoVA Study trial
| Metric | AFib | CAD | T2D | BrCa | CRCa | PrCa |
|---|---|---|---|---|---|---|
| Total | ||||||
| Screened participants | 1,077 | 1,077 | 1,077 | 259 | 1,077 | 818 |
| Mispredicted as negative* | 13 | 20 | 15 | 2 | 2 | 4 |
| NPV-RW | 98.8% | 98.2% | 98.6% | 99.2% | 99.9% | 99.5% |
| Male | ||||||
| Screened participants | 818 | 818 | 818 | - | 818 | 818 |
| Mispredicted as negative | 11 | 19 | 13 | - | 1 | 4 |
| NPV-RW | 98.7% | 97.7% | 98.4% | - | 99.9% | 99.5% |
| Female | ||||||
| Screened participants | 259 | 259 | 259 | 259 | 259 | - |
| Mispredicted as negative | 2 | 1 | 2 | 2 | 1 | - |
| NPV-RW | 99.2% | 99.6% | 99.2% | 99,2% | 99.6% | - |
| Non-Hispanic White | ||||||
| Screened participants | 592 | 592 | 592 | 186 | 592 | 406 |
| Mispredicted as negative | 8 | 10 | 4 | 1 | 1 | 2 |
| NPV-RW | 98.7% | 98.3% | 99.3% | 99.5% | 99.8% | 99.5% |
| Non-Hispanic Other Races | ||||||
| Screened participants | 380 | 380 | 380 | 65 | 380 | 315 |
| Mispredicted as negative | 3 | 7 | 7 | 1 | 1 | 1 |
| NPV-RW | 99.2% | 98.2% | 98.2% | 98.5% | 99.7% | 99.7% |
| Hispanic White | ||||||
| Screened participants | 66 | 66 | 66 | 1 | 66 | 65 |
| Mispredicted as negative | 2 | 2 | 2 | 0 | 0 | 1 |
| NPV-RW | 97.1% | 97.1% | 97.1% | 100% | 100% | 98.5% |
| Hispanic Other Races | ||||||
| Screened participants | 39 | 39 | 39 | 7 | 39 | 32 |
| Mispredicted as negative | 0 | 1 | 2 | 0 | 0 | 0 |
| NPV-RW | 100% | 97.5% | 95.1% | 100% | 100% | 100% |
54 participants self-reported a diagnosis of exactly 1 disease, and 2 participants self-reported diagnoses for 2 separate diseases each.
Figure 2. Real-world negative predictive value of six disease classifiers versus disease prevalence in target population.
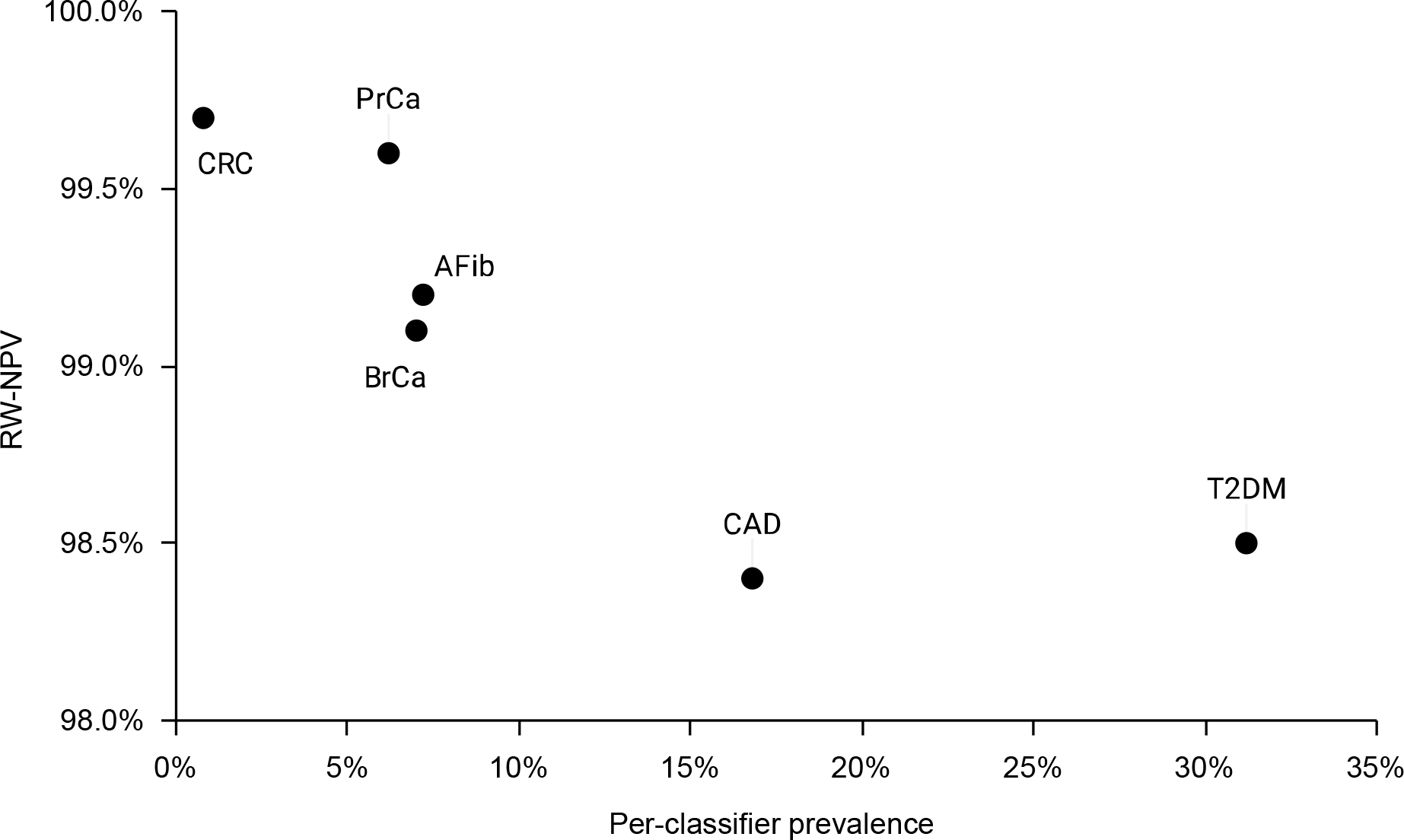
Data are the NPV-RW of the disease classifiers during the first 16 months of recruitment for the GenoVA Study trial, plotted versus the per-classifier disease prevalence among the 20,518 VA Boston patients aged 50–70 years meeting insurance and PCP relationship criteria.
DISCUSSION
Data from EHRs have become a staple for efficient cohort selection, subject recruitment, and outcomes collection in clinical trials.19–21 We sought to implement validated EHR-based classifiers as exclusion criteria for a clinical trial of polygenic risk scoring for six common diseases. We found that simple classifiers consisting of structured data such as diagnosis and procedure codes, prescriptions, and cancer registry entries achieved the desired performance, either as published or with minimal manual review and iterative optimization. During the first 16 months of the GenoVA Study trial, the implementation of these classifiers optimized the efficiency of recruitment, misclassifying only 5% of screened individuals as negative for the six diseases. These results confirm the utility of EHR-based disease classifiers for facilitating pragmatic and other types of clinical trials.
For the GenoVA Study, our primary objective was to exclude patients with a known diagnosis of the six target diseases. A classifier that incorrectly identifies large proportions of patients as having the conditions would unnecessarily decrease the size of the patient population deemed eligible for the trial. This result would have hampered recruitment efforts and yielded an enrolled sample non-representative of the population for whom the polygenic risk score intervention is intended. However, the resulting threat to trial validity would not have been as consequential as that resulting from classifiers with the reverse bias, which would have let into the trial participants already experiencing the primary outcome (diagnoses of the target diseases). Moreover, each FN classification reduces recruitment efficiency, wasting the personnel time and effort to recruit and screen an ultimately ineligible participant. Therefore, we developed our classifiers with the primary objective of maximizing TNR, tolerating some misclassification of patients who lack a disease of interest (FP). Depending on the study objectives, other trials might prioritize maximizing TPR instead; for example, a treatment trial would want to minimize the number of recruited participants without the target disease for which the treatment is intended. Future work should evaluate whether our method generalizes to inclusion criteria.
Even though all 6 disease classifiers achieved a NPV-RW >98% among the first 1,077 participants screened for the GenoVA Study, we still observed an inverse correlation between NPV-RW and disease prevalence. This observation likely reflects the greater accuracy of registry data for rarer conditions (in this case, breast, colorectal, and prostate cancer) and the lower sensitivity of multi-component classifiers for more common conditions. Disease prevalence may be important for trialists to consider in determining the optimal balance between TPR and TNR for efficiency and accuracy in study recruitment.
Our findings support key recommendations for trialists looking to use EHR classifiers in clinical trial screening. First, the provenance of diagnosis codes, or the “context of evidence,” impacts their accuracy.18,22 Professionally managed cancer registries, such as the VA Central Cancer Registry used in our study, proved highly accurate in identifying prevalent diagnoses of the three target malignancies in our study, without further modification. Trials should leverage such well curated data, if available. In contrast, non-standardized coding practices across different providers or care settings may reduce the accuracy of routinely collected EHR data for use in clinical trials.23 For instance in the GenoVA Study, diagnosis codes originating from personnel not specifically tasked with making definitive diagnoses were more susceptible to FP misclassifications, as in our observed pharmacist use of AFib diagnostic codes in anticoagulation clinics. In our chart reviews, we also identified FP diagnoses of T2D from emergency care providers administering one dose of insulin. Trialists may want to consider incorporating design patterns such as the “credentials of the actor” and “context of evidence” in optimizing disease classifiers.18
Second, trialists may want to consider using combinations of disease-specific diagnosis codes, medication prescriptions, and/or temporality, rather than a single diagnostic or procedure code, to more accurately identify disease cases. Such approaches might need to be developed and validated locally, tailored to the specific patient population, local practice, and the goals of the study.24 Prior to using the VA Central Cancer Registry to define CRCa, we noted instances where endoscopists used CRCa codes either prior to a colonoscopy or before the pathology report confirmed absence of malignancy, likely to indicate that the purpose of the procedure was to rule out cancer. Similarly, we observed CAD diagnosis codes at the time a cardiac stress test was ordered to rule out CAD. In the case of CAD, we chose to correct these errors by imposing the easily computable requirement of 2 CAD codes, as has been recommended for other diagnoses.25 However, this requirement of a second encounter will miss very new diagnoses, which we observed among some FN cases recruited in the GenoVA Study.
Our work has some limitations to note. First, because we considered this work to be a validation and optimization of existing published classifiers, as opposed to new classifier development, we employed only one clinical expert reviewer and samples of 20 records for each round of review. Larger record samples and use of more than one reviewer would increase the precision and rigor, respectively, of the findings. Second, our cohort is composed of military Veterans who may receive some proportion of healthcare services outside of VHA. As a result, and given the importance of excluding prevalent diagnoses of the 6 target diseases, we could not fully automate eligibility screening with the disease classifiers alone. However, the necessary telephone eligibility screen afforded the opportunity to perform the present analysis on the classifiers’ real-world performance. Third, although our classifiers achieved 100% true positive rates during development, we cannot comment on their real-world true positive rates, since participants meeting case status were not contacted for recruitment and additional screening. Finally, we did not consider unstructured data such as images or clinical notes in developing our disease classifiers, nor did we consider more advanced computational methods such as machine learning. Such approaches may improve the performance of EHR-based classifiers but may require additional time and computational resources out of reach for most pragmatic trials.
In conclusion, evaluating previously published disease classifiers and, when necessary, using simple heuristics to optimize their performance resulted in computable trial eligibility criteria that greatly improved the efficiency of recruitment. Our approach serves as a model for other trialists implementing EHR-based disease classifiers in participant screening.
Supplementary Material
Funding
This work was supported by the National Human Genome Research Institute (R35 HG01706).
Footnotes
Declaration of Competing Interest
The authors declare they have no competing interests.
Appendix A. Supplementary data
Supplementary data to this article can be found online at TBD
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary material, with the exception of subject-level data that may allow identification of individual participants.
REFERENCES
- 1.Brunette CA et al. Pragmatic Trials in Genomic Medicine: The Integrating Pharmacogenetics In Clinical Care (I-PICC) Study. Clin. Transl. Sci. 13, 381–390 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.NIH Pragmatic Trials Collaboratory. NIH Collaboratory Rethinking Clinical Trials - The Living Textbook. Rethinking Clinical Trials https://rethinkingclinicaltrials.org/. [Google Scholar]
- 3.Cowie MR et al. Electronic health records to facilitate clinical research. Clin. Res. Cardiol. 106, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogers JR et al. Contemporary use of real-world data for clinical trial conduct in the United States: a scoping review. J. Am. Med. Inform. Assoc. 28, 144–154 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weng C Optimizing Clinical Research Participant Selection with Informatics. Trends Pharmacol. Sci. 36, 706–709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan KS, Fowles JB & Weiner JP Review: electronic health records and the reliability and validity of quality measures: a review of the literature. Med. Care Res. Rev. MCRR 67, 503–527 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Hao L et al. Development of a clinical polygenic risk score assay and reporting workflow. Nat. Med. 28, 1006–1013 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torkamani A, Wineinger NE & Topol EJ The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 19, 581–590 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Donahue M et al. Veterans Health Information Exchange: Successes and Challenges of Nationwide Interoperability. AMIA. Annu. Symp. Proc. 2018, 385–394 (2018). [PMC free article] [PubMed] [Google Scholar]
- 10.National Center for Veterans Analysis and Statistics. VA Benefits and Health Care Utilization: At-A-Glance Card, Q1, 2022. (2022).
- 11.Department of Veterans Affairs. Veterans Health Administration Directive 1601A.01. (2020). [Google Scholar]
- 12.Boston Healthcare System. About us. Veteran Affairs https://www.va.gov/boston-healthcare/about-us/ (2021).
- 13.Price LE, Shea K & Gephart S The Veterans Affairs’s Corporate Data Warehouse: Uses and Implications for Nursing Research and Practice. Nurs. Adm. Q. 39, 311–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peltzman T, Rice K, Jones KT, Washington DL & Shiner B Optimizing Data on Race and Ethnicity for Veterans Affairs Patients. Mil. Med. 187, e955–e962 (2022). [DOI] [PubMed] [Google Scholar]
- 15.Cheng D et al. Updating and Validating the U.S. Veterans Affairs Frailty Index: Transitioning From ICD-9 to ICD-10. J. Gerontol. A. Biol. Sci. Med. Sci. 76, 1318–1325 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centers for Medicare & Medicaid Services. 2018 ICD-10 CM and GEMs. Centers for Medicare & Medicaid Services https://www.cms.gov/Medicare/Coding/ICD10/2018-ICD-10-CM-and-GEMs.
- 17.R Core Team. R: A language and environment for statistical computing. (2021).
- 18.Rasmussen LV et al. Design patterns for the development of electronic health record-driven phenotype extraction algorithms. J. Biomed. Inform. 51, 280–286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller HN et al. Electronic medical record–based cohort selection and direct-to-patient, targeted recruitment: early efficacy and lessons learned. J. Am. Med. Inform. Assoc. 26, 1209–1217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richesson RL et al. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory. J. Am. Med. Inform. Assoc. JAMIA 20, e226–231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richesson RL et al. Enhancing the use of EHR systems for pragmatic embedded research: lessons from the NIH Health Care Systems Research Collaboratory. J. Am. Med. Inform. Assoc. JAMIA 28, 2626–2640 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Divney AA et al. Research-grade data in the real world: challenges and opportunities in data quality from a pragmatic trial in community-based practices. J. Am. Med. Inform. Assoc. 26, 847–854 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powell GA et al. Using routinely recorded data in a UK RCT: a comparison to standard prospective data collection methods. Trials 22, 429 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad FS et al. Computable Phenotype Implementation for a National, Multicenter Pragmatic Clinical Trial. Circ. Cardiovasc. Qual. Outcomes 13, e006292 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chamberlain AM et al. Identification of Incident Atrial Fibrillation From Electronic Medical Records. J. Am. Heart Assoc. 11, e023237 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary material, with the exception of subject-level data that may allow identification of individual participants.


