Abstract
Communication between CD4 T cells and cognate B cells is key for the former to fully mature into germinal center T follicular helper (GC-Tfh) cells, and for the latter to mount a CD4 T cell-dependent (TD) humoral immune response. While this interaction occurs in a B:T synapse dependent manner, how CD4 T cells transcriptionally regulate B:T synapse formation remains largely unknown. Here we report that Mef2d, an isoform of the myocyte enhancer factor 2 (Mef2) transcription factor family, is a critical regulator of this process. In CD4 T Cells Mef2d negatively regulates expression of Sh2d1a, which encodes SLAM-associated protein (SAP), a critical regulator of B:T synapses. We found that Mef2d regulates Sh2d1a expression via DNA binding-dependent transcriptional repression, inhibiting SAP-dependent B:T synapse formation, and preventing antigen-specific CD4 T cells from differentiating into GC-Tfh cells. Mef2d also impeded IL-21 production by CD4 T cells, an important B cell help signaling molecule, via direct repression of the Il21 gene. In contrast, CD4 T cell-specific disruption of Mef2d led to a significant increase in GC-Tfh differentiation in response to protein immunization, concurrent with enhanced SAP expression. MEF2D mRNA expression inversely correlates with human SLE patient autoimmune parameters, including circulating Tfh-like cell frequencies, autoantibodies, and SLEDAI scores. These findings highlight Mef2d as a pivotal rheostat in CD4 T cells for controlling GC formation and antibody production by B cells.
One Sentence Summary
Mef2d controls GC-Tfh differentiation and TD humoral immunity via transcriptional repression of Sh2d1a and Il21 in CD4 T cells.
INTRODUCTION
The immune and nervous systems have developed specialized adhesive structures or synapses to monitor and properly respond to internal or external changes (1, 2). While the neuronal synapse functions as a key framework for the delivery of neuronal information from the axon terminal of one neuron to the dendrite of the next (3), the immune synapse ensures the stable interaction of CD4 T cells with professional antigen-presenting cells (APCs), through which appropriate immune responses could be elicited in antigen-specific CD4 T cells (4). These systems exhibit substantial similarities in establishing synapses with structural and functional domains (2, 5). Synapses are formed and stabilized by the structural domain, whereas triggering of intracellular signaling pathways occurs through the functional domain, via molecular interactions with corresponding ligands such as neurotransmitters and ions in the nervous system, antigen/MHC complexes, signaling ligands, and co-stimulatory or co-inhibitory molecules in the immune system (4, 6).
While the immune system has developed molecules to regulate synapse formation and convey signals downstream of the synapse (4), in order to support these processes efficiently, it has also adapted molecular machinery that operates during corresponding processes in the nervous system. The N-methyl-D-aspartate receptor (NMDAR), one of the ion channels that plays a pivotal role in the regulation of synaptic functions in neurons (7), is also highly expressed by developing T cell precursors in the thymus. NMDAR contributes to the negative selection of auto-reactive thymocytes by sustaining Ca2+ influx during interaction with autoantigen-presenting thymic dendritic cells (DCs) (8). Dlgh1, a member of the membrane-associated guanylate kinase (MAGUK) protein family that functions as a neuronal synaptic scaffolding molecule (9), was found to co-localize with T cell receptors (TCRs) at the interface between CD4 T cells and APCs, and to play indispensable roles in triggering immune responses of CD4 T cells (10). Furthermore, dopamine, a well-known neurotransmitter, controls proliferation and cytokine production in human and murine CD4 T cells (11).
The formation of immune synapses with DCs or other monocyte-derived APCs in the T cell zone of lymphoid organs leads to the activation of intracellular signaling molecules in CD4 T cells, which are ultimately converted into nuclear cues (i.e., activation of transcription factors such as NFAT, AP-1, and NF-kB) that lead to the proliferation and activation of antigen-specific CD4 T cells (12). Activated CD4 T cells then differentiate into diverse effectors (Th1, Th2, Th17, Tfh, and iTreg), depending on the instructive signals provided by APCs and the surrounding environment (13). Among effector CD4 T cells, T follicular helper (Tfh) cells are highly dependent on the subsequent formation of cognate interactions with B cells at the B:T border for full maturation into germinal center Tfh (GC-Tfh) cells, which in turn provide signals for cognate B cells to form and undergo GC reactions (14, 15). This feedback regulation, a fundamental process in GC-dependent humoral immunity, occurs in a B:T synapse-dependent manner. Alterations in the genes encoding molecules including CD40 and ICOS that support stable B:T interactions or blocking antibodies against these molecules impede the generation of long-lived plasma cells and memory B cells (16, 17), the effector B cells that predominantly develop from GC responses (18).
Hypogammaglobulinemia and severe deficiency of isotype-switched memory B cells are clinical manifestations associated with X-linked lymphoproliferative (XLP) disease (19, 20), a fatal primary immunodeficiency condition caused by mutations in the SH2D1A gene in humans (21, 22). Sh2d1a encodes SAP (SLAM-associated protein), a key adaptor molecule that functions downstream of the signaling lymphocyte activation molecule (SLAM) family receptors. Sh2d1a gene deficiency in mice recapitulated profound defects in GC-dependent humoral immunity in XLP patients (23, 24). Sh2d1a in CD4 T cells is indispensable for B:T synapse formation (25, 26), the lack of which resulted in an almost complete block of GC-Tfh differentiation and GC formation in response to exogenous antigens (27, 28) as well as to self-antigens (29). Despite its critical roles in B:T synapse formation, subsequent GC-Tfh maturation, and GC-dependent humoral immunity, little information is available to delineate how SH2D1A (Sh2d1a) gene expression is regulated in CD4 T cells.
Here, we examined the Mef2d transcription factor, an isoform that was reported to function downstream of TCR signaling in CD4 T cells (30), of the Mef2 transcription factor family that negatively regulates neuronal synaptic plasticity and memory formation (31–33), in the context of synapse formation of CD4 T cells with cognate B cells and investigated its roles in Tfh differentiation and T cell-dependent humoral immunity. Mef2d directly repressed Sh2d1a gene expression in murine CD4 T cells and negatively regulated SAP-dependent B:T synapse formation. In protein immunization, the Mef2d transcription factor governs GC formation and antigen-specific antibody production by B cells via negative regulation of GC-Tfh differentiation and inhibition of IL-21 production by CD4 T cells. Moreover, MEF2D expression in CD4 T cells exhibited profound inverse relationships with humoral autoimmune conditions [cTfh cell frequency, SLEDAI, and autoantibodies] in SLE patients, indicating critical physiological roles of Mef2d mediated transcriptional repression of the Sh2d1a and Il21 genes of CD4 T cells in GC-dependent humoral (auto)immunity.
RESULTS
Mef2D negatively regulates germinal center follicular T helper differentiation
The myocyte enhancer factor-2 (MEF2) family of transcription factors play a critical role in transcriptional regulation of neuronal synapse formation (31–33). T follicular helper (Tfh) cells, to properly regulate synapse formation with cognate B cells (14, 15), employ molecules of the nervous system (34, 35), leading us to hypothesize that MEF2 transcription factors might function during Tfh differentiation of CD4 T cells. We first assessed the expression of MEF2 transcription factors in tonsillar GC-Tfh (PD-1hiCXCR5+) and non-Tfh (PD-1−CXCR5−) cells (fig. S1A), where BCL6 and PRDM1 mRNA was robustly expressed (fig. S1B), as reported (36). While only MEF2A and MEF2D are highly expressed in human peripheral blood CD4 T cells (fig. S1C), MEF2B was also abundantly expressed by the effector CD4 T cells in human tonsils (fig. S1D). In contrast to comparable mRNA expression of MEF2A and MEF2B genes by GC-Tfh and non-Tfh cells (fig. S1D), MEF2D expression was significantly reduced in GC-Tfh cells compared to non-Tfh effectors and naïve CD4 T cells at both mRNA (fig. S1E) and protein (fig. S1F) levels. In murine CD4 T cells, we found that Mef2a and Mef2d mRNA were highly expressed by non-Tfh and Tfh cells in comparison to the other isoforms (fig. S1G–H) and that only Mef2d transcripts were significantly curtailed by Tfh cells (fig. S1H).
A similar expression pattern of MEF2D (Mef2d) in non-Tfh and GC-Tfh cells to that of BLIMP-1, a well-known transcription factor that antagonizes Tfh differentiation (37, 38), led us to speculate that MEF2D (Mef2d) might play an inhibitory role in Tfh (GC-Tfh) differentiation of CD4 T cells. This point was tested using a murine system, given the similar expression profiles of the MEF2D (Mef2d) isoform in both species (fig. S1, E and H). Ectopic expression of Mef2d in OTII CD4 T cells (Fig. 1A) did not alter the frequency of OTII CD4 T cells in the popliteal lymph nodes (popLNs) after NP-OVA immunization (Fig. 1B–C), whilst GC-Tfh cells were significantly reduced, and non-Tfh cells were expanded (Fig. 1C–D). Surface expression of PD-1 and CXCR5 was significantly reduced on the Mef2d-RV OTII CD4 T cells compared to the controls (Fig. 1E–F). Formation of Bcl6+CXCR5+ GC-Tfh cells was significantly lower in Mef2d-RV OTII CD4 T cells than in control OTII CD4 T cells (Fig. 1G).
Fig. 1. Mef2d inhibits germinal center Tfh (GC-Tfh) formation of OTII TCRtg CD4 T cells.
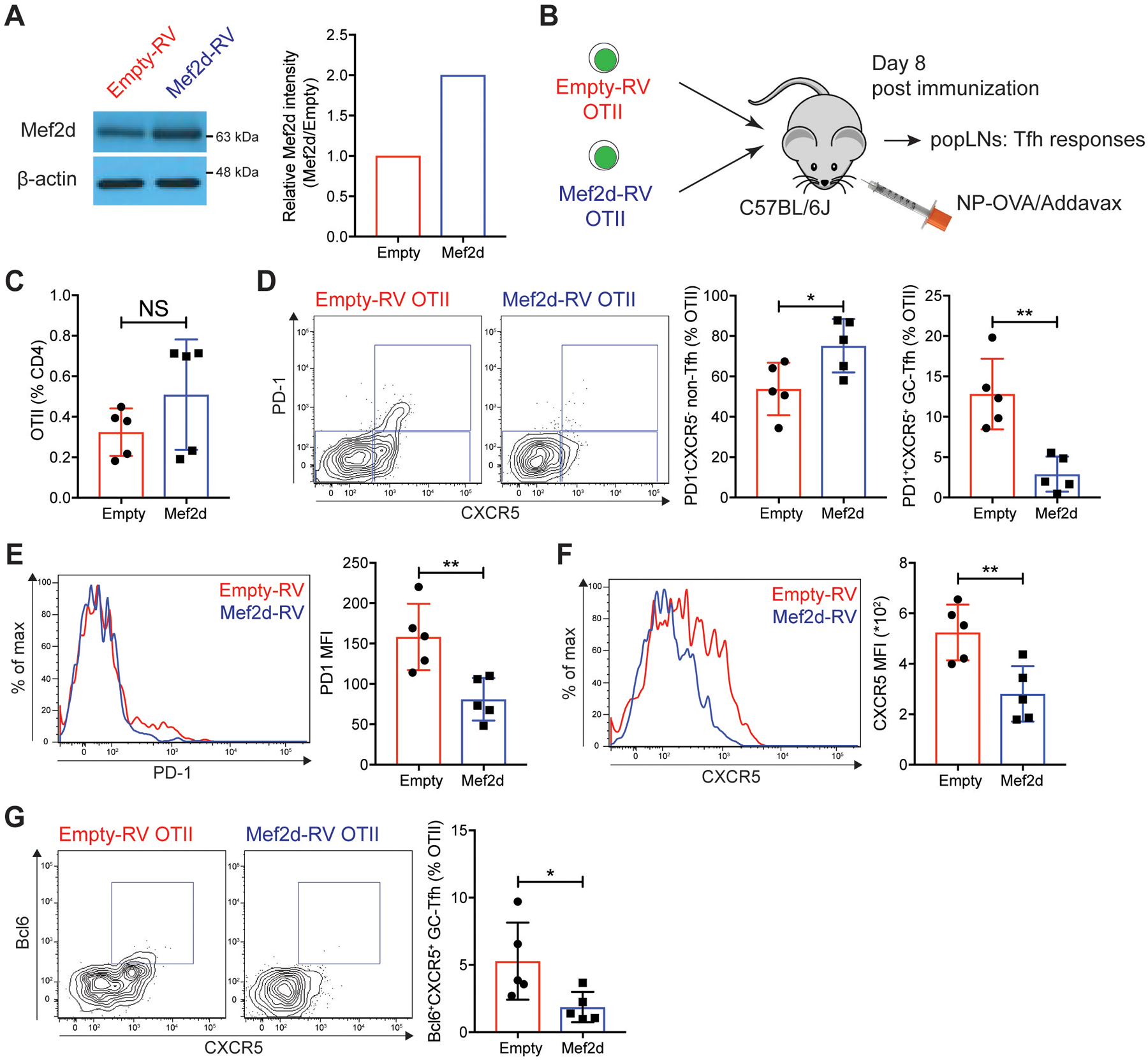
(A) Immunoblots of β-actin and Mef2d protein. Relative Mef2d band intensity of Mef2d-RV cells to those of empty-RV cells.
(B) Experimental scheme. Equal numbers of empty-RV or Mef2d-RV OTII CD4 T cells were transferred to C57BL/6J mice. Eight days after subcutaneous footpad immunization with NP-OVA, popLNs were examined for GC-Tfh differentiation of GFP+ OTII CD4 T cells.
(C) The frequencies of GFP+ OTII CD4 T cells calculated as the percentage of total CD4 T cells.
(D) Flow cytometry plots of GFP+ OTII CD4 T cells. Gates indicate PD-1−CXCR5− non-Tfh, PD-1−CXCR5+ Tfh, and PD-1+CXCR5+ GC-Tfh cells. Quantified frequencies of PD-1−CXCR5− non-Tfh and PD-1+CXCR5+ GC-Tfh cells among GFP+ OTII CD4 T cells.
(E and F) Overlaid histograms of PD-1 (E) and CXCR5 (F) of empty-RV (red) or Mef2d-RV (blue) OTII CD4 T cells. MFIs were calculated.
(G) Flow cytometry plots of GFP+ OTII CD4 T cells with gates indicating Bcl6+CXCR5+ GC-Tfh cells. The frequencies of Bcl6+CXCR5+ GC-Tfh cells were calculated.
Representative of two independent experiments with n=4–5 per group.
Error bars indicate mean with SD. Statistical significance values were determined using two-tailed Student’s t-test. NS, statistically non-significant; * p <0.05; ** p <0.01.
Significant defects in GC-Tfh differentiation by Mef2d were also observed when Sm TCRtg CD4 T cells, specific to the gp66–77 peptide of the LCMV virus (39), were analyzed in the presence or absence of ectopic Mef2d expression. Eight days after gp61-KLH immunization (fig. S2A), Mef2d-RV Sm CD4 T cells formed significantly fewer PD-1+CXCR5+ GC-Tfh cells compared to empty-RV Sm CD4 T cells (fig. S2B), with curtailed surface expression of PD-1 and CXCR5 (fig. S2C–D). Our data demonstrate that Mef2d negatively regulates GC-Tfh differentiation of CD4 T cells in the context of acute protein immunization.
Mef2d regulates germinal center formation and antigen-specific antibody production by B cells via inhibition of GC-Tfh maturation
As Mef2d inhibited GC-Tfh differentiation of CD4 T cells in the draining LNs after protein immunization (Fig. 1 and fig. S2), we next investigated whether Mef2d in CD4 T cells regulates the formation of GCs and the production of high-affinity antibodies by B cells. Significantly fewer Mef2d-RV OTII CD4 T cells were localized in the GC areas of the popLNs after NP-OVA immunization (Fig. 2A–B). We used Cd4CreBcl6fl/fl mice (Bcl6 CKO mice hereafter) whose endogenous CD4 T cells could not help B cells to form GCs (40) as recipients for adoptive transfer experiments (Fig. 2C). Seven days after immunization, transferred Mef2d-RV OTII CD4 T cells formed significantly less PSGL-1loCXCR5+ GC-Tfh cells compared to control (Fig. 2D). Total CD19 B cell frequencies were comparable (Fig. 2E), whilst Fas+PNA+ GC B cells were barely detectable in the Bcl6 CKO mice (Fig. 2F) which received Mef2d-RV OTII CD4 T cells. Moreover, the magnitude of high-affinity (NP8-bound) IgG production of Bcl6 CKO mice was significantly reduced by Mef2d-RV OTII CD4 T cells compared with the control OTII CD4 T cells (Fig. 2G). B cell production of NP49-bound (low-affinity) IgG antibodies also decreased by ectopic Mef2d expression (Fig. 2G), which suggests that the Mef2d transcription factor might also play a role in the GC-independent antibody response. Nonetheless, the impaired ability of B cells to produce antigen-specific IgGs by Mef2d-RV OTII CD4 T cells lasted until 28 days after immunization (Fig. 2H and fig. S3A). These data demonstrate that GC development and CD4 T cell-dependent production of antigen-specific antibodies are controlled by CD4 T cell intrinsic functions of Mef2d transcription factor.
Fig. 2. Germinal center formation and antigen-specific antibody production by B cells are negatively regulated by Mef2d function in CD4 T cells.
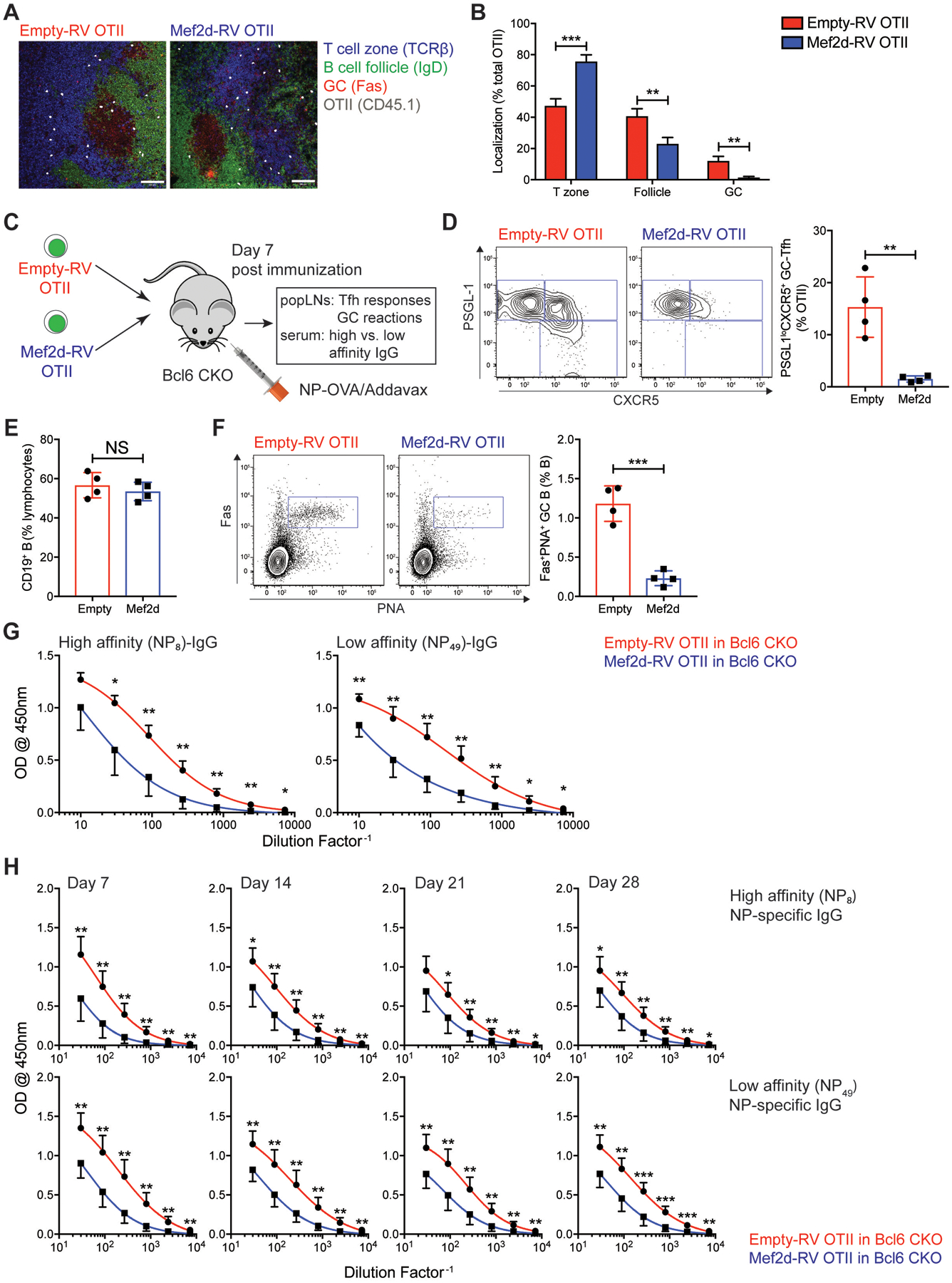
(A) Empty-RV (left) or Mef2d-RV (right) CD45.1 OTII CD4 T cell were transferred into C57BL/6J mice. Seven days after NP-OVA immunization, popLNs were taken for IF analysis of CD45.1 OTII CD4 T cell localization (T cell zone, B cell follicle, or GC area). CD45.1 OTII CD4 T cells were shown in large dots to clarify. Scale bar = 100μm.
(B) The frequencies of CD45.1 OTII CD4 T cells positioned in each location were calculated.
(C to G) Empty-RV or Mef2d-RV OTII CD4 T cells were transferred into Bcl6 CKO (Cd4CreBcl6fl/fl) mice, which were immunized subcutaneously with NP-OVA. Seven days after NP-OVA immunization, popLNs and serum were examined for GC-Tfh differentiation and GC formation and NP-specific IgG production, respectively.
(C) Experimental scheme for investigating the regulation of humoral immune response by the Mef2d function of CD4 T cells.
(D) Flow cytometry plots of GFP+ OTII CD4 T cells present in popLNs of Bcl6 CKO mice. Gates indicate PSGL-1hiCXCR5− non-Tfh, PSGL-1hiCXCR5+ Tfh, and PSGL-1loCXCR5+ GC-Tfh cells. The frequencies of PSGL-1loCXCR5+ GC-Tfh cells were quantified.
(E) The frequencies of CD19 B cells. Calculated as the percentage of total lymphocytes.
(F) Flow cytometry plots of CD19 B cells with gates indicating Fas+PNA+ GC B cells. The GC B cell frequencies among the total B cells were calculated.
(G) Amounts of high- (NP8-BSA) and low- (NP49-BSA) affinity NP-specific IgG antibodies measured in serum from Bcl6 CKO mice adoptively transferred with empty-RV (red) or Mef2d-RV (blue) OTII CD4 T cells.
(H) Empty-RV (red) or Mef2d-RV (blue) OTII CD4 T cells were adoptively transferred into Bcl6 CKO mice, which were bled at days 7, 14, 21 and 28 after NP-OVA immunization. The high- and low-affinity NP-specific IgG production was measured by ELISA.
Representative of two independent experiments (n=4–5 per group) (C to G) and composite data from two independent experiments using n=6 recipient mice per group (H).
Error bars indicate mean with SEM (B) and mean with SD (D to H). Statistical significance values were determined using two-tailed Student’s t-test. NS, statistically non-significant; * p <0.05; ** p <0.01; *** p <0.001.
Early Tfh differentiation is inhibited by Mef2d function in CD4 T cells
Profound defects in the formation of GC-Tfh cells by Mef2d expression led us to investigate whether it might play inhibitory roles during the early stage of Tfh differentiation of CD4 T cells prior to GC-Tfh maturation (14, 15). The frequency of OTII CD4 T cells in the popLNs was not altered by ectopic Mef2d expression three days after NP-OVA immunization (Fig. 3A–B), which suggests that Mef2d, in contrast to its pro-apoptotic functions downstream of TCR-mediated Ca2+ influx during T cell development (30), does not lead to apoptosis of mature CD4 T cells during the immune response to acute protein immunization. The frequencies of PD-1+CXCR5+ and PSGL-1loCXCR5+ Tfh cells which differentiated from OTII CD4 T cells were significantly curtailed by Mef2d overexpression (Fig. 3C–D), with reduced expression of canonical Tfh cell markers (PD-1 and CXCR5) (Fig. 3E–F), and reciprocal increase in the surface expression of PSGL-1 (Fig. 3G), a molecule involved in CD4 T cell positioning in the T cell zone (41). Equivalent expression of CD25 (Fig. 3H), a high-affinity IL-2 receptor subunit (42), as well as similar IL-2 production, was observed between empty-RV and Mef2d-RV OTII CD4 T cells. Furthermore, IFN-γ− and IL-17A-production was comparable (fig. S3B), indicating that the overall activation and Th1 or Th17 differentiation of CD4 T cells was not modulated by ectopic Mef2d expression.
Fig. 3. Mef2d inhibits the early Tfh differentiation of antigen-specific CD4 T cells.
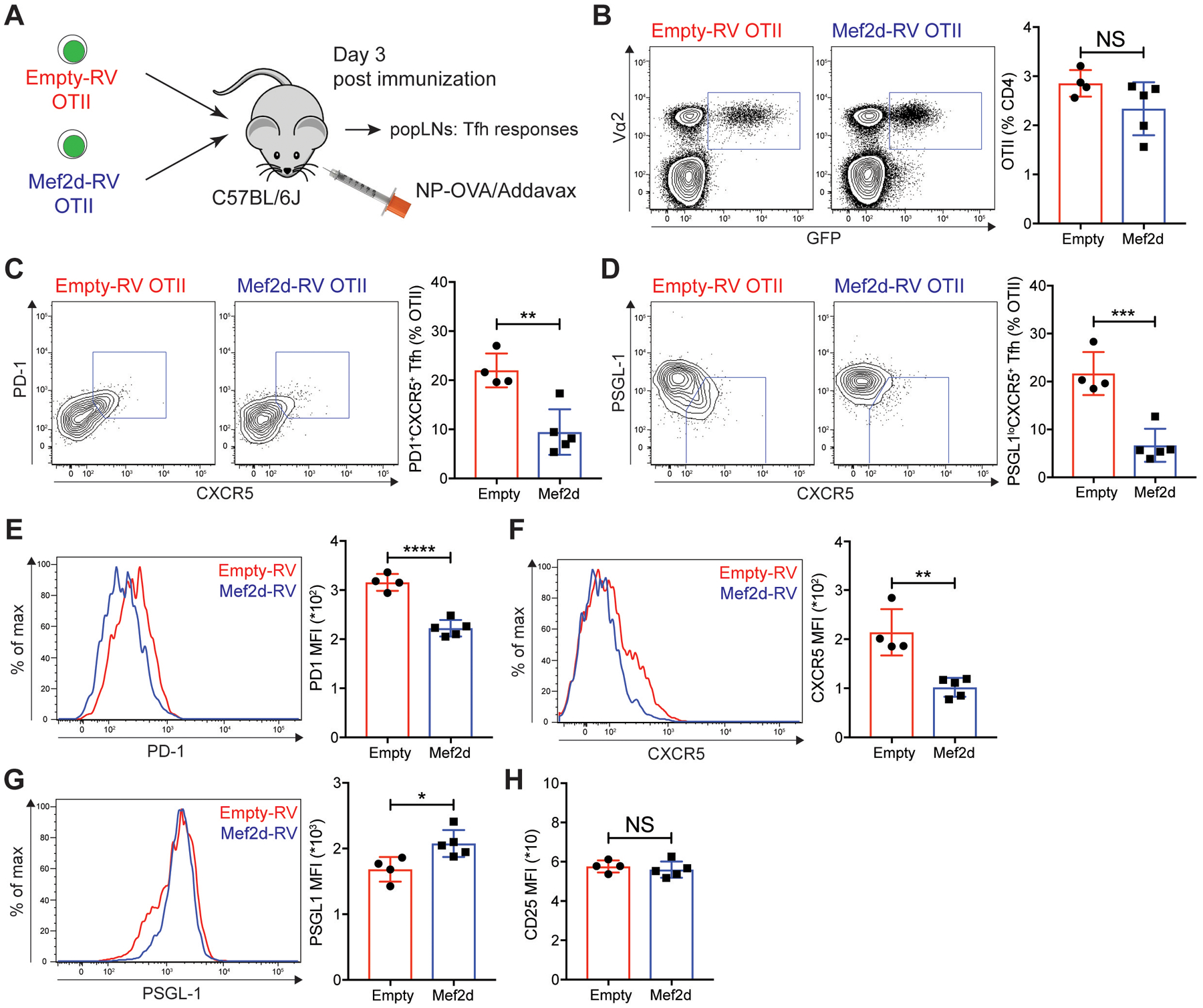
(A) Experimental scheme to analyze Mef2d functions during early Tfh differentiation of CD4 T cells. The same number of empty-RV or Mef2d-RV OTII CD4 T cells were transferred into C57BL/6J mice. Three days after subcutaneous NP-OVA immunization, popLNs were examined for Tfh differentiation of GFP+ OTII CD4 T cells.
(B) Flow cytometry plots of CD4 T cells in the popLNs. Vα2+GFP+ OTII CD4 T cells were gated and quantified.
(C and D) Flow cytometry plots of the transferred GFP+ OTII CD4 T cells with gates indicating PD-1+CXCR5+ (C) and PSGL-1loCXCR5+ (D) Tfh cells. The frequencies of PD-1+CXCR5+ and PSGL-1loCXCR5+ Tfh cells among the GFP+ OTII CD4 T cells were calculated.
(E to G) Overlaid histograms of PD-1 (E), CXCR5 (F), and PSGL-1 (G) of empty-RV (red) and Mef2d-RV (blue) OTII CD4 T cells. MFIs were calculated.
(H) Quantification of CD25 MFIs from empty-RV and Mef2d-RV OTII CD4 T cells.
Representative of two independent experiments with n=4–5 mice per group.
Error bars indicate mean with SD. Statistical significance values were determined using two-tailed Student’s t-test. NS, statistically non-significant; * p <0.05; ** p <0.01; *** p <0.001; **** p <0.0001.
We previously reported that cognate B cells function as major APCs in the Tfh differentiation between 72 and 96 hours after acute viral infection (43). In the context of protein immunization, antigen-specific CD4 T cells exhibited a slightly earlier requirement for cognate B cells for Tfh differentiation (fig. S3C–E). 72 hours after NP-OVA immunization (fig. S3C), generation of PD-1+CXCR5+ and PSGL-1loCXCR5+ Tfh cells from OTII CD4 T cells was significantly impaired in B cell deficient μMT mice compared to C57BL/6J mice (fig. S3D–E). Collectively, these data imply that Mef2d might function during cognate B cell-dependent Tfh differentiation.
Mef2d transcriptionally represses Sh2d1a in CD4 T cells
We next aimed to understand how Mef2d negatively regulates cognate B cell-dependent Tfh and GC-Tfh differentiation. RNA-seq was performed with PSGL-1loCXCR5+ Tfh, and PSGL-1hiCXCR5− non-Tfh cells, which developed from empty-RV (control Tfh and control non-Tfh) or Mef2d-RV (Mef2d Tfh and Mef2d non-Tfh) OTII CD4 T cells three days after NP-OVA immunization (fig. S4A). DE-seq analyses revealed that genes associated with Tfh differentiation in positive (Cxcr5, Bcl6, Tox2, Sh2d1a, Tcf7, and Pdcd1) or negative (Ccr7, Klf2, Id2, Bach2, and Selplg) ways were highly expressed by control Tfh and control non-Tfh cells, respectively (fig. S4B), which were subsequently categorized in heatmaps based on the previously reported functions [transcription factors, B:T interaction, and migration] (Fig. 4A). Control Tfh cells exhibited a strong enrichment for Tfh gene signatures, which were previously reported by transcriptomic analyses of Tfh and non-Tfh cells that formed in the context of various immunological stimuli (28, 44, 45) (fig. S4C), confirming the quality of the datasets generated by our RNA-seq analysis pipeline. We then compared the expression of genes in control Tfh cells vs. Mef2d Tfh cells and control non-Tfh cells vs. Mef2d non-Tfh cells to screen for potential targets of Mef2d. Approximately 320 genes were differentially expressed (padj < 0.05; more than 1.5-fold) in Tfh (175 genes) and non-Tfh (142 genes) compartments that developed from Mef2d-RV OTII CD4 T cells compared with the corresponding cells formed by empty-RV OTII CD4 T cells (fig. S4D). While a role for Mef2d could not be ruled out in directly or indirectly regulating the expression of genes whose functions are related to Tfh (Bcl6, Tcf7, and Cxcr5) or non-Tfh (Prdm1, Bach2, Klf2, and Ccr7) differentiation (Fig. 4A and fig. S4E), we were interested in the genes that exhibited RPKM alterations in the same direction (upregulated or downregulated) in both Tfh and non-Tfh compartments by increased Mef2d function. Among the 317 genes, 21 and 17 genes met this condition (table S2), which we considered as potential target genes of Mef2d in the regulation of Tfh and GC-Tfh differentiation of CD4 T cells.
Fig. 4. Mef2d impedes Tfh differentiation of CD4 T cells via direct repression of the Sh2d1a expression.
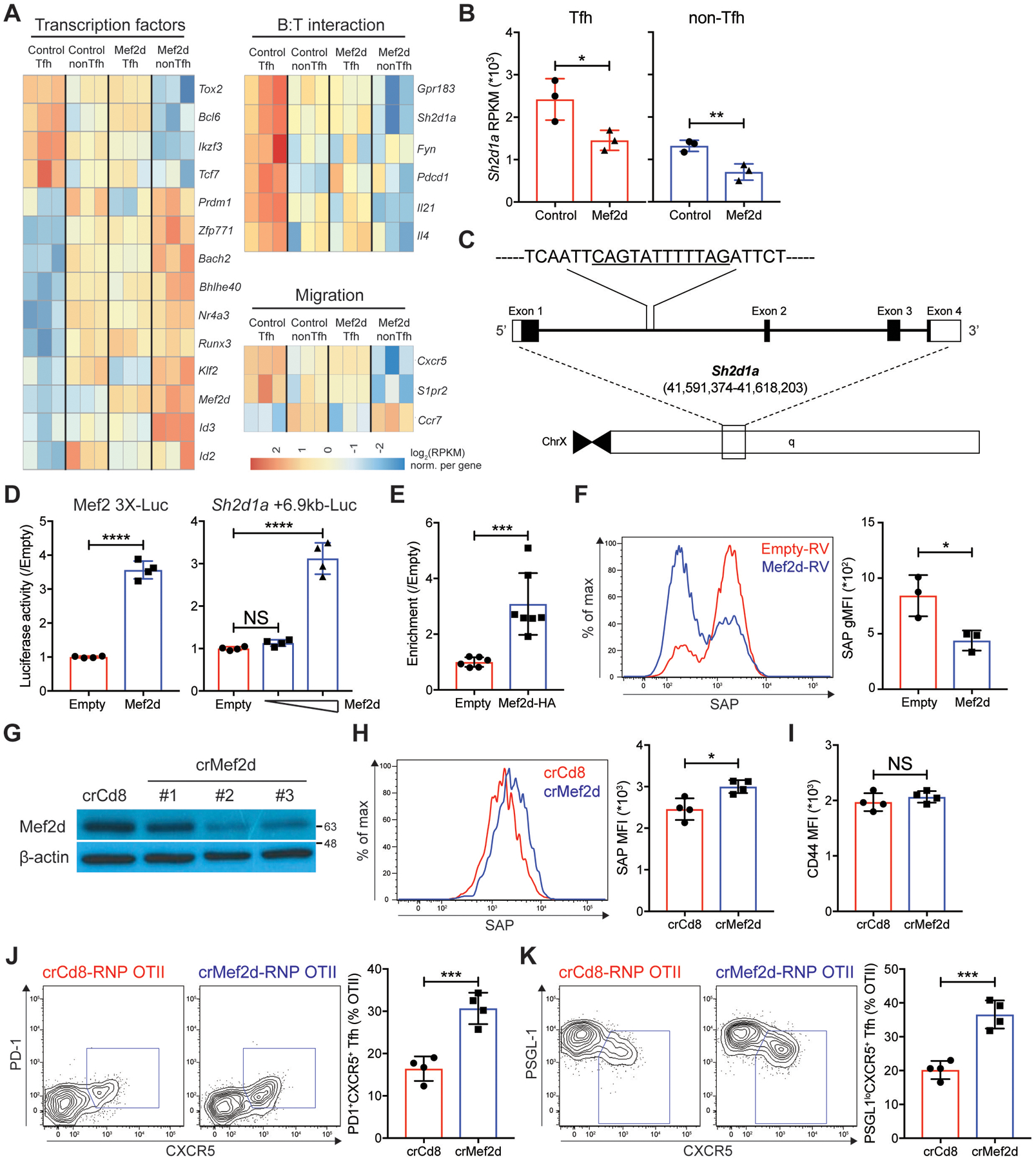
(A and B) RNA-seq was performed with three biological replicates of Tfh cells and non-Tfh cells that developed from empty-RV or Mef2d-RV OTII CD4 T cells. Each biological sample was obtained by sorting GFP+ Tfh and GFP+ non-Tfh cells from 8–12 recipient C57BL/6J mice per group.
(A) Differentially expressed genes by control Tfh, control non-Tfh, Mef2d Tfh, and Mef2d non-Tfh cells, which were categorized based on reported gene functions and presented as high (red) to low (blue) expression levels.
(B) Sh2d1a gene RPKMs of Tfh (red) and non-Tfh (blue) cells developed from empty-RV (circles) or Mef2d-RV (triangles) OTII CD4 T cells.
(C) The genomic region of the murine Sh2d1a gene with a putative Mef2d binding site (CAGTATTTTTAG) at +6.9 kb from the Sh2d1a gene transcription start site.
(D) Luciferase activities of the pGL4.10 plasmids with three Mef2 binding sites (Mef2 3X-Luc) or with a 1.5 kb region of the Sh2d1a locus containing CAGTATTTTTAG (Sh2d1a +6.9kb-Luc) in the presence or absence of Mef2d co-expression (0.05 μg for Mef2 3X-Luc and 0.05 and 0.8 μg for Sh2d1a +6.9kb-Luc). Data from two independent experiments with duplicate wells per each condition.
(E) Shown is the fold enrichment of the Sh2d1a +6.9kb DNA region bound by HA-tagged Mef2d in HEK293T cells transfected with empty-pCMV or Mef2d-HA-pCMV. Data from three independent experiments with duplicate samples for each condition.
(F) Intracellular SAP expression of in vitro stimulated polyclonal CD4 T cells, which were transduced with empty-RV (red) or Mef2d-RV (blue). Overlaid SAP histograms. Geometric SAP MFIs of GFP+ CD4 T cells from three independent experiments were calculated.
(G to K) SAP expression and early Tfh differentiation of Mef2d gene disrupted OTII CD4 T cells were examined. CD45.1 OTII CD4 T cells were transfected with either crCd8-RNP or crMef2d-RNP (#2), which were then adoptively transferred into C57BL/6J mice. Three days after NP-OVA immunization, the donor cells were examined for Tfh differentiation and SAP expression.
(G) β-actin and Mef2d immunoblots with cell lysates obtained from crCd8-RNP+ or crMef2d-RNP+ OTII CD4 T cells.
(H) Overlaid histograms of SAP of the donor cells. SAP MFIs were quantified.
(I) CD44 MFIs of the donor OTII CD4 T cells were calculated.
(J and K) Flow cytometry plots of the donor OTII CD4 T cells with gates indicating PD-1+CXCR5+ (J) and PSGL-1loCXCR5+ (K) Tfh cells. The frequencies of PD-1+CXCR5+ and PSGL-1loCXCR5+ Tfh cells among the donor OTII CD4 T cells were calculated.
Representative of two independent experiments with n=4 mice per group.
Error bars indicate mean with SD. Statistical significance values were determined using two-tailed Student’s t-test [B, D (left) to F, and H to K] and one-way ANOVA with Tukey’s multiple comparisons test [D (right)]. NS, statistically non-significant; * p <0.05; ** p <0.01; *** p <0.001; **** p <0.0001.
One of the 17 genes whose expression was curtailed in both Tfh and non-Tfh cells by ectopic Mef2d expression was Sh2d1a (table S2). The Sh2d1a RPKM of Mef2d Tfh cells was as low as that of control non-Tfh cells, the effector CD4 T cells that do not need to interact with cognate B cells, and further declined in Mef2d non-Tfh cells (Fig. 4B). This gene encodes SAP, a key adaptor molecule required for CD4 T cells to form synapse with B cognate cells (25, 26), and thus plays an indispensable role in cognate B cell-dependent Tfh differentiation and full GC-Tfh maturation (14, 15). Thus, we hypothesized that Mef2d might control Tfh and GC-Tfh differentiation of CD4 T cells via transcriptional repression of the Sh2d1a gene. To address this point, we first scanned the genomic DNA region of the murine Sh2d1a gene for potential DNA-binding sites ([C/T]TA[T/A]4TA[G/A]) of the Mef2 transcription factor family (46). The putative Mef2 binding sequence (TATTTTTAG) exists around the +6.9kb region from the transcription start site (TSS) of the Sh2d1a gene (Fig. 4C). To determine whether Mef2d could directly bind to and regulate Sh2d1a expression, the DNA region that spans 1.5kb, including the putative Mef2 binding site (TATTTTTAG), was cloned into the pGL4.10 luciferase plasmid (Sh2d1a +6.9kb-Luc). The pGL4.10 luciferase plasmid with a DNA sequence harboring three Mef2 binding sites (Mef2 3X-Luc) served as a positive control (Fig. 4D). Luciferase activity of the Sh2d1a +6.9kb-Luc plasmid was increased by Mef2d co-expression in a dose-dependent manner (Fig. 4D), indicating that Mef2d binds to this regulatory region of the Sh2d1a gene. To further validate the direct binding of the Mef2d transcription factor to this site, we performed ChIP-qPCR using an anti-HA98–106 antibody to pull down DNA regions bound by the Mef2d transcription factor, whose N-terminal domain was tagged with influenza hemagglutinin (HA98–106) molecule (30). The DNA region harboring the corresponding Mef2 binding site within the +6.9kb region on the Sh2d1a gene was enriched about three-fold in the presence of Mef2d-HA co-transfection (Fig. 4E). Collectively, our data demonstrated that Mef2d directly binds to the regulatory region of the Sh2d1a gene.
We then investigated whether Mef2d could inhibit SAP protein expression in CD4 T cells. Polyclonal CD4 T cells isolated from naïve mice were stimulated in vitro with plate-bound anti-CD3 and anti-CD28 antibodies, transduced with either empty-RV or Mef2d-RV, and stained for intracellular SAP protein. While SAP expression increased profoundly in CD4 T cells upon TCR- and CD28-mediated stimulation, it was significantly reduced by ectopic Mef2d expression (Fig. 4F), strongly indicating that Mef2d is able to directly modulate SAP expression in murine CD4 T cells. The negative regulatory role of Mef2d in SAP expression of CD4 T cells was further substantiated using RNP mediated Mef2d gene disruption (47). Nucleofection of Cas9 protein complexed with a crRNA targeting the murine Mef2d gene (crMef2d #2) greatly reduced Mef2d protein expression in naïve OTII CD4 T cells (Fig. 4G). We examined SAP expression and the cognate B cell-dependent Tfh differentiation of CD4 T cells in the popLNs three days after NP-OVA immunization, the time point when the RNA-seq analysis was performed (fig. S4). crMef2d-RNP+ OTII CD4 T cells expressed higher levels of SAP protein than control cells (Fig. 4H). The difference in SAP expression was not due to incongruent activation status of CD4 T cells, as CD44 was comparably expressed regardless of Mef2d gene editing (Fig. 4I). Increased SAP expression in crMef2d-RNP+ OTII CD4 T cells was accompanied by increased formation of PD-1+CXCR5+ and PSGL-1loCXCR5+ Tfh cells compared to controls (Fig. 4J–K). Collectively, these data indicate that the Mef2d transcription factor functions as a transcriptional repressor of the Sh2d1a gene in CD4 T cells.
Mef2d negatively regulates GC-Tfh differentiation via transcriptional regulation of SAP expression in CD4 T cells
MEF2D-mediated negative regulation of SAP expression also appears to operate in human effector CD4 T cells. In human tonsils, BCL6+CXCR5+ GC-Tfh cells expressed SAP protein more strongly than BCL6−CXCR5− non-Tfh cells (fig. S5A–B). The magnitude of SAP expression was inversely related to MEF2D mRNA expression in these two compartments (fig. S5C). Moreover, there was a negative correlation between mRNA expression of the MEF2D and SH2D1A genes in tonsillar GC-Tfh cells (fig. S5D), implying that along with the ability of Mef2d to bind to the Sh2d1a loci and to repress its expression in CD4 T cells, Mef2d might control GC-Tfh differentiation via transcriptional control of SAP expression.
To address this point, we first examined whether GC-Tfh differentiation and SAP expression in CD4 T cells are modulated by Mef2d in a DNA binding-dependent manner. As the N-terminal MADS-box and Mef2 domain are highly conserved among the Mef2 isoforms (46, 48, 49), we tested a point mutation at the 24th amino acid position in the MADS-box domain (R to L, R24L hereafter). R24L compromised the DNA binding of Mef2c (50), so we aimed to determine if this could abolish DNA binding of Mef2d. Luciferase activity of the Mef2 3X-Luc plasmid, which was induced by wild-type Mef2d (Fig. 4D), was not triggered by the R24L mutant (fig. S5E). We then explored whether (1) the R24L Mef2d failed to inhibit GC-Tfh differentiation and (2) it was unable to repress SAP expression. OTII CD4 T cells, which had been transduced with empty-RV, wild-type Mef2d-RV, or R24L Mef2d-RV, were examined for GC-Tfh differentiation and SAP expression seven days after NP-OVA immunization. The ectopic expression of wild-type Mef2d inhibited OTII CD4 T cells from differentiating into PD-1+CXCR5+ and PSGL-1loCXCR5+ GC-Tfh cells (Fig. 5A–B). The R24L point mutation almost completely abrogated this inhibitory role of Mef2d, as the R24L Mef2d-RV OTII CD4 T cells not only formed GC-Tfh cells (Fig. 5A–B) but also expressed PD-1 and CXCR5 at comparable levels to the controls (Fig. 5C–D).
Fig. 5. Mef2d negatively regulates GC-Tfh differentiation of antigen-specific CD4 T cells via DNA binding-dependent regulation of SAP expression.
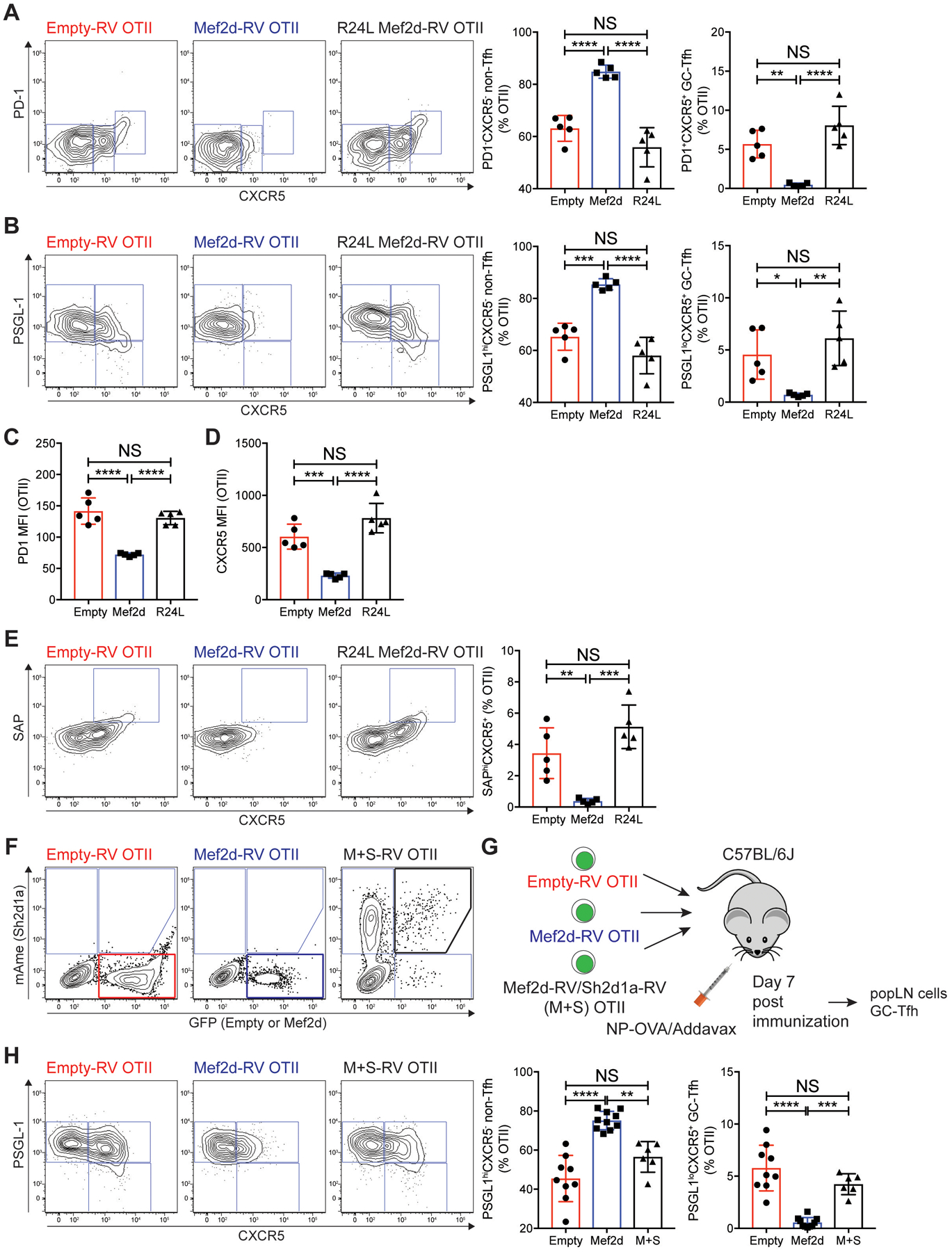
(A to E) Empty-RV (red), Mef2d-RV (blue), or R24L Mef2d-RV (black) CD45.1 OTII CD4 T cells were adoptively transferred into C57BL/6J mice, which were immunized subcutaneously with NP-OVA. Seven days after NP-OVA immunization, the transferred GFP+ CD45.1 OTII CD4 T cells in popLNs were analyzed.
(A and B) Flow cytometry plots of GFP+ CD45.1 OTII CD4 T cells. Gates indicate non-Tfh (PD-1−CXCR5− or PSGL-1hiCXCR5−), Tfh (PD-1−CXCR5+ or PSGL-1hiCXCR5+), and GC-Tfh (PD-1+CXCR5+ or PSGL-1loCXCR5+) cells. The frequencies of non-Tfh and GC-Tfh cells developed from the respective donor cells were calculated.
(C and D) MFIs of PD-1 (C) and CXCR5 (D) of the donor cells were quantified.
(E) Flow cytometry plots of the donor cells. Gates indicate the SAPhiCXCR5+ compartment. The SAPhiCXCR5+ cell frequencies were calculated.
(F to H) GC-Tfh differentiation of Mef2d-RV OTII CD4 T cells was examined in the presence or absence of ectopic co-expression of SAP.
(F) Flow cytometry plots CD45.1 OTII CD4 T cells transduced with empty-RVGFP or Mef2d-RVGFP with or without Sh2d1a-RVmAmetrine. Empty-RVGFP+ (red box), Mef2d-RVGFP+ (blue box), or Mef2d+Sh2d1a-RVGFP+mAmetrine+ (black box) OTII CD4 T cells were highlighted.
(G) The respective RV+ OTII CD4 T cells were sorted and transferred into C57BL/6J mice. Seven days after NP-OVA immunization, popLNs were examined for GC-Tfh differentiation and IL-21 production (fig. S7, C–D) of the donor OTII CD4 T cells were examined.
(H) Flow cytometry plots of RV+ OTII CD4 T cells with gates indicating non-Tfh (PSGL-1hiCXCR5−), Tfh (PSGL-1hiCXCR5+), and GC-Tfh (PSGL-1loCXCR5+) cells. The frequencies of non-Tfh and GC-Tfh cells were calculated.
Representative of three independent experiments with n=5 mice per group (A to E) and composite data from two independent experiments using n=6–10 recipient mice per group (F to H).
Error bars indicate mean with SD. Statistical significance values were determined using one-way ANOVA with Tukey’s multiple comparisons test. NS, statistically non-significant; * p <0.05; ** p <0.01; *** p <0.001; **** p <0.0001.
In CD4 T cells, SAP expression was dynamically regulated depending on activation status (fig. S5F) and during differentiation into effector cells (fig. S5G). In the popLNs after NP-OVA immunization, SAP expression was most highest in PD-1+CXCR5+ GC-Tfh cells, followed by PD-1−CXCR5+ Tfh cells, whereas the lowest SAP expression was found in PD-1−CXCR5− non-Tfh cells (fig. S5G). As such, the SAPhi compartment could be observed among the high CXCR5 expressors (mostly PD1+CXCR5+ GC-Tfh cells) (fig. S5H). Mef2d-RV OTII CD4 T cells failed to form the SAPhi compartment (Fig. 5E), consistent with their inability to differentiate into GC-Tfh cells. Importantly, when Mef2d R24L was ectopically expressed, formation of the SAPhi compartment in OTII CD4 T cells was rescued (Fig. 5E). These data demonstrate that GC-Tfh differentiation and SAP expression of CD4 T cells are controlled together by DNA binding-dependent Mef2d function.
The question remained whether Mef2d negatively regulates GC-Tfh differentiation via transcriptional control of SAP expression. To directly test this, we examined whether Mef2d-dependent impairment of GC-Tfh differentiation could be reversed by restoring SAP expression in Mef2d-RV OTII CD4 T cells. OTII CD4 T cells transduced with empty-RV, Mef2d-RV, or Mef2d-Sh2d1a-RV (M+S) (Fig. 5F), in the popLNs were examined for GC-Tfh differentiation seven days after NP-OVA immunization (Fig. 5G). Whilst forced Mef2d expression led to a significant loss of PSGL-1loCXCR5+ GC-Tfh differentiation (Fig. 5H), Mef2d-RV OTII CD4 T cells, when SAP was co-expressed, were able to differentiate into PSGL-1loCXCR5+ GC-Tfh cells as comparable as the controls (Fig. 5H). Taken together, these data demonstrate that Mef2d negatively regulates GC-Tfh differentiation of CD4 T cells via transcriptional repression of the Sh2d1a gene.
The B:T immune synapse is negatively regulated by Mef2d in CD4 T cells
Given the inhibitory roles of Mef2d in transcriptional regulation of SAP expression, and during SAP-dependent GC-Tfh differentiation, we examined whether B:T synapse formation might be modulated by Mef2d. For this, an in vitro B:T conjugation assay was performed (25, 26). Equal numbers of empty-RV (CD45.11) and Mef2d-RV (CD45.12) OTII CD4 T cells were co-cultured with splenic B cells pulsed with OVA323–339 peptide (1 or 10 μg/mL) that were pre-stimulated with LPS (fig. S6A–B). The background B:T conjugate formation was determined with OTII CD4 T cells after co-culture with pre-activated B cells in the absence of OVA323–339 peptide. The antigen-specific B:T synapse (Δ conjugate, hereafter), measured by subtracting [CD4+CD19+ % among the total CD4+]− Ag from [CD4+CD19+ % among the total CD4+]+ Ag, dramatically increased in an OVA323–339 peptide dose-dependent manner (fig. S6A). In the Δ conjugate, Mef2d-RV OTII CD4 T cells were present at significantly lower frequencies than the control OTII CD4 T cells (fig. S6B), indicating that Mef2d inhibits CD4 T cells from establishing antigen-specific B:T synapse. We assessed expression of Vα2, the TCR alpha chain of the OTII TCRtg CD4 T cells (51), and CD18 (β2 integrin subunit of LFA-1), the lack of which compromises LFA-1 function in CD4 T cells (52). Expression of both Vα2 and CD18 was comparable between Mef2d-RV and control OTII CD4 T cells (fig. S6C), implying that Mef2d regulated B:T synapse formation of CD4 T cells in a TCR- and LFA-1-independent manner.
RPKMs of the SLAM family members (Slamf5 and Slamf6 genes encoding CD84 and Ly108, respectively) and Cd40lg that function in CD4 T cell’s synapse formation with antigen presenting APCs (53, 54) were not altered in Tfh and non-Tfh cells by Mef2d (fig. S4E), indicating that Mef2d might function to specifically modulate synapse formation of CD4 T cells with B cells. Indeed, we found that Mef2d-RV OTII CD4 T cells formed immune synapse with antigen-presenting DCs at similar magnitudes to the controls (fig. S6D–E).
Mef2d negatively regulates IL-21 production of CD4 T cells
In our RNA-seq analysis, Il21 was also listed as a potential target of Mef2d (table S2). The amount of Il21 mRNA was significantly reduced in Mef2d Tfh cells, with levels similar to non-Tfh cells, and expression further diminished in Mef2d non-Tfh cells (Fig. 6A). Moreover, potential Mef2 binding sites were found at around +3.2kb (TATAAATAG) and +6.6 kb (CTAAAAATAG) regions in the Il21 locus (Fig. 6B). Luciferase assays using plasmids harboring either +3.2kb or +6.6kb region of the Il21 gene revealed that Mef2d was able to bind to these sites, with more robust binding to the +3.2kb region (Fig. 6C), indicating that IL-21 production of CD4 T cells could be modulated by Mef2d. To test this hypothesis, polyclonal CD4 T cells isolated from naïve mice were stimulated under a culture condition that induces IL-21 production by in vitro activated CD4 T cells (55). The frequency of IL-21 producing CD4 T cells was significantly reduced by ectopic Mef2d expression (fig. S7A–B), indicating Mef2d as a potential transcriptional repressor of the Il21 gene.
Fig. 6. Mef2d hinders IL-21 production of antigen-specific CD4 T cells via DNA binding dependent repression of the Il21 gene.
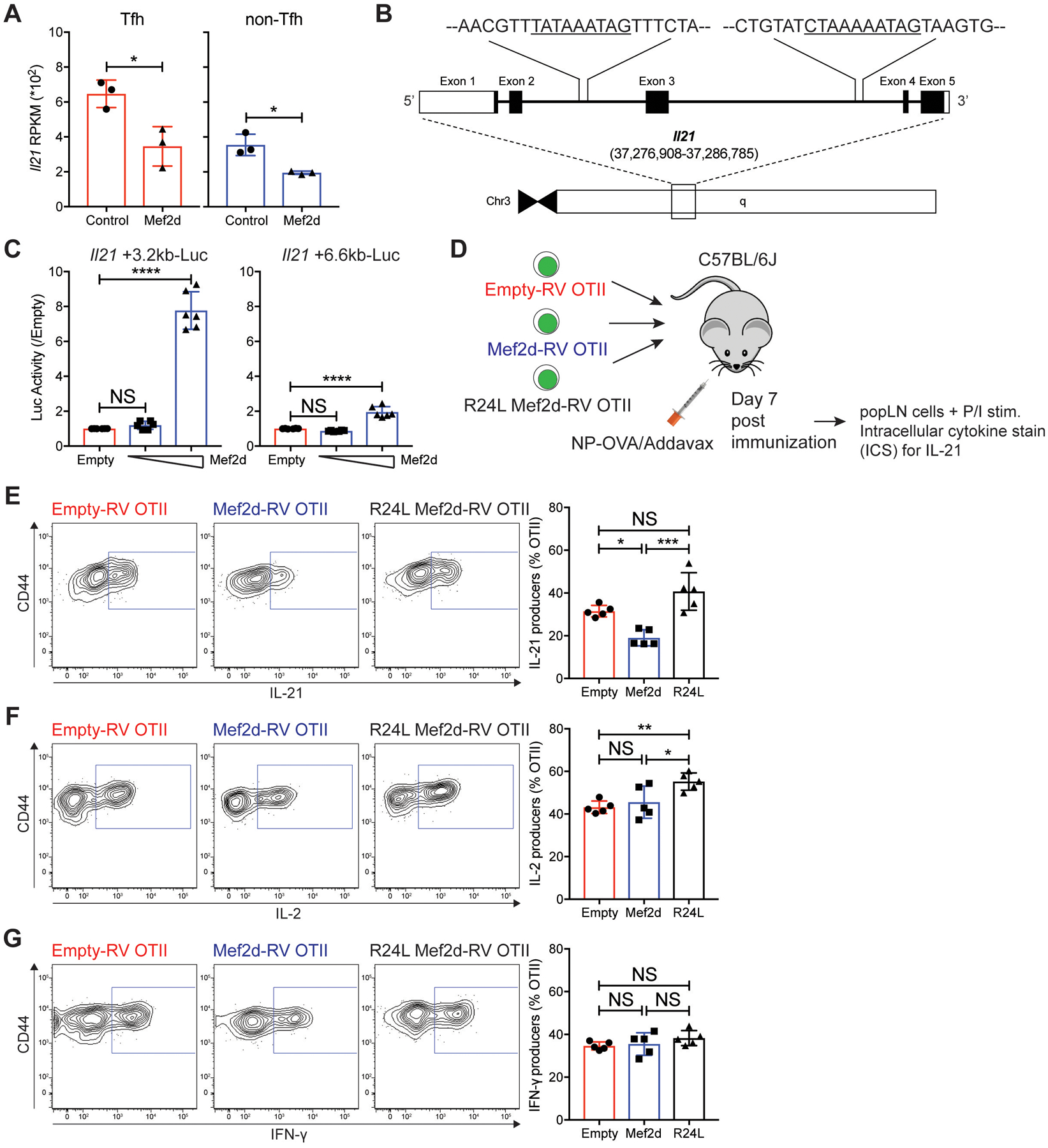
(A) RPKMs of the Il21 gene of Tfh (red) and non-Tfh (blue) cells developed from empty-RV (circles) or Mef2d-RV (triangles) OTII CD4 T cells.
(B) The genomic region of the murine Il21 gene with putative Mef2d binding sites at +3.2 kb (TATAAATAG) and +6.6 kb (CTAAAAATAG) from the Il21 gene transcription start site.
(C) Luciferase activities of the pGL4.10 plasmids with +3.2 kb or +6.6 kb region of Il21 locus (~1.5kb long) containing the potential Mef2d binding sites in the presence or absence of Mef2d co-expression (0.05 and 0.8 μg for respective luciferase plasmids).
(D to G) CD45.1 OTII CD4 T cells, transduced with empty-RV, Mef2d-RV, or R24L Mef2d-RV, were transferred into C57BL/6J mice. Seven days after NP-OVA immunization, the transferred cells were analyzed for production of IL-21, IL-2 and IFN-γ.
(D) Experimental scheme to investigate the regulation of IL-21 production of CD4 T cells by Mef2d.
(E to G) Flow cytometry plots of empty-RV, Mef2d-RV, and R24L Mef2d-RV OTII CD4 T cells, as shown in Fig. 5 A–E. Gates indicate IL-21 (E), IL-2 (F), and IFN-γ (G) producers. The frequencies of respective populations among GFP+ OTII CD4 T cells were calculated.
Analysis was performed with the data obtained from three independent experiments (two replicate samples/experiment) (C) and data are representative of three independent experiments with n=5 mice per group (D to G).
Error bars indicate mean with SD. Statistical significance values were determined using two-tailed Student’s t-test (A) and one-way ANOVA with Tukey’s multiple comparisons test (C, E to G). NS, statistically non-significant; * p <0.05; ** p <0.01; *** p <0.001; **** p <0.0001.
Next, we investigated whether Mef2d-mediated regulation of IL-21 production occurs in a DNA binding-dependent manner (Fig. 6D). IL-21 ICS (intracellular cytokine staining) was performed with empty-RV, wild-type Mef2d-RV or R24L Mef2d-RV OTII CD4 T cells from popLNs (Fig. 5A–E). IL-21 production in OTII CD4 T cells was significantly reduced by ectopic expression of wild-type Mef2d (Fig. 6E), while their ability to produce IL-2 or IFN-γ remained unchanged (Fig. 6F–G). The R24L mutant Mef2d failed to inhibit IL-21 production (Fig. 6E), consistent with its inability to suppress GC-Tfh differentiation.
IL-21 is a major B cell-help cytokine produced mostly by Tfh and GC-Tfh cells (15). Therefore, it is plausible that the impaired IL-21 production might be secondary to the inherent defects of the Mef2d-RV OTII CD4 T cells undergoing SAP-dependent Tfh and GC-Tfh differentiation (Fig. 4 and Fig. 5), and not a direct consequence of transcriptional repression of the Il21 gene. To directly delineate the “Mef2d – Il21” axis, we examined the IL-21 production of M+S OTII CD4 T cells, whose GC-Tfh differentiation defects by the forced Mef2d expression were almost fully restored by recovering SAP expression (Fig. 5F–H). Defective IL-21 production was, however, not completely rescued by SAP co-expression (fig. S7C). The partial restoration of IL-21 production was not due to potential issues in general CD4 T cell activation, as IL-2 production of OTII CD4 T cells was comparable irrespective of ectopic expression of Mef2d or Sh2d1a gene, or both. Collectively, these data delineate Mef2d as a transcriptional repressor of the Il21 gene.
The Mef2d gene controls GC-Tfh differentiation of CD4 T cells
Given its ability to repress SAP dependent B:T synapse formation and GC-Tfh differentiation, we hypothesized that Mef2d deficiency might lead to strong GC-Tfh differentiation via de-repressed SAP expression. This point was addressed by analyzing SAP expression and GC-Tfh differentiation in CD4 T cells, whose Mef2d gene was disrupted by CRISPR/Cas9 system (56). Cd4CreROSA26CAG-Cas9-EGFP/Wt mice (57) were crossed with OTII TCRtg mice to produce Cas9-expressing OTII (Cas9+ OTII, hereafter) CD4 T cells. Two sgRNA sequences targeting the murine Mef2d gene were tested for Mef2d gene disruption (fig. S8A). Cas9+ OTII CD4 T cells transduced with a retrovirus expressing sgCd8 or sgMef2d (#1), which resulted in a substantial knockdown of Mef2d protein expression (fig. S8B), were adoptively transferred into Cd4CreROSA26CAG-Cas9-EGFP mice (fig. S8C) to rule out a potential challenge of Cas9+ CD4 T cells being rejected in C57BL/6J mice (58). The donor cells were examined for GC-Tfh differentiation seven days after NP-OVA immunization. While Mef2d gene disruption did not alter OTII CD4 T cell expansion (fig. S8D), it strongly induced PSGL-1loCXCR5+ GC-Tfh differentiation (fig. S8E) with a significant increase in CXCR5 expression (fig. S8F). Most importantly, sgMef2d+Cas9+ OTII CD4 T cells formed the SAPhiCXCR5+ compartment more robustly than sgCd8+Cas9+ OTII CD4 T cells (fig. S8G), which likely results from a lack of the transcriptional repression of the Sh2d1a gene.
We further studied Mef2d gene function using CD4 T cell-specific Mef2d knockout (Mef2d CKO, hereafter) mice (fig. S9) using the Mef2dfl/fl line (59). CD4 T cell-specific deficiency of Mef2d protein expression, validated by Mef2d immunoblot (fig. S9A), did not compromise thymic T cell development (fig. S9B) or CD4 or CD8 T cell maturation (fig. S9C–E), consistent with previous findings (60). WT and Mef2d CKO CD45.2 OTII CD4 T cells were adoptively transferred into CD45.1 B6.SJL mice. Seven days after NP-OVA immunization, the donor cells in the popLNs were examined for GC-Tfh differentiation, intracellular SAP expression and IL-21 production. Comparable CD44 expression indicates that antigen-specific CD4 T cell activation was not perturbed due to a lack of the Mef2d gene (Fig. 7A). Mef2d gene deficiency, however, resulted in profound increases in GC-Tfh differentiation of OTII CD4 T cells compared to control OTII CD4 T cells (Fig. 7B–C), confirming our findings with CRISPR/Cas9 mediated Mef2d gene disruption (fig. S8). We also observed enhanced surface expression of canonical Tfh markers (PD-1 and CXCR5) (Fig. 7D) and reduced expression of a T cell zone homing PSGL-1 (Fig. 7E) in Mef2d CKO OTII CD4 T cells, highlighting the suppressive functions of Mef2d in Tfh and GC-Tfh differentiation. Mef2d CKO OTII CD4 T cells formed the SAPhiCXCR5+ compartment and expressed higher levels of intracellular SAP protein than control OTII CD4 T cells (Fig. 7F). IL-21 production in antigen-specific CD4 T cells was also increased by Mef2d gene deficiency (Fig. 7G). Collectively, this data identifies that the Mef2d transcription factor is a a key transcriptional regulator of SAP dependent Tfh and GC-Tfh differentiation from CD4 T cells, and its subsequent role in B cell help.
Fig. 7. Mef2d deficiency leads to enhanced SAP expression, IL-21 production and GC-Tfh differentiation of antigen-specific CD4 T cells.
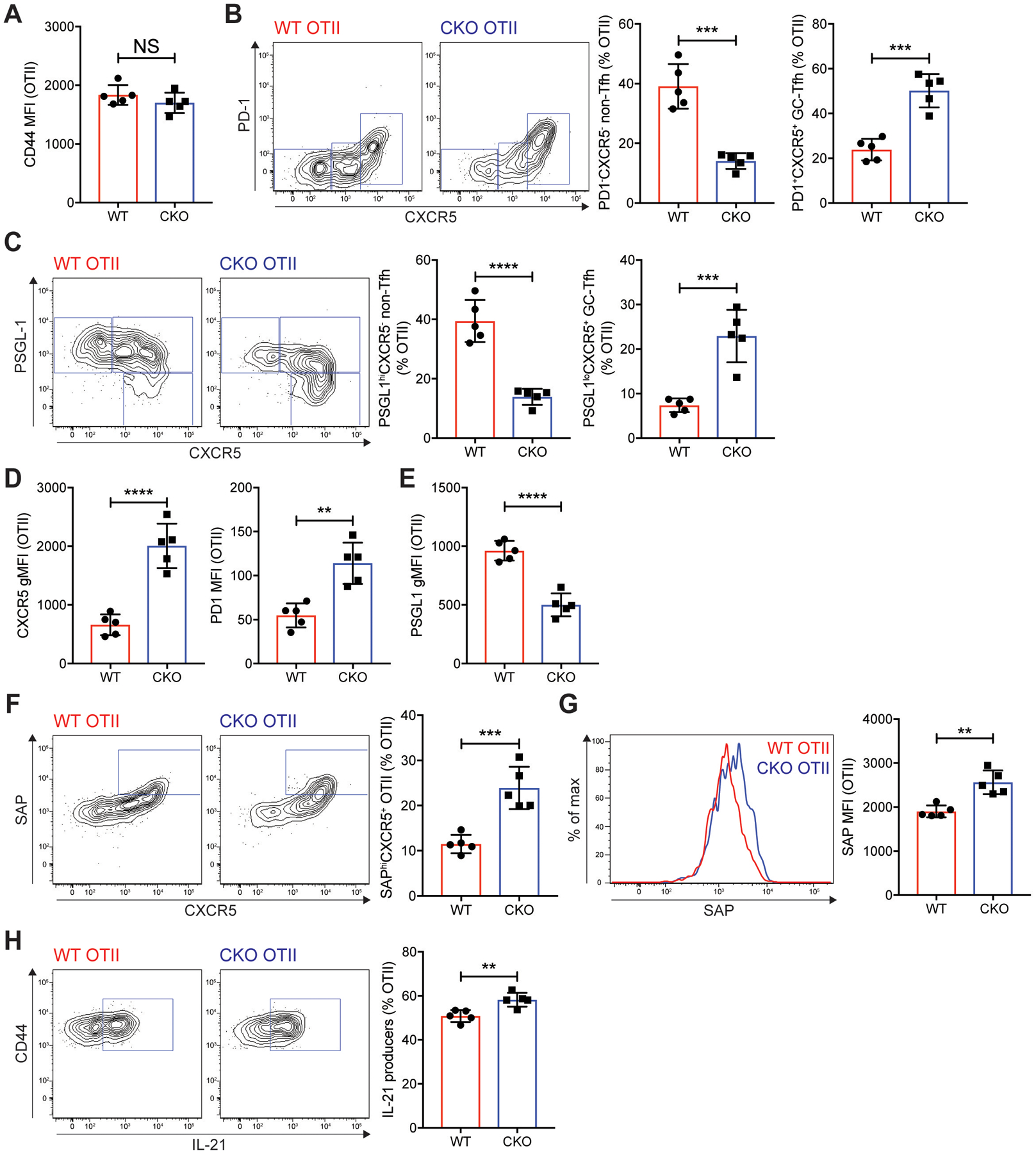
WT control and Mef2d CKO (Cd4CreMef2dfl/fl) CD45.2 OTII CD4 T cells were transferred into CD45.1 B6.SJL mice, which were immunized subcutaneously with NP-OVA. Seven days later, popLNs were examined for GC-Tfh differentiation, SAP expression, and IL-21 production.
(A) CD44 MFIs of the donor OTII CD4 T cells in the popLNs.
(B and C) Flow cytometry plots of the transferred OTII CD4 T cells. Gates indicate non-Tfh (PD-1−CXCR5− or PSGL-1hiCXCR5−), Tfh (PD-1−CXCR5+ or PSGL-1hiCXCR5+), and GC-Tfh (PD-1+CXCR5+ or PSGL-1loCXCR5+) cells. The frequencies of non-Tfh and GC-Tfh cells were calculated.
(D) CXCR5 gMFIs and PD-1 MFIs of the donor OTII CD4 T cells were quantified.
(E) PSGL-1 gMFIs of the donor cells were calculated.
(F and G) SAP expression of the WT control and Mef2d CKO OTII CD4 T cells were examined. Flow cytometry plots of the donor cells with gates indicating SAPhiCXCR5+ compartment (F). Overlaid SAP histograms (G). SAPhiCXCR5+ cell frequencies and intracellular SAP MFIs were quantified.
(H) Flow cytometry plots of the transferred OTII CD4 T cells. Gates indicate IL-21 producing cells. The frequencies of IL-21 producers were quantified.
Representative of two independent experiments with n=5 mice per group.
Error bars indicate mean with SD. Statistical significance values were determined using two-tailed Student’s t-test. NS, statistically non-significant; ** p <0.01; *** p <0.001; **** p <0.0001.
Reduced MEF2D expression in CD4 T cells is associated with humoral autoimmune conditions in SLE patients
The regulation of SAP-dependent GC-Tfh differentiation and IL-21 production by CD4 T cells is crucial to control CD4 T cell-dependent humoral immunity, the dysregulation of which is highly associated with the pathophysiology of autoimmune diseases such as systemic lupus erythematosus (61). Recently, a novel genetic variant (rs200395694: G>T) was found in the regulatory region of the MEF2D gene in a subgroup of Swedish SLE patients, which exhibits strong associations with the magnitude of autoantibodies (62). Although its impact on MEF2D expression in CD4 T cells was not examined, the variant, due partly to its location within the transcription factor binding sites, led to a reduced luciferase activity compared with the reference allele in Jurkat cells (62), suggesting this variant might negatively impact on transcriptional activation of the MEF2D gene in CD4 T cells. Healthy individuals and SLE patients (table S3a) were examined for MEF2D expression in CD4 T cells, which was further analyzed for a potential association with autoimmune features of SLE patients. Phenotypic examination of the peripheral blood mononuclear cells (PBMCs) (fig. S10A) revealed that the frequencies of CD4 T cells (Fig. 8A) and CD45RA− (activated/memory) CD4 T cells (fig. S10B) were reduced and increased, respectively, in the SLE patients compared to those in healthy individuals. As previously reported by others (63), the frequency of the circulating Tfh (cTfh)-like cells (ICOS+PD-1+ cells) among the CD45RA−CXCR5+ compartment was significantly increased in the SLE patients in comparison to the controls (Fig. 8B) with strong positive correlations with SLEDAI and the amount of the serum autoantibodies (anti-dsDNA Ig) (Fig. 8C).
Fig. 8. Reduced MEF2D expression in CD4 T cells is associated with autoimmune SLE conditions.
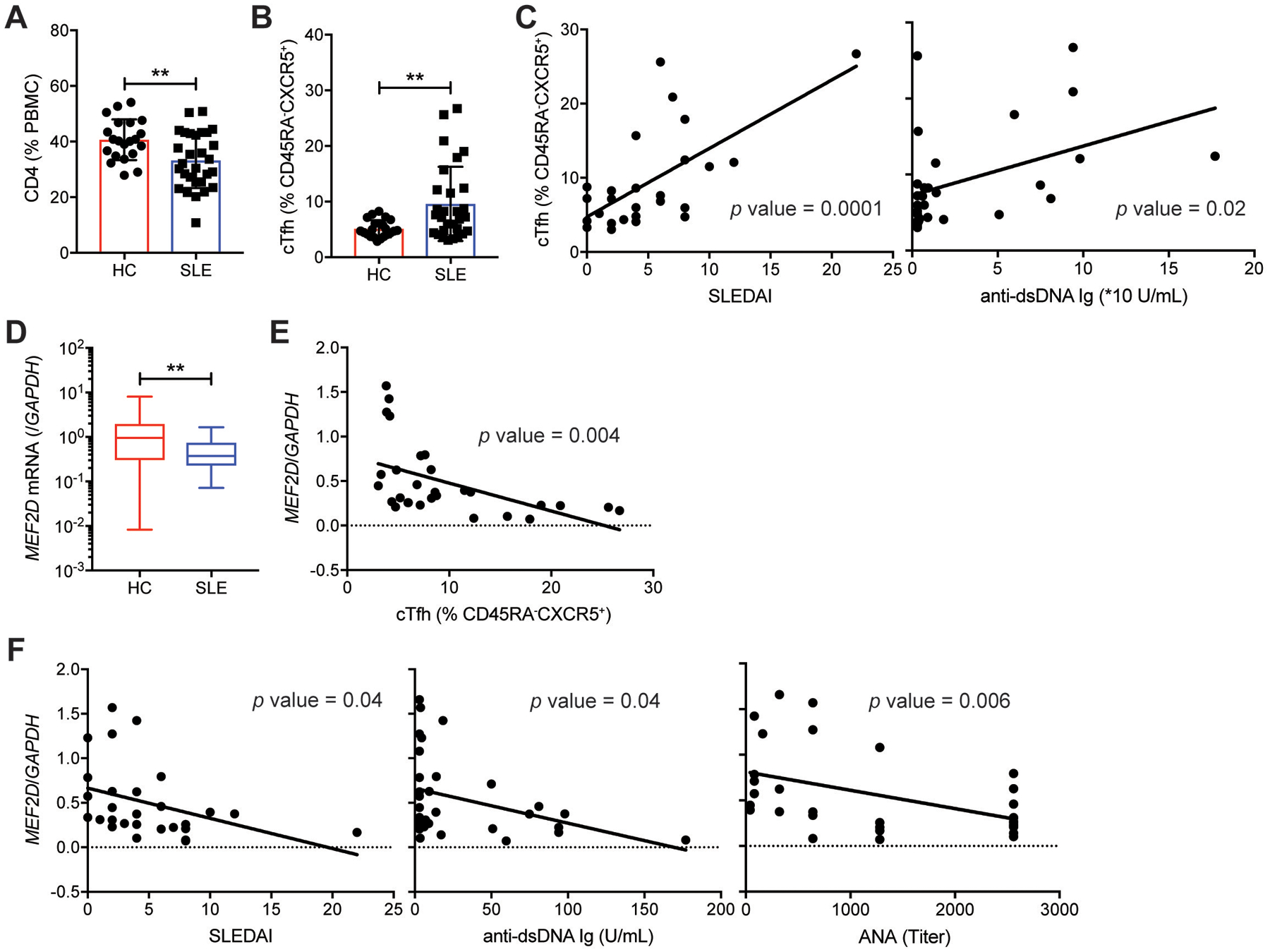
(A) Frequencies of CD4 T cells in PBMC from healthy controls and SLE patients (table S3a).
(B) Frequencies of ICOS+PD-1+ circulating Tfh (cTfh) cells (gating strategy shown in fig. S10A) among peripheral blood CD45RA−CXCR5+ CD4 T cells of healthy controls and SLE patients (table S3a).
(C) Correlations of ICOS+PD-1+ cTfh cell frequency with SLEDAI and anti-dsDNA Ig level in the SLE patients.
(D) MEF2D mRNA measured by qPCR in peripheral blood CD4 T cells obtained from healthy controls and from the SLE patients (table S3a).
(E) Correlation of the MEF2D expression of CD4 T cells with the ICOS+PD-1+ cTfh cell frequency in the SLE patients.
(F) Correlations of the MEF2D expression of the SLE CD4 T cells with SLEDAI, anti-dsDNA Ig, and ANA.
Error bars indicate mean with SD (A and B) and mean with min to max (D). Statistical significance values were determined using two-tailed Student’s t-test (A, B, and D) and Pearson’s correlation analysis (C, E, and F). ** p <0.01.
We then measured the MEF2D mRNA of the peripheral blood CD4 T cells obtained from the SLE patients and those of the healthy individuals by qPCR. The patients’ CD4 T cells expressed approximately three-fold less MEF2D mRNA than those of the healthy individuals (Fig. 8D). The reduction might have resulted from the increased cTfh cell frequency in the patients (Fig. 8B), given the lower MEF2D expression by GC-Tfh cells in comparison to non-Tfh effector CD4 T cells in tonsils (fig. S1E–F). Possible pathophysiological roles of reduced MEF2D expression in the patients’ CD4 T cells were subsequently examined by quantifying MEF2D mRNA expression of naïve CD4 T cells of healthy individuals and the patients (table S3b). CD45RA+ naïve CD4 T cells were FACS-sorted from PBMCs, whose MEF2D mRNA amount was measured by qPCR. CD45RA+ naïve CD4 T cells in the patients expressed significantly lower MEF2D mRNA than those in the controls (fig. S10C).
Interestingly, the MEF2D mRNA quantity exhibited a significant negative association (p=0.004) with the cTfh cell frequency in the SLE patients (Fig. 8E). Moreover, it was inversely correlated with the SLEDAI (p=0.04) and the amounts of the serum autoantibody [anti-dsDNA (p=0.04) and ANA (p=0.006]) of the patients (Fig. 8F), implying a potential physiological link between reduced MEF2D expression in CD4 T cells and dysregulated humoral autoimmunity. While we cannot fully rule out other pathological factors associated with reduced MEF2D expression, the magnitude of CD4 T cell activation (fig. S10D) or the frequencies of other effector CD4 T cells (CXCR3+CCR6− Th1, CXCR3−CCR6− Th2, and CXCR3−CCR6+ Th17 cells) did not show a significant inverse correlation with the MEF2D mRNA level (fig. S10E). Given that the reduced Mef2d expression in CD4 T cells (by crMef2d-RNP nucleofection or sgMef2d-Cas9+) led to a significant increase in Tfh and GC-Tfh differentiation (Fig. 4 and fig. S8), out study suggests a possible pathophysiologic role of the “Mef2d – Sh2d1a/Il21 gene” axis in the control of T-dependent humoral autoimmunity.
DISCUSSION
Spatiotemporal coordination of cognate interactions between B cells and CD4 T cells is fundamental to the development of Tfh (GC-Tfh) cells, as well as to the generation of CD4 T cell-dependent (TD) humoral immunity (14, 15, 64). It is becoming apparent that B cells and CD4 T cells have adopted components of the central nervous system for rapid and effective exchange of immunological information at the interface (35, 65, 66). Here, we provide evidence to further substantiate this concept. We report that Mef2d, a transcriptional inhibitor of neuronal synapse formation (31, 32), negatively controlled (1) SAP-dependent immune synapse between B cells and CD4 T cells, and (2) IL-21 production, a key signaling molecule produced at the B:T synaptic junction, via transcriptional repression of the Sh2d1a and Il21 genes in CD4 T cells. Mef2d thus functions as a crucial regulator of Tfh and GC-Tfh differentiation of CD4 T cells and subsequent antigen-specific antibody production of B cells in germinal centers.
Stable B:T interaction guarantees cognate B cells are selected by CD4 T cells to trigger humoral immunity. In this context, Mef2d mediated SAP regulation could function as a critical checkpoint to assure bystander (or non-cognate) B cells at the B:T border do not enter the GC, and to prevent B cell clones with impaired (unwanted, sometimes autoreactive) antigen specificities that might have been acquired by random somatic hypermutation from being selected during GCs (67). Our findings highlight the importance of Mef2d in controlling the B:T synapse dependent checkpoint, as Mef2d gene disruption or deletion in CD4 T cells leads to a robust GC-Tfh differentiation via unrestrained SAP expression and autoimmune features of the SLE patients are associated with reduced MEF2D mRNA expression in CD4 T cells. Our study also suggests that MEF2D expression needs to be tightly regulated in CD4 T cells, and a breach in this process might contribute to autoimmunity via uncontrolled autoantibody production. It is important to determine whether dysregulated MEF2D expression (Fig. 8D and fig. S10C), caused in some instances by the rs200395694: G>T alteration (62), might trigger the onset of SLE via unchecked SAP expression. However, given the Mef2d roles to positively regulate Treg cell functions (68), we cannot rule out a possibility that the autoimmune features of the SLE patients might result from improper control of autoimmune cells by Treg cells. Nonetheless, minor autoimmune features of Foxp3CreMef2dfl/fl mice (68) and profound SAP dependent GC-Tfh differentiation (Fig. 7) seen in Cd4CreMef2dfl/fl OTII CD4 T cells raises the question of whether Cd4CreMef2dfl/fl mice might develop SLE like autoimmune features via dysregulated SAP expression and spontaneous GC formation.
It is interesting that Mef2a, while it is also highly expressed in CD4 T cells in both humans and mice (fig. S1), did not play redundant roles to Mef2d in transcriptional regulation of SAP and IL-21 (fig. S12). Due to the conserved DNA binding domain (50), Mef2 transcription factor isoforms could function in concert. Mef2a and Mef2d play important roles in modulating apoptosis of the cerebellar granule neurons in the nervous system (69), whereas Mef2c works together with Mef2d and Mef2b in supervising the large pre-B to the small pre-B stage transition (70) and in controlling GC formation (71), respectively. Mef2a gene disruption, however, was unable to phenocopy the increased SAP expression and SAP-dependent Tfh/GC-Tfh differentiation observed by Mef2d gene disruption or deletion. It might be thus interesting to examine whether these isoforms are differentially regulated in CD4 T cells or during Tfh differentiation processes, since functions of Mef2 transcription factor isoforms are under control of post-transcriptional and post-translational modifications in the nervous system (72).
Another enigmatic aspect of the Mef2 transcription factors is that their target genes appear to be regulated in a cell-type and stage specific manner. While Mef2d was previously shown to activate Nur77 induction in autoreactive thymocytes, and function as a pro-apoptotic factor (30, 73), the Nr4a1 gene was not transcriptionally modulated by Mef2d in mature CD4 T cells (fig. S4F). Moreover, Mef2b appears to bind to target genes and to regulate their expression differently in B cells and CD4 T cells. While MEF2B (Mef2b) is a well-known transcriptional activator of BCL6 (Bcl6) in human and mouse GC B cells (71, 74), even after ectopically expressed, it was not able to induce Bcl6 expression in CD4 T cells (fig. S11A–B). Instead, Mef2b-RV OTII CD4 T cells were severely impaired in their ability to undergo Tfh differentiation, similar to Mef2d-RV OTII CD4 T cells, in the context of NP-OVA immunization.
BCL6 is a transcriptional activator of SH2D1A gene in human CD4 T cells (75), and there are BCL6 binding motifs in the promoter and distal regions of the MEF2D gene in human GC-Tfh cells (76). In human lymphoid non-Tfh and GC-Tfh cells, the striking similarity of MEF2D mRNA expression pattern to that of the PRDM1 gene (fig. S1), whose transcriptional activation is repressed by BCL6 (37), suggests that MEF2D-dependent regulation of SH2D1A expression might occur via BCL6, as recently proposed by Crotty and colleagues (56). In the study, Bcl6 was revealed to regulate the Tfh differentiation program of murine CD4 T cells, at least in part by transcriptional repression of genes that encode transcriptional repressors (i.e., Id2, Runx2, and Klf2, among others) of Tfh differentiation. While our preliminary investigation suggests that Mef2d might not be targeted by Bcl6 (fig. S11D), this potential “Bcl6 –| Mef2d –| Sh2d1a axis” needs more careful investigations with physiologically relevant samples, as the forced Bcl6 expression by itself is not able to instruct human and murine CD4 T cells to undergo Tfh differentiation during in vitro stimulation (38, 75).
IL-21 is a major B cell help cytokine produced by Tfh/GC-Tfh cells and plays important roles for B cells to form GCs, undergo isotype switching, and differentiate into plasma cells. Our study revealed that Mef2d also controls the TD humoral immunity via transcriptional regulation of B cell help function of CD4 T cells. This conclusion is supported by our findings; (1) Icos-independent IL-21 production of CD4 T cells was shown to be negatively regulated by ectopic Mef2d expression (fig. S7A–B) and (2) the impaired IL-21 production was not fully restored by rescuing SAP expression in the Mef2d-RV OTII CD4 T cells (fig. S7C). Thus, our findings provide mechanistic insights into the Mef2d-mediated transcriptional modulation of B:T synapse formation and B cell-help function of CD4 T cells, highlighting Mef2d as a double-layer control switch of the TD humoral (auto)immunity.
MATERIALS AND METHODS
Study design
This study aimed to examine the function of the Mef2d transcription factor in the regulation of Tfh differentiation and humoral immunity. Retroviral expression, conditional knockout and CRISPR/Cas9 mediated gene disruption systems were used for gain- and loss-of-Mef2d function studies with primary murine (TCRtg) CD4 T cells. RNA-seq was performed to screen for potential target genes of Mef2d, and molecular and biochemical assays were used to validate Sh2d1a and Il21 genes as direct targets of Mef2d in murine CD4 T cells. An in vitro conjugation assay was performed to examine the Mef2d roles in SAP-dependent B:T synapse formation. Lymphoid tissues or peripheral blood of healthy individuals and SLE patients were analyzed for physiological relevance of the MEF2D expression of CD4 T cells in human diseases. All experiments were performed in at least two independent experiments, with three or more mice per group.
Mice
C57BL/6J mice were purchased from DBL (Seongnam, Republic of Korea). μMT and ROSA26CAG-Cas9-EGFP mice were obtained from Jackson Laboratory. Mef2dfl/fl mice (RIKEN BRC) were kindly provided by Dr. Takahisa Furukawa at Osaka University (59). Cd4CreBcl6fl/fl, OTII, and SMARTA (TCRtg specific for LCMV glycoprotein amino acids 66–77 presented by I-Ab) mice were kindly provided by Dr. Shane Crotty. All mouse strains on a C57BL/6J background were bred under specific pathogen-free conditions in accordance with the Institute for Experimental Animals at Seoul National University. TCRtg mice (6–12 weeks of age) were used for the adoptive transfer experiments with sex- and age-matched recipient mice. All animal experiments were performed in accordance with the guidelines of the Seoul National University Institutional Animal Care and Use Committee.
Human Studies
Healthy donors were recruited from the Department of Otorhinolaryngology and the Department of Pathology at Seoul National University Hospital. The detailed demographic information is provided in table S1. All studies were approved by the SNUH Institutional Review Board (SNUH IRB # 1609-129-795 and 1504-059-664), and healthy donors were enrolled after providing written informed consent.
Systemic lupus erythematosus (SLE) patients and healthy individuals were recruited with informed consent after IRB approval from the Department of Rheumatology at Ajou University Hospital (demographic information in table S4). All studies were reviewed and approved by the Ajou Institutional Review Board (IRB # AJIRB-BMR-SMP-17-155).
Adoptive cell transfer and immunization
Naïve, retrovirus-transduced, or RNP-transfected TCRtg CD4 T cells were transferred intravenously via the retro-orbital sinus. 0.1–2 × 106 cells/mouse were transferred to examine the donor cells on days 3 and 7 (or 8) post immunization (77). Following the adoptive transfer, mice were immunized with NP-OVA (Biosearch Technologies) or gp61-KLH (GenScript) emulsified in Addavax (InvivoGen) or CFA (Sigma Aldrich) via subcutaneous footpad injection.
Plasmids and retrovirus production
The amplified murine Mef2d, Mef2b, or Bcl6 coding sequences were cloned into a retroviral expression vector (pMIG). R24L Mef2d-pMIG vector was generated using a site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions. The murine Sh2d1a plasmid was kindly provided by Dr. Hai Qi at Tsinghua University. The Sh2d1a coding sequence was cloned into pMIG-mAmetrine (pMIA) vector. Single-guide RNA (sgRNA) sequences (listed in table S5) (https://chopchop.cbu.uib.no/) were cloned into pLsg-mAmetrine (pLsgA) plasmid (kindly provided by Dr. Shane Crotty) (56). For retrovirus production, Plat-E cells were transfected with the plasmids of interest. Culture supernatants were collected at 24 and 48 hours after transfection, filtered through a 0.45 μm syringe filter and stored at 4 °C until use.
In vitro polarization of IL-21 producing CD4 T cells
Polyclonal CD4 T cells were isolated and transduced with empty-retroviral (empty-RV) or Mef2d-retroviral (Mef2d-RV) as described in supplementary materials and methods. During and after transduction, cells were cultured in IL-21 polarizing medium [D10 with 50 μM BME, 10 ng/mL recombinant mouse IL-21 (BioLegend), 10 μg/mL anti-IFNγ (XMG1.2; BioXcell), and 10 μg/mL anti-IL-4 (11B11; BioXcell)] for an additional 4 days.
Flow cytometry
For mouse samples, single-cell suspensions were prepared from draining LNs or spleens or after in vitro stimulation. The cells were stained with fluorochrome-conjugated antibodies against CD4 (RM4–5), CD45.1 (A20), CD18 (M18/2), and CD19 (6D5) from BioLegend; CD4 (RM4–5), B220 (RA3–6B2), ICOS (7E.17G9), and CD44 (IM7) from eBioscience; PD-1 (J43) from Invitrogen; CD8 (53–6.7), B220 (RA3–6B2), TCR Vα2 (B20.1), CD45.1 (A20), CD45.2 (104), PSGL-1 (2PH1), PD-1 (J43), and Fas (Jo2) from BD Biosciences; PNA (FL-1071) from Vector Laboratories. Two-step and three-step CXCR5 staining were used for FACS analysis of the transferred cells on day 3 after immunization with biotinylated anti-CXCR5 (2G8) antibody and days 7–8 post-immunization with purified anti-CXCR5 (2G8) antibody (from BD Bioscience), respectively (78). For intracellular cytokine staining, cells were stimulated with 40 ng/mL PMA (Merck) and 1.5 μg/mL ionomycin (Abcam) in the presence of 2 μg/mL brefeldin A (Merck) for 3 hours. After surface staining, cells were fixed, permeabilized, and stained with antibodies against IL-2 (JES6–5H4), IFNγ (XMG1.2), IL-17A (TC11–18H10.1) (from Biolegend), and recombinant mouse IL-21 receptor Fc (R&D Systems) followed by anti-human IgG (Jackson Immunoresearch).
Human mononuclear cells were stained with the following fluorochrome-conjugated antibodies: CD4 (RPA-T4), CD8 (HIT8a), CD45RO (UCHL1) (BioLegend), Fixable Viability Dye, CXCR5 (MU5UBEE) (eBioscience), CD19 (HIB19) (Invitrogen), CXCR5 (RF8B2), and PD-1 (EH12.1) (BD Biosciences).
Intracellular SAP or Bcl6 staining was conducted with fluorochrome-conjugated antibodies [anti-SAP (1A9) and anti-Bcl6 (K112–91) from BD Biosciences] after fixation and permeabilization using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience).
All stained cells were acquired using LSRII, LSRFortessa, or LSRFortessa X-20 (BD Biosciences) and analyzed using FlowJo software v.9.6 (FlowJo).
Luciferase assay
An approximately 1.5kb region of the Sh2d1a or Il21 gene, encompassing potential Mef2d binding sites, was amplified from mouse genomic DNA by PCR and cloned into the pGL4.10 vector (Sh2d1a +6.9kb-Luc or Il21 +3.2kb-Luc and Il21 +6.6kb-Luc, respectively). A vector including the 3X Mef2 binding sequence amplified from the 3XMEF2-Luc plasmid (a gift from Dr. Ron Prywes at Columbia University; Addgene plasmid # 32967) was used as a positive control. HEK293T cells were transfected with luciferase plasmids and a β-galactosidase plasmid as an internal control in the presence of empty-pCMV, pCMV-Mef2d, or pCMV-R24L Mef2d plasmids. Three days after transfection, cells were lysed with Luciferase Cell Culture Lysis 5X Reagent buffer (Promega). Luminescent signals were measured with the cell lysates using the Luciferase Assay System (Promega), following the manufacturer’s instructions. Luciferase activity was normalized with β-galactosidase activity quantified using o-nitrophenyl-β-D-galactopyranoside (ONPG) substrate.
In vitro conjugation assay
The B:T conjugation assay (26, 79) was performed as follows. Empty-GFP (CD45.11) and Mef2d-GFP (CD45.12) OTII CD4 T cells were prepared. Splenic B cells, isolated from C57BL/6J mice, were stimulated with LPS (1 μg/mL) for two days. Activated B cells were labeled with 50 μM CMF2HC (Invitrogen) for 30 minutes at room temperature, followed by a brief pulse with OVA323–339 peptide (0, 1, or 10 μg/mL) for 30 minutes at 37 °C. 1.25 × 105 empty-GFP and 1.25 × 105 Mef2d-GFP OTII CD4 T cells were co-cultured with 5.0 × 105 B cells for 20 or 30 minutes at 37 °C.
The DC:T conjugation assay was done in similar manners to the B:T conjugation assay with following modifications. Splenic DCs, after isolation with CD11c microbeads (Miltenyi Biotec) and subsequent staining with CD11c antibody for 20 minutes at 4 °C, were pulsed with OVA323–339 peptide for 2 hours at 37 °C. 2.5 × 105 DCs were co-cultured with empty-GFP and Mef2d-mAmetrine OTII CD4 T cells.
Conjugate formation of the respective OTII CD4 T cells with B cells or DCs was analyzed based on congenic molecule (CD45.11 vs. CD45.12) expression or based on GFP and mAmetrine expression among CD4 T cells in the conjugates.
Investigation of human peripheral blood mononuclear cells
Sample preparation:
PBMCs from SLE patients and healthy donors were isolated as previously described (80). The first cohort’s PBMC samples were used for FACS and quantitative RT-PCR (qPCR) analyses. PBMCs obtained from the second cohort were used to isolate naïve (CD45RA+) CD4 T cells for qPCR analysis.
FACS analysis:
PBMCs were stained with Fixable Viability Dye (Invitrogen) in PBS, followed by staining with fluorochrome-conjugated antibodies against CD4 (RPA-T4), CD45RA (HI100), CXCR5 (J252D4), CXCR3 (G025H7), CCR6 (11A9), PD-1 (EH12.2H7), and ICOS (C398.4A) (from BioLegend and BD Biosciences). Cells were stained in MACS buffer (0.5 % FBS, 1 % penicillin and streptomycin, and 2 mM EDTA in PBS) for 30 min at 4°C.
qPCR analysis:
Total or naïve CD4 T cells were isolated from PBMCs using the Human CD4 T cell Isolation Kit (Miltenyi Biotec) or by FACS-sort. RNA was extracted using Tri Reagent (MRC) according to the manufacturer’s instructions. Oligo(dT) primers (Promega) and the Improm-II Reverse Transcriptase kit (Promega) were used for cDNA synthesis following the manufacturer’s instructions. qPCR was performed using SYBR Premix Ex Taq (Takara) on a Rotor-Gene Q (Qiagen). The primer sequences for MEF2D and GAPDH are listed in table S5.
Quantification of autoantibody production:
Anti-nuclear and anti-dsDNA antibodies were measured in serum samples obtained from SLE patients. Anti-nuclear antibodies (ANA) were measured using ANA HEp-2 Plus (Generic Assays), and anti-dsDNA was measured using the Amerlex Anti-dsDNA Kit (Trinity Biotech) according to the manufacturer’s instructions.
Statistical analysis
All experiments were independently performed two or three times. Data sets were analyzed with the two-tailed unpaired or paired Student’s t-test, one-way ANOVA with Tukey’s multiple comparisons test, two-way ANOVA with Bonferroni’s multiple comparisons test, or Pearson’s correlation using Prism 7.0 software (GraphPad). Differences were considered statistically significant at P < 0.05.
Supplementary Material
Fig. S1. Expression of the MEF2 (Mef2) transcription factor family members in human and murine CD4 T cell subsets.
Fig. S2. Impaired GC-Tfh differentiation of Sm TCRtg CD4 T cells by ectopic Mef2d expression following gp61-KLH immunization.
Fig. S3. Cognate B cell-dependent Tfh differentiation of CD4 T cells.
Fig. S4. Gene expression analyses for screening potential Mef2d target genes in the regulation of Tfh/GC-Tfh differentiation of CD4 T cells.
Fig. S5. Intracellular SAP expression by human and mouse CD4 T cell subsets.
Fig. S6. Mef2d-mediated negative regulation of antigen-specific B:T conjugate formation.
Fig. S7. IL-21 production of CD4 T cells is impeded by the Mef2d transcription factor.
Fig. S8. CRISPR/Cas9 mediated Mef2d gene disruption promotes GC-Tfh differentiation of OTII TCRtg CD4 T cells with de-repressed SAP expression.
Fig. S9. T cell development and maturation in the Mef2d CKO mice.
Fig. S10. Phenotypic identification of naïve and activated effector CD4 T cells in human peripheral blood and correlation analyses with MEF2D mRNA expression.
Fig. S11. Mef2d- and Mef2b-mediated inhibition of Tfh differentiation may occur in a Bcl6-independent manner.
Fig. S12. Mef2a does not play redundant roles to Mef2d in the regulation of SAP-dependent GC-Tfh differentiation of antigen-specific CD4 T cells after NP-OVA immunization.
Table S1. Information from tonsil donors.
Table S2. Genes regulated by Mef2d in Tfh and non-Tfh cells.
Table S3. Clinical information from SLE patients and healthy individuals.
Table S4. Demographic information from SLE patients and healthy individuals.
Table S5. sgRNA and crRNA sequences and primer sequences used in qPCR and ChIP assays.
Supplementary data file S1. Raw data excel file
Acknowledgments:
We thank GC Han and CS Kim for assisting with the experiments and RNA-seq analysis. We also thank Dr. Hai Qi for kindly providing the In vitro conjugation assay protocol.
Funding:
This research was supported by scholarships from the Fellowship for Fundamental Academic Fields provided by Seoul National University, and from the BK21 FOUR education program to Y.-J.K., by a grant from the US National Institutes of Health (NIH) (NIAID P01 AI145815) to S.C., and by the Basic Science Research Program through a National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (NRF-2017R1A2B2008507 and NRF-2019M3C7A1032653), by the Creative-Pioneering Researchers Program through Seoul National University, and by the SNUH Research Fund (Grant No. 320200270) to Y.S.C.
Footnotes
Competing interests: The authors, including S.-H.I, the CEO and major shareholder of ImmunoBiome Inc., declare no competing financial interests.
Data and materials availability:
RNA sequencing data for this study are available in the Gene Expression Omnibus (GEO) repository under accession number GSE220287. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. Requests for research materials generated in this study should be addressed to Y.S.C. (younsoo94@snu.ac.kr).
REFERENCES AND NOTES
- 1.Donnadieu E, Revy P, Trautmann A, Imaging T-cell antigen recognition and comparing immunological and neuronal synapses. Immunology 103, 417–425 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dustin ML, Colman DR, Neural and immunological synaptic relations. Science 298, 785–789 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Yamada S, Nelson WJ, Synapses: sites of cell recognition, adhesion, and functional specification. Annu. Rev. Biochem 76, 267–294 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fooksman DR, Vardhana S, Vasiliver-Shamis G, Liese J, Blair DA, Waite J, Sacristán C, Victora GD, Zanin-Zhorov A, Dustin ML, Functional anatomy of T cell activation and synapse formation. Annu. Rev. Immunol 28, 79–105 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dustin ML, Signaling at neuro/immune synapses. J. Clin. Investig 122, 1149–1155 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalva MB, McClelland AC, Kayser MS, Cell adhesion molecules: signalling functions at the synapse. Nat. Rev. Neurosci 8, 206–220 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt DL, Castillo PE, Synaptic plasticity of NMDA receptors: mechanisms and functional implications. Curr. Opin. Neurobiol 22, 496–508 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Affaticati P, Mignen O, Jambou F, Potier MC, Klingel-Schmitt I, Degrouard J, Peineau S, Gouadon E, Collingridge GL, Liblau R, Capiod T, Cohen-Kaminsky S, Sustained calcium signalling and caspase-3 activation involve NMDA receptors in thymocytes in contact with dendritic cells. Cell Death Differ. 18, 99–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng C-Y, Seabold GK, Horak M, Petralia RS, MAGUKs, synaptic development, and synaptic plasticity. Neuroscientist 17, 493–512 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Round JL, Tomassian T, Zhang M, Patel V, Schoenberger SP, Miceli MC, Dlgh1 coordinates actin polymerization, synaptic T cell receptor and lipid raft aggregation, and effector function in t cells. J. Exp. Med 201, 419–430 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarkar C, Basu B, Chakroborty D, Dasgupta PS, Basu S, The immunoregulatory role of dopamine: an update. Brain Behav. Immun 24, 525–528 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Padhan K, Varma R, Immunological synapse: a multi-protein signalling cellular apparatus for controlling gene expression. Immunology 129, 322–328 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Shea JJ, Paul WE, Mechanisms underlying lineage commitment and plasticity of helper CD4+ t cells. Science 327, 1098–1102 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crotty S, T follicular helper cell biology: a decade of discovery and diseases. Immunity 50, 1132–1148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinuesa CG, Linterman MA, Yu D, MacLennan ICM, Follicular helper T cells. Annu. Rev. Immunol 34, 335–368 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ, Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol. Rev 229, 152–172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimbacher B, Hutloff A, Schlesier M, Glocker E, Warnatz K, Dräger R, Eibel H, Fischer B, Schäffer AA, Mages HW, Kroczek RA, Peter HH, Homozygous loss of ICOS is associated with adult-onset common variable immunodeficiency. Nat. Immunol 4, 261–268 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Victora GD, Nussenzweig MC, Germinal centers. Annu. Rev. Immunol 30, 429–457 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Hügle B, Suchowerskyj P, Hellebrand H, Adler B, Borte M, Sack U, Overberg-Schmidt US, Strnad N, Otto J, Meindl A, Schuster V, Persistent hypogammaglobulinemia following mononucleosis in boys is highly suggestive of X-linked lymphoproliferative disease--report of three cases. J. Clin. Immunol 24, 515–522 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Ma CS, Hare NJ, Nichols KE, Dupré L, Andolfi G, Roncarolo M-G, Adelstein S, Hodgkin PD, Tangye SG, Impaired humoral immunity in X-linked lymphoproliferative disease is associated with defective IL-10 production by CD4+ t cells. J. Clin. Invest 115, 1049–1059 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nichols KE, Harkin DP, Levitz S, Krainer M, Kolquist KA, Genovese C, Bernard A, Ferguson M, Zuo L, Snyder E, Buckler AJ, Wise C, Ashley J, Lovett M, Valentine MB, Look AT, Gerald W, Housman DE, Haber DA, Inactivating mutations in an SH2 domain-encoding gene in X-linked lymphoproliferative syndrome. Proc. Natl. Acad. Sci. U.S.A 95, 13765–13770 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, van Schaik S, Notarangelo L, Geha R, Roncarolo MG, Oettgen H, De Vries JE, Aversa G, Terhorst C, The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature 395, 462–469 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Crotty S, Kersh EN, Cannons J, Schwartzberg PL, Ahmed R, SAP is required for generating long-term humoral immunity. Nature 421, 282–287 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Hron JD, Caplan L, Gerth AJ, Schwartzberg PL, Peng SL, SH2D1A regulates T-dependent humoral autoimmunity. J. Exp. Med 200, 261–266 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cannons JL, Qi H, Lu KT, Dutta M, Gomez-Rodriguez J, Cheng J, Wakeland EK, Germain RN, Schwartzberg PL, Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity 32, 253–265 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN, SAP-controlled T-B cell interactions underlie germinal centre formation. Nature 455, 764–769 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, Schwartzberg PL, Crotty S, The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity 36, 986–1002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, Crotty S, Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J. Immunol 185, 190–202 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linterman MA, Rigby RJ, Wong RK, Yu D, Brink R, Cannons JL, Schwartzberg PL, Cook MC, Walters GD, Vinuesa CG, Follicular helper T cells are required for systemic autoimmunity. J. Exp. Med 206, 561–576 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Youn HD, Sun L, Prywes R, Liu JO, Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science 286, 790–793 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Cole CJ, Mercaldo V, Restivo L, Yiu AP, Sekeres MJ, Han J-H, Vetere G, Pekar T, Ross PJ, Neve RL, Frankland PW, Josselyn SA, MEF2 negatively regulates learning-induced structural plasticity and memory formation. Nat. Neurosci 15, 1255–1264 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Flavell SW, Cowan CW, Kim T-K, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME, Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311, 1008–1012 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Yap E-L, Greenberg ME, Activity-regulated transcription: Bridging the gap between neural activity and behavior. Neuron 100, 330–348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bush JO, Soriano P, Ephrin-B1 regulates axon guidance by reverse signaling through a PDZ-dependent mechanism. Genes Dev. 23, 1586–1599 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papa I, Saliba D, Ponzoni M, Bustamante S, Canete PF, Gonzalez-Figueroa P, McNamara HA, Valvo S, Grimbaldeston M, Sweet RA, Vohra H, Cockburn IA, Meyer-Hermann M, Dustin ML, Doglioni C, Vinuesa CG, TFH-derived dopamine accelerates productive synapses in germinal centres. Nature 547, 318–323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinstein JS, Lezon-Geyda K, Maksimova Y, Craft S, Zhang Y, Su M, Schulz VP, Craft J, Gallagher PG, Global transcriptome analysis and enhancer landscape of human primary T follicular helper and T effector lymphocytes. Blood 124, 3719–3729 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crotty S, Johnston RJ, Schoenberger SP, Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol 11, 114–120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnston RJ, Poholek AC, Ditoro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S, Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325, 1006–1010 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oxenius A, Bachmann MF, Zinkernagel RM, Hengartner H, Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur. J. Immunol 28, 390–400 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Kaji T, Ishige A, Hikida M, Taka J, Hijikata A, Kubo M, Nagashima T, Takahashi Y, Kurosaki T, Okada M, Ohara O, Rajewsky K, Takemori T, Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J. Exp. Med 209, 2079–2097 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, Dong X, Odegard JM, Kaech SM, Dent AL, Crotty S, Craft J, In vivo regulation of Bcl6 and T follicular helper cell development. J. Immunol 185, 313–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malek TR, Castro I, Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity 33, 153–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, Crotty S, ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 34, 932–946 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, Flavell RA, Tian Q, Dong C, Bcl6 expression specifies the T follicular helper cell program in vivo. J. Exp. Med 209, 1841–1852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang Y.-h., Watowich SS, Jetten AM, Tian Q, Dong C, Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity 29, 138–149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKinsey TA, Zhang CL, Olson EN, MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci 27, 40–47 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Seki A, Rutz S, Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J. Exp. Med 215, 985–997 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Black BL, Olson EN, Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol 14, 167–196 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Potthoff MJ, Olson EN, MEF2: a central regulator of diverse developmental programs. Development 134, 4131–4140 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Molkentin JD, Black BL, Martin JF, Olson EN, Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol 16, 2627–2636 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barnden MJ, Allison J, Heath WR, Carbone FR, Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol 76, 34–40 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Koboziev I, Karlsson F, Ostanin DV, Gray L, Davidson M, Zhang S, Grisham MB, Role of LFA-1 in the activation and trafficking of T cells: Implications in the induction of chronic colitis. Inflamm. Bowel Dis 18, 2360–2370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cannons JL, Tangye SG, Schwartzberg PL, SLAM family receptors and SAP adaptors in immunity. Annu. Rev. Immunol 29, 665–705 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Han S, Hathcock K, Zheng B, Kepler TB, Hodes R, Kelsoe G, Cellular interaction in germinal centers. Roles of CD40 ligand and B7–2 in established germinal centers. J. Immunol 155, 556–567 (1995). [PubMed] [Google Scholar]
- 55.Suto A, Kashiwakuma D, Kagami S.-i., Hirose K, Watanabe N, Yokote K, Saito Y, Nakayama T, Grusby MJ, Iwamoto I, Nakajima H, Development and characterization of IL-21-producing CD4+ T cells. J. Exp. Med 205, 1369–1379 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi J, Diao H, Faliti CE, Truong J, Rossi M, Bélanger S, Yu B, Goldrath AW, Pipkin ME, Crotty S, Bcl-6 is the nexus transcription factor of T follicular helper cells via repressor-of-repressor circuits. Nat. Immunol 21, 777–789 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F, CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chew WL, Tabebordbar M, Cheng JKW, Mali P, Wu EY, Ng AHM, Zhu K, Wagers AJ, Church GM, A multifunctional AAV-CRISPR-Cas9 and its host response. Nat. Methods 13, 868–874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Omori Y, Kitamura T, Yoshida S, Kuwahara R, Chaya T, Irie S, Furukawa T, Mef2d is essential for the maturation and integrity of retinal photoreceptor and bipolar cells. Genes Cells 20, 408–426 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Pattison MJ, Naik RJ, Reyskens KMSE, Arthur JSC, Loss of Mef2D function enhances TLR induced IL-10 production in macrophages. Biosci. Rep 40, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dörner T, Giesecke C, Lipsky PE, Mechanisms of B cell autoimmunity in SLE. Arthritis Res. Ther 13, 243–212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farias FHG, Dahlqvist J, Kozyrev SV, Leonard D, Wilbe M, Abramov SN, Alexsson A, Pielberg GR, Hansson-Hamlin H, Andersson G, Tandre K, Bengtsson AA, Sjöwall C, Svenungsson E, Gunnarsson I, Rantapää-Dahlqvist S, Syvänen A-C, Sandling JK, Eloranta M-L, Rönnblom L, Lindblad-Toh K, A rare regulatory variant in the MEF2D gene affects gene regulation and splicing and is associated with a SLE sub-phenotype in Swedish cohorts. Eur. J. Hum. Genet 27, 432–441 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi J-Y, Ho J. H.-e., Pasoto SG, Bunin V, Kim ST, Carrasco S, Borba EF, Gonçalves CR, Costa PR, Kallas EG, Bonfa E, Craft J, Circulating follicular helper-like T cells in systemic lupus erythematosus: association with disease activity. Arthritis Rheumatol. 67, 988–999 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mesin L, Ersching J, Victora GD, Germinal center B cell dynamics. Immunity 45, 471–482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu P, Shih C, Qi H, Ephrin B1-mediated repulsion and signaling control germinal center T cell territoriality and function. Science 356, eaai9264 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Papa I, Vinuesa CG, Synaptic interactions in germinal centers. Front. Immunol 9, 1858 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mayer CT, Gazumyan A, Kara EE, Gitlin AD, Golijanin J, Viant C, Pai J, Oliveira TY, Wang Q, Escolano A, Medina-Ramirez M, Sanders RW, Nussenzweig MC, The microanatomic segregation of selection by apoptosis in the germinal center. Science 358, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Di Giorgio E, Wang L, Xiong Y, Akimova T, Christensen LM, Han R, Samanta A, Trevisanut M, Bhatti TR, Beier UH, Hancock WW, MEF2D sustains activation of effector Foxp3+ Tregs during transplant survival and anticancer immunity. J. Clin. Invest 19, 183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li M, Linseman DA, Allen MP, Meintzer MK, Wang X, Laessig T, Wierman ME, Heidenreich KA, Myocyte enhancer factor 2A and 2D undergo phosphorylation and caspase-mediated degradation during apoptosis of rat cerebellar granule neurons. J. Neurosci 21, 6544–6552 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herglotz J, Unrau L, Hauschildt F, Fischer M, Kriebitzsch N, Alawi M, Indenbirken D, Spohn M, Müller U, Ziegler M, Schuh W, Jäck H-M, Stocking C, Essential control of early B-cell development by Mef2 transcription factors. Blood 127, 572–581 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Brescia P, Schneider C, Holmes AB, Shen Q, Hussein S, Pasqualucci L, Basso K, Dalla-Favera R, MEF2B instructs germinal center development and acts as an oncogene in B cell lymphomagenesis. Cancer Cell 34, 453–465.e459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Assali A, Harrington AJ, Cowan CW, Emerging roles for MEF2 in brain development and mental disorders. Curr. Opin. Neurobiol 59, 49–58 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Youn HD, Liu JO, Cabin1 represses MEF2-dependent Nur77 expression and T cell apoptosis by controlling association of histone deacetylases and acetylases with MEF2. Immunity 13, 85–94 (2000). [DOI] [PubMed] [Google Scholar]
- 74.Ying CY, Dominguez-Sola D, Fabi M, Lorenz IC, Hussein S, Bansal M, Califano A, Pasqualucci L, Basso K, Dalla-Favera R, MEF2B mutations lead to deregulated expression of the oncogene BCL6 in diffuse large B cell lymphoma. Nat. Immunol 14, 1084–1092 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, Crotty S, Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J. Immunol 188, 3734–3744 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hatzi K, Nance JP, Kroenke MA, Bothwell M, Haddad EK, Melnick A, Crotty S, BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J. Exp. Med 212, 539–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Choi YS, Crotty S, Retroviral vector expression in TCR transgenic CD4(+) T cells. Methods Mol. Biol 1291, 49–61 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, Love PE, Peng W, Xue H-H, Crotty S, LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat. Immunol 16, 980–990 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chu C, Wang Y, Zhang X, Ni X, Cao J, Xu W, Dong Z, Yuan P, Wei W, Ma Y, Zhang L, Wu L, Qi H, SAP-regulated T cell-APC adhesion and ligation-dependent and -independent Ly108-CD3ζ interactions. J. Immunol 193, 3860–3871 (2014). [DOI] [PubMed] [Google Scholar]
- 80.Kim CJ, Lee C-G, Jung J-Y, Ghosh A, Hasan SN, Hwang S-M, Kang H, Lee C, Kim G-C, Rudra D, Suh C-H, Im S-H, The transcription factor Ets1 suppresses T follicular helper type 2 cell differentiation to halt the onset of systemic lupus erythematosus. Immunity 49, 1034–1048.e1038 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Expression of the MEF2 (Mef2) transcription factor family members in human and murine CD4 T cell subsets.
Fig. S2. Impaired GC-Tfh differentiation of Sm TCRtg CD4 T cells by ectopic Mef2d expression following gp61-KLH immunization.
Fig. S3. Cognate B cell-dependent Tfh differentiation of CD4 T cells.
Fig. S4. Gene expression analyses for screening potential Mef2d target genes in the regulation of Tfh/GC-Tfh differentiation of CD4 T cells.
Fig. S5. Intracellular SAP expression by human and mouse CD4 T cell subsets.
Fig. S6. Mef2d-mediated negative regulation of antigen-specific B:T conjugate formation.
Fig. S7. IL-21 production of CD4 T cells is impeded by the Mef2d transcription factor.
Fig. S8. CRISPR/Cas9 mediated Mef2d gene disruption promotes GC-Tfh differentiation of OTII TCRtg CD4 T cells with de-repressed SAP expression.
Fig. S9. T cell development and maturation in the Mef2d CKO mice.
Fig. S10. Phenotypic identification of naïve and activated effector CD4 T cells in human peripheral blood and correlation analyses with MEF2D mRNA expression.
Fig. S11. Mef2d- and Mef2b-mediated inhibition of Tfh differentiation may occur in a Bcl6-independent manner.
Fig. S12. Mef2a does not play redundant roles to Mef2d in the regulation of SAP-dependent GC-Tfh differentiation of antigen-specific CD4 T cells after NP-OVA immunization.
Table S1. Information from tonsil donors.
Table S2. Genes regulated by Mef2d in Tfh and non-Tfh cells.
Table S3. Clinical information from SLE patients and healthy individuals.
Table S4. Demographic information from SLE patients and healthy individuals.
Table S5. sgRNA and crRNA sequences and primer sequences used in qPCR and ChIP assays.
Supplementary data file S1. Raw data excel file
Data Availability Statement
RNA sequencing data for this study are available in the Gene Expression Omnibus (GEO) repository under accession number GSE220287. All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. Requests for research materials generated in this study should be addressed to Y.S.C. (younsoo94@snu.ac.kr).


