Abstract
Biomarkers are commonly used in pediatric medicine to identify disease and guide clinical management for children. Biomarkers can be used to predict risk of disease, provide diagnostic clarification, and offer prognostic expectations. Specimens for biomarker testing might require noninvasive collection (eg, urine, exhaled breath) or invasive procedures (eg, blood, bronchoalveolar lavage) and testing might use various methodologies (eg, genomics, transcriptomics, proteomics, metabolomics). Specimen type and testing methodology depends on the disease of interest, ability to obtain sample, and availability of biomarker testing. To develop a new biomarker, researchers must first identify and validate the target, and then determine the test characteristics of the biomarker. Once it has undergone initial development and testing, a new biomarker is then tested in the clinical setting before being implemented into practice. An ideal biomarker is one that is feasible to obtain, readily quantifiable, and offers meaningful information that impacts care. Learning how to reliably interpret the performance and clinical application of a new biomarker is an important skillset for all pediatricians in the hospital setting. Here we provide a high-level overview of the process from biomarker discovery to application. In addition, we provide an example for the real-world application of biomarkers as an opportunity for clinicians to build on their ability to critically evaluate, interpret, and implement biomarkers in clinical practice.
Biomarkers are commonly used in pediatric medicine to assist in managing and targeting care for children. A biomarker is a measure of a patient’s individual biochemical, physiologic, or behavioral characteristics that is associated with a person’s health or disease status.1 Biomarkers are typically used as indicators of abnormalities and can be obtained at a single time point or sequentially. Ideal biomarkers are readily quantifiable, economical, reproducible, sensitive, specific, and, when possible, can be detected early to guide interventions.2 Biomarkers can be used for different purposes such as diagnosis, prognosis, or to predict the risk, exposure, or disease of interest. Biomarkers can greatly influence care directly by providers ordering and interpreting biomarkers to help guide patient care or indirectly by influencing decisions based on clinical pathway or risk-stratification schema. Biomarkers may be sampled from noninvasive sources such as urine, exhaled breath, breast milk, hair, nails, saliva, and meconium, or can be more invasive such as blood, tissue samples, or bronchoalveolar fluid. This article will provide a general overview for the basic steps in biomarker discovery (Table 1), describe a clinical example in which biomarkers are currently used, and in which they are being further developed to improve the care of children.
TABLE 1.
Process for Biomarker Development from Identification to Implementation
| Phase | Description | Measure of Success |
|---|---|---|
| Phase 1: Identify the biomarker | • Choose type of biomarker: diagnostic, prognostic, and/or predictive • Identify the correct specimen type • Use genomic, transcriptomics, proteomics, metabolomics tools (Fig 1) |
• The biomarker and specimen type for the biomarker chosen must match the physiologic process being measured. |
| Phase 2: Validate the biomarker | • Content • Construct • Criterion |
• Biomarkers must be successfully validated before being used clinically. |
| Phase 3: Evaluate the biomarker10 | • Sensitivity: Ability of a particular biomarker to identify a true positive • Specificity: Ability of a particular biomarker to identify a true negative • Positive predictive value: Proportion of patients with a positive test who actually have the disease • Negative predictive value: Proportion of patients with a negative test who do not have the disease • AUC ROC: Overall diagnostic accuracy of a biomarker to correctly identify patients with and without the disease |
• Test characteristics of the new biomarker must demonstrate accurate and reliable performance when compared with a thoughtfully chosen gold standard before implementing the biomarker clinically. |
| Phase 4: Test the biomarker in the clinical setting | • Use dissemination and implementation methodology to test the ability to use the biomarker in a clinical setting. • Identify high-yield clinical scenarios in which the biomarker improves patient outcomes or increases the value of care. |
• Biomarkers must be readily available and easily interpretable for clinicians to use in the clinical setting. |
AUC ROC, area under the curve receiver operating curve.
Phases of Biomarker Development
Identify the Biomarker
The first phase of biomarker development requires identifying potential biomarkers that offer clinical promise while considering the practical purpose of that potential biomarker. Identifying novel biomarkers requires a fundamental understanding of the disease pathway in question and can be done using different fields and accompanying technologic approaches such as genomics (eg, sequencing), transcriptomics (eg, microarray, gene expression), proteomics (eg, mass spectrometry, protein chips), or metabolomics (eg, mass spectrometry, nuclear magnetic resonance) (Fig 1). Genomics can be useful in determining the genetic predisposition to disease, whereas proteomics may be informative of the host’s response to disease or the environment. Potential targets might be those most likely to be aberrant in disease or might be identified simultaneously, comparing many biomarkers at once using nontargeted methods to identify hidden patterns of a combination of biomarkers. Nontargeted methods are useful in the discovery of novel biomarkers as all potential biomarkers in a sample are investigated simultaneously. Promising targets worthy of study are often identified from smaller cohort studies or biospecimen repositories from patients with specific diagnoses or clinical phenotypes.
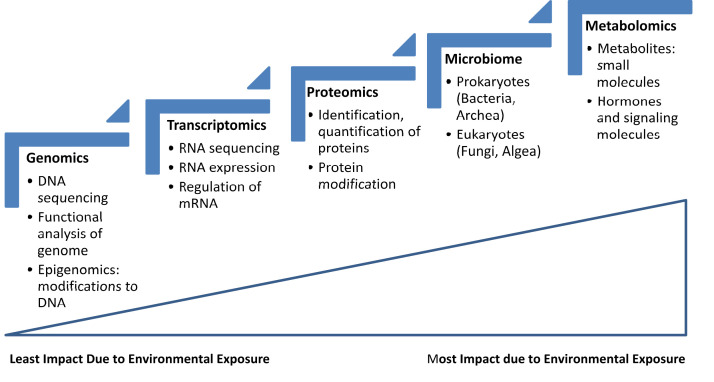
FIGURE 1.
“Omics” approaches and environmental influence. Environmental exposure will change one’s metabolome to a greater extent than one’s genome. Therefore, choosing the biomarker (eg, a gene vs protein) will depend on whether the biomarker is supposed to detect a genetic change (eg, DNA, RNA) versus a physiologic change (eg, protein, metabolite). Also, the microbiome is heavily influenced by the environment and represents the host’s underlying microbiota. mRNA, messenger RNA.
Validate the Biomarker
The second phase is to validate the potential biomarker’s content validity, construct validity, and criterion validity.3 Content validity is the degree to which a biomarker reflects the biological phenomenon studied (eg, degree to which hemoglobin A1c is representative of a diagnosis of diabetes). Construct validity pertains to the ability of the biomarker to align with the other relevant characteristics of the disease or trait (eg, degree to which hemoglobin A1c values align with other characteristics such as glucose levels or presence of diabetic neuropathy). Criterion validity is the extent to which the biomarker correlates with the specific exposure or disease of interest (eg, how accurate hemoglobin A1c is as a measure of severity of hyperglycemia).4 Once the biomarker is identified and meets these validity criteria, determining whether a dose-response relationship exists between the candidate biomarker and disease (eg, higher levels of biomarker represent more severe disease) can help to further establish a causal relationship between the 2. These validations are routinely performed in larger cohort studies to support the clinical plausibility of future use of the biomarker.
Evaluate the Biomarker
To support implementation of biomarkers into clinical practice, test characteristics including sensitivity, specificity, positive predictive values, and negative predictive values are calculated based on the ability of the biomarker to correctly identify the risk, exposure, or disease of interest (Table 1). For instance, when considering the ability of C-reactive protein (CRP) to identify children with acute bacterial infections, the sensitivity of CRP is quite high because it is often elevated in children with true bacterial disease. However, the specificity of CRP for bacterial infection is less robust because many other conditions can elevate CRP levels. Thus, CRP will not reliably distinguish children without bacterial infection. It is important to recognize that positive and negative predictive values are dynamic metrics that vary based on pretest probability, which is in turn influenced by disease prevalence. For instance, the positive predictive value of CRP to identify bacterial infections may be increased when used in a population with a very high prevalence of bacterial infection (eg, children with high fevers, dysuria, flank pain) compared with a population with lower prevalence of bacterial infection (eg, children with fever, mild upper respiratory infection symptoms).
These measures of biomarker performance cannot be interpreted in isolation. Instead, biomarkers must be evaluated collectively and in the context of what the biomarker’s purported use will be. In addition to these frequently referenced measures of test performance, other metrics may be used to interpret a biomarker’s performance. For instance, area under the curve receiver operating curve can be used to precisely define ideal cutoff values for a particular biomarker that optimizes sensitivity and specificity (Fig 1).
Appraisal of the gold or reference standard used when determining the test characteristics of a novel biomarker is essential for clinicians when critically reviewing the evidence. For example, the reference standard of “community-acquired bacterial pneumonia” may take various forms including clinical and radiographic criteria, clinical consensus from a panel of providers, or microbiological confirmation via pathogen identification. The use of a gold standard that itself is not specific to a disease state can have profound effects, both positive and negative, on a biomarker’s described performance. Therefore, the standard used to derive these measures must be considered when evaluating a biomarker’s test performance.
Test the Biomarker in a Clinical Setting
Once the biomarker achieves clinically acceptable performance characteristics, which can vary significantly based on the intended purpose of the biomarker, the next challenge is to implement the biomarker into clinical practice. Dissemination and implementation methodologies are often used to ensure successful adoption and sustained implementation of biomarkers in the clinical setting. Practical issues to consider when implementing a biomarker include the limit of detection, variability, half-life of the biomarker, methods needed to collect the biomarker, specimen storage, and turnaround time for results. The last phase in biomarker development is to test the biomarker in a clinical setting, typically using a randomized controlled study with clear endpoints that demonstrate change in clinical management or earlier diagnosis of a disease process. Defining a clear endpoint can vary significantly based on the biomarker being studied; some studies easily define a clear outcome of interest, whereas others must rely on more ambiguous endpoints. For instance, presence of a serious bacterial infection may be the clear endpoint used for a study evaluating the utility of procalcitonin in critically ill children, whereas more nebulous measures for quality-of-life metrics may be used alongside serial hemoglobin A1c levels for children with type 1 diabetes.
A Shift Toward a Personalized Approach
Asthma is a disease process that provides an ideal example of how biomarkers can be identified, validated, and used to improve clinical care. Asthma is challenging to manage because it is complex and heterogenous with substantial variability in clinical phenotypes and treatment response.5 Traditional guidelines for asthma management followed a 1-size-fits-all approach that considered asthma a singular clinical entity. However, recognition of the diversity in asthma phenotypes helped identify distinct disease pathways and revealed the need for a personalized approach to advance asthma care and improve morbidity in children. Personalized asthma management has progressed through identification of biomarkers that are effective in differentiating asthma phenotypes and subsequently predicting treatment response. These identified biomarkers have also served as targets for the development of novel therapeutics.
Biomarkers currently used to identify the type 2 asthma phenotype, and to predict and monitor treatment response in children with severe type 2 asthma, are an example of diagnostic and predictive biomarkers that have been developed, validated, and implemented into clinical practice. The type 2 asthma phenotype is characterized by significant airway inflammation and driven by type 2 inflammatory cytokines, including eosinophils. Blood and sputum eosinophil counts, serum immunoglobulin E levels, and fraction of exhaled nitric oxide values have been shown to be diagnostic and predictive biomarkers with established cutoffs that can reliably identify children with severe type 2 asthma who might benefit from biologic agents targeting type 2 cytokines.6 Recently, the MUPPITS-2 trial demonstrated that phenotype-directed therapy with mepolizumab, a monoclonal antibody directed at interleukin-5 that mediates eosinophil production, reduced the number of asthma exacerbations in children with type 2 asthma.7
The MUPPITS-2 trial also provides an example of novel biomarkers that may offer more precise prediction of treatment response. Although blood eosinophil counts helped to identify the subset of children with exacerbation-prone type 2 asthma who might benefit most from mepolizumab therapy, the relative reduction in blood eosinophil counts after mepolizumab therapy was not associated with treatment efficacy. Instead, investigators of the MUPPITS-2 trial demonstrated that nasal transcriptomic profiling identified transcriptomic modules (networks of functionally related genes) that more precisely described the mechanisms underlying the variable response to mepolizumab therapy.7 A previous impediment to airway transcriptomic analysis in children was the need to perform invasive bronchoscopies to collect airway samples. However, performing transcriptomic analysis of nasal epithelia collected by minimally invasive nasal swabs has been shown to be an excellent surrogate for bronchial brushings and is feasible to obtain in children.8 Thus, with further validation and advances in technology allowing for real-time results, airway transcriptomic modules have potential as more precise biomarkers of treatment response to advance asthma management.
Although substantial progress has been made in the development of biomarkers for the type 2 asthma phenotype, further advancement is needed to expand biomarker representation of broader asthma phenotypes (eg, the non–type 2 asthma phenotype). In addition, most progress has occurred in the outpatient setting for management of chronic asthma. In the acute care and inpatient settings, management of children with acute asthma continues to follow a 1-size-fits-all approach with variable response to standard acute asthma therapies.9 Thus, there is a critical need to develop biomarkers for the acute care and inpatient settings to predict treatment response, drive treatment decisions, and identify potential targets for novel therapeutics. Last, although there is a need for continued identification of asthma biomarkers, there is an equally important need for integration and application of our knowledge to transition toward individualized asthma care.
Conclusions
Conventional and novel biomarkers hold promise to better predict, diagnose, and prognosticate both common and rare diseases among children. Clinical scientists and bench researchers must continue to uncover mechanistic pathways and think creatively to identify biomarkers with promise for clinical applications. Similarly, because these diagnostic tools represent a constantly evolving field, clinicians face a Sisyphean task of continuous exposure to and interpretation of newly implemented biomarkers in the patient care setting. Leveraging available biomarkers for individual patients requires a specific skillset for clinicians to successfully interpret a given result and then take actionable steps to improve care based on the biomarker’s result. As experts in practical decision-making at the bedside, pediatric hospitalists should continue to critically evaluate newly proposed biomarkers in the literature to both implement those that improve care value and deimplement those that reduce care value.
Footnotes
FUNDING: This study was supported by the National Institutes of Health National Heart Lung Blood Institute (K23HL161354 to N.N.) and from National Institute of Allergy and Infectious Diseases (R21AI154239 and K01AI125413 to L.A.). The funders did not have any role in study design, data collection, statistical analysis, or manuscript preparation. Funded by the National Institutes of Health (NIH).
CONFLICT OF INTEREST DISCLOSURES: Dr Ambroggio has an ongoing study supported by Pfizer Inc. investigating pneumococcal urine antigen diagnostics. Drs Navanandan and Searns have indicated they have no potential financial conflicts to disclose.
Drs Navanandan, Searns, and Ambroggio conceptualized the manuscript, drafted the initial manuscript, and reviewed, revised, and approved the final manuscript as submitted.
References
- 1. Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69(3):89–95 [DOI] [PubMed] [Google Scholar]
- 2. Florin TA, Ambroggio L. Biomarkers for community-acquired pneumonia in the emergency department. Curr Infect Dis Rep. 2014;16(12):451. [DOI] [PubMed] [Google Scholar]
- 3. Mayeux R. Biomarkers: potential uses and limitations. NeuroRx. 2004;1(2):182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lyons TJ, Basu A. Biomarkers in diabetes: hemoglobin A1c, vascular and tissue markers. Transl Res. 2012;159(4):303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zahran HS, Bailey CM, Damon SA, Garbe PL, Breysse PN. Vital signs: asthma in children - United States, 2001–2016. MMWR Morb Mortal Wkly Rep. 2018;67(5):149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Global Initiative for Asthma. Global strategy for asthma management and prevention, 2020. Available at: https://ginasthma.org/wp-content/uploads/2020/06/GINA-2020-report_20_06_04-1-wms.pdf. Accessed March 23, 2023
- 7. Jackson DJ, Bacharier LB, Gergen PJ, et al. Mepolizumab for urban children with exacerbation-prone eosinophilic asthma in the USA (MUPPITS-2): a randomised, double-blind, placebo-controlled, parallel-group trial. Lancet. 2022;400(10351):502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poole A, Urbanek C, Eng C, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol. 2014;133(3):670–8.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O’Connor MG, Saville BR, Hartert TV, Arnold DH. Treatment variability of asthma exacerbations in a pediatric emergency department using a severity-based management protocol. Clin Pediatr (Phila). 2014;53(13):1288–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Szklo M, Nieto FJ. Epidemiology Beyond the Basics, 2nd ed. Burlington, MA: Jones and Bartlett Publishers; 2007 [Google Scholar]