Abstract
Over the last two decades, cancer researchers have taken the promise offered by the Human Genome Project and have expanded its capacity to use sequencing to identify the genomic alterations that give rise to and sustain individual tumors. This expansion has allowed researchers to identify and target highly recurrent alterations in specific cancer contexts, such as EGFR mutations in non-small cell lung cancer (Lynch et al, N Engl J Med 350:2129–2139, 2004; Sharifnia et al., Proc Natl Acad Sci U S A 111:18661–18666, 2014), BCR-ABL translocations in chronic myeloid leukemia (Deininger, Pharmacol Rev 55:401–423. https://doi.org/10.1124/pr.55.3.4, 2003; Druker et al, N Engl J Med 344. 1038–1042, 2001; Druker et al, N Engl J Med 344:1031–1037. https://doi.org/10.1056/NEJM200104053441401, 2001), or HER2 amplifications in breast cancer (Slamon et al, N Engl J Med 344:783–792. https://doi.org/10.1056/NEJM200103153441101, 2001; Solca et al, Beyond trastuzumab: second-generation targeted therapies for HER-2-positive breast cancer. In: Sibilia M, Zielinski CC, Bartsch R, Grunt TW (eds) Drugs for HER-2-positive breast cancer. Springer, Basel, pp 91–107, 2011). Despite these advances in our capacity to identify the genetic alterations that drive tumor initiation, survival, and proliferation, our ability to target these alterations to provide effective treatment options for patients in need, particularly those with rare or advanced cancers, remains limited (Gould et al, Nat Med 21:431–439. https://doi.org/10.1038/nm.3853, 2015). Patient-derived models of cancer offer one potential mechanism to overcome this barrier between the bench and bedside. Through the development and testing of patient-derived models of cancer, functional genomics efforts can identify tumor-specific drug sensitivities and thereby provide a connection between tumor genetics and effective therapeutics for patients in need of treatment options.
Recognizing that cancer is a multifaceted set of disease states, the development of personalized models of cancer that can be used to compare treatment options, identify tumor-specific vulnerabilities, and guide clinical decision-making has tremendous potential for improving patient outcomes. This chapter will describe a representative set of patient-derived models of cancer, reviewing each of their strengths and weaknesses and highlighting how selecting a model to suit a specific question or context is critical. Each model comes with a unique set of pros and cons, making them more or less appropriate for each specific research or clinical question. As each model can be leveraged to gain new insights into cancer biology, the key to their deployment is to identify the most appropriate model for a specific context, while carefully considering the strengths and limitations of the selected model. When used appropriately, patient-derived models may prove to be the missing link needed to bring the promise of personalized oncology to fruition in the clinic.
Introduction: Why Are Patient-Derived Models Important and Useful?
Most cancer subtypes are complex and heterogeneous in histological presentation, genetic variation, and prognostic outcomes. For the most part, engineered models fail to recapitulate this diversity, and natural processes that underpin this diversity, ultimately creating models that fail to recreate the complexity of the disease states. Patient-derived cancer models were developed in order to more closely recapitulate patient tumors and allow us to capture the specifics of individual tumors. They can be loosely defined as any model of cancer that is developed from patient samples. Cancerous patient tissues and/or cells, as compared to genetically engineered cells or animal models, provide the benefit of having evolved in a patient and thereby having the full complement of genomic alterations acquired over time and driven by unique, environmental pressures present in that patient. Every tumor consists of a unique ratio of tumor cells, immune cells, fibroblasts, extracellular scaffolding, and endothelial cells, all of which interact both physically and molecularly. Unlike traditional two-dimensional cancer cell lines, modern patient-derived models of cancer seek to preserve elements of the genetic profile, cell-cell interactions, and physical components of a given tumor.
Patient-derived models of cancer provide tractable platforms with which researchers can test specific hypotheses. No model fully recapitulates the unique context of a patient’s tumor and, as a general rule, the greater the complexity of a model, the more limited is the number of ways one can perturb and/or evaluate it (Fig. 12.1). For example, two-dimensional cell line models can be plated in hundreds to thousands of replicates for high-throughput assays, but generating the equivalent number of mouse models is not practically feasible. Even the most complex cancer models do not fully recapitulate the native state of tumors, so when using these models, it is critical to account for the ways in which they do and do not faithfully represent the tumor from which they were derived. Additionally, evaluating agents that target tumor-extrinsic factors, such as the vasculature or immune system, is largely futile in simple systems that lack the complexity to evaluate multicellular (not to mention multisystem) therapeutic responses. Interpretation of the data generated in a specific model and any clinically relevant conclusions thereof are limited to the capacity of said model to recapitulate the tumor from which they were derived. Furthermore, limitations can be present in a variety of different elements within a model system as well as in how it is being assessed. In short, it is critical to recognize the strengths and weaknesses of each model, to evaluate the data generated in each model within the context of its specific capacity and limitations, and to design experiments and workflows accordingly [1].
Fig. 12.1.
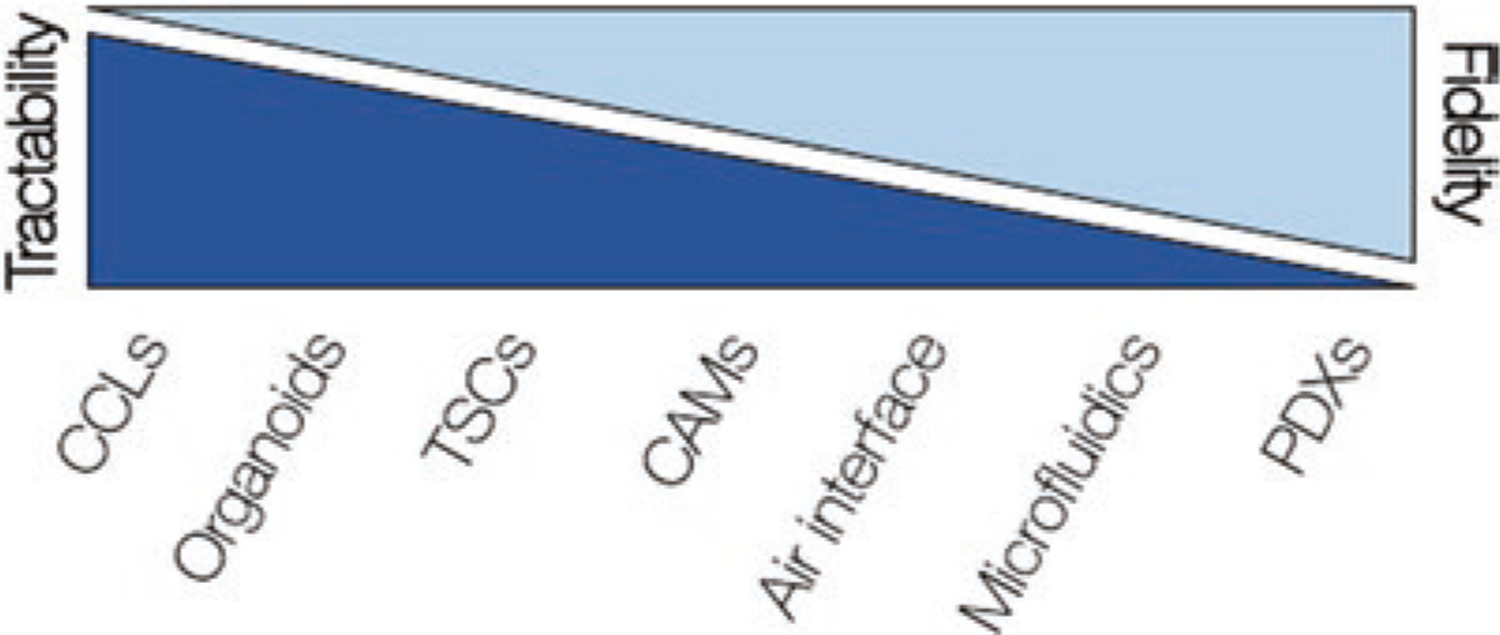
Relative relationship between ease of use in a laboratory setting and fidelity to the original patient across select patient-derived models of cancer. CCLs Cancer cell lines, TSCs tumor slice cultures, CAMs chorioallantoic membrane models, PDXs patient-derived xenografts
For decades and at present, cancer modeling has been dominated by the use of two-dimensional cell lines as representations of different tumor types. For much of this time, the focus was warranted since site of origin and pathology were the best available methods for determining diagnosis and treatment. There are many benefits to working with two-dimensional cell lines, since they provide a relatively stable and largely reproducible platform for experimentation and analysis. Unfortunately, they fail to recapitulate many tumor elements that are critical to therapeutic response, e.g., tumor-stromal interactions and microenvironment [2], restricting researchers’ capacity to directly translate findings from these models into the clinic. Moreover, over time, cell lines evolve to their culture conditions, losing the heterogeneity and features of the cancer of which they were derived.
While patient-derived models can be powerful tools to study individual tumors, our capacity to use them to study the specific impact of a given gene alteration may be limited since models do not have naturally occurring isogenic controls and, rather, represent the accumulation of all of the alterations in a cell rather than an isolated few. Conversely, however, the genetic complexity of these models may be critical for gaining insight into the tumor’s signaling or metabolism or other interactions that play critical role in its sensitivity to therapeutics. One could supplement a patient-derived model with engineered cell lines, e.g., those with targeted oncogenic mutations in genes such as a KRAS G12D [3] or deletions of tumor suppressors such as PTEN [4], which is beneficial when attempting to demonstrate the relationship between a specific alteration and a given phenotype. Recognizing that each tumor is the result of its own unique environment and the selective pressures to which it was exposed, patient-derived models provide a means to assess each tumor individually. When paired with genomics, this information may prove vital to elucidating the complex interplay between genomics and therapeutic response.
With the advent of rapid next-generation sequencing technologies, there has been a shift in the clinic from a singular focus on tissue of origin toward using molecular diagnostics to inform therapeutic strategies. This shift has allowed for the development and use of agents not focused on tumor type but on targeting the genetic events that drive and sustain individual tumors [5–12] and even agents that target recurrence [13]. While actionable mutations can be predictive of therapeutic response, their predictive power is often context-dependent. To better capture how inter-patient physiological and genetic variation contributes to therapeutic responses, it is imperative to both generate new cancer models and expand upon existing models to more accurately recapitulate what is observed in vivo on a molecular, genetic, histologic, and patient level. This will not only allow for discovery of novel interactions but will help in elucidating drug efficacy and possibly even stratifying patients with similar cancer profiles into new therapeutic groups. In order to continue developing agents that target these tumor-specific alterations, there is a need for the development and use of increasingly personalized and higher-fidelity models. In the next decade, diagnostic approaches that incorporate functional genomics have the potential to become a routine part of patient care, whereby direct assessments of drug sensitivity in patient-derived models could provide an avenue for rapid comparisons and personalization of therapeutics prescribed in the clinic [14–16]. Taken together with our increased capacity to sequence tumors to understand the genetic alterations driving and sustaining tumor growth, development, and treatment response, there has been an expeditious development of myriad patient-derived modeling platforms that allow investigators to assess the effects of different treatment modalities on various aspects of patient tumors (Fig. 12.2). Combining physiologically relevant information from complimentary model systems could be the key to designing functional pipelines that allow for personalized therapeutic decision-making in the clinic.
Fig. 12.2.
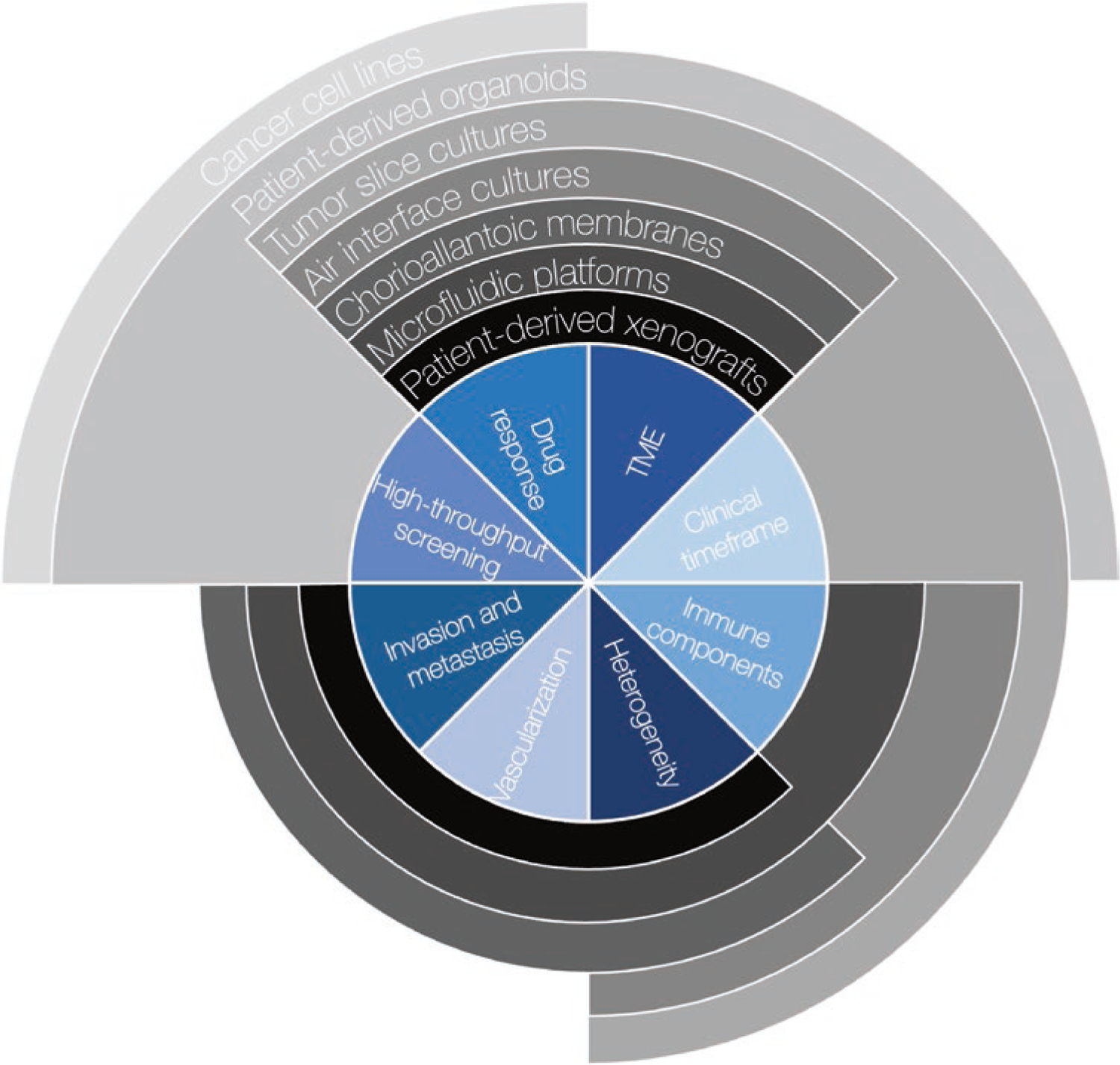
Relative capacity of select patient-derived models of cancer to recapitulate two elements directly related to patient care, high-throughput screening and assessment on a clinical timeframe, and a select six core elements of tumor architecture and proliferation: Ability to quantify response to drugs or other perturbations, tumor microenvironment (TME) factors like stromal cells or extrinsic growth factors, various immune components, intra-tumoral cellular and/or molecular heterogeneity, vascular organization and angiogenesis paradigms, and the ability to invade surrounding tissue or matrix and to metastasize
Types of Models
This section will review several patient-derived models of cancer. It is key to note that each model has its own set of strengths and weaknesses and that their use is predicated on understanding and controlling for these system-specific considerations. For example, patient-derived xenografts in humanized mice may recapitulate more elements of the patient condition, but the feasibility, high cost, long timeline, and variable stability of these models limit the applications of their use. Conversely, patient-derived, two-dimensional tumor cell lines are relatively cheap and tractable but fail to reconstitute many of the elements of the tumor and its microenvironment that are targeted by therapeutics, thus limiting the scope and therefore the questions that can be asked with them.
Two-Dimensional Cancer Cell Lines
Two-dimensional cancer cell lines (CCLs) are a well-established model system for testing small molecules, such as the National Cancer Institute 60 (NCI60) project [17, 18]. Over the decades, thousands of commercially available cancer cell lines have been generated [19] and assessed using high-throughput drug and genetic screens. CCLs can also be established directly from dissociated patient tissue. While not always easy to establish, the strength of these models is in the ease of their propagation and culture. Hundreds to thousands of copies of these lines can be generated in very short timeframes, making them ideal for high-throughput screening. They have been used in pseudo-unselected trials for multiple compounds targeting similar pathways [20] and for pseudo-enrichment trials for drug efficacy in breast cancer [21, 22]. There have also been large-scale efforts by the Dependency Map Consortium (DepMap) to conduct RNAi and CRISPR screens in genomically characterized CCLs to identify cancer type-specific and pan-cancer dependencies [23–26] in conjunction with small molecule screening via the PRISM method that utilizes DNA barcoding to pool cell lines for high-efficiency drug screening [27]. Multiple other groups have also worked to generate publicly available pharmacogenomics datasets, such as the Cancer Cell Line Encyclopedia (CCLE) [28, 29], Genomics of Drug Sensitivity in Cancer (GDSC) [30, 31], and the Cancer Therapeutics Response Portal (CTRP) [32–34].
Established CCLs have been shown to be inconsistent across different laboratories, e.g., in a study of 27 cell lines all labeled as MCF7 (breast cancer), wide genetic variation and response to anticancer therapeutics were recorded [35], but this is an issue inherent in divergent evolution during propagation and variable maintenance practices, which is seen in other in vitro culture systems. As the most broadly used models of cancer, CCLs have been at the center of debates concerning consistency across different datasets, namely, the CCLE and the GDSC. Studies have shown that drug-gene interactions matched between CCLE and GDSC exhibited poor correlation and inconsistencies [36, 37], prompting other groups to join the debate on how best correct for experimental and methodological variation between the original drug screens and subsequent computational analysis [29, 38–43]. In response to these issues, multiple patient-derived tumor modeling platforms, e.g., a customizable Functional Genomics Pipeline [44] and the National Cancer Institute’s Patient-Derived Models Repository [45–47], have both implemented fidelity checks that use a combination of genomics and pathology to ensure model fidelity, and also produced datasets that explain the observed differences in drug sensitivity.
As patient-derived culture systems have been developed for the full gambit of tumor types, it is interesting to note that some tumor types, e.g., liver, are highly amenable to growth in two-dimensional culture while being resistant to growth in three dimensions. As such, two-dimensional culture of patient-derived models still has the potential to play a critical role in both precision oncology and cancer research at large. Besides known issues with inconsistent cell line nomenclature and contamination [48], a key outstanding question for the implementation of patient-derived models is how well they model tumor dynamics or recapitulate clinical cancer vulnerabilities. Despite many large-scale grants and clinical trials being fundamentally anchored by results from screens conducted in CCLs, these models tend to require further validation, as their simplicity, which is key to their broad use, also limits their fidelity. Moreover, cancer cell cultures grown in a monolayer lose their three-dimensional architecture and the resultant intercellular interactions. These changes induce changes in gene and protein expression. Thus, patient-derived models from heterogeneous cancers undergo in vitro selection, potentially altering their fidelity to the tumors from which they were derived.
Patient-Derived Organoids
Patient-derived organoids (PDOs) are generated via the dissociation and subsequent expansion of patient tumor samples. Unlike traditional two-dimensional cultures, PDOs are grown in the context of a matrix, e.g., laminin, to generate three-dimensional models of the tumors from which they were derived. Because these cells are grown in a three-dimensional matrix, the models retain some of the structural features and cellular diversity of the tumors from which they were derived (Table 12.1). By culturing PDOs in conditions that mimic the native environment, investigators are able to recapitulate elements of the primary tumor that are lost in two-dimensional culture, including preserving cell-cell and cell-extracellular matrix interactions.
Table 12.1.
Select publications demonstrating patient-derived organoid development for specific cancer types
The PDO system is a middle-ground approach between more complex models and two-dimensional cultures in that it has the capacity for layered complexity through the addition of other cell types (such as T cells) while still allowing investigators to generate thousands of replicates, which can be evaluated in shorter time frames and across more conditions than complex models. PDOs are also amenable to application in other model systems in which the tumor organoids serve as the base patient-derived model that is then made increasingly complex by the addition of alternative platforms, e.g., air interface models, tumor-immune co-cultures, and chorioallantoic membrane models. Furthermore, one can run concurrent-specific and high-throughput drug screens in mice and in three-dimensional culture (with or without co-cultures), respectively. When PDOs are used in co-cultures and other more complex models, one can assess the intricate interplay between tumors and their micro-and macroenvironments, making them a useful tool for the development of both clinical and pre-clinical pipelines.
Air Interface Cultures
Cultures with an air-liquid interface (ALI) expose three-dimensional organoids to the air rather than encapsulating them in media and allow for long-term propagation of organoids. ALI organoids have been used to study differentiation programs [72], as well as paracrine signaling, and architecture of oncogenically transformed gastrointestinal tissues [73]. This model allows for in vitro profiling of primary tumor epithelium and immune and stromal components from patient biopsies, and it has been shown to accurately model the effects of immunotherapy on endogenous tumor-infiltrating lymphocytes [74]. Non-small cell lung cancer ALI cultures have been used to successfully screen aerosolized drugs, suggesting ALI cultures as a viable alternative to animal studies in regard to studying anticancer drug effects in the respiratory tract and inhalation delivery [75, 76]. These features make ALI models a good fit for studies assessing tumor-immune interactions, but the added complexity of the model increases the barrier to using them at large scale.
Chorioallontoic Membrane Models
Chorioallontoic membrane (CAM) models use the developing chicken embryo as the host into which patient-derived tumor models can be implanted in order to evaluate growth interactions with the vasculature and microenvironment. In contrast to the classic in vivo rodent PDX model, CAMs provide a more tractable and cheaper option that is naturally immunodeficient [78], provide an easily manipulated vascular environment, has relatively reduced maintenance requirements, and requires shorter experimental timelines [79]. CAMs have been used to evaluate nanoparticle drug delivery in ovarian cancer [80], in vivo perineural invasion of head and neck squamous cell carcinoma [81], cancer-associated autophagy programs [82], metastatic capacity of non-small cell lung and prostate cancer cells and screening for putative anti-metastatic drugs [83], and the effects of tumor cell invasion on vascular network structure and stability [83]. While CAMs are relatively tractable compared to PDXs, they are not as easily genetically manipulated, and many reagents, e.g., antibodies or cytokines, are incompatible with the avian model. Despite the aforementioned pitfalls of this model, CAMs can be used to provide insight to invasion studies, elucidate molecular and drug mechanisms, and drug pharmacokinetic and pharmacodynamic studies.
Tumor Slice Cultures
Tumor slice cultures (TSCs) are generated by isolating tumors from patients and creating ex vivo cultures that maintain the composition and orientation of the native tumor microenvironment and extracellular matrix. They are particularly well suited for assessment of the extent to which inter- and intra-tumoral heterogeneity affects the tumor response to therapies. In a TSC, the myriad cell types, vascular networks, and tissue organization from the original tumor are maintained and can be easily imaged and visualized. TSCs are also easily monitored over time, allowing researchers to examine the temporal dynamics of perturbation at the cellular level, a feat that is increasingly complicated in murine models, which cannot be as readily imaged. For example, Minami et al. used real-time and serial imaging and immunohistochemical analysis of drug-treated TSCs from a mouse model of malignant glioma to inform the ways in which organotypic brain slice cultures could be used in testing anti-glioma drugs [84]. Since TSCs maintain the complexity and heterogeneity of the tumor from which they are derived and are rapidly culturable following biopsy, they have potential as personalized preclinical models to stratify individual patients for treatment on a diagnostic timeline. Patient tissue is the limiting reagent in this model, rendering TSCs a low-throughput model. Furthermore, using certain media conditions for each tumor may select for tumor growth over sustaining the growth of other cells (i.e., immune cell populations). Unlike PDXs or cell-based models, these cultures cannot be propagated, only remain viable for limited time windows, and develop abnormal growth kinetics and signaling after approximately 6 days in culture, depending on the tumor type and level of optimization of the culture conditions.
Over time, protocols for slice cultures have evolved to standardize TSC volume and surface area, permitting these models to be used for measuring metabolic activity across TSCs within and across patients, ultimately allowing for quantitative measurements of different biologic activities [85]. Vaira et al. showed that slices taken from human colon, lung, and prostate tumors maintained proliferative capacity and native morphology in culture, and demonstrated reduced proliferation upon treatment with targeted inhibition of Mdm2 and PI3K [86]. Merz et al. demonstrated that patient-derived glioblastoma TSCs recapitulated clinical responses to X-ray, spread-out Bragg-peak carbon irradiation, and temozolomide, suggesting their use in understanding therapeutic effects in glioblastoma and dissecting resistance mechanisms [87]. Similarly, Martin et al. showed that patient-derived TSCs of liver metastases of colorectal cancer could be screened with cetuximab, oxaliplatin, and pembrolizumab in order to identify patient-specific response to these standard of care regimens, suggesting a potential utility for TSCs in personalized oncology [88]. TSCs (ex vivo and from PDX) have also been shown to be a viable system for testing drug efficacy as shown in Table 12.2.
Table 12.2.
Select drug screening experiments in tumor slice culture models
| Cancer type | Tumor source | Drugs tested | Reference |
|---|---|---|---|
| Pancreatic ductal adenocarcinoma | Patient | Staurosporine, cycloheximide | [89] |
| Pancreatic ductal adenocarcinoma | Patient | Rapamycin | [90] |
| Colon, triple-negative breast cancer | PDX PDX |
Staurosporine Panels of FDA-approved drugs |
[91] |
| Colorectal cancer | Patient | Cetuximab, oxaliplatin, and pembrolizumab | [88] |
| Colorectal cancer | Patient | 5-fluorouracil (5-FU) and FOLFOX (5-FU and oxaliplatin) | [92] |
| Breast | Patient | FAC (5-FU, doxorubicin, 4-HC or preactivated cyclophosphamide) | [93] |
| Glioma | Mouse | Cisplatin, temozolomide, paclitaxel, tranilast | [84] |
| Colon, lung, prostate | Human | LY294002 (PI3K inhibitor), Nutlin-3 | [86] |
In addition to their potential use in evaluating tumor-specific drug sensitivities, TSCs have the capacity to stably model the tumor micro- and immune environment. Naipal et al. developed culture methods for breast cancer TSCs that maintained tumor and stromal cell morphological and viability characteristics for up to 7 days [93]. TSC treatment with FAC in decreasing dilutions revealed variation in sensitivity to the chemotherapeutic regimen due to morphological and proliferative capacities as well as prior exposure to neo-adjuvant therapy in vivo [93]. Jiang et al. showed that TSCs of pancreatic ductal adenocarcinoma (PDAC) maintained staining for the stromal component α-smooth muscle actin and infiltrating T cells and macrophages between day 1 and 6 of culture [89], while Misra et al. showed that these PDAC models stably preserve the tumor micro- and immune environment, and cancerous cells maintained their proliferative capacity and recapitulated the differentiation grade of the primary tumor, allowing for pharmacologic screening of heterogeneous patient-derived tissue [90]. Varying dose responses for 5-FU and FOLFOX were evaluated in patient-derived colorectal cancer TSCs, further underscoring the ability of TSCs to capture inter-patient heterogeneity in drug sensitivity [92]. Sivakumar et al. determined that the immune cell composition in TSCs from syngeneic mouse models of pancreatic, breast, and colon cancer, melanoma, and a primary liver tumor sample remained stable over 7 days in culture and that TSCs from PDX models were valuable models for pharmacologic screening [91]. Taken together, TSC is a high-fidelity model that can be used to address drug sensitivity and mechanisms-of-action, metabolic studies, as well as monitor cell-cell molecular and physical interactions within a patient’s tumor, but their use is limited to a relatively small scale and the short time interval for which these cultures remain viable.
Microfluidic Platforms
In microfluidic platforms, synthetic scaffolds made of glass or polymers provide substrate onto which three-dimensional models can be seeded. Three-dimensional microfluidic systems have been used to mimic vascular biology and physiological conditions by flowing fluid through chambers seeded with the cells of interest and monitoring phenotypes of interests [94]. In the cancer context, perfusable microfluidic systems have been shown to recapitulate expected drug toxicities in hepatoblastoma [95], triple-negative breast cancer [96], and head and neck cancer [97], to name a few. These platforms can also be customized to mimic in vivo tumor microenvironment by repopulating decellularized matrix [98], which is an ideal system for pharmacologic screening. Another use case for microfluidic platforms is to test candidate therapeutics against patient-derived single-cell suspensions, which can increase throughput from limited patient sample volume [99]. Lung adenocarcinoma PDX biopsies have been successfully screened with staurosporine in a microfluidic platform, indicating the potential of these systems to maintain physical interactions between cancers and their native tumor microenvironment [100].
Immune checkpoint blockade (ICB) treatment has been tested in murine- and patient-derived organotypic spheroids suspended in perfusable collagen hydrogels, which allows for monitoring of immune cell compositions and profiling of the secretome in response to ICB [101], though this model is restricted to tumor-infiltrating cells and does not address using appropriate immune cell ratios found in the parent patient tissue. Deng et al. showed that patient-derived tumor spheroids in a 3D microfluidic device treated with CDK4/6 inhibitors palbociclib and trilaciclib released increased T-helper 1 cytokines, supporting this model system as a future direction for studying the tumor immune microenvironment ex vivo [102]. Microfluidics have been used to identify factors that influence TCR-engineered T cell efficacy against cancerous hepatocytes [103, 104].
More complex microfluidic “organ-on-a-chip” (OOAC) systems can be used to assess how vascularization affects therapeutic efficacy and delivery in co-cultures of endothelial cells, fibroblasts, and tumor cells [105] and have been used to assess dynamics of nanoparticle intravasation from vessels into tumors [106]. Colorectal and breast cancer cells have been shown to grow around and utilize synthetic vasculature and display clinically relevant responses to anticancer therapies, suggesting that these vascularized micro-organs and micro-tumors could be a promising model for therapeutic testing of angiogenesis inhibitors [107]. However, tumor vasculature has been shown to have varying effects on drug delivery (and even anti-angiogenic medications) due to poorly executed neo-angiogenesis and remodeling [108], which may alter drug efficacy modeled in synthetic vasculature. A breast cancer OOAC system successfully modeled ductal carcinoma in situ and mammary tissue layers that recapitulated clinical response to paclitaxel treatment [109]. In another OOAC study, cancer growth and invasion of non-small cell lung cancer was faithfully modeled, as were the effect of mechanical breathing on vascularization and cancer cell response to tyrosine kinase inhibitor rociletinib [110].
While these microfluidic platforms can be difficult and expensive to develop and maintain, the capacity to customize three-dimensional microfluidic platforms makes them ideal for modeling the complex interactions between cell populations and highlights them as a powerful tool for dissecting the molecular mechanisms underpinning treatment efficacy.
Patient-Derived Xenografts
Patient-derived xenografts (PDXs) are generated through the implantation of patient tumor tissue into mice, thus creating a mammalian model of the patient’s tumor [111]. Once implanted, these tumors have the capacity to grow, establish a blood supply, and interact with the murine host, thereby providing a living mammalian system in which to run analyses. Once established, PDX models can be propagated; however, much like other patient derived models, they can lose their heterogeneity and be subject to further evolution in their new hosts over time as dominant clones take over and the tumor endures subsequent passaging. While early PDXs can retain elements of the tumor microenvironment, e.g., cancer-associated fibroblasts [112, 113], these elements can be lost over time as the tumors continue to evolve in their new environment. In a study of over a thousand PDXs across more than 18 tumor types, it was found that the selective environment of passaging xenografts in the mouse leads to the accumulation of copy number alterations, which puts evolutionary distance between the primary patient and the model [114]. This supports the notion that passage number may be a critical factor in retaining the complexity of PDX models and underscores the need to evaluate model fidelity not only at the time of development but also at the time of use to ensure that key components of the tumor are being recapitulated as expected.
Increased fidelity to the patient of origin may be obtained for some tumor types by altering the site of xenograft implantation. Traditionally, all PDXs regardless of tumor type have been implanted in the flank of their murine hosts, but through orthotopic implantation into the tissue of origin, researchers may produce a more accurate tumor model [115, 116]. By placing the PDX in a context that more closely resembles its native site of initiation, the model can provide contextual feedbacks specific to the site of origin, e.g., air interface in the lung or microbes in the gut. The type of mouse, e.g., athymic nude or NOD-SCID [111], used can also significantly alter the fidelity of these models and the types of questions that can be addressed. While PDXs have been traditionally generated in immune-compromised mice, in the last decade multiple groups have generated increasingly complex “humanized mice” that are engineered to express human genes (or mice transplanted with human monocytes) to allow for modeling of interactions between the tumor and the immune system [117]. Adding yet more complexity, other groups are advancing toward being able to generate personalized murine avatars that have patient-matched tumor and immune components [118], though these models are not stable for extended periods, which limits their capacity to model long-term therapeutic responses.
There are several strengths associated with PDX models for drug testing. They represent the gold standard for preclinical models by providing a system where researchers can evaluate the impact of therapies or other perturbations on patient tumors while also evaluating their effects on other organs in a mammalian system. PDXs can be generated in genetically modified animals to evaluate the role of a specific host gene or protein upon tumor development, progression, or drug response [116]. PDX models can be used concurrently with clinical trials to evaluate the impact of drugs on anti-tumor efficacy and to generate a readout of potential toxicities in critical organs [119–121]. Though PDXs can be incredibly powerful in both model capacity and level of fidelity, it is impractical to generate sizeable cohorts of these models to exhaustively test larger drug libraries. Furthermore, their clinical use is limited by factors such as differences in surgical techniques, take rates (i.e., the fraction of tumors that successfully implant to generate a PDX, which is dependent upon multiple factors such as tumor type and implantation site), and timing (the time to generate a cohort of PDXs from a single patient tumor adequate for testing can be months or years depending on growth rates). One could use PDXs to address the efficacy and toxicity of drugs on a patient tumor and organ systems, and supplement this experiment with both genetically similar murine allografts and human xenografts to incorporate the immune system effects and increase the speed and power of the drug study. All together, these features limit the speed at which data from PDX models can be generated and used to identify treatment options for the patient from which they were derived but make them a useful tool for the development of preclinical data sets that can be used to support clinical trials (Box 12.1).
Box 12.1.
While not directly derived from patient tissue, it is worth noting that the power of fly genetics allows researchers to design complex Drosophila (fly) avatars that represent the individual genetics of a given patient with >15 putative driver mutations. While fly avatars do not provide a mammalian environment, the genomic drivers present in a patient’s cancer are modeled in Drosophila hindguts, which can then be screened with candidate therapeutics to assess drug toxicity and efficacy in vivo and to quantify animal survival rates in a clinically relevant timeline of 3 weeks [122, 123]. While the fidelity of these models to the initial patient tumor is much lower than that of PDXs, this avatar system is similar in that it can be read out in terms of survival, allowing researchers to gauge tumor-specific treatment efficacy as compared to generic toxicity. Unlike PDXs, these models can be generated and evaluated in clinically relevant time frames and on a sufficiently large scale to allow for broader testing of drug libraries, and ultimately, generating data that can be used to guide clinical decision-making [124].
Conclusion
Patient-derived models of cancer have the potential to inform novel therapeutic options and elucidate the complex genetics and multifactorial intercellular interactions underlying cancer phenotypes. Depending on the specific research question, clinical context, or use case, and desired time frame, all patient-derived models have the potential to be the “correct” model, especially when more than one model is used in an orthogonal way or to approach different aspects of the same question. Key features to consider that were touched on in this chapter are time, cost, scale, and fidelity (Table 12.3). While genomics has rapidly expanded our understanding of tumorigenesis and tumor maintenance, the careful application of appropriate patient-derived models could provide a path to truly personalized oncology by providing platforms to understand the complex interplay between tumors, their environment, and therapeutic sensitivities.
Table 12.3.
Comparison of features of different patient-derived models of cancer
| Model | Heterogeneity | Model complexity | Tumor microenvironment | Immune system components | Renewability | Screening capacity | Screening throughput |
|---|---|---|---|---|---|---|---|
| 2-D cell lines | Little to none unless co-cultured | Large preexisting banks can be established from other models or directly form patients | None | None | Can be passaged and banked for future use | Single and combination agent | High |
| Organoids | Is lost over time, cellular diversity can be enforced through co-culture approaches | Establishment requires customization of culture conditions | TME can be engineered | None | Can be passaged and banked for future use | Single and combination agent | High |
| Air interface | Low to high | Complex experimental system that requires specialized equipment | TME from patient is maintained | Patient tumor-infiltrating immune cells are maintained | Can be passaged and banked for future use | ICB, single and combination agent | Low |
| CAM | Low to high | Easy to establish | TME from patient is maintained | Patient tumor-infiltrating immune cells are maintained | Can be regenerated with new eggs and cells from culture | Single and combination agent | Medium |
| TSC | High | Easy to establish | TME from patient is maintained | Patient tumor-infiltrating immune cells are maintained | Non-renewable | Single and combination agent | Low |
| Microfluidic co-cultures | Low to high | Complex experimental system that requires specialized equipment | TME is engineered | Immune complexity is engineered | Non-renewable | ICB, single and combination agent | Medium |
| PDX | High | Labor intensive to establish and grow out | TME from patient is partially maintained | None | Renewable as long as you have mice | ICB, CAR-T, single and combination agent | Low |
References
- 1.Gould SE, Junttila MR, de Sauvage FJ. Translational value of mouse models in oncology drug development. Nat Med. 2015;21:431–9. 10.1038/nm.3853. [DOI] [PubMed] [Google Scholar]
- 2.Klemm F, Joyce JA. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol. 2015;25:198–213. 10.1016/j.tcb.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, Frese KK, DeNicola G, Feig C, Combs C, Winter SP, Ireland-Zecchini H, Reichelt S, Howat WJ, Chang A, Dhara M, Wang L, Rückert F, Grützmann R, Pilarsky C, Izeradjene K, Hingorani SR, Huang P, Davies SE, Plunkett W, Egorin M, Hruban RH, Whitebread N, McGovern K, Adams J, Iacobuzio-Donahue C, Griffiths J, Tuveson DA. Inhibition of hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–61. 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, Cordon-Cardo C, Catoretti G, Fisher PE, Parsons R. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci. 1999;96:1563–8. 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sos ML, Michel K, Zander T, Weiss J, Frommolt P, Peifer M, Li D, Ullrich R, Koker M, Fischer F, Shimamura T, Rauh D, Mermel C, Fischer S, Stückrath I, Heynck S, Beroukhim R, Lin W, Winckler W, Shah K, LaFramboise T, Moriarty WF, Hanna M, Tolosi L, Rahnenführer J, Verhaak R, Chiang D, Getz G, Hellmich M, Wolf J, Girard L, Peyton M, Weir BA, Chen T-H, Greulich H, Barretina J, Shapiro GI, Garraway LA, Gazdar AF, Minna JD, Meyerson M, Wong K-K, Thomas RK. Predicting drug susceptibility of non–small cell lung cancers based on genetic lesions. J Clin Invest. 2009;119:1727–40. 10.1172/JCI37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch TJ, Well DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 7.Sharifnia T, Rusu V, Piccioni F, Bagul M, Imielinski M, Cherniack AD, Pedamallu CS, Wong B, Wilson FH, Garraway LA, Altshuler D, Golub TR, Root DE, Subramanian A, Meyerson M. Genetic modifiers of EGFR dependence in non-small cell lung cancer. Proc Natl Acad Sci U S A. 2014;111:18661–6. 10.1073/pnas.1412228112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deininger MWN. Specific targeted therapy of chronic myelogenous leukemia with imatinib. Pharmacol Rev. 2003;55:401–23. 10.1124/pr.55.3.4. [DOI] [PubMed] [Google Scholar]
- 9.Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, Capdeville R, Talpaz M. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–42. 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 10.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, Sawyers CL. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–7. 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 11.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–92. 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 12.Solca FF, Adolf GR, Jones H, Uttenreuther-Fischer MM. Beyond trastuzumab: second-generation targeted therapies for HER-2-positive breast cancer. In: Sibilia M, Zielinski CC, Bartsch R, Grunt TW, editors. Drugs for HER-2-positive breast cancer. Basel: Springer; 2011. p. 91–107. 10.1007/978-3-0346-0094-1_6. [DOI] [Google Scholar]
- 13.Shaw AT, Friboulet L, Leshchiner I, Gainor JF, Bergqvist S, Brooun A, Burke BJ, Deng Y-L, Liu W, Dardaei L, Frias RL, Schultz KR, Logan J, James LP, Smeal T, Timofeevski S, Katayama R, Iafrate AJ, Le L, McTigue M, Getz G, Johnson TW, Engelman JA. Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N Engl J Med. 2016;374:54–61. 10.1056/NEJMoa1508887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johannessen CM, Boehm JS. Progress towards precision functional genomics in cancer. Curr Opin Syst Biol. 2017;2:74–83. 10.1016/j.coisb.2017.02.002. [DOI] [Google Scholar]
- 15.Letai A. Functional precision cancer medicine—moving beyond pure genomics. Nat Med. 2017;23:1028–35. 10.1038/nm.4389. [DOI] [PubMed] [Google Scholar]
- 16.Tyner JW. Integrating functional genomics to accelerate mechanistic personalized medicine. Cold Spring Harb Mol Case Stud. 2017;3:a001370. 10.1101/mcs.a001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alley MC, Scudiere DA, Monks A, Hursey ML, Czerwinski MJ, Fine DL, Abbott BJ, Mayo JG, Shoemaker RH, Boyd MR. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988;48:589–601. [PubMed] [Google Scholar]
- 18.Stinson S, Alley M, Kopp W, Fiebig H, Mullendore L, Pittman A, Kenney S, Keller J, Boyd M. Morphological and immunocytochemical characteristics of human tumor cell lines for use in a disease-oriented anticancer drug screen. Anticancer Res. 1992;12:1035–53. [PubMed] [Google Scholar]
- 19.Bairoch A. The cellosaurus, a cell-line knowledge resource. J Biomol Tech. 2018;29:25–38. 10.7171/jbt.18-2902-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greshock J, Bachman KE, Degenhardt YY, Jing J, Wen YH, Eastman S, McNeil E, Moy C, Wegrzyn R, Auger K, Hardwicke MA, Wooster R. Molecular target class is predictive of in vitro response profile. Cancer Res. 2010;70:3677–86. 10.1158/0008-5472.CAN-09-3788. [DOI] [PubMed] [Google Scholar]
- 21.Heiser LM, Sadanandam A, Kuo W-L, Benz SC, Goldstein TC, Ng S, Gibb WJ, Wang NJ, Ziyad S, Tong F, Bayani N, Hu Z, Billig JI, Dueregger A, Lewis S, Jakkula L, Korkola JE, Durinck S, Pepin F, Guan Y, Purdom E, Neuvial P, Bengtsson H, Wood KW, Smith PG, Vassilev LT, Hennessy BT, Greshock J, Bachman KE, Hardwicke MA, Park JW, Marton LJ, Wolf DM, Collisson EA, Neve RM, Mills GB, Speed TP, Feiler HS, Wooster RF, Haussler D, Stuart JM, Gray JW, Spellman PT. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A. 2012;109:2724–9. 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, Virtanen C, Bradner JE, Bader GD, Mills GB, Pe’er D, Moffat J, Neel BG. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 2016;164:293–309. 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald ER, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, Belur D, Castelletti D, Frias E, Gampa K, Golji J, Kao I, Li L, Megel P, Perkins TA, Ramadan N, Ruddy DA, Silver SJ, Sovath S, Stump M, Weber O, Widmer R, Yu J, Yu K, Yue Y, Abramowski D, Ackley E, Barrett R, Berger J, Bernard JL, Billig R, Brachmann SM, Buxton F, Caothien R, Caushi JX, Chung FS, Cortés-Cros M, deBeaumont RS, Delaunay C, Desplat A, Duong W, Dwoske DA, Eldridge RS, Farsidjani A, Feng F, Feng J, Flemming D, Forrester W, Galli GG, Gao Z, Gauter F, Gibaja V, Haas K, Hattenberger M, Hood T, Hurov KE, Jagani Z, Jenal M, Johnson JA, Jones MD, Kapoor A, Korn J, Liu J, Liu Q, Liu S, Liu Y, Loo AT, Macchi KJ, Martin T, McAllister G, Meyer A, Mollé S, Pagliarini RA, Phadke T, Repko B, Schouwey T, Shanahan F, Shen Q, Stamm C, Stephan C, Stucke VM, Tiedt R, Varadarajan M, Venkatesan K, Vitari AC, Wallroth M, Weiler J, Zhang J, Mickanin C, Myer VE, Porter JA, Lai A, Bitter H, Lees E, Keen N, Kauffmann A, Stegmeier F, Hofmann F, Schmelzle T, Sellers WR. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell. 2017;170:577–592.e10. 10.1016/j.cell.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 24.McFarland JM, Ho ZV, Kugener G, Dempster JM, Montgomery PG, Bryan JG, Krill-Burger JM, Green TM, Vazquez F, Boehm JS, Golub TR, Hahn WC, Root DE, Tsherniak A. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun. 2018;9:1–13. 10.1038/s41467-018-06916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, Goodale A, Lee Y, Ali LD, Jiang G, Lubonja R, Harrington WF, Strickland M, Wu T, Hawes DC, Zhivich VA, Wyatt MR, Kalani Z, Chang JJ, Okamoto M, Stegmaier K, Golub TR, Boehm JS, Vazquez F, Root DE, Hahn WC, Tsherniak A. Computational correction of copy-number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49:1779–84. 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, Meyers RM, Ali L, Goodale A, Lee Y, Jiang G, Hsiao J, Gerath WFJ, Howell S, Merkel E, Ghandi M, Garraway LA, Root DE, Golub TR, Boehm JS, Hahn WC. Defining a cancer dependency map. Cell. 2017;170:564–576. e16. 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu C, Mannan AM, Yvone GM, Ross KN, Zhang Y-L, Marton MA, Taylor BR, Crenshaw A, Gould JZ, Tamayo P, Weir BA, Tsherniak A, Wong B, Garraway LA, Shamji AF, Palmer MA, Foley MA, Winckler W, Schreiber SL, Kung AL, Golub TR. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat Biotechnol. 2016;34:419–23. 10.1038/nbt.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jané-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Cell Line Encyclopedia Consortium, Genomics of Drug Sensitivity in Cancer Consortium. Pharmacogenomic agreement between two cancer cell line data sets. Nature. 2015;528:84–7. 10.1038/nature15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, Iorio F, Surdez D, Chen L, Milano RJ, Bignell GR, Tam AT, Davies H, Stevenson JA, Barthorpe S, Lutz SR, Kogera F, Lawrence K, McLaren-Douglas A, Mitropoulos X, Mironenko T, Thi H, Richardson L, Zhou W, Jewitt F, Zhang T, O’Brien P, Boisvert JL, Price S, Hur W, Yang W, Deng X, Butler A, Choi HG, Chang JW, Baselga J, Stamenkovic I, Engelman JA, Sharma SV, Delattre O, Saez-Rodriguez J, Gray NS, Settleman J, Futreal PA, Haber DA, Stratton MR, Ramaswamy S, McDermott U, Benes CH. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, Ramaswamy S, Futreal PA, Haber DA, Stratton MR, Benes C, McDermott U, Garnett MJ. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2012;41:D955–61. 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, Ebright RY, Stewart ML, Ito D, Wang S, Bracha AL, Liefeld T, Wawer M, Gilbert JC, Wilson AJ, Stransky N, Kryukov GV, Dancik V, Barretina J, Garraway LA, Hon CS, Munoz B, Bittker JA, Stockwell BR, Khabele D, Stern AM, Clemons PA, Shamji AF, Schreiber SL. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–61. 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S, Javaid S, Coletti ME, Jones VL, Bodycombe NE, Soule CK, Alexander B, Li A, Montgomery P, Kotz JD, Hon CS-Y, Munoz B, Liefeld T, Dančík V, Haber DA, Clish CB, Bittker JA, Palmer M, Wagner BK, Clemons PA, Shamji AF, Schreiber SL. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016;12:109–16. 10.1038/nchembio.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, Jones V, Bodycombe NE, Soule CK, Gould J, Alexander B, Li A, Montgomery P, Wawer MJ, Kuru N, Kotz JD, Hon CS, Munoz B, Liefeld T, Dančík V, Bittker JA, Palmer M, Bradner JE, Shamji AF, Clemons PA, Schreiber SL. Harnessing connectivity in a large-scale small-molecule sensitivity dataset. Cancer Discov. 2015;5:1210–23. 10.1158/2159-8290.CD-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben-David U, Siranosian B, Ha G, Tang H, Oren Y, Hinohara K, Strathdee CA, Dempster J, Lyons NJ, Burns R, Nag A, Kugener G, Cimini B, Tsvetkov P, Maruvka YE, O’Rourke R, Garrity A, Tubelli AA, Bandopadhayay P, Tsherniak A, Vazquez F, Wong B, Birger C, Ghandi M, Thorner AR, Bittker JA, Meyerson M, Getz G, Beroukhim R, Golub TR. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560:325–30. 10.1038/s41586-018-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJWL, Quackenbush J. Inconsistency in large pharmacogenomic studies. Nature. 2013;504:389–93. 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jang IS, Neto EC, Guinney J, Friend SH, Margolin AA. Systematic assessment of analytical methods for drug sensitivity prediction from cancer cell line data. Pac Symp Biocomput. 2014:63–74. [PMC free article] [PubMed] [Google Scholar]
- 38.Geeleher P, Cox NJ, Huang RS. Cancer biomarker discovery is improved by accounting for variability in general levels of drug sensitivity in pre-clinical models. Genome Biol. 2016;17:190. 10.1186/s13059-016-1050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geeleher P, Gamazon ER, Seoighe C, Cox NJ, Huang RS. Consistency in large pharmacogenomic studies. Nature. 2016;540:E1–2. 10.1038/nature19838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hatzis C, Bedard PL, Juul Birkbak N, Beck AH, Aerts HJWL, Stern DF, Shi L, Clarke R, Quackenbush J, Haibe-Kains B. Enhancing reproducibility in cancer drug screening: how do we move forward? Cancer Res. 2014;74:4016–23. 10.1158/0008-5472.CAN-14-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haverty PM, Lin E, Tan J, Yu Y, Lam B, Lianoglou S, Neve RM, Martin S, Settleman J, Yauch RL, Bourgon R. Reproducible pharmacogenomic profiling of cancer cell line panels. Nature. 2016;533:333–7. 10.1038/nature17987. [DOI] [PubMed] [Google Scholar]
- 42.Safikhani Z, El-Hachem N, Smirnov P, Freeman M, Goldenberg A, Birkbak NJ, Beck AH, Aerts HJWL, Quackenbush J, Haibe-Kains B. Safikhani et al. reply. Nature. 2016;540:E2–4. 10.1038/nature19839. [DOI] [PubMed] [Google Scholar]
- 43.Safikhani Z, Smirnov P, Freeman M, El-Hachem N, She A, Rene Q, Goldenberg A, Birkbak NJ, Hatzis C, Shi L, Beck AH, Aerts HJWL, Quackenbush J, Haibe-Kains B. Revisiting inconsistency in large pharma cogenomic studies. F1000Research. 2017;5:2333. 10.12688/f1000research.9611.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pauli C, Hopkins BD, Prandi D, Shaw R, Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, McNary TJ, Churakova Y, Cheung C, Triscott J, Pisapia D, Rao R, Mosquera JM, Robinson B, Faltas BM, Emerling BE, Gadi VK, Bernard B, Elemento O, Beltran H, Demichelis F, Kemp CJ, Grandori C, Cantley LC, Rubin MA. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7:462–77. 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evrard YA, Srivastava A, Randjelovic J, Doroshow JH, Dean DA, Morris JS, Chuang JH. Systematic establishment of robustness and standards in patient-derived xenograft experiments and analysis. Cancer Res. 2020;80:2286–97. 10.1158/0008-5472.CAN-19-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meehan TF, Conte N, Goldstein T, Inghirami G, Murakami MA, Brabetz S, Gu Z, Wiser JA, Dunn P, Begley DA, Krupke DM, Bertotti A, Bruna A, Brush MH, Byrne AT, Caldas C, Christie AL, Clark DA, Dowst H, Dry JR, Doroshow JH, Duchamp O, Evrard YA, Ferretti S, Frese KK, Goodwin NC, Greenawalt D, Haendel MA, Hermans E, Houghton PJ, Jonkers J, Kemper K, Khor TO, Lewis MT, Lloyd KCK, Mason J, Medico E, Neuhauser SB, Olson JM, Peeper DS, Rueda OM, Seong JK, Trusolino L, Vinolo E, Wechsler-Reya RJ, Weinstock DM, Welm A, Weroha SJ, Amant F, Pfister SM, Kool M, Parkinson H, Butte AJ, Bult CJ. PDX-MI: minimal information for patient-derived tumor xenograft models. Cancer Res. 2017;77:e62–6. 10.1158/0008-5472.CAN-17-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tatum JL, Kalen JD, Jacobs PM, Ileva LV, Riffle LA, Hollingshead MG, Doroshow JH. A spontaneously metastatic model of bladder cancer: imaging characterization. J Transl Med. 2019;17:425. 10.1186/s12967-019-02177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu M, Selvaraj SK, Liang-Chu MMY, Aghajani S, Busse M, Yuan J, Lee G, Peale F, Klijn C, Bourgon R, Kaminker JS, Neve RM. A resource for cell line authentication, annotation and quality control. Nature. 2015;520:307–11. 10.1038/nature14397. [DOI] [PubMed] [Google Scholar]
- 49.Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, Chua CW, Barlow LJ, Kandoth C, Williams AB, Bergren SK, Pietzak EJ, Anderson CB, Benson MC, Coleman JA, Taylor BS, Abate-Shen C, McKiernan JM, Al-Ahmadie H, Solit DB, Shen MM. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell. 2018;173:515–528.e17. 10.1016/j.cell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullenders J, de Jongh E, Brousali A, Roosen M, Blom JPA, Begthel H, Korving J, Jonges T, Kranenburg O, Meijer R, Clevers HC. Mouse and human urothelial cancer organoids: a tool for bladder cancer research. Proc Natl Acad Sci. 2019;116:4567–74. 10.1073/pnas.1803595116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ballabio C, Anderle M, Gianesello M, Lago C, Miele E, Cardano M, Aiello G, Piazza S, Caron D, Gianno F, Ciolfi A, Pedace L, Mastronuzzi A, Tartaglia M, Locatelli F, Ferretti E, Giangaspero F, Tiberi L. Modeling medulloblastoma in vivo and with human cerebellar organoids. Nat Commun. 2020;11:583. 10.1038/s41467-019-13989-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linkous A, Balamatsias D, Snuderl M, Edwards L, Miyaguchi K, Milner T, Reich B, Cohen-Gould L, Storaska A, Nakayama Y, Schenkein E, Singhania R, Cirigliano S, Magdeldin T, Lin Y, Nanjangud G, Chadalavada K, Pisapia D, Liston C, Fine HA. Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep. 2019;26:3203–3211. e5. 10.1016/j.celrep.2019.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H, Korving J, van Boxtel R, Duarte AA, Lelieveld D, van Hoeck A, Ernst RF, Blokzijl F, Nijman IJ, Hoogstraat M, van de Ven M, Egan DA, Zinzalla V, Moll J, Boj SF, Voest EE, Wessels L, van Diest PJ, Rottenberg S, Vries RGJ, Cuppen E, Clevers H. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–386.e10. 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 54.Mazzucchelli S, Piccotti F, Allevi R, Truffi M, Sorrentino L, Russo L, Agozzino M, Signati L, Bonizzi A, Villani L, Corsi F. Establishment and morphological characterization of patient-derived organoids from breast cancer. Biol Proced Online. 2019;21:1–3. 10.1186/s12575-019-0099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldhammer N, Kim J, Timmermans-Wielenga V, Petersen OW. Characterization of organoid cultured human breast cancer. Breast Cancer Res. 2019;21:141. 10.1186/s13058-019-1233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, Ohta Y, Toshimitsu K, Nakazato Y, Kawasaki K, Uraoka T, Watanabe T, Kanai T, Sato T. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18:827–38. 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Narasimhan V, Wright JA, Churchill M, Wang T, Rosati R, Lannagan TRM, Vrbanac L, Richardson AB, Kobayashi H, Price T, Tye GXY, Marker J, Hewett PJ, Flood MP, Pereira S, Whitney GA, Michael M, Tie J, Mukherjee S, Grandori C, Heriot AG, Worthley DL, Ramsay RG, Woods SL. Medium-throughput drug screening of patient-derived organoids from colorectal peritoneal metastases to direct personalized therapy. Clin Cancer Res. 2020;26:3662–70. 10.1158/1078-0432.CCR-20-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otte J, Dizdar L, Behrens B, Goering W, Knoefel WT, Wruck W, Stoecklein NH, Adjaye J. FGF signalling in the self-renewal of colon cancer organoids. Sci Rep. 2019;9:17365. 10.1038/s41598-019-53907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roerink SF, Sasaki N, Lee-Six H, Young MD, Alexandrov LB, Behjati S, Mitchell TJ, Grossmann S, Lightfoot H, Egan DA, Pronk A, Smakman N, van Gorp J, Anderson E, Gamble SJ, Alder C, van de Wetering M, Campbell PJ, Stratton MR, Clevers H. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. 2018;556:457–62. 10.1038/s41586-018-0024-3. [DOI] [PubMed] [Google Scholar]
- 60.Verissimo CS, Overmeer RM, Ponsioen B, Drost J, Mertens S, Verlaan-Klink I, van Gerwen B, van der Ven M, van de Wetering M, Egan DA, Bernards R, Clevers H, Bos JL, Snippert HJ. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. eLife. 2016;5:e18489. 10.7554/eLife.18489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R, Rata M, Koh D-M, Tunariu N, Collins D, Hulkki-Wilson S, Ragulan C, Spiteri I, Moorcraft SY, Chau I, Rao S, Watkins D, Fotiadis N, Bali M, Darvish-Damavandi M, Lote H, Eltahir Z, Smyth EC, Begum R, Clarke PA, Hahne JC, Dowsett M, de Bono J, Workman P, Sadanandam A, Fassan M, Sansom OJ, Eccles S, Starling N, Braconi C, Sottoriva A, Robinson SP, Cunningham D, Valeri N. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–6. 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan HHN, Siu HC, Law S, Ho SL, Yue SSK, Tsui WY, Chan D, Chan AS, Ma S, Lam KO, Bartfeld S, Man AHY, Lee BCH, Chan ASY, Wong JWH, Cheng PSW, Chan AKW, Zhang J, Shi J, Fan X, Kwong DLW, Mak TW, Yuen ST, Clevers H, Leung SY. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. 2018;23:882–897.e11. 10.1016/j.stem.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 63.Schutgens F, Rookmaaker MB, Margaritis T, Rios A, Ammerlaan C, Jansen J, Gijzen L, Vormann M, Vonk A, Viveen M, Yengej FY, Derakhshan S, de Winter-de Groot KM, Artegiani B, van Boxtel R, Cuppen E, Hendrickx APA, van den Heuvel-Eibrink MM, Heitzer E, Lanz H, Beekman J, Murk J-L, Masereeuw R, Holstege F, Drost J, Verhaar MC, Clevers H. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat Biotechnol. 2019;37:303–13. 10.1038/s41587-019-0048-8. [DOI] [PubMed] [Google Scholar]
- 64.Kim M, Mun H, Sung CO, Cho EJ, Jeon H-J, Chun S-M, Jung DJ, Shin TH, Jeong GS, Kim DK, Choi EK, Jeong S-Y, Taylor AM, Jain S, Meyerson M, Jang SJ. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat Commun. 2019;10:3991. 10.1038/s41467-019-11867-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z, Qian Y, Li W, Liu L, Yu L, Liu X, Wu G, Wang Y, Luo W, Fang F, Liu Y, Song F, Cai Z, Chen W, Huang W. Human lung adenocarcinoma-derived organoid models for drug screening. iScience. 2020;23:101411. 10.1016/j.isci.2020.101411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hill SJ, Decker B, Roberts EA, Horowitz NS, Muto MG, Worley MJ, Feltmate CM, Nucci MR, Swisher EM, Nguyen H, Yang C, Morizane R, Kochupurakkal BS, Do KT, Konstantinopoulos PA, Liu JF, Bonventre JV, Matulonis UA, Shapiro GI, Berkowitz RS, Crum CP, D’Andrea AD. Prediction of DNA repair inhibitor response in short-term patient-derived ovarian cancer organoids. Cancer Discov. 2018;8:1404–21. 10.1158/2159-8290.CD-18-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kopper O, de Witte CJ, Lõhmussaar K, Valle-Inclan JE, Hami N, Kester L, Balgobind AV, Korving J, Proost N, Begthel H, van Wijk LM, Revilla SA, Theeuwsen R, van de Ven M, van Roosmalen MJ, Ponsioen B, Ho VWH, Neel BG, Bosse T, Gaarenstroom KN, Vrieling H, Vreeswijk MPG, van Diest PJ, Witteveen PO, Jonges T, Bos JL, van Oudenaarden A, Zweemer RP, Snippert HJG, Kloosterman WP, Clevers H. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat Med. 2019;25:838–49. 10.1038/s41591-019-0422-6. [DOI] [PubMed] [Google Scholar]
- 68.Nelson SR, Zhang C, Roche S, O’Neill F, Swan N, Luo Y, Larkin A, Crown J, Walsh N. Modelling of pancreatic cancer biology: transcriptomic signature for 3D PDX-derived organoids and primary cell line organoid development. Sci Rep. 2020;10:2778. 10.1038/s41598-020-59368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tiriac H, Belleau P, Engle DD, Plenker D, Deschênes A, Somerville TDD, Froeling FEM, Burkhart RA, Denroche RE, Jang G-H, Miyabayashi K, Young CM, Patel H, Ma M, LaComb JF, Palmaira RLD, Javed AA, Huynh JC, Johnson M, Arora K, Robine N, Shah M, Sanghvi R, Goetz AB, Lowder CY, Martello L, Driehuis E, LeComte N, Askan G, Iacobuzio-Donahue CA, Clevers H, Wood LD, Hruban RH, Thompson E, Aguirre AJ, Wolpin BM, Sasson A, Kim J, Wu M, Bucobo JC, Allen P, Sejpal DV, Nealon W, Sullivan JD, Winter JM, Gimotty PA, Grem JL, DiMaio DJ, Buscaglia JM, Grandgenett PM, Brody JR, Hollingsworth MA, O’Kane GM, Notta F, Kim E, Crawford JM, Devoe C, Ocean A, Wolfgang CL, Yu KH, Li E, Vakoc CR, Hubert B, Fischer SE, Wilson JM, Moffitt R, Knox J, Krasnitz A, Gallinger S, Tuveson DA. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Res. 2018;19:1112–29. 10.1158/2159-8290.CD-18-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, Clevers H. Organoid culture systems for prostate epithelial tissue and prostate cancer tissue. Nat Protoc. 2016;11:347–58. 10.1038/nprot.2016.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Puca L, Bareja R, Prandi D, Shaw R, Benelli M, Karthaus WR, Hess J, Sigouros M, Donoghue A, Kossai M, Gao D, Cyrta J, Sailer V, Vosoughi A, Pauli C, Churakova Y, Cheung C, Deonarine LD, McNary TJ, Rosati R, Tagawa ST, Nanus DM, Mosquera JM, Sawyers CL, Chen Y, Inghirami G, Rao RA, Grandori C, Elemento O, Sboner A, Demichelis F, Rubin MA, Beltran H. Patient derived organoids to model rare prostate cancer phenotypes. Nat Commun. 2018;9:2404. 10.1038/s41467-018-04495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR, Kuo CJ. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–6. 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li X, Nadauld L, Ootani A, Corney DC, Pai RK, Gevaert O, Cantrell MA, Rack PG, Neal JT, Chan CW-M, Yeung T, Gong X, Yuan J, Wilhelmy J, Robine S, Attardi LD, Plevritis SK, Hung KE, Chen C-Z, Ji HP, Kuo CJ. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat Med. 2014;20:769–77. 10.1038/nm.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, Liu IH, Chiou S-H, Salahudeen AA, Smith AR, Deutsch BC, Liao L, Zemek AJ, Zhao F, Karlsson K, Schultz LM, Metzner TJ, Nadauld LD, Tseng Y-Y, Alkhairy S, Oh C, Keskula P, Mendoza-Villanueva D, De La Vega FM, Kunz PL, Liao JC, Leppert JT, Sunwoo JB, Sabatti C, Boehm JS, Hahn WC, Zheng GXY, Davis MM, Kuo CJ. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988.e16. 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Movia D, Bazou D, Volkov Y, Prina-Mello A. Multilayered cultures of NSCLC cells grown at the air-liquid interface allow the efficacy testing of inhaled anti-cancer drugs. Sci Rep. 2018;8:12920. 10.1038/s41598-018-31332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Movia D, Bazou D, Prina-Mello A. ALI multilayered co-cultures mimic biochemical mechanisms of the cancer cell-fibroblast cross-talk involved in NSCLC MultiDrug Resistance. BMC Cancer. 2019;19:1–21. 10.1186/s12885-019-6038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leene W, Duyzings MJ, van Steeg C. Lymphoid stem cell identification in the developing thymus and bursa of Fabricius of the chick. Z Zellforsch Mikrosk Anat Vienna Austria 1948. 1973;136:521–33. 10.1007/BF00307368. [DOI] [PubMed] [Google Scholar]
- 78.Nowak-Sliwinska P, Segura T, Iruela-Arispe ML. The chicken chorioallantoic membrane model in biology, medicine and bioengineering. Angiogenesis. 2014;17:779–804. 10.1007/s10456-014-9440-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vu BT, Shahin SA, Croissant J, Fatieiev Y, Matsumoto K, Le-Hoang Doan T, Yik T, Simargi S, Conteras A, Ratliff L, Jimenez CM, Raehm L, Khashab N, Durand J-O, Glackin C, Tamanoi F. Chick chorioallantoic membrane assay as an in vivo model to study the effect of nanoparticle-based anticancer drugs in ovarian cancer. Sci Rep. 2018;8:8524. 10.1038/s41598-018-25573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schmitd LB, Liu M, Scanlon CS, Banerjee R, D’Silva NJ. The chick chorioallantoic membrane in vivo model to assess perineural invasion in head and neck cancer. JoVE J Vis Exp. 2019:e59296. 10.3791/59296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Janser FA, Ney P, Pinto MT, Langer R, Tschan MP. The Chick Chorioallantoic Membrane (CAM) assay as a three-dimensional model to study autophagy in cancer cells. Bio-Protoc. 2019;9:e3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pawlikowska P, Tayoun T, Oulhen M, Faugeroux V, Rouffiac V, Aberlenc A, Pommier AL, Honore A, Marty V, Bawa O, Lacroix L, Scoazec JY, Chauchereau A, Laplace-Builhe C, Farace F. Exploitation of the chick embryo chorioallantoic membrane (CAM) as a platform for anti-metastatic drug testing. Sci Rep. 2020;10:16876. 10.1038/s41598-020-73632-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mangir N, Raza A, Haycock JW, Chapple C, Macneil S. An improved in vivo methodology to visualise tumour induced changes in vasculature using the chick chorionic allantoic membrane assay. In Vivo. 2018;32:461–72. 10.21873/invivo.11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Minami N, Maeda Y, Shibao S, Arima Y, Ohka F, Kondo Y, Maruyama K, Kusuhara M, Sasayama T, Kohmura E, Saya H, Sampetrean O. Organotypic brain explant culture as a drug evaluation system for malignant brain tumors. Cancer Med. 2017;6:2635–45. 10.1002/cam4.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kenerson HL, Sullivan KM, Seo YD, Stadeli KM, Ussakli C, Yan X, Lausted C, Pillarisetty VG, Park JO, Riehle KJ, Yeh M, Tian Q, Yeung RS. Tumor slice culture as a biologic surrogate of human cancer. Ann Transl Med. 2020;8:114. 10.21037/atm.2019.12.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vaira V, Fedele G, Pyne S, Fasoli E, Zadra G, Bailey D, Snyder E, Faversani A, Coggi G, Flavin R, Bosari S, Loda M. Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc Natl Acad Sci. 2010;107:8352–6. 10.1073/pnas.0907676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Merz F, Gaunitz F, Dehghani F, Renner C, Meixensberger J, Gutenberg A, Giese A, Schopow K, Hellwig C, Schäfer M, Bauer M, Stöcker H, Taucher-Scholz G, Durante M, Bechmann I. Organotypic slice cultures of human glioblastoma reveal different susceptibilities to treatments. Neuro-Oncol. 2013;15:670–81. 10.1093/neuonc/not003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martin SZ, Wagner DC, Hörner N, Horst D, Lang H, Tagscherer KE, Roth W. Ex vivo tissue slice culture system to measure drug-response rates of hepatic metastatic colorectal cancer. BMC Cancer. 2019;19:1–14. 10.1186/s12885-019-6270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jiang X, Seo YD, Chang JH, Coveler A, Nigjeh EN, Pan S, Jalikis F, Yeung RS, Crispe IN, Pillarisetty VG. Long-lived pancreatic ductal adenocarci noma slice cultures enable precise study of the immune microenvironment. Onco Targets Ther. 2017;6:e1333210. 10.1080/2162402X.2017.1333210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Misra S, Moro CF, Del Chiaro M, Pouso S, Sebestyén A, Löhr M, Björnstedt M, Verbeke CS. Ex vivo organotypic culture system of precision-cut slices of human pancreatic ductal adenocarcinoma. Sci Rep. 2019;9:2133. 10.1038/s41598-019-38603-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sivakumar R, Chan M, Shin JS, Nishida-Aoki N, Kenerson HL, Elemento O, Beltran H, Yeung R, Gujral TS. Organotypic tumor slice cultures provide a versatile platform for immuno-oncology and drug discovery. OncoImmunology. 2019;8:e1670019. 10.1080/2162402X.2019.1670019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sönnichsen R, Hennig L, Blaschke V, Winter K, Körfer J, Hähnel S, Monecke A, Wittekind C, Jansen-Winkeln B, Thieme R, Gockel I, Grosser K, Weimann A, Kubick C, Wiechmann V, Aigner A, Bechmann I, Lordick F, Kallendrusch S. Individual susceptibility analysis using patient-derived slice cultures of colorectal carcinoma. Clin Colorectal Cancer. 2018;17:e189–99. 10.1016/j.clcc.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 93.Naipal KAT, Verkaik NS, Sánchez H, van Deurzen CHM, den Bakker MA, Hoeijmakers JHJ, Kanaar R, Vreeswijk MPG, Jager A, van Gent DC. Tumor slice culture system to assess drug response of primary breast cancer. BMC Cancer. 2016;16:78. 10.1186/s12885-016-2119-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol. 2014;32:760–72. 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 95.Bhise NS, Manoharan V, Massa S, Tamayol A, Ghaderi M, Miscuglio M, Lang Q, Shrike Zhang Y, Shin SR, Calzone G, Annabi N, Shupe TD, Bishop CE, Atala A, Dokmeci MR, Khademhosseini A. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication. 2016;8:014101. 10.1088/1758-5090/8/1/014101. [DOI] [PubMed] [Google Scholar]
- 96.Lanz HL, Saleh A, Kramer B, Cairns J, Ng CP, Yu J, Trietsch SJ, Hankemeier T, Joore J, Vulto P, Weinshilboum R, Wang L. Therapy response testing of breast cancer in a 3D high-throughput perfused microfluidic platform. BMC Cancer. 2017;17:1–11. 10.1186/s12885-017-3709-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jin D, Ma X, Luo Y, Fang S, Xie Z, Li X, Qi D, Zhang F, Kong J, Li J, Lin B, Liu T. Application of a microfluidic-based perivascular tumor model for testing drug sensitivity in head and neck cancers and toxicity in endothelium. RSC Adv. 2016;6:29598–607. 10.1039/C6RA01456A. [DOI] [Google Scholar]
- 98.Lu S, Cuzzucoli F, Jiang J, Liang L-G, Wang Y, Kong M, Zhao X, Cui W, Li J, Wang S. Development of a biomimetic liver tumor-on-a-chip model based on decellularized liver matrix for toxicity testing. Lab Chip. 2018;18:3379–92. 10.1039/C8LC00852C. [DOI] [PubMed] [Google Scholar]
- 99.Eduati F, Utharala R, Madhavan D, Neumann UP, Longerich T, Cramer T, Saez-Rodriguez J, Merten CA. A microfluidics platform for combinatorial drug screening on cancer biopsies. Nat Commun. 2018;9:2434. 10.1038/s41467-018-04919-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Holton AB, Sinatra FL, Kreahling J, Conway AJ, Landis DA, Altiok S. Microfluidic biopsy trapping device for the real-time monitoring of tumor microenvironment. PLoS One. 2017;12:e0169797. 10.1371/journal.pone.0169797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, Bowden M, Deng J, Liu H, Miao D, He MX, Walker W, Zhang G, Tian T, Cheng C, Wei Z, Palakurthi S, Bittinger M, Vitzthum H, Kim JW, Merlino A, Quinn M, Venkataramani C, Kaplan JA, Portell A, Gokhale PC, Phillips B, Smart A, Rotem A, Jones RE, Keogh L, Anguiano M, Stapleton L, Jia Z, Barzily-Rokni M, Cañadas I, Thai TC, Hammond MR, Vlahos R, Wang ES, Zhang H, Li S, Hanna GJ, Huang W, Hoang MP, Piris A, Eliane J-P, Stemmer-Rachamimov AO, Cameron L, Su M-J, Shah P, Izar B, Thakuria M, LeBoeuf NR, Rabinowits G, Gunda V, Parangi S, Cleary JM, Miller BC, Kitajima S, Thummalapalli R, Miao B, Barbie TU, Sivathanu V, Wong J, Richards WG, Bueno R, Yoon CH, Miret J, Herlyn M, Garraway LA, Allen EMV, Freeman GJ, Kirschmeier PT, Lorch JH, Ott PA, Hodi FS, Flaherty KT, Kamm RD, Boland GM, Wong K-K, Dornan D, Paweletz CP, Barbie DA. Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discov. 2018;8:196–215. 10.1158/2159-8290.CD-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Deng J, Wang ES, Jenkins RW, Li S, Dries R, Yates K, Chhabra S, Huang W, Liu H, Aref AR, Ivanova E, Paweletz CP, Bowden M, Zhou CW, Herter-Sprie GS, Sorrentino JA, Bisi JE, Lizotte PH, Merlino AA, Quinn MM, Bufe LE, Yang A, Zhang Y, Zhang H, Gao P, Chen T, Cavanaugh ME, Rode AJ, Haines E, Roberts PJ, Strum JC, Richards WG, Lorch JH, Parangi S, Gunda V, Boland GM, Bueno R, Palakurthi S, Freeman GJ, Ritz J, Nicholas Haining W, Sharpless NE, Arthanari H, Shapiro GI, Barbie DA, Gray NS, Wong K-K. CDK4/6 inhibition augments antitumor immunity by enhancing T-cell activation. Cancer Discov. 2018;8:216–33. 10.1158/2159-8290.CD-17-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee SWL, Adriani G, Ceccarello E, Pavesi A, Tan AT, Bertoletti A, Kamm RD, Wong SC. Characterizing the role of monocytes in T cell cancer immunotherapy using a 3D microfluidic model. Front Immunol. 2018;9:416. 10.3389/fimmu.2018.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pavesi A, Tan AT, Koh S, Chia A, Colombo M, Antonecchia E, Miccolis C, Ceccarello E, Adriani G, Raimondi MT, Kamm RD, Bertoletti A. A 3D microfluidic model for preclinical evaluation of TCR-engineered T cells against solid tumors. JCI Insight. 2017;2:e89762. 10.1172/jci.insight.89762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Paek J, Park SE, Lu Q, Park K-T, Cho M, Oh JM, Kwon KW, Yi Y, Song JW, Edelstein HI, Ishibashi J, Yang W, Myerson JW, Kiseleva RY, Aprelev P, Hood ED, Stambolian D, Seale P, Muzykantov VR, Huh D. Microphysiological engineering of self-assembled and perfusable microvascular beds for the production of vascularized three-dimensional human microtissues. ACS Nano. 2019;13:7627–43. 10.1021/acsnano.9b00686. [DOI] [PubMed] [Google Scholar]
- 106.Jarvis M, Arnold M, Ott J, Pant K, Prabhakarpandian B, Mitragotri S. Microfluidic co-culture devices to assess penetration of nanoparticles into cancer cell mass. Bioeng Transl Med. 2017;2:268–77. 10.1002/btm2.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sobrino A, Phan DTT, Datta R, Wang X, Hachey SJ, Romero-López M, Gratton E, Lee AP, George SC, Hughes CCW. 3D microtumors in vitro supported by perfused vascular networks. Sci Rep. 2016;6:31589. 10.1038/srep31589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Seo BR, DelNero P, Fischbach C. In vitro models of tumor vessels and matrix: engineering approaches to investigate transport limitations and drug delivery in cancer. Adv Drug Deliv Rev. 2014;69–70:205–16. 10.1016/j.addr.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Choi Y, Hyun E, Seo J, Blundell C, Kim HC, Lee E, Lee SH, Moon A, Moon WK, Huh D. A microengineered pathophysiological model of early-stage breast cancer. Lab Chip. 2015;15:3350–7. 10.1039/C5LC00514K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hassell BA, Goyal G, Lee E, Sontheimer-Phelps A, Levy O, Chen CS, Ingber DE. Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro. Cell Rep. 2017;21:508–16. 10.1016/j.celrep.2017.09.043. [DOI] [PubMed] [Google Scholar]
- 111.Hidalgo M, Amant F, Biankin AV, Budinská E, Byrne AT, Caldas C, Clarke RB, de Jong S, Jonkers J, Mælandsmo GM, Roman-Roman S, Seoane J, Trusolino L, Villanueva A. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014;4:998–1013. 10.1158/2159-8290.CD-14-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Invrea F, Rovito R, Torchiaro E, Petti C, Isella C, Medico E. Patient-derived xenografts (PDXs) as model systems for human cancer. Curr Opin Biotechnol. 2020;63:151–6. 10.1016/j.copbio.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 113.Linxweiler J, Hajili T, Körbel C, Berchem C, Zeuschner P, Müller A, Stöckle M, Menger MD, Junker K, Saar M. Cancer-associated fibroblasts stimulate primary tumor growth and metastatic spread in an orthotopic prostate cancer xenograft model. Sci Rep. 2020;10:12575. 10.1038/s41598-020-69424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ben-David U, Ha G, Tseng Y-Y, Greenwald NF, Oh C, Shih J, McFarland JM, Wong B, Boehm JS, Beroukhim R, Golub TR. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet. 2017;49:1567–75. 10.1038/ng.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Erstad DJ, Sojoodi M, Taylor MS, Ghoshal S, Razavi AA, Graham-O’Regan KA, Bardeesy N, Ferrone CR, Lanuti M, Caravan P, Tanabe KK, Fuchs BC. Orthotopic and heterotopic murine models of pancreatic cancer and their different responses to FOLFIRINOX chemotherapy. Dis Model Mech. 2018;11:dmm034793. 10.1242/dmm.034793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lai Y, Wei X, Lin S, Qin L, Cheng L, Li P. Current status and perspectives of patient-derived xenograft models in cancer research. J Hematol Oncol. 2017;10:106. 10.1186/s13045-017-0470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tian H, Lyu Y, Yang Y-G, Hu Z. Humanized rodent models for cancer research. Front Oncol. 2020;10:1696. 10.3389/fonc.2020.01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jespersen H, Lindberg MF, Donia M, Söderberg EMV, Andersen R, Keller U, Ny L, Svane IM, Nilsson LM, Nilsson JA. Clinical responses to adoptive T-cell transfer can be modeled in an autologous immune-humanized mouse model. Nat Commun. 2017;8:707. 10.1038/s41467-017-00786-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Clohessy JG, Pandolfi PP. Mouse hospital and co-clinical trial project—from bench to bedside. Nat Rev Clin Oncol. 2015;12:491–8. 10.1038/nrclinonc.2015.62. [DOI] [PubMed] [Google Scholar]
- 120.Clohessy JG, Pandolfi PP. The mouse hospital and its integration in ultra-precision approaches to cancer care. Front Oncol. 2018;8:340. 10.3389/fonc.2018.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Koga Y, Ochiai A. Systematic review of patient-derived xenograft models for preclinical studies of anti-cancer drugs in solid tumors. Cells. 2019;8:418. 10.3390/cells8050418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bangi E, Murgia C, Teague AGS, Sansom OJ, Cagan RL. Functional exploration of colorectal cancer genomes using Drosophila. Nat Commun. 2016;7:13615. 10.1038/ncomms13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Das TK, Esernio J, Cagan RL. Restraining network response to targeted cancer therapies improves efficacy and reduces cellular resistance. Cancer Res. 2018;78:4344–59. 10.1158/0008-5472.CAN-17-2001. [DOI] [PubMed] [Google Scholar]
- 124.Bangi E, Ang C, Smibert P, Uzilov AV, Teague AG, Antipin Y, Chen R, Hecht C, Gruszczynski N, Yon WJ, Malyshev D, Laspina D, Selkridge I, Rainey H, Moe AS, Lau CY, Taik P, Wilck E, Bhardwaj A, Sung M, Kim S, Yum K, Sebra R, Donovan M, Misiukiewicz K, Schadt EE, Posner MR, Cagan RL. A personalized platform identifies trametinib plus zoledronate for a patient with KRAS-mutant metastatic colorectal cancer. Sci Adv. 2019;5:eaav6528. 10.1126/sciadv.aav6528. [DOI] [PMC free article] [PubMed] [Google Scholar]


