Abstract
Antibodies are critical reagents to detect and characterize human proteins. The biomedical research community aspires to have at least one potent and selective renewable antibody for each human protein, and for each of the most common applications. To quantify progress toward this goal, a standardized characterization approach to assess the performance of 614 commercial antibodies for 65 neuroscience-related proteins in Western blot, immunoprecipitation, and immunofluorescence applications, using isogenic knockout cells as controls. Side-by-side comparisons of the antibodies demonstrated that: i) for each application tested, approximately half of this protein set was covered by at least one high-performing antibody, suggesting that coverage of human proteins by commercial antibodies is significant but gaps are still present; ii) on average recombinant antibodies performed better than monoclonal or polyclonal antibodies; iii) hundreds of underperforming antibodies have been used in published articles. Encouragingly, more than half of the commercial antibodies that did not perform as expected were removed from the market by manufacturers or had alterations made to their recommended usage based on this body of work. This work suggests an efficient strategy toward achieving coverage of the human proteome with selective renewable antibodies is to mine the existing commercial antibody repertoire, using this knowledge to focus new renewable antibody generation efforts.
Keywords: antibody, antibody validation, antibody characterization, open science
Introduction
Antibodies as reagents enable the identification and quantification of proteins for a range of applications. The community aspires to have at least one, and ideally two or more, potent and selective renewable (monoclonal or recombinant monoclonal) antibodies for each human protein and each essential application [1]. Unfortunately, although there are over 1.5 million reagent antibodies on the market that target human proteins [2], there is no agreed-upon mechanism to determine, validate or compare their performance. It is thus difficult to assess progress toward the objective of well-validated antibodies for each human protein, or to design a strategy to accomplish this aim.
To help quantify the status of the coverage of human proteins with high-quality antibodies, we created a partnership of academics, funders and commercial antibody manufacturers. This includes 10 companies representing approximately 30% of antibody manufacturing worldwide. The goal is to systematically compare the performance of commercial antibodies using knockout (KO) cell lines and isogenic parental cells as controls. For each protein target, we test selected commercial antibodies in parallel using standardized protocols, agreed upon by all parties, in Western blot (WB), immunoprecipitation (IP), and immunofluorescence (IF) applications. All data are shared rapidly and openly on ZENODO, a preprint server. One long-term goal is to build a database of antibody characterization data that can serve as a valuable resource for antibody users, saving them time and money, reducing erroneous data, and improving rigour and reproducibility.
Thus far we have tested 614 commercially available antibodies targeted to 65 proteins for WB, IP and IF, and found that approximately two thirds of this protein set was covered by at least one high-performing antibody and half covered by at least one high-performing renewable antibody, suggesting that coverage of human proteins by commercial antibodies is significant. This sample size is also large enough to observe several trends in antibody performance across various parameters. First, our study clearly demonstrates the robustness of a KO-based approach to evaluate antibody specificity. Second, we found that approximately half of all antibodies do not perform to the expected standard as per manufacturer recommendations, with some variability by application. Third, we observed that recombinant antibodies perform better than traditional antibodies. Finally, we have also observed that suboptimal antibodies have been used, and continue to be used, in numerous peer-reviewed research articles.
Results
Assembling KO cell lines and antibodies
Our goal is to enable the biomedical research enterprise to have access to at least one potent and selective renewable antibody for each human protein, and for each of the common applications [3], initially for WB, IP and IF. Our first objective was to assess the current coverage of the human proteome by antibodies that are commercially available. Here, we report the analysis of 614 commercially available antibodies to the first 65 protein targets, which were chosen by disease charities, academia, and industry without consideration of antibody coverage. Most targets are related to neurodegenerative diseases, including Alzheimer’s disease (AD), amyotrophic lateral sclerosis (ALS), and Parkinson’s disease (PD). The list comprises 32 AD-related proteins that were community-nominated through an NIH-funded project on dark AD genes (https://agora.adknowledgeportal.org/), 22 proteins that were nominated within the ALS Reproducible Antibody Platform (ALS-RAP) project, 5 PD-linked proteins that were nominated by the Michael J. Fox Foundation (MJFF) and 6 proteins nominated by industry (Figure 1A). Within the 65 target proteins, 56 are intracellular and 9 are secreted. The description of each protein target is indicated in Table 1.
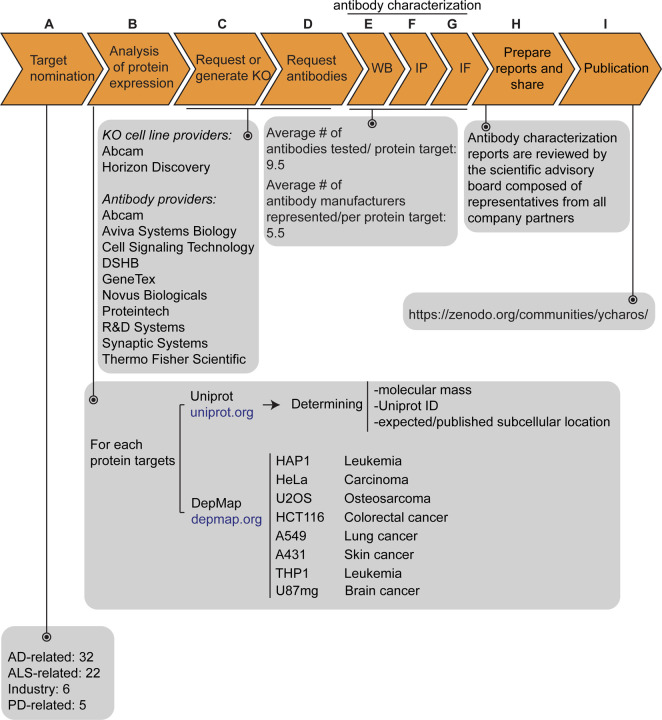
Figure 1: Antibody characterization platform.
(A) The funders of the targets analyzed in this study and the number of targets proposed by each are indicated. (B) Bioinformatic analyses of nominated proteins using Uniprot to determine their molecular mass, unique Uniprot ID and published/expected subcellular distribution. In parallel, analyses of the Cancer Dependency Map (“DepMap”) portal provided RNA sequencing data for the designated target, which guided our selection of cell lines with adequate expression for the generation of custom KO cell lines. A subset of cell lines amenable for genome engineering were prioritized. (C) Receive relevant KO cell lines or generate custom KO lines and (D) receive antibodies from manufacturing partners. All contributed antibodies were tested in parallel by (E) WB using WT and KO cell lysates ran side-by-side, (F) IP followed by WB using a KO-validated antibody identified in E and by (G) IF using a mosaic strategy to avoid imaging and analysis biases. (H) Antibody characterization data for all tested antibodies were presented in a form of a protein target report. All reports were shared with participating manufacturers for their review. (I) Reviewed reports were published on ZENODO, an open access repository. ALS-RAP=amyotrophic lateral sclerosis-reproducible antibody platform, AD=Alzheimer’s disease, MJFF=Michael J. Fox Foundation. KO=knockout cell line
Each protein was searched through Uniprot to determine the Uniprot identifier and the molecular mass, and to predict whether the protein is secreted or intracellular (Figure 1B). Our strategy to compare antibody performance from KO cell lines to isogenic parental controls was predicated on identifying a parental cell line(s) that expressed sufficient levels of the target protein to be able to be detected by an antibody with a binding affinity in the range of 1–50 nM. To identify candidate lines, we searched the Cancer Dependency Map Portal (DepMap) using the “Expression 22Q1” database, which houses the RNA-level analysis of >1800 cancer cell lines [4] (Figure 1B). After our initial experience with a few dozen targets comparing RNA expression and the ability to detect a clear signal, we selected 2.5 log2(TPM+1) as an RNA-level threshold to select a candidate cell line to create a KO. Among the cell lines showing expression above this level, we prioritized a group of 8 common cell line backgrounds representing different cell/tissue types because their doubling time is short, and they are amenable to CRISPR-Cas9 technology (Figure 1B). These 8 cell lines were used in 62 out of the 65 antibody characterization studies (Table 1).
After identifying candidate cell lines for each target, we either obtained KO cell lines from our industry consortium partners or generated them in-house (Figure 1C). Antibodies were provided from antibody manufacturers, who were responsible for selecting antibodies to be tested from their collections (Figure 1D). Most antibody manufacturers in the collaboration prioritized renewable antibodies, either monoclonal antibodies derived from hybridoma cultures, or recombinant monoclonals generated in vitro from cloned immunoglobulins. The highest priority was given to recombinant antibodies as they represent the ultimate renewable reagent [1] and have advantages in terms of adaptability, such as switching IgG subclass [5] or using molecular engineering to achieve higher affinity binding than B-cell generated antibodies [6].
All available antibodies from all companies were tested side-by-side in parental and KO lines. The protocols used were established by our previous work [7] and refined in collaboration with antibody manufacturers and on occasion differed from the protocols they used for their internal characterization. All antibodies were tested for all 3 applications, independently of the antibody manufacturers’ recommendations. We received on average 9.5 antibodies per protein target contributed from an average of at least 5 different antibody manufacturers (Figure 1E, F, G). Companies often contributed more than one antibody per target (Table 1).
Antibody and cell line characterization
Antibodies were tested by WB against parental and either KO lysates (intracellular proteins) or cell media (secreted proteins) to verify their performance (Figure 1E). For 86% of target proteins (56/65), we identified one or more antibodies that successfully immunodetected their cognate protein. This experiment not only identified well-performing antibodies, but also validates the efficacy of the KO line. For the remaining 14% of targets (9/65), we were able to identify at least one specific, non-selective antibody that detects the cognate protein by WB, but also recognizes unrelated proteins (there were non-specific bands). All 614 antibodies were tested by immunoprecipitation on non-denaturing cell lysates (intracellular proteins) or cell media (secreted proteins), using WB with a successful antibody from the previous step to evaluate the immunocapture (Figure 1F). All antibodies against intracellular proteins were then tested for IF using a strategy that imaged a mosaic of parental and KO cells in the same visual field to reduce imaging and analysis biases (Figure 1G).
For each protein target, we consolidated all screening data into a report, which we make available without restriction on ZENODO, a data-sharing website operated by CERN. On ZENODO, all 65 reports are gathered together under the Antibody Characterization through Open Science (YCharOS) community: https://ZENODO.org/communities/ycharos/ (Figure 1). Prior to its release, each antibody characterization report underwent a technical review by a group of scientific advisors from academia and industry (Figure 1). All 65 reports are currently available (Table 1), and new reports are regularly uploaded on the YCharOS community at a current rate of approximately 4 per week.
Coverage of human proteins by renewable antibodies
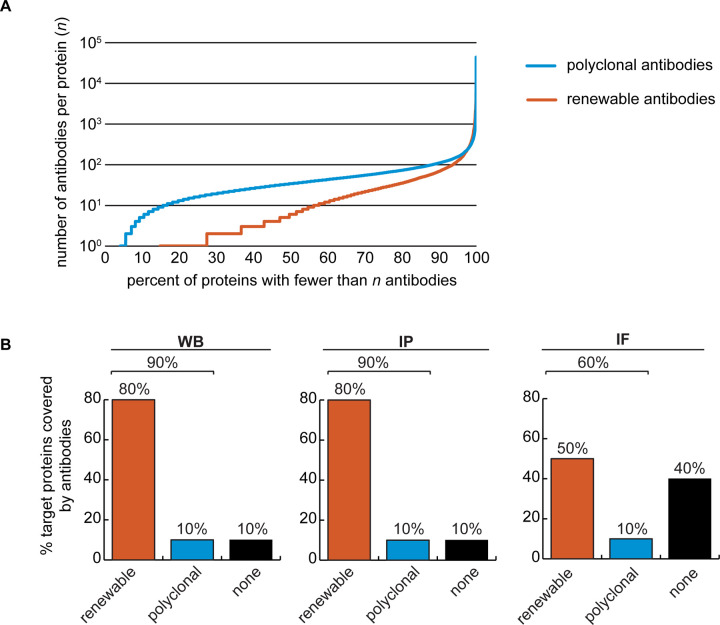
The Antibody Registry (www.antibodyregistry.org, SCR_006397) indicates that there are ~1.6 million antibodies covering ~96% of human proteins [2], with 53% of human proteins covered by at least five renewable antibodies (Figure 2A, Supplementary Data 1). 21% of human proteins are covered by only one or two renewable antibodies, and ~15% have no renewable antibodies available (Figure 2A). In our set of 65 proteins, and from the manufacturers represented, 49 were covered by at least 3 renewable antibodies, 15 proteins are covered by one or two renewable antibodies, and one protein was not covered by any renewables (Table 1).
Figure 2: Analysis of human protein coverage by antibodies.
(A) Cumulative plot showing the percentage of the human proteome that is covered by polyclonal antibodies (blue line) and renewable antibodies (monoclonal + recombinant; orange line). The number of antibodies per protein was extracted from the Antibody Registry database. (B) Percentage of target proteins covered by minimally one renewable successful antibody (orange column) or covered by only successful polyclonal antibodies (blue column) is showed for each indicated applications using a bar graph. Lack of successful antibody (“none”) is also shown (black column).
After testing all 614 antibodies against the 65 targets, we found a well-performing renewable antibody for 50 targets in WB application (Figure 2B, left bar graph), for 49 targets in IP (Figure 2B, middle bar graph), and for 30 targets in IF (Figure 2B, right bar graph). This coverage was encouragingly high; there is a well-performing commercially available renewable antibody for a large fraction of the proteins in our test set, in each application. For some proteins lacking coverage by renewable antibodies or lacking successful renewable antibodies, well-performing polyclonal antibodies were identified (Figure 2B). Lastly, some proteins were not covered by any successful antibodies; notable ~40% of our protein set lacked a successful antibody for IF (Figure 2B, right bar graph).
Recombinant antibody performance
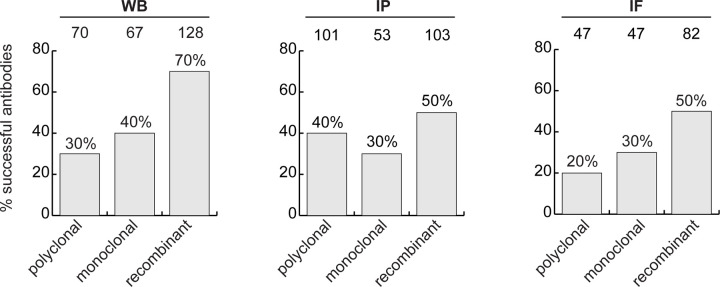
Our data demonstrate that there may be well-performing renewable antibodies for WB, IP and IF that are already available for many human proteins. To inform a strategy to generate reagents for the remaining human proteins or applications, we compared the performance of the various antibody types (polyclonal, monoclonal, and recombinant mono- or polyclonal) in our dataset, in each of the three applications. The set constituted 258 polyclonal antibodies, 165 monoclonal antibodies and 191 recombinants.
For WB, 27% of the polyclonal antibodies, 41% of the monoclonal antibodies and 67% of the recombinant antibodies immunodetected their target protein (Figure 3, left bar graph). For IP, the trends were similar: 39%, 32% and 54% of polyclonal, monoclonal and recombinants, respectively, immunodepleted their cognate antigen from lysates or medium (Figure 3, middle bar graph). We tested 529 antibodies against the set of intracellular proteins for IF, and 22% of polyclonal antibodies, 31% of monoclonal antibodies, and 48% of recombinant antibodies generated selective fluorescence signals in images of parental versus KO cells (Figure 3, right bar graph).
Figure 3: Analysis of antibody performance by antibody types.
The percentage of successful antibodies based on their clonality is shown using a bar graph, for each indicated application. The percentage corresponding to each column is shown in the corresponding column bar, and the number of antibodies represented in each category is indicated above the corresponding bar.
In sum, in our set and for all applications, recombinant antibodies are on average better performers than polyclonal or monoclonal antibodies in each of the applications. It should be noted that recombinant antibodies are newer protein reagents compared to polyclonal and monoclonal hybridomas and their superior performance could be a consequence of enhanced internal characterization by the commercial suppliers. That said, recombinant antibodies perform well and have significant advantages from the perspective of reproducible science.
Optimizing an antibody characterization strategy
We characterized the potency and selectivity of the antibodies in our dataset by comparing signals from parental and KO cells. While this method is clearly preferred [7–10], not all antibodies on the market are characterized this way, largely due to cost and the range of alternative methods that are employed [11]. To assess whether the cost of “KO characterization” is justified, we compared the performance of antibodies in our dataset with our standardized KO characterization protocol to the performance predicted by KO and other characterization methods used in the companies.
We first compared antibody performance based on the manufacturer’s recommendations. In all, 578 of the 614 antibodies we tested were recommended for WB by the manufacturers. Of these, 44% were successful by WB and 56% failed (Supplemental Figure 2A, left bar graph). Most antibodies are not recommended for IP by the suppliers, perhaps because they are not tested. Of the 143 of 614 that were recommended for IP, 58% enriched their cognate antigen from cell extracts. Interestingly, of the 471 remaining antibodies that presumably never been tested for IP, 37% were able to enrich their cognate antigen (Supplemental Figure 2A, middle bar graph). Of the 529 antibodies tested in IF, 293 were recommended for this application by the suppliers and 236 were not. Only 39% of the antibodies recommended for IF were successful (Supplemental Figure 2A, right bar graph).
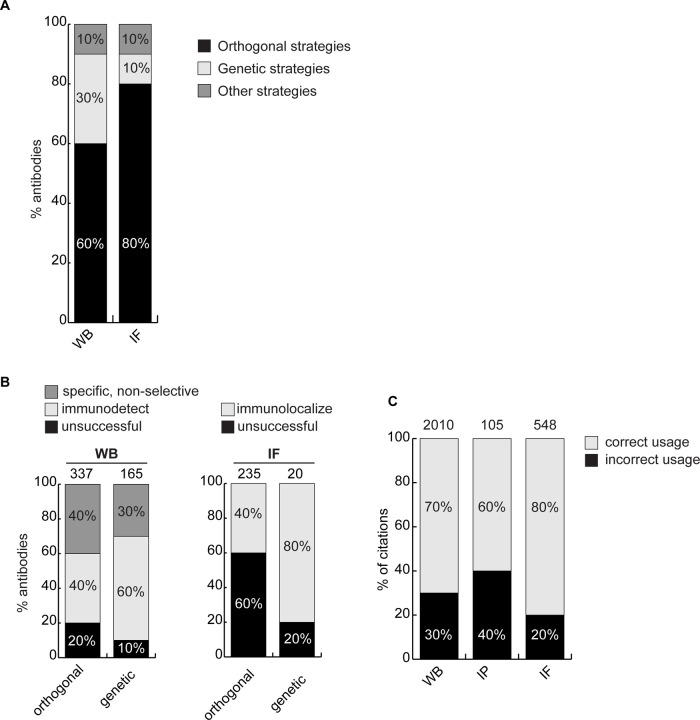
We next investigated if antibody validation strategies have equal scientific value. Broadly, antibodies are characterized either using genetic approaches, which exploit KO or knockdown (KD) samples as controls, or using orthogonal approaches, which use known information about the target protein of interest as a correlate to validate the performance of antibodies. Orthogonal and genetic strategies represent the most common characterization strategies used in industry. For WB, 61% of antibodies were recommended based on orthogonal approaches, 30% using genetic approaches and 9% using other strategies. For IF, 83% of the antibodies were recommended based on orthogonal approaches, 7% using genetic approaches and 10% using other strategies (Figure 4A).
Figure 4: Scientific value of antibody characterization methods and research usage.
(A) Percentage of antibodies validated by suppliers using one of the indicated methods for WB or IF showed using a bar graph with stacked columns. The percentage corresponding to each section of the bar graph is shown in the corresponding column. Orthogonal= orthogonal strategies, genetic= genetic strategies. (B) Percentage of successful (gray) and unsuccessful (black) antibodies according to the validation method used by the manufacturer for WB and IF as compared to the KO strategy used in this study. Data is shown using a bar graph with stacked columns. The percentage corresponding to each section of the bar graph is shown in the corresponding column. The number of antibodies analyzed corresponding to each condition is shown above each bar. (C) Percentage of publications that used antibodies that successfully passed validation (correct usage) or to antibodies that were unsuccessful in validation (incorrect usage) showed using a bar graph with stacked columns. The number of publications was found by searching CiteAb. The percentage corresponding to each section of the bar graph is shown in the corresponding column and the number of publications represented in each category is shown above the corresponding bar.
For WB, ~80% of the antibodies recommended by the manufacturer based on orthogonal strategies and ~90% of antibodies recommended based on genetic strategies could detect the intended target protein (Figure 4B, left bar graph). For IF, 38% of the antibodies recommended by the manufacturer based on orthogonal strategies were confirmed using KO cells as controls, and 80% of antibodies validated with genetic strategies succeeded to confirm in our hands (Figure 4B, right bar graph). While orthogonal strategies are somewhat suitable for WB, genetic strategies generate far more robust characterization data for IF.
Antibodies and reproducible science
The availability of renewable, well-characterized antibodies would be expected to increase the reproducibility of research. To assess the bibliometric impact of underperforming antibodies, we used the reagent search engine CiteAb (https://www.citeab.com/) to quantify how antibodies in our dataset have been used in the literature.
We identified 2010 publications that employed one of the 180 antibodies we tested for WB. Of those, 69% (1378 publications) used an antibody that specifically immunodetected its target protein by WB but 31% (632 publications) citations used an unsuccessful antibody (Figure 4C). For IP, 105 publications employed 41 of our tested antibodies. 65% of citations (68 publications) used a well-performing antibody but 35% (37 publications) employed an antibody unable to immunocapture its target protein (Figure 4C). For IF, we found 548 publications that employed 80 of the antibodies we tested. 22% (123 publications) of these studies used an antibody unable to immunolocalize its target protein (Figure 4C). If our results are representative, this suggests that 20–30% of figures in the literature are generated using antibodies that do not recognize their intended target, and that more effort in antibody characterization is highly justified.
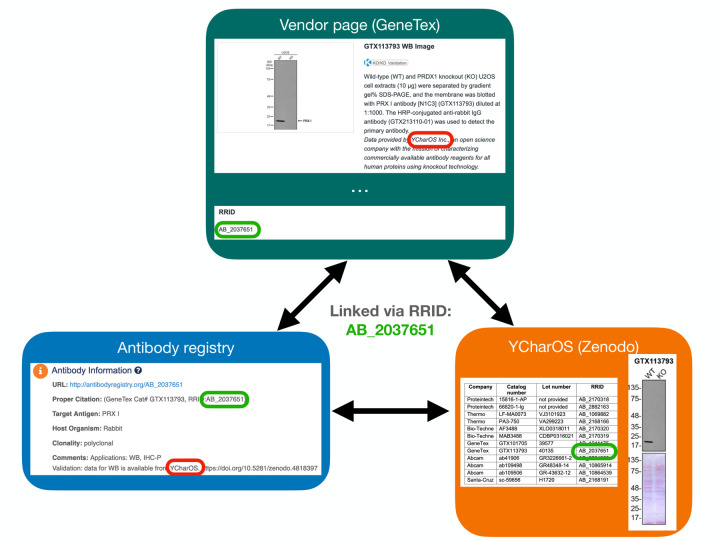
A Research Resource Identification (RRID) was assigned to each 614 antibody tested, indicated in each 65 antibody characterization reports available on ZENODO (Figure 5, bottom right image). Antibody characterization data generated by this organization are being disseminated by the RRID community and are directly connected through the Antibody Registry, or the RRID Portal (Figure 5, bottom left image) and participating antibody manufacturers’ websites (Figure 5, top image).
Figure 5: Accessing antibody characterization data using the RRID portal.
An antibody RRID can be used to search characterization studies across various databases, such as vendor page, the Antibody Registry and on the YCharOS community page on ZENODO. AB_2037651 is given as an example.
Discussion
Here we present a comprehensive and standardized analysis of the performance of commercially available antibodies using isogenic KO cell lines as controls. Our analyses involved side-by-side comparison of 614 antibodies by WB, IP and IF against 65 human proteins. All raw data are openly available (https://ZENODO.org/communities/ycharos/), identifiable on the RRID portal and on participating antibody manufacturers’ websites. Our studies provide an unbiased and scalable analytical framework for the representation and comparison of antibody performance, an estimate of the coverage of human proteins with renewable antibodies and an assessment of the scientific value of common antibody characterization methods.
Life scientists tend to focus on a small subset of human proteins, leading to an imbalance between a small percentage of well-studied proteins and a much higher percentage remaining poorly characterized [12]. Our set of 65 funder-designated proteins is an unbiased sample representative of the heterogeneity of knowledge of the human proteome; a search of the NIH protein database revealed that 15 proteins (23% of our protein sample) are well studied with more than 500 publications and 50 proteins (77% of our protein sample) have corresponding publications ranging from 37 to 498 (Table 1). Although we observed that there are more commercial antibodies available for the best-studied proteins (Table 1), an encouraging result of our work is that more than half of our protein targets are covered by well-performing, renewable antibodies for WB, IP and IF - including both well characterized and more poorly studied proteins. Within the antibodies we tested, we found a successful renewable antibody for WB for 77% of proteins (50/65), for IP for 75% of proteins (49/65) and for IF for 54% of proteins (30/56) examined. Extrapolation of our findings to the human proteome would suggest that it might be possible to identify well-performing renewable reagents for half the human proteome, including poorly characterized proteins, simply by mining commercial collections. Indeed, it is likely that the coverage is greater because our corporate partners only represent 27% of the antibody production worldwide. A practical conclusion of our study is that the first step toward covering the human proteome with renewable antibodies should be to take stock of the existing repertoire, and use this information to guide new antibody generation efforts.
The research market is heavily dominated by polyclonal antibodies, and their use lies at the heart of the reproducibility crisis observed in biomedical research [13, 14] and present important ethical concerns [1]. From a scientific perspective, polyclonal antibodies suffer from batch-to-batch variation and are thus in conflict with the scientific community desire to use and provide only renewable reagents. From an ethical perspective, the generation of polyclonal antibodies requires large number of animals yearly [15]. While recombinant antibodies rely on the use of animals for the initiation of an antibody generation program, animal-free in vitro biochemical strategies are used for the latter steps of production, and to generate new batches of these antibodies [6]. As of today, the uptake of recombinant antibodies by the scientific community has not been satisfactory. For example, while leading antibody manufacturers are converting top-cited polyclonal antibodies into recombinant antibodies and removing underperforming antibodies from their catalogues, scientists tend to stay away from newer recombinant technologies. This situation has also been observed and described by the EU Reference Laboratory for Alternatives to Animal Testing and a lack of understanding in the use of recombinant methods has been suggested by authors of a recent correspondence to the editors of neuter Biotechnology [6]. One reason for this confusion could be the absence of large-scale performance data comparing the various antibody generation technologies. In our dataset, recombinant antibodies performed well in all applications tested, arguing there is no reason not to adopt the recombinant technology. Moreover, our study strongly supports the use of recombinant idea that future antibody generation programs should focus on recombinant technologies.
Our analyses also inform the characterization pipelines to use for newly generated antibodies. Currently, it is common to use WB as the initial screen [16]. However, we find that success in IF is the best predictor of performance in WB and IP (Supplemental Figure 3). Given that it is difficult to imagine a process dependent on IF, we suggest that using knockout (or knock-down in the case of an essential gene) strategies to screen antibodies for the intended application will provide the most effective approach to identify selective antibodies. Currently, one of the main barriers to large-scale production of high-quality antibodies is the lack of availability of KO lines derived from cells express detectable levels of each human protein. Creation of a broadly accessible biobank of bespoke KO cells for each human gene should be a priority for the community.
Our studies are rapidly shared via the open platform ZENODO, and selected studies were published on the F1000 publication platform (https://f1000research.com/ycharos). Approximately 4 studies are being uploaded on ZENODO every month. This data generation and dissemination is intended to benefit the global life science community, but its impact depends on the real-world uptake of the data. Thus, we have partnered with the RRID Portal Community to improve our dissemination strategies. The Antibody Registry is a comprehensive repository of over 2.5 million commercial antibodies that have been assigned with RRIDs to ensure proper reagent identification [2]. Our data can be searched in the AntibodyRegistry.org and other portals that display this data such as the RRID.site portal and dkNet.org. The search term “ycharos” will bring back all of the currently available antibodies that have been characterized and searching for the target or the catalogue number of the antibody in any of these portals will also bring back the YCharOs information. In the RRID.site portal and dkNet there will also be a green star, tagging the antibodies to further highlight the contribution of YCharOS. The project is also being promoted through large international bioimaging networks including Canada BioImaging (CBI - https://www.canadabioimaging.org/), BioImaging North America (BINA - https://www.bioimagingnorthamerica.org/) and Global BioImaging (GBI - https://globalbioimaging.org/).
The potential impacts of poorly performing antibodies are well documented [17–20]; our analyses of the use of a few hundred antibodies begins to provide insight into the magnitude of the problem. While we acknowledge that our platform has clear limitations in selecting the best antibody for an application, such as using universal protocols for all tested antibodies or testing in a single cell line, the process can robustly identify antibodies that do not recognize their intended antigen. Removal of these products from the market is essential in that hundreds of published papers report the use of these poor-quality antibodies. Importantly, from a total of 409 antibodies that presented conflicting data between our characterization data and antibody supplier’s recommendations, participating companies informed us that based on our work they have withdrawn 73 antibodies from the market and changed recommendations for 153 antibodies (Supplemental Figure 4). In turn, high quality antibodies are being promoted. We expect to see additional changes and an overall improvement in the general quality of commercial reagents as more antibody characterization reports are generated.
Overall, this project provides the global life sciences community with a tremendous resource for the study of human proteins and will result in significant improvements in rigour and reproducibility in antibody-based assays and scientific discovery.
Materials and methods
Data analysis
Performance of each antibody was retrieved from the corresponding ZENODO report or publication (Table 1), for WB, IP and IF, and analyzed following the performance criteria described in Box 1. Antibody properties, application recommendations and antibody characterization strategies were taken from the manufacturers’ datasheets. Throughout the manuscript, renewable antibodies refer to monoclonal antibodies from hybridomas or recombinant antibodies (monoclonal and polyclonal recombinant antibodies) generated in vitro.
Box 1: Key definitions.
| Definition | |
|---|---|
| Successful antibody for Western blot | A successful primary antibody immunodetects the target protein, and the signal observed in the WT lysate is lost in the KO lysates (Supplemental Figure 1A). The antibody does not recognize any other protein under the conditions tested. |
| Specific, non-selective antibody for Western blot | The primary antibody specifically recognize the target protein, as well as unrelated protein(s) (Supplemental Figure 1A). |
| Successful antibody for immunoprecipitation | Under the conditions used, a successful primary antibody immunocaptures the target protein to at least 10% of the starting material (Supplemental Figure 1B), which leads to an observable depletion of the target protein from the starting material. |
| Successful antibody for immunofluorescence | A successful primary antibody immunolocalizes the target protein by generating a fluorescence signal in WT cells that is at least 1.5-fold higher than the signal in KO cells (Supplemental Figure 1C). Signal provided by such antibody staining can be easily distinguished from unspecific background and noise. |
For Figure 2A, the analysis of the antibody coverage of human proteins was performed as previously described [2] and antibodies were divided into polyclonal and renewable categories.
The correlation of antibody performance between two applications were evaluated by the McNemar test, followed by the chi-square statistic (Supplemental Figure 3). The number of antibodies was reported in each corresponding cell of the 2×2 contingency tables, and chi-square statistic was computed as follows: . The null hypothesis is (where p is the population proportion). Note that these hypotheses relate only for the cells that assess change in status, that is cell b which contains the number of antibodies which passed application #2, but failed application #1, whereas cell c contains the number of antibodies which passed application #1, but failed application #2. The test measures the effectiveness of antibodies for one application (from fail to pass) against the other application (change from pass to fail). If , the performance of one application is not correlated with the performance of another application, whereas if pb<or>pC, then antibody performance from one application can inform on the performance of the other application. The computed value is compared to the chi-square probability table to identify the p-value (degree of freedom is 1). The percentage of antibodies indicated in the double y-axis graph was computed by dividing the number of antibodies in the corresponding cell to the total number of antibodies (sum of cell a, b, c and d).
We asked participating antibody suppliers to indicate the number of antibodies eliminated from the market, and the number of antibodies for which there was a change in recommendation due to their evaluation of our characterization data (Supplemental Figure 5).
To evaluate the number of citations corresponding to each antibody (Figure 4C), we searched CiteAb (between November 2022 and March 2023) and used the provided analysis of citations per application.
The number of articles corresponding to each human target protein was assessed by searching the NIH protein database (https://www.ncbi.nlm.nih.gov/protein/) on May 4, 2023.
Resource information (alphabetical order)
Supplementary Material
Supplemental Figure 1: Schematic representations of antibody performance
(A) Schematic representations of a successful antibody (left schematic) or antibodies that fail (middle and right schematic) WB. (B) Schematic representations of a successful antibody (left schematic) or antibodies that fail (middle and right schematics) IP. (C) Schematic representation of the mosaic strategy used (left schematic). WT cells are labelled with a fluorescent cell dye (green), and KO cells are labelled with a different fluorescent cell dye (magenta) plated together as a mosaic. Schematic representations of a successful antibody (antibody #1) and a non-successful (antibody #2) for IF are shown.
Supplemental Figure 2: Analysis of antibody performance by manufacturer’s catalogue recommendation
Percentage of successful (gray) or unsuccessful (black) antibodies for the indicated applications are shown using a bar graph with stacked columns. Antibodies were divided according to whether they were recommended or not recommended by the manufacturers for the indicated applications. The percentage corresponding to each section of the bar graph is shown in the graph, and the total number of antibodies represented in each category is indicated above the corresponding bar.
Supplemental Figure 3: Correlation of antibody performance between applications
(A) Representation of a 2 × 2 contingency table used to apply the McNemar Test as well as the equation of the chi-square (Χ2) statistic used. Analysis of antibody performance correlation, represented as a contingency table and as a double y-axis graph between (B) WB and IP, (C) IF and IP and (D) IF and WB. n/s = non-significant
Supplemental Figure 4: Actions taken from participating companies
The percentage of antibodies removed from the market, or for which catalogue recommendations were modified following assessment of our data by our antibody manufacturing partners. The number of antibodies represented in each category is indicated above the corresponding bar.
| Name of the resource | RRID | Website |
|---|---|---|
| Antibody Registry | SCR_006397 | https://antibodyregistry.org |
| Cancer Dependency Map Portal (DepMap) | SCR_017655 | https://depmap.org/portal/ |
| CiteAb | SCR_009653 | https://www.citeab.com |
| F1000research (YCharOS Gateway) | - | https://f1000research.com/ycharos |
| NIH protein database | - | https://www.ncbi.nlm.nih.gov/protein/ |
| Universal Protein Resource (Uniprot) | SCR_002380 | https://www.uniprot.org/ |
| ZENODO (YCharOS community) | - | https://zenodo.org/communities/ycharos |
Acknowledgement
This work was supported by the Emory-Sage-SGC TREAT-AD center established by the National Institutes of Aging (NIA) U54AG065187 grant and additional support by RF1AG057443, by a grant from the Michael J. Fox Foundation for Parkinson’s Research (no. 18331), by a grant from the Motor Neurone Disease Association (UK), The ALS Association (USA) and ALS Canada to develop ALS-RAP, by a Canadian Institutes of Health Research Foundation Grant (FDN154305) and by the Government of Canada through Genome Canada, Genome Quebec and Ontario Genomics (OGI-210). The Structural Genomics Consortium is a registered charity (no. 1097737) that receives funds from Bayer AG, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Genome Canada through Ontario Genomics Institute (grant no. OGI-196), the EU and EFPIA through the Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant no. 875510), Janssen, Merck KGaA (also known as EMD in Canada and the United States), Pfizer and Takeda. RA is supported by a Mitacs postdoctoral followship. Images were collected and/or image processing and analysis for this manuscript was performed in the McGill University Advanced BioImaging Facility (ABIF), RRID:SCR_017697. We thank Chetan Raina (YCharOS Inc.) for his important contribution to the creation of an open scientific ecosystem of antibody manufacturers and knockout cell line suppliers.
Abbreviations:
- DepMap
Cancer Dependency Map Portal
- KO
knockout
- KD
knockdown
- ID
identifier
- IP
immunoprecipitation
- IF
immunofluorescence
- MWM
molecular weight mass
- RRID
Research Resource Identifier
- SOP
Standard Operating Procedure
- TPM
Transcripts Per Million
- WB
Western blot
Footnotes
Competing interests
The laboratory of Peter S McPherson was awarded a Genomic Applications Partnership Program grant from Genome Canada in 2021. For this project, the laboratory of Peter McPherson developed partnerships with high-quality antibody manufacturers and knockout cell lines providers. The partners provide antibodies and knockout lines to the McPherson laboratory at no cost. Partners are: Abcam, Aviva Systems Biology, Cell Signaling Technology, Developmental Studies Hybridoma Bank, GeneTex, Novus Biologicals, Horizon Discovery, Proteintech, R&D Systems, Synaptic Systems and Thermo Fisher Scientific.
References
- 1.Marx V., Change-makers bring on recombinant antibodies. Nat Methods, 2020. 17(8): p. 763–766 DOI: 10.1038/s41592-020-0915-8. [DOI] [PubMed] [Google Scholar]
- 2.Bandrowski A., et al. , The Antibody Registry: ten years of registering antibodies. Nucleic Acids Res, 2023. 51(D1): p. D358–D367 DOI: 10.1093/nar/gkac927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laflamme C., et al. , Opinion: Independent third-party entities as a model for validation of commercial antibodies. N Biotechnol, 2021. 65: p. 1–8 DOI: 10.1016/j.nbt.2021.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Ghandi M., et al. , Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature, 2019. 569(7757): p. 503–508 DOI: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews N.P., et al. , A toolbox of IgG subclass-switched recombinant monoclonal antibodies for enhanced multiplex immunolabeling of brain. Elife, 2019. 8 DOI: 10.7554/eLife.43322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray A., et al. , Animal-free alternatives and the antibody iceberg. Nat Biotechnol, 2020. 38(11): p. 1234–1239 DOI: 10.1038/s41587-020-0687-9. [DOI] [PubMed] [Google Scholar]
- 7.Laflamme C., et al. , Implementation of an antibody characterization procedure and application to the major ALS/FTD disease gene C9ORF72. Elife, 2019. 8 DOI: 10.7554/eLife.48363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis M.J., et al. , Validation of Tau Antibodies for Use in Western Blotting and Immunohistochemistry. bioRxiv, 2023: p. 2023.04.13.536711 DOI: 10.1101/2023.04.13.536711. [DOI]
- 9.Lutz A.K., et al. , SHANK3 Antibody Validation: Differential Performance in Western Blotting, Immunocyto- and Immunohistochemistry. Front Synaptic Neurosci, 2022. 14: p. 890231 DOI: 10.3389/fnsyn.2022.890231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies P., et al. , Comprehensive characterization and optimization of anti-LRRK2 (leucine-rich repeat kinase 2) monoclonal antibodies. Biochem J, 2013. 453(1): p. 101–13 DOI: 10.1042/BJ20121742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uhlen M., et al. , A proposal for validation of antibodies. Nat Methods, 2016. 13(10): p. 823–7 DOI: 10.1038/nmeth.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter A.J., et al. , Target 2035: probing the human proteome. Drug Discov Today, 2019. 24(11): p. 2111–2115 DOI: 10.1016/j.drudis.2019.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Baker M., Reproducibility crisis: Blame it on the antibodies. Nature, 2015. 521(7552): p. 274–6 DOI: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- 14.Baker M., When antibodies mislead: the quest for validation. Nature, 2020. 585(7824): p. 313–314 DOI: 10.1038/d41586-020-02549-1. [DOI] [PubMed] [Google Scholar]
- 15.Gray A.C., et al. , Animal-Friendly Affinity Reagents: Replacing the Needless in the Haystack. Trends Biotechnol, 2016. 34(12): p. 960–969 DOI: 10.1016/j.tibtech.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 16.Lund-Johansen F. and Browning M.D., Should we ignore western blots when selecting antibodies for other applications? Nat Methods, 2017. 14(3): p. 215 DOI: 10.1038/nmeth.4192. [DOI] [PubMed] [Google Scholar]
- 17.Voskuil J.L.A., et al. , The Antibody Society’s antibody validation webinar series. MAbs, 2020. 12(1): p. 1794421 DOI: 10.1080/19420862.2020.1794421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aponte Santiago N., et al. , Tales of the unexpected. Elife, 2023. 12 DOI: 10.7554/eLife.87444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freedman L.P., et al. , [Letter to the Editor] The need for improved education and training in research antibody usage and validation practices. Biotechniques, 2016. 61(1): p. 16–8 DOI: 10.2144/000114431. [DOI] [PubMed] [Google Scholar]
- 20.Sato Y., et al. , Nonspecific binding of common anti-CFTR antibodies in ciliated cells of human airway epithelium. Sci Rep, 2021. 11(1): p. 23256 DOI: 10.1038/s41598-021-02420-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Schematic representations of antibody performance
(A) Schematic representations of a successful antibody (left schematic) or antibodies that fail (middle and right schematic) WB. (B) Schematic representations of a successful antibody (left schematic) or antibodies that fail (middle and right schematics) IP. (C) Schematic representation of the mosaic strategy used (left schematic). WT cells are labelled with a fluorescent cell dye (green), and KO cells are labelled with a different fluorescent cell dye (magenta) plated together as a mosaic. Schematic representations of a successful antibody (antibody #1) and a non-successful (antibody #2) for IF are shown.
Supplemental Figure 2: Analysis of antibody performance by manufacturer’s catalogue recommendation
Percentage of successful (gray) or unsuccessful (black) antibodies for the indicated applications are shown using a bar graph with stacked columns. Antibodies were divided according to whether they were recommended or not recommended by the manufacturers for the indicated applications. The percentage corresponding to each section of the bar graph is shown in the graph, and the total number of antibodies represented in each category is indicated above the corresponding bar.
Supplemental Figure 3: Correlation of antibody performance between applications
(A) Representation of a 2 × 2 contingency table used to apply the McNemar Test as well as the equation of the chi-square (Χ2) statistic used. Analysis of antibody performance correlation, represented as a contingency table and as a double y-axis graph between (B) WB and IP, (C) IF and IP and (D) IF and WB. n/s = non-significant
Supplemental Figure 4: Actions taken from participating companies
The percentage of antibodies removed from the market, or for which catalogue recommendations were modified following assessment of our data by our antibody manufacturing partners. The number of antibodies represented in each category is indicated above the corresponding bar.