Abstract
The emergence of synthetic cannabinoid receptor agonists (SCRAs) as illicit psychoactive substances has posed considerable public health risks that include fatalities. Many SCRAs exhibit much higher efficacy and potency, compared with the phytocannabinoid Δ9-tetrahydrocannabinol (THC), at the cannabinoid receptor 1 (CB1R), a G protein-coupled receptor involved in modulating neurotransmitter release. In this study, we investigated structure activity relationships (SAR) of aminoalkylindole SCRAs at CB1Rs, focusing on 5F-pentylindoles containing an amide linker attached to different head moieties. Using in vitro bioluminescence resonance energy transfer (BRET) assays, we identified a few of SCRAs exhibiting significantly higher efficacy in engaging the Gi protein and recruiting β-arrestin than the reference CB1R full agonist CP55940. Importantly, adding a methyl group at the head moiety of 5F-MMB-PICA yielded 5F-MDMB-PICA, an agonist exhibiting a large increase in efficacy and potency at the CB1R. This pharmacological observation was supported by a functional assay of the effects of these SCRAs on glutamate field potentials recorded in hippocampal slices. Molecular modeling and simulations of the CB1R bound with either of the SCRAs revealed critical structural determinants contributing to the higher efficacy of 5F-MDMB-PICA, and how these subtle differences propagated to the receptor-G protein interface. Thus, we find that apparently minor structural changes in the head moiety of SCRAs can cause major changes in efficacy. Our results highlight the need for close monitoring of structural modifications of newly emerging SCRAs and their potential for toxic drug responses in humans.
Keywords: synthetic cannabinoids, cannabinoid receptor 1, bioluminescence resonance energy transfer, molecular dynamics
INTRODUCTION
Synthetic cannabinoid receptor agonists (SCRAs) were originally developed as research compounds for studying the endocannabinoid system and subsequently repurposed as nausea suppressants, appetite stimulants, and for neuropathic pain relief [1, 2]. Unfortunately, since their introduction in medicinal chemistry, SCRAs have extensively infiltrated the recreational drug market as new psychoactive substances (NPS), leading to significant public health threat due to their elevated potential to cause psychiatric and medically serious disorders as well as mortality in humans. Over the past decade, the relatively widespread use of these unregulated NPS has led to numerous cases of mass overdoses, accidental intoxications, and fatalities [3–5]. Moreover, the increased efficacy and potency of SCRAs synthesized in illicit laboratories is associated to increased risks of their adverse and toxic effects [6]. A striking example of the danger posed by SCRAs is a mass intoxication event that occurred in New York City in 2016, commonly referred to as a “zombie outbreak”. This event resulted from the use of the molecule MMB-FUBINACA, which resulted in symptoms including catatonia, seizures, and psychosis [4]. Since this event, numerous additional reports have emerged describing deaths from the use of 5F-MDMB-PINACA [3, 7]. Where investigated, it was observed that these SCRAs exhibit high potency at CB1Rs and CB2Rs. Additionally, these drugs act as full agonists at CB1Rs, which distinguishes them from endogenous cannabinoids and the well-known psychoactive constituent of cannabis, THC [8–10].
The CB1R is one of the most abundant G protein-coupled receptors in the brain that primarily activates Gi/o proteins to inhibit adenylyl cyclases to inhibit cAMP production leading to the inhibition of gene transcription and synaptic remodeling [11–13]. CB1Rs are widely expressed on axon terminals where they can be activated by endocannabinoids. CB1Rs can also activate G protein-coupled inwardly-rectifying potassium channels (GIRKs) to hyperpolarize neurons and inhibit voltage-gated calcium channels (VGCCs), primarily through liberation of G protein β and γ subunits [14–17]. The inhibition of neurotransmitter release by CB1Rs has been identified in many neuronal pathways, including neurotransmission mediated by glutamate and GABA, acetylcholine, and monoamines, such as serotonin [14–17]. In addition, CB1Rs expressed on astrocytes are reported to regulate the release of gliotransmitters that can further modulate neuronal activity [18, 19]. Together, through their ability to suppress neurotransmission in many neuronal circuits, CB1Rs and the endocannabinoid system regulate a wide variety of physiological functions, including learning and memory, motivation, pain, and anxiety [10, 12].
It is noteworthy that SCRAs induce a distinct subset of adverse effects such as nausea, anxiety, hypothermia, hallucination, catalepsy, tachycardia, hypertension, and withdrawal, which are unlike those associated with phytocannabinoids [20–24]. Many SCRAs share an aminoalkylindole scaffold, which is divided into four connected moieties known as the “head, linker, core, and tail” [1]. Structure-activity studies have identified several high efficacy agonists at the CB1R, including 4CN-CUMYL-BUTINACA, 5F-MMB-PINACA, and 5F-CUMYL-PINACA, among others. All of these exhibit much higher potency and efficacy at CB1Rs, compared to phytocannabinoids and many first generation SCRAs, and this has been associated with much higher incidences of neurological symptomology [25]. Additionally, some adverse effects of these SCRAs may also be attributed to off-site interactions with non-canonical receptor targets [26].
Crystallographic and cryo-EM studies of CB1Rs have greatly advanced our mechanistic understanding of their function, revealing both active and inactive conformational states at the atomistic level [27–32]. Notably, the active molecular structures bind to a variety of CB1R agonists, not only the widely used reference SCRA CP55940, but also a few other SCRAs such as AM11542, AM841, and MDMB-FUBINACA. Importantly, the active CB1R structures in complex with Gi protein can serve as excellent templates for investigating the SAR of SCRAs.
Synthetic orthosteric agonists that share binding sites yet elicit a higher maximum response than the endogenous agonist, resulting in Emax greater 100%, are considered as superagonists for the target receptor [33, 34]. However, this concept is often contentious as the readouts used to compare efficacies may be influenced by signal amplification, which leads to higher apparent Emax values. Despite the complexities in assay interpretation and subject receptor system, there are instances where synthetic ligands confer superagonism [35–37].
5F-pentylindoles, a subgroup of highly potent and efficacious SCRAs, have been associated with a surge in human use and coinciding hospital visits [24]. In this study, we aim to investigate 5F-pentylindoles and analyze their analogues especially within their “head” moiety that manifests as efficacy above the full agonist. Here, we provide molecular mechanisms that may explain superagonism at the CB1R.
RESULTS
Subtle changes in the head moiety of 5F-pentylindoles result in distinct pharmacological profiles of Gi1 engagement
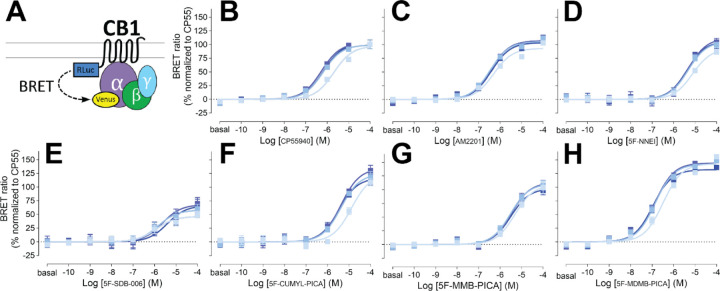
Aminoalkylindole SCRAs, such as JWH018 and AM2201, which exhibit high potency and efficacy at CB1Rs, were widely abused in early 2010’s [38–40]. In this study, we investigated the SAR of aminoalkylindole SCRAs specifically with a 5F-pentylindole scaffold and focused on several molecules containing an amide linker attached to different head moieties. To study CB1R-Gi1 coupling, we used G protein engagement BRET in heterologous HEK293 cells [41]. This method has the advantage over other functional assays such as adenylyl cyclase inhibition, as it provides a direct measurement of Gi1 coupling and is not confounded by potential off-target receptor activation of SCRAs. We collected readouts at four time points (2, 16, 30, 44 min) of ligand incubation to capture kinetic profiles of receptor G-protein coupling, as lipophilic ligands are known to have binding kinetics different from hydrophilic ones [42, 43]. Results obtained at the 16-min time point were utilized in the analysis as the responses of most agonists reached a stable plateau by this time point that did not change.
Our results show that AM2201 displays a similar potency and efficacy profile to a commonly used reference full agonist CP55940, with a slightly higher Emax, although the difference is not significant (Table 1). Conversion of the ketone moiety of AM2201 to an amide linker results in a less potent compound (N-1-naphthyl, 5F-NNEI) compared to AM2201 (1-naphthyl) (see Figure 1, Supplementary Figure 1, Table 1). Substitution of 1-naphthyl of 5F-NNEI with a less bulky benzyl group (N-1-benzyl, 5F-SDB-006) substantially lowers the efficacy but slightly increases the potency relative to 5F-NNEI.
Table 1.
Pharmacological comparison of 5-fluoropentylindole ligands at CB1R.
| CB1R-Gi1 engagement at 16 min | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CP55940 | AM2201 | 5F-NNEI | 5F-SDB-006 | 5F-CUMYL-PICA | 5F-MMB-PICA | 5F-MDMB-PICA | WIN55-212-2 | ∆9-THC | AEA | 2AG | |
| Emax (%) | 100.0±3.2 | 105.8±2.3 | 106.2±6.5 | 57.9±6.2**** | 124.2±6.7* | 111.2±5.2 | 142.1±2.7**** | 98.8±5.2 | 36.1±4.1****# | 57.8±3.1****# | 61.2±2.7****# |
|
| |||||||||||
| pEC50 (M) | 6.19±0.08 | 6.47±0.06 | 5.33±0.12* | 5.38±0.25 | 5.38±0.11 | 5.53±0.10 | 6.85±0.06 | 5.00±0.09**** | ND | 4.92±0.49 | 5.21±0.16 |
| CB1R-βarrestin2 recruitment at 16 min | |||||||||||
| CP55940 | AM2201 | 5F-NNEI | 5F-SDB-006 | 5F-CUMYL-PICA | 5F-MMB-PICA | 5F-MDMB-PICA | WIN55-212-2 | ∆9-THC | AEA | 2AG | |
| Emax (%) | 100.0±3.1 | 106.5±1.9 | 110.3±2.9 | 46..0±2.5**** | 91.0±3.8 | 99.9±4.1 | 147.7±1.6**** | 121.3±4.7# | 36.0±7.1****# | 49.9±3.0****# | 85.3±4.9# |
|
| |||||||||||
| pEC50 (M) | 5.19±0.06 | 6.12±0.04* | 5.28±0.05 | 5.61±0.12 | 5.86±0.10* | 5.44±0.09 | 6.24±0.03* | 4.72±0.07 | ND | 4.95±0.11 | 4.55±0.92 |
Mean ± SEM values for Emax and pEC50 are reported. Emax is normalized against CP55940.
Y-axis cut-off value at the highest concentration is shown. ND – EC50 cannot be determined. One-way ANOVA followed by Tukey test
p<0.05
p<0.0001 against corresponding CP55940 in the same assay.
Figure 1. Gi1 engagement BRET.
Drug-induced engagement BRET between CB1R-Rluc and Gi1-Venus (A) measured in response to CP55940 (B), AM2201 (C), 5F-NNEI (D), 5F-SDB-006 (E), 5F-CUMYL-PICA (F), 5F-MMB-PICA (G), 5F-MDMB-PICA (H) at various time points (2, 16, 30, 44 min light to dark blue). Concentration-response curves are plotted as a percentage of maximal response by CP55940 at each time point and presented as means ± SEM of n ≥ 3 independent experiments.
Replacing the benzyl group of 5F-SDB-006 with a cumyl group, which effectively adds two methyl groups and results in 5F-CUMYL-PICA, substantially increased Emax to 124% of that of the reference CP55940. Substituting the benzyl group of 5F-SDB-006 with a methyl 3-methylbutanoate group, resulting in 5F-MMB-PICA, also increased the efficacy, which was comparable to 5F-NNEI. Most strikingly, adding an extra methyl group at the 3 position of 5F-MMB-PICA, resulting in 5F-MDMB-PICA, dramatically increased both the efficacy and potency of the compound. For comparison, we also included concentration response curves for additional structurally unrelated cannabinoid agonists (see Supplementary Figure 2, Table 1). Among them, Δ9-THC, anandamide, and 2-arachidonoylglycerol showed a lower potency and efficacy than CP55940.
SAR for β-arrestin 2 recruitment by 5F-pentylindoles is similar to that for G protein engagement
β-arrestins play a crucial role in both desensitizing receptors and initiating their own signaling cascades [44]. Moreover, β-arrestin 2 is also involved in CB1R desensitization and internalization [45–49]. Therefore, we compared G protein engagement and β-arrestin recruitment by CB1Rs using 5F-pentylindole agonists. To ensure an unbiased comparison in a heterologous system, the same CB1R Rluc-fusion construct used for the CB1R-Gi1 coupling was also used for β-arrestin 2 recruitment (see Methods).
It has been reported that agonists have lower potencies for β-arrestin recruitment compared to G protein coupling [50]. As expected, most of the agonists tested in this study demonstrated ~3-fold weaker potencies for β-arrestin 2 recruitment than for Gi1 engagement (Figure 2, Table 1). Nonetheless, the potency ranking among SCRAs for β-arrestin 2 recruitment was similar to that observed for Gi1 engagement. Specifically, the ketone-to-amide linker change resulted in decreased potency, as observed in the comparison between AM2201 and 5F-NNEI. Interestingly, among all the tested 5F-pentylindoles with amide-linker, only 5F-MDMB-PICA showed a higher potency than AM2201. Furthermore, similar to the Gi1 engagement assay, the addition of one extra methyl group from 5F-MMB-PICA to 5F-MDMB-PICA led to a more than 6-fold increase in potency. The 5F-SDB-006 ligand had the lowest efficacy among the tested 5F-pentylindoles, while 5F-MDMB-PICA showed a much higher efficacy than 5F-MMB-PICA. Additionally, dose response profiles for other structurally unrelated cannabinoid agonists are displayed for comparison (Supplementary Figure 3, Table 1). Among them, THC, anandamide, and 2-arachidonoylglycerol showed lower potency and efficacy than CP55940.
Figure 2. β-arrestin 2 recruitment BRET.
Drug-induced recruitment BRET between CB1R-Rluc and β-arrestin 2-Venus (A) measured in response to CP55940 (B), AM2201 (C), 5F-NNEI (D), 5F-SDB-006 (E), 5F-CUMYL-PICA (F), 5F-MMB-PICA (G), 5F-MDMB-PICA (H) at various time points (2, 16, 30, 44 min light to dark blue). Concentration-response curves are plotted as a percentage of maximal response by CP55940 at each time point and presented as means ± SEM of n ≥ 3 independent experiments.
No significant bias was observed between G protein engagement and β-arrestin recruitment
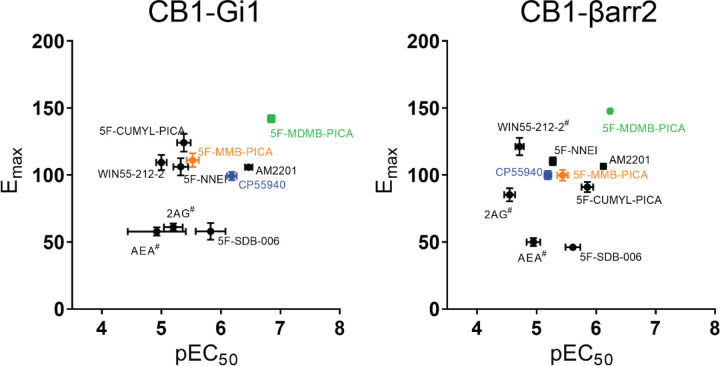
Signaling bias towards G protein activation or β-arrestin recruitment can result in downstream responses that deviate from those of balanced endogenous agonists. These differences could underlie the unique physiological outcomes reported for the use of SCRA in humans. We plotted the efficacies and potencies of various SCRAs and reference agonists for Gi1 engagement and β-arrestin 2 recruitment (Figure 3). Many of the 5F-pentylindoles showed similar profiles for both signaling pathways, including 5F-MMB-PICA and 5F-MDMB-PICA (orange and green symbols in Figure 3). Of these, 5F-MDMB-PICA showed the highest efficacy and potency for both pathways. CP55940 noticeably showed a lower potency for β-arrestin 2 recruitment relative to other compounds (blue symbol, Figure 3). None of the agonists showed significant bias towards either pathway (i.e., with a bias factor >2.00 or <−2.00; Table 2).
Figure 3. Emax-pEC50 plot.
Emax-pEC50 comparison between CB1R-Gi1 engagement and β-arrestin 2 recruitment
# Y-axis cut-off value at the highest concentration is shown for its Emax.
Table 2.
Coupling bias between Gi1 and β-arrestin 2 among the tested CB1R agonists
| Bias factor | |
|---|---|
| CP55940 | 0.00 |
| AM2201 | −0.65 |
| 5F-NNEI | −0.97 |
| 5F-SDB-006 | −0.68 |
| 5F-CUMYL-PICA | −1.34 |
| 5F-MMB-PICA | −0.86 |
| 5F-MDMB-PICA | −0.41 |
| WIN55-212-2 | −0.76 |
In both the Gi1 engagement and β-arrestin 2 recruitment assays, 5F-MDMB-PICA demonstrated the highest efficacy among all the compounds tested. This represented an increase of more than 40% compared to CP55940 and 5F-MMB-PICA, which has only one less methyl group in the head moiety. Therefore, to further characterize the actions of SCRAs at the CB1R, we focused on comparing 5F-MDMB-PICA and 5F-MMB-PICA in subsequent ex vivo recording and molecular simulation studies.
Significant efficacy difference was observed between 5F-MDMB-PICA and 5F-MMB-PICA at presynaptic CB1Rs in the hippocampus
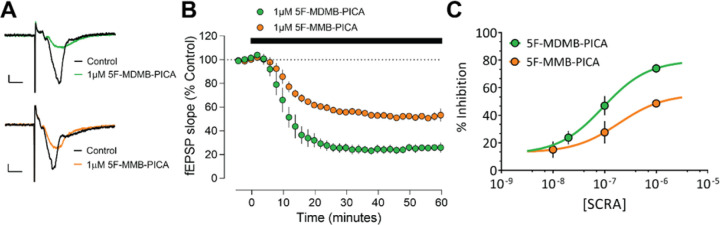
CB1Rs are expressed at the presynaptic terminals of neurons to regulate neurotransmitter release in the brain. In the hippocampus, CB1Rs are found on pyramidal neuron axonal terminals, known as Schaffer collateral/commissural (Scc) fibers projecting from the CA3 to CA1 regions [51]. Activation of CB1Rs results in Gi/o protein-mediated inhibition of voltage-dependent Ca2+ channels, leading to a reduction in glutamate release from Scc fibers in CA1 [51–53]. We utilized extracellular recordings of the rising slopes of field excitatory postsynaptic potentials (fEPSPs), evoked by electrical stimulation of Scc fibers in hippocampal brain slices, to compare the effects of 5F-MDMB-PICA and 5F-MMB-PICA on glutamate release [54].
As previously demonstrated with other synthetic CB1R agonists [55], fEPSPs were significantly reduced by bath application of either 5F-MDMB-PICA or 5F-MMB-PICA (1 µM, Figure 4A). As depicted in a time-course plot of fEPSP slope, the effects of both agonists are apparent within 10 min of application with calculated tau values of 6.34 and 8.80 min for 5F-MDMB-PICA and 5F-MMB-PICA, respectively (Figure 4B). However, the maximal inhibition produced by 5F-MDMB-PICA was significantly greater than that observed with 5F-MMB-PICA. Thus, concentration-response curves revealed significantly different Emax values for the two SCRAs (Figure 4C, t-test P = 0.0255). In order to confirm the CB1R-dependence of these effects, we applied the CB1R-selective antagonist AM251 after the effects of the agonists were stable. We found that AM251 fully reversed the effects of both agonists (Supplementary Figure 4).
Figure 4. Brain slice electrophysiology.
Concentration‐dependent inhibition of hippocampal glutamate release by 5F-MDMB-PICA and 5F-MMB-PICA. (A) Representative averaged traces for 5F-MDMB-PICA (upper) and 5F-MMB-PICA (lower), (B) Summary time course of recordings (n ≥ 3 recordings) demonstrating the effect of SCRAs. (C) Concentration–response curve for 5F-MDMB-PICA and 5F-MMB-PICA (n ≥ 3 slices per concentration). The pEC50 was calculated to be 7.20 and 6.72 for 5F-MDMB-PICA and 5F-MMB-PICA respectively. Data points are presented as means ± SEM.
Adding an extra methyl group to the ligand head moiety changes the dynamics of the agonist binding pocket
The in vitro and ex vivo assays demonstrated that 5F-MDMB-PICA is significantly more efficacious than 5F-MMB-PICA, with the latter’s efficacy being comparable to that of the reference full agonist CP55940 (Figure 3). This observation led us to investigate the molecular mechanism underlying 5F-MDMB-PICA’s higher efficacy at the CB1R. That only the addition of a single methyl group to the head moiety of 5F-MMB-PICA could cause such a large increase in 5F-MDMB-PICA’s efficacy permitted us to investigate how this affects ligand-receptor interactions, and the propagation to receptor/G-protein coupling. To do this, we conducted comparative molecular modeling and simulations of the CB1R bound to each of these SCRAs.
We used the CB1R/Gαi protein complex bound with MDMB-FUBINACA (Figure 5A; PDB 6N4B), as the template to construct the CB1R/5F-MDMB-PICA and CB1R/5F-MMB-PICA complex models (see Methods). In the structure of 6N4B, the head moiety and linker of MDMB-FUBINACA occupy a pocket that is enclosed by residues from transmembrane helices (TMs) 1, 2, and 7, while its p-fluorobenzyl tail is enclosed by TMs 3, 5, 6, and second extracellular loop (ECL2). Notably, 5F-MDMB-PICA and MDMB-FUBINACA share the same head moiety, while 5F-MMB-PICA has one less methyl group than these two ligands (Supplementary Figure 5). Therefore, we assumed that 5F-MDMB-PICA or 5F-MMB-PICA would have a similar head-tail orientation as MDMB-FUBINACA in the ligand binding pocket (Supplementary Figure 5), and therefore selected their corresponding docking poses accordingly to construct the complex models. As a control, we also simulated the CB1R/MDMB-FUBINACA model (Table 3). During our simulations, 5F-MMB-PICA and 5F-MDMB-PICA maintained their binding poses in the same orientation as MDMB-FUBINACA (Figure 5C) but exhibited some variations in both the local interactions within the binding pocket and the resulting global receptor conformation.
Figure 5. Ligands’ dihedral angle and binding pocket.
(A) The zoom-out view of the receptor (grey) bound with Gαi (dark grey) and MDMB-FUBINACA (magenta). (B) The binding mode of 5F-MDMB-PICA (green), MDMB-FUBINACA (magenta), and 5F-MMB-PICA (yellow and orange) and their associated binding site residues that show differences in contact frequency between 5F-MDMB-PICA, MDMB-FUBINACA, and 5F-MMB-PICA. The residues that show differences are color coded correspondingly. (C) The zoom-in view of the superimposed ligands in the binding site. They occupy the same region in binding pocket but 5F-MMB-PICA picks two distinct conformers for head moiety. (D) The 2D map of head moiety dihedral angles (Supplementary Figure 1).
Table 3.
Summary of simulated conditions and simulation lengths.
| Ligand | Number of trajectories | Length of trajectories (μs) |
|---|---|---|
| 5F-MDMB-PICA | 5 | 14.5 |
| 5F-MMB-PICA | 6 | 14.5 |
| MDMB-FUBINACA | 3 | 6.6 |
Based on the equilibrated simulation results, we first assessed the impact of the extra methyl group on the ligand binding poses and measured two dihedral angles in the head moiety for each bound ligand (designated as and , Supplementary Figure 5). The distribution of revealed that 5F-MMB-PICA had two distinct modes in the binding site, with (~50 ≤ peak 1 ≤ ~115 and ~135 ≤ peak 2 ≤ ~180), resulting from the rotation of the head moiety around the N-C bond (Figure 5D). In contrast, there appeared to be minimal energy barriers for the head moieties of the bound 5F-MDMB-PICA and MDMB-FUBINACA to transition between these two modes. The dihedral angle did not exhibit any noticeable differences among the three ligands.
We hypothesized that the distinct dynamics of the ligand head moiety in the binding pocket might impact their interactions with the receptor. Consequently, to evaluate the effects of the extra methyl group on the dynamics of the binding site, we calculated the contact frequency and residues’ sidechain dihedral angles for each ligand (see Methods). The outcomes revealed that contact residues were present in TM2, TM3, ECL2, TM5, TM6, and TM7. TM2 and TM3 had a more significant role in ligand binding interactions than other segments (Supplementary Table 1). Notably, our studies revealed clear distinctions among the three simulated conditions investigated. The contact frequency and sidechain dihedral angle analyses suggested that 5F-MDMB-PICA and MDMB-FUBINACA, with the same head moiety, shared similarities for most of their contacting residues, and their differences with 5F-MMB-PICA were similar (Supplementary Table 1). Specifically, several residues from TM2 (I1692.56, V1712.58, S1732.60, and H1782.65), TM3 (G1943.30), and ECL2 (I267ECL2, P269ECL2, and H270ECL2), which all interact with the ligand head moieties, showed similar differences.
In particular, our analysis shows that H1782.65 plays a crucial role in ligand interactions, with both 5F-MDMB-PICA and MDMB-FUBINACA interacting more frequently (~100%) with this residue than 5F-MMB-PICA (46%). In addition, our results reveal that 5F-MDMB-PICA interacts more with I267ECL2, I269ECL2, and H270ECL2 compared to 5F-MMB-PICA, as do I267ECL2 and H270ECL2 in the case of MDMB-FUBINACA (Supplementary Table 1, Figure 5B). Furthermore, we observed that the dynamics of the ligands in the binding pocket can induce changes in the sidechain conformations of certain residues, as highlighted in Supplementary Table 1. For example, the rotamer of S1732.60 for 5F-MDMB-PICA and MDMB-FUBINCA has more than 70° difference from that 5F-MMB-PICA. Additionally, there are noticeable differences in the sidechain conformation of P269ECL2 between 5F-MDMB-PICA/5F-MMB-PICA and MDMB-FUBINACA/5F-MMB-PICA, (Supplementary Table 1) which could be related to their different puckering states.
The additional methyl group in head moiety causes conformational changes in TM1 and TM2
To investigate the conformational changes beyond the ligand pocket, we first compared the available CB1R structures in the active and inactive states. The CB1R shares activation features with other class A GPCRs, including outward movement of the intracellular portion of TM6 (TM6i) and inward movement of TM7i, accompanied by changes in conserved motifs [56]. Our quantitative analysis, shown in Supplementary Figure 6, demonstrates these movements between the active CB1R-Gαi structures (PDB: 6N4B, 6KPG, and 7WV9) [28–30] and the inactive structures (PDB: 5U09 [31] and PDB: 5TGZ [32]). In addition to these features, CB1R also exhibits inward movements of the extracellular portion of TM1 (TM1e) and TM2e with the displacement of the N terminus upon activation [28].TM2 plays a distinct role in CB1R activation by engaging the ligand in hydrophobic/polar interactions with TM2e and stabilizing the agonist binding conformation[28, 32]. However, 6N4B shows another interesting feature at the extracellular domain which is the inward movement of TM6e towards TM1e and TM2e (Supplementary Figure 6).
For our simulation results, after performing RMSD-based clustering of CB1R TM domain and conducting the PIA-GPCR analysis, we observed conformational differences in the TM1e and TM2e region between CB1R/5F-MDMB-PICA and CB1R/5F-MMB-PICA (Figure 6A). Specifically, we noted that in CB1R/5F-MDMB-PICA, TM1e and TM2e moved inward towards the center of the ligand binding pocket, in contrast to their configuration in CB1R/5F-MMB-PICA. As a result of the inward movement of TM2 in CB1R/5F-MDMB-PICA, key residues in TM2e, such as H1782.65, had more frequent interactions with the head moiety of 5F-MDMB-PICA in the binding pocket (Supplementary Table 1), thus stabilizing the bound 5F-MDMB-PICA.
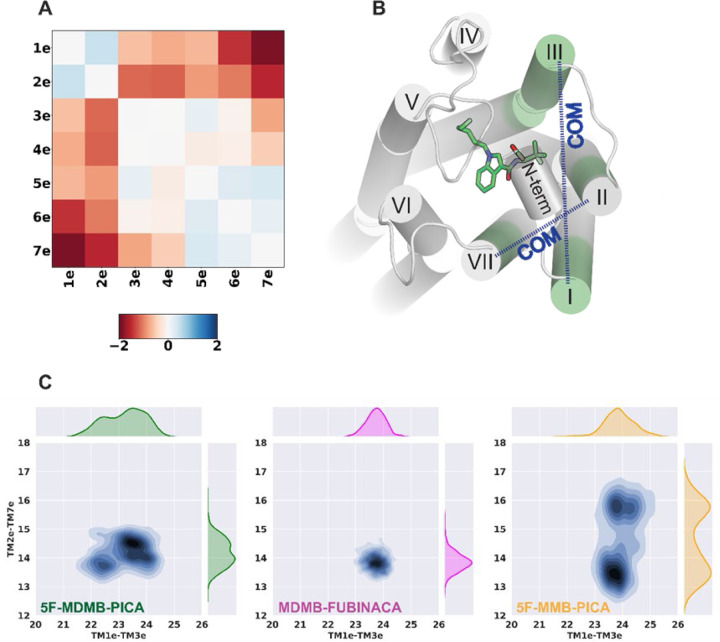
Figure 6. PIA-GPCR analysis and TMs’ COM distribution.
(A) PIA-GPCR COM results for TMs’ extracellular domain (ΔTMe(COM) = TMe(COM)5F-MDMB-PICA – TMe(COM)5F MMB-PICA). (B) The 5F-MDMB-PICA in the binding pocket. The highlighted transmembrane sub-segments are extracellular part of TM1, TM2, TM3, and TM7 (denoted as TM1e, TM2e, TM3e, and TM7e). The blue dashed arrows show the COM distance between TM1e – TM3e and TM2e – TM7e. (C) The 2D distribution of the COM distances. 5F-MMB-PICA shows two distinct distributions for TM2e – TM7e COM distance which seems to be the result of the changes in head moiety seen for this ligand.
To further characterize the conformational changes in this receptor region that accommodate the head groups of the SCRAs, we analyzed the center-of-mass (COM) distances between two pairs of transmembrane subsegments: TM1e – TM3e and TM2e – TM7e (Figure 6B). The COM distance distributions between TM2e – TM7e shows two distinct peaks for 5F-MMB-PICA compared to 5F-MDMB-PICA and MDMB-FUBINCA (Figure 6C). This could be due to the two conformations that the head moiety of 5F-MMB-PICA can adopt. The extra methyl group of 5F-MDMB-PICA results in a more stable conformation for its head moiety, which is associated with the changes in the dynamics of the extracellular regions of the receptor surrounding the head moiety.
Divergence in the allosteric interaction network between 5F-MDMB-PICA and 5F-MMB-PICA
We then carried out a correlation-based network analysis (see Methods) to compare how the bindings of 5F-MDMB-PICA and 5F-MMB-PICA may induce different conformational changes from the ligand binding pocket to the receptor-G protein interface. Our results identified several significant differences between CB1R/5F-MDMB-PICA and CB1R/5F-MMB-PICA conditions, as illustrated in Figure 7A.
Figure 7. Correlation-based network pathway.
(A) The CB1R structure bound with 5F-MDMB-PICA with final correlated pairs mapped on the structure. The green pairs are representative pairs for 5F-MDMB-PICA and the yellow pairs are representative for 5F-MMB-PICA. There is a noticeable difference between these two ligands signaling pathway. The CB1R/5F-MDMB-PICA network includes a pathway consisting of eight strongly correlated pairs (solid green lines) which connect the E1331.49-L3997.55 (TM1i – TM7i) changes to the changes at the extracellular-middle domain, F1742.61-D1762.63 (TM2m – TM2e), the intracellular domain, A1602.47-A3987.54 (TM2i – TM7i), H1431.59-F408H8 (TM1i – H8), T3446.36-L3997.55 (TM6i – TM7i), H406H8-R409H8 (H8 – H8), and the changes between the intracellular domain and Gα,T313ICL3-S265Gα (ICL3 – Gα), T313 ICL3-E318Gα (ICL3 – Gα), and R4007.56- K349Gα (TM7i – Gα). E1331.49-L3997.55 (TM1i – TM7i) is at the core of these correlated pairs. There is no such pathway for CB1R/5F-MMB-PICA. (B) The TM2e key residue H1782.65 contributes to the 5F-MDMB-PICA signaling pathway. (C) The TM2i residue (F1552.42) and TM7i NPxxY residues (P3947.50 and Y3977.53) are unique to 5F-MDMB-PICA network pathway
Specifically, TMs 1, 2, and 7 are more engaged in the allosteric network for CB1R/5F-MDMB-PICA, while TMs 3,4, and 5 are more involved for the CB1R/5F-MMB-PICA. The analysis shows that ~45% of TM2 residues are part of the strongly correlated network for CB1R/5F-MDMB-PICA while only 14% of TM2 residues are involved in the network for CB1R/5F-MMB-PICA. Notably, the two key residues in CB1R binding (H1782.65) (Figure 7B) and activation (F1552.42) (Figure 7C) [29] contribute to the network for CB1R/5F-MDMB-PICA but not for CB1R/5F-MMB-PICA. Similarly, for TM7, the contribution of residues for CB1R/5F-MDMB-PICA and CB1R/5F-MMB-PICA is 44% and 15%, respectively. Among these residues, P3947.50 and Y3977.53 from the NPxxY motif are unique to the CB1R/5F-MDMB-PICA network (Figure 7C). As reviewed and described above, TM2 and TM7 have been shown to play critical roles in CB1R activation. Therefore, such distinct TM and residue involvements between these two conditions may be associated with the different efficacies of 5F-MDMB-PICA and 5F-MMB-PICA at CB1R.
Additionally, the number of correlated residue pairs for CB1R/5F-MDMB-PICA is almost twice that of CB1R/5F-MMB-PICA. The results showed pairs with strong correlations that connect the changes in binding pocket between the 5F-MDMB-PICA and receptor residues to other pairs (receptor-receptor, receptor-Gαi). While these residue pairs do not form a complete pathway for either CB1R/5F-MDMB-PICA or CB1R/5F-MMB-PICA, the rigid body movement of the receptor regions between them likely propagate the impact.
Thus, our results underscore the contributions of the highly conserved NpxxY motif [56], as well as the key residues H2.65 and F2.42 [29] in the allosteric network that may be associated with receptor activation.
DISCUSSION
Here we demonstrated substantial increase in the efficacy and potency of a 5-fluoropentylindole SCRA through the addition of a single methyl group to the head moiety (i.e., 5F-MDMB-PICA). This modification led to superagonism, which refers to the phenomenon where agonist efficacy surpasses that of endogenous agonists [33]. Although “superagonism” has a clear definition, its interpretation can often be complicated when applied to assays that not directly detect receptor activation status. These results can be confounded by signal amplification when large receptor reserves are present, which can lead to inaccurate representation of the intrinsic efficacy of the agonist [57]. The BRET assays utilized in this study rely on the direct coupling between a receptor and a transducer, which can effectively avoid signal amplification and detect transducer engagement or recruitment resulting from receptor activation. However, compared to the cAMP assay, the use of fusion constructs in BRET assays, which enable specificity and direct coupling, may lead to comparatively lower potencies. Nonetheless, a comparison between the test ligands and a reference CP55940 ensures proper calibration of the relative potencies of SCRAs. Our finding of 5F-MDMB-PICA as a superagonist at the CB1R is supported not only by our own electrophysiological results but also by previous studies in cellular functional assays measuring membrane potential [8, 40], and the cannabinoid triad behavioral assays [58]. Notably, the presynaptic inhibition measured using field recordings in brain slices is known to be sensitive to differences in pharmacological properties of CB1R ligands that possess an adequate “ceiling” to characterize high efficacy agonists.
Addition of a methyl substituent, although not a dramatic structural change, is known to yield profound ligand-receptor interactions in many cases [59, 60], as observed here with the methyl-3-methylbutanoate to methyl-3,3-dimethylbutanoate modification (i.e., MMB- to MDMB- change). Extensive interactions between the methyl-3,3-dimethylbutanoate head moiety and the TM2e region were demonstrated in the cryo-EM structure [28], and our computational modeling demonstrated how these interactions can significantly affect the potency and efficacy of 5F-MDMB-PICA compared to 5F-MMB-PICA. Our simulation results also indicated additional interactions of 5F-MDMB-PICA with ECL2 residues, compared to 5F-MMB-PICA, which maybe also relate to its higher potency and efficacy. This finding aligns with previous mutagenesis work of the CB1R, which have highlighted the critical role of ECL2 in ligand binding [28, 61], G protein coupling, and receptor trafficking [61]. In particular, alanine scanning mutations on both N- and C-terminal regions of ECL2 have consistently resulted in more than a 10-fold increase in EC50 with significantly reduced Emax values, suggesting the indispensability of these residues for both ligand binding and receptor activation [61].
Since the initial reports of illicit usage around 2010, there has been an increasing recognition of the association between the potency and efficacy of SCRAs at CB1Rs and their clinically adverse effects [62]. Several studies have focused on assessing the pharmacological properties of the head, linker, core, and tail moieties comprising SCRAs [63, 64]. Consistent with the findings of our current study, previous data have also indicated that even minor structural variations within SCRAs can lead to significant efficacy changes, thereby potentially influencing clinical toxicity [6, 25, 65]. Among them, 5F-MDMB-PICA, a schedule-I drug, has been identified as one of the most prevalent SCRAs in United States, as of 2019 [66]. Fatal events have been associated with the use of this drug [67, 68], as well as reports of notably strong depressant effects [69], and other toxic interactions [70]. Our present findings suggest that even minor structural modifications in newly emerging SCRAs, particularly in relation to the presence of substituted head moieties, can strongly increase SCRA efficacy, and potentially contribute to the adverse effects occurring with human consumption of these illicit compounds. Our study can be used to further our understanding of how specific alterations in cannabinoid receptor ligands may contribute to their toxicity in humans and the potential life threatening consequences of their use.
METHODS AND MATERIALS
Cell culture, constructs, BRET, analysis
Variations of bioluminescence resonance energy transfer (BRET) assay were performed to detect ligand-induced receptor-signaling protein coupling events. A constant amount of plasmid cDNA (15 μg) was transfected into human embryonic kidney cells 293 T (HEK-293T) using polyethylenimine (PEI; Sigma) in a 1:2 weight ratio in 10 cm plates. Cells were maintained in culture with Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Atlanta), 2 mM l-glutamine (Gibco), and 1% penicillin streptomycin (Gibco) and kept in an incubator at 37 °C and 5% CO2. The transfected amount and ratio among the receptor and heterotrimeric G proteins were tested for the optimized dynamic range in drug-induced BRET. Experiments were performed approximately 48 h post-transfection. As reported previously [71], cells were collected, washed, and resuspended in phosphate-buffered saline (PBS). Approximately 200,000 cells/well were distributed in 96-well plates, and 5 μM coelenterazine H (luciferase substrate) was added to each well. One minute after the addition of coelenterazine, CB1R ligands were added to each well. Two different configurations of BRET were used: (i) Gαi1-engagement and (ii) β-arrestin-2 recruitment. (i) Gi1 protein engagement assay uses Rluc-fused receptor and mVenus-fused Gαi1 for a resonance energy transfer (RET) pair. Untagged Gβ1 and Gγ2constructs were cotransfected. (ii) β-arrestin-2 recruitment uses D2R-Rluc-β-arrestin-2-Venus for a RET pair. GRK2 was cotransfected to assist an enhanced phosphorylation required for the β-arrestin-2 recruitment. The donor luminescence as well as the acceptor fluorescence were always quantified for consistent expression levels across different experiments such that no significant expression differences. For fluorescence measurement, Venus was excited at 500 nm and measured at an emission wavelength of 530 nm over 1 s, using a Pherastar FSX plate reader (BMG Labtech, Cary, NC, USA). For kinetic experiments, cells were incubated at 37 °C within the Pherastar FSX plate reader (BMG Labtech, Cary, NC, USA) with BRET signal measurements taken at various time-points ranging from 2–46 min. The BRET1 signal from the same batch of cells was calculated as the ratio of the light emitted by Venus (530 nm) over that emitted by coelenterazine H (485 nm). BRET change was defined as BRET ratio for the corresponding drug minus BRET ratio in the absence of the drug. Emax values are expressed as the basal subtracted BRET change and in the dose–response graphs. Data and statistical analysis were performed with Prism 9 (GraphPad Software). Bias factors between Gi1-engagement and β-arrestin-2 were calculated as previously described [71, 72].
Brain Slice Electrophysiology
Experiments were performed as described previously, with additional modifications [55, 71]. Brain slices were prepared from adult wildtype C57BL/6J mice (male and female) that were anesthetized with isoflurane and euthanized by decapitation. Prior to decapitation, mice were intracardially perfused with modified artificial cerebral spinal fluid (mACSF) containing (in mM): 92 NMDG, 20 HEPES, 25 glucose, 30 NaHCO3, 1.2 NaH2PO4, 2.5 KCl, 5 sodium ascorbate, 3 sodium pyruvate, 2 thiourea, 10 MgSO4, 0.5 CaCl2, 300–310 mOsm, at pH 7.3–7.4. The brains were sectioned in cold mACSF, saturated with 95% O2 and 5% CO2 (carbogen), and transverse hippocampal slices (220 µm) were obtained using a vibrating tissue slicer. The slices were immediately placed in the same buffer and maintained at 32 °C for 10 min, before moving them to a holding chamber filled with carbogen saturated ACSF (holding ACSF) containing, in mM: 92 NaCl, 20 HEPES, 25 glucose, 30 NaHCO3, 1.2 NaH2PO4, 2.5 KCl, 5 sodium ascorbate, 3 sodium pyruvate, 2 thiourea, 1 MgSO4, 2 CaCl2, 300–310 mOsm, at pH 7.3–7.4. During electrophysiological recordings, slices were continuously perfused at 2 mL/min with carbogen-saturated ACSF containing (in mM): 124 NaCl, 2.5 KCl, 1.25 NaH2PO4, 1 MgCl2, 26 NaHCO3, 11 glucose, 2.4 CaCl2, 300–310 mOsm, at pH 7.3–7.4. The temperature of the recording chamber was maintained at 31–32 °C. Brain slices were visualized using an upright microscope to confirm placement of an extracellular recording electrodes (3–5 MΩ), backfilled with ACSF, within the stratum radiatum of area CA1. A bipolar stimulating wire was positioned in area CA3A to activate Scc fibers and evoke fEPSPs. Extracellular recordings were made using a MultiClamp 700B amplifier (10 kHz low-pass Bessel filter) and Digidata 1550B (20 kHz digitization) with pClamp 11 software (Molecular Devices).
As we have previously found that endogenous adenosine can disrupt CB1R‐mediated inhibition of glutamate release in the hippocampus [71], we included the adenosine A1 receptor antagonist, caffeine (50 μM) in the ACSF throughout incubation and recordings.
The following numbers of slices were recorded for the drug perfusion conditions listed: 2×108, 10−7, 10−6 M 5F-MDMB-PICA (6 slice recordings from 4 animals); 10−8, 10−7, 10−6 M 5F-MMB-PICA (5 slice recordings from 4 animals); 2×10−8 M 5F-MDMB-PICA + 10−6 M AM251 (5 slice recordings from 5 animals); 10−6 M 5F-MMB-PICA + 3×10−6 M AM251 (6 slice recordings from 6 animals). All data are reported as mean ± SEM. Data were analyzed in Clampex and statistically compared with Prism 9 (GraphPad Software) by paired t-test.
Molecular dynamics simulations
We used the (MDMB-FUBINACA)-CB1R-Gαi crystal structure (PDB 6N4B) [28] as the starting point for our modeling and molecular dynamics (MD) simulations. The CB1R missing residues on the N-terminus (residues 104–108), extracellular loop 2 (ECL2) (residues 258–263), and helix-8 (H8) (residues 412–414) were added using MODELER (version 9.24) with CB1R structure (PDB 5XRA) [27] as the template.
The structure was further processed and refined using the Protein Preparation Wizard and Ligand Preparation implemented in Maestro (Schrödinger suite 2019–4). In addition, the residues D1632.50 and D2133.49 were protonated to their neutral forms as assumed in the active state of rhodopsin-like GPCRs [73–75].
Initial poses of 5F-MDMB-PICA and 5F-MMB-PICA were selected by analyzing the results of docking the ligands into the prepared (MDMB-FUBINACA)-CB1R binding site using the Induced-Fit Docking (IFD) protocol [76] (Schrödinger suite 2019–4). Using Desmond System Builder (Schrödinger suite 2019–4), CB1R models were placed into explicit 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine lipid bilayer (POPC) with the standard membrane-protein orientation provided by the Orientation of Proteins in Membranes (OPM) database [77]. Simple point charge (SPC) water model [78] was used to solvate the system, charges were neutralized, and 0.15 M NaCl was added. The total system size was ~177000 atoms. The OPLS3e force field (PMID: 30768902) was used throughout this study. The initial parameters for 5F-MDMB-PICA and MDMB-FUBINACA based on the default atom typing of OPLS3e was further optimized by the force field builder (Schrödinger suite 2019–4). Desmond MD system (D. E. Shaw Research, New York, NY) [79] was used for the unbiased all-atom MD simulations.
We applied similar simulation protocols for GPCRs [80] to minimize and equilibrate the system. The initial energy minimization was followed by equilibration with the restraints on the ligand heavy atoms and protein backbone atoms. In the production runs, we used the NPT ensemble with constant temperature at 310 K maintained with Langevin dynamics and 1 atm constant pressure was achieved by hybrid Nosé-Hoover Langevin dynamics on an anisotropic flexible periodic cell with a constant surface tension (x-y plane). In production runs, all restraints on the CB1R were released. However, restraints on and N-terminus segments of Gα were kept for 1.2 μs and then released.
Clustering analysis
We performed root-mean-square deviation (RMSD)-based clustering of our aggregated MD simulation trajectories to study the conformational dynamics of protein domains. The clustering is based on all-to-all RMSD of the backbone of transmembrane segments (TMs). The computed all-to-all RMSD matrix was then subjected to the K-means clustering algorithm [81]. For each ligand, we combined frames collected from all independent trajectories, then sampled 10000 frames with replacement. Finally, we merged the sampled frames from each ligand and used them to calculate the all-to-all RMSD.
To estimate the number of clusters, we used the Gap Statistic method (1) [81]. In this technique the change in within-cluster sum of squares error is compared with the expected value under an appropriate null distribution.
| (1) |
where is the expectation of and is the size of the sample from the reference distribution. can be estimated as follows: 1) Uniformly generate the reference distribution 2) Draw number of Monte Carlo samples from the reference distribution 3) Determine the for each sample 4) Estimate by an average of copies . We can then calculate the gap statistic by following two steps: 1), change the number of clusters form , cluster the observed data, and for each calculate the within-sum of squares of error 2) for each , generate reference data sets from the uniform distribution and measure within-sum of squares of error . The gap statistic can be calculated as (2):
| (2) |
and using , which is the simulation error in, the number of clusters is the smallest value for such that
Frames from selected clusters (clustered frames*) were then mapped to the original frames and saved to perform further analyses (e.g., RMSD, dihedral angles, contact frequency, Protein Interaction Analyzer (PIA-GPCR) [82], and correlation-based network analysis).
Contact frequency:
We considered the heavy atoms of binding site residues and each ligand to find the residues that were within 6 Å of each ligand to calculate the contact frequency for each ligand. If any of the ligands showed 50% or more frequency for any of the binding site residues, we tabulated the results for that residue for all studied ligands. We also calculated the differences between CB1R/5F-MDMB-PICA or CB1R/MDMB-FUBINACA, and CB1R/5F-MMB-PICA, respectively. The differences of 25% and more were highlighted.
PIA-GPCR:
The following structural elements were defined for the analysis of coarse-grained interaction network of the hCB1. The “e”,” m”, and “i” symbols define the extracellular, middle, and intracellular section of TM region, respectively. TM1e (residues 1131.29-1201.36), TM1m (residues 1211.37-1291.45), TM1i (residues 1301.46-1441.60), TM2e (residues 1752.62-1792.66), TM2m (residues 1652.52-1742.61), TM2i (residues 1512.38-1642.51), TM3e (residues 1863.22-1953.31), TM3m (residues 1963.32-2023.38), TM3i (residues 2033.39-2203.56), TM4e (residues 2474.56-2534.62), TM4m (residues 2414.50-2464.55), TM4i (residues 2294.38-2404.49), TM5e (residues 2725.36-2805.44), TM5m (residues 2815.45-2885.52). TM5i (residues 2895.53-3115.75), TM6e (residues 3636.55-3686.60), TM6m (residues 3556.47-3626.54), TM6i (residues 3356.27-3546.46), TM7e (residues 3747.30-3827.38), TM7m (residues 3837.39-3907.46), TM7i (residues 3917.47-4007.56).
For PIA-GPCR sidechain rotamer analysis, the residues extracted from contact frequency analysis along with the residues reported in the experimental studies [27, 28] but not identified in contact frequency results were included.
RMSD, dihedral angles, and contact frequency were calculated with VMD (version 1.9.3), PIA-GPCR with our in-house scripts, and the Cα-Cα distances for network analysis with MDAnalysis (version 1.1.1).
Correlation-based network analysis
We performed Pearson correlation analysis to identify the unique signaling pathways for ligands studied in our work. We divided the signaling pathway into three regions and for each region systematically scanned and included distance/dihedral measurements in our initial data set for network analysis based on the following considerations:
Ligand-binding site:
Ligand polar groups play a significant role in ligand binding. In both 5F-MDMB-PICA and 5F-MMB-PICA the polar groups are distributed in head, linker, core and tail moieties. Three oxygen atoms in the head moiety, two nitrogen atoms in the linker and core, respectively, and one fluorine atom in the tail. Therefore, the dynamics of the ligand in the binding site can be characterized using the distances between these atoms and receptor residues. Clustered frames* were used to calculate the contact frequency between ligand and binding site residues. The residues within 6 Å of each polar atom that have 10% contact frequency were selected and the distances between those residues and the corresponding polar atom were included in network analysis. The dihedral angles of the ligands’ head moiety were also included in the analysis.
TMs site:
Clustered frames* were used to measure the distances between the Cα-Cα of all 7TM residues. We then filtered out the Cα-Cα distances that their average value was larger than 12 Å and the rest were included in the final set of measurements for network analysis. The dihedral angles of the conserved motifs that play a significant role in GPCRs activation (C6.47W6.48×6.49P6.50, D3.49R3.50Y3.51, P5.50I3.40F6.44, and N7.49P7.50×7.51×7.52Y7.53) were also measured and included in the analyses.
CB1R-Gαi site:
Clustered frames* were used to systematically scan the distances between receptor residues (TMs + intracellular loops) and the Gα residues. The Cα-Cα distances that their average value was larger than 12 Å were filtered out and the rest were included in the final set of measurements.
The sample Pearson correlation coefficient (r) was calculated for all distance/dihedral pairs. We then applied three filtering criteria on Pearson correlation coefficient matrix
Threshold for r and only pairs with r ≥ 0.85 were kept.
Let’s show each correlated pair of distances as (distance ) and (distance ) where and are the residue numbers for pairs and . If that pairs were kept.
For each set of correlated pairs that share one common distance, and the other distances include residues from the same region of TMs, only one pair with the maximum difference between residue numbers either or or both was kept.
The clustering and network analyses were repeated 10 times (bootstrap sampling with replacement). The final correlated pairs resulted from each bootstrap sampling which followed by clustering and network analysis were combined, and the common pairs were extracted for each ligand. The common pairs for each ligand were then mapped to the structure for visualization. These final pairs are considered as a hot region of allosteric pathway at the selected correlation threshold (0.85) for each ligand. To make sure the correlation between each pair is real (the change in variable m(t) (e.g., distance or dihedral angle) at time and is accompanied by the change in variable n(t) at the corresponding times), the time series for each final correlated pair were evaluated by plotting their distribution as a 2D map.
Supplementary Material
Acknowledgements
The work is supported by Brain & Behavior Research Foundation Young Investigator Grant (H.Y.) Support for this research was provided by the National Institute on Drug Abuse–Intramural Research Program (Z1A DA000606 to L.S., Z1A DA000487 to CRL, Z1A DA000523 to MHB). This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Footnotes
Competing Interest Statement
The authors declare no competing financial interests.
REFERENCES
- 1.Huffman JW, Dai D., Martin B.R., Compton D.R.: Design, synthesis and pharmacology of cannabimimetic indoles. Bioorganic & Medicinal Chemistry Letters 1994, 4(4):563–566. [Google Scholar]
- 2.Wiley JL, Compton DR, Dai D, Lainton JA, Phillips M, Huffman JW, Martin BR: Structure-activity relationships of indole- and pyrrole-derived cannabinoids. J Pharmacol Exp Ther 1998, 285(3):995–1004. [PubMed] [Google Scholar]
- 3.Usui K, Fujita Y, Kamijo Y, Kokaji T, Funayama M: Identification of 5-Fluoro ADB in Human Whole Blood in Four Death Cases. J Anal Toxicol 2018, 42(2):e21–e25. [DOI] [PubMed] [Google Scholar]
- 4.Adams AJ, Banister SD, Irizarry L, Trecki J, Schwartz M, Gerona R: “Zombie” Outbreak Caused by the Synthetic Cannabinoid AMB-FUBINACA in New York. N Engl J Med 2017, 376(3):235–242. [DOI] [PubMed] [Google Scholar]
- 5.Notes from the field: Severe illness associated with synthetic cannabinoid use - Brunswick, Georgia, 2013. MMWR Morb Mortal Wkly Rep 2013, 62(46):939. [PMC free article] [PubMed] [Google Scholar]
- 6.Lie W, Cheong EJY, Goh EML, Moy HY, Cannaert A, Stove CP, Chan ECY: Diagnosing intake and rationalizing toxicities associated with 5F-MDMB-PINACA and 4F-MDMB-BINACA abuse. Arch Toxicol 2021, 95(2):489–508. [DOI] [PubMed] [Google Scholar]
- 7.Boland DM, Reidy LJ, Seither JM, Radtke JM, Lew EO: Forty-Three Fatalities Involving the Synthetic Cannabinoid, 5-Fluoro-ADB: Forensic Pathology and Toxicology Implications. J Forensic Sci 2020, 65(1):170–182. [DOI] [PubMed] [Google Scholar]
- 8.Banister SD, Moir M, Stuart J, Kevin RC, Wood KE, Longworth M, Wilkinson SM, Beinat C, Buchanan AS, Glass M et al. : Pharmacology of Indole and Indazole Synthetic Cannabinoid Designer Drugs AB-FUBINACA, ADB-FUBINACA, AB-PINACA, ADB-PINACA, 5F-AB-PINACA, 5F-ADB-PINACA, ADBICA, and 5F-ADBICA. ACS Chem Neurosci 2015, 6(9):1546–1559. [DOI] [PubMed] [Google Scholar]
- 9.Antonides LH, Cannaert A, Norman C, Vives L, Harrison A, Costello A, Nic Daeid N, Stove CP, Sutcliffe OB, McKenzie C: Enantiospecific Synthesis, Chiral Separation, and Biological Activity of Four Indazole-3-Carboxamide-Type Synthetic Cannabinoid Receptor Agonists and Their Detection in Seized Drug Samples. Front Chem 2019, 7:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Marzo V, Stella N, Zimmer A: Endocannabinoid signalling and the deteriorating brain. Nat Rev Neurosci 2015, 16(1):30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heifets BD, Castillo PE: Endocannabinoid signaling and long-term synaptic plasticity. Annu Rev Physiol 2009, 71:283–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piomelli D: The molecular logic of endocannabinoid signalling. Nat Rev Neurosci 2003, 4(11):873–884. [DOI] [PubMed] [Google Scholar]
- 13.Marsicano G, Lutz B: Expression of the cannabinoid receptor CB1 in distinct neuronal subpopulations in the adult mouse forebrain. Eur J Neurosci 1999, 11(12):4213–4225. [DOI] [PubMed] [Google Scholar]
- 14.Kano M, Ohno-Shosaku T, Maejima T: Retrograde signaling at central synapses via endogenous cannabinoids. Mol Psychiatry 2002, 7(3):234–235. [DOI] [PubMed] [Google Scholar]
- 15.Wilson RI, Nicoll RA: Endogenous cannabinoids mediate retrograde signalling at hippocampal synapses. Nature 2001, 410(6828):588–592. [DOI] [PubMed] [Google Scholar]
- 16.Guo J, Ikeda SR: Endocannabinoids modulate N-type calcium channels and G-protein-coupled inwardly rectifying potassium channels via CB1 cannabinoid receptors heterologously expressed in mammalian neurons. Mol Pharmacol 2004, 65(3):665–674. [DOI] [PubMed] [Google Scholar]
- 17.Hoffman AF, Lupica CR: Mechanisms of Cannabinoid Inhibition of GABAASynaptic Transmission in the Hippocampus. The Journal of Neuroscience 2000, 20(7):2470–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernández-Moncada I, Marsicano G: Astroglial CB1 receptors, energy metabolism, and gliotransmission: an integrated signaling system? Essays Biochem 2023, 67(1):49–61. [DOI] [PubMed] [Google Scholar]
- 19.Navarrete M, Araque A: Endocannabinoids mediate neuron-astrocyte communication. Neuron 2008, 57(6):883–893. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz MD, Trecki J, Edison LA, Steck AR, Arnold JK, Gerona RR: A Common Source Outbreak of Severe Delirium Associated with Exposure to the Novel Synthetic Cannabinoid ADB-PINACA. J Emerg Med 2015, 48(5):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ukaigwe A, Karmacharya P, Donato A: A Gut Gone to Pot: A Case of Cannabinoid Hyperemesis Syndrome due to K2, a Synthetic Cannabinoid. Case Rep Emerg Med 2014, 2014:167098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takematsu M, Hoffman RS, Nelson LS, Schechter JM, Moran JH, Wiener SW: A case of acute cerebral ischemia following inhalation of a synthetic cannabinoid. Clin Toxicol (Phila) 2014, 52(9):973–975. [DOI] [PubMed] [Google Scholar]
- 23.Kazory A, Aiyer R: Synthetic marijuana and acute kidney injury: an unforeseen association. Clin Kidney J 2013, 6(3):330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ford BM, Tai S, Fantegrossi WE, Prather PL: Synthetic Pot: Not Your Grandfather’s Marijuana. Trends Pharmacol Sci 2017, 38(3):257–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel M, Manning JJ, Finlay DB, Javitch JA, Banister SD, Grimsey NL, Glass M: Signalling profiles of a structurally diverse panel of synthetic cannabinoid receptor agonists. Biochem Pharmacol 2020, 175:113871. [DOI] [PubMed] [Google Scholar]
- 26.Yano H, Adhikari P, Naing S, Hoffman AF, Baumann MH, Lupica CR, Shi L: Positive Allosteric Modulation of the 5-HT(1A) Receptor by Indole-Based Synthetic Cannabinoids Abused by Humans. ACS Chem Neurosci 2020, 11(10):1400–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hua T, Vemuri K, Nikas SP, Laprairie RB, Wu Y, Qu L, Pu M, Korde A, Jiang S, Ho JH et al. : Crystal structures of agonist-bound human cannabinoid receptor CB(1). Nature 2017, 547(7664):468–471. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Krishna Kumar K, Shalev-Benami M, Robertson MJ, Hu H, Banister SD, Hollingsworth SA, Latorraca NR, Kato HE, Hilger D, Maeda S et al. : Structure of a Signaling Cannabinoid Receptor 1-G Protein Complex. Cell 2019, 176(3):448–458.e412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua T, Li X, Wu L, Iliopoulos-Tsoutsouvas C, Wang Y, Wu M, Shen L, Brust CA, Nikas SP, Song F et al. : Activation and Signaling Mechanism Revealed by Cannabinoid Receptor-G(i) Complex Structures. Cell 2020, 180(4):655–665.e618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X, Wang X, Xu Z, Wu C, Zhou Y, Wang Y, Lin G, Li K, Wu M, Xia A et al. : Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1. Nat Chem Biol 2022, 18(8):831–840. [DOI] [PubMed] [Google Scholar]
- 31.Shao Z, Yin J, Chapman K, Grzemska M, Clark L, Wang J, Rosenbaum DM: High-resolution crystal structure of the human CB1 cannabinoid receptor. Nature 2016, 540(7634):602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hua T, Vemuri K, Pu M, Qu L, Han GW, Wu Y, Zhao S, Shui W, Li S, Korde A et al. : Crystal Structure of the Human Cannabinoid Receptor CB(1). Cell 2016, 167(3):750–762.e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrage R, De Min A, Hochheiser K, Kostenis E, Mohr K: Superagonism at G protein-coupled receptors and beyond. Br J Pharmacol 2016, 173(20):3018–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith NJ, Bennett KA, Milligan G: When simple agonism is not enough: emerging modalities of GPCR ligands. Mol Cell Endocrinol 2011, 331(2):241–247. [DOI] [PubMed] [Google Scholar]
- 35.Tan CM, Wilson MH, MacMillan LB, Kobilka BK, Limbird LE: Heterozygous alpha 2A-adrenergic receptor mice unveil unique therapeutic benefits of partial agonists. Proc Natl Acad Sci U S A 2002, 99(19):12471–12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holst B, Brandt E, Bach A, Heding A, Schwartz TW: Nonpeptide and peptide growth hormone secretagogues act both as ghrelin receptor agonist and as positive or negative allosteric modulators of ghrelin signaling. Mol Endocrinol 2005, 19(9):2400–2411. [DOI] [PubMed] [Google Scholar]
- 37.Schrage R, Seemann WK, Klöckner J, Dallanoce C, Racké K, Kostenis E, De Amici M, Holzgrabe U, Mohr K: Agonists with supraphysiological efficacy at the muscarinic M2 ACh receptor. Br J Pharmacol 2013, 169(2):357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seely KA, Patton AL, Moran CL, Womack ML, Prather PL, Fantegrossi WE, Radominska-Pandya A, Endres GW, Channell KB, Smith NH et al. : Forensic investigation of K2, Spice, and “bath salt” commercial preparations: a three-year study of new designer drug products containing synthetic cannabinoid, stimulant, and hallucinogenic compounds. Forensic Sci Int 2013, 233(1–3):416–422. [DOI] [PubMed] [Google Scholar]
- 39.Wilson CD, Tai S, Ewing L, Crane J, Lockhart T, Fujiwara R, Radominska-Pandya A, Fantegrossi WE: Convulsant Effects of Abused Synthetic Cannabinoids JWH-018 and 5F-AB-PINACA Are Mediated by Agonist Actions at CB1 Receptors in Mice. J Pharmacol Exp Ther 2019, 368(2):146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sachdev S, Vemuri K, Banister SD, Longworth M, Kassiou M, Santiago M, Makriyannis A, Connor M: In vitro determination of the efficacy of illicit synthetic cannabinoids at CB(1) receptors. Br J Pharmacol 2019, 176(24):4653–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yano H, Sánchez-Soto M, Ferré S: Bioluminescence Resonance Energy Transfer Assay to Characterize Gi-Like G Protein Subtype-Dependent Functional Selectivity. Curr Protoc Neurosci 2017, 81:5.33.31–35.33.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sykes DA, Parry C, Reilly J, Wright P, Fairhurst RA, Charlton SJ: Observed Drug-Receptor Association Rates Are Governed by Membrane Affinity: The Importance of Establishing “Micro-Pharmacokinetic/Pharmacodynamic Relationships” at the β2-Adrenoceptor. Molecular Pharmacology 2014, 85(4):608–617. [DOI] [PubMed] [Google Scholar]
- 43.Sykes DA, Stoddart LA, Kilpatrick LE, Hill SJ: Binding kinetics of ligands acting at GPCRs. Mol Cell Endocrinol 2019, 485:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei H, Ahn S, Shenoy SK, Karnik SS, Hunyady L, Luttrell LM, Lefkowitz RJ: Independent beta-arrestin 2 and G protein-mediated pathways for angiotensin II activation of extracellular signal-regulated kinases 1 and 2. Proc Natl Acad Sci U S A 2003, 100(19):10782–10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Daigle TL, Kearn CS, Mackie K: Rapid CB1 cannabinoid receptor desensitization defines the time course of ERK1/2 MAP kinase signaling. Neuropharmacology 2008, 54(1):36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daigle TL, Kwok ML, Mackie K: Regulation of CB1 cannabinoid receptor internalization by a promiscuous phosphorylation-dependent mechanism. J Neurochem 2008, 106(1):70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin W, Brown S, Roche JP, Hsieh C, Celver JP, Kovoor A, Chavkin C, Mackie K: Distinct domains of the CB1 cannabinoid receptor mediate desensitization and internalization. J Neurosci 1999, 19(10):3773–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laprairie RB, Bagher AM, Kelly ME, Dupré DJ, Denovan-Wright EM: Type 1 cannabinoid receptor ligands display functional selectivity in a cell culture model of striatal medium spiny projection neurons. J Biol Chem 2014, 289(36):24845–24862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zagzoog A, Brandt AL, Black T, Kim ED, Burkart R, Patel M, Jin Z, Nikolaeva M, Laprairie RB: Assessment of select synthetic cannabinoid receptor agonist bias and selectivity between the type 1 and type 2 cannabinoid receptor. Sci Rep 2021, 11(1):10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manning JJ, Rawcliffe G, Finlay DB, Glass M: Cannabinoid 1 (CB(1) ) receptor arrestin subtype-selectivity and phosphorylation dependence. Br J Pharmacol 2023, 180(3):369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffman AF, Laaris N, Kawamura M, Masino SA, Lupica CR: Control of cannabinoid CB1 receptor function on glutamate axon terminals by endogenous adenosine acting at A1 receptors. J Neurosci 2010, 30(2):545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Godino MC, Torres M, Sánchez-Prieto J: Inhibition of N- and P/Q-type Ca2+ channels by cannabinoid receptors in single cerebrocortical nerve terminals. FEBS Lett 2005, 579(3):768–772. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan JM: Mechanisms of cannabinoid-receptor-mediated inhibition of synaptic transmission in cultured hippocampal pyramidal neurons. J Neurophysiol 1999, 82(3):1286–1294. [DOI] [PubMed] [Google Scholar]
- 54.Lupica CR, Hu Y, Devinsky O, Hoffman AF: Cannabinoids as hippocampal network administrators. Neuropharmacology 2017, 124:25–37. [DOI] [PubMed] [Google Scholar]
- 55.Hoffman AF, Lycas MD, Kaczmarzyk JR, Spivak CE, Baumann MH, Lupica CR: Disruption of hippocampal synaptic transmission and long-term potentiation by psychoactive synthetic cannabinoid ‘Spice’ compounds: comparison with Δ(9) - tetrahydrocannabinol. Addict Biol 2017, 22(2):390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Q, Yang D, Wu M, Guo Y, Guo W, Zhong L, Cai X, Dai A, Jang W, Shakhnovich EI et al. : Common activation mechanism of class A GPCRs. Elife 2019, 8. [DOI] [PMC free article] [PubMed]
- 57.Gillis A, Kliewer A, Kelly E, Henderson G, Christie MJ, Schulz S, Canals M: Critical Assessment of G Protein-Biased Agonism at the μ-Opioid Receptor. Trends Pharmacol Sci 2020, 41(12):947–959. [DOI] [PubMed] [Google Scholar]
- 58.Glatfelter GC, Partilla JS, Baumann MH: Structure-activity relationships for 5F-MDMB-PICA and its 5F-pentylindole analogs to induce cannabinoid-like effects in mice. Neuropsychopharmacology 2022, 47(4):924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schönherr H, Cernak T: Profound methyl effects in drug discovery and a call for new C-H methylation reactions. Angew Chem Int Ed Engl 2013, 52(47):12256–12267. [DOI] [PubMed] [Google Scholar]
- 60.Leung CS, Leung SS, Tirado-Rives J, Jorgensen WL: Methyl effects on protein-ligand binding. J Med Chem 2012, 55(9):4489–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahn KH, Bertalovitz AC, Mierke DF, Kendall DA: Dual role of the second extracellular loop of the cannabinoid receptor 1: ligand binding and receptor localization. Mol Pharmacol 2009, 76(4):833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alves VL, Gonçalves JL, Aguiar J, Teixeira HM, Câmara JS: The synthetic cannabinoids phenomenon: from structure to toxicological properties. A review. Crit Rev Toxicol 2020, 50(5):359–382. [DOI] [PubMed] [Google Scholar]
- 63.Banister SD, Connor M: The Chemistry and Pharmacology of Synthetic Cannabinoid Receptor Agonists as New Psychoactive Substances: Origins. Handb Exp Pharmacol 2018, 252:165–190. [DOI] [PubMed] [Google Scholar]
- 64.Banister SD, Connor M: The Chemistry and Pharmacology of Synthetic Cannabinoid Receptor Agonist New Psychoactive Substances: Evolution. Handb Exp Pharmacol 2018, 252:191–226. [DOI] [PubMed] [Google Scholar]
- 65.Banister SD, Longworth M, Kevin R, Sachdev S, Santiago M, Stuart J, Mack JB, Glass M, McGregor IS, Connor M et al. : Pharmacology of Valinate and tert-Leucinate Synthetic Cannabinoids 5F-AMBICA, 5F-AMB, 5F-ADB, AMB-FUBINACA, MDMB-FUBINACA, MDMB-CHMICA, and Their Analogues. ACS Chem Neurosci 2016, 7(9):1241–1254. [DOI] [PubMed] [Google Scholar]
- 66.Krotulski AJ, Mohr ALA, Logan BK: Emerging Synthetic Cannabinoids: Development and Validation of a Novel Liquid Chromatography Quadrupole Time-of-Flight Mass Spectrometry Assay for Real-Time Detection. Journal of Analytical Toxicology 2019, 44(3):207–217. [DOI] [PubMed] [Google Scholar]
- 67.Tokarczyk B, Jurczyk A, Krupińska J, Adamowicz P: Fatal intoxication with new synthetic cannabinoids 5F-MDMB-PICA and 4F-MDMB-BINACA-parent compounds and metabolite identification in blood, urine and cerebrospinal fluid. Forensic Sci Med Pathol 2022, 18(4):393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Groth O, Roider G, Angerer V, Schäper J, Graw M, Musshoff F, Auwärter V: “Spice”-related deaths in and around Munich, Germany: A retrospective look at the role of synthetic cannabinoid receptor agonists in our post-mortem cases over a seven-year period (2014–2020). Int J Legal Med 2023. [DOI] [PMC free article] [PubMed]
- 69.Janssens LK, Hudson S, Wood DM, Wolfe C, Dargan PI, Stove CP: Linking in vitro and ex vivo CB(1) activity with serum concentrations and clinical features in 5F-MDMB-PICA users to better understand SCRAs and their metabolites. Arch Toxicol 2022, 96(11):2935–2945. [DOI] [PubMed] [Google Scholar]
- 70.Kleis J, Germerott T, Halter S, Héroux V, Roehrich J, Schwarz CS, Hess C: The synthetic cannabinoid 5F-MDMB-PICA: A case series. Forensic Sci Int 2020, 314:110410. [DOI] [PubMed] [Google Scholar]
- 71.Bonifazi A, Yano H, Guerrero AM, Kumar V, Hoffman AF, Lupica CR, Shi L, Newman AH: Novel and Potent Dopamine D(2) Receptor Go-Protein Biased Agonists. ACS Pharmacol Transl Sci 2019, 2(1):52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Free RB, Chun LS, Moritz AE, Miller BN, Doyle TB, Conroy JL, Padron A, Meade JA, Xiao J, Hu X et al. : Discovery and characterization of a G protein-biased agonist that inhibits β-arrestin recruitment to the D2 dopamine receptor. Mol Pharmacol 2014, 86(1):96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dror RO, Arlow DH, Maragakis P, Mildorf TJ, Pan AC, Xu H, Borhani DW, Shaw DE: Activation mechanism of the β2-adrenergic receptor. Proc Natl Acad Sci U S A 2011, 108(46):18684–18689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ghanouni P, Schambye H, Seifert R, Lee TW, Rasmussen SG, Gether U, Kobilka BK: The effect of pH on beta(2) adrenoceptor function. Evidence for protonation-dependent activation. J Biol Chem 2000, 275(5):3121–3127. [DOI] [PubMed] [Google Scholar]
- 75.Ranganathan A, Dror RO, Carlsson J: Insights into the role of Asp79(2.50) in β2 adrenergic receptor activation from molecular dynamics simulations. Biochemistry 2014, 53(46):7283–7296. [DOI] [PubMed] [Google Scholar]
- 76.Sherman W, Beard HS, Farid R: Use of an induced fit receptor structure in virtual screening. Chem Biol Drug Des 2006, 67(1):83–84. [DOI] [PubMed] [Google Scholar]
- 77.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI: OPM: orientations of proteins in membranes database. Bioinformatics 2006, 22(5):623–625. [DOI] [PubMed] [Google Scholar]
- 78.Berendsen H, Postma JPM, van Gunsteren W, Hermans J: Interaction Models for Water in Relation to Protein Hydration. In., vol. 11; 1981: 331–342. [Google Scholar]
- 79.Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD et al. : Scalable algorithms for molecular dynamics simulations on commodity clusters. In: Proceedings of the 2006 ACM/IEEE conference on Supercomputing; Tampa, Florida. Association for Computing Machinery; 2006: 84–es. [Google Scholar]
- 80.Lane JR, Abramyan AM, Adhikari P, Keen AC, Lee KH, Sanchez J, Verma RK, Lim HD, Yano H, Javitch JA et al. : Distinct inactive conformations of the dopamine D2 and D3 receptors correspond to different extents of inverse agonism. Elife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lloyd S: Least squares quantization in PCM. IEEE Transactions on Information Theory 1982, 28(2):129–137. [Google Scholar]
- 82.Stolzenberg S, Michino M, LeVine MV, Weinstein H, Shi L: Computational approaches to detect allosteric pathways in transmembrane molecular machines. Biochim Biophys Acta 2016, 1858(7 Pt B):1652–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ballesteros JA, Weinstein H: [19] Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. In: Methods in Neurosciences. Edited by Sealfon SC, vol. 25: Academic Press; 1995: 366–428. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.